Abstract
This study addresses how visual food cues are encoded in reward related brain areas and whether this encoding might provide information to differentiate between patients suffering from eating disorders [binge‐eating disorder (BED) and bulimia nervosa (BN)], overweight controls (C‐OW), and normal‐weight controls (C‐NW). Participants passively viewed pictures of food stimuli and neutral stimuli in a cue reactivity design. Two classification analyses were conducted. First, we used multivariate pattern recognition techniques to decode the category of a currently viewed picture from local brain activity patterns. In the second analysis, we applied an ensemble classifier to predict the clinical status of subjects (BED, BN, C‐OW, and C‐NW) based on food‐related brain response patterns. The left insular cortex separated between food and neutral contents in all four groups. Patterns in the right insular cortex provided a maximum diagnostic accuracy for the separation of BED patients and C‐NW (86% accuracy, P < 10−5, 82% sensitivity, and 90% specificity) as well as BN patients and C‐NW (78% accuracy, P = 0.001, 86% sensitivity, and 70% specificity). The right ventral striatum separated maximally between BED patients and C‐OW (71% accuracy, P = 0.013, 59% sensitivity, and 82% specificity). The right lateral orbitofrontal cortex separated maximally between BN patients and C‐OW (86% accuracy, P < 10−4, 79% sensitivity, and 94% specificity). The best differential diagnostic separation between BED and BN patients was obtained in the left ventral striatum (84% accuracy, P < 10−3, 82% sensitivity, and 86% specificity). Our results indicate that pattern recognition techniques are able to contribute to a reliable differential diagnosis of BN and BED. Hum Brain Mapp 33:2135–2146, 2012. © 2011 Wiley Periodicals, Inc.
Keywords: eating disorders, bulimia nervosa, cue reactivity, classification, functional magnetic resonance imaging
INTRODUCTION
Bulimia nervosa (BN) is a common eating disorder that affects 1–3% of the population. The cardinal symptom of BN is the occurrence of eating attacks (“binge‐eating”), during which a person experiences loss of control. In the majority of bulimic patients [80–90%; American Psychiatric Association,1994], a binge‐eating episode is followed by vomiting to prevent weight gain. Moreover, other compensatory strategies might be employed such as use of laxatives/diuretics, phases of controlled eating/dieting, and physical exercise. Recently, it has been discussed in the DSM‐IV [American Psychiatric Association,1994; criteria sets provided for further study] whether a second, similar eating disorder known as binge‐eating disorder (BED) should be differentiated from BN as an autonomous disorder. It shares binge eating with BN as a major symptom. However, in contrast to BN, it does not involve regular behaviors to counteract weight gain. Both syndromes are characterized by comparable levels of body dissatisfaction and fear of weight gain [e.g., Barry et al.,2003; please see also section Participants for further details]. Because patients suffering from BED and BN share such a variety of clinical symptoms, it is unclear whether BED is an autonomous diagnostic category or a variant of BN.
On the neuronal level, two recent neuroimaging studies investigated differential brain activation to visual food stimuli in patients suffering from binge‐eating syndromes [Karhunen et al.,2000; Schienle et al.,2009]. For BN patients they found increased activation of frontal regions involved in selective attention [anterior cingulate cortex (ACC)] and subcortical areas involved in interoception (insula). BED patients showed enhanced recruitment of prefrontal regions involved in the processing of the hedonic value of primary reinforcers [orbitofrontal cortex (OFC)].
However, these studies were based on more conventional neuroimaging analyses that focus on activation differences across extended regions of cortex. Although conventional methods provide insights into differential neuronal processes, they are not optimally suited to investigate the diagnostic separability of two disorders based on neuroimaging data. They leave out lot of potential sources of information because they analyze activation differences for each position of the brain independently. For optimal classification it is important to include the information encoded in conjoint activation in distributed patterns of activity in the brain [Haynes and Rees,2006]. This can be achieved using multivariate pattern recognition techniques (“classifiers” or “decoders”) that have higher sensitivity compared to more traditional voxel‐based analysis approaches [Pereira et al.,2009] and that are specifically designed to categorize data patterns into distinct groups. Thus, it is important to assess whether such techniques might allow diagnosing the clinical status of individuals based on their distributed patterns of brain activity.
In line with this, we and others recently demonstrated the relevance and power of pattern‐based techniques for cognitive paradigms in healthy subjects, for example, in vision [Haxby et al.,2001; Haynes and Rees,2005; Haynes et al.,2005], language [Chen et al.,2006; Formisano et al.,2008], motor tasks [Strother et al.,2004], emotion [Mourão‐Miranda et al.,2007], decision making [Soon et al.,2008], attention [Mourão‐Miranda et al.,2005], and lie‐detection [Davatzikos et al.,2005]. See [Haynes and Rees,2006; Norman et al.,2006; Pereira et al.,2009] for an overview of pattern recognition in MRI. In clinical neuroimaging, classifiers have been applied to the diagnosis of several disorders based on neuroimaging signals, for example, in depression [Fu et al.,2008; Marquand et al.,2008], substance abuse [Zhang et al.,2005], schizophrenia [Koutsouleris et al.,2009; Meyer‐Lindenberg et al.,2001], and dementia [Kloeppel et al.,2008; McEvoy et al.,2009], however not yet to eating disorders.
In this study, we investigated how different visual food cues (e.g., french fries, ice cream, cake, chips, etc.) are encoded in the brain and how this encoding differs between patients suffering from BED, BN, overweight controls (C‐OW), and normal‐weight controls (C‐NW). For that, we conducted two pattern recognition analyses based on data from Schienle et al. [2009]. In the first analysis, we evaluated whether pictures depicting food cues can be separated from pictures depicting neutral content based on brain activation patterns of the gustatory system and the reward system. In the second analysis, we investigated whether it is possible to separate different eating disorders based on spatial brain activation patterns. Here, the discriminability of groups exhibiting binge‐eating syndromes was of central interest, as this analysis can provide additional information concerning the diagnostic autonomy of BED.
MATERIALS AND METHODS
Participants
Women suffering from BED (n = 17) according to DSM‐IV research criteria [American Psychiatric Association,1994], from BN (purging type; n = 14) according to DSM‐IV and healthy controls with no previous history of eating disorders [normal‐weight (C‐NW; n = 19), overweight (C‐OW; n = 17)] gave written informed consent to participate in this study. All women were non‐medicated, right‐handed, Caucasian, and did not smoke. Participants with clinically relevant depression were excluded from the study. Further, other current psychiatric comorbidities (e.g., substance abuse, mood disorders, anxiety disorders, etc.) led to exclusion. An exception was made for specific phobias (two participants). Patients interested in treatment were assisted with referrals. The study was approved by the ethics committee of the German Society for Psychology.
Stimuli and Design
The participants passively viewed a total of 45 pictures from the categories food (e.g., french fries, ice cream, cake, chips, etc), disgust (e.g., dirty toilets, maggots, etc), and neutral (household articles), which had been matched for complexity, brightness, and color composition. For further details, see [Schienle et al.,2009]. For this investigation, we focus on food and neutral pictures.
Brain Imaging
The results presented here are based on a reanalysis of data originally collected in [Schienle et al.,2009]. There, brain images were acquired using a 1.5 Tesla whole‐body tomograph (Magnetom Symphony, Siemens, Erlangen, Germany) with a standard head coil. For the functional imaging, 380 volumes were measured using a T2*‐weighted gradient echo‐planar imaging sequence with 30 slices covering the whole brain (slice thickness = 4 mm; 1 mm gap, interleaved, TR = 3,000 ms; TE = 50 ms, flip angle = 90°, field of view = 192 × 192 mm2; matrix size = 64 × 64).
Data Preprocessing and Analysis
Preprocessing
Image preprocessing was performed using SPM2 and included slice time correction, realignment, normalization to the Montreal Neurological Institute (MNI) brain template, smoothing (isotropic three‐dimensional Gaussian filter, full width at half maximum = 8 mm), and application of a high pass filter (128 s). Activation parameter maps for the experimental conditions (food, neutral, and disgust) were calculated for each subject using a general linear model (GLM) for the coordinates of several bilaterally defined regions of interest (ROIs): ACC, amygdala, insula, lateral and medial OFC, and the ventral striatum (i.e., 12 ROIs in total). The regions were selected based on previous literature on the processing of food cues [e.g., Pelchat et al.,2004; Schienle et al.,2009; Wang et al.,2004)] and are associated with gustatory and attentional processing as well as the hedonic value or the incentive salience of food stimuli or associated cues. The ROIs were created with MARINA [Walter et al.,2003]. Except for the ventral striatum, they are based on the parcellation of the brain proposed by Tzourio‐Mazoyer et al. [2002]. The mask for the ventral striatum was defined manually in accordance with [O'Dougherty et al.,2004]. The volume of the ROIs for the left/right hemisphere is (mm3): ACC: 11,200/10,505; amygdala: 1,760/1,984; insula: 14,864/14,160; lateral OFC: 20,624/21,776; medial OFC: 14,520/13,936; ventral striatum: 2,176/2,024. The three conditions were modeled as a boxcar function convolved with a hemodynamic response function and entered into the model as effects of interest (the condition disgust was modeled as a parameter of no interest). The resulting activation parameter maps were used in Decoding Analysis 1: Picture Categories (see below). Finally, difference contrast maps were calculated for the ROI coordinates for each subject separately based on the activation parameter maps (GLM activation parameter estimates for food cue presentation minus activation parameter estimates for neutral picture presentation). These difference contrast maps were used in Decoding Analysis 2: Ensemble Decoding of Groups (see below).
Decoding Analysis 1: Picture Categories
In a first step, we assessed to which degree brain activity of the four different participant groups (BED, BN, C‐OW, and C‐NW) encodes information about the difference between food and neutral stimuli. For this, we used a so‐called “searchlight” approach [Haynes et al.,2007; Kriegeskorte et al.,2006] that searches across the brain for local activity patterns that are informative about the stimulus category. For a given “center” voxel cvi, in the brain, the searchlight is defined as a spherical cluster with a radius of three voxels surrounding a given center coordinate. Within this cluster of voxels, we extracted the spatial pattern of responses from the activation parameter maps of the two stimulus conditions (food, neutral) of each subject in a given group. Then a classifier was trained and tested based on the patterns of each searchlight position separately using leave‐one‐out (LOO) cross‐validation. As accuracy measure, we calculated the percentage of correctly classified brain responses. By iterating this procedure across all ROI coordinates, we obtained an accuracy map depicting the local discriminative information for the decoding of stimuli for each of the participant groups. For details see Figure 1.
Figure 1.
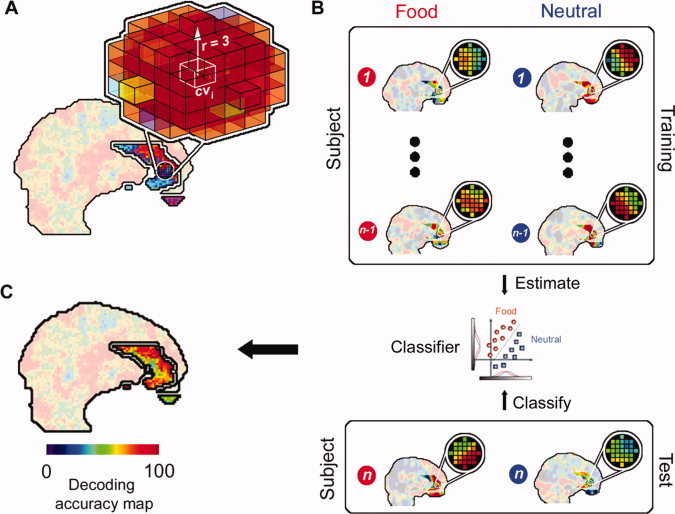
Decoding of food versus neutral images. (A) “Searchlight” approach that searches across the brain for local activity patterns that are informative about the stimulus category (food, neutral). For a given “center” voxel cvi in the brain, the searchlight is defined as a spherical cluster with a radius of three voxels surrounding the center coordinate. (B) Within this cluster of voxels, the spatial pattern of responses for the activation maps is extracted for each stimulus category separately. The data from all but one subject (1…n − 1) are used as a “training dataset” to train a classifier (linear Support Vector Machine [Fung and Mangasarian,2003]) to distinguish between patterns from the two conditions. The classifier is then tested by applying it to the data from the remaining “test” subject (n). This leave‐one‐out (LOO) cross‐validation procedure was then repeated n‐times by leaving out the data of one subject at a time from the training data set. LOO cross‐validation avoids the circular inference that has recently been heavily debated in neuroimaging [Kriegeskorte et al.,2009]. The success of the classifier is an estimate of the local information at that position in the brain. (C) The resulting accuracy was then noted at the coordinate cvi as the local information related to the stimulus category. This was repeated for each center position cvi thus yielding a 3‐dimensional map of local information. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The classification algorithm we used was a linear support vector machine (SVM) classifier [Fung and Mangasarian,2003; http://www.cs.wisc.edu/dmi/svm/nsvm/nsvm.m]. The algorithm attempts to find a linear decision boundary that separates the patterns of two classes (here: searchlight activity patterns emerging during presentation of food and neutral images, respectively). During identification of the decision boundary the algorithm optimizes a free parameter that determines the tradeoff between classifier complexity and number of non‐separable patterns [please see Schölkopf and Smola2002] for further details). Instead of explicitly using cross‐validation to optimize this parameter, the algorithm approximates the cross‐validation rate as this procedure saves computational cost.
The decoding analysis was performed independently for the four different participant groups. The results report searchlight center coordinates cvi that exhibit a significant accuracy on a family‐wise‐error (FWE) corrected level of P < 0.05. Probabilities were calculated using the χ2‐distribution. Additionally, we also separately report coordinates that exhibit a trend towards significance (P uncorrected = 0.001). To reduce the risk of false positive findings, we defined a cluster size criterion for these coordinates, that is, a number (k) of neighboring searchlights yielding significant accuracies and set this threshold to k = 5.
To additionally assess whether local separability coincides with regions of activation or deactivation we computed a conventional t‐contrast based on the GLM results for each subject for each voxel underlying an above‐chance searchlight classifier (parameter estimates for food minus neutral condition). Results report the percentage of voxels located within the radius of (a cluster of) significant searchlights that showed a significant activation difference (P uncorrected = 0.001, no cluster size criterion, and two‐sided). Moreover, we report the mean t‐values for these voxels.
Decoding Analysis 2: Ensemble Decoding of Groups
In a second decoding analysis, we proceeded to the crucial clinical analysis and investigated whether it is possible to distinguish between the four participant groups (BED, BN, C‐OW, and C‐NW). The decoding was conducted for all pairs of eating disordered and control groups and the pair of both eating disordered groups separately (BED & C‐OW; BED & C‐NW; BN & C‐OW; BN & C‐NW; BED & BN). The difference contrast maps (activation parameter estimates for food cue presentation minus activation parameter estimates for neutral picture presentation) of the n subjects of a pair of groups entered each pairwise analysis. Then, we divided the data into training and test datasets by leaving the contrast map of one subject out at a time in a first level LOO loop. The training dataset (size: n − 1) was then again subdivided into training and test data by leaving another subject out (size: n − 2) in a second level or nested LOO loop. For each coordinate cvi a linear SVM classifier [Fung and Mangasarian,2003] was now trained based on n − 2 contrast maps and predicted the group of the contrast map left out in each second level iteration. The procedure was repeated n − 1 times for each coordinate cvi and results in a decoding accuracy map that is independent of the contrast map removed in the first level loop. Once this map was determined, we selected the five most predictive searchlight positions cvi, separately for each of the 12 ROIs, to construct 12 “ensemble” classifiers (one per ROI) that were applied to the remaining test dataset from the first level.
Finally, the ensemble classification was made on a per‐ROI basis based on the average decision of the five selected classifiers in that ROI. The procedure was repeated n times and resulted in one ensemble decoding accuracy per ROI. The ensemble decoding approach was chosen, as it was recently shown to have better generalization abilities and robustness compared to individual classifiers [Martinez‐Ramon et al.,2006]. For further details, see Figure 2.
Figure 2.
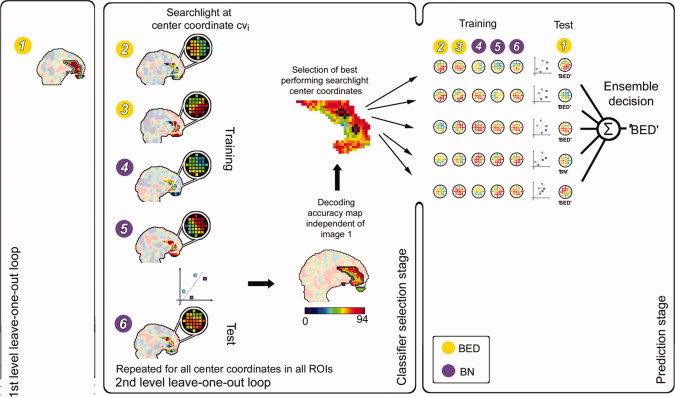
Decoding of eating disorders. Example for a hypothetical data set (three binge‐eating disorder [BED, yellow] and three bulimia nervosa [BN, purple] patients, i.e. n = 6). Preceding the classification analysis, we extracted the contrast map for food minus neutral for each subject n. The decoding of clinical conditions starts with subject one. In a first level leave‐one‐out (LOO) loop, this subject's contrast map is removed from the data set. The remaining contrast maps are then used as training dataset (size n − 1) that is again subdivided into training and test data in the second level LOO loop by leaving another subject out (size n − 2). Searchlight classifiers are now trained with the training data set of size n − 2 and the resulting classification model is used to classify the test map of the second level LOO iteration. This two layer approach is commonly known as a nested cross validation and it is used to further optimize the classification while at the same time avoiding circular inferences because the second level separation into training and test is used to find the optimal classification and then this is tested by applying it to the first level test dataset. The procedure is repeated n − 1 times for each coordinate cvi and results in a decoding accuracy map that is independent of the data of subject one. Once this map is determined, the five best performing searchlights of each ROI (shown: ACC) are selected and trained on all contrast maps except the map of the initially removed subject. Each of the selected classifiers is now tested on its respective pattern from the map of subject one and comes to a decision individually. Subsequently, the final decision is given by the average decision of the individual classifiers, that is, the classifier‐ensemble. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Probabilities of observed accuracies were calculated using the χ2‐distribution. We report ROIs exhibiting FWE‐corrected results (Bonferroni‐correction, i.e., P FWE = 0.05 [α]/12 [number of ROIs] = 0.004) as well as results that have a trend to significance (P uncorrected = 0.05). For each of these ROIs, we report the sensitivity, specificity, and the mean of sensitivity and specificity. The latter is reported as diagnostic accuracy measure instead of the percentage of correctly classified patterns, as it is more robust towards biases in the output of classifiers that could arise from an unbalanced number of subjects in the two groups of a pair.
To additionally assess whether local separability of groups coincides with regions of relative activation or deactivation, we computed a between‐group t‐contrast (BED − C‐OW, BED − C‐NW, BN − C‐OW, BN − C‐NW, and finally BED − BN; P uncorrected = 0.001, no cluster size criterion, two‐sided) based on the contrast maps of the subjects. Results report for each ROI separately the percentage of voxels located within the radius of searchlight classifiers selected across all first level LOO iterations that showed a significant difference (P uncorrected = 0.001, no cluster size criterion, two‐sided). Moreover, we report the mean t‐values for these voxels.
Please note that the base classifiers of each ROI ensemble classifier were selected separately for each iteration of the first level LOO loop following their performance in the second level loop. Therefore, we do not report coordinates in MNI‐space as the five base classifiers of maximal accuracy in each ROI may have varied their position across iterations.
Finally, we used a large‐scale decoding procedure to evaluate the performance of our ensemble method. For that, we merged all abovementioned ROIs to define a single search space containing brain areas involved in food cue processing. Then, we trained and tested a single SVM classifier [Fung and Mangasarian,2003] on patterns drawn from this space in a standard LOO cross‐validation approach.
RESULTS
Participant Characteristics
The groups had a comparable mean age (years): BED: 26.4 (SD = 6.4), BN: 23.1 (SD = 3.8), C‐NW: 22.3 (SD = 2.6), C‐OW: 25.0, (SD = 4.7); [F(3,63) = 2.6, and P = 0.07]. They were comparable with regard to mean years of education [BED: 13.0 (SD = 1.5); BN: 12.7 (SD = 0.8); C‐NW: 13.2 (SD = 0.9); C‐OW: 12.5 (SD = 1.9); F(3,63) = 0.7; P = 0.56]. The majority of participants were students. The patients reported a comparable illness duration (BN: M = 7.3 years, SD = 3.6; BED: M = 6.8 years, SD = 4.0; t(29) = 0.5, P = 0.66) and degree of binge‐eating as indicated by self report (subscale “binging” of the Eating Disorder Inventory [EDI; Diehl and Staufenbiehl,1994]; BED: 14.4 (SD = 2.0), BN: 15.8 (SD = 2.0); t(29) = 1.95, P = 0.06). Bulimic patients all used vomiting as the main method of weight control. Their self‐reported vomiting (means for subscale “vomiting,” EDI) was pronounced [11.8 (SD = 2.5), whereas it was minimal in BED patients (3.0, SD = 2.5; t(29) = 9, P < 0.001]. Four patients in the BN group had previously suffered from anorexia nervosa (lifetime diagnosis). BED and BN patients did not differ in further EDI subscales: “body dissatisfaction”; BED: 16 (3.8), BN: 15.9 (2.7); t(29) = 0.1, P = 0.91 and “fear of weight gain”; BED: 13.4 (3.3), BN: 15.4 (2.2); t(29) = 1.9, P = 0.07. The mean body mass index (BMI) for the four groups were as follows: BED: 32.2 (SD = 4.0), BN: 22.1 (SD = 2.5), C‐NW: 21.7 (SD = 1.4), C‐OW: 31.6 (4.7). The BED patients and C‐OW subjects were obese and had a comparable mean body mass index (BMI; t(32) = 0.3, P = 0.76). Bulimic and C‐NW subjects had a comparable mean BMI [t(31) = 0.6, P = 0.54].
Decoding Analysis 1: Picture Categories
We were able to correctly classify the stimulus category (food, neutral) for all four participant groups, however, there were important differences in informative brain regions between the patients and controls (see Table I for full details). For all four groups, we were able to decode the stimulus category from the insula. Except for C‐OW, we were also able to decode the stimulus type from the OFC in all groups. In patients, but not in controls, we were able to decode the stimulus category also from two additional regions, the amygdala and the ACC. Furthermore, decoding was above chance in the ventral striatum but only in BED patients, whereas in BN patients the information in this region had a clear trend that failed to reach statistical significance. On average, only 2% of all voxels underlying significant searchlight classifiers showed significant activation differences between stimuli in BED patients, 5% in BN patients, 3% in C‐OW, and 12% in C‐NW (Table I; Fig. 3). This suggests that multivariate classifiers did not mainly rely on information from voxels with strong activity differences, but included information from voxels that show weak differential effects.
Table I.
Cross‐validation results for the decoding of picture categories in binge‐eating patients, bulimic patients, and healthy control subjects
| Group/Region | H | CS | x | y | z | DA (%) | P | Mean t‐value | Mean beta‐values | Vox* (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| BED | ||||||||||
| ACC | L | 16 | −10 | 42 | −4 | 88.2 | <10−5 | 1.7 | −0.2/−0.7 | 0 |
| 13 | 0 | 50 | 0 | 82.4 | <10−3 | 1.3 | −0.3/−0.8 | 0 | ||
| R | 60 | 10 | 42 | 12 | 88.2 | <10−5 | 1.1 | −0.1/−0.4 | 3 | |
| Amygdala | R | 3 | 28 | 4 | −16 | 88.2 | <10 −5 | 1.0 | 0.5/0.2 | 0 |
| Insula | L | 34 | −40 | −10 | 6 | 88.2 | <10−5 | 0.1 | 0.1/0.1 | 0 |
| lat. OFC | L | 7 | −26 | 38 | −16 | 88.2 | <10−5 | 1.9 | 0.5/−0.1 | 10 |
| R | 9 | 32 | 26 | −8 | 85.3 | <10−4 | 0.3 | 0.3/0.2 | 0 | |
| v. Striatum | L | 2 | −4 | 6 | −8 | 82.4 | <10−3 | 1.2 | 0.1/−0.3 | 0 |
| BUL | ||||||||||
| ACC | L | 7 | −6 | 38 | 20 | 85.7 | <10−3 | 1.3 | 0.1/−0.3 | 0 |
| 7 | 0 | 24 | 30 | 85.7 | <10−3 | 3.0 | 0.5/−0.5 | 16 | ||
| 6 | −6 | 32 | 26 | 89.3 | <10−4 | 2.8 | 0.4/−0.4 | 10 | ||
| R | 6 | 10 | 48 | 10 | 82.1 | <10−3 | 1.0 | 0.0/−0.3 | 0 | |
| 5 | 4 | 28 | 14 | 82.1 | <10−3 | 1.8 | 0.1/−0.4 | 0 | ||
| Amygdala | L | 46 | −26 | −2 | −20 | 96.4 | <10−6 | 2.1 | 0.5/0.0 | 2 |
| Insula | L | 19 | −38 | −4 | −2 | 92.9 | <10−5 | 2.6 | 0.5/−0.1 | 17 |
| 18 | −38 | 4 | −2 | 89.3 | <10−4 | 1.6 | 0.3/−0.1 | 1 | ||
| R | 7 | 38 | −12 | −6 | 89.3 | <10−4 | 1.4 | 0.3/−0.1 | 4 | |
| 9 | 42 | −12 | 6 | 89.3 | <10−4 | 1.0 | 0.1/−0.2 | 1 | ||
| lat. OFC | L | 99 | −22 | 44 | −20 | 92.9 | <10−5 | 1.6 | 0.5/0.1 | 13 |
| 5 | −36 | 56 | −2 | 92.9 | <10−5 | 0.9 | 0.3/0.0 | 0 | ||
| med. OFC | L | 1 | −20 | 44 | −20 | 89.3 | <10−4 | 0.2 | 0.3/0.3 | 0 |
| 5 | −24 | 60 | −6 | 85.7 | <10−3 | 1.9 | 0.0/−0.6 | 0 | ||
| C‐OW | ||||||||||
| Insula | L | 20 | −44 | −6 | −6 | 85.3 | <10−4 | 0.8 | 0.3/0.1 | 2 |
| R | 24 | 38 | 18 | −14 | 85.3 | <10−4 | 1.5 | 0.7/0.3 | 4 | |
| 5 | 42 | 4 | −6 | 82.4 | <10−3 | 1.8 | 0.5/−0.1 | 4 | ||
| C‐NW | ||||||||||
| Insula | L | 21 | −40 | −4 | 6 | 85 | <10−5 | 1.4 | 0.2/−0.1 | 2 |
| R | 6 | 46 | −6 | −2 | 82.5 | <10−4 | −0.2 | −0.3/−0.3 | 0 | |
| lat. OFC | L | 11 | −24 | 30 | −20 | 82.5 | <10−4 | 3.4 | 1.0/0.1 | 40 |
| med. OFC | L | 1 | −16 | 26 | −14 | 85 | <10−5 | 1.9 | 0.2/−0.1 | 5 |
H: hemisphere; CS: cluster size, that is the number of neighboring significant searchlights; x, y, and z: Montreal Neurological Institute coordinate of the center of the searchlight classifier with the peak accuracy; DA(%): decoding accuracy; P: probability of the accuracy according to χ2‐distribution; Mean t‐value for the contrast food minus neutral for voxels underlying (a cluster of) significant searchlight classifier(s); mean beta‐values: mean beta‐regression coefficients for voxels underlying a (cluster of) significant searchlight classifier(s) for food and neutral picture presentation; Vox*(%): percentage of these voxels showing significant results for the contrast (puncorrected = 0.001, no cluster size criterion, two‐sided). ACC: anterior cingulate cortex; OFC: orbitofrontal cortex; lat.: lateral; med.: medial; v: ventral. Non‐bold text indicates a significant decoding accuracy not corrected for multiple comparisons (puncorrected = 0.001, k = 5). Bold text indicates that the accuracy of the classifier with the peak accuracy in a cluster is significant on a FWE‐corrected level (Bonferroni‐correction, i.e., pFWE = 0.05 [α]/total number of searchlight centers in a given ROI).
Figure 3.
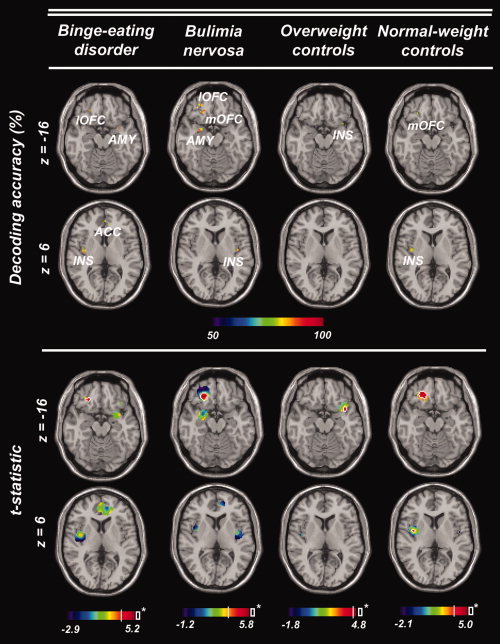
Brain regions encoding visual food cues in BED patients, BN patients, overweight, and normal‐weight control subjects. Top: Center coordinates of searchlight classifiers showing above‐chance decoding accuracy in the separation of visual food cues and neutral stimuli. Axial slices were chosen to show maximally informative regions in the amygdala and the orbitofrontal cortex (AMY; OFC; l, lateral; m, medial; z = −16), or the anterior cingulate cortex and the insula (ACC; INS; z = 6) respectively. Bottom: t‐statistic (food minus neutral) for each voxel underlying a significant searchlight classifier based on the GLM results for each subject. White contour lines highlight voxels showing significant activation differences (P uncorrected = 0.001, no cluster size criterion, two‐sided). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Decoding Analysis 2: Ensemble Decoding of Groups
We were also able to correctly classify the participant group (BED, BN, C‐OW, and C‐NW). Ensemble classifiers located in the right insula and the left lateral OFC separated between BED patients and C‐NW. Classifiers in the right ACC, the left insula and medial OFC, and the right ventral striatum separated between BED patients and C‐OW. Classifiers in the left ACC, the right insula, and the left ventral striatum separated between BN patients and C‐NW and classifiers in the right lateral OFC separated between bulimic patients and C‐OW. Finally, searchlights within the right ACC, the insular cortices of the left and right hemisphere and the left ventral striatum separated between BED and BN patients. Maximal accuracy for the separation of BED patients and C‐NW as well as for BN patients and C‐NW was obtained in the right insula (BED vs. C‐NW: 86% accuracy, P < 10−5, corr., 82% sensitivity, 90% specificity; BN vs. C‐NW: 78% accuracy, P = 0.001, corr., 86% sensitivity, and 70% specificity). The best separation between BED and C‐OW was obtained in the right ventral striatum (71% accuracy, P = 0.013, uncorr., 59% sensitivity, and 82% specificity). Maximal accuracy for the separation of BN patients and C‐OW was obtained in the right lateral OFC (86% accuracy, P < 10−4, corr., 79% sensitivity, and 94% specificity). The best differential diagnostic separation between BED and BN patients was obtained in the left ventral striatum (84% accuracy, P < 10−3, corr., 82% sensitivity, and 86% specificity). Again, the contribution of pronounced activation differences in individual voxels to the separation of groups was very small. On average, only 2% of all voxels underlying selected searchlight classifiers showed significant activation differences between BED patients and C‐NW, 1% in the case of separation of BN patients and C‐NW. Significant activation differences in individual voxels did contribute neither to the separation of BED patients and overweight‐controls nor to the separation of BN patients and overweight‐controls. Only 2% of all voxels underlying selected searchlight classifiers showed significant activation differences in the case of the separation of BED and BN patients. Interestingly, BN patients had higher contrast values than BED patients in the corresponding voxels in all ROIs separating between both eating disorder groups (ACC, insula, and ventral striatum), as far as the mean of t‐values of voxels is concerned (Table II).
Table II.
Cross‐validation results for the decoding of eating disorders
| Groups/Region | H | DA (%) | P | SEN | SPE | Mean t‐value | Mean con‐values | Vox* (%) |
|---|---|---|---|---|---|---|---|---|
| BED and C‐OW | ||||||||
| ACC | R | 67.6 | 0.031 | 82.4 | 52.9 | −0.3 | 0.16/0.30 | 0 |
| Insula | L | 70.6 | 0.016 | 76.5 | 64.7 | −0.9 | −0.15/0.07 | 1 |
| med. OFC | L | 67.7 | 0.039 | 70.6 | 64.7 | 0.1 | 0.29/0.22 | 0 |
| v. Striatum | R | 70.6 | 0.013 | 58.8 | 82.4 | −1.5 | −0.02/0.52 | 0 |
| BED and C‐NW | ||||||||
| Insula | R | 86.2 | <10−5 | 82.4 | 90.0 | 0.5 | −0.04/−0.12 | 0 |
| lat. OFC | L | 70.3 | 0.014 | 70.6 | 70.0 | −1.0 | −0.03/0.21 | 4 |
| BN and C‐OW | ||||||||
| lat. OFC | R | 86.3 | <10−4 | 78.6 | 94.1 | −0.1 | −0.05/−0.03 | 0 |
| BN and C‐NW | ||||||||
| ACC | L | 67.1 | 0.040 | 64.3 | 70.0 | 0.8 | 0.51/0.28 | 1 |
| Insula | R | 77.9 | 0.001 | 85.7 | 70.0 | 1.4 | 0.18/−0.14 | 3 |
| v. Striatum | L | 68.2 | 0.037 | 71.4 | 65.0 | 0.1 | 0.44/0.40 | 0 |
| BED and BN | ||||||||
| ACC | R | 71.0 | 0.019 | 70.6 | 71.4 | −0.6 | 0.29/0.50 | 0 |
| Insula | R | 70.4 | 0.022 | 76.4 | 64.3 | −1.6 | −0.12/0.35 | 5 |
| Insula | L | 68.1 | 0.040 | 64.7 | 71.4 | −0.9 | 0.00/0.26 | 1 |
| v. Striatum | L | 84.0 | <10−3 | 82.4 | 85.7 | −1.1 | 0.21/0.63 | 1 |
H: Hemisphere; DA (%): decoding accuracy calculated as the mean of sensitivity and specificity. Thereby, in each pair‐wise analysis (BED vs. C‐OW; BED vs. C‐NW; BN vs. C‐OW; BN vs. CNW; BED vs. BN) a correctly identified subject of the group named first was considered as a true positive; P: probability of observed DA according to χ2‐distribution; SEN: sensitivity; SPE: specificity; mean t‐value for the difference of contrast maps between groups (BED minus C‐OW, BED minus C‐NW, BN minus C‐OW, BN minus C‐NW, and BED minus BN) in voxels underlying searchlight classifiers selected across all first level LOO iterations for a given ROI; mean con‐values: mean of contrast values for both groups of a pair in voxels underlying searchlight classifiers selected across all first level LOO iterations for a given ROI; Vox*(%): percentage of voxels underlying selected searchlight classifiers showing significant t‐contrasts (P uncorrected = 0.001, no cluster size criterion, two‐sided); lat.: lateral; v: ventral. Bold text indicates a significant decoding accuracy on a FWE‐corrected level (Bonferroni‐correction, i.e., P FWE = 0.05 [α]/12 [number of ROIs] = 0.004). Non‐bold text indicates a significant decoding accuracy according to an uncorrected threshold (P uncorrected = 0.05).
Using the large‐scale decoding approach, we obtained a significant result in case of the separation of BED patients and C‐OW (71% accuracy, P = 0.013, 82% sensitivity, and 59% specificity). For this pair of groups, the large‐scale approach reached the same accuracy as the ensemble approach. For all other pairs of groups, the results were worse and not significant.
DISCUSSION
In this study, we were able to demonstrate that the information encoded about food stimuli as well as subjects' clinical condition can be decoded accurately from local spatial brain patterns in gustatory and reward‐related brain regions. In contrast to several other studies that only addressed the separability of patients from healthy controls [e.g., Fu et al.,2008; Marquand et al.,2008; Zhang et al.,2005], we could separate two similar syndromes, BED versus BN, from the same disorder spectrum based on brain activation patterns.
We conducted two pattern recognition analyses. In the first analysis, we investigated the encoding of visual food cues. Across all patient and control groups spatial activation patterns in the insula differentiated between food and neutral pictures. Similarly, the OFC separated between stimuli in all groups except for the C‐OW. For that group it showed a clear trend that however did not reach the required cluster size criterion. Another set of regions, that is, the amygdala, the ventral striatum, as well as the ACC separated between food and non‐food stimuli exclusively in eating disordered patients.
The stimulus‐related information in the insula is in line with the fact that the anterior insula is the primary gustatory cortex [Augustine,1996]. Gustatory sensations can be elicited by electrical stimulation of insular neurons, whereas insular lesions have been linked to deficits in flavor recognition and reduced taste intensity [Pritchard et al.,1999]. Convergent evidence for the role of the insula in the processing of visual food cues has been obtained in several neuroimaging studies [e.g., Schienle et al.,2009; Wang et al.,2004]. Decoding in the OFC is compatible with the fact that gustatory information is relayed from the insula to the OFC, which is the secondary gustatory cortex [Baylis et al.,1995]. Previous research has indicated that the OFC represents the hedonic value of food stimuli and it plays a central role in reward processing [e.g., Kringelbach,2004]. In line with these findings, the consistent involvement of the insula across groups in this study could be understood as a basic brain response pattern to food cues in healthy as well as disordered subjects reflecting gustatory properties of food.
Subregions of the amygdala, the ventral striatum, and the ACC separated between stimuli only in patients with eating disorders. These structures are known to be involved in the processing of the incentive salience of reward‐related cues [Berridge,2009; Mahler and Berridge,2009]. Correspondingly, a recent study [Mahler and Berridge,2009] showed that the amygdala converts learning into motivation in rats, and amplifies and focuses learned incentive salience onto a particular cue thus making the cue more attractive and in turn triggering consummatory behavior. The ACC has been identified in a variety of studies as being part of a brain network involved in food‐cue processing [e.g., Berridge,2009; Mahler and Berridge,2009; Pelchat et al.,2004; St‐Onge et al.,2005]. It is assumed that activity in the ACC reflects attentional processing of salient food‐cues [Pelchat et al.,2004]. Finally, the finding of discriminative information in the ventral striatum is in line with studies showing that this structure is central for the processing of the incentive value of reward‐related cues [Diekhof et al.,2008]. A direct connection between striatal activity and increased incentive salience of food cues has been demonstrated recently [e.g., Farooqi et al.,2007; Kelley 2004]. Salient food cues can elicit food “wanting” [Berridge,2009] that is a potential trigger of food consumption.
In the second analysis, we addressed the clinically crucial question whether it is possible to distinguish between the four participant groups based on brain response patterns. In this analysis, the insular cortex turned out as a key structure that was involved in the discrimination of each participant group. Insular involvement in the separation of groups is in line with findings of previous studies [Rothemund et al.,2007; Schienle et al.,2009] showing that subjects suffering from several eating disorders and obese subjects differed in their insular response to high‐caloric food cues. Garavan [2010] suggests that such group differences might be explained by differences in food craving.
Besides insular patterns, spatial brain activity patterns in the ventral striatum, and the ACC separated between BED patients and C‐OW, BN patients and C‐NW, as well as BED patients and BN patients. In line with the findings for the decoding of stimuli, these results could be explained by differences of incentive salience and food wanting between groups. Interestingly, striatal patterns and patterns in the ACC were characterized by slightly stronger responses to food‐cues as compared to neutral stimuli in BN patients and obese control subjects compared to patterns of these regions in BED patients. This is indicated by mean contrast values determined for these areas (see Table II). Thus, our findings suggest that relative to BED patients, food‐cues are more attractive and attention grabbing for BN patients and obese subjects and exert a stronger impact to seek and consume food.
Importantly, this interpretation does not interfere with the results for the decoding of stimuli. Although it is true that separation of stimuli is possible in these regions for the BED group but is not possible for the C‐OW group, mean beta regression coefficients depicted in Table I show that food‐cue evoked patterns do not separate from neutral patterns based on pronounced positive activation processes in the BED group. For the ventral striatum, patterns of both stimulus types have a mean β‐value close to zero. Patterns in the ACC even show a latent deactivation for food and a somewhat stronger deactivation for neutral picture presentation. Thus, it is quite possible that moderate food‐specific activation in these regions in the C‐OW group (as is indicated in Table II) induces separability from the BED group whereas it is not sufficient to separate between stimuli within the C‐OW group. The reasons why patterns in the striatum do not separate between stimuli in the C‐OW group remain speculative. There are studies reporting a striatal involvement in food‐cue processing of obese subjects in terms of differential activation on the voxel level [e.g. Farooqi et al.,2007; Rothemund et al.,2007]. However, in line with the results of the present study that clearly show that areas of diagnostic information are not (necessarily) areas of pronounced activation differences on the voxel‐level, processes observed in these studies may not be sufficiently reliable on the pattern level to separate between stimuli.
Besides differences in food reward processing specific differences in eating disorder behaviors between BED and BN patients such as vomiting may account for the striatal involvement in the separation of these groups. This view is also supported by the analysis of the structural MRI data of these groups [Schäfer et al.,2010]. In this study, it could be shown that BN patients had a greater grey matter volume of the ventral striatum compared to BED patients and controls. Moreover, the striatal volume was negatively correlated with the BMI of the bulimic patients and positively with their degree of vomiting. Thus, BN patients with a larger striatum vomited more frequently and by this executed more efficient weight control. This finding can be interpreted in the framework of instrumental reward conditioning, which is related to striatal functions as it is known that the ventral striatum facilitates the acquisition of behaviors related to obtaining a reward [e.g., Diekhof et al.,2008]. In this sense, BN patients purge to reduce stress and weight, which can be labeled as negative reinforcement. Taken together these results strengthen the assumption of the diagnostic autonomy of BED, as it is possible to diagnose BED and BN accurately based on brain patterns drawn from regions that are associated to specific eating‐disordered behaviors and that are coding for the incentive salience of disorder‐related stimuli.
This is the first study investigating the neural or hemodynamic correlates of food‐cue processing in patients suffering from different binge‐eating disorders and controls using multivariate classification. This technique has several advantages. As opposed to methods investigating differential activation [as e.g. used in Schienle et al.,2009], it allows investigating the diagnostic information contained in food‐cue elicited activation patterns for the separation of patients and controls as it assesses the separability of individual data exemplars belonging to different categories. This is not possible using methods analyzing processes of differential activation as these techniques evaluate the relation of data distributions and not individual exemplars. Furthermore, multivariate pattern recognition techniques have higher sensitivity compared to the conventional techniques as they analyze the information contained in distributed patterns of activity. This is underlined by the fact that the classification is successful even without strong overall activation differences between groups. To put it differently, the diagnostic information is not only contained in regions with significant univariate effects. A drawback of the study is that it still lacks replication. To confirm the diagnostic relevance of the identified brain regions and the performance of the classification framework proposed a prospective blind study has to be conducted in the future. If it will be possible to predict diagnoses of unknown subjects in this future study based on the classification models derived from the samples in this study with high accuracy, this might justify the belief in the diagnostic significance of brain regions identified and the classification setup proposed.
To summarize, we were able to demonstrate that the information encoded about food stimuli as well as subject's clinical condition can be decoded accurately from local spatial brain patterns. In the decoding of stimuli, the insula separated between food stimuli in patients as well as controls. The involvement of this region could be understood as a basic brain response pattern to food cues reflecting gustatory properties of food. In patients, the amygdala, the ACC and the ventral striatum separated between stimuli additionally. This suggests that eating disordered patients exhibit a deviant motivational and attentional processing of visual food cues, which could trigger binge‐eating attacks. In the decoding of groups the insula, the amygdala, the OFC, the ACC, and the ventral striatum separated between groups. The fact that regions of the incentive salience network contained diagnostically relevant information for the separation of BED from BN patients supports the assumption of the diagnostic autonomy of BED.
Acknowledgements
The authors thank Carlo Reverberi, Jakob Heinzle, Kerstin Hackmack, Kai Goergen, Andrea Hermann, Dieter Vaitl, and Rudolf Stark for helpful discussions and comments.
REFERENCES
- American Psychiatric Association ( 1994): Diagnostic and statistical manual of mental disorders, 4th ed. Washington, DC: American Psychiatric Association. [Google Scholar]
- Augustine JR ( 1996): Circuitry and functional aspects of the insular lobe in primates including humans. Brain Res Rev 22: 229–244. [DOI] [PubMed] [Google Scholar]
- Barry DT, Grilo CM, Masheb RM ( 2003): Comparison of patients with bulimia nervosa, obese patients with binge eating disorder, and non‐obese patients with binge eating disorder. J Nerv Ment Dis 9: 598–595. [DOI] [PubMed] [Google Scholar]
- Baylis LL, Rolls ET, Baylis GC ( 1995): Afferent connections of the caudolateral orbitofrontal cortex taste area of the primate. Neuroscience 64: 801–812. [DOI] [PubMed] [Google Scholar]
- Berridge KC ( 2009): ‘Liking’ and ‘wanting’ food rewards: Brain substrates and roles in eating disorders. Physiol Behav 97: 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Pereira F, Lee W, Strother S, Mitchell T ( 2006): Exploring predictive and reproducible modeling with the single‐subject FIAC dataset. Hum Brain Mapp 27: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Ruparel K, Fan Y, Shen DG, Acharyya M, Loughead JW, Gur RC, Langleben DD ( 2005): Classifying spatial patterns of brain activity with machine learning methods: Application to lie detection. Neuroimage 28: 663–668. [DOI] [PubMed] [Google Scholar]
- Diehl JM, Staufenbiel T ( 1994): Essstörungs‐inventar [eating disorder inventory]. Supplement zum IEG. Eschborn: Verlag Dietmar Klotz. [Google Scholar]
- Diekhof EK, Falkai P, Gruber O ( 2008): Functional neuroimaging of reward processing and decision‐making: A review of aberrant motivational and affective processing in addiction and mood disorders. Brain Res Rev 59: 164–184. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, Bullmore E, Keogh J, Gillard J, O'Rahilly S, Fletcher PC ( 2007): Leptin regulates striatal regions and human eating behavior. Science 317: 1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano E, De Martino F, Bonte M, Goebel R ( 2008): Who is saying what? Brain‐based decoding of human voice and speech. Science 322: 970–973. [DOI] [PubMed] [Google Scholar]
- Fu CHY, Mourao‐Miranda J, Costafreda SG, Khanna A, Marquand AF, Williams SCR, Brammer MJ ( 2008): Pattern classification of sad facial processing: Toward the development of neurobiological markers in depression. Biol Psychiatry 63: 656–662. [DOI] [PubMed] [Google Scholar]
- Fung G, Mangasarian OL ( 2003): Finite Newton method for lagrangian support vector machine classification. Data mining institute. Technical Report 02–01, Neurocomputing 55: 39–55. [Google Scholar]
- Garavan H ( 2010): Insula and drug cravings. Brain Struct Funct 214: 593–601. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Rees G ( 2005): Predicting the stream of consciousness from activity in early visual cortex. Curr Biol 15: 1301–1307. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Rees G ( 2006): Decoding mental states from brain activity in humans. Nat Rev Neurosci 7: 523–534. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Deichmann R, Rees GE ( 2005): Eye‐specific suppression in human LGN reflects perceptual dominance during binocular rivalry. Nature 438: 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes JD, Sakai K, Rees G, Gilbert S, Frith C, Passingham RE ( 2007): Reading hidden intentions in the human brain. Curr Biol 17: 323–328. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P ( 2001): Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293: 2425–2430. [DOI] [PubMed] [Google Scholar]
- Heinz A, Siessmeier T, Wrase J, Hermann D, Klein S, Grüsser‐Sinopoli SM, Flor H, Braus DF, Buchholz HG, Gründer G, Schreckenberger M, Smolka MN, Rösch F, Mann K, Bartenstein P ( 2004): Correlation between dopamine D2 receptors in the ventral striatum and central processing of alcohol cues and craving. Am J Psychiatry 161: 1783–1789. [DOI] [PubMed] [Google Scholar]
- Karhunen LJ, Vanninen EJ, Kuikka JT, Lappalainen RI, Tiihonen J, Uusitupa MJI ( 2000): Regional cerebral blood flow during exposure to food in obese binge eating women. Psychiatry Res 99: 29–42. [DOI] [PubMed] [Google Scholar]
- Kloeppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ ( 2008): Automatic classification of MR scans in Alzheimer's disease. Brain 131: 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsouleris N, Meisenzahl EM, Davatzikos C, Bottlender R, Frodl T, Scheuerecker J, Schmitt G; Zetzsche T, Decker D, Reiser M, Möller HJ, Gaser C ( 2009): Use of neuroanatomical pattern classification to identify subjects in at‐risk mental states of psychosis and predict disease transition. Gen Arch Psychiatry 66: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Goebel R, Bandettini P ( 2006): Information‐based functional brain mapping. Proc Natl Acad Sci 103: 3863–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PSF, Baker CI ( 2009): Circular analysis in systems neuroscience: The dangers of double dipping. Nat Neurosci 12: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelbach ML ( 2004): Food for thought: Hedonic experience beyond homeostasis in the human brain. Neuroscience 126: 807–819. [DOI] [PubMed] [Google Scholar]
- Mahler SV, Berridge KC ( 2009): Which cue to “want?” Central amygdala opioid activation enhances and focuses incentive salience on a prepotent reward cue J Neurosci 29: 6500–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquand AF, Mourao‐Miranda J, Brammer MJ, Cleare AJ, Fu CHY ( 2008): Neuroanatomy of verbal working memory as a diagnostic biomarker for depression. Neuroreport 19: 1507–1511. [DOI] [PubMed] [Google Scholar]
- Martinez‐Ramon M, Koltchinskii V, Heileman GL, Posse S ( 2006): fMRI pattern classification using neuroanatomically constrained boosting. Neuroimage 31: 1129–1141. [DOI] [PubMed] [Google Scholar]
- McEvoy LK, Fennema‐Notestine C, Roddey JC, Hagler DJ Jr, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM ( 2009): Alzheimer disease: Quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology 251: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer‐Lindenberg A, Poline JB, Kohn PD, Holt JL, Egan MF, Weinberger DR, Berman KF ( 2001): Evidence for abnormal cortical functional connectivity during working memory in schizophrenia. Am J Psychiatry 158: 1809–1817. [DOI] [PubMed] [Google Scholar]
- Mourão‐Miranda J, Bokde ALW, Born C, Hampel H, Stetter S ( 2005): Classifying brain states and determining the discriminating activation patterns: Support vector machine on functional MRI data. Neuroimage 28: 980–995. [DOI] [PubMed] [Google Scholar]
- Mourão‐Miranda J, Friston KJ, Brammer M ( 2007): Dynamic discrimination analysis: A spatial‐temporal SVM. Neuroimage 36: 88–99. [DOI] [PubMed] [Google Scholar]
- Norman KA, Polyn SM, Detre GJ, Haxby JV ( 2006): Beyond mind‐reading: Multi‐voxel pattern analysis of fMRI data. Trends Cogn Sci 10: 424–430. [DOI] [PubMed] [Google Scholar]
- O'Dougherty J, Dayan P, Schultz J, Deichmann R, Friston K, Dolan RJ ( 2004): Dissociable roles of the ventral and dorsal striatum in instrumental conditioning. Science 304: 452–454. [DOI] [PubMed] [Google Scholar]
- Pelchat ML, Johnson A, Chan R, Valdez J, Ragland JD ( 2004): Images of desire: Food‐craving activation during fMRI. Neuroimage 23: 1486–1493. [DOI] [PubMed] [Google Scholar]
- Pereira F, Mitchell T, Botvinick M ( 2009): Machine learning classifiers and fMRI: A tutorial overview. Neuroimage 45: S199–S209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard TC, Macaluso DA, Eslinger PJ ( 1999): Taste perception in patients with insular cortex lesions. Behav Neurosci 113: 663–671. [PubMed] [Google Scholar]
- Rothemund Y, Preuschhof C, Bohner G, Bauknecht HC, Klingebiel R, Flor H, Klapp BF ( 2007): Differential activation of the dorsal striatum by high‐caloric visual food stimuli in obese individuals. Neuroimage 37: 410–421. [DOI] [PubMed] [Google Scholar]
- Schäfer A, Vaitl D, Schienle A ( 2010): Regional grey matter volume abnormalities in bulimia nervosa and binge‐eating disorder. Neuroimage 50: 639–643. [DOI] [PubMed] [Google Scholar]
- Schienle A, Schaefer A, Hermann A, Vailt D ( 2009): Binge‐eating disorder: Reward sensitivity and brain activation to images of food. Biol Psychiatry 65: 654–661. [DOI] [PubMed] [Google Scholar]
- Schölkopf B, Smola A ( 2002): Learning with Kernels. MIT Press, Cambridge, MA.
- Soon CS, Brass M, Heinze HJ, Haynes JD ( 2008): Unconscious determinants of free decisions in the human brain. Nat Neurosci 11: 543–545. [DOI] [PubMed] [Google Scholar]
- St‐Onge MP, Sy M, Heymsfield SB, Hirsch J ( 2005): Human cortical specialization for food: A functional magnetic resonance imaging investigation. J Nutr 135: 1014–1018. [DOI] [PubMed] [Google Scholar]
- Strother S, La Conte S, Hansen LK, Anderson J, Zhang J, Pulapura S, Rottenberg D ( 2004): Optimizing the fMRI data‐processing pipeline using prediction and reproducibility performance metrics: I. A preliminary group analysis. Neuroimage 23: S196–S207. [DOI] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N ( 2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. Neuroimage 15: 273–289. [DOI] [PubMed] [Google Scholar]
- Walter B, Blecker C, Kirsch P, Sammer G, Schienle A, Stark R, Vaitl D (2003). MARINA: An easy to use tool for the creation of MAsks for Region of INterest Analyses [abstract]. Presented at the 9th International Conference on Functional Mapping of the Human Brain, June 19–22, 2003, New York, NY. Available on CD‐Rom in NeuroImage 19.
- Wang GJ, Volkow ND, Telang F, Jayne M, Ma J, Rao M, Zhu W, Wong CT, Pappas NR, Geliebter A, Fowler JS ( 2004): Exposure to appetitive food stimuli markedly activates the human brain. Neuroimage 21: 1790–1797. [DOI] [PubMed] [Google Scholar]
- Zhang L, Samaras D, Tomasi D, Volkow N, Goldstein R ( 2005): Machine learning for clinical diagnosis from functional magnetic resonance imaging. Comp Vis Pat Rec. IEEE Proc of CVPR 1: 1211–1217. [Google Scholar]


