ABSTRACT
One natural recombinant avian leukosis virus (ALV) strain GX14DJ3–18 was isolated from a native gamecock by DF-1 cell culture and identified with Polymerase Chain Reaction (PCR), immunofluorescence assay and the viral genome's nucleotide sequencing. This strain was revealed as a novel recombinant virus with nucleotide sequence similarities of 95.4% Long Terminal Repeated (LTR), 95.8% 5′, UTR, 97.9% gag, and 92.9% 3′untranslated regions (UTR) in ALV-J. Also we found sequence similarities of 99.3% pol and 99.0% gp37 in ALV-E, and 89.9% gp85 in ALV-A. The simulated congenital infection with GX14DJ3–18 in Three-Yellow chickens exhibited a significant negative effect on the development of immune organs (P < 0.05). Also, lower antibody responses were found to vaccinations with the commercial vaccines of Newcastle disease virus and with subtypes H5 and H9 of avian influenza virus (P < 0.05). The incidence of tumor or tumor-like lesions in the challenged birds was 14.28% (5/35), while none were observed in the un-challenged control group (0/35). These results suggested that GX14DJ3–18 is a novel recombinant ALV that can induce pathogenicity in the commercial Three-Yellow chickens. We speculated that cross-provincial sales of gamecocks in which ALVs have not been eradicated thoroughly might be a potential route for the transmission of ALVs to commercial chickens.
Keywords: avian leukosis virus, recombinant, genome sequence, pathogenicity, transmission
INTRODUCTION
Avian leukosis viruses (ALVs) are known as a variety of infectious retroviral agents that cause severe economic loss to the poultry industry worldwide (Payne and Nair, 2012). Avian leukosis viruses that infect chickens comprise 7 envelope subgroups designated A to E, J and K (Wang et al., 2018a). Subgroups A, B, C, D, J, and K are exogenous ALVs while subgroup E is endogenous (Su et al., 2018). Among these subgroups, subgroup J (ALV-J) is the most prevalent in chickens (Lin et al., 2017) and causes multiple malignant tumors, such as myelocytoma and hemangioma (Zhang et al., 2018). Subgroups A, B, and K are recognized as common pathogenic viruses that induce lymphoid leukosis and sarcoma (Wang et al., 2017). Subgroups C and D infections have been reported rarely (Li et al., 2018a).
In China, owing to the initiation of a Nationwide Eradication Program (NEP) from 2008, the infection of the exogenous ALVs was significantly reduced in big representative breeding farms (Wang et al., 2018b). However, various ALV field strains are still circulating in numerous small-scale native chicken breeds in which strict and effective eradication programs have not been used (Su et al., 2018). The Guangxi gamecock is a rare bird known for its good fighting ability, competition, and entertainment value. About 80,000 gamecocks per year were sold to other parts of Guangxi and other provinces in China like Guizhou, Henan, Anhui, Xinjiang (Wei et al., 2019).
In this study, a unique ALV strain was isolated from a Guangxi gamecock. The isolate's genome was characterized by complete genome sequencing and comparing with other ALV reference strains. The pathogenicity study of the strain was carried out using day-old commercial Three-Yellow chickens through intra-abdominal injection. We describe the effect on the infected flock, including the effect on the development of immune organs, the immune-responses to the inactivated vaccines and tumor incidence of the birds.
MATERIAL AND METHODS
Recently, a total of five thousand five hundred 17- to 20-week-old gamecocks showed mental depression, stunted growth, and other production problems as well as clinical tumor lesions. Mortality rates for certain flocks reached 4 to 10% (He et al., 2016). To understand the epidemiology of ALV infections in gamecocks, 3 flocks from the local gamecock farms in Pingxiang county of Guangxi, China were tested for ALVs infection and a total of 276 birds (20 to 22 wk of age) were included in the analysis. Serum samples were collected for detection of ALV antibodies (Avian Leukosis Virus Antibody Test Kit, IDEXX), and plasma samples were collected for virus isolation. The results showed that a total of 22.46% (62/276) of the sampled birds were positive for ALV-J reactive antibodies, 7.25% (20/276) were positive for A/B subgroup specific antibodies, and 12.68% (35/276) were positive for the virus.
Virus Isolation and Identification
DF-1 cells were used to exclude the possibility of replicating endogenous viruses interfering with virus isolation as previously reported (Wang et al., 2017). Briefly, 2 × 107 DF-1 cells were plated in each of 24-well plates. Twenty four hours later, positive serum were inoculated into the DF-1 cell cultures for 1 h, and then replenished with Dulbecco's modified Eagle's medium containing 2% fetal calf serum (Life, Australia). After 3 serial passages (7 D for each passage) (Maas et al., 2006), cell cultures were used for the identifications of the viruses by Polymerase Chain Reaction (PCR) and immunofluorescence assay (IFA). Specific monoclonal antibodies (McAb) against ALV-A, ALV-B, and ALV-J were applied separately to detect the viral gp85 proteins in the infected DF-1 as previously reported (Qin et al., 2001). Also the deoxyribonucleic acid (DNA) from the cell culture samples was prepared using a commercial DNA extraction kit (TIANGEN, Beijing, China), and further used for the PCR detection of different ALV subgroups. The primers used for the PCR detection are presented in Table 1. The conditions for PCR are as follows: 95°C 4 min; 95°C 30 s, 50 to 60°C 30 s, 72°C, 30 s/kb (32 cycles); and 72°C 10 min. The PCR products were analyzed by electrophoresis using 1.5% agarose in a tris-Ethylene Diamine Tetraacetic Acid buffer gel containing 0.6 mg/mL ethidium bromide. If the PCR product's band-size in the gel is the same as the target band-size, it indicates that the corresponding virus is positive.
Table 1.
Oligonucleotides synthesized for amplifying the entire proviral genome of the isolate.
| Primersa | Sequences (5′-3′) | Annealing temperature (°C) | Size of PCR products |
|---|---|---|---|
| ALV-J-F | GGATGAGGTGACTAAGAAAG | 56 | 545 bp |
| ALV-J-R | CGAACCAAAGGTAACACACG | ||
| ALV-A-F | GGATGAGGTGACTAAGAAAG | 50 | 692 bp |
| ALV-A-R | AGAGAAAGAGGGGTGTCTAAGGAG | ||
| ALV-B-F | GGATGAGGTGACTAAGAAAG | 50 | 847 bp |
| ALV-B-R | ATGGACCAATTCTGACTCATT | ||
| ALV-C-F | GGATGAGGTGACTAAGAAAG | 50 | 860 bp |
| ALV-C-R | GAGGCCAGTACCTCCCACG | ||
| ALV-D-F | GGATGAGGTGACTAAGAAAG | 50 | 797 bp |
| ALV-D-R | ATCCATACGCACCACAGTATTCG | ||
| ALV-K-F | TCTGCCTCTCTACACAGTCAGCCAC | 60 | 2.0 kb |
| ALV-K-R | CTGCTTCATTCAGGTGTTCGCAATC | ||
| ALV-1F | GCCATTTGACCATTCACCAC | 50 | 2.2 kb |
| ALV-1R | ATCCCGCCTTCTGCACTGTTTAGC | ||
| ALV-2F | GGGATGGACAAACTGGGTCGGGTG | 50 | 1.7 kb |
| ALV-2R | TGGGTTGGGTGGAGAATA | ||
| ALV-3F | GTATGTAGCACCCGTAGG | 62 | 1.6 kb |
| ALV-3R | ACAAGACCAGGACACCAAT | ||
| ALV-4F | GGATGAGGTGACTAAGAAAG | 58 | 2 kb |
| ALV-4R | ACACTACATTTCCCCCTCCCTAT | ||
| ALV-5F | TGAAAGGACTGCTTTTGGGGCTTGTAGT | 58 | 0.6 kb |
| ALV-5R | TGTGGTGAATGGTCAAATGGCGTTTA | ||
| ALV-6F | TGAAAGGACTGCTTTTGGGGCTTGTAGT | 56 | 1.2 kb |
| ALV-6R | TTTTCCCGCAATAGGTTTTACAC |
F and R denote the forward and reverse primers for a specific fragment, respectively.
Whole Genome Sequencing and Alignments
A total of 6 primer pairs were designed to amplify 6 fragments that overlapped with each other and covered the whole genomic sequences of ALV isolates (Table 1) (Cai et al., 2013). The sequence alignments were compared with other ALVs reference strains from GenBank (ALV-A strain RSA, M37980; ALV-B strain Schmidt-Ruppin B, AF052428; ALV-C strain Prague C, J02342; ALV-D strain Schmidt-Ruppin D, D10652; ALV-E strain ev-1, AY013303; ALV-J strain HPRS-103, Z46390; ALV-K strain JS11X1, KF746200). The genomic nucleotide sequences were aligned using the Clustal W method in MegAlign program of DNASTAR software. Phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis version 5.0 with the maximum likelihood method with 1000 bootstrap replicates. Transcriptional regulatory elements in U3 were analyzed by the online service system of NSITE from Soft Berry.
Animal Experiment
Eighty day-old commercial Three-Yellow chicks without a history of vaccination were purchased from a local farm that was accredited by authorities for the elimination of specific avian diseases including AL. Upon arrival, the birds were tested again to make sure they were free from ALV infection by ELISA using ALV Antigen Kit (IDEXX, America) against p27 antigen in meconium according to the manufacturer's instructions. In the infected group, 35 birds were inoculated intra-abdominally with 100 μL of the isolated virus stock containing 104TCID50/0.1 mL. In the control group, 35 birds were injected with the supernates of uninfected DF-1 cell cultures via the same route. Birds of different groups were maintained separately in isolators for 21 wk with ad lib access to commercial feed and drinking water. At the corresponding detection time, 10 birds (5 from experimental group and 5 from control group) were euthanatized humanely and the corresponding tissue was collected for testing.
At 7 D of age, the birds of all the groups were intramuscularly injected in the left leg with 0.2 mL avian influenza virus (AIV)-H5 inactivated oil-emulsion vaccine (OEV) and subcutaneously injected in the neck with 0.2 mL Newcastle disease virus (NDV) + AIV-H9 bivalent inactivated OEV (Merial Nanjing Animal Health Co., Ltd Strain La Sota+ F). Sera of each group (n = 35/each) were sampled before the vaccination and at 3, 4, and 5 wk post-vaccination (wpv). All sera were frozen at −20°C until the haemagglutination inhabitation test was carried out.
RESULTS
Virus Identification
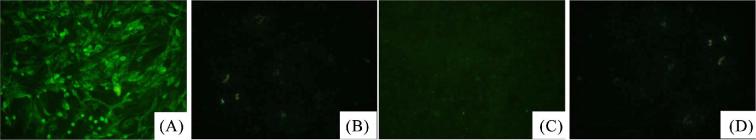
In one of the samples the lysate of the cell culture was ALV-p27 positive by the ELISA test and the positive PCR product was obtained using the primers specific for ALV-A gp85 and not with the primers for ALV-B, C, D, J, or K. The IFA results showed that the isolate-infected cells reacted with the ALV-A subgroup-specific McAb but not with the ALV-B or the ALV-J subgroup-specific McAb (Figure 1). The data indicated that a mono-infection ALV strain containing the gp85 gene of subgroup A (named GX14DJ3–18) was isolated.
Figure 1.
(A, B, C) DF-1 cells infected with GX14DJ3–18 were detected with immunofluorescence assay mediated by monoclonal antibodies specific for ALV-A, ALV-B, and ALV-J; (D) Uninoculated DF-1cells served as a negative control.
Whole Genome Sequence Analysis
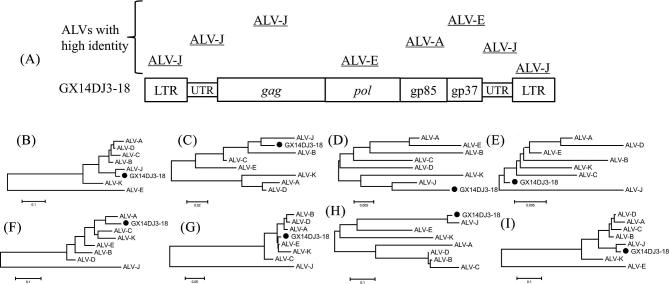
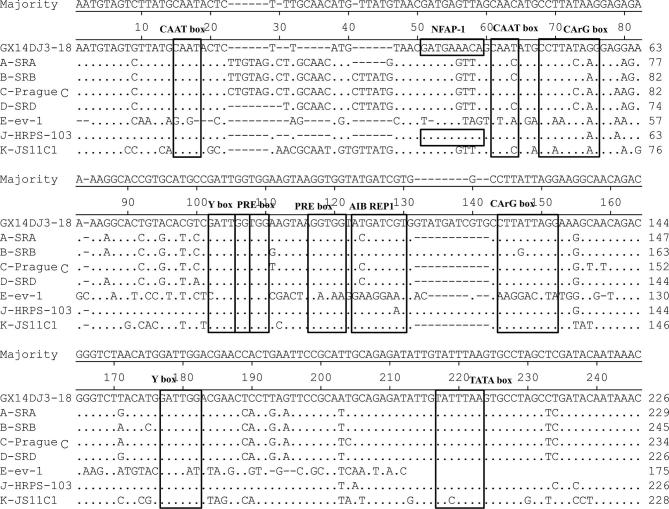
The full-length proviral genome of GX14DJ3–18 was 7605 nucleotides (nt) in length (GenBank Accession Number: MH213216), with the typical arrangement of retroviral genes being Long Terminal Repeated (LTR)-leader-gag-pol-env-LTR. The structure of the isolate is exhibited in Figure 2 with a comparison with other ALVs. The isolate displayed the highest similarity with the LTR (95.4%), 5′3′untranslated regions (UTR) (95.8%), gag (97.9%), and 3′UTR (92.9%) nucleotide sequences of ALV-J. Also the highest similarity was demonstrated with the pol (99.3%) and gp37 (99.0%) of ALV-E. Finally, the highest similarity was demonstrated with the gp85 (89.9%) of ALV-A (Table 2). In addition, the U3 region of GX14DJ3–18 revealed high identity to that of the ALV-J prototype strain HPRS103 (Figure 3). Transcriptional regulation elements of GX14DJ3–18 were identified in the U3 region, including 2 CAAT, CArG, Y, and PRE box, 1 NFAP-1, AIB REP1, and TATA box.
Figure 2.
Sequence comparison between the field isolate and the reference ALVs. (A) The genome structure of GX14DJ3–18. The boxed sequence line shows the structure of the entire proviral DNA of GX14DJ3–18. The lines over the schematic of the genome of GX14DJ3–18 indicate regions exhibiting high homology with the strains listed above. (B), (C), (D), (E), (F), (G), (H), and (I) Phylogenetic relationship of GX14DJ3–18 to reference ALV strains, based on the 5′LTR, 5′UTR, gag, pol, gp85, gp37, 3′UTR, and 3′LTR genome sequence, respectively. Black circle (●) indicates ALV isolates in this study.
Table 2.
The percentages of similarities between the GX14DJ3–18 and the 7 subgroups for each of the segments.
| 5′LTR | 5′UTR | gag | pol | gp85 | gp37 | 3′UTR | 3′LTR | |
|---|---|---|---|---|---|---|---|---|
| ALV-A | 89.3% | 86.4% | 95.3% | 98.8% | 89.9% | 95.6% | 88.1% | 89.3% |
| ALV-B | 88.5% | 93.7% | 94.8% | 98.4% | 83.4% | 93.9% | 82.3% | 88.5% |
| ALV-C | 86.6% | 95.0% | 96.4% | 98.7% | 86.8% | 92.4% | 67.5% | 86.6% |
| ALV-D | 89.7% | 84.6% | 95.6% | 98.1% | 84.8% | 94.9% | 81.8% | 89.7% |
| ALV-E | 55.1% | 89.2% | 94.5% | 99.3% | 87.1% | 99.0% | 74.0% | 55.1% |
| ALV-J | 95.4% | 95.8% | 97.9% | 97.9% | 50.1% | 59.5% | 92.9% | 95.4% |
| ALV-K | 81.5% | 81.3% | 95.4% | 98.9% | 86.7% | 95.7% | 84.4% | 81.5% |
Figure 3.
Comparative analysis of the U3 region nucleotide sequences of ALV strains. Dots indicate identical residues, while letters indicate base substitutions. Dashes indicate gaps in the alignment. Motifs were indicated in panes.
Influences of GX14DJ3-18 Infection on the Development of Immune Organs in the Experiment Birds
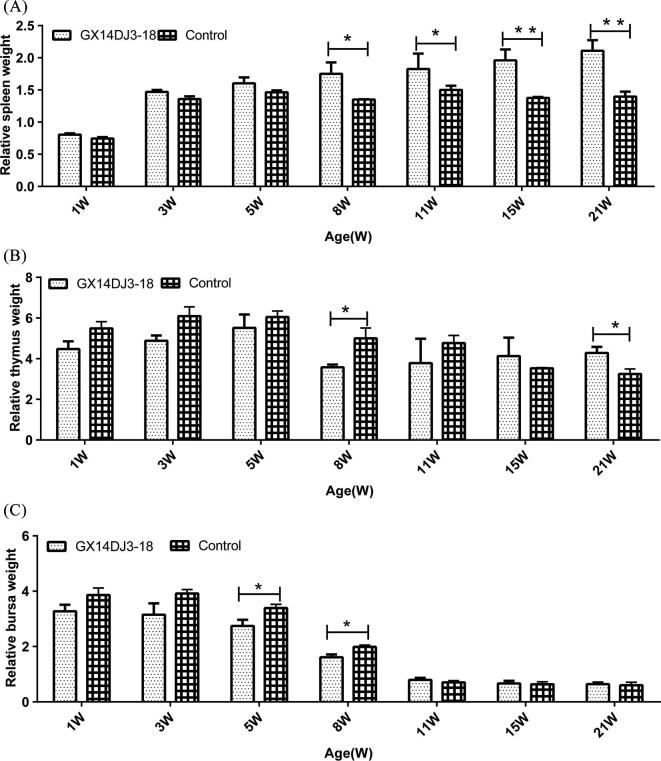
For the spleen (Figure 4A), the mean Relative Weight of Immune Organs (RWIO) (RWIO = immune organ weight (g)/body weight (kg) of birds in the infected group was significantly higher than that of the uninfected control group at 8 and 11 wpi (P < 0.05). In addition at 15 wpi and 21 wpi, the mean RWIO of the infected group was very significantly higher than that of the uninfected control group (P < 0.01). The mean RWIO for the thymus in the infected group at 8 wpi was significantly lower than that of the uninfected control group (P < 0.05), and at 21 wpi, the mean RWIO of the infected group was significantly higher than that of the uninfected control group (P < 0.05) (Figure 4B). For the bursa (Figure 4C), the mean RWIO of birds in the infected group at 5 wpi and 8 wpi were significantly lower than that of the uninfected control group (P < 0.05).
Figure 4.
Dynamic change of relative weight of immune organs. (A) Spleen relative weight change. (B) Thymus relative weight change. (C) Bursa relative weight change. The chicks were weighed on weeks 1, 3, 5, 8, 11, 15, and 21 (n = 5/group). Data shown are the means ± SD. (*P < 0.05, statistically significant; **P < 0.01 very significant).
Inhibition of the Antibody Response to the Vaccinations with Commercial NDV and AIV-H5/H9 Vaccines
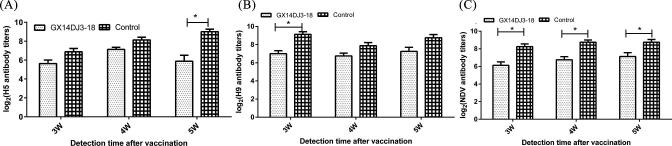
As shown in Figure 5, the GX14DJ3–18 infected birds exhibited significantly lower antibody responses to the OEVs of AIV-H5 and AIV-H9 when compared to those of the uninfected control group at 5 and 3 wpv, respectively (P < 0.05). Significant differences were observed for antibody responses to NDV vaccination between the control and GX14DJ3–18 infected groups at 3, 4, and 5 wpv (P < 0.05).
Figure 5.
Influence of GX14DJ3–18 on the inhibition of antibody titers against Newcastle disease virus and AIV-H5/H9 after vaccination. (A) AIV-H5 haemagglutination inhabitation (HI) antibody titers changes. (B) AIV-H9 HI antibody titers changes. (C) NDV HI antibody titers changes. Data shown are the means ± SE.
Gross and Histological Lesions
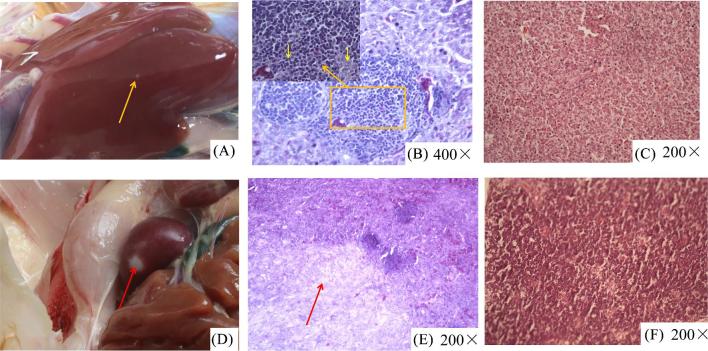
During the whole experiment, no deaths were found in the infected birds or in the un-infected control group. The incidence of tumor or tumor-like lesions in the infected birds were 14.28% (5/35) while none were observed in the un-infected control group (0/35). The first case of clinical tumor was observed at 11 wpi in the infected group and presented as white tumor nodules, approximately 2 to 3 mm in diameter, on the liver surface (Figure 6A). At 21 wpi, following necropsy some of the infected birds had gross tumor or tumor-like lesions in the spleen (Figure 6D). Microscopically, the hepatocytes were compressed by trachychromatic lymphocytes (Figure 6B), and a large number of irregular fibrous cells were infiltrated in the spleen tissue (Figure 6E). No histological lesions were observed with chickens in the negative control group, and tumors were notably absent in the livers and spleens of chickens in the control group (Figures 6C and 6F).
Figure 6.
Anatomical and histological lesions. (A) White tumor nodules, approximately 2 to 3 mm in diameter, on the liver surface. (B) Lymphocytic proliferation foci were found in the livers. (C) Normal liver tissues. (D) Sarcoma in the spleens. (E) A large number of irregular fibrous cells infiltrated into spleen tissues. (F) Normal spleen tissues.
DISCUSSION
By the early 2000s ALV-J had spread into China and had caused severe losses in the poultry industry (Li et al., 2018b). In addition, with the spread of ALV-J in local breeds of chickens in China, ALV-J and other subpopulations of recombinant viruses have emerged one after another. For example FJ15HT0 (Wu et al., 2017), consisting of an ALV-B gp85 gene, an ALV-E gp37 and a J subgroup LTR region induced significantly reduced growth and immune suppression in SPF chickens. Also, HLJ13SH01 (Gao et al., 2015) and GX14FF03 (Wang et al., 2017) and LC110515–5, were identified. These data show that ALV recombination is currently a relatively common phenomenon. However, there is rarely a report of the recombination of viruses ALV-A, ALV-J, and ALV-E.
In the present study, one natural recombinant virus named GX14DJ3–18 was isolated from gamecocks. As other articles have reported, the LTR, UTR, and env gene are all virulence factors of ALVs (Wang et al., 2012), the recombination of GX14DJ3–18 inevitably results in altered clinical symptoms from classical ALV-A and ALV-J strains. The unique strain GX14DJ3–18 supplies an extremely important natural model virus to study the function of different genomic regions of ALVs. The Three-Yellow chicken is a major and popular local breed of broiler chickens in southern China and favored by the domestic markets. In order to study whether the virus isolated from gamecocks is pathogenic to commercial chicken, we examined the pathogenic effects of GX14DJ3–18 by challenging the Three-Yellow birds.
Animal experiments showed that GX14DJ3–18 caused severe immunosuppression in chickens. The trend of RWIO for the spleens in infected birds was higher than in the uninfected controls (P < 0.05), while for the bursa the trend in infected birds was lower than in the uninfected controls (P < 0.05), and for the thymus the trend in the infected group was lower than in the uninfected controls during the earlier periods of the experiment and became higher at the later stage (P < 0.05). It was also demonstrated that the infected birds had lower antibody titer responses to the vaccinations of NDV, AVI-H5 and H9 vaccines than the uninfected control group birds (P < 0.05). These data indicated that GX14DJ3–18 induced significant immune suppression in Three-Yellow birds.
The unique isolate GX14DJ3–18 containing ALV-A gp85 and ALV-E gp37 gene induced fibrosarcoma and lymphocytoma that are commonly induced by ALV-A. The tumor phenotype was determinated by ALV's viral envelope and ALV-E has little or no oncogenicity (Payne and Nair, 2012). Based on the above reasons, the fibrosarcoma and lymphocytoma were mainly determined by the gp85 gene of the recombinant virus. The ALVs lacking oncogenes induce neoplasms in chickens by integrating LTR enhancer sequences into or near the tumor genes of the host and perturbing their expression. The incidence of tumor in the birds induced by GX14DJ3–18 (containing an ALV-J-like LTR region) were 14.28%. These data suggest that the enhancer activity of the LTR contributed to the high tumor incidence rate (Justice et al., 2015).
ALVs can be transmitted vertically or horizontally (Lin et al., 2013) and the commercial live vaccine might also be a potential route for ALVs transmission (Wang et al., 2018a). In this study, a strain that isolated from gamecocks was found to cause severe immunosuppression and tumor incidence in the commercial Three-Yellow chicken. Previous studies showed gamecock translocation has the risk of spreading ND and AI (Lefrancois et al., 2010). The infected gamecocks in Pingxiang (a small town in southwest China neighboring Vietnam) are descendants of cross-breeding of a foreign male line with the local female line of gamecock. Therefore, there is the potential that ALV will infect and/or be carried by the parent birds that might allow the virus to spread vertically and/or horizontally to the offspring gamecocks. As a result ALV might have a risk of spreading to other parts of the country through the commercial flow of the birds from Pingxiang and pose a potential threat to the commercial chickens in other locations.
CONCLUSION
A naturally recombinant strain of ALV consisting of ALV-J's LTR, UTR and gag, ALV-E's pol and gp37, and ALV-A's gp85 was isolated from a gamecock flock in China. Commercial Three-Yellow chickens infected with the recombinant strain showed severe immunosuppression and tumors. The cross-provincial sales of gamecocks in which ALVs have not been eradicated thoroughly might be a potential route of ALV transmission and could be a threat to the commercial poultry industry in general.
ACKNOWLEDGMENTS
This work was supported by the Guangxi Special Funding on Science and Technology Research [AA17204057], the Guangxi Program for Modern Agricultural Industry Technical System Construction-Chicken Industry [nycytxgxcxtd-19–03], the Shandong Provincial Natural Science Foundation, China [ZR201807070202]. The manuscript was kindly reviewed by Richard Roberts, Aurora, CO 80014, USA.
Conflict of interest statement
The authors declare that they have no competing interests.
REFERENCES
- Cai L., Shen Y., Wang G., Guo H., Liu J., Cheng Z.. 2013. Identification of two novel multiple recombinant avian leukosis viruses in two different lines of layer chicken. J. Gen. Virol. 94:2278–2286. [DOI] [PubMed] [Google Scholar]
- Gao Y., Guan X., Liu Y., Li X., Yun B., Qi X., Wang Y., Gao H., Cui H., Liu C., Zhang Y., Wang X., Gao Y.. 2015. An avian leukosis virus subgroup J isolate with a Rous sarcoma virus-like 5'-LTR shows enhanced replication capability. J. Gen. Virol. 96:150–158. [DOI] [PubMed] [Google Scholar]
- He C. W., Wang P. K., Qin L. L., Bi Y. Y., Zhou G. Z., Yang Y. L., Peng H., Wei P.. 2016. Detection of Avian leukosis virus in Game Fowl fo Pingxiang, Guangxi and env gene analysis of isolates. Prog. Vet. Med. 37:23–26. [Google Scholar]
- Justice J. F., Morgan R. W., Beemon K. L.. 2015. Common viral integration sites identified in avian leukosis virus-induced b-cell lymphomas. mBio 6:e01863–01815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois T., Hendrikx P., Vachiéry N., Ehrhardt N., Millien M., Gomez L., Gouyet L., Gerbier G., Gongora V., Shaw J. J. T., Diseases E.. 2010. Interaction between research and diagnosis and surveillance of avian influenza within the Caribbean animal health network (CaribVET). Transbound. Emerg. Dis. 57:11–14. [DOI] [PubMed] [Google Scholar]
- Li H., Wang P., Lin L., Shi M., Gu Z., Huang T., Mo M. L., Wei T., Zhang H., Wei P.. 2018a. The emergence of the infection of subgroup J avian leucosis virus escalated the tumour incidence in commercial Yellow chickens in Southern China in recent years. Transbound. Emerg. Dis. 66: 312–316. [DOI] [PubMed] [Google Scholar]
- Li J., Meng F., Li W., Wang Y., Chang S., Zhao P., Cui Z. J.. 2018b. Characterization of avian leukosis virus subgroup J isolated between 1999 and 2013 in China. Poult. Sci. 97:3532–3539. [DOI] [PubMed] [Google Scholar]
- Lin L., Wang P., Yang Y., Li H., Huang T., Wei P.. 2017. Full-length genome sequence analysis of four subgroup J avian leukosis virus strains isolated from chickens with clinical hemangioma. Virus Genes 53:868–875. [DOI] [PubMed] [Google Scholar]
- Lin Y., Xia J., Zhao Y., Wang F., Yu S., Zou N., Wen X., Cao S., Huang Y.. 2013. Reproduction of hemangioma by infection with subgroup J avian leukosis virus: the vertical transmission is more hazardous than the horizontal way. Virol. J 10:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas R., Zoelen D. V., Oei H., Claassen I. J. B.. 2006. Replacement of primary chicken embryonic fibroblasts (CEF) by the DF-1 cell line for detection of avian leucosis viruses. Biologicals 34:177–181. [DOI] [PubMed] [Google Scholar]
- Payne L. N., Nair V.. 2012. The long view: 40 years of avian leukosis research. Avian Pathol. 41:11–19. [DOI] [PubMed] [Google Scholar]
- Qin A. J., Lee L. F., Fadly A., Cui Z.. 2001. Development and characterization of monoclonal antibodies to subgroup J avian leukosis virus. Chinese Journal of Animal and Veterinary Sciences 32:556–562. [PubMed] [Google Scholar]
- Su Q., Li Y., Li W., Cui S., Tian S., Cui Z., Zhao P., Chang S.. 2018. Molecular characteristics of avian leukosis viruses isolated from indigenous chicken breeds in China. Poult. Sci. 97:2917–2925. [DOI] [PubMed] [Google Scholar]
- Wang P., Lin L., Li H., Shi M., Gu Z., Wei P.. 2018a. Full-length genome sequence analysis of an avian leukosis virus subgroup J (ALV-J) as contaminant in live poultry vaccine: the commercial live vaccines might be a potential route for ALV-J transmission. Transbound. Emerg. Dis. 65:1103–1106. [DOI] [PubMed] [Google Scholar]
- Wang P., Lin L., Li H., Yang Y., Huang T., Wei P.. 2018b. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989-2016: coexistence of five extremely different clusters. Arch. Virol. 163:377–389. [DOI] [PubMed] [Google Scholar]
- Wang P., Yang Y., Lin L., Li H., Wei P.. 2017. Complete genome sequencing and characterization revealed a recombinant subgroup B isolate of avian leukosis virus with a subgroup J-like U3 region. Virus Genes 53:927–930. [DOI] [PubMed] [Google Scholar]
- Wang Q., Gao Y., Wang Y., Qin L., Qi X., Qu Y., Gao H., Wang X.. 2012. A 205-nucleotide deletion in the 3' untranslated region of avian leukosis virus subgroup J, currently emergent in China, contributes to its pathogenicity. J. Virol. 86:12849–12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Deng Q., Zhai G., He C., Li H., Zhang Y., Zeng R., Mo M., Huang T., Wei P.. 2019. Re-emergence of a genotype VIII virulent Newcastle disease virus isolated from Chinese game fowl after 13 years. Transbound. Emerg. Dis. 66:1077–1084. [DOI] [PubMed] [Google Scholar]
- Wu X., Zhao J., Zeng Y., Wu Y., Wang Q., Wu B., Huang Y.. 2017. A novel avian retrovirus associated with lymphocytoma isolated from a local Chinese flock induced significantly reduced growth and immune suppression in SPF chickens. Vet. Microbiol. 205:34–38. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Guan X., Chen Z., Cao D., Kang Z., Shen Q., Lei Q., Li F., Li H., Leghari M. F., Wang Y., Qi X., Wang X., Gao Y.. 2018. The high conserved cellular receptors of avian leukosis virus subgroup J in Chinese local chickens contributes to its wide host range. Poult. Sci. 97:4187–4192. [DOI] [PubMed] [Google Scholar]