Abstract
Neurofibromatosis Type 1 (NF1) is a common genetic condition associated with cognitive dysfunction. However, the pathophysiology of the NF1 cognitive deficits is not well understood. Abnormal brain structure, including increased total brain volume, white matter (WM) and grey matter (GM) abnormalities have been reported in the NF1 brain. These previous studies employed univariate model‐driven methods preventing detection of subtle and spatially distributed differences in brain anatomy. Multivariate pattern analysis allows the combination of information from multiple spatial locations yielding a discriminative power beyond that of single voxels. Here we investigated for the first time subtle anomalies in the NF1 brain, using a multivariate data‐driven classification approach. We used support vector machines (SVM) to classify whole‐brain GM and WM segments of structural T 1‐weighted MRI scans from 39 participants with NF1 and 60 non‐affected individuals, divided in children/adolescents and adults groups. We also employed voxel‐based morphometry (VBM) as a univariate gold standard to study brain structural differences. SVM classifiers correctly classified 94% of cases (sensitivity 92%; specificity 96%) revealing the existence of brain structural anomalies that discriminate NF1 individuals from controls. Accordingly, VBM analysis revealed structural differences in agreement with the SVM weight maps representing the most relevant brain regions for group discrimination. These included the hippocampus, basal ganglia, thalamus, and visual cortex. This multivariate data‐driven analysis thus identified subtle anomalies in brain structure in the absence of visible pathology. Our results provide further insight into the neuroanatomical correlates of known features of the cognitive phenotype of NF1. Hum Brain Mapp 35:89–106, 2014. © 2012 Wiley Periodicals, Inc.
Keywords: neurofibromatosis type 1, support vector machine (SVM), neuroimaging, multivariate pattern classification, MRI, brain structure
INTRODUCTION
The brain changes during development as a function of genetic and environmental conditions, as well as in the context of neurodevelopmental disorders. To understand abnormal neurodevelopment and its consequences it is fundamental to investigate the structural phenotype and its possible role in cognition [Hoeft et al., 2008].
Neurofibromatosis Type 1 (NF1) is a common, single gene, developmental disorder with an incidence of 1 in 3,500, characterized by increased predisposition for tumor development and cognitive deficits [Kayl and Moore, 2000]. NF1 features are detectable in infancy or early childhood, suggesting a role for the NF1 gene in normal development [Daston and Ratner, 1992]. Previous studies indicated that the NF1 gene is expressed throughout the brain during development and in adulthood, both in neurons and glia [Daston and Ratner, 1992; Zhu et al., 2005]. While during development the protein product of the NF1 gene, neurofibromin, is expressed in all organ systems, in adult tissues expression predominates in the nervous system (including the cerebral cortex, cerebellum, and brainstem) [Daston and Ratner, 1992; Gutmann et al., 1995]. Neurofibromin is involved in cell proliferation and differentiation [Lee et al., 2010], suggesting that brain structure might be affected in NF1. Indeed, an increase in total brain volume is a widely reported structural abnormality [Cutting et al., 2002; Payne et al., 2010]. Other studies show evidence for more specific localized structural deficits, e.g., the thalamus and the corpus callosum [Cutting et al., 2000; Dubovsky et al., 2001; Greenwood et al., 2005; Kayl et al., 2000; Moore et al., 2000; Payne et al., 2010; Steen et al., 2001].
Importantly, individuals with NF1 and no brain tumors show impaired cognitive abilities suggesting anomalous brain function or structure independently of focal lesions [De Winter et al., 1999; Moore et al., 1994; Schrimsher et al., 2003]. Furthermore, visuospatial, memory, and motor deficits have so far been described in individuals with NF1 without clearly identified brain mechanisms [Hyman et al., 2005, 2006; Levine et al., 2006; North, 2000]. The prominent visuospatial deficits reported in these patients suggest parietal dysfunction [Billingsley et al., 2002; Clements‐Stephens, et al., 2008] while studies using NF1 animal models indicate abnormal function of the hippocampus, prefrontal cortex and striatum, related to visuospatial memory, working memory, and attentional deficits, respectively [Costa et al., 2002; Park et al., 2009].
Magnetic resonance imaging (MRI) is a standard method for studying brain structure, with studies mainly focusing either on volume changes in particular anatomical structures, lobes, or on the whole brain [Bray et al., 2009; Hoeft et al., 2008; Mietchen and Gaser, 2009]. To date, only univariate analysis methods have been applied to study brain anatomical alterations in patients with NF1 [Payne et al., 2010]. In contrast, multivariate methods enable the identification of subtle neuroanatomical discriminative patterns encoded across brain regions, in the absence of a priori hypotheses. This method determines group differences from a large number of simultaneously evaluated spatial locations (voxels). Notably, individual voxels might not be significantly different between groups but can still contribute to a significantly different overall spatial pattern [Ecker et al., 2010; Marzelli et al., 2011; Pereira et al., 2009]. In NF1, as in other clinical populations, multivariate pattern analysis techniques are of particular interest for studying the brain anatomy, as studies have shown that these disorders rarely affect single brain structures [Bray et al., 2009]. We hypothesize that structural brain alterations would involve multiple and distributed anatomical structures of the brain, composing a complex spatial pattern that may not be easily observed using univariate approaches. Here we used a data‐driven multivariate classifier, employing a support vector machine (SVM) algorithm, which has previously been employed to investigate group differences with structural MRI [Ecker et al., 2010; Klöppel et al., 2008; Marzelli et al., 2011]. We used linear SVM to classify whole‐brain high‐resolution anatomical images. We employed a leave‐two‐out cross‐validation procedure to discriminate participants with NF1 from non‐affected controls and to identify differences in neuroanatomical spatial patterns. Furthermore, we also used the standard univariate voxel‐based morphometry (VBM) analysis, commonly used for whole brain structural analysis. This method is not directly comparable with the multivariate classification technique, but allowed us to better interpret the meaning of the weights identified by SVM analysis while at the same time confirming whether the distributed patterns highlighted with SVM included locations identified by VBM. The identified spatial patterns and their functional significance provide novel insights on current pathophysiological hypotheses in NF1.
METHODS
Participants
For this study, we recruited 112 individuals, 26 NF1 children/adolescents, 31 control children/adolescents, 20 NF1 adults, and 35 control adults. Although NF1 is an autosomal dominant genetic disorder, novel mutations are common and therefore diagnosis does not rely solely on genetic testing. All participants with NF1 were recruited and diagnosed in collaboration with the Clinical Genetics Department of the Pediatrics Hospital of Coimbra according to the NIH clinical criteria for NF1 [Neurofibromatosis. Conference statement. National Institutes of Health Consensus Development Conference, 1988]. The presence of two or more of the following criteria constitutes definitive diagnosis:
Six or more “cafe‐au‐lait” spots greater than 5 mm in diameter in prepubertal children or greater than 15 mm in diameter in postpubertal individuals.
Two or more neurofibromas of any form or one plexiform neurofibroma.
Freckling in the axillary or inguinal regions.
Optic glioma.
Two or more Lisch nodules (iris hamartomas).
A distinctive osseous lesion such as sphenoid dysplasia or thinning of long bone cortex with or without pseudoarthrosis.
A first‐degree relative with NF1 by the above criteria.
Symptom severity is variable from mild symptoms in most individuals diagnosed with NF1 to some debilitating cases, characterized by severe cognitive impairments and serious medical complications [Tonsgard, 2006]. Here, we investigated individuals without brain tumors to prove that a phenotype is still present in NF1 in the absence of severe structural abnormalities.
Children and adolescent controls were recruited among unaffected siblings and from a local school. Adult controls were recruited among the unaffected parents or from an adult education school. Neuroradiological assessments of FLAIR and T 1‐weighted MRI scans were carried out by an experienced neuroradiologist in order to detect central nervous system pathologies. We excluded participants with a clinically significant intracranial abnormality on MRI, as intracranial tumor, optic glioma, or other imaging abnormalities. T2‐hyperintensities were not considered an exclusion criterion. T2‐hyperintensities are the most commonly identified abnormalities on T 2‐weighted MR images in participants with NF1. T2‐hyperintensities tend to resolve with adulthood and the hypothesis that these lesions are associated cognitive impairment in children with NF1 remains controversial [Payne et al., 2010]. None of the participants had a psychiatric illness and individuals with other neurological problems (e.g., epilepsy) were excluded. We excluded 13 participants based on these criteria.
T 1‐weighted MRI brain scans from 99 participants divided in two groups were analyzed: a group of children/adolescents (7–17 years old), 21 participants diagnosed with NF1 and 29 age‐ and gender‐matched controls; and a group of adults (19–50 years old), 18 participants with NF1 and 31 age‐ and gender‐matched controls. Overall, 95 participants were right handed and only 4 participants were left handed. Demographic data for the included participants are reported in Table 1.
Table 1.
Demographic data on study cohort
| Group | Children/adolescents | Adults | ||
|---|---|---|---|---|
| NF1 | Controls | NF1 | Controls | |
| n | 21 | 29 | 18 | 31 |
| Age | 11.12 (2.35) | 12.08 (2.41) | 33.05 (5.42) | 34.97 (7.93) |
| Gender (F/M) | 14/7 | 16/13 | 17/1 | 25/6 |
Data expressed as mean (SD). There were no significant statistical differences in age (using independent samples t‐tests) and gender ratios (using Chi‐square test) between the clinical groups.
In order to correlate the level of cognitive impairments with the output of SVM classification, we performed the neuropsychological characterization of the participants. In the group of children/adolescents, we applied the Portuguese adapted version of the Wechsler Intelligence Scale for children (WISC‐III) to measure the IQ [Wechsler, 2003]. For adult participants, we applied the first set of the Raven Advanced Progressive Matrices [Raven, 1947] as an indication of non‐verbal intelligence. Participants in both age groups also performed the Benton's Judgment of Line Orientation (JLO) test, a standardized measure of visuospatial judgment [Benton et al., 1978]. The neuropsychological characterization of a subgroup of these patients has already been reported before in our previous study [Ribeiro et al., 2012]. Neuropsychological differences between participants with NF1 and controls were evaluated using independent samples t‐tests, after testing for normality of the data. In all cases we failed to reject the null hypothesis that the samples came from a normal distribution (except for JLO, in which case non parametric tests were used). Neuropsychological characterization is reported in Table 2.
Table 2.
Volumetric and neuropsychological measures on patients and control groups
| Group | Children/adolescents | Adults | ||
|---|---|---|---|---|
| NF1 | Controls | NF1 | Controls | |
| Grey matter (ml) | 701.63 (111.73) | 740.79 (88.85) | 654.97* (55.50) | 616.51* (51.06) |
| White matter (ml) | 498.33* (59.44) | 456.76* (54.08) | 556.02** (51.62) | 475.50** (56.75) |
| Cerebral spinal Fluid (ml) | 237.82 (79.94) | 210.76 (30.29) | 206.79 (34.94) | 195.71 (19.12) |
| Brain total (ml) | 1437.78 (136.96) | 1408.31 (141.82) | 1417.78** (113.37) | 1287.72** (111.82) |
| Full‐scale IQ | 96.60* (15.21)a | 114.42* (20.53)a | — | — |
| Raven score | — | — | 0.50* (0.23)a | 0.66* (0.23)a |
| JLO score | 16.89** (6.29)a | 24.26** (3.97)a | 17.94* (4.22)a | 22.43* (6.05)a |
Data expressed as mean (SD). Significant group differences (NF1 vs. Controls) are marked: *P < 0.05, **P < 0.001.
JLO: Judgment of Line Orientation test.
n = 21, 24, 18, 22, 19, 23, 18.
Standard Protocol Approvals, Registrations, and Patient Consents
The study was approved by the Ethics Committees of the Faculty of Medicine of the University of Coimbra and of the Pediatrics Hospital of the University of Coimbra. Written informed consent was obtained from the adult participants and from the legal representative in the case of participants under 18 years old. Oral or written consent was also obtained from participants under 18 years old.
MR Image Acquisition and Preprocessing
All subjects were scanned at the Portuguese Brain Imaging Network facilities in Coimbra, Portugal, on a 3T research scanner (Magnetom TIM Trio, Siemens) using a 12‐channel birdcage head coil. Two high‐resolution whole‐brain anatomical T 1‐weighted MR images (MPRAGE sequence, 1 × 1 × 1 mm3 voxel size, repetition time (TR) 2.3 s, echo time (TE) 2.98 ms, flip angle (FA) 9°, field of view (FOV) 256 × 256, 160 slices) were acquired for each participant, as well as a T 2‐weighted MR image (FLAIR sequence, 1 × 1 × 1 mm3 voxel size, TR 5 s, TE 2.98 ms, inversion time (TI) 1.8 s, FOV 250 × 250, 160 slices).
Data were pre‐processed using SPM8 software (Wellcome Trust Centre for Neuroimaging, Institute of Neurology, UCL, London, UK, http://www.fil.ion.ucl.ac.uk/spm) and VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm8/) in the Matlab computing environment (version 7.6.0 R2008a, The Mathworks, MA). Each T 1‐weighted native image volume was manually aligned onto the axis of the anterior and posterior commisures. Images were automatically corrected for inhomogeneity of the magnetic field and segmented into grey matter (GM), white matter (WM), and cerebrospinal fluid, with the value at each voxel representing the proportion of the corresponding tissue type [Ashburner and Friston, 2005]. For the adults' group, we used the standard MNI template for spatial normalization and segmentation. For the children/adolescents' group, we created a custom template with the TOM toolbox [Wilke et al., 2008]. We used the high‐dimensional registration DARTEL algorithm in SPM [Ashburner, 2007] to spatially align each subject's image with the corresponding template. This high‐dimensional normalization procedure in SPM includes both linear (affine) and nonlinear components. The affine scaling matches the subject brain to the template in overall shape and the nonlinear component expands and contracts some brain regions on a locally specific basis. To correct for the effects of spatial normalization in our study, we used a non‐linear only “modulation” step. This accounts for local amount of expansion or contraction of brain structures, so that the total amount of GM/WM in the modulated images remains the same as it would be in the original images. For instance, if spatial normalization doubles the volume of a certain structure, then the correction will halve the intensity of the signal in this region. With the adjustment, we can compare the total volume of tissue in each structure corrected for individual brain size (tissue volume per unit volume of spatially normalized image) [Ashburner, 2009; Ashburner and Friston, 2000], which we will denominate as relative volume. If the linear (affine) component of the modulation had been included, through the determinant of the affine matrix [see Buckner et al., 2004 for a complete review of the method] the same corrections would have been applied on a global scale: i.e., larger brains would show a globally higher intensity due to the necessary global contraction to fit the template, and vice‐versa. We have chosen to ignore the affine modulation step, so the GM/WM maps are also corrected for overall global size and shape and there is no need to include global measures as nuisance covariates in subsequent statistical models [Ashburner, 2009].
We also generated custom GM and WM templates for each group (children/adolescents and adults) from GM and WM images of each group's subjects, for optimization of the segmentation procedure and for presentation of the results. Global brain volumes were determined through these automatic segmentation procedures. Differences between participants with NF1 and controls were evaluated using independent samples t‐tests. Volumetric measures for the included participants are reported in Table 2.
Support Vector Pattern Classification
As a supervised learning method, the SVM algorithm determines a map between features of the data and the associated label. In the context of our study, the features of the data are the intensity values in each voxel of the segmented and modulated volume images (GM or WM), which represent the relative volume of tissue (as discussed earlier) and the label of each image is the group to which it belongs (NF1 or Control). Aiming at distinguishing the images into two discrete classes, we perform a binary classification task, in which each image is classified individually as “NF1” or “Control.” Prior to classification, each feature is scaled to a z‐score in the matrix containing all the participants. The z‐score is computed for each voxel (corresponding to a column in the data matrix while each image of an individual is a row of the matrix) using the mean and standard deviation along the column. After scaling, each voxel will have mean 0 and standard deviation 1, compensating for a possible wider variation in signal amplitude in some voxels than others [Pereira et al., 2009]. A set of images is used to train the classifier to learn the neuroanatomical relationships between the relative volume across voxels and the group membership. In the training of the classifier, a mathematical decision function that best distinguishes the images of the two groups is established. Once the decision function is learned, the model can be applied to predict the group assignment of a previously unseen test image [Pereira et al., 2009]. We used a linear SVM due to its simplicity, interpretability, and generally good performance [Pereira et al., 2009]. The linear classifier predicts the class of the test image based on a linear combination of the features.
In high‐dimensional problems, as is the case of the whole‐brain approach in our study, it is likely that only a subset of voxels will actually provide enough information for classification. In order to reduce the number of voxels used in the classification task, we used a simple filtering approach, the Fisher score, which allows choosing the voxels that are most discriminative between classes. The Fisher criterion score is a simple filtering approach, independent of the classifier that uses inter‐ and intra‐class variance to measure the power of the feature in discriminating the examples' labels [Chang and Lin, 2008]. The higher the F‐score the more discriminative is the feature. The voxels were ranked by discriminative power regarding the class and we selected the n voxels with highest discriminative power for classification. We analyzed images from the different age groups (children/adolescents or adults) and tissue type (GM or WM) separately, using 100, 1,000, 10,000, 50,000, 100,000, or 150,000 voxels. Importantly, the feature selection step was performed on the training set only.
To extract more general conclusions about the structural patterns distinguishing patients with NF1 from controls we used a Leave‐Two‐Out Cross‐Validation (L2OCV) method [Bray et al., 2009; Pereira et al., 2009]. In the L2OCV method, pairs of images (one from each class) are left out for testing the model in each L2OCV iteration. In our procedure, each image of one class was tested paired with each image of the other class and this procedure was repeated 100 times. Given the larger number of controls than participants with NF1 in our data set, a number of control participants (8 in the children/adolescents and 13 in the adults) were randomly excluded in each of the 100 L2OCV repetitions, ensuring that the classification was performed with the same number of participants for each clinical group. After constructing the training set of each cross‐validation fold, the F‐score ranking of all the voxels was computed only in this set. Then the highest n ranked voxels were selected to form the reduced training set and the same voxels were selected in the test set to test the examples in the test fold of the cross‐validation iteration. We repeated the L2OCV 100 times, an arbitrary value, to ensure that each participant would be tested several times and to build a sample of classification accuracy values to compare with the sample of accuracy values obtained with permuted labels. Each participant was included in L2OCV procedure 84 times, on average. We then computed the mean sensitivity (measuring correct identification of NF1 images), specificity (measuring correct identification of control images), and classification accuracy (the proportion of correct predictions) over all iterations of the procedure.
To evaluate the statistical significance of the classification results we needed to determine the probability of obtaining the observed classification results if the null hypothesis was true (that there is no information about the label in the data). Due to the large number of voxels and training/testing trials it was not computationally feasible to generate the null distribution by permutation for each cross‐validation fold in each repetition [Golland and Fischl, 2003; Pereira et al., 2009]. We rather repeated 200 times the same L2OCV procedure with a different shuffling of the labels each time. Over many repetitions, this yields a sample of classification results under the null hypothesis that there is no class information in the data [Pereira and Botvinick, 2011]. Normality of the distribution was assessed with the Shapiro–Wilk test (in all cases failed to reject the null hypothesis) and we used the t distribution to assess the significance of the classification results. We performed independent sample t‐tests to compare the mean classification performance values (accuracy, sensitivity and specificity) between the real tests distribution and the distribution from the tests with randomized labels to extract the corresponding P‐values [Kaplan and Meyer, 2012].
Discriminative Patterns
Linear SVM determines the weights of the classification model during training. The weights, one for each voxel, can be used to characterize the specific pattern of brain differences between NF1 and control brains. Although not in all cases, the absolute values of the weights often indicate which voxels are more important for classification [Cristianini and Shawe‐Taylor, 2000]. A higher absolute weight indicates that the voxel is considered more in determining the group assignment than another voxel with lower weight. This helps to identify the brain regions that strongly contributed, albeit not alone, to the discrimination between individuals with NF1 and controls in the GM or WM tissues. Note that the interpretation of the voxels' weights of a linear SVM classifier must be done with care, because although the sign and strength of the weight of a voxel may often imply the sign and strength of the correlation that a feature has with the labels, this does not always hold true, given the multivariate nature of the decision boundary [Pereira and Botvinick, 2011]. In principle, a positive/negative weight means higher/lower tissue relative volume in patients than in controls [Ecker et al., 2010], respectively, but it can really only be determined from univariate assessment (which our VBM approach provides). Thus, we examined how the combined relationships between the voxels contribute to the discrimination by analyzing the distribution of the voxels' weights, and used VBM results to inform the meaning of SVM patterns. We mapped the voxels' weights in the custom GM and WM templates for each group. As the cross‐validation method comprises the training of many SVM models, we averaged the voxels' weights across iterations to determine the spatial discriminative pattern of tissue relative volume differences. We also extracted the discriminative map from an SVM model trained with the entire datasets (children/adolescents and adults separately), which resulted in a pattern very similar to the weight vector of each L2OCV iteration. We discuss the discriminative maps in terms of the regions with strong weights, contained within the identified distributed patterns, and its probable functional significance to the cognitive phenotype of NF1. Note that these regions should be seen as components of a complex and distributed network of subtle differences in the NF1 brain.
Voxel‐Based Morphometry Analysis
Univariate VBM is a traditionally used univariate method to study whole brain morphometry, when no a priori hypotheses are available [Ashburner and Friston, 2000]. Thus, we performed VBM in the same GM/WM segments used in the classification analysis in order to gain information about voxel‐wise local volumetric differences that can help the interpretation of the SVM findings. We performed VBM analysis comparing NF1 and control brains in each group, adults and children/adolescents. First, we smoothed the modulated normalized GM and WM volume images with 3‐dimensional 8‐mm full‐width‐at‐half‐maximum (FWHM) isotropic Gaussian kernels. Smoothing in VBM is required to guarantee normality of the data for subsequent statistical analysis. We then applied the general linear model (GLM) at each voxel using SPM8 and VBM8 toolbox to investigate between‐group differences in GM or WM regional volume. Statistical inference of significant clusters of volumetric differences (NF1 vs. controls) in GM and WM was performed using a voxel‐wise two‐sample t‐test corrected for multiple comparisons. We used a voxel level P‐value < 0.05 corrected employing the family wise error (FWE) rate and accounting for non‐uniform smoothness of the data [Hayasaka et al., 2004; Worsley et al., 1999]. When there were no suprathreshold voxels, we lowered the statistical stringency to uncorrected P‐value < 0.001 to provide heuristic information on the meaning of the sign of SVM weight maps. To display the regions showing changes that might be involved in NF1, the output maps were overlaid onto custom GM and WM templates for each group.
RESULTS
Neuropsychological Characterization and Volumetric Measurements
Volumetric and neuropsychological measures for the included participants are reported in Table 2.
Children and adolescents with NF1 had significantly higher WM volumes than controls (P < 0.05). In contrast, GM volume and total intracranial volume were not significantly different from control levels. In adult participants, statistically significant volumetric differences were found in the global volume of GM (P < 0.05), WM (P < 0.001), and whole brain (P < 0.05). In both children/adolescents and adults, we found statistically significant differences between NF1 and controls in intelligence measures [full‐scale IQ (P < 0.05) for children and adolescents and Raven test score (P < 0.05) for adults] and visuospatial abilities (JLO test score: P < 0.05 in children/adolescents, P < 0.001 in adults). For all these measures the scores of the individuals with NF1 were lower than the scores of the control participants.
SVM Classification Performance
The results of the classification between individuals with NF1 and controls using GM and WM images are shown in Table 3. The best classification accuracy was obtained in the adult's group using the GM images. Here, individuals with NF1 were correctly classified in 93.60% of all cases (sensitivity = 91.65%; specificity = 95.56%). Slightly lower classification accuracies were observed on the basis of the WM images, which resulted in correct classification in 91.96% of adults with sensitivity of 89.64% and specificity of 94.28%. In the children/adolescents group, the classification accuracy using GM was 89.91% (sensitivity = 90.54%; specificity = 89.29%) and using WM was 87.10% (sensitivity = 88.99%; specificity = 85.22%). The performance (accuracy, sensitivity and specificity) of the classification was always significantly higher than that derived with the null distribution obtained with permuted labels (P < 0.001) for both age groups (children/adolescents and adults), number of voxels used and tissue types (GM and WM).
Table 3.
Results of support vector machine pattern classification of whole‐brain grey and white matter images of NF1 patients and Controls
| Number of voxels | Grey matter | White matter | ||||
|---|---|---|---|---|---|---|
| Accuracy (%) | Sensitivity (%)a | Specificity (%)a | Accuracy (%) | Sensitivity (%)a | Specificity (%)a | |
| Children | ||||||
| 100 | 83.12 | 81.41 | 84.82 | 78.60 | 75.75 | 81.46 |
| 1,000 | 86.40 | 85.29 | 87.51 | 81.18 | 77.92 | 84.44 |
| 10,000 | 88.70 | 89.61 | 87.79 | 84.75 | 83.86 | 85.63 |
| 50,000 | 88.35 | 90.36 | 86.34 | 86.32 | 87.13 | 85.51 |
| 100,000 | 89.38 | 90.62 | 88.13 | 86.94 | 88.19 | 85.68 |
| 150,000 | 89.91 | 90.54 | 89.29 | 87.10 | 88.99 | 85.22 |
| Adults | ||||||
| 100 | 78.62 | 76.80 | 80.44 | 79.56 | 80.17 | 78.95 |
| 1,000 | 86.50 | 84.80 | 88.19 | 86.52 | 87.23 | 85.81 |
| 10,000 | 90.15 | 86.89 | 93.41 | 91.69 | 90.92 | 92.47 |
| 50,000 | 92.35 | 89.36 | 95.33 | 92.09 | 89.96 | 94.22 |
| 100,000 | 93.20 | 90.82 | 95.58 | 92.07 | 89.69 | 94.44 |
| 150,000 | 93.60 | 91.65 | 95.56 | 91.96 | 89.64 | 94.28 |
Results of SVM classification using different number of voxels containing grey matter or white matter, selected from the whole brain for image analysis.
We consider a correctly identified NF1 case a true positive.
Multivariate SVM Classifier Weight Vectors
The WM discriminative maps showing spatially distributed patterns of differences between NF1 and controls are displayed in Figures 1 and 2 for children/adolescents and adults, respectively, while the GM discriminative maps are shown in Figures 3 and 4, for children/adolescents and adults, respectively. Due to the multivariate nature of SVM, the discriminative maps should be interpreted as spatially distributed patterns of local volumetric differences. Nonetheless, we have highlighted the regions that contributed with higher absolute weights to the discrimination between patients with NF1 and controls. Despite the fact that the relationship between the sign and strength of the voxels' weights and the increase/decrease in relative volume is not necessarily direct, it usually holds true [Pereira and Botvinick, 2011]. A positive weight in the discriminative map suggests relatively higher tissue volume in patients than in controls and a negative weight means relatively lower tissue volume in patients. As changes in relative volumes can only be definitely determined by univariate assessment, we also employed the VBM approach. We found an overall agreement between the directions of the local volumetric differences in VBM and the voxels' weights in SVM discriminative maps [e.g. in the thalamus, with VBM we observed higher relative volume in NF1 images (see Supporting Information Figs. 1 and 2) and positive weights in the same voxels of the SVM multivariate maps (see Figs. 3 and 4)]. Notably, the discriminative patterns further covered regions with differences that were not discernible at voxel level using P corrected VBM analysis. Still, the multivariate discriminative maps should always be considered as so rather than claiming one single region to influence a phenotypic trait.
Figure 1.
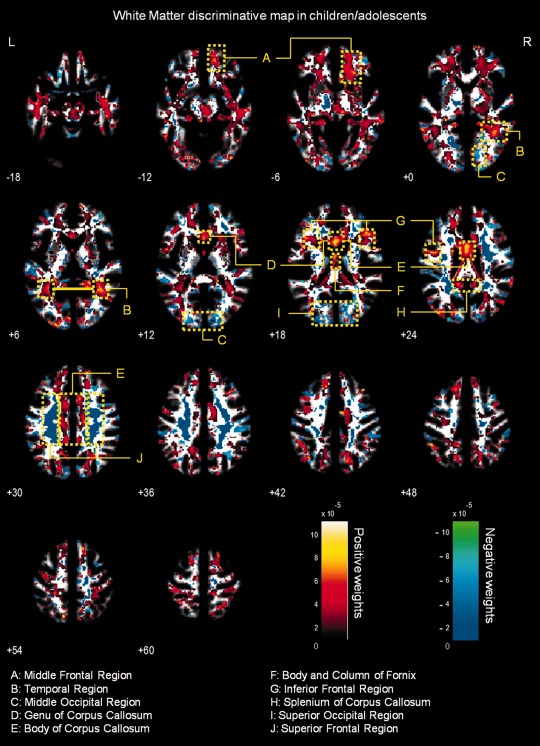
Whole‐brain representation of the discriminative map for WM relative volume classification in children/adolescents. The weight vectors are displayed from a leave‐two‐out linear SVM using 150,000 voxels. Positively weighted voxels in NF1 vs. controls are displayed in red/yellow, while negatively weighted voxels are displayed in blue/green. Regions with relatively stronger classification absolute weights are identified. The map is overlayed on the group WM template from all subjects. The z‐coordinate for each axial slice in the standard MNI space is given.
Figure 2.
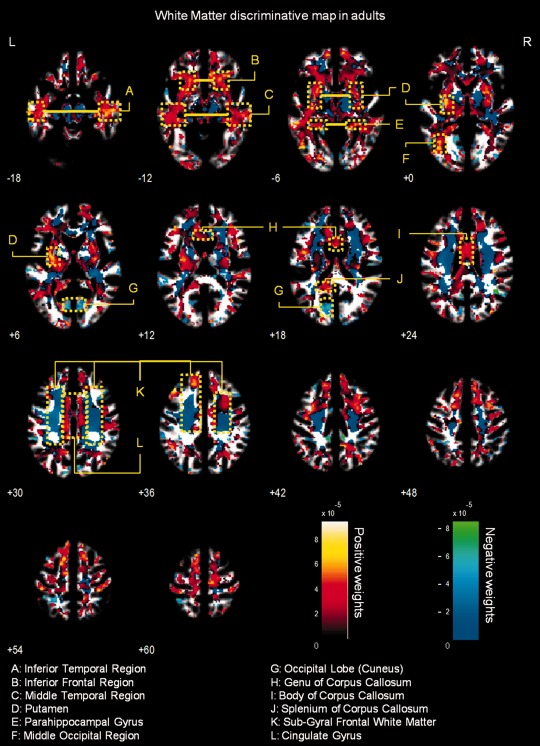
Whole‐brain representation of the discriminative map for WM relative volume classification in adults. The weight vectors are displayed from a leave‐two‐out linear SVM using 150,000 voxels. Positively weighted voxels in NF1 vs. controls are displayed in red/yellow, while negatively weighted voxels are displayed in blue/green. Regions with relatively stronger classification absolute weights are identified. The map is overlayed on the group WM template from all subjects. The z‐coordinate for each axial slice in the standard MNI space is given.
Figure 3.
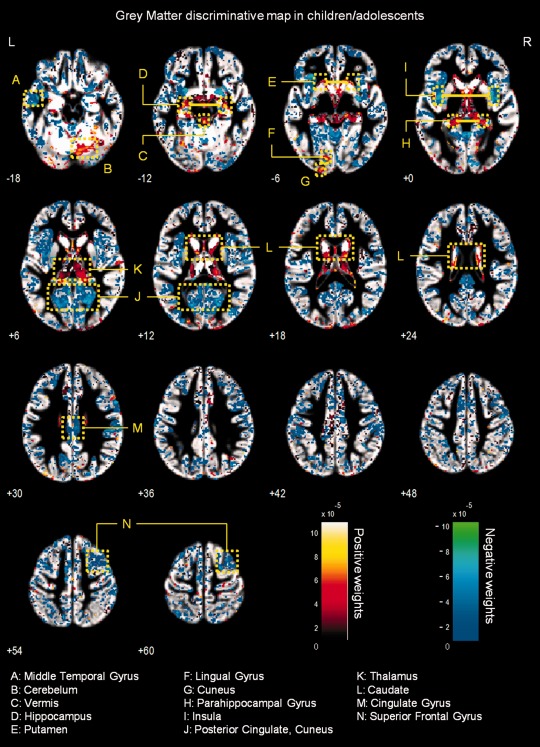
Whole‐brain representation of the discriminative map for GM relative volume classification in children/adolescents. The weight vectors are displayed from a leave‐two‐out linear SVM using 150,000 voxels. Positively weighted voxels in NF1 vs. controls are displayed in red/yellow, while negatively weighted voxels are displayed in blue/green. Regions with relatively stronger classification absolute weights are identified. The map is overlayed on the group GM template from all subjects. The z‐coordinate for each axial slice in the standard MNI space is given.
Figure 4.
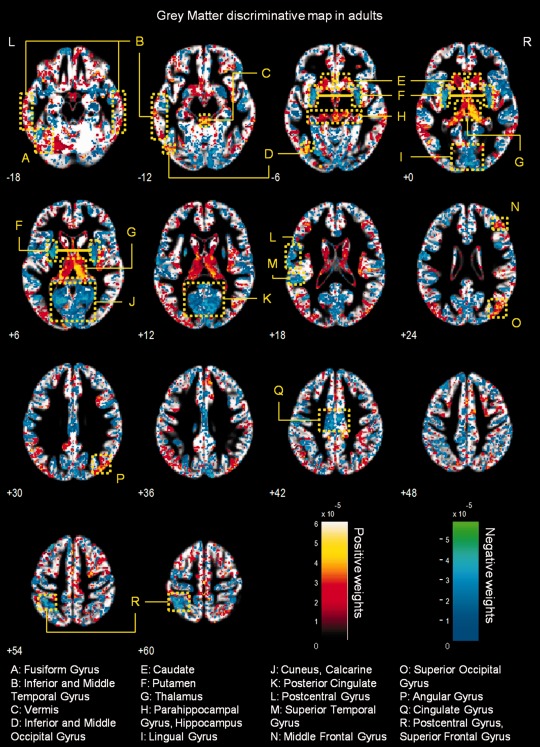
Whole‐brain representation of the discriminative map for GM relative volume classification in adults. The weight vectors are displayed from a leave‐two‐out linear SVM using 150,000 GM voxels. Positively weighted voxels in NF1 vs. controls are displayed in red/yellow, while negatively weighted voxels are displayed in blue/green. Regions with relatively stronger classification absolute weights are identified. The map is overlayed on the group GM template from all subjects. The z‐coordinate for each axial slice in the standard MNI space is given.
White matter multivariate differences
The WM discriminative maps, for both adults and children/adolescents, show a similar pattern displaying a predominance of positive weights in participants with NF1 relative to controls (see Figs. 1 and 2). Differences were mainly observed in anterior frontal and temporal WM regions and the corpus callosum. Few voxels showed negative weights in WM, and those identified had small absolute values.
Grey matter multivariate differences
Discriminative GM patterns contain positive and negative discriminant weights, in both children/adolescents and adults (see Figs. 3 and 4). Voxels with strongly positive weights were mainly found in subcortical structures: the caudate nuclei, the hippocampus, the vermis, and the thalamus. Other strongly positive weighted voxels contributing to the discrimination pattern were located in temporal and occipital regions. In NF1 children/adolescents but not in adults, we could also observe positively weighted GM voxels in the cerebellum. On the other hand, negatively weighted GM voxels were found in both NF1 clinical groups mostly in the occipital cortex, including the calcarine fissure, the cuneus, and the lingual gyrus. We also found voxels with negative weights in other cortical regions such as the temporal gyrus and the cingulate cortex. In NF1 adults (but not in the younger age group), we observed GM voxels with negative weights in the putamen, whilst only in children/adolescents we observed negative weights in GM voxels located bilaterally in the insula.
Univariate VBM Analyses
Using the VBM approach, we found similar maps of differences in WM relative volume as observed using the SVM approach. The statistical maps of VBM analysis of WM with significant tissue relative volume differences, corrected for multiple comparisons, are reported in Figures 5 and 6 for children/adolescents and adults, respectively. In Supporting Information, Table SI provides the identification of the regions that are significantly different between the groups. We did not find any statistically significant differences in univariate VBM analysis of GM tissue, when using corrected thresholds (data not shown). To compare the results obtained with VBM and SVM, we used VBM threshold at P < 0.001 (uncorrected). Interestingly, these VBM maps highlighted group differences in regions also identified in the SVM weights' maps. The output maps are presented in Supporting Information Figures 1 and 2 for children/adolescents and adults, respectively.
Figure 5.
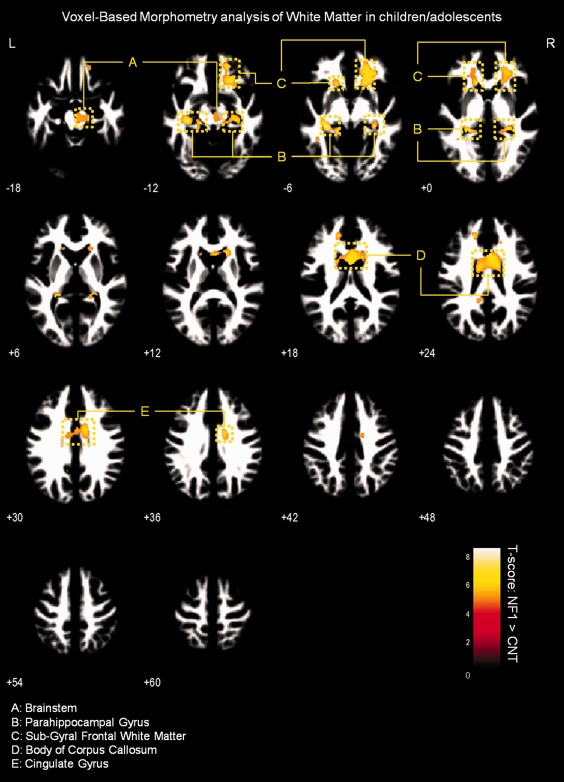
Results of VBM analysis of WM in children/adolescents. Results are presented at a voxel‐level P‐value < 0.05, corrected for FWE and non‐stationary smoothness. Voxels showing significant WM relative volume differences are overlaid on group‐customized WM template.
Figure 6.
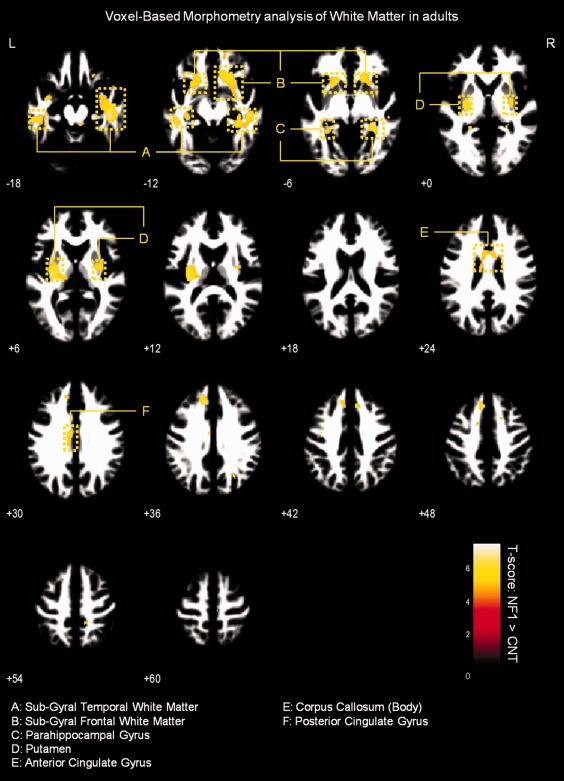
Results of VBM analysis of WM in adults. Results are presented at a voxel‐level P‐value < 0.05, corrected for FWE and non‐stationary smoothness. Voxels showing significant WM relative volume differences are overlaid on group‐customized WM template.
White matter VBM results
We observed significantly higher WM relative volume in individuals with NF1 compared to controls in the regions adjacent to the lingual gyrus, cingulate gyrus, temporal gyrus, parahippocampal gyrus, and in the body of the corpus callosum (voxel‐level FWE corrected P < 0.05). We also found WM relative volume increases in brainstem of NF1 children/adolescents and near the putamen of NF1 adults (see Figs. 5 and 6 and Supporting Information Table SI).
Grey Matter VBM results
No significant between‐group (NF1 vs. Control) differences in GM relative volume of either children/adolescents or adults were observed when correcting for multiple comparisons at the voxel‐level (FWE P < 0.05). Nonetheless, the exploratory analysis of uncorrected univariate VBM maps of GM also revealed regions, mainly with GM relative volume deficits in the NF1 groups, covered by the multivariate SVM discriminative maps. These were the vermis, hippocampus, lingual gyrus, and the cuneus. Lower relative GM volume was also detectable bilaterally in the insula of children/adolescents with NF1. On the other hand, in accordance with the multivariate analysis, the thalamus was identified with higher GM relative volume in NF1 children/adolescents and adult participants (see Supporting Information Figs. 1 and 2).
Correlation of SVM output with neuropsychological measures
We computed the correlation of the SVM test margins with Full‐scale IQ (for children/adolescents), Raven matrices scores (an indication of non‐verbal intelligence for adults), and JLO scores (a measure of visuospatial performance consistently impaired in patients with NF1). For all these measures, we found univariate significant differences between clinical groups (NF1 vs. Controls, P < 0.05). We computed the correlations including the examples from both classes (NF1 and Controls) and we did not find any significant correlation between the neuropsychological measurements and the output of the SVM (distance of each example to the separating boundary).
DISCUSSION
This study is, to our knowledge, the first to use multivariate pattern analysis of whole‐brain GM and WM volumetric segments of structural MRI scans in individuals with NF1. Linear SVM were able to correctly discriminate between NF1 and control images with high accuracy based on T 1‐weighted anatomical images in two different populations, children/adolescents and adults. The high classification performance demonstrates that the SVM is a powerful method to predict group separation. Additionally, it confirms the presence of neuroanatomical differences between individuals with NF1 and controls.
Macrocephaly, associated with bigger brain global volumes, is commonly found in individuals with NF1 [Payne et al., 2010]. Indeed, in our cohorts, we also found differences in global brain volume measures (Table 2). However, our aim was to determine whether patients with NF1 could be discriminated based on local relative volume anomalies that could be related to the cognitive profile of these patients. Thus, we discarded global brain volume differences between the groups, by only applying nonlinear modulation to the data. Note that, in the preprocessing of the data, the normalization applied to the whole brain does not introduce a biased intensification of the local effects [Ashburner, 2009; Ashburner and Friston, 2000]. Thus, global volume differences are not a likely explanation for the distributed discriminative patterns found (in fact GM global differences are even absent in our children cohort) suggesting that different brain regions are affected by the disease in different ways.
Our data driven analysis allowed the identification of abnormal structural patterns that may be relevant to understanding disease mechanisms. In our sample, the traditional VBM analysis detected no significant between‐group differences in GM relative volumes after correcting for multiple comparisons. Furthermore, only few significant differences were identified in WM maps. This supports the idea that the univariate nature of the VBM method makes subtle or distributed differences hard to find [Ecker et al., 2010; Marzelli et al., 2011], possibly as a consequence of the necessary smoothing step in VBM. This step ensures the normality of the data for statistical analysis and it inevitably produces loss of spatial resolution. Nonetheless, the spatial maps of WM as well as the uncorrected exploratory analysis of GM were in agreement with the multivariate discrimination results.
Unlike VBM, which considers each voxel as a spatially independent unit, SVM is a multivariate technique. Due to the method inputs and preprocessing steps, the discriminative maps should be interpreted as spatially distributed networks of local relative volume differences rather than strong claims about effects in individual regions. The SVM discriminative map contains information about the relative importance of each voxel to the classification but its interpretation needs to be done with caution. We do not claim any specific region to be individually responsible for a certain phenotypic feature but we note its contribution to the multi‐part neuroanatomical pattern. Voxels with positive/negative high weights in the multivariate analysis are likely to present true higher/lower relative tissue volume in NF1 group but other scenarios are also possible. The combination with VBM analysis is thus useful to confirm whether local volumetric differences are consistent with the discriminative pattern.
It is worth to note, as a limitation of this study, that, due to the nature of the MR signal, besides pure volumetric differences our results could also reflect differences in tissue content that might give rise to differences in signal T1 properties. These differences could lead the segmentation algorithm to interpret signal intensity differences as volumetric differences and miss‐assign these as more/less tissue volume. Thus, the differences in discriminative maps could also arise, at least in part, from different water content in GM and WM, different myelin composition or myelin thickness, different neural or neuropil density, or different neural size between groups. Recent diffusion weighted imaging and diffusion tensor imaging (DWI/DTI) data in NF1 is consistent with this notion [Wignall et al., 2010; Zamboni et al., 2007]. Further investigation is needed to elucidate this issue. Nevertheless, even if the discriminative voxels do not represent true differences in relative volume they still point out regions with structural alterations and provide starting points for localized investigations (e.g., ROI ‐ Region of Interest ‐ analysis). Accordingly, we discuss the plausible influence of these regions, as constituents of a complex network, to the phenotypic profile in NF1.
Distributed Neuroanatomical Networks Discriminating NF1 Brains
Regarding WM, discriminative maps show mainly positively weighted voxels discriminating between NF1 and controls mostly located in frontal, temporal, and occipital regions and were detected also with voxel‐wise VBM, suggesting that these differences are more pronounced than GM anomalies. Our data span regions such as corpus callosum (genu, body, and splenium) in children/adolescents which corroborates other morphometric findings in children with NF1 [Cutting et al., 2000; Dubovsky et al., 2001; Kayl et al., 2000; Moore et al., 2000; Steen et al., 2001]. Also our VBM results of higher local relative WM volume are in accordance with the sign of the weights attributed by SVM to these regions. Increased callosal volume has been found to be related to low IQ, impaired visuospatial, and motor skills and learning problems in NF1 children [Moore et al., 2000; Pride et al., 2010]. In line with our results, it is also known that besides being larger than normal, the NF1 corpus callosum has abnormal microstructure as measured by DTI, most likely compromising callosal function [Wignall et al., 2010; Zamboni et al., 2007]. Our data revealed abnormal structure in other strongly weighted WM voxels, such as premotor and frontal WM regions. We further detected increased relative volume of frontal WM in NF1 brain with VBM. These findings are also in accordance with other morphometric NF1 studies reporting significant increases in cerebral WM volume, predominantly in frontal regions of the brain [Cutting et al., 2000; Greenwood et al., 2005]. The frontal cortex is involved in executive function, abnormal in patients with NF1 [Levine et al., 2006; Roy et al., 2010]. Thus, this deficit might be related to abnormal connectivity in the frontal lobe. Noteworthy, frontal WM was also found to have abnormal microstructure in a previous DTI study [Zamboni et al., 2007]. The concordance of our findings with previous reports using different approaches, suggest consistent frontal WM structural anomalies in NF1.
We could also identify positive and negative weighted WM discriminative voxels in the temporal lobe, in a region crossed by the superior longitudinal fasciculus, a major associative intrahemispheric fiber tract that connects parieto‐temporal association areas with the frontal lobe and vice versa [Makris et al., 2005]. Structural abnormalities in this pathway suggest associative memory deficits. In addition, we observed positively weighted WM voxels in a region of the temporal lobe that might comprise part of the posterior thalamic radiation, a projection fiber from the posterior part of the thalamus to the occipital cortex, which also includes the optic radiation [Cheon et al., 2011; Wakana et al., 2004]. These results are consistent with visual and/or memory deficits found in individuals with NF1.
We have identified more complex discriminative patterns of differences in GM voxels. As discussed before, their interpretation benefits from joint consideration of other methods. However, in our study we did not find statistically significant voxel‐wise relative volume differences with VBM analysis of GM relative volume. Nevertheless, with pattern recognition we have identified regions, as part of a complex network important for patient classification, which could potentially underlie the cognitive problems that are commonly found in NF1, such as deficient visuospatial function, memory, executive function, and motor function.
Visuospatial deficits are considered the hallmark of the cognitive profile of NF1 [Levine et al., 2006]. Notably, we found between‐group differences in discriminative patterns including the visual cortex; the parahippocampal gyrus, involved in visual navigation [Epstein, 2008]; the angular gyrus, involved in high‐level vision [Seghier et al., 2010]; and the pulvinar nucleus of the thalamus, which was also detected with uncorrected exploratory VBM analysis, involved in visual attention [Smith et al., 2009]. Concurrent alteration of these structures might therefore underlie the visuospatial deficits observed in the NF1 population. Memory deficits in NF1+/− mice are related to abnormal physiology of the hippocampus [Costa and Silva, 2003; Costa et al., 2002; Cui et al., 2008; Donarum et al., 2006; Park et al., 2009]. Notably, in our study we observed positively weighted GM discriminative voxels in this structure in participants with NF1. In addition, VBM also revealed (below corrected statistical significance) univariate local differences in this region with patients with NF1 showing higher relative GM volume than controls. As the hippocampus plays an important role in visual and spatial learning and memory function [Morgado‐Bernal, 2011], affected in patients with NF1 [Levine et al., 2006], our results suggest a structural hippocampal abnormality underlying these deficits. Thus, our findings of abnormal structure in the hippocampus are suggestive of its non‐optimal functioning and implicate for the first time this structure in the human NF1, in line with the findings from the animal model.
Our findings of generalized discriminant contribution of voxels in the thalamus are in line with previous studies reporting abnormal thalamic function and metabolic content in patients with NF1 [Barbier et al., 2011; Kaplan et al., 1997; Wang et al., 2000]. The thalamus, acting as a relay station, receives sensory information and projects to several areas of the cortex, including the frontal cortex (involved in executive function) and the visual cortex. Indeed, global thalamic dysfunction can lead to memory problems, sensory perception deficits, and executive dysfunction, all being features of NF1 [Barbier et al., 2011; Hyman et al., 2007; Levine et al., 2006; Moore et al., 1996; Roy et al., 2010]. Furthermore, we observed between‐group differences in the caudate nucleus and the putamen, the former involved in goal oriented and executive function and the latter in motor planning and implementation of automatic routines. Interestingly, reduced striatal dopamine levels have been linked to attentional deficits in an NF1 mouse model [Brown et al., 2010]. In addition, a human study reported clusters of dysplastic cells in the caudate nucleus and putamen of individuals with NF1 [Yokota et al., 2008]. Our results further support the hypothesis that these structures have abnormal constitution and might contribute to the executive and motor dysfunction in NF1. Also parts of the cerebellum were observed to contribute to the discriminative pattern of GM differences in the children with NF1 involved in this study. Our findings thus suggest that the basis of the motor deficits in NF1 population could be an abnormal structural network including this brain region [Puttemans et al., 2005]. However, the interpretation of such complex patterns in terms of region‐specific differences must be cautious, particularly regarding heterogeneous samples.
In the present study, we did not find a significant correlation of cognitive impairment measures with the SVM output (distance from each example and the separating boundary). On the one hand, this lack of correlation might reflect the fact that neuropsychological measures are generally associated with large coefficients of variation. Furthermore, the association of brain tissue volumes with IQ is still controversial, even in studies with large samples [see e.g. Brain Development Cooperative Group, 2012]. On the other hand, cognitive deficits in patients with NF1 might be more closely related with abnormal function than with structural anomalies. Finally, it should be taken into account that care must be taken when using the test margin to correlate with behavioral variables given that the test margin depends crucially on the training margin [Marquand et al., 2010]. The SVM algorithm employed in our study was based on the binary separation between patients with NF1 and the control group. In this type of approach, the test margins therefore ignore specific cognitive profiles [Ecker et al., 2010].
CONCLUSION
This work showed that SVMs, in the context of a multivariate pattern analysis of whole‐brain T 1‐weighted structural images, can be used to distinguish between individuals diagnosed with NF1 and control participants on the basis of neuroanatomical differences. Furthermore, this data‐driven analysis indicated biologically plausible, spatially distributed networks of brain regions with abnormal structure in individuals with NF1 providing important clues for the pathophysiology of the cognitive phenotype associated with this disorder. A causal link still remains however to be established as it could also be that abnormal function of these areas lead to abnormal development of structure and not the other way around.
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information
Supporting Information Tables.
ACKNOWLEDGMENTS
Authors thank the NF1 participants and their families, as well as all the control subjects that participated in this study. Authors also thank Carlos Ferreira and João Marques for help with MRI scanning.
REFERENCES
- Ashburner J (2007): A fast diffeomorphic image registration algorithm. NeuroImage 38:95–113. [DOI] [PubMed] [Google Scholar]
- Ashburner J (2009): Computational anatomy with the SPM software. Magn Reson Imaging 27:1163–1174. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ (2000): Voxel‐based morphometry—The methods. NeuroImage 11:805–821. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ (2005): Unified segmentation. NeuroImage 26:839–851. [DOI] [PubMed] [Google Scholar]
- Barbier C, Chabernaud C, Barantin L, Bertrand P, Sembely C, Sirinelli D, Castelnau P, Cottier JP (2011): Proton MR spectroscopic imaging of basal ganglia and thalamus in neurofibromatosis type 1: Correlation with T2 hyperintensities. Neuroradiology 53:141–148. [DOI] [PubMed] [Google Scholar]
- Benton AL, Varney NR, Hamsher KD (1978): Visuospatial judgment. A clinical test. Arch Neurol 35:364–367. [DOI] [PubMed] [Google Scholar]
- Billingsley RL, Schrimsher GW, Jackson EF, Slopis JM, Moore BD III (2002): Significance of planum temporale and planum parietale morphologic features in neurofibromatosis type 1. Arch Neurol 59:616–622. [DOI] [PubMed] [Google Scholar]
- Brain Development Cooperative Group (2012): Total and regional brain volumes in a population‐based normative sample from 4 to 18 years: The NIH MRI study of normal brain development. Cereb Cortex:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Chang C, Hoeft F (2009): Applications of multivariate pattern classification analyses in developmental neuroimaging of healthy and clinical populations. Front Hum Neurosci 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Emnett RJ, White CR, Yuede CM, Conyers SB, O'Malley KL, Wozniak DF, Gutmann DH (2010): Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis‐1 mutant mice. Hum Mol Genet 19:4515–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ (2004): A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas‐based head size normalization: Reliability and validation against manual measurement of total intracranial volume. Neuroimage 23:724–738. [DOI] [PubMed] [Google Scholar]
- Chang YW, Lin CJ (2008): Feature ranking using linear SVM InJMLR: Workshop and Conference Proceedings,Hong‐Kong,3:53–64. [Google Scholar]
- Cheon KA, Kim YS, Oh SH, Park SY, Yoon HW, Herrington J, Nair A, Koh YJ, Jang DP, Kim YB, Leventhal BL, Cho ZH, Castellanos FX, Schultz RT (2011): Involvement of the anterior thalamic radiation in boys with high functioning autism spectrum disorders: A Diffusion Tensor Imaging study. Brain Res 1417:77–86. [DOI] [PubMed] [Google Scholar]
- Clements‐Stephens AM, Rimrodt SL, Gaur P, Cutting LE (2008): Visuospatial processing in children with neurofibromatosis type 1. Neuropsychologia 46:690–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Silva AJ (2003): Mouse models of neurofibromatosis type I: Bridging the GAP. Trends Mol Med 9:19–23. [DOI] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ (2002): Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415:526–530. [DOI] [PubMed] [Google Scholar]
- Cristianini N, Shawe‐Taylor J (2000): An introduction to Support Vector Machines and Other Kernel‐Based Learning Methods.Cambridge: Cambridge University Press. [Google Scholar]
- Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, Parada LF, Mody I, Silva AJ (2008): Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 135:549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting LE, Koth CW, Burnette CP, Abrams MT, Kaufmann WE, Denckla MB (2000): Megalencephaly in NF1: Predominantly white matter contribution and mitigation by ADHD. J Child Neurol 15:157–160. [DOI] [PubMed] [Google Scholar]
- Cutting LE, Cooper KL, Koth CW, Mostofsky SH, Kates WR, Denckla MB, Kaufmann WE (2002): Megalencephaly in NF1: Predominantly white matter contribution and mitigation by ADHD. Neurology 59:1388–1394. [DOI] [PubMed] [Google Scholar]
- Daston MM, Ratner N (1992): Neurofibromin, a predominantly neuronal GTPase activating protein in the adult, is ubiquitously expressed during development. Dev Dyn 195:216–226. [DOI] [PubMed] [Google Scholar]
- De Winter AE, Moore BD III, Slopis JM, Ater JL, Copeland DR (1999): Brain tumors in children with neurofibromatosis: Additional neuropsychological morbidity? Neuro Oncol 1:275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donarum EA, Halperin RF, Stephan DA, Narayanan V (2006): Cognitive dysfunction in NF1 knock‐out mice may result from altered vesicular trafficking of APP/DRD3 complex. BMC Neurosci 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovsky EC, Booth TN, Vezina G, Samango‐Sprouse CA, Palmer KM, Brasseux CO (2001): MR imaging of the corpus callosum in pediatric patients with neurofibromatosis type 1. AJNR Am J Neuroradiol 22:190–195. [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Rocha‐Rego V, Johnston P, Mourão‐Miranda J, Marquand A, Daly EM, Brammer MJ, Murphy C, Murphy DG, MRC AIMS Consortium (2010): Investigating the predictive value of whole‐brain structural MR scans in autism: A pattern classification approach. NeuroImage 49:44–56. [DOI] [PubMed] [Google Scholar]
- Epstein RA (2008): Parahippocampal and retrosplenial contributions to human spatial navigation. Trends Cogn Sci 12:388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golland P, Fischl B (2003): Permutation tests for classification: Towards statistical significance in image‐based studies. Lect Notes Comput Sci 2732:330–341. [DOI] [PubMed] [Google Scholar]
- Greenwood RS, Tupler LA, Whitt JK, Buu A, Dombeck CB, Harp AG, Payne ME, Eastwood JD, Krishnan KRR, MacFall JR (2005): Brain morphometry, T2‐weighted hyperintensities, and IQ in children with neurofibromatosis type 1. Arch Neurol 62:1904–1908. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Geist RT, Wright DE, Snider WD (1995): Expression of the neurofibromatosis 1 (NF1) isoforms in developing and adult rat tissues. Cell Growth Differ 6:315–323. [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE (2004): Nonstationary cluster‐size inference with random field and permutation methods. NeuroImage 22:676–687. [DOI] [PubMed] [Google Scholar]
- Hoeft F, Lightbody AA, Hazlett H, Patnaik S, Piven J, Reiss AL (2008): Morphometric spatial patterns differentiate fragile X syndrome, typical developing and developmentally delayed boys of ages one to three. Arch Gen Psychiatry 65:1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SL, Shores EA, North KN (2005): The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology 65:1037–1044. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Shores EA, North KN (2006): Learning disabilities in children with neurofibromatosis type 1: Subtypes, cognitive profile, and attention‐deficit‐hyperactivity disorder. Dev Med Child Neurol 48:973–977. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Gill DS, Shores EA, Steinberg A, North KN (2007): T2 hyperintensities in children with neurofibromatosis type 1 and their relationship to cognitive functioning. J Neurol Neurosurg Psychiatry 78:1088–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J, Meyer K (2012): Multivariate pattern analysis reveals common neural patterns across individuals during touch observation. Neuroimage 60:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan AM, Chen K, Lawson MA, Wodrich DL, Bonstelle CT, Reiman EM (1997): Positron emission tomography in children with neurofibromatosis‐1. J Child Neurol 12:499–506. [DOI] [PubMed] [Google Scholar]
- Kayl AE, Moore BD III (2000): Behavioral phenotype of neurofibromatosis, type 1. Ment Retard Dev Disabil 6:117–124. [DOI] [PubMed] [Google Scholar]
- Kayl AE, Moore BD III, Slopis JM, Jackson EF, Leeds NE (2000): Quantitative morphology of the corpus callosum in children with neurofibromatosis and attention‐deficit hyperactivity disorder. J Child Neurol 15:90–96. [DOI] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR Jr, Ashburner J, Frackowiak RSJ (2008): Automatic classification of MR scans in Alzheimer's disease. Brain 131:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Yeh TH, Emnett RJ, White CR, Gutmann DH (2010): Neurofibromatosis‐1 regulates neuroglial progenitor proliferation and glial differentiation in a brain region‐specific manner. Genes Dev 24:2317–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TM, Materek A, Abel J, O'Donnell M, Cutting LE (2006): Cognitive profile of neurofibromatosis type 1. Semin Pediatr Neurol 13:8–20. [DOI] [PubMed] [Google Scholar]
- Makris N, Kennedy DN, McInerney S, Sorensen AG, Wang R, Caviness VS, Pandya DN (2005): Segmentation of subcomponents within the superior longitudinal fascicle in humans: A quantitative, in vivo, DT‐MRI study. Cereb Cortex 15:854–869. [DOI] [PubMed] [Google Scholar]
- Marquand A, Howard M, Brammer M, Chu C, Coen S, Mourão‐Miranda J (2010): Quantitative prediction of subjective pain intensity from whole‐brain fMRI data using Gaussian processes. Neuroimage 49:2178–2189. [DOI] [PubMed] [Google Scholar]
- Marzelli MJ, Hoeft F, Hong DS, Reiss AL (2011): Neuroanatomical spatial patterns in Turner syndrome. NeuroImage 55:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietchen D, Gaser C (2009): Computational morphometry for detecting changes in brain structure due to development, aging, learning, disease and evolution. Front Neuroinform 3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BD III, Ater JL, Needle MN, Slopis J, Copeland DR (1994): Neuropsychological profile of children with neurofibromatosis, brain tumor, or both. J Child Neurol 9:368–377. [DOI] [PubMed] [Google Scholar]
- Moore BD III, Slopis JM, Schomer D, Jackson EF, Levy BM (1996): Neuropsychological significance of areas of high signal intensity on brain MRIs of children with neurofibromatosis. Neurology 46:1660–1668. [DOI] [PubMed] [Google Scholar]
- Moore BD III, Slopis JM, Jackson EF, De Winter AE, Leeds NE (2000): Brain volume in children with neurofibromatosis type 1: Relation to neuropsychological status. Neurology 54:914–920. [DOI] [PubMed] [Google Scholar]
- Morgado‐Bernal I (2011): Learning and memory consolidation: Linking molecular and behavioral data. Neuroscience 176:12–19. [DOI] [PubMed] [Google Scholar]
- Neurofibromatosis .Conference statement. National Institutes of Health Consensus Development Conference (1988):Arch Neurol 45:575–578. [PubMed] [Google Scholar]
- North KN (2000): Neurofibromatosis type 1. Am J Med Genet 97:119–127. [DOI] [PubMed] [Google Scholar]
- Park CS, Zhong L, Tang SJ (2009): Aberrant expression of synaptic plasticity‐related genes in the NF1+/− mouse hippocampus. J Neurosci Res 87:3107–3119. [DOI] [PubMed] [Google Scholar]
- Payne JM, Moharir MD, Webster R, North KN (2010): Brain structure and function in neurofibromatosis type 1: Current concepts and future directions. J Neurol Neurosurg Psychiatry 81:304–309. [DOI] [PubMed] [Google Scholar]
- Pereira F, Botvinick M (2011): Information mapping with pattern classifiers: A comparative study. Neuroimage 56:476–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Mitchell T, Botvinick M (2009): Machine learning classifiers and fMRI: A tutorial overview. NeuroImage 45:S199–S209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride N, Payne JM, Webster R, Shores EA, Rae C, North KN (2010): Corpus callosum morphology and its relationship to cognitive function in neurofibromatosis type 1. J Child Neurol 25:834–841. [DOI] [PubMed] [Google Scholar]
- Puttemans V, Wenderoth N, Swinnen SP (2005): Changes in brain activation during the acquisition of a multifrequency bimanual coordination task: From the cognitive stage to advanced levels of automaticity. J Neurosci 25:4270–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JC (1947): Raven's Advanced Progressive Matrices APM Set I.Oxford:Oxford Psychologists Press. [Google Scholar]
- Ribeiro MJ, Violante IR, Bernardino I, Ramos F, Saraiva J, Reviriego P, Upadhyaya M, Silva ED, Castelo‐Branco M (2012): Abnormal achromatic and chromatic contrast sensitivity in neurofibromatosis type 1. Invest Ophthalmol Vis Sci 53:287–293. [DOI] [PubMed] [Google Scholar]
- Roy A, Roulin JL, Charbonnier V, Allain P, Fasotti L, Barbarot S, Stalder JF, Terrien A, Gall DL (2010): Executive dysfunction in children with neurofibromatosis type 1: A study of action planning. J Int Neuropsychol Soc 16:1056–1063. [DOI] [PubMed] [Google Scholar]
- Schrimsher GW, Billingsley RL, Slopis JM, Moore BD III (2003): Visual‐spatial performance deficits in children with neurofibromatosis type‐1. Am J Med Genet A 120:326–330. [DOI] [PubMed] [Google Scholar]
- Seghier ML, Fagan E, Price CJ (2010): Functional subdivisions in the left angular gyrus where the semantic system meets and diverges from the default network. J Neurosci 30:16809–16817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AT, Cotton PL, Bruno A, Moutsiana C (2009): Dissociating vision and visual attention in the human pulvinar. J Neurophysiol 101:917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen RG, Taylor JS, Langston JW, Glass JO, Brewer VR, Reddick WE, Mages R, Pivnick EK (2001): Prospective evaluation of the brain in asymptomatic children with neurofibromatosis type 1: Relationship of macrocephaly to T1 relaxation changes and structural brain abnormalities. AJNR Am J Neuroradiol 22:810–817. [PMC free article] [PubMed] [Google Scholar]
- Tonsgard JH (2006): Clinical manifestations and management of neurofibromatosis type 1. Semin Pediatr Neurol 13:2–7. [DOI] [PubMed] [Google Scholar]
- Wakana S, Jiang H, van Zijl PCM, Mori S (2004): Fiber tract‐based atlas of human white matter anatomy. Radiology 230:77–87. [DOI] [PubMed] [Google Scholar]
- Wang PY, Kaufmann WE, Koth CW, Denckla MB, Barker PB (2000): Thalamic involvement in neurofibromatosis type 1: Evaluation with proton magnetic resonance spectroscopic imaging. Ann Neurol 47:477–484. [PubMed] [Google Scholar]
- Wechsler D (2003): Escala de Inteligência de Wechsler para Crianças—Terceira Edição (WISC‐III): Manual.Lisboa:CEGOC‐Tea. [Google Scholar]
- Wignall EL, Griffiths PD, Papadakis NG, Wilkinson ID, Wallis LI, Bandmann O, Cowell PEE, Hoggard N (2010): Corpus callosum morphology and microstructure assessed using structural MR imaging and diffusion tensor imaging: Initial findings in adults with neurofibromatosis type 1. AJNR Am J Neuroradiol 31:856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C (2008): Template‐O‐Matic: A toolbox for creating customized pediatric templates. NeuroImage 41:903–913. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC (1999): Detecting changes in nonisotropic images. Hum Brain Mapp 8:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota O, Tsuchiya K, Hayashi M, Kakita A, Ohwada K, Ishizu H, Takahashi H, Akiyama H (2008): Glial clusters and perineuronal glial satellitosis in the basal ganglia of neurofibromatosis type 1. Acta Neuropathol 116:57–66. [DOI] [PubMed] [Google Scholar]
- Zamboni SL, Loenneker T, Boltshauser E, Martin E, Il'yasov KA (2007): Contribution of diffusion tensor MR imaging in detecting cerebral microstructural changes in adults with neurofibromatosis type 1. AJNR Am J Neuroradiol 28:773–776. [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Harada T, Liu L, Lush ME, Guignard F, Harada C, Burns DK, Bajenaru ML, Gutmann DH, Parada LF (2005): Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development 132:5577–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information
Supporting Information Tables.


