ABSTRACT
Centers for Disease Control and Prevention (CDC), health departments, and other state and federal partners have linked contact with live poultry to 70 human Salmonella outbreaks in the United States from 2000 to 2017, which resulted in a total of 4,794 illnesses, 894 hospitalizations, and 7 deaths. During human salmonellosis outbreaks environmental sampling is rarely conducted as part of the outbreak investigation. CDC was contacted by state health officials on June 12, 2018, to provide support during an investigation of risk factors for Salmonella infections linked to live poultry originating at a mail-order hatchery. From January 1, 2018, to June 15, 2018, 13 human Salmonella infections in multiple states were attributed to exposure to live poultry from a single hatchery. Two serotypes of Salmonella were associated with these infections, Salmonella Enteritidis and Salmonella Litchfield. Molecular subtyping of the S. Enteritidis clinical isolates revealed they were closely related genetically (within 0 to 9 alleles) by core genome multi-locus sequence typing (cgMLST) to isolates obtained from environmental samples taken from hatchery shipping containers received at retail outlets. Environmental sampling and onsite investigation of practices was conducted at the mail-order hatchery during an investigation on June 19, 2018. A total of 45 environmental samples were collected, and 4 (9%) grew Salmonella. A chick box liner from a box in the pre-shipping area yielded an isolate closely related to the S. Enteritidis outbreak strain (within 1 to 9 alleles by cgMLST). The onsite investigation revealed lapses in biosecurity, sanitation, quality assurance, and education of consumers. Review of Salmonella serotype testing performed by the hatchery revealed that the number of samples and type of samples collected monthly varied. Also, S. Enteritidis was identified at the hatchery every year since testing began in 2016. Recommendations to the hatchery for biosecurity, testing, and sanitation measures were made to help reduce burden of Salmonella in the hatchery and breeding flocks, thereby reducing the occurrence of human illness.
Keywords: Salmonella, hatchery management, environment
INTRODUCTION
The Centers for Disease Control and Prevention (CDC) estimates non-typhoidal Salmonella causes approximately 1.2 million illnesses, 23,000 hospitalizations, and 450 deaths in the United States annually. Non-typhoidal Salmonella can be transmitted by food, water, direct animal contact, or person-to-person contact, (Pires et al., 2014). Approximately 1 million illnesses result from eating contaminated food, and 127,000 are attributed to contact with animals (Hale et al., 2012). This includes illnesses linked to contact with live poultry. Salmonella is a Gram-negative rod-shaped bacterium that causes gastrointestinal illness in humans and is most often acquired by the fecal-oral route. People infected with Salmonella who become symptomatic might develop diarrhea, fever, and abdominal cramps between 12 and 72 h after infection. Most infected individuals recover without treatment, yet, severe illness may occur especially in older adults, infants, and those who are immunocompromised (Lund and O'Brien, 2011).
From 2000 to 2017, 70 human Salmonella outbreaks were linked with live poultry in the United States, resulting in 4,794 illnesses, 894 hospitalizations, and 7 deaths (personal communication). Poultry become infected with Salmonella after contact with contaminated litter, feces, feed, water, shavings, equipment, or after contact with other infected animals (domestic and wild) or personnel (Poppe, 2000). Infected hens can also produce eggs that are internally contaminated with Salmonella (Poppe, 2000). Once infected, poultry can continue to intermittently shed Salmonella in their feces, even if they appear healthy. (Brownell et al., 1969; Nakamura et al., 1993; Gast and Holt, 1998). In attempts to reduce Salmonella shedding, vaccinations are often used; however, research has shown that vaccines do not eliminate Salmonella from the environment (Sharma et al., 2018), thus, a multifactor approach is required for Salmonella reduction in poultry (Barton-Behravesh et al., 2014).
According to the American Veterinary Medical Association (2018) and the United States Department of Agriculture (2013) keeping live poultry in backyard flocks is a growing trend in the United States. There are approximately 20 US mail-order hatcheries that produce over 582 million birds annually (USDA, 2018). Regulatory oversight authority regarding zoonotic disease control in these hatcheries is minimal. Consumers purchase poultry directly from hatcheries or from agriculture feed stores. Agriculture feed stores typically purchase poultry directly from mail-order hatcheries; however, industry practices such as drop-shipping often result in poultry originating from a different hatchery than the hatchery from which poultry were originally ordered. Drop-shipping, is a practice in which the initial hatchery relies on a different hatchery to supply and ship a customer's order, while using the initial hatchery's name (Nichols et al., 2018). Drop-shipping can make it difficult for public health investigators to establish an epidemiologic link between patients and a source hatchery during outbreak investigations. To help reduce Salmonella incidence at the hatchery level, mail-order hatcheries can voluntarily participate in the United States Department of Agriculture National Poultry Improvement Plan (NPIP) for Salmonella reduction (2014a). Participating hatcheries should employ enhanced biosecurity, sanitation and quality assurance, and are expected to work only with other hatcheries and suppliers that also participating in the program.
This report describes the investigation of a multistate outbreak of human Salmonella Enteritidis and Salmonella Litchfield infections linked to contact with multiple species of live poultry originating from one mail-order hatchery in the United States (Hatchery A). Routine, ongoing surveillance was key to detecting this outbreak of human illnesses. The investigation included interviews with ill people, traceback of implicated poultry to identify the mail-order hatchery source, and a site assessment of Hatchery A, including: environmental sampling, a survey of the facilities and work flow processes, and a review of Salmonella control measures. At the conclusion of the investigation, we made recommendations to Hatchery A. Implementation of interventions at Hatchery A to reduce the risk of human Salmonella infections are ongoing.
METHODS AND MATERIALS
Surveillance
As part of ongoing surveillance, state and local public health laboratories characterized Salmonella isolates from ill people by using serotyping, pulse-field gel electrophoresis (PFGE), and whole genome sequencing. Whole genome sequencing in this investigation included whole genome multi-locus sequence typing (wgMLST), high-quality single nucleotide polymorphism analysis, and core genome multi-locus sequence typing (cgMLST). Sequences were submitted by public health laboratories to PulseNet, the national molecular subtyping network for foodborne pathogens. During this investigation, PulseNet underwent a planned transition in molecular subtyping methods from wgMLST and high-quality single nucleotide polymorphism analysis to cgMLST. From 2016 to 2018 the state public health agency, in the state which Hatchery A is located, also collected environmental samples from poultry shipping containers arriving at agricultural feed stores (Sidge et al., 2019). Initially, a case was defined as isolation of S. Enteritidis PFGE XbaI pattern JEGX01.0004 or S. Litchfield PFGE XbaI pattern JGXX01.0009 (outbreak strains) from a person with illness onset from January 1, 2018, to June 15, 2018, with either exposure to live poultry or an isolate closely related by wgMLST or high-quality single nucleotide polymorphism analysis. This case definition was later refined to be a Salmonella infection with one of the outbreak strains and corresponding PFGE patterns, with illness onset from January 1, 2018, to June 15, 2018, and an isolate closely related genetically to one of the strains by cgMLST. Antibiotic resistance was predicted from whole genome sequencing data by the National Antimicrobial Resistance Monitoring System using established methods (McDermott et al., 2016).
State and local public health officials interviewed case-patients or their guardians using standard questionnaires developed by state and local health departments and CDC (Nakao et al., 2015). These initial questionnaires asked about foods eaten and food purchase locations, water exposures, and animal contact. If live poultry contact was reported on initial interview, an additional CDC-developed questionnaire was administered which included more specific questions about live poultry exposure, including: type of exposure and degree of contact, species of live poultry, and purchase information.
Traceback
Retail locations where ill people reported purchasing live poultry were contacted to obtain information on where the live poultry were sourced, tracing them back to a hatchery of origin. In addition, source hatchery information was obtained for live poultry sampled in feed store locations by state and local health officials (Sidge et al., 2019). CDC, state, and local public health personnel conducted the traceback investigations.
Site Assessment
Environmental Sampling
Environmental sampling for Salmonella was conducted at Hatchery A by state and local health departments and CDC staff on June 19, 2018 (Hardy et al., 2019). All environmental sampling was done on a single day and was conducted in a manner which represented movement of eggs and chicks through the hatchery during production. Based on epidemiologic evidence and recognized potential risk factors for Salmonella transmission, a prioritized list of sampling areas was prepared. Sampled areas in priority order were: chick environment (including liners inside egg-hatching incubators and inside and outside egg-hatching incubators); pre-shipping area; adult bird environment (including nest and laying boxes, bedding, and food and water containers); and trucks used for live poultry and egg transportation onsite and offsite. Shoe covers worn by the sampling team inside the hatchery buildings were also tested after sampling was complete.
Sample collectors were briefed on priorities and techniques on the day of sampling to ensure consistency. The sampling team followed best practices for biosecurity on poultry farms (USDA, 2014b). Three different swabbing techniques were used: sterile polyurethane culture swabs in liquid Amies agar gel, sterile wooden swabs, and sterile gauze squares (USDA, 2014b). Additional samples were collected from chick box liners and bedding then placed in sterile whirl pack bags and sterile collection cups, respectively.
Using chain-of-custody protocols, samples were transported at ambient temperature in sealed containers and delivered to the state public health laboratory within 6 h of the start of sampling. Samples were cultured and screened by polymerase chain reaction; presumptive Salmonella colonies were biochemically identified and analyzed by matrix assisted laser desorption ionization-time of flight mass spectrometry and serotyped. Salmonella isolates were characterized through PFGE and wgMLST. Salmonella isolates were retrospectively characterized by cgMLST to further examine genetic relatedness.
Survey
Recommended hatchery facility and workflow processes are available in the NPIP Best Management Practice Handbook (USDA, 2014a). Recommendations are made in 4 categories: biosecurity, sanitation, quality assurance, and education of consumers. Hatchery A processes were assessed using a hatchery questionnaire provided by NPIP, and augmented with a CDC hatchery-specific questionnaire (Nakao et al., 2015). This questionnaire was developed to facilitate the traceback investigation, identify the original poultry source, and better characterize industry practices such as drop-shipping, comingling, and multiplying that may complicate outbreak investigations. The number of NPIP recommendations that were implemented on the day of assessment within each category were tabulated.
Record Review
We reviewed Hatchery A's records, including the measures they were taking to reduce the burden of Salmonella in the hatchery environment at the time of the outbreak. These measures were developed in 2016 by a veterinary consultant retained by Hatchery A. These measures were compared to those recommended by the NPIP to determine percent agreement. Salmonella test results of hatchery facility environmental and carcass samples, reported by Hatchery A from January 2016 to May 2018 were also reviewed. These results included the number of samples tested, sample type, sample location, and Salmonella serotype identified per sample. Number of Salmonella positive samples by sample type, date, and facility location were assessed. Commercial operations from where the hatchery obtained eggs and chicks for the 2018 season were reviewed to determine if eggs and chicks were obtained from hatcheries and suppliers that participated in the NPIP Salmonella monitoring and control program. Finally, a list of hatcheries for which Hatchery A performed drop-shipping, including quantity, breeds, and time period over which drop-shipping occurred was reviewed.
RESULTS
Surveillance
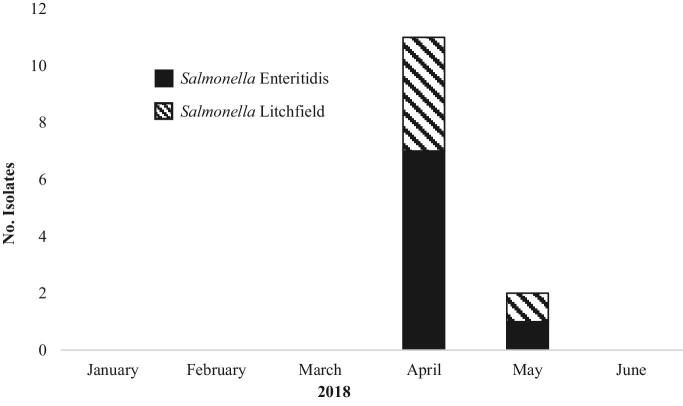
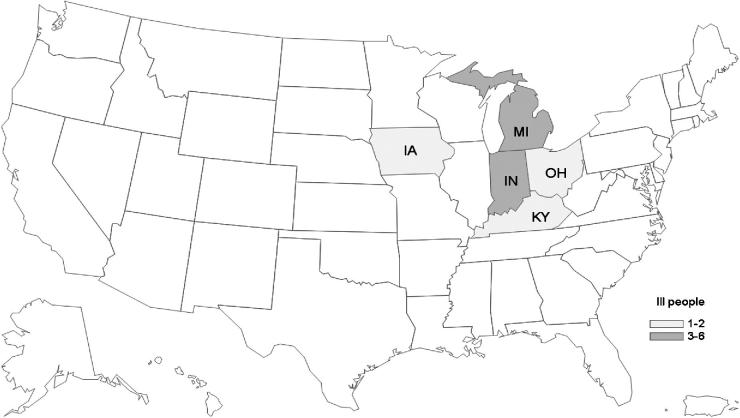
From January 1, 2018 to June 15, 2018, 24 cases were identified from 11 states; however, when the refined case definition was applied, and genetic relatedness was evaluated by cgMLST, the number of cases was reduced to 13 from 5 states (Table 1, and Figures 1 and 2). The median age of case-patients was 22 yr with a range of less than 1 to 73 yr; 4 case-patients were 5 yr of age or younger. Nine of 13 case-patients were female, 3 of 7 with information available were hospitalized, and no deaths were reported. No isolates had predicted resistance to the National Antimicrobial Resistance Monitoring System standard panel of antimicrobial agents. Information on contact with live poultry was available for 8 patients, 6 of whom reported they had contact with live poultry in the week preceding illness onset (Table 1). The outbreak strain of S. Enteritidis was also isolated from 8 live poultry shipping container liners (Sidge et., al. 2019). Sequence analysis revealed that the S. Enteritidis clinical isolates were closely related to one another and to the 8 environmental isolates obtained from live poultry shipping containers (0 to 9 alleles by cgMLST, Figure 3); the S. Litchfield clinical isolates were also closely related to one another (0 to 7 alleles by cgMLST, Figure 3).
Table 1.
Selected demographic and clinical information of ill people infected with outbreak strains of S. Enteritidis and S. Litchfield, January 1, 2018 to June 15, 2018.1
| Variable | S. enteritidis | S. litchfield | Total |
|---|---|---|---|
| No. of states with cases reporting to PulseNet | 4 | 3 | 52 |
| No. of ill people | 8 | 5 | 13 |
| Age range in years (median) | 2 to 60 (29) | <1 to 73 (11) | <1 to 73 (22) |
| Females—no. (n3) | 6 (n = 8) | 3 (n = 5) | 9 (n = 13) |
| No. hospitalized patients (n3) | 1 (n = 4) | 2 (n = 3) | 3 (n = 7) |
| No. patients with live poultry exposure (n3) | 4 (n = 6) | 2 (n = 2) | 6 (n = 8) |
Information was obtained from the PulseNet database.
Three states had both S. Enteritidis and S. Litchfield cases.
n is the number of patients with information available.
Figure 1.
Number of clinical isolates with one of the Salmonella outbreak strains, by isolation date as reported to PulseNet database, January 1, 2018 to June 15, 2018 (n = 13).
Figure 2.
Number of S. Enteritidis and S. Litchfield illnesses linked to live poultry, by state of residence, January 1, 2018 to June 15, 2018 (n = 13). The data are shown according to the state that tested the patient for the outbreak strains.
Figure 3.
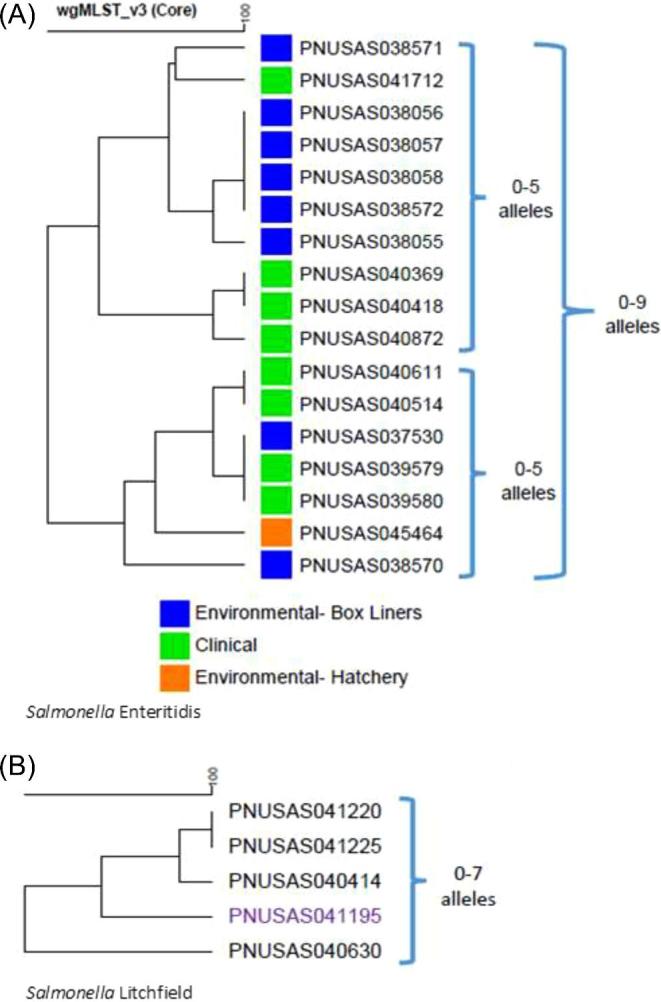
Dendrogram of clinical and environmental isolates in this outbreak by core genome multi-locus sequence typing (n = 22). A. Salmonella Enteritidis. B. Salmonella Litchfield.
Traceback
Patients infected with S. Enteritidis reported purchasing poultry from agricultural feed stores that sourced poultry from Hatchery A. Additionally, shipping containers at feed stores from which samples were taken as part of ongoing surveillance, and yielded the outbreak strain of S. Enteritidis, were labeled as originating from Hatchery A. During patient interviews, people infected with the outbreak strains of S. Litchfield indicated they had purchased poultry from stores that reported sourcing poultry from Hatchery A.
Site Assessment
Environmental Sampling
A total of 45 environmental samples were collected at Hatchery A, and 4 (9%) grew Salmonella. A chick box liner collected in the pre-shipping area yielded an isolate closely related genetically to the S. Enteritidis outbreak strain (1 to 9 alleles by cgMLST, Figure 3). Three environmental samples collected from an outbuilding used to house breeder stock birds, grew Salmonella Typhimurium. Initial screening was performed by the state public health laboratory, and sequencing found them to be closely related genetically (0 to 1 alleles) by cgMLST. Sequence analysis of these isolates performed by the United States Department of Agriculture's Center for Veterinary Biologics determined them to be closely related to the strain used in the modified-live Poulvac ST vaccine (Zoetis, USA) used to vaccinate breeder birds at Hatchery A (personal communication). PulseNet was queried for any clinical isolates matching these environmental S. Typhimurium isolates by PFGE. Three clinical isolates were identified, but based on cgMLST, these isolates were up to 305 alleles from the environmental isolates and were classified as not closely related.
Survey
The onsite investigation revealed lapses in biosecurity, sanitation, quality assurance, and education of consumers. Facility observations and answers to the NPIP survey and CDC hatchery-specific questionnaire were used to evaluate Hatchery A's adherence to NPIP best practices. Adherence to recommendations intended to reduce Salmonella in the hatchery environment was lower than expected for a large, commercial mail-order hatchery for the categories of sanitation (15%), biosecurity (39%), quality assurance (40%), and education of consumers (50%). Some of these finding are also lower than what was previously noted in a survey of other hatcheries (Nakao et al., 2015; Sharma et al., 2018). Significant sanitation findings included the observation that beverages intended for human consumption were stored in the hatchery; employees consumed food and beverages on the table used for sexing chicks; the incubators are constructed of unsealed wood which cannot be sterilized; and the potable water source is an unchlorinated and unfiltered well. Key biosecurity findings included the observation that employees wore casual attire during work, and no shower or coveralls were provided; no employee identification system was in place; no wheel disinfection procedures were present; and employee foot baths were dry. Regarding quality assurance, Salmonella serotype testing was performed as recommended (USDA, 2014b), but the number of samples obtained per month was highly variable. Finally, it was observed that consumer materials for education of Salmonella health risks and prevention measures were inconsistently provided in chick shipping boxes.
Record Review
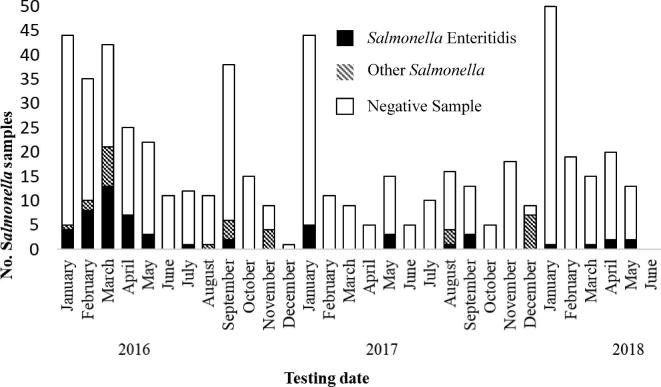
Hatchery A provided a record of Salmonella mitigation measures recommended by a paid consultant veterinarian. A comparison of the consultant's recommendations to the NPIP recommendations revealed that the consultant made recommendations in 10 of the 12 major categories provided by NPIP (USDA, 2014a).
Review of Hatchery A Salmonella serotype testing performed by a commercial laboratory, revealed that the monthly sample collection process for Salmonella testing was inconsistent, with the number of samples submitted ranging from 1 to 50 per month, with a median of 15 per month (Figure 4). Salmonella Enteritidis was identified at the hatchery every year since testing began in 2016; it was found in every month tested in 2018 with the exception of February.
Figure 4.
Hatchery A Salmonella testing results by month. Hatchery A collects a combination of chicks and environmental samples; sample testing and isolate serotyping is performed by a commercial laboratory. Other Salmonella serotypes included Infantis, Kentucky, Memphis, Muenster, and Saintpaul.
Testing records indicated that Hatchery A sampled from 2 general locations, the breeder coops and inside the hatchery. Breeder coop samples predominantly consisted of litter, and the percent of samples positive for Salmonella decreased over the years of sampling as follows: 2016 (19%), 2017 (9%), and through May 8, 2018 (0%). Hatchery samples consisted predominately of hatch debris, live chicks, and environmental swabs. As seen in Table 2, the greatest number of samples were live chicks (269), followed by environmental swabs (100), and hatch debris (33). The sample type with the greatest number of samples positive for Salmonella was environmental swabs, followed by live chicks, and hatch debris.
Table 2.
Hatchery A Salmonella testing results by year and sample type, 2016 to 2018.
| Sample type | Year | No. samples | No. Salmonella positive samples | Percent positive for Salmonella |
|---|---|---|---|---|
| Hatch debris | 2016 | 15 | 2 | 13% |
| 2017 | 4 | 0 | 0% | |
| 20181 | 14 | 1 | 7% | |
| Live chick | 2016 | 116 | 34 | 29% |
| 2017 | 78 | 5 | 6% | |
| 20181 | 75 | 0 | 0% | |
| Environmental swab | 2016 | 29 | 9 | 31% |
| 2017 | 36 | 16 | 44% | |
| 20181 | 35 | 5 | 14% |
Includes sample information from January 1, 2018 to May 8, 2018.
A review of commercial and non-commercial operations that were sources of eggs and chicks for the 2018 season revealed that some eggs were sourced from suppliers that do not participate in the USDA-NPIP Salmonella monitoring and control program. Finally, records indicated that Hatchery A, drop-shipped to states known to have clinical isolates of the outbreak strain but not previously linked to Hatchery A through traceback.
DISCUSSION
Live-poultry associated infections have increased concurrently with an increase in the number of backyard poultry flocks in the United States since 1990 (Basler et al., 2016). Backyard flocks are most frequently stocked with poultry acquired from agricultural retail stores, or more rarely, are purchased directly from a hatchery. Every year, over 50 million chicks are distributed nationally from a core group of approximately 20 mail-order hatcheries (Gaffga et al., 2012). Whether mailed directly to consumers or to an agricultural retail store, baby poultry are shipped through the U.S. Postal Service. When shipped, groupings of baby poultry consisting of up to 120 chicks, 60 ducklings, 32 goslings, or 80 turkey poults (personal communication) are packaged into cardboard boxes and a single box might contain multiple species. This concentration of baby poultry within a shipping container may increase the risk for cross contamination and sharing of Salmonella strains between birds. Shipping in this manner has been shown to increase animal stress, which can lead to increased Salmonella shedding (Gast and Holt, 1998). The nationwide distribution of poultry shipping combined with the stress of shipping and the opportunity for cross contamination may explain, in part, the multistate distribution of recent live-poultry associated Salmonella outbreaks (Nichols et al., 2018).
As indicated by the surveillance and traceback efforts during this 2018 investigation, the Salmonella outbreak strains (S. Enteritidis and S. Litchfield) were isolated from all ill people in the outbreak. In all 6 of 8 patients reported contact with live poultry in the week preceding illness, with 2 of these individuals reporting purchasing the poultry from agricultural feed stores that sourced their birds from Hatchery A. This was not the first year that Hatchery A was linked to human Salmonella illness outbreaks. Historic human Salmonella outbreaks linked to Hatchery A date back from 1999 (Bidol et al., 2000). Additional links were made between human Salmonella outbreak strains and Hatchery A in 2000, 2006, and from 2015 to 2018 (Wilkins et al., 2002; Bidol et al., 2007). State public health officials worked with animal health and NPIP partners to provide recommendations and interventions to mitigate and control Salmonella at Hatchery A. In 2018, state and local public health officials together with CDC, developed plans for a joint site assessment and sampling strategy at Hatchery A.
Salmonella Enteritidis is a common serotype in outbreaks linked to live poultry exposure (Bäumler et al., 2000 and Velge et al., 2005), therefore it is not surprising that it was the predominant serotype identified in this outbreak. Salmonella Litchfield however, historically has not been found to be associated with outbreaks linked to live poultry exposure, but was linked to Hatchery A in 2017. The Salmonella testing Hatchery A performed from 2016 until the site assessment in 2018 identified a multitude of Salmonella serotypes including Infantis, Kentucky, Memphis, Muenster, and Saintpaul; however, S. Enteritidis was the predominant serotype, representing 65% of the positive samples. Salmonella Enteritidis was identified within the hatchery in 2016, 2017, and in almost every month in 2018, up to the site assessment, suggesting long-standing colonization, or repeated introduction of this serotype into the hatchery. It is possible that S. Litchfield was not identified at Hatchery A during this investigation because it was likely at a lower level of environmental contamination at the facility compared to S. Enteritidis.
Hatchery A's routine Salmonella testing focused on hatch debris, live chicks, and environmental samples. From 2016 to 2018, the percent of samples positive for Salmonella decreased for both hatch debris and live chicks, but not for environmental samples. Environmental samples consistently produced the highest percent of Salmonella positives every year and increased 13% from 2016 to 2017 (Table 2). These results are higher than the expected isolation rates for the sample types taken (Sharma et al., 2017). In addition, the 2018 S. Enteritidis positive samples from the hatchery were isolated from the floor in both the egg and hatch rooms, as well as the top of the incubators. These results indicate that S. Enteritidis may be resident within the hatchery and not continually imported from eggs and chicks sourced from breeders. However, as Hatchery A sources birds from non-NPIP participating suppliers, the initial contamination might have come from a supplier.
The findings of this investigation are subject to several limitations. Case ascertainment relied on laboratory data, and additional cases might have been missed if they were not detected by PulseNet during the timeframe of the investigation. The site assessment at Hatchery A was conducted on a single day and thus is constrained by the limitations of cross-sectional observations. However, based on findings from the review of Hatchery A's records and from answers to the NPIP and CDC hatchery-specific questionnaire provided by Hatchery A, it is unlikely that findings would have changed if multiple assessments had been conducted over multiple days. Also, the NPIP hatchery manager survey was largely dependent on the recall ability of Hatchery A, and consequently subject to recall bias. Many of the answers provided by Hatchery A were corroborated by visual inspection and review of records. Other answers however, contrasted with our observations and favored processes that aligned with NPIP best practices. In addition, because routine Salmonella testing by the hatchery was performed at a commercial laboratory, only serotype was identified, and no isolates were available for additional analysis. Consequently, it is not possible to determine with certainty if the S. Enteritidis samples identified by Hatchery A are the outbreak strain. Finally, drop-shipping by Hatchery A made it difficult for public health investigators to establish a connection, between some patients and Hatchery A in states with clinical isolates but no epidemiologic link to Hatchery A.
The U.S. Department of Agriculture's National Poultry Improvement Plan (USDA, 2014a) has developed biosecurity, sanitation, quality assurance, and consumer education standards “to assist hatchery operators in mitigating Salmonella contamination of birds to be sold through the mail, feed stores, or other retail outlets.” Preventing the spread of Salmonella transmitted by eggs and disseminated at hatcheries, as well as promoting the proper handling and husbandry of live poultry by consumers, will reduce the number of human illnesses linked to contact with live poultry. However, compliance with NPIP recommendations by mail-order hatcheries is voluntary, and adherence is variable among the mail-order hatcheries. Comprehensive control programs based on NPIP recommendations can be developed in collaboration with a consulting veterinarian. When developing control programs, hatcheries and consulting veterinarians should consider routine microbiologic monitoring and effective sanitation procedures, including procedures targeted to address results of microbiologic monitoring. However, the high cost of implementation of all recommendations might be cost prohibitive for some smaller hatcheries.
We encourage more collaboration between NPIP, industry partners (including hatcheries and agricultural feed stores), and animal and public health agencies at the local, state, and federal level. More direct communication between hatcheries, and health and regulatory agencies might expedite public health officials’ awareness of potential disease risks before an outbreak occurs, and might also alert a hatchery to a disease contaminant that could be eliminated. Efforts to promote microbiologic monitoring through state laboratories with the capacity to conduct sequencing and perform comparisons between human, animal, and environmental samples, will allow for more efficient identification of outbreaks through molecular subtyping of isolates, thereby facilitating a faster, more focused response. Finally, information regarding the potential risk of Salmonella infection associated with live poultry and measures consumers can take to reduce risk should be consistent between all partners within the distribution network. This outbreak has prompted regular direct communication, monitoring, and a corrective action timeline for Hatchery A by local public health officials.
ACKNOWLEDGMENTS
We wish to thank the United States Department of Agriculture's Center for Veterinary Biologics, Ames, Iowa for their assistance in identifying the 3 Hatchery A environmental samples collected from an outbuilding used to house breeder stock birds, as the strain used in the modified-live Poulvac ST vaccine (Zoetis US). Also, we wish to thank the Michigan Department of Health and Human Services, Lansing, Michigan, and the Ottawa County Department of Public Health, Holland, Michigan for their significant contribution to this investigation.
Notes
The Salmonella nucleotide sequence data reported in this paper have been uploaded to the PulseNet Salmonella bioproject (PRJNA230403) at the National Center for Biotechnology Information, U.S. National Library of Medicine, Bethesda MD, 20894
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- American Veterinary Medical Association (AVMA). 2018. AVMA pet ownership and demographics sourcebook executive summary, 2017–2018 edition. AVMA. Accessed Jan. 2019. https://www.avma.org/PracticeManagement/Documents/AVMA-Pet-Demographics-Executive-Summary.pdf. [Google Scholar]
- Basler C., Nguyen T., Anderson T., Hancock T., Barton-Behravesh C.. 2016. Outbreaks of human Salmonella infections associated with live poultry, United States, 1990–2014. Emerg. Infect Dis. 22:1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Behravesh C., Briston D., Hopkins B., Gomez T.. 2014. Backyard poultry flocks and salmonellosis: a recurring, yet preventable public health challenge. CID. 58:1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler A. J., Hargis B. M., Tsolis R. M.. 2000. Tracing the origins of Salmonella outbreaks. Science. 287:50–52. [DOI] [PubMed] [Google Scholar]
- Bidol S., Stobierski M. G., Robinson-Dunn B., Massey J., Hall W., Boulton M., Warwick M., Borst M., Wiedlocher S.. 2000. Salmonellosis associated with chicks and ducklings-Michigan and Missouri, Spring 1999. MMWR Morb. Mortal Wkly. Rep. 49:297–299. [PubMed] [Google Scholar]
- Bidol S., Stobierski M. G., Leschinsky D., Ettestad P., Smelser C., Sena-Johnson D., Jungk J., Tafoya N., Torres P., Taylor F., Keene W., Plantenga M., Progulske B., TenEyck R., Rada R., Effinger L., Lockett J., Patel N., Angulo F., Bair-Breake H., Gaffga N.. 2007. Three outbreaks of salmonellosis associated with baby poultry from three hatcheries—United States. MMWR Morb. Mortal Wkly. Rep. 56:273–276. [PubMed] [Google Scholar]
- Brownell J. R., Sadler W. W., Fanelli M. J.. 1969. Factors influencing the intestinal infection of chickens with Salmonella Typhimurium. Avian Dis. 13:804–816. [PubMed] [Google Scholar]
- Gaffga N., Barton-Behravesh C., Ettestad P., Smelser C., Rhorer A., Cronquist A., Comstock N., Bidol S., Patel N., Gerner-Smidt P., Keene W., Gomez T.. 2012. Outbreak of salmonellosis linked to live poultry from a mail-order hatchery. N. Engl. J. Med. 366:2065–2073. [DOI] [PubMed] [Google Scholar]
- Gast R. K., Holt P. S.. 1998. Persistence of Salmonella Enteritidis from one day of age until maturity in experimentally infected layer chickens. Poult. Sci. 77:1759–1762. [DOI] [PubMed] [Google Scholar]
- Hale C., Scallan E., Cronquist A., Dunn J., Smith K., Robinson T., Lathrop S., Tobin-D'Angelo M., Clogher P.. 2012. Estimates of enteric illness attributable to contact with animals and their environments in the United States. CID. 54:S472–S479. [DOI] [PubMed] [Google Scholar]
- Hardy M., Robertson S., Sidge J., Signs K., Stobierski M. G., Jones K., Soehnlen M., Stefanovsky L., Hambley A., Brandenburg J., Martin H., Lauer A. C., Fields P., Koski L., Stevenson L., Pabilonia K., Nichols M., Basler C., Ribot E., Hise K.. 2019. Notes from the field: environmental investigation of a multistate salmonellosis outbreak linked to live backyard poultry from a mail-order hatchery — Michigan, 2018. MMWR Morb. Mortal Wkly. Rep. 67:1430–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund B., O'Brien S. J.. 2011. The occurrence and prevention of foodborne disease in vulnerable people. Foodborne Pathog. Dis. 8:961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott P., Tyson G., Kabera C., Chen Y., Li C., Folster J., Ayers S., Lam C., Tate H., Zhao S.. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60:5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Nagamine N., Suzuki S., Norimatsu M., Oishi K., Kijima M., Tamura Y., Sato S.. 1993. Long-term shedding of Salmonella Enteritidis in chickens which received a contact exposure within 24 hrs of hatching. J. Vet. Med. Sci. 55:649–653. [DOI] [PubMed] [Google Scholar]
- Nakao J., Pringle J., Jones R., Nix B., Borders J., Heseltine G., Gomez T., McCluskey B., Roney C., Brinson D., Erdman M., McDaniel A., Barton-Behravesh C.. 2015. ‘One health’ investigation: outbreak of human Salmonella Braenderup infections traced to a mail-order hatchery – United States, 2012–2013. Epidemiol. Infect. 143:2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols M., Stevenson L., Whitlock L., Pabilonia K., Robyn M., Basler C., Gomez T., Barton Behravesh C.. 2018. Preventing human Salmonella infections resulting from live poultry contact through interventions at retail stores. J. Agric. Saf. Health. 24:155–166. [DOI] [PubMed] [Google Scholar]
- Pires S., Vieira A., Hald T., Cole D.. 2014. Source attribution of human salmonellosis: an overview of methods and estimates. Foodborne Pathog. Dis. 11:667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C. 2000. Salmonella infections in the domestic fowl. Pages107–132 in Salmonella in domestic animals. Wray C., Wray A., ed. CAB International, New York, NY. [Google Scholar]
- Sharma A., Erdman M., Muñoz‐Vargas L., Mollenkopf D., Habing G.. 2017. Changes in the prevalence, genotypes and antimicrobial resistance phenotypes of non-typhoidal Salmonella recovered from mail-order hatchling poultry sold at US feed stores, 2013–2015. Zoonoses. 65:102–112. [DOI] [PubMed] [Google Scholar]
- Sharma P., Caraguel C., Sexton M., McWhorter A., Underwood G., Holden K., Chousalkar K.. 2018. Shedding of Salmonella Typhimurium in vaccinated and unvaccinated hens during early lay in field conditions: a randomized controlled trial. BMC Micro. 18:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidge J., Signs K., Bidol S., Jones K., Robeson S., Soehnlen M., Stobierski M.. 2019. Live poultry shipment box sampling at feed stores as an indicator for human Salmonella infections. MMWR Morb. Mortal Wkly. Rep. 68:407–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Department of Agriculture (USDA)-Animal and Plant Health Inspection Service-Veterinary Services-National Animal Health Monitoring System. 2013. Urban chicken ownership in four U.S. cities. USDA. Accessed Jul. 2018. https://www.aphis.usda.gov/animal_health/nahms/poultry/downloads/poultry10/Poultry10_dr_Urban_Chicken_Four.pdf. [Google Scholar]
- United States Department of Agriculture (USDA)-Animal and Plant Health Inspection Service-Veterinary Services-National Poultry Improvement Plan. 2014a. Best management practices handbook: a guide to the mitigation of Salmonella contamination at poultry hatcheries. USDA. Accessed Jul. 2018. http://poultryimprovement.org/documents/BestManagementPracticesHatcheries.pdf. [Google Scholar]
- United States Department of Agriculture (USDA)-Animal and Plant Health Inspection Service-Veterinary Services. 2014b. National poultry improvement plan standards. USDA. Accessed Jul. 2018. https://www.poultryimprovement.org/documents/ProgramStandardsAugust2014.pdf. [Google Scholar]
- United States Department of Agriculture (USDA)-National Agricultural Statistics Service. 2018. Hatchery production 2017 summary. USDA. Accessed Jul. 2019. https://www.nass.usda.gov/Publications/Todays_Reports/reports/htpdan18.pdf [Google Scholar]
- Velge P., Cloeckaert A., Barrow P.. 2005. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 36:267–288. [DOI] [PubMed] [Google Scholar]
- Wilkins M. J., Bidol S. A., Boulton M. L., Stobierski M. G., Massey J. P., Robinson-Dunn B.. 2002. Human salmonellosis associated with young poultry from a contaminated hatchery in Michigan and the resulting public health interventions, 1999 and 2000. Epidemiol. Infect. 129:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]