Abstract
Voxel‐based morphometry (VBM) has been used repeatedly in single‐center studies to investigate regional gray matter (GM) atrophy in multiple sclerosis (MS). In multi‐center trials, across‐scanner variations might interfere with the detection of disease‐specific structural abnormalities, thereby potentially limiting the use of VBM. Here we evaluated longitudinally inter‐site differences and inter‐site comparability of regional GM in MS using VBM. Baseline and follow up 3D T1‐weighted magnetic resonance imaging (MRI) data of 248 relapsing‐remitting (RR) MS patients, recruited in two clinical centers, (center1/2: n = 129/119; mean age 42.6 ± 10.7/43.3 ± 9.3; male:female 33:96/44:75; median disease duration 150 [72–222]/116 [60–156]) were acquired on two different 1.5T MR scanners. GM volume changes between baseline and year 2 while controlling for age, gender, disease duration, and global GM volume were analyzed. The main effect of time on regional GM volume was larger in data of center two as compared to center one in most of the brain regions. Differential effects of GM volume reductions occured in a number of GM regions of both hemispheres, in particular in the fronto‐temporal and limbic cortex (cluster P corrected <0.05). Overall disease‐related effects were found bilaterally in the cerebellum, uncus, inferior orbital gyrus, paracentral lobule, precuneus, inferior parietal lobule, and medial frontal gyrus (cluster P corrected <0.05). The differential effects were smaller as compared to the overall effects in these regions. These results suggest that the effects of different scanners on longitudinal GM volume differences were rather small and thus allow pooling of MR data and subsequent combined image analysis. Hum Brain Mapp, 2011. © 2011 Wiley‐Liss, Inc.
Keywords: gray matter, MRI, multiple sclerosis, scanner, voxel‐based morphometry, longitudinal
INTRODUCTION
Previous longitudinal single‐site studies have shown progressive regional gray matter (GM) atrophy in relapsing‐remitting multiple sclerosis (RRMS) using voxel‐based morphometry (VBM) [Audoin et al., 2006; Battaglini et al., 2009; Bendfeldt et al., 2009; Bodini et al., 2009; Pagani et al., 2005; Sepulcre et al., 2006]. Providing automated measures of highly localized regional differences in the concentration/volume of GM or white matter (WM) [Ashburner and Friston, 2000; Good et al., 2001], the VBM technique can be used as an indirect measure of regional pathology in MS. Regional measures in principle are more sensitive to subtle inhomogenously distributed cerebral volume changes than global measures, and thus may have the potential to facilitate the assessment and monitoring of MS disease. In contrast to WM lesions, however, the majority of regional GM changes are not visible on conventional magnetic resonance imaging (MRI) [Pirko et al., 2007].
Therefore, large numbers of subjects are required, necessitating pooling of data from different centers to increase statistical power. In longitudinal MS‐trials, it is common to pool1 scanning resources from multiple centers. Comparison of VBM data derived from different MRI scanners, however, has been critically discussed because the potential confound introduced by different scanners [Ashburner and Friston, 2000; Stonnington et al., 2008] might reduce or wholly offset any gain in power for detecting group differences [Schnack et al., 2010].
A number of studies have been conducted to investigate the reliability of multicenter VBM (Table I). This previous work covers (a) phantom tests [Ewers et al., 2006], (b) studies of healthy volunteers [Ewers et al., 2006; Huppertz et al., 2010; Moorhead et al., 2009; Tardif et al., 2009], and (c) studies of the brain in different states of function and dysfunction, the latter comparing either patients with Alzheimer's disease and patients with Mild Cognitive Impairment [Ewers et al., 2006] or with cognitively normal elderly controls [Stonnington et al., 2008], Childhood Absence Epilepsy subjects with healthy controls [Pardoe et al., 2008], and groups of patients with psychiatric diseases and twins [Schnack et al., 2010].
Table I.
Neuroimaging studies on the reliability of multicenter VBM
| Design | Subjects | Sequence/imaging analysis tools | Outcome | Findings | ||
|---|---|---|---|---|---|---|
| Phantom studies | (Ewers et al., 2006) | –eleven 1.5T scanners | (n = 1) | –T1‐weighted scans/MPRAGE | multicenter variability | –nine of eleven centers met the reliability criteria of the phantom test, whereas two centers showed aberrations in spatial resolution, slice thickness and slice position |
| Healthy volunteers studies | (Tardif et al., 2009) | –two time‐points; –two different scanners (1.5 and 3T) | HV (n = 8) | –MPRAGE; ‐MNI image processing tools; ‐SPM5 | (1) image quality (SNR/image uniformity); (2) GM density; (3) power analysis for longitudinal and cross‐sectional VBM study | (1) SNR and Image non‐uniformity increased significantly at 3 T; (2) regional biases between protocols in the VBM results, in particular at 3 T; (3) smaller number of subjects required in a longitudinal study to detect a difference in GM density at 3 T for MP‐RAGE |
| (Moorhead et al., 2009) | –three 1.5T scanners; –two time‐points | HV (n = 14) | –T1‐weighted MRI scans; ‐SPM5 (separate sets of tissue priors for each scanner) | (1) intra‐ and inter‐scanner variability | (1) inter‐scanner variability not reduced to the level of intra‐scanner variability (scanner specific priors for SPM assist in pooling of data from different sites) | |
| (Huppertz et al., 2010) | –six different sites; –different vendors; –different field strengths (1.5 and 3T); –three time‐points | HV (n = 1) | –MPRAGE; ‐SPM5 (predefined masks derived from a probabilistic whole‐brain atlas) | (1) intra‐scanner variability; (2) inter‐scanner variability; (3) MPVD | (1) CV per brain structure: median 0.89%; (2) CV: median 4.74% (combined variability: median, 4.80%); (3) MPVD: (for CV results 0.50, 3.78, and 3.80%): 1.4% for the same scanner, 10.5% for different scanners | |
| Patient studies (different states of function and dysfunction) | (Stonnington et al., 2008) | –one site; −10 years; −6 scanners (1.5 T; same platform); ‐multiple upgrades over time | – AD (n = 62); –cognitively normal elderly controls (n = 74) | –T1‐weighted MRI scans; ‐whole‐brain voxel‐wise analysis; –SPM5 | (1) effect of disease; (2) effect of scanner; (3) interaction of scanner and disease | (1) reduction of GM in medial temporal lobe; (2) less than group differences and only significant in thalamus; (3) no significant interaction of scanner with disease group; → results not confounded by scanner |
| (Pardoe et al., 2008) | –three different sites; –3T and 1.5T scanners | –CAE; ‐ HV; n (CAE/controls) = site A) 10/213 site B) 15/33 site C) 19/11 | – T1‐weighted MRI scans; ‐optimized VBM | (1) comparisons of CAE subjects and controls stratified by site; (2) inter‐site comparison of controls from each site; (3) factorial analysis of all data with site and disease status as factors | (1) consistent regions of structural change in the thalamic nuclei; (2) site‐specific differences between controls, which requires adjustment for site in the combined analyses; (3) thalamic atrophy in CAE cases; → combined VBM: consistent patterns of structural change in CAE when site factor in statistical analysis | |
| (Ewers et al., 2006)a | –ten of eleven 1.5T scanners; –six different 1.5T scanners | –HV (n = 1); –AD (n = 73); –MCI (n = 76) | –T1‐weighted MRI scans or MPRAGE; –manual hippocampal volumetry; ‐automatic segmentation of brain compartments; –SPM2/VBM | (1) multisite variability; (2) Power analysis for detection of a difference in GMV between AD and MCI patients across centers | (1) CV: 3.55% (hippocampus); 5.02% (grey matter) 4.87% (white matter); 4.66% (cerebrospinal fluid); 12.81% (± 9.06) voxel intensities GM; 8.19% (± 6.9) WM; (2) (d = 0.42): N = 180; → good reliability across centers | |
| (Schnack et al., 2010) | –four 1.5T scanners, one 1.0T scanner (four vendors); –different acquisition protocols | – HV (n = 6) | −3D‐FFE, SPGR, 3D‐FLASH, MPRAGE; –MNI image processing tools, both for VBM and CORT; ‐development of methods to detect reproducibility of VBM/CORT to detect group differences | (1) group effect; (2) heritability | (1) reliability maps showed an overall good comparability between the sites; (2) scan pooling improved heritability estimates |
AD, Alzheimer's disease; CAE, childhood absence epilepsy; CORT, cortical thickness measurement; CV, coefficient of variation; GM, gray matter; GMV, gray matter volume; HV, healthy volunteer; MCI, mild cognitive impairment; MPRAGE, magnetization‐prepared rapid gradient echo; MPVD, minimum percentage volume difference for detecting a significant volume change between two volume measurements in the same subject calculated for each substructure; MNI, Montreal Neurological Institute; MRI, magnetic resonance imaging; VBM voxel‐based morphometry; SNR, signal:noise ratio, SPM, statistical parametric mapping; 3D‐FFE, three‐dimensional T1‐weighted coronal spoiled gradient echo scan WM white matter.
Same publication reports the phantom study above.
To date, however, VBM studies focusing on reliability of multicenter MRI in MS are missing. Therefore, the goal of the present study was to investigate whether GM volume changes can be elucidated in multi‐center studies in the context of MS. In particular, we investigated inter site differences and inter site comparability of GM changes in a large sample of RRMS patients recruited and scanned in two different clinical centers on two different 1.5T scanners.
On the basis of the previous literature, we hypothesized that differential effects of time on regional GM volume changes between the two centers would occur. We also hypothesized that the interaction of scanner‐and disease‐related effects would be rather small in this longitudinal combined dataset and that this would allow pooling of MR data.
Over and above these methodological aspects, on the basis of previouslongitudinal MRI studies of MS using either smaller samples or different methodology [Audoin et al. 2006; Battaglini et al. 2009; Bendfeldt et al. 2009; Chen et al. 2004; Pagani et al. 2005], we hypothesized a general predominance of GM volume changes in fronto‐temporal cortical regions in RRMS patients in this combined multi‐site dataset.
MATERIALS AND METHODS
Patients
We analyzed pairs of MRI data from 248 Caucasian patients (77 men, 171 women) with a diagnosis of clinically definite relapsing remitting MS [Polman et al., 2005] of the case‐controlled study for genotype‐phenotype associations in MS (GeneMSA; GSK, UK) recruited in two clinical centers (center one: n = 129, center two: n = 119) participating in the GeneMSA consortium. Patients with a clinical relapse or glucocorticosteroid treatment within the month previous to baseline or follow‐up scan were excluded, whereas the concomitant use of disease modifying therapies for MS was permitted. 127 patients (center one: 80, center two: 47) received immunomodulatory‐immunosuppressive drugs (interferon‐β‐1a, interferon‐β‐1b, glatiramer acetate) during the entire study; no change of these medications occurred between baseline and follow‐up scan at 2 years. During follow‐up 75 RRMS patients had received corticosteroid therapy to treat acute relapse. At the time of baseline and follow up MR scan, all patients had been relapse‐free and interacted with steroids for at least 1 month. The study was approved by the local ethical standards committee and written informed consent was obtained from each subject.
There is overlap between subjects in this study and those used in previous MR structural imaging studies [Bendfeldt et al., 2009, 2010c]. Previously we have searched for GM volume changes in RRMS patients from center one. In those studies, between‐group differences (baseline vs. 1‐year follow‐up) in GM volume were estimated by fitting an analysis of covariance (ANCOVA) model at each intracerebral voxel in standard space. For each subject follow‐up minus baseline difference images were created, and then analyzed with a regression model with an intercept (parameter of interest) and centered covariates of age, gender and disease duration. Within the RRMS group, we specifically focused on those patients with increasing T2 and T1 lesion burden (n = 45) and patients lacking an increase in WM lesion burden (n = 44). The former studies provided evidence of an association between the progression of regional GM volume reductions in specific fronto‐temporal cortical areas and WM lesion volume progression on the one hand [Bendfeldt et al., 2009; Nakamura and Fisher, 2009] and lesion location on the other hand [Kappos et al., 2006].
In the present study we analysed subsets of 129 patients from center one and 119 patients from center two (baseline vs. 2‐year follow‐up). The study focuses (a) on the differential effects potentially occurring in patients from different sites/scanners and (b) on the overall disease related effects in the whole dataset of the pooled samples rather then on associations of GM and WM changes. In contrast to the former analyses, here, we have included T2‐ and T1 lesion volumes and scanner as additional covariates.
MR Image Acquisition
All subjects were scanned twice (baseline and 2‐year follow‐up) using either one of two 1.5T MR systems (center one: Siemens Avanto; center two: Siemens Vision) with similar protocols. For VBM analysis, 3D‐heavily T1‐weighted gradient echo images were acquired (TR: 7–20.8 ms; TE: 2–4 ms; TI: 300–400 ms), consisting of isotropic 1 × 1 × 1 mm3 voxels. Additionally, dual echo‐T2‐weighted images (magnetization‐prepared rapid gradient echo “MP‐RAGE”; TR: 2,000–4,000 ms; TE: 14–20/80–108 ms), with interleaved axial 3.0‐mm‐thick slices and an in‐plane resolution of 1.0 × 1.0 mm2 were acquired. Lastly, post‐contrast T1‐weighted spin‐echo images (TR: 467–650 ms; TE: 8–17 ms; axial 3.0‐mm‐thick slices with an in‐plane resolution of 1.0 × 1.0 mm2) were obtained. The same image acquisition parameters were used between timepoint no. 1 and timepoint no. 2.
MR Imaging Data Analysis
We analyzed MR images for all subjects on a commercially available Intel‐based workstation running Debian Linux 3.1 using VBM. Images were processed with Statistical Parametric Mapping software (SPM5, Wellcome Department of Imaging Neurosciences, University College London, [http://www.fil.ion.ucl.ac.uk/spm] version 958, last updated December 13, 2007) running under the MATLAB 7.00 (R14) environment.
The images were processed using the VBM toolbox v1.03 (http://dbm.neuro.uni-jena.de/vbm/, last updated December 6, 2006) as described before [Bendfeldt et al., 2009, 2010a].
In brief, the method was modified to reduce the influence of MS lesions in the process, which could alter the normalization and segmentation procedures. To prevent WM lesions from being misclassified as GM, lesions identified on T2 images were masked from the three‐dimensional MP‐RAGE images [Nakamura and Fisher, 2009]. MS lesions were outlined on the proton density scans (to calculate 3D binary masks and quantify the areas of previously identified brain lesions) using the commercial semi‐automatic thresholding contour software AMIRA 3.1.1 (Mercury Computer Systems), [Kappos et al., 2006]. The 3D binary masks were then co‐registered to the MP‐RAGE to remove the MS lesions. All VBM input images were controlled carefully regarding WM lesions adjacent to the cortex or deep gray matter to estimate the amount of false positive or negative GM volume changes, respectively.
In the segmentation step, images were spatially normalized into the same stereotactic space. In SPM5, prior probability maps that are relevant to tissue segmentation are warped to the individual brains, making the creation of a customized template unnecessary. The normalization was performed by first estimating the optimum 12‐variable affine transformation for matching images and then optimizing the normalization using 16 nonlinear iterations using 6 × 8 × 6 basis functions to account for global non‐linear shape differences [Ashburner and Friston, 1999]. To preserve the total within‐voxel volume, which may have been affected by the nonlinear transformation, every voxel's signal intensity in the segmented GM images was multiplied by the Jacobian determinants derived from the spatial normalization.
When using unsupervised clustering methods like SPM5 in combination with a lesion mask, simple changes in the lesion mask (such as increases in T2 lesion volume) could have an effect on the clustering results. As the set of voxels used for parameter estimation changes, the segmentation could change as a result.
The potential bias coming from errors in registration has been minimized by visually checking all GM and WM lesion registrations analyses to ensure that there were no failures of alignment and consequent misclassification of tissues. Segmentation accuracy was assessed by examining axial slices of each subject's GM, WM, and cerebrospinal fluid (CSF) image in the individual's space. The warping accuracy was assessed by displaying axial slices from each subject with edges from the atlas image.
To preserve the total within‐voxel volume, which may have been affected by the nonlinear transformation, every voxel's signal intensity in the segmented GM images was multiplied by the Jacobian determinants derived from the spatial normalization. The analysis of these modulated datasets was used to detect regional differences in absolute tissue volume. Finally, in order to increase the signal‐to‐noise ratio and to account for variations in normal gyral anatomy all images were smoothed using a 5‐mm full‐width‐at half‐maximum isotropic Gaussian kernel as done before [Bendfeldt et al., 2009; Borgwardt et al., 2007a, b, 2008; Fusar‐Poli et al., 2007]. On the basis of the expected subtle regional differences [Ashburner and Friston, 2000], we have chosen a small smoothing kernel, because it allows us to detect a greater number of regions with small structures as the medial temporal lobes, parahippocampal gyrus, and anterior cingulate cortex. Also, according to the matched filter theorem, the width of the smoothing kernel determines the scale at which morphological changes are most sensitively detected [White et al., 2001].
Statistical Analysis
Demographic data
The median and interquartile range, or the mean and standard deviation were used to describe clinical and MRI characteristics. We used chi‐squared test, paired t test, Mann‐Whitney‐U and Wilcoxon test for nonparametric data to compare demographic and clinical variables. For these tests, a significance level of P < 0.05 was considered. Statistical analysis was performed with SPSS software, version 15 (SPSS, Chicago, IL).
MRI data
Between‐group differences (baseline vs. 2‐year follow‐up) in gray matter volume were estimated by fitting an analysis of covariance (ANCOVA) model at each intracerebral voxel in standard space. We have chosen a full‐factorial design with centered covariates age, gender, disease duration, T2‐ and T1 lesion volume, and scanner. Before entering the linear regression models, T2 and T1 lesion volume was transformed to reduce skew of lesion volume and reduce the impact of outlier lesion volumes using the logarithm with base 10 (logT2LV, log T1LV). To assess additional nuisance variation due to head size differences the analysis was adjusted for each subject's global GM volume (GMV) by entering the global values as additional covariate. GMV (mean value) was calculated by SPM5. We performed F‐tests to investigate whether there were any longitudinal GM volume changes in each of the two centers followed by subsequent T‐tests contrasting each center against the other.
Statistical maps were assessed for significance with cluster size inference adjusted for non‐stationarity [Hayasaka et al., 2004; Moorhead et al., 2005] (http://dbm.neuro.uni-jena.de/vbm/non-stationary-cluster-extent-correction/). A cluster‐defining threshold of P = 0.001 uncorrected was used, and clusters were considered significant at P < 0.05 cluster level, corrected for a whole‐brain search (though for completeness our tables also report family‐wise error (FWE)‐corrected voxel‐wise P‐values as well). Significant clusters were anatomically localized using the atlas of Talairach and Tournoux, except for foci in and close to the cerebellum, which were localized using the atlas of [Schmahmann et al., 1999].
All the potentially confounding covariates were included in the original analysis. However, to confirm the results in a more stringent way, we also investigated two matched groups of patients of center one (n = 73) and center two (n = 73) (Table II). These patients were sex‐matched on top of being age‐matched. Then each subject of center one was assigned a patient of center two which did not differ by more than ±5 years in age at baseline and by more than ±2 years in disease duration.
Table II.
Clinical and MRI characteristics
| “Center one” | “Center two” | Statistics | |
|---|---|---|---|
| N = 129 | N = 119 | Center comparison | |
| N = 73 | N = 73 | (P value) | |
| Age at bs in years, mean (SD) | 42.6 (10.7) | 43.3 (9.3) | 0.587 |
| 41.2 (9.0) | 41.1 (8.7) | 0.966 | |
| Male/female (ratio) | 33/96 (1:2.9) | 44/75 (1:1.7) | 0.052 |
| 22/51 (1:2.3) | 22/51 (1:2.3) | 1 | |
| Disease duration: Time since first symptoms at bs in months, median (IQR) | 150 (72–222) | 116 (60–156) | 0.024 |
| 96 (48–186) | 96 (60–186) | 0.895 | |
| Scan‐interval, months (SD) | 24.5 (1) | 25.4 (1.6) | <0.001 |
| 24.5 (1) | 25.5 (1.6) | <0.001 | |
| Drug treatmenta (T/NT) | 80/47 | 47/71 | <0.001 |
| 41/32 | 41/32 | 0.966 | |
| EDSS at bs, median (IQR) | 2.5 (1.5–3.0) | 3.0 (2.0–4.0) | <0.001 |
| 2.0 (1.5–2.5) | 3.0 (2.0–4.0) | <0.001 | |
| EDSS at y2, median (IQR) | 2.5 (1.5–3.5) | 3.5 (2.5–4.0) | <0.001 |
| 2.0 (1.5–3.0) | 3.5 (2.5–4.0) | <0.001 | |
| Statistics: EDSS change, bs versus y2 (P value) | 0.162 | 0.005 | |
| 0.169 | 0.006 | ||
| GMV in cm3 (mean SD) at bs | 635 (74) | 624 (68) | 0.210 |
| 644 (75) | 624 (65) | 0.108 | |
| GMV in cm3 (mean SD) at y2 | 631 (75) | 625 (67) | 0.550 |
| 641 (79) | 628 (65) | 0.406 | |
| Statistics: GMVC (bs versus y2, P‐value) | 0.018 | 0.314 | |
| 0.168 | 0.396 | ||
| T2 lesion load in ml, median (IQR) at bs | 2.9 (1.0–8.0) | 2.6 (1.0–7.8) | 0.583 |
| 2.6 (0.9–7.3) | 2.6 (1.2–8.2) | 0.81 | |
| T2 lesion load in ml, median (IQR) at y2 | 3.5 (1.0–8.5) | 2.8 (1.0–7.9) | 0.546 |
| 2.9 (0.9–7.5) | 2.6 (1.0–8.1) | 0.862 | |
| Statistics: T2 lesion volume change (increase), bs versus y2: P‐value | 0.016 | 0.010 | |
| 0.079 | 0.011 | ||
| T1 lesion load in ml, median (IQR) at bs | 0.7 (0.1–2.7) | 0.5 (0.1–2.5) | 0.416 |
| 0.5 (0.1–2.7) | 0.5 (0.1–2.2) | 0.883 | |
| T1 lesion load in ml, median (IQR) at y2 | 0.9 (0.1–2.6) | 0.6 (0.2–2.4) | 0.410 |
| 0.6 (0.1–2.6) | 0.6 (0.1–2.3) | 0.810 | |
| Statistics: T1 lesion volume change, bs versus y2: P‐value | 0.928 | 0.808 | |
| 0.795 | 0.795 | ||
| New T2‐Lesions at y2 [count, median (IQR)] | 0 (0–1) | 0 (0–1) | 0.092 |
| 0 (0–1) | 0 (0–2) | 0.304 | |
| New gadoliniumd‐enhancing lesions at y2 [count, median (IQR)] | 0 (0–0) | 0 (0–0) | 0.038 |
| 0 (0–0) | 0 (0–0) | 0.056 |
T, treated; NT, not treated; SD, standard deviation; IQR, interquartile range; EDSS, expanded disability status scale; bs, baseline; y2, year2; GMV, global gray matter volume; GMVC, gray matter volume change. Results of a subgroup with optimally pair wise matched subjects of n = 73 (center one) vs. n = 73 (center two) is presented in italic letters.
No changes in medication during follow‐up.
RESULTS
Clinical and MRI (non‐VBM) Characteristics
The clinical and MRI characteristics of the 248 RRMS patients are reported in Table II. Cross‐sectionally, subjects from both centers did not differ significantly with respect to age, gender, GMV, T1, and T2 lesion volumes and number of new T2 lesions neither at baseline nor at follow‐up. Scan‐interval, disease duration, EDSS, number of new gadolinium‐enhancing lesions, and the proportion of patients without/with immunomodulatory treatment were significantly different between the centers. Gray matter volume decreased slightly over time in center one and T2 lesion volumes increased in both centers. To account for these differences, we also report the results from the matched samples (Table II, italic letters).
Any Longitudinal Gray Matter Volume Change: Main Effect of Time
We investigated the main effect of time on GM volume within each of the two centers. Contrast estimates and 90% confidence intervals, which are reflective of the standard deviations, are shown in Table II. Generally, the effect of time on GM volume was rather small. It was larger, however, in most of the significant clusters of the limbic, frontal, and occipital cortices in center two as compared to center one. In these regions, confidence intervals were larger relative to the contrast estimates in center one, and similar between the two centers. Smaller effects in center one occurred in particular in regions of the the left parahippocampal, cingulate, rectal, medial frontal gyrus, paracentral lobule and in the left claustrum, as well as in the right precentral and lingual gyrus and in the right precuneus. Larger effects in center one occurred bilaterally in regions of the medial frontal gyrus and in the left precuneus. Effects of similar magnitude occurred in the left postcentral gyrus and cuneus, as well as in the right temporal gyrus (Table III).
Table III.
Any longitudinal GM volume change
| Area | MNI coordinates of cluster maximum (x y z) | Contrast estimate: mean (SD)Center 1 | Contrast estimate: mean (SD)Center 2 | F | |
|---|---|---|---|---|---|
| Contrast estimatecenter 1 < center 2 | |||||
| Limbic lobe | Claustrum | (−36/−9/−2) | 0.0039 (0.0041) | 0.0093 (0.0043) | 15.48 |
| Parahippocampal gyrus | (−29/−31/−10) | 0.0060 (0.005) | 0.0120 (0.0052) | 18.77 | |
| Cingulate gyrus | (−1/−42/28) | 0.0024 (0.0053) | 0.0124 (0.0055) | 14.26 | |
| Frontal lobe | Rectal gyrus | (−9/26/−24) | 0.0083 (0.0056) | 0.0176 (0.0058) | 30.80 |
| Paracentral lobule | (−5/−37/67) | 0.0038 (0.0068) | 0.0184 (0.0070) | 19.36 | |
| Medial frontal gyrus | (−35/7/55) | 0.0093 (0.0073) | 0.0131 (0.0073) | 12.39 | |
| Precentral gyrus | (13/−33/64) | 0.0033 (0.0048) | 0.0148 (0.0050) | 24.72 | |
| Occipital lobe | Lingual gyrus | (16/−93/−12) | 0.0031 (0.0075) | 0.0189 (0.0077) | 16.65 |
| Precuneus | (7/−65/48) | 0.0078 (0.0055) | 0.0105 (0.0057) | 14.93 | |
| Contrast estimatecenter 1 > center 2 | |||||
| Frontal lobe | Medial frontal gyrus | (26/29/46) | 0.0151 (0.0059) | 0.0021 (0.0061) | 18.21 |
| (−24/37/41) | 0.0177 (0.0060) | 0.0008 (0.0062) | 24.10 | ||
| Occipital lobe | Precuneus | (−29/−63/48) | 0.0140 (0.0060) | 0.0054 (0.0062) | 17.01 |
| Contrast estimatecenter 1 ∼ center 2 | |||||
| Parietal lobe | Postcentral gyrus | (−49/−21/53) | 0.0075 (0.0051) | 0.0070 (0.0053) | 15.68 |
| Temporal lobe | Superior temporal gyrus | (59/−53/18) | 0.0083 (0.0053) | 0.0087 (0.0055) | 13.41 |
| (60/−40/12) | 0.0075 (0.0051) | 0.0070 (0.0053) | 10.63 | ||
| Occipital lobe | Cuneus | (−24/−80/31) | 0.0089 (0.0061) | 0.0074 (0.0063) | 9.47 |
Comparison of main effects of time in MR data from center one (n = 129) and center two (n = 119) (Cluster P corrected < 0.05). Coordinates (x, y, and z) refer to the point of maximal change in each cluster in stereotactic space as defined in the MNI atlas.
Inter‐Site Differences
To look for inter‐site differences of regional GM volume reductions, we analyzed the interaction of time and center, while correcting for multiple comparisons across the brain. First, to increase statistical power, we analyzed the whole samples. Then, to reduce the confounding effect of age, gender, disease duration, and medication with GM volume, we repeated the analysis in a subsample of optimally‐matched pairs of patients from both scanners (n = 73 each).
Differential effects in the complete data
Statistically significant differences occurred bilaterally in the precuneus, superior temporal, and medial frontal gyrus. In the left hemisphere, additional differences occur in the rectal, postcentral, parahippocampal, and cingulate gyrus, as well as in the cuneus, paracentral lobule, and claustrum. In the right hemisphere, differences occur in the precentral and lingual gyrus and in the superior parietal lobule (Table IV, Fig. 1).
Table IV.
Differential effects: Interaction of center and time
| Area | MNI coordinates of cluster maximum (x y z) | T | Cluster size k E(voxels) | Cluster P corrected | Voxel P FWE‐corrected | |
|---|---|---|---|---|---|---|
| Left hemisphere | ||||||
| Limbic lobe | Claustrum | −36/−9/−2 | 5.19 | 1,326 | <0.001 | 0.032 |
| Parahippocampal gyrus | −29/−31/−10 | 5.85 | 3,573 | <0.001 | 0.001 | |
| Cingulate gyrus | −1/−42/28 | 4.50 | 2,796 | <0.001 | 0.410 | |
| Frontal lobe | Rectal gyrus | −9/26/−24 | 7.46 | 10,522 | <0.001 | <0.001 |
| Paracentral lobule | −5/−37/67 | 5.26 | 3,355 | <0.001 | 0.024 | |
| Medial frontal gyrus | −37/41/27 | 4.79 | 3,332 | <0.001 | 0.150 | |
| −35/7/55 | 4.92 | 2,677 | <0.001 | 0.104 | ||
| −24/37/41 | 5.02 | 1,465 | <0.001 | 0.069 | ||
| Temporal lobe | Superior temporal gyrus | −54/−58/30 | 4.88 | 1,436 | <0.001 | 0.120 |
| Parietal lobe | Postcentral gyrus | −49/−21/53 | 4.94 | 1,221 | <0.001 | 0.095 |
| Occipital lobe | Precuneus | −29/−63/48 | 5.24 | 3,371 | <0.001 | 0.026 |
| Cuneus | −24/−80/31 | 4.32 | 647 | 0.014 | <0.001 | |
| Right hemisphere | ||||||
| Frontal lobe | Medial frontal gyrus | 27/55/15 | 5.56 | 5,737 | <0.001 | 0.006 |
| 26/29/46 | 4.73 | 564 | 0.029 | 0.191 | ||
| Precentral gyrus | 13/−33/64 | 6.02 | 3,099 | <0.001 | <0.001 | |
| Temporal lobe | Superior temporal gyrus | 59/−53/18 | 5.16 | 5,665 | <0.001 | 0.037 |
| 60/−40/12 | 4.60 | 572 | 0.027 | 0.299 | ||
| Parietal lobe | Superior parietal lobule | 17/−48/63 | 4.09 | 771 | 0.005 | 0.906 |
| Occipital lobe | Precuneus | 7/−65/48 | 5.41 | 1,765 | <0.001 | 0.012 |
| Lingual gyrus | 16/−93/−12 | 4.77 | 666 | 0.012 | 0.165 | |
Results refer to comparison of MR data from center one (n = 129) and center two (n = 119) (Cluster P corrected < 0.05). Coordinates (x, y, and z) refer to the point of maximal change in each cluster in stereotactic space as defined in the MNI atlas.
Figure 1.
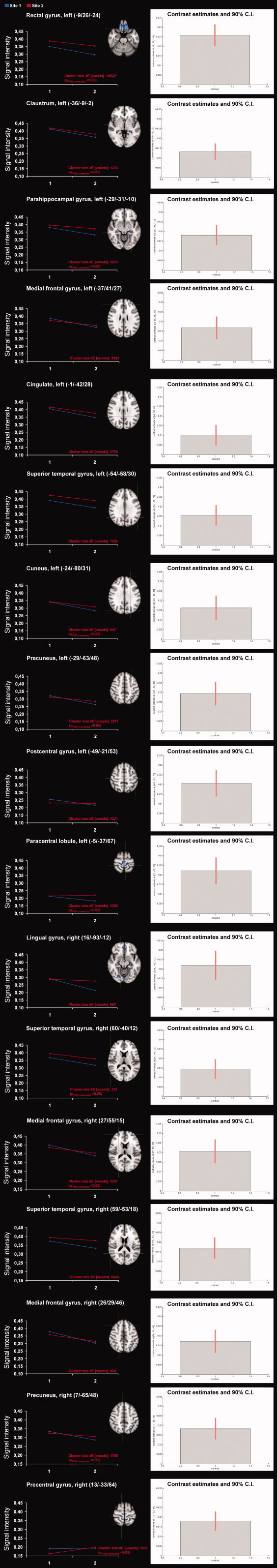
Comparison of longitudinal regional GM volume changes from two different 1.5T scanners in patients with RRMS For each individual region from left to right: (a) Superimposed image of the significant GM volume differences (P < 0.01 corrected) onto an MNI‐template. (b) Mean signal intensities (“eigenvalues”) for the interaction between the effects of site (center one and center two) and time (baseline vs. follow‐up scan) in a variety of cortical regions. * for more details see also Table III) Contrast estimates and 90% confidence intervals for the differential effects. Images are presented in standard radiological fashion, with the right hemisphere shown on the left of the figure, and vice versa. The crosshairs show the focus of the cluster and refer to the MNI coordinates. The X/Y/Z coordinates show the position of each slice with respect to MNI atlas. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Confidence intervals were small relative to the contrast estimates and similar between the different brain regions (Fig. 1).
Differential effects in the matched groups
Statistically significant differences between the matched groups occurred bilaterally in a number of fronto‐temporal and parietal cortical regions and in the cerebellum. In contrast to the whole samples, the limbic and occipital lobes were not involved (Table V).
Table V.
Differential effects: Interaction of center and time (matched groups)
| Area | MNI coordinates of cluster maximum (x y z) | T | Cluster size k E (voxels) | Cluster P corrected | Voxel P FWE‐corrected | |
|---|---|---|---|---|---|---|
| Left hemisphere | ||||||
| Frontal lobe | Rectal gyrus | −9/24/−27 | 6.19 | 1,480 | <0.001 | <0.001 |
| Precentral gyrus | −12/−29/68 | 5.53 | 6,852 | <0.001 | 0.007 | |
| MFG | −35/10/−21 | 4.83 | 1,463 | <0.001 | 0.133 | |
| −29/36/40 | 4.74 | 942 | 0.004 | 0.184 | ||
| Temporal lobe | Fusiform gyrus | −50/−37/−23 | 5.24 | 1,448 | <0.001 | 0.025 |
| Hippocampus | −30/−31/−9 | 4.43 | 822 | 0.008 | 0.485 | |
| Parietal lobe | Angular gyrus | −54/−56/35 | 4.45 | 1,239 | 0.001 | 0.460 |
| Right hemisphere | ||||||
| Frontal lobe | Paracentral lobule | 7/−30/70 | 5.35 | 3,570 | <0.001 | 0.015 |
| Orbital gyrus | 17/36/−26 | 5.22 | 1,560 | <0.001 | 0.027 | |
| MFG | 39741/22 | 3.99 | 594 | 0.042 | 0.987 | |
| Temporal lobe | Hippocampus | 27/−34/−2 | 4.27 | 1,039 | 0.002 | 0.694 |
| Supramarginal gyrus | 55/−51/28 | 4.37 | 775 | 0.011 | 0.570 | |
| Parietal lobe | Superior parietal lobule | 29/−51/62 | 4.37 | 915 | 0.004 | 0.564 |
| Cerebellum | 25/−68/−21 | 5.13 | 918 | 0.004 | 0.039 | |
Results refer to comparison of the optimally pair‐wise matched groups from center one (n = 73) and center two (n = 73) (Cluster P corrected < 0.05). Coordinates (x, y, and z) refer to the point of maximal change in each cluster in stereotactic space as defined in the MNI atlas.
Disease‐Related Versus Differential Effects (Matched Groups)
We also looked for GM volume reductions between baseline and follow‐up in the complete data of the matched groups. Significant GM volume reductions in patients with RRMS were found bilaterally in the cerebellum, in the left inferior temporal gyrus and insula, and in the right superior frontal gyrus and uncus (Table VI). Figure 2 shows that the differential effects of the two centers/scanners were smaller than the disease‐related effects in these regions.
Table VI.
Overall longitudinal GM volume reductions in the complete data (matched groups)
| Area | MNI coordinates of cluster maximum (x y z) | T | Cluster size k E(voxels) | Cluster P corrected | Voxel P FWE‐corrected | |
|---|---|---|---|---|---|---|
| Left hemisphere | ||||||
| Frontal lobe | MFG | −44/33/34 | 3.89 | 619 | 0.035 | 0.986 |
| Parietal lobe | Inferior parietal lobule | −42/−54/55 | 4.07 | 1,038 | 0.002 | 0.911 |
| Cerebellum | −39/−40/−36 | 7.13 | 47,528 | <0.001 | <0.001 | |
| Right hemisphere | ||||||
| Frontal lobe | Inferior orbital gyrus | 40/−89/−3 | 5.87 | 864 | 0.006 | 0.001 |
| Paracentral lobule | 4/−32/48 | 4.95 | 1,530 | <0.001 | 0.081 | |
| Temporal lobe | Uncus | 20/−4/−33 | 5.63 | 3,172 | <0.001 | 0.004 |
| Occipital lobe | Precuneus | 6/−71/44 | 4.58 | 887 | 0.005 | 0.313 |
| Cerebellum | 42/−39/−31 | 7.07 | 15,980 | <0.001 | <0.001 | |
Results refer to the complete MR data (matched groups) from center one (n = 73) and center two (n = 73) (FWE corrected <0.05). Coordinates (x, y, and z) refer to the point of maximal change in each cluster in stereotactic space as defined in the MNI atlas.
Figure 2.
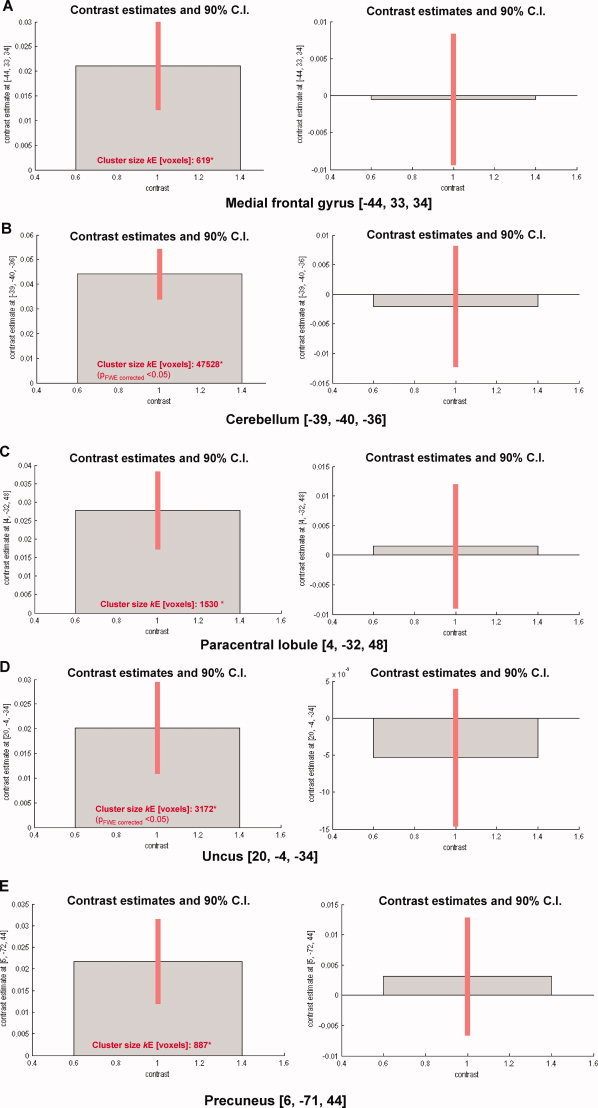
Contrast estimates and 90% confidence intervals for disease‐related effects and center/scanner effects (matched groups) Disease‐related effect in the complete data in the (a) left medial frontal gyrus at [−44, 33, 34; x, y, z], (c) left cerebellum [−39, −40, −36], (e) right paracentral lobule [4, −32, 48], (g) right uncus [20, −4, −33], and (J) right precuneus [6, −71, 44]. Differential effects of center/scanner in the (b) left medial frontal gyrus at [−44, 33,34], (d) left cerebellum [−39, −40, −36], (f) right paracentral lobule [4, −32, 48], (h) right uncus [20, −4, −33], and (k) right precuneus [6, −71, 44]. * cluster sizes refer both to the left and right panels; for more details see also Table III. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
The principal aim of the current study was to investigate longitudinal GM volume changes in a large sample of RRMS patients (n = 248) recruited and scanned at two different sites. It is the first VBM study looking for the effects of sites/scanners on the detection of subtle longitudinal GM volume differences in MS in a comparative manner.
The results suggest larger effects of time on GM volume change in data of scanner two as compared to scanner one in most of the brain regions. Additionally, differential longitudinal GM volume reductions were observed bilaterally in a number of fronto‐temporal, parietal, limbic and occipital cortical regions of both hemispheres as well as in the left claustrum. Differential effects in the fronto‐temporal and parietal cortex were also confirmed with the analysis of the matched groups. Longitudinal GM volume reductions in the complete data of the matched groups were found bilaterally in the cerebellum, in the left inferior temporal gyrus and insula, and in the right superior frontal gyrus and uncus. The center/scanner‐related effects were smaller as compared to the disease‐related effects in these regions.
Interindividual and Between‐Scanner Effects
In principle, several factors may affect the capacity of VBM to detect regional GM loss, including physiological and pathological interindividual heterogeneity as well as scanner effects that may have introduced systematic error [Ashburner and Friston, 2000; Stonnington et al., 2008], thus making the interpretation of results difficult. Scanner effects due to partial volume effects [Li et al., 2005], noise of the electronics of the MRI system, imaging gradient non‐linearity [Jovicich et al., 2006], and/or differential scanner drift over time may all contribute to image intensity inhomogeneity. Furthermore, differences in subject positioning between sites can occur and images can vary as a function of protocol differences between a baseline and a later scan or with drifts in instrument signal to noise over time [Preboske et al., 2006]. The interaction of scanner differences with segmentation remains a particular concern and potential cause of varied measures of regional tissue volume in the brain [Ashburner and Friston, 2000].
In the current study interindividual heterogeneity was compensated for by using a large sample of 248 patients scanned twice. The sample sizes of the two centers (129 and 119, respectively) provided adequate power for detecting even subtle changes in regional GM volume. Although the effect sizes of the longitudinal regional GM volume reductions were rather small, confidence intervals, which are reflective of the standard deviations, for the contrast estimates were small relative to the effect sizes and similar between the different regions and scanners, indirectly suggestive of relatively little variance across the different regions and scanners.
Generally, as compared to cross‐sectional comparisons, for longitudinal VBM analyses most of the above mentioned factors should be of minor relevance. With respect to the clinical and MRI data, except for the slight longitudinal changes of GMV in center one and EDSS score in center two, none of these variables changed significantly in the course of time. To account for potentially confounding group‐specific factors, however, we included age, gender, global GM volume, disease duration and WM lesion volumes as covariates in the SPM regression model.
Additionally, given the intercenter differences in disease duration and drug treatment we created two groups of strictly matched subjects from each center (n = 73, each group). Interestingly, the comparison of these two well‐matched groups revealed similar results in a variety of cortical regions. Except for the EDSS scores, which are slightly different even after matching, matching minimized the possibility that the unequal covariates may have biased the result. Thus, the changes in regional GM volume that we observed in RRMS patients are unlikely to be related to one of these factors.
To account for potential scanner effects in the analyses we used “optimized” and modulated VBM with linear and non‐linear modulation [Good et al., 2001], thus minimizing the potentially confounding effects of errors in stereotactic normalization, global brain shape, head size differences, and differences in subject positioning as well as for image intensity variability. It is, however, necessary to rule out any possible interaction between scanner and effect of interest and/or account for the effects of different scanners in a principle manner [Mikol et al., 2008; O'Connor et al., 2009; Stonnington et al., 2008]. Therefore, in the statistical model, we have additionally corrected for center.
The misclassification of lesions as GM is a potential problem, in particular in longitudinal studies, because MS lesions are highly dynamic and the misclassified lesion volume changes may be even greater than the true GM volume change [Bendfeldt et al., 2009]. Thus, to avoid misclassification, lesions identified on T2w images were masked from the three‐dimensional MP‐RAGE images [Battaglini et al., 2009; Bendfeldt et al., 2009; Pagani et al., 2005; Sepulcre et al., 2006]. As recently demonstrated [Bendfeldt et al., 2009] this might be insufficient to produce an accurate GM segmentation. To minimize the potential bias coming from errors in registration, we visually checked all GM and WM lesion registration analyses to ensure that there were no failures of alignment and consequent misclassification of tissues.
Early accelerated loss of brain volume and a rapid decrease in the number of gadolinium‐enhancing lesions has been discussed as “pseudoatrophy” related to the anti‐inflammatory effects of medication [Barkhof et al., 2009]. We have therefore accounted for this effect by excluding patients who changed from one to another immunomodulatory drug less than 6 months before baseline MRI or during follow‐up.
Relevance of Regional GM Changes in Relation to Disease
Recently, research has focused on the tissue compartments and regions within which brain atrophy occurs [Chard et al., 2002; Chen et al., 2004; Dalton et al., 2004; De Stefano et al., 2003; Jasperse et al., 2007; Pagani et al., 2005; Prinster et al., 2006; Quarantelli et al., 2003; Sailer et al., 2003; Sepulcre et al., 2006; Tedeschi et al., 2005; Tiberio et al., 2005]. The topography of GM involvement differs among patients with different clinical phenotypes, with a prominent involvement of the thalamus in the early stages and an extensive and diffuse cortical GM loss in the progressive forms [Ceccarelli et al., 2008].
The results of the present study are broadly consistent with previous longitudinal VBM studies, as well as studies using other image analysis methods, which report progressive GM atrophy in MS patients in both the fronto‐temporal cortices and the deep GM regions [Audoin et al., 2006; Battaglini et al., 2009; Bendfeldt et al., 2009; Chen et al., 2004; Pagani et al., 2005; Sepulcre et al., 2006] (Table VII).
Table VII.
Longitudinal neuroimaging studies on regional GM involvement in MS
| Author and year of publication | n | Disease course | Age; mediana (range) | Sex (m/f) | Disease duration (ys/mthsa) | EDSS | “Scaninterval” (monthsa, yearsb) | MR sequence | Field strength/ scanner | Method of image analysis (smoothing kernel) | Statistics | Cluter‐defining threshold, P‐value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Chen et al., 2004) | 20 | stable progressive | 40 (22–54) | 9/11 | 8 (2–15) | 2 (1.5–5) | 0.8 (0.5–1.1)b | T1‐w (3 mm) | 1.5‐T | automated quantification of GM thickness: SIENA‐based | Multivariate linear model (SPSS) | 0.05 |
| 10 | RRMS or SPMS | 42 (26–54) | 2/8 | 10 (5–16) | 3.5 (1.4–4.5) | 0.9 (0.5–2.5)b | Philips Gyroscan | |||||
| (Pagani et al., 2005) | 20 | RRMS | 49.9 (25–68) | 8/12 | 10 (0–34) | 5 (0–8) | 15 ± 0.5a | T1‐w (3mm) | 1.5T | SIENA/SPM99 | GLM | 0.001 |
| 19 | SPMS | 35.9 (25–43) | 6/13 | 5 (0–20) | 3 (0–5) | Siemens Vision | (10‐mm FWHM) | |||||
| 31 | PPMS | 49.3 (35–58) | 12/19 | 16 (6–34) | 6 (4–8) | |||||||
| (Audoin et al., 2006) | 21 | RRMS | 36 (27–55) | 5/16 | 25.8a (14.4–45.5) | 1.0 (0–3) | 2b | FSPGR, 3D | 1.5T | SPM2 | Two‐sample t tests | 0.05 |
| 10 | NC | 37 (31–52) | 6/4 | GE Signa | ||||||||
| (Sepulcre et al., 2006) | 31 | PPMS | 43.7 | 22/39 | 3 (2–5) | 4.5 | 1b | FSPGR, 3D | 1.5T | VBM/SPM2 (12‐mm FWHM) | 1‐way ANOVA (within subjects) | 0.05 |
| 15 | NC | 43.2 | GE Signa | |||||||||
| (Battaglini et al., 2009) | 59 | RRMS | 41.0 (10.6) | 23/36 | 1.8 (0.1–17) | bs: 1.5 (0–5) | 2 (3–4.8)b | T1‐w (3 mm) | 1.5T | FSL‐VBM | GLM, permutation testing | 0.05 |
| fu: 1.5 (1–5) | Philips Gyroscan | SIENAr (10‐mm FWHM) | ||||||||||
| (Bendfeldt et al., 2009) | 151 | RRMS | 35 (17–60) | 36/112 | 10 (6–17) | bs: 2.0 (1.5–3.0) | 12.6a | MP‐RAGE (1 mm) | 1.5T | SPM5‐VBM (5‐mm FWHM) | GLM, paired t‐tests | 0.001 |
| fu: 2.3 (1.5–3.0) | Siemens Avanto | |||||||||||
| (Bendfeldt et al., present study) | 119 | RRMS | 42.6 (10.7) | 33/96 | 150 (72–222) | bs: 2.5 (1.5–3.0) | 24.5 (1)a | MP‐RAGE (1mm) | 1.5T | SPM5‐VBM | GLM, | 0.001 |
| fu: 2.5 (1.5–3.5) | 25.4 (1.6)a | Siemens Avanto/ | (5 mm FWHM) | Full‐factorial design | ||||||||
| 119 | 43.3 (9.3) | 44/75 | 116 (60–156) | bs: 3.0 (2.0–4.0) | ||||||||
| fu: 3.5 (2.5–4.0) | Siemens Vision |
ANOVA, analysis of variance; FSPGR, fast spoiled gradient echo; FWHM, full width at half maximum; GLM, general linear model; GM, gray matter; MP‐RAGE, magnetization‐prepared rapid gradient echo; NC, normal controls; PPMS, primary progressive MS; RRMS, relapsing‐remitting MS; SIENA, structural image evaluation, using normalization of atrophy; SPMS, secondary progressive MS; SPM, statistical parametric mapping; SPSS, statistical analysis software; T, Tesla; 3D, three‐dimensional; T1‐w, T1‐weighted; VBM, voxel based morphometry.
In our former longitudinal study of 151 patients with RRMS from center one followed up for 1 year, we showed significant cortical GM volume reductions in the anterior and posterior cingulate, the temporal cortex, and cerebellum [Giorgio et al., 2010], while GM volume reductions in primary sensory, visual, or motor areas were not evident. Another recent publication based on a rather small cohort of 20 patients with RRMS followed‐up for 15 months, which combined Structural Image Evaluation Using Normalization of Atrophy (SIENA) and SPM analysis [Pagani et al., 2005], reported brain atrophy development in the insula, cingulate sulcus, as well as in frontal, parietal, and temporal regions.
In the present study, we have investigated GM changes between baseline and Year 2. Overall longitudinal GM volume reductions in the complete data of the matched groups occurred bilaterally in the cerebellum, in the left inferior temporal gyrus and insula, and in the right superior frontal gyrus and uncus. In contrast to the separate analyses (data not shown), the pooled dataset provided significant brain volume reductions in brain areas that were consistent with previous longitudinal single‐site 1.5T MR studies in RRMS with different sample sizes, different clinical data, and different morphometry tools (see parameters in Table VII). These results support the notion that a pooled analysis gives brain areas with significant longitudinal effects that are not detectable in single‐site datasets.
Contrary to previous studies [Bendfeldt et al., 2009; Pagani et al., 2005], significant effects in the anterior and posterior cingulate were not found in this pooled analysis. This could reflect either demographical or clinical sample differences, scanner effects, or statistical power of the analysis. Short‐term fluctuations of GM volume during the 2‐year time interval of 2 years could also play a role.
Furthermore, we have shown that the development of GM reductions is closely associated with the concurrent progression of T2 and T1 lesion volumes [Bendfeldt et al., 2009]. Therefore, although we have included lesion load as a covariate in the model, the different proportions of patients with “progressive” and “non‐progressive” WM lesion load in the different samples, might have influenced the pattern of GM atrophy differentially.
Finally, the locations of the WM lesions per se might have influenced the pattern of atrophy. This is in part supported by the finding that WM lesions are mainly located in the periventricular regions in patients with clinically isolated syndrome (CIS), RRMS, and secondary progressive (SP) MS [Ceccarelli et al., 2008]. Increasing degeneration of those lesions could interrupt tracts that originate from or project to prefrontal, cingulate, and association areas. The anterior cingulate, e.g., has extensive cortico‐cortical connections with highly inter‐connected cerebral regions, such as the insula, which in turn has numerous connections with other parts of the limbic system, e.g., the hippocampus, parahippocampal gyrus, as well as the frontal, parietal and temporal cortices. Those interconnected areas might prone to be affected by axonal degeneration in the cerebral white matter [Charil et al., 2007]. This could help explain why highly interconnected cortical areas might be more vulnerable to atrophy than regions with relatively fewer connections. Furthermore, histopathologic studies in MS have also demonstrated that the cingulate gyrus, temporal lobe, and insula generally show a higher prevalence of cortical demyelinated lesions than other areas [Kutzelnigg and Lassmann, 2005].
Limitations
Very few methodological approaches have been described to establish the reliability of multicenter VBM [Clark et al., 2006; Ewers et al., 2006; Moorhead et al., 2009; Pardoe et al., 2008; Schnack et al., 2010; Stonnington et al., 2008; Tardif et al., 2009]. Ewers et al. [ 2006] calculated voxelwise coefficients of variance from a single subject scanned on 10 scanners while Schnack et al. [ 2010] applied a multicenter calibration study with six healthy volunteers scanned at five sites with scanners from four different manufacturers, each running different acquisition protocols. The resulting reliability maps showed good comparability between the four sites, showing a reasonable gain in sensitivity in most parts of the brain whereas in some brain areas, e.g., around the thalamus, scan pooling was difficult. Clark et al. [ 2006], investigating scanner/post‐processing combinations showed that due to partial voluming effects the thalamic region was susceptible to voxelwise segmentation errors. In patients, [Pardoe et al., 2008] carried out a multicenter study on childhood absence epilepsy and [Stonnington et al., 2008] analyzed multicenter Alzheimer's disease data.
Longitudinal studies using other approaches then VBM also show that the reliability of MRI across centers was relatively good even when scanners with different field strength were used. In a longitudinal aging study of healthy adults using different scanners did not affect measured intracranial volume with a manual tracing method [Raz et al., 2005], and manual hippocampal measurements performed on both 1.5 T and 3.0 T scanners were not affected by field strength [Briellmann et al., 2001]. Multi‐center data from three different 1.5T scanners have also been used to explore the validity and the variability of some of the freely available automated methods currently being used to segment GM and to estimate GM atrophy in MS [Derakhshan et al., 2010].
The current literature, however, is devoid of VBM studies that describe the analysis of longitudinal MS data acquired on different scanners with regard to the interaction of scanner with effects of interest.
To compare the longitudinal similarities and/or differences between regional GM volumes of patients scanned at two different sites, in the present study, we used a whole‐brain imaging method. VBM is a widely used method for assessing differences in regional volume or tissue “concentration” across subjects in conventional MR images. The procedure is relatively straightforward and is most commonly carried out using the statistical parametric mapping (SPM) software package. SPM5/VBM (Statistical Parametric Mapping software), was already used in our prior longitudinal VBM studies of regional GM volume changes in RRMS [Bendfeldt et al., 2009, 2010a, b].
In terms of the differential effects of disease and of scanner, a factorial design would have been favorable, with scanner as one factor (e.g., Center 1 and 2) and group (e.g., the longitudinal GM volume changes of patients and longitudinal GM volume changes of controls) as the other. Presumably, the effects of scanner would be present regardless of disease, so this would have been another way to assess the effect of disease and the effect of scanner and also potentially allow pooling of the data. In the GeneMSA study, however, although a series of clinical data from healthy relatives were collected, MRI data were not recorded, so that no controls were available for our retrospective study. A previous test–retest study performed with healthy volunteers [Han et al., 2006] has shown that cortical thickness is comparable across 1.5T sites (even from different MR vendors), indirectly supporting our finding of low scanner‐related variation in longitudinal GM volumes. Therefore, although MRI data used here has rather low heterogeneity from the point of view of a multi‐site MR study—two sites, with MR systems of the same field strength and from the same vendor—the results might benefit planning of multisite VBM‐MS‐MRI studies to reduce the sources of heterogeneity in the future. In the present study, we have shown that multicenter VBM data can be used in terms of reliability and expected gain obtained from pooling the data. Additionally, VBM techniques may be extended to multi‐center studies involving other imaging modalities as well.
It is rather unlikely that the observed differences between centers were related to WM lesion load, because T2‐ as well as T1 lesion volumes did not differ between the centers. A significant impact of new lesions in relevant WM tracts is unlikely as well, because the differences between the numbers of new lesions in the two centers were rather small and it is known from clinical trials, that the different immunomodulatory drugs used in the current study are equally good at preventing the occurrence of new inflammatory lesions. Lesions in the internal capsule and periventricular WM are critical for functional disability as measured by the Expanded Disability Status Scale (EDSS). Furthermore, in patients with EDSS scores below 4, the specific pathways interrupted by lesions seem to be important in explaining cognitive deficits [Giorgio et al., 2010]. EDSS scores in center two were about 0.5 to one point larger than in center one and further increased during follow‐up.
Medication might have influenced the trajectory of regional GM volumes differentially in the two centers. Whereas there is clear evidence demonstrating the impact of medication on disease activity as measured by clinical and MRI parameters, its impact on regional GM atrophy in particular is widely unclear. Although in treatment trials with different agents, no significant effects of medication on global GM volume were reported, the different drugs used in the current study (i.e., interferon‐β‐1a, interferon‐β‐1b, glatiramer acetate) as well as the proportion of immunomodulatory‐treated and untreated patients in both centers might have influenced regional GM volume development differentially. The results from a previous analysis [Bendfeldt et al., 2010b] assessing the effect of immunomodulatory medication indeed suggest differences in the dynamics of regional GM volume atrophy in differentially treated (Interferon 1a/b or glatiramer acetate) or untreated MS patients. Differences in the insula and hippocampus were found in patients treated with either interferons or glatiramer acetate, but not between treated and untreated patients (treatment allocation was non‐randomized). Although we accounted for potentially confounding covariates in the analysis of the complete data, we can not rule out an effect on the results of the present study. However, matching the patient groups of both centers for disease duration and the proportion of immunomodulatory treated and untreated patients, revealed similar results.
Statistically significant differences between the matched groups occurred bilaterally in a number of fronto‐temporal and parietal cortical regions and in the cerebellum. In contrast to the whole samples, however, the limbic and occipital lobes were not involved. This might have been caused by reduced power of the subset analysis on the one hand or better matching of the samples on the other hand. Furthermore, as discussed earlier, the location of WM lesions might have influenced the pattern of atrophy differentially.
There are a few regions with greater effects in center one than center two or vice versa. This could reflect either heterogeneity of the sample, systematic change due to scanner variation or protocol disparity. Because the brain regions are not influenced consistently, it is rather unlikely, however, that these differences are due to systematic change alone. Because the scan‐intervals were significantly different between the two centers, they were considered in the statistical model as well (data not shown). The results show slightly reduced effect sizes in some of the significant regions, which has no impact on the main conclusions of this study.
Another limiting factor might be the fact, that the effect of aging is not well modeled by a linear fit across the age range in this study. This is of particular importance because aging‐related structural changes might have interfered with disease‐specific effects. Although we have included age as a covariate in the statistical model, and it is a relatively small longitudinal age change (2 years), we cannot completely exclude a potential confound of aging in the current study [Sowell et al., 2007].
CONCLUSION
To our knowledge, this is the first longitudinal large‐scale study to provide evidence that, among certain brain areas, VBM studies using data from more than one clinical center in MS offer similar results. This is of great interest, as differences in GM volumes between studies using different MR scanners are not comparable a priori. We showed that the effects of the different sites/scanners on the detection of longitudinal GM volume differences were rather small. It is, however, still recommended to rule out possible interactions between time point and effect of interest for each individual study and/or account for the effects of different scanners in a principle manner.
Future studies exploring the comparability of different scanners are necessary to reduce the sources of heterogeneity of VBM studies, and to sustain the ongoing research in clinical neurology.
Footnotes
Note that “pooling” is used here in the sense of “combining” data; not in the sense of “collapsing of factor levels when combining data” in which it is often used in multicenter trials [e.g., Schwemer, 2000].
REFERENCES
- Ashburner J, Friston KJ ( 1999): Nonlinear spatial normalization using basis functions. Hum Brain Mapp 7: 254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ ( 2000): Voxel‐based morphometry—The methods. NeuroImage 11: 805–821. [DOI] [PubMed] [Google Scholar]
- Audoin B, Davies GR, Finisku L, Chard DT, Thompson AJ, Miller DH ( 2006): Localization of grey matter atrophy in early RRMS. J Neurol 253: 1495–1501. [DOI] [PubMed] [Google Scholar]
- Barkhof F, Calabresi PA, Miller DH, Reingold SC ( 2009): Imaging outcomes for neuroprotection and repair in multiple sclerosis trials. Nat Rev Neurol 5: 256–266. [DOI] [PubMed] [Google Scholar]
- Battaglini M, Giorgio A, Stromillo ML, Bartolozzi ML, Guidi L, Federico A, De Stefano N ( 2009): Voxel‐wise assessment of progression of regional brain atrophy in relapsing‐remitting multiple sclerosis. J Neurol Sci 282: 55–60. [DOI] [PubMed] [Google Scholar]
- Bendfeldt K, Kuster P, Traud S, Egger H, Winklhofer S, Mueller‐Lenke N, Naegelin Y, Gass A, Kappos L, Matthews PM, Nichols, Thomas E, Radue Ernst‐Wilhelm, Borgwardt Stefan J. ( 2009): Association of regional gray matter volume loss and progression of white matter lesions in multiple sclerosis—A longitudinal voxel‐based morphometry study. NeuroImage 45: 60–67. [DOI] [PubMed] [Google Scholar]
- Bendfeldt K, Blumhagen JO, Egger H, Loetscher P, Denier N, Kuster P, Traud S, Mueller‐Lenke N, Naegelin Y, Gass A, Hirsch J, Kappos L, Nichols TE, Radue EW, Borgwardt SJ. ( 2010a): Spatiotemporal distribution pattern of white matter lesion volumes and their association with regional grey matter volume reductions in relapsing‐remitting multiple sclerosis. Hum Brain Mapp 31: 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendfeldt K, Egger H, Nichols TE, Loetscher P, Denier N, Kuster P, Traud S, Mueller‐Lenke N, Naegelin Y, Gass A, Kappos L, Radue EW, Borgwardt SJ. ( 2010b): Effect of immunomodulatory medication on regional gray matter loss in relapsing‐remitting multiple sclerosis—A longitudinal MRI study. Brain Res 1325: 174–182. [DOI] [PubMed] [Google Scholar]
- Bendfeldt K, Kappos L, Radue EW, Borgwardt S ( 2010c): Longitudinal spatiotemporal distribution of gray and white matter pathology in multiple sclerosis. AJNR Am J Neuroradiol 31: E45; author reply E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodini B, Khaleeli Z, Cercignani MH, Miller D, Thompson AJ, Ciccarelli O ( 2009): Exploring the relationship between white matter and gray matter damage in early primary progressive multiple sclerosis: An in vivo study with TBSS and VBM. Hum Brain Mapp 30: 2852–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgwardt SJ, McGuire PK, Aston J, Berger G, Dazzan P, Gschwandtner U, Pfluger M, D'Souza M, Radue EW, Riecher‐Rossler A ( 2007a): Structural brain abnormalities in individuals with an at‐risk mental state who later develop psychosis. Br J Psychiatry Suppl 51: s69–s75. [DOI] [PubMed] [Google Scholar]
- Borgwardt SJ, Riecher‐Rossler A, Dazzan P, Chitnis X, Aston J, Drewe M, Gschwandtner U, Haller S, Pfluger M, Rechsteiner E, et al. ( 2007b): Regional gray matter volume abnormalities in the at risk mental state. Biol Psychiatry 61: 1148–1156. [DOI] [PubMed] [Google Scholar]
- Borgwardt SJ, McGuire P, Fusar‐Poli P, Radue EW, Riecher‐Rossler A ( 2008): Anterior cingulate pathology in the prodromal stage of schizophrenia. Neuroimage 39: 553–554. [DOI] [PubMed] [Google Scholar]
- Briellmann RS, Syngeniotis A, Jackson GD ( 2001): Comparison of hippocampal volumetry at 1.5 tesla and at 3 tesla. Epilepsia 42: 1021–1024. [DOI] [PubMed] [Google Scholar]
- Ceccarelli A, Rocca MA, Pagani E, Colombo B, Martinelli V, Comi G, Filippi M ( 2008): A voxel‐based morphometry study of grey matter loss in MS patients with different clinical phenotypes. NeuroImage 42: 315–322. [DOI] [PubMed] [Google Scholar]
- Chard DT, Griffin CM, Parker GJM, Kapoor R, Thompson AJ, Miller DH ( 2002): Brain atrophy in clinically early relapsing‐remitting multiple sclerosis. Brain 125: 327–337. [DOI] [PubMed] [Google Scholar]
- Charil A, Dagher A, Lerch JP, Zijdenbos AP, Worsley KJ, Evans AC ( 2007): Focal cortical atrophy in multiple sclerosis: Relation to lesion load and disability. NeuroImage 34: 509–517. [DOI] [PubMed] [Google Scholar]
- Chen JT, Narayanan S, Collins DL, Smith SM, Matthews PM, Arnold DL ( 2004): Relating neocortical pathology to disability progression in multiple sclerosis using MRI. NeuroImage 23: 1168–1175. [DOI] [PubMed] [Google Scholar]
- Clark KA, Woods RP, Rottenberg DA, Toga AW, Mazziotta JC ( 2006): Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images. NeuroImage 29: 185–202. [DOI] [PubMed] [Google Scholar]
- Dalton CM, Chard DT, Davies GR, Miszkiel KA, Altmann DR, Fernando K, Plant GT, Thompson AJ, Miller DH ( 2004): Early development of multiple sclerosis is associated with progressive grey matter atrophy in patients presenting with clinically isolated syndromes. Brain 127: 1101–1107. [DOI] [PubMed] [Google Scholar]
- De Stefano N, Matthews PM, Filippi M, Agosta F, De Luca M, Bartolozzi ML, Guidi L, Ghezzi A, Montanari E, Cifelli A, et al. ( 2003): Evidence of early cortical atrophy in MS: Relevance to white matter changes and disability. Neurology 60: 1157–1162. [DOI] [PubMed] [Google Scholar]
- Derakhshan M, Caramanos Z, Giacomini PS, Narayanan S, Maranzano J, Francis SJ, Arnold DL, Collins DL ( 2010): Evaluation of automated techniques for the quantification of grey matter atrophy in patients with multiple sclerosis. NeuroImage 52: 1261–1267. [DOI] [PubMed] [Google Scholar]
- Ewers M, Teipel SJ, Dietrich O, Schönberg SO, Jessen F, Heun R, Scheltens P, Pol Lvd, Freymann NR, Moeller HJ, et al. ( 2006): Multicenter assessment of reliability of cranial MRI. Neurobiol Aging 27: 1051–1059. [DOI] [PubMed] [Google Scholar]
- Fusar‐Poli P, Perez J, Broome M, Borgwardt S, Placentino A, Caverzasi E, Cortesi M, Veggiotti P, Politi P, Barale F, et al. ( 2007): Neurofunctional correlates of vulnerability to psychosis: A systematic review and meta‐analysis. Neurosci Biobehav Rev 31: 465–484. [DOI] [PubMed] [Google Scholar]
- Giorgio A, Palace J, Johansen‐Berg H, Smith SM, Ropele S, Fuchs S, Wallner‐Blazek M, Enzinger C, Fazekas F ( 2010): Relationships of brain white matter microstructure with clinical and MR measures in relapsing‐remitting multiple sclerosis. J Magn Reson Imaging 31: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RNA, Friston KJ, Frackowiak RSJ ( 2001): A voxel‐based morphometric study of ageing in 465 normal adult human brains. NeuroImage 14: 21–36. [DOI] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Busa E, Pacheco J, Albert M, Killiany R, et al. ( 2006): Reliability of MRI‐derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. Neuroimage 32: 180–194. [DOI] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE ( 2004): Nonstationary cluster‐size inference with random field and permutation methods. NeuroImage 22: 676–687. [DOI] [PubMed] [Google Scholar]
- Huppertz HJ, Kroll‐Seger J, Kloppel S, Ganz RE, Kassubek J ( 2010): Intra‐ and interscanner variability of automated voxel‐based volumetry based on a 3D probabilistic atlas of human cerebral structures. NeuroImage 49: 2216–2224. [DOI] [PubMed] [Google Scholar]
- Jasperse B, Vrenken H, Sanz‐Arigita E, de Groot V, Smith SM, Polman CH, Barkhof F ( 2007): Regional brain atrophy development is related to specific aspects of clinical dysfunction in multiple sclerosis. NeuroImage 38: 529–537. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, MacFall J, et al. ( 2006): Reliability in multi‐site structural MRI studies: Effects of gradient non‐linearity correction on phantom and human data. NeuroImage 30: 436–443. [DOI] [PubMed] [Google Scholar]
- Kappos L, Antel J, Comi G, Montalban X, O'Connor P, Polman CH, Haas T, Korn AA, Karlsson G, Radue EW, et al. ( 2006): Oral fingolimod (FTY720) for relapsing multiple sclerosis. N Engl J Med 355: 1124–1140. [DOI] [PubMed] [Google Scholar]
- Kutzelnigg A, Lassmann H ( 2005): Cortical lesions and brain atrophy in MS. J Neurol Sci 233: 55–59. [DOI] [PubMed] [Google Scholar]
- Li X, Li L, Lu H, Liang Z ( 2005): Partial volume segmentation of brain magnetic resonance images based on maximum a posteriori probability. Med Phys 32: 2337–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikol DD, Barkhof F, Chang P, Coyle PK, Jeffery DR, Schwid SR, Stubinski B, Uitdehaag BMJ ( 2008): Comparison of subcutaneous interferon beta‐1a with glatiramer acetate in patients with relapsing multiple sclerosis (the REbif vs. Glatiramer acetate in relapsing MS disease [REGARD] study): A multicentre, randomized, parallel, open‐label trial. Lancet Neurol 7: 903–914. [DOI] [PubMed] [Google Scholar]
- Moorhead TWJ, Job DE, Spencer MD, Whalley HC, Johnstone EC, Lawrie SM ( 2005): Empirical comparison of maximal voxel and non‐isotropic adjusted cluster extent results in a voxel‐based morphometry study of comorbid learning disability with schizophrenia. NeuroImage 28: 544–552. [DOI] [PubMed] [Google Scholar]
- Moorhead TWJ, Gountouna VE, Job DE, McIntosh AM, Romaniuk L, Lymer GK, Whalley HC, Waiter GD, Brennan D, Ahearn TS, et al. ( 2009): Prospective multi‐centre voxel based morphometry study employing scanner specific segmentations: Procedure development using CaliBrain structural MRI data. BMC Med Imaging 9: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Fisher E ( 2009): Segmentation of brain magnetic resonance images for measurement of gray matter atrophy in multiple sclerosis patients. NeuroImage 44: 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor P, Filippi M, Arnason B, Comi G, Cook S, Goodin D, Hartung H‐P, Jeffery D, Kappos L, Boateng F, et al. ( 2009): 250 μg or 500 μg interferon beta‐1b versus 20 mg glatiramer acetate in relapsing‐remitting multiple sclerosis: A prospective, randomized, multicentre study. Lancet Neurol 8: 889–897. [DOI] [PubMed] [Google Scholar]
- Pagani E, Rocca MA, Gallo A, Rovaris M, Martinelli V, Comi G, Filippi M ( 2005): Regional brain atrophy evolves differently in patients with multiple sclerosis according to clinical phenotype. AJNR Am J Neuroradiol 26: 341–346. [PMC free article] [PubMed] [Google Scholar]
- Pardoe H, Pell GS, Abbott DF, Berg AT, Jackson GD ( 2008): Multi‐site voxel‐based morphometry: Methods and a feasibility demonstration with childhood absence epilepsy. NeuroImage 42: 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirko I, Lucchinetti CF, Sriram S, Bakshi R ( 2007): Gray matter involvement in multiple sclerosis. Neurology 68: 634–642. [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Edan G, Filippi M, Hartung H‐P, Kappos L, Lublin F, Metz L, McFarland H, O'Connor P, et al. ( 2005): Diagnostic criteria for multiple sclerosis: 2005 revisions to the ldquoMcDonald Criteriardquo. Ann Neurol 58: 840–846. [DOI] [PubMed] [Google Scholar]
- Preboske GM, Gunter JL, Ward CP, Jack CR Jr ( 2006): Common MRI acquisition non‐idealities significantly impact the output of the boundary shift integral method of measuring brain atrophy on serial MRI. Neuroimage 30: 1196–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinster A, Quarantelli M, Orefice G, Lanzillo R, Brunetti A, Mollica C, Salvatore E, Morra VB, Coppola G, Vacca G, et al. ( 2006): Grey matter loss in relapsing‐remitting multiple sclerosis: A voxel‐based morphometry study. NeuroImage 29: 859–867. [DOI] [PubMed] [Google Scholar]
- Quarantelli M, Ciarmiello A, Morra VB, Orefice G, Larobina M, Lanzillo R, Schiavone V, Salvatore E, Alfano B, Brunetti A ( 2003): Brain tissue volume changes in relapsing‐remitting multiple sclerosis: Correlation with lesion load. NeuroImage 18: 360–366. [DOI] [PubMed] [Google Scholar]
- Raz N, Lindenberger U, Rodrigue KM, Kennedy KM, Head D, Williamson A, Dahle C, Gerstorf D, Acker JD ( 2005): Regional brain changes in aging healthy adults: General trends, individual differences and modifiers. Cereb Cortex 15: 1676–1689. [DOI] [PubMed] [Google Scholar]
- Sailer M, Fischl B, Salat D, Tempelmann C, Schonfeld MA, Busa E, Bodammer N, Heinze H‐J, Dale A ( 2003): Focal thinning of the cerebral cortex in multiple sclerosis. Brain 126: 1734–1744. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Doyon J, McDonald D, Holmes C, Lavoie K, Hurwitz AS, Kabani N, Toga A, Evans A, Petrides M ( 1999): Three‐dimensional MRI atlas of the human cerebellum in proportional stereotaxic space. NeuroImage 10: 233–260. [DOI] [PubMed] [Google Scholar]
- Schnack HG, van Haren NE, Brouwer RM, van Baal GC, Picchioni M, Weisbrod M, Sauer H, Cannon TD, Huttunen M, Lepage C, et al. ( 2010): Mapping reliability in multicenter MRI: Voxel‐based morphometry and cortical thickness. Hum Brain Mapp 31: 1967–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemer G ( 2000): General linear models for multicenter clinical trials. Control Clin Trials 21: 21–29. [DOI] [PubMed] [Google Scholar]
- Sepulcre J, Sastre‐Garriga J, Cercignani M, Ingle GT, Miller DH, Thompson AJ ( 2006): Regional gray matter atrophy in early primary progressive multiple sclerosis: A voxel‐based morphometry study. Arch Neurol 63: 1175–1180. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Kan E, Woods RP, Yoshii J, Bansal R, Xu D, Zhu H, Thompson PM, Toga AW ( 2007): Sex differences in cortical thickness mapped in 176 healthy individuals between 7 and 87 years of age. Cereb Cortex 17: 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonnington CM, Tan G, Klöppel S, Chu C, Draganski B, Jack CR Jr, Chen K, Ashburner J, Frackowiak RSJ ( 2008): Interpreting scan data acquired from multiple scanners: A study with Alzheimer's disease. NeuroImage 39: 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif CL, Collins DL, Pike GB ( 2010): Regional impact of field strength on voxel‐based morphometry results. Hum Brain Mapp 31: 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi G, Lavorgna L, Russo P, Prinster A, Dinacci D, Savettieri G, Quattrone A, Livrea P, Messina C, Reggio A, et al. ( 2005): Brain atrophy and lesion load in a large population of patients with multiple sclerosis. Neurology 65: 280–285. [DOI] [PubMed] [Google Scholar]
- Tiberio M, Chard DT, Altmann DR, Davies G, Griffin CM, Rashid W, Sastre‐Garriga J, Thompson AJ, Miller DH ( 2005): Gray and white matter volume changes in early RRMS: A 2‐year longitudinal study. Neurology 64: 1001–1007. [DOI] [PubMed] [Google Scholar]
- White T, O'Leary D, Magnotta V, Arndt S, Flaum M, Andreasen NC ( 2001): Anatomic and functional variability: The effects of filter size in group fMRI data analysis. NeuroImage 13: 577–588. [DOI] [PubMed] [Google Scholar]


