SUMMARY
The incidence of human papilloma virus (HPV)-associated head and neck squamous cell carcinoma (HNSCC) is increasing and implicated in more than 60% of all oropharyngeal carcinomas (OPSCCs). Although whole-genome, transcriptome, and proteome analyses have identified altered signaling pathways in HPV-induced HNSCCs, additional tools are needed to investigate the unique pathobiology of OPSCC. Herein, bioinformatics analyses of human HPV(+) HNSCCs revealed that all tumors express full-length E6 and identified molecular subtypes based on relative E6 and E7 expression levels. To recapitulate the levels, stoichiometric ratios, and anatomic location of E6/E7 expression, we generated a genetically engineered mouse model whereby balanced expression of E6/E7 is directed to the oropharyngeal epithelium. The addition of a mutant PIK3CAE545K allele leads to the rapid development of pre-malignant lesions marked by immune cell accumulation, and a subset of these lesions progress to OPSCC. This mouse provides a faithful immunocompetent model for testing treatments and investigating mechanisms of immuno- suppression.
Graphical Abstract

In Brief
Carper et al. present the ‘‘iKHP’’ mouse, in which HPV16 oncogenes are inducibly activated in vivo in a tissue-specific and temporal manner. Oropharyngeal- specific expression of E6/E7 with PIK3CAE545K in these mice promotes the development of premalignant lesions marked by immune cell infiltration, but only a subset spontaneously convert to OPSCC.
INTRODUCTION
Head and neck squamous cell carcinomas (HNSCCs) are the 6th most common cancer worldwide, with nearly 65,000 people diagnosed annually in the United States alone (Global Burden of Disease Cancer Collaboration et al., 2017). Oral cavity squamous cell carcinoma (OSCC), oropharyngeal squamous cell carcinoma (OPSCC), and laryngeal squamous cell carcinoma (LSCC) represent distinct anatomic subsites of HNSCCs that account for the majority of cases (~90%), with the remaining ~10% including other locations, such as the thyroid and nasopharynx. Nearly 50% of HNSCC patients who present with lymph-node- positive disease (relapsed/metastatic) have a dismal 5-year survival of <50%, despite current therapeutic strategies that include radiation, surgery, chemotherapy, and monoclonal antibodies. Hence, an urgent need exists to better understand the pathobiology of these cancers to facilitate the development of new targeted therapies.
Known risk factors for the development of HNSCC include tobacco and alcohol consumption and/or viral infections from high-risk human papillomavirus (HPV) (Stein et al., 2014; Suh et al., 2014). Although carcinogen-induced models exist, human HNSCCs caused by tobacco and alcohol consumption have been on the decline in recent decades. In stark contrast, the global incidence of HPV-associated (HPV(+)) HNSCC is steadily increasing irrespective of sex and ethnicity and currently accounts for 30% of all HNSCCs but up to 80% of all oropharyngeal cancers (OPSCCs) (Chaturvedi et al., 2011; D’Souza et al., 2016). Although the HPV family is comprised of over 120 known genotypes that can infect the basal layer of mucosal or cutaneous epithelia in humans (https://pave.niaid.nih.gov) (Bernard et al., 2010; Van Doorslaer et al., 2017), a subset of six high-risk HPVs are associated with cancer (Ha and Califano, 2016; Moody and Laimins, 2010; Münger et al., 2004). Specifically, HPV16 is associated with the majority of all HPV(+) cancers, including 90% of head and neck malignancies wherein the expression of two oncogenes, namely, E6 and E7, is a universal feature (Gillison et al., 2018; Leemans et al., 2018; Cancer Genome Atlas Network, 2015). Interestingly, HPV16 transcribes E6 and E7 as a polycistronic pre-mRNA that undergoes posttranscriptional processing by the host RNA splicing machinery, resulting in multiple splice isoforms, including the E6*I variant whose accumulation is critical for efficient translation of E7 (Ajiro et al., 2012; Graham and Faizo, 2017; Tang et al., 2006). Compared to HPV-negative HNSCCs, HPV(+) cases have distinct molecular and clinical features, such as age of onset, prognosis, and immunologic profile (D’Souza et al., 2016; Mandal et al., 2016). More importantly, patients with HPV(+) OPSCC are almost universally p53 wild type and have superior treatment responses compared to their generally p53-mutant HPV(—) OSCC counterparts (Leemans et al., 2018). Consequently, several ongoing clinical trials are attempting to de-escalate standard therapy for HPV(+) OPSCC (Blitzer et al., 2014; Gabani et al., 2019) and, yet, few preclinical genetically engineered mouse models (GEMMs) of HPV(+) OPSCC exist and none faithfully recapitulate these unique features.
To address these limitations, we generated and characterized an inducible GEMM of HPV16-driven oropharyngeal cancer. Our efforts establish an autochthonous, immunocompetent HPV(+) GEMM wherein E6 and E7 expression combined with tissue-specific expression of mutant PIK3CAE545K faithfully phenocopies the histologic and molecular features in developing human HPV(+) oropharyngeal carcinoma. Consequently, this model provides a valuable tool for studying the underlying mechanisms that HPV16-E6/E7 and mutant PIK3CA use to induce malignancy and may reveal insights that can lead to new therapies that offer similar oncologic outcomes to radiation, with less treatment morbidity. Moreover, these data highlight the strength of our immunocompetent GEMM for modeling field cancerization and HPV(+) OPSCC development and progression.
RESULTS
Unspliced E6 and Alternatively Spliced E6*I Transcript Expression Are Hallmarks of HPV(+) HNSCC
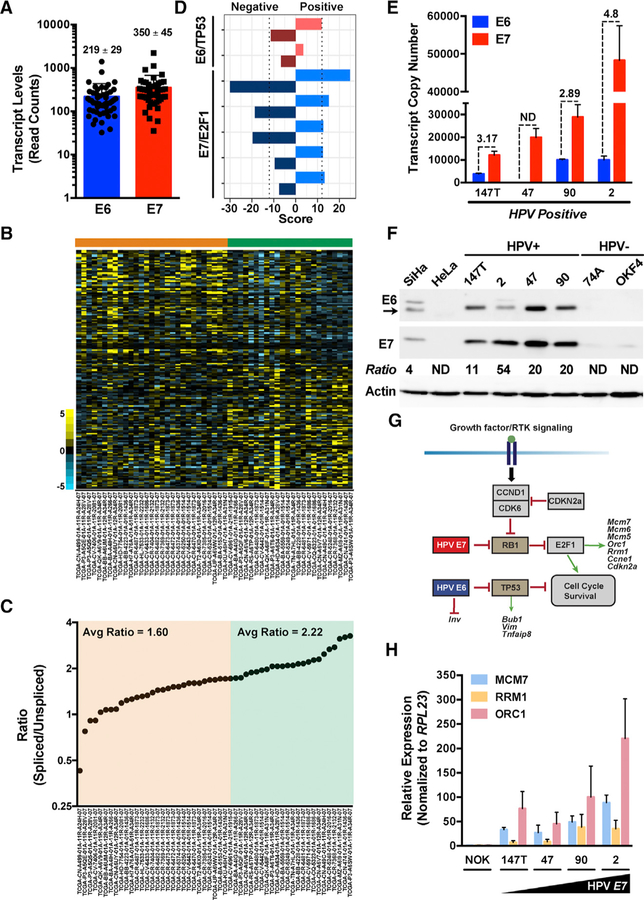
Although several studies have identified E7 as the dominant viral oncogene responsible for driving HPV-associated tumors, cooperation between E7 and E6 is essential for inducing and promoting tumorigenesis, respectively (Halbert et al., 1991; Jabbar et al., 2010; Riley et al., 2003; Strati and Lambert, 2007). Due to the limited number of nucleotides present between the HPV16 E6 and E7 open reading frames (ORFs), the accumulation of alternatively spliced E6*I transcripts is critical for efficient translation of E7 (Ajiro et al., 2012; Tang et al., 2006). Thus, we analyzed RNA sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA; n = 530) HNSCC Research Network (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) to investigate the relative abundance of E7 (E6*I) and E6 in HPV(+) HNSCC. An analysis of read alignments identified 53 cases with median coverage of >100 for the total, unspliced HPV16 E6 full-length transcript (Figure S1A). The apparent decrease in mapped reads between nucleotides 226 and 409 suggested that E6*I, and consequently its translated product E7, is the predominant variant expressed in HPV(+) HNSCC, as reported by others (Gillison et al., 2018; Nulton et al., 2017). However, reads mapping to positions 1–225 and 409–880 represent the sum of E6 and E6*I transcripts. To deconvolute these transcript variant data and to determine relative E7 versus E6 expression levels, we focused on reads mapping to splice junctions unique to each transcript variant, summed the total read counts, and calculated the ratio of spliced/unspliced (E6*I/E6) transcripts (Figure S1B; Table S1). Remarkably, E6 and the E6*I variant (i.e., E7) are expressed to near equivalent levels in HPV(+) HNSCC, whereas the alternate E6*II variant commonly expressed in HPV(+) cervical carcinoma (Pastuszak-Lewandoska et al., 2014) is poorly expressed (Figure 1A; Figures S2A–S2C; Table S1).
Figure 1. HPV16 E6:E7 Stoichiometry Impacts Gene Expression in HPV(+) HNSCCs.
(A) Raw RNA-seq read counts for full-length, unspliced HPV16 E6 and alternatively spliced E6*I (translated to E7) transcripts.
(B) Heatmap of differentially expressed genes (rows) ordered using the HPV16 E7/E6 ratio as a continuous variable. Samples (columns, n = 53) are arranged according to increasing ratio from left to right.
(C) Spliced to unspliced ratios of HPV16 E7/E6 transcripts plotted against a sorted rank of the ratios from left to right.
(D) Gene set enrichment analysis (GSEA) of E6/TP53 and E7/E2F1 pathway genes. Separate scores were calculated for each pathway listed to identify genes that positively correlate with the E7/E6 ratio versus genes that negatively correlate with the E7/E6 ratio.
(E) Quantitative real-time PCR analysis of HPV16 E6 and E7 transcript copy numbers calculated using respective standard curves established with plasmid DNA encoding E6 and E7. Quantitative real-time PCR was performed using 10 ng of indicated cDNA samples as input (n = 3, mean ± SEM). ND, not detected.
(F) Representative immunoblots of the HPV16 E6 and E7 levels in HPV-positive HNSCC cell lines relative to control HPV16(+) SiHa and HPV18(+) HeLa or HPV(—) HNSCC and normal oral keratinocyte (NOK) cell lines. Densitometry quantification was performed by normalizing the E6 and E7 band intensities to the actin loading control band intensity for each respective sample prior to calculating the E7/E6 ratio.
(G) Schematic of the cell cycle, survival, and differentiation signaling pathways and genes regulated by high-risk HPV16 oncogenes. E6 targets the tumor suppressor TP53, whereas E7 targets the tumor suppressor pRb, relieving E2F1 target gene repression.
(H) Quantitative real-time PCR analysis of select E7/ E2F1 host pathway genes in HPV-positive HNSCC cell lines relative to control HPV(−) NOK cells.
Relative fold expression is shown normalized to RPL23 mRNA levels (n = 3, mean ± SEM). Related to Figures S1 and S2 and Tables S1, S2, and S3.
To assess if variations in E6/E7 stoichiometry impact host gene expression, we analyzed mRNA expression of the 53 HPV(+) tumors by ranking the samples as a continuous variable according to the spliced/unspliced ratio and qualitatively identified two groups with a ratio cutoff at 2.0 (Figure 1B). The spliced/ unspliced ratio varied from 0.43 to 3.26, with the majority (n = 30) of cases in group 1 (orange) displaying near equivalent 1:1 ratios (average [avg] = 1.60) and cases in group 2 (n = 23) displaying near 2:1 ratios (avg = 2.22) (Figure 1C). Gene set enrichment analysis (GSEA) revealed that top signatures associated with these groups include TP53 and E2F1/pRB, targets of E6 and E7, respectively (Figure 1D; Figure S2D; Tables S2 and S3). Quantification of E6 and E7 transcript levels within established HPV16(+) HNSCC cell lines revealed that spliced/unspliced ratios are 1.81- to >3.0-fold higher than the average ratio observed in group 1 samples and to 1.3- to 2.2-fold higher in group 2 samples (Figure 1E). Interestingly, western blot analysis confirmed that E7 levels are much higher than E6 and densitometry measurements revealed that E7/E6 ratios range from 11 to 54, suggesting that modest differences in E7/E6 mRNA expression levels can have profound effects at the protein level (Figure 1F). HPV16 E6 and E7 deregulate a host of signaling pathways and transcription factors by TP53 and E2F1/pRB, respectively, which target genes involved in cell cycle and proliferation (Figure 1G). We next tested the consequence of higher E7/E6 ratios and observed that higher E7 expression correlates with higher E2F1/pRB target gene expression (Figure 1H). Collectively, these data indicate that the E7/E6 ratio impacts the pathobiology of HPV(+) tumors.
Generation of an Inducible HPV16 Mouse Model with Balanced E6 and E7 Oncogene Stoichiometry
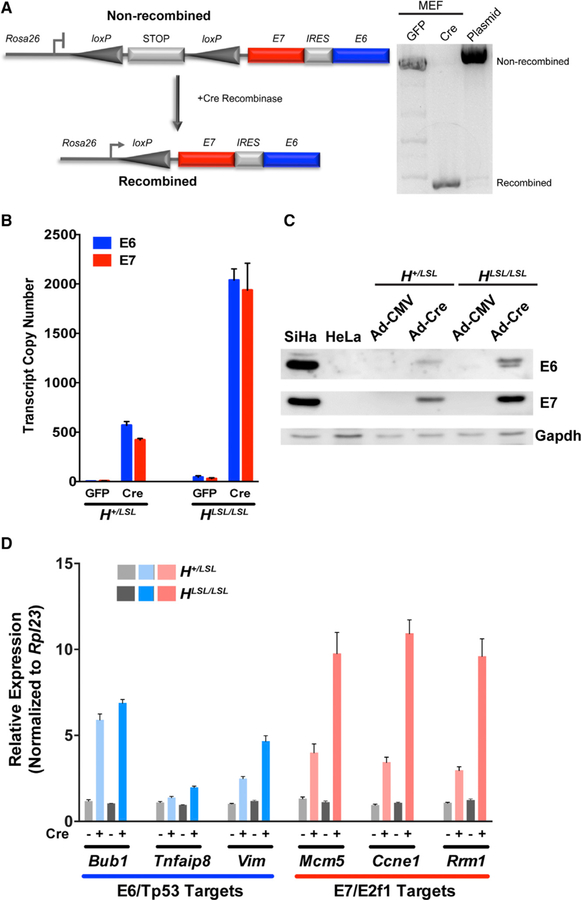
Previous HPV16-driven GEMMs employed either constiutive or inducible expression of the E6 and/or E7 oncogenes (Table S4). However, currently available inducible HPV16 GEMMs were generated using HPV16 genomic regions that encompass the E6/E7 ORFs. Unfortunately, E7 expression depends on viral and host factors that regulate alternative splicing of the E6 transcript, which makes E6 and E7 levels difficult to predict. To develop a conditional and inducible HPV16 GEMM that recapitulates the E6:E7 expression stoichiometry characteristic of most HPV(+) HNSCCs, we generated knockin transgenic mice harboring a cassette with the E7 and E6 ORFs separated by an internal ribosomal entry site (IRES) regulated by a LoxP-STOP-LoxP element at the constitutively active Rosa26 locus (Figure 2A). Mouse embryonic fibroblasts (MEFs) isolated from Rosa26-LSL-E7iresE6 mice were infected with an adenovirus expressing GFP or Cre recombinase (Ad-Cre), leading to recombination and to the excision of the transcriptional stop element resulting in the expression of the polycistronic E7-IRES-E6 message driven by the Rosa26 promoter. The comparison of E6 and E7 expression levels in heterozygous (H+/LSL) versus homozygous (HLSL/LSL) MEFs confirmed the expected increase in expression with near-equivalent E6:E7 stoichiometry, and these RNA levels are reflected in the relative protein levels (Figures 2B and 2C). Notably, E6:E7 expression ratios observed in H+/ LSL MEFs are comparable to that seen in HPV(+) HNSCC. We next analyzed the effects of E6 and E7 expression on canonical targets downstream of Tp53 and E2f1/pRb and observed expected changes in the expression profiles of select cell cycle, cell proliferation, DNA replication and repair, and differentiation (Figure 2D; Figures S3A and S3B). These data confirm that E6 and E7 are both inducibly expressed and functional.
Figure 2. Generation and Characterization of an Inducible Knockin Mouse Model for Investigating E6:E7-Driven Oropharyngeal Tumorigenesis.
(A) Schematic depicting the high-risk HPV16 cassette introduced into the Rosa26 locus (left). A transcriptional STOP element flanked by LoxP sites is located upstream of the E7 ORF, an internal ribosomal entry site (IRES), and the E6 ORF (Rosa26-LSL-E7iresE6). Primary mouse embryonic fibroblasts (MEFs) transduced with either GFP or Cre adenoviruses (MOI = 20) were collected 72 h later and recombination confirmed by PCR-gel electrophoresis (right).
(B) Quantitative real-time PCR analysis of E6 and E7 transcript copy numbers from heterozygous or homozygous Rosa26-LSL-E7iresE6 primary MEFs (H+/LSL or HLSL/LSL) were calculated using respective standard curves (n = 3, mean ± SEM).
(C) Representative immunoblots of the E6 and E7 levels in H+/LSL or HLSL/LSL MEFs transduced with either GFP or Cre adenoviruses relative to control HPV16(+) SiHa and HPV18(+) HeLa cells.
(D) Quantitative real-time PCR analysis of select E6/ Tp53 and E7/E2F1 host pathway genes in H+/LSL or HLSL/LSL MEFs transduced with either GFP or Cre adenoviruses. Relative fold expression is shown normalized to Ubb mRNA levels (n = 3, mean ± SEM).
Related to Figure S3.
Conditional Expression of HPV16 E7iresE6 Disrupts Epithelial Homeostasis and Promotes Tumorigenesis
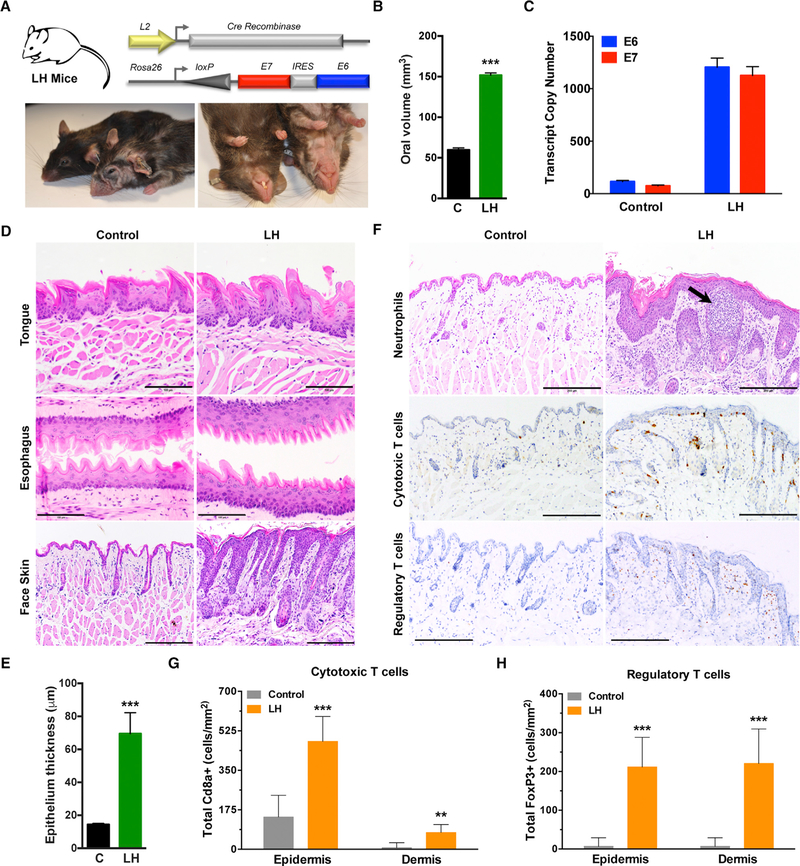
To validate the effects of balanced E6 and E7 oncogene expression in vivo, we spatially targeted the expression of E7-IRES-E6 mRNAs to squamous epithelia lining the oral cavity, pharynx, esophagus, and forestomach by crossing the Rosa26-LSL-E7iresE6 mice to L2-Cre mice, which express Cre-recombinase postnatally within the oro-esophageal epithelia (Nakagawa et al., 1997; Opitz et al., 2002; Stairs et al., 2011). The L2-Cre;Rosa26-LSL-E7iresE6 mice (LH mice) were born with normal Mendelian distribution but developed several overt phenotypes, including hair loss; wrinkling and thickening of the skin, ears, paws, and muzzle; as well as occasional ulcerative lesions (Figure 3A; Figure S4A). Gross macroscopic examination revealed enlarged oral tissues with significant changes in oral volume in LH versus control littermate mice and qRT-PCR confirmed E6 and E7 expression (Figures 3B and 3C). Histologic analysis at 8–10 months of age demonstrated fully penetrant epithelial hyperplasia and mild dysplasia, primarily within esophageal and cutaneous (face skin) epithelia but with variable penetrance of mild dysplasia within the tongue (Figure 3D). Compared to control littermates, LH mice displayed a significant increase in epithelium thickness but rarely (1 tumor, n = 16) developed tumors (Figure 3E). Moreover, addition of the chemical carcinogen 4NQO, frequently used to induce oral tumors (Vitale-Cross et al., 2009), failed to accelerate or increase the frequency of oropharyngeal tumor development at low doses (10 µg/ml) (Figure S4B). Quantification of epithelium thickness from different anatomic locations of the face skin with or without inclusion of the keratin layer did not differ between control and 4NQO-treated animals (Figures S4C and S4D). An analysis of subcutaneous face skin revealed the striking infiltration of several different immune cell populations, including polymorphonuclear cells (Ly6g+ neutrophils), cytotoxic T cells (Cd8+ T cells), and regulatory T cells (FoxP3+ T cells), supporting the presence of an immunologic response to viral antigen expression (Figures 3F–3H; Figure S4E).
Figure 3. Conditional Oral-Esophageal Expression of E6 and E7 Induces Hyperplasia and Immune Cell Infiltration.
(A) General strategy for generating LH mice by crossing a Cre recombinase strain driven by the Epstein-Barr virus ED-L2 promoter (L) to our knockin HPV16 strain (H; top). Representative photographs of control littermates and the LH animals at 8–10 months (bottom).
(B) Quantification of oral volume calculated based on the formula for an ellipsoid volume less the oral aperture (n = 3, ***p < 0.0001).
(C) Quantitative real-time PCR analysis of E6 and E7 transcript copy numbers from dissected whole tongue of LH and control littermate mice calculated using respective standard curves (n = 3, mean ± SEM).
(D) Histologic analysis of epithelial tissues. Representative sagittal sections from adult tongue, esophagus, and cutaneous epithelia were formalin fixed and embedded, and sections were stained with H&E. Magnification, 200×; bar, 200 µm.
(E) Quantification of cutaneous epithelial thickness calculated in adult LH and control littermate mice (n = 3, ***p < 0.0001).
(F) Histologic and immunohistochemical analysis of immune cell infiltration. Representative sagittal sections from adult cutaneous epithelia were formalin fixed, embedded, and H&E stained (top) and reveal morphologic and histologic abnormalities, such as hyperplasia, dysplasia, and pockets of neutrophil infiltrates. IHC staining is shown for Cd8a+ cytotoxic T cells (middle) and FoxP3+ regulatory T cells (bottom). Magnification, 200×; bar, 200 µm.
(G) Quantification of epidermal versus dermal Cd8a+ cytotoxic T cell infiltration (n = 3, ***p < 0.0001, **p < 0.001).
(H) Quantification of epidermal versus dermal FoxP3+ regulatory T cell infiltration (n = 3, ***p < 0.0001).
Data presented in (B), (E), (G), and (H) are shown as mean ± SEM. Student’s t test (2 tailed) was used to determine significance. Related to Figure S4.
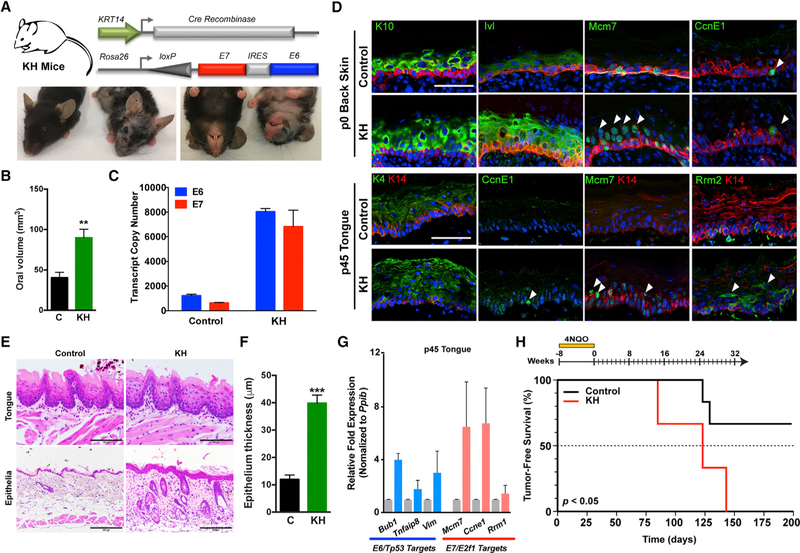
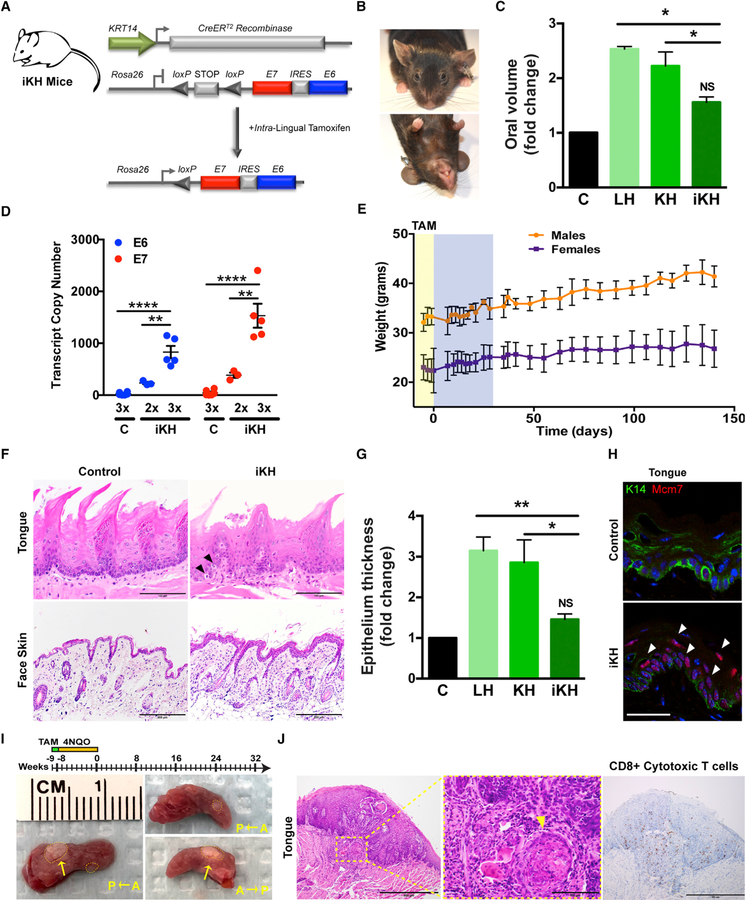
To more specifically target progenitor cells within the basal epithelia, we bred the Rosa26-LSL-E7iresE6 mice to KRT14-Cre, which is expressed in the basal epithelia as early as E12. The offspring of these crosses (KH mice) displayed similar overt phenotypes to the LH mice (Figure 4A). Macroscopic examination revealed hair loss; wrinkling and thickening of the skin, ears, paws, and muzzle; but also disruption of hair follicle bulge stem cells during anagen, as evidenced by patches of new hair growth, similar to that previously described for HPV16 E6 and E7 (Michael et al., 2013) (Figure S5A). Similar to LH mice, we observed significant changes in oral volume in KH versus control littermate mice and E6 and E7 expression, although transcript levels were 6- to 7-fold higher in KH epithelia (Figures 4B and 4C).
Figure 4. Conditional Basal Epithelial Expression of E6 and E7 Induces Canonical Tp53/pRb-E2f1 Pathway Genes and Epithelial Tumorigenesis.
(A) General strategy for generating KH mice by crossing a Cre recombinase strain driven by the human keratin 14 (KRT14) promoter (K) to our knockin HPV16 strain (H; top). Representative photographs of control littermates and the KH animals at 1–2 months (bottom).
(B) Quantification of oral volume calculated based on the formula for an ellipsoid volume less the oral aperture (n = 3, mean ± SEM; **p < 0.001).
(C) Quantitative real-time PCR analysis of E6 and E7 transcript copy numbers from dissected whole tongue of KH and control littermate mice calculated using respective standard curves (n = 3, mean ± SEM).
(D) Immunofluorescence (IF) analysis of neonatal and adult epithelial tissues. Representative sections from neonatal back skin (p0) and adult tongue (p45) were fixed in paraformaldehyde (PFA) and embedded, and sections were stained for epithelial differentiation markers or E7/ E2F1 targets. White arrowheads denote suprabasal events. Magnification, 400×; bar, 50 µm.
(E) Histologic analysis of epithelial tissues. Representative sagittal sections from adult tongue and cutaneous epithelia were formalin fixed and embedded, and sections were H&E stained. Magnification, 200×; bar, 200 µm.
(F) Quantification of cutaneous epithelial thickness calculated in adult KH and control littermate mice (n = 3, ***p < 0.0001).
(G) Quantitative real-time PCR analysis of select E6/Tp53 and E7/E2F1 host pathway genes in adult KH and control littermate mice. Relative fold expression is shown normalized to Ppib mRNA levels (n = 3, mean ± SEM).
(H) Epithelial E6 and E7 expression together with 4NQO treatment increases oral tumor development. Kaplan-Meier analysis of KH and control littermate mice provided with 4NQO (20 mg/ml) in the drinking water for 8 weeks followed by weekly oral cavity examination for gross pathologic changes. Tumor onset was significantly reduced (p = 0.0499) for KH mice (median tumor-free survival = 123 days) compared to control littermates.
Data presented in (B) and (F) are shown as mean ± SEM. Student’s t test (2 tailed) was used to determine significance. Related to Figure S5.
Because KRT14 drives Cre expression early in fetal development, we next examined the impact of E6:E7 expression on epithelial proliferation and differentiation in KH mice at select developmental time points. Compared to control littermates, KH mice displayed increased suprabasal expression of several differentiation markers, including keratins (K4 and K10) and involucrin (Ivl), in addition to the cell cycle and proliferation marker Ccne1, and DNA replication and repair markers Mcm7 and Rrm2 (Figure 4D; Figures S5B and S5C). A significant increase (p < 0.05) in suprabasal mitoses was also observed in KH mice compared to control littermates (Figures S5B and S5C). Histologic analysis of adult lingual and cutaneous epithelia at 1–2 months of age demonstrated fully penetrant epithelial hyperplasia, dysplasia, and hyperkeratosis in KH animals compared to control littermates (Figures 4E and 4F). Analysis of adult (p45) tongues versus embryonic day 16.5 (E16.5) back skin confirmed induced and sustained expression of E6/ Tp53 and E7/E2f1 pathway target genes in basal epithelia (Figure 4G; Figure S5D). However, similar to LH mice, KH mice rarely presented with oral cavity or oropharyngeal tumors despite frequently developing thymic tumors, as described with other HPV16 E6/E7 models (Arbeit et al., 1994; Carraresi et al., 2001) (Figure S5E). Therefore, to assess the effects of E6:E7 expression on tumorigenesis, we administered a moderate 4NQO dose (20 µg/ml) and observed a significant decrease (p < 0.05) in oral tumor-free survival for 4NQO-treated KH animals compared to control littermates (Figure 4H). Collectively, these data confirm the effects of conditional HPV16 E6 and E7 on epithelial proliferation and differentiation and validate the ability of stoichiometrically balanced E6 and E7 oncogene expression to drive tumorigenesis in a head and neck carcinogenesis model.
Intralingual E7iresE6 Activation with Chemical Carcinogenesis Induces Oropharyngeal Carcinoma
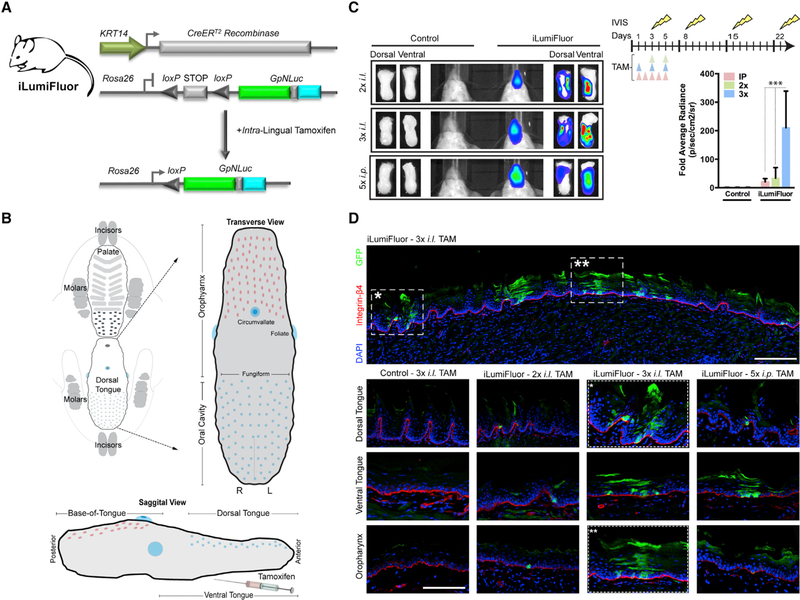
Mounting evidence has supported a role for field cancerization in the development and progression of HNSCCs (Califano et al., 1996; Slaughter et al., 1953). Previous efforts to inducibly and conditionally activate the HPV16 E6/E7 oncogenes in adult mice used tamoxifen-regulated Cre (CreERtam), but these ubiquitously expressed E6 and E7 within all epithelial tissues (Zhong et al., 2014). Thus, we sought to develop an autochthonous method that drives mosaic transgene activation for modeling field cancerization. We recently reported the creation of bioluminescent-fluorescent fusion reporters (LumiFluor) (Schaub et al., 2015) composed of eGFP and nanoluciferase that provide enhanced optical properties and signal penetration through tissues in vivo due to bioluminescence resonance energy transfer (BRET) (De et al., 2013). Therefore, we generated a knockin Rosa26-LSL-LumiFluor mouse strain and bred this animal to KRT14-CreERtam mice to establish iLumiFluor offspring (Figure 5A). To spatially restrict E6 and E7 expression at anatomically relevant tissues where HPV-induced HNSCCs arise, we delivered tamoxifen directly to the tongue to specifically activate the inducible transgenes only within the oropharyngeal regions of the lingual epithelia (Figure 5B). Arbitrary landmarks separate the divisions of the human pharynx, such that the oropharynx corresponds to the retromolar trigone posterior to the oral cavity and includes the lateral posterior one-third of the tongue. However, this does not translate to the same anatomic region in the mouse due to the elongated head of rodents and a relatively straight pharynx (Treuting and Dintzis, 2012). Rather, the posterior two-thirds of the mouse tongue encompasses the murine equivalent of an oropharyngeal region and includes intermediate epithelial transition zones where lesions are known to be manifest in humans. Different routes of administration (intraperitoneal [i.p.] or intralingual [i.l.]) and varying doses of tamoxifen were evaluated in a longitudinal manner using non-invasive IVIS bioluminescent imaging (BLI) over a 3-week time-course (Figure 5C). Non-invasive BLI revealed robust signal detection from oropharyngeal tissues with as few as two i.l. doses but a significant >100-fold increase (p = 0.001) with three i.l. doses compared to five i.p. doses, and these differences were confirmed to arise from the tongue upon end-point resection. Notably, i.l. tamoxifen delivery significantly reduced (>5-fold; p < 0.05) off-target reporter expression in cutaneous epithelia compared to conventional i.p. administration routes (Figure S6). To assess the levels and geographic locations of induced transgene expression, labeled cells were analyzed in frozen sagittal sections taken from tongues resected at end-point (Figure 5D). Patches of LumiFluor-labeled cells (GFP+) were observed throughout the basal epithelial layers of the dorsal, ventral, and base of tongue, establishing that i.l. tamoxifen delivery provides a robust and reproducible method for inducing mosaic transgene expression within lingual epithelia and supporting the utility of this methodology for modeling field cancerization.
Figure 5. Generation and Application of Inducible Reporter Knockin Mice (iLumiFluor) for Developing Methods to Target Lingual and Oropharyngeal Epithelia.
(A) General strategy for testing the efficacy of intralingual (i.l.) tamoxifen (TAM) administration to activate the estrogen receptor Cre recombinase fusion (KRT14-CreERtam) and induce eGFP-NanoLuc (GpNLuc LumiFluor) transgene expression within the tongue following Cre-mediated recombination and excision of the STOP cassette.
(B) Schematic of transverse and sagittal views highlighting key lingual anatomic features including the oropharynx, dorsal tongue, and ventral tongue. TAM was delivered by i.l. injection to the posterior, dorsal-lateral tongue to minimize dorsal tongue swelling and disruption to eating or drinking.
(C) Targeted induction of bioluminescent signal in adult tongues. Bioluminescent imaging (BLI) of iLumiFluor and control littermate mice injected either 2× or 3× i.l. versus 5× i.p. with TAM. Live animal, non-invasive BLI was performed temporally and signal levels graphed relative to baseline. Representative images for both live, non-invasive BLI and end-point resected tongues 16 days post-TAM are presented (n = 3, ***p < 0.0001).
(D) Immunofluorescence analysis reveals mosaic reporter expression throughout the adult tongue. Representative sections of adult tongues fixed in paraformaldehyde (PFA) and embedded, and sections were stained for epithelial basement membrane (Integrin-β4; red), LumiFluor (eGFP; green), and nuclei (DAPI; blue). Tile scan of 3× i.l. TAM (top). Comparison of TAM treatment regimen effects at various anatomic locations in iLumiFluor and control littermate mice (bottom). Tile scan of magnification at 20× with 1.25 optical zoom.
Data presented in (C) is shown as mean ± SEM of a representative experiment (n = 3 experiments). Two-way ANOVA with Bonferroni’s post hoc tests were used to determine significance. Related to Figure S6.
Next, we generated mice that enable conditional and inducible E7iresE6 expression within tongues of adult animals by breeding KRT14-CreERtam and Rosa26-LSL-E7iresE6 mice. As expected, the offspring of these crosses (iKH mice) displayed no overt phenotypes in the absence of tamoxifen (Figures 6A and 6B). Notably, in stark contrast to LH and KH mice, there is a significant reduction in the oral volumes of i.l. tamoxifen-treated iKH animals, indicating that i.l. tamoxifen delivery minimizes expression in off-target tissues despite robust lingual E6 and E7 expression (Figures 6C and 6D). To assess if direct injection of tamoxifen into the tongues has any adverse effects on feeding behaviors, we closely monitored animals over the course of 5 months and found no significant change in body weight for male or female animals (Figure 6E). Histologic analysis at 8–10 months of age demonstrated fully penetrant lingual hyperplasia and dysplasia but a significant reduction in cutaneous (face skin) epithelial thickness compared to LH and KH mice (Figures 6F and 6G). Moreover, iKH mice displayed increased lingual expression of the E7/E2f1-regulated target Mcm7 within the suprabasal layers compared to control littermates (Figure 6H). Notably, the addition of 4NQO to iKH mice promoted the development of oropharyngeal squamous cell carcinomas (Figure 6I; Figure S7A). Histologic and immunohistochemical (IHC) analysis of tongue sections confirmed recruitment of immune infiltrates, including neutrophils, cytotoxic T cells, and regulatory T cells (Figure 6J; Figure S7A). Collectively, these data validate the utility of this method for generating an autochthonous model of HPV16-associated oropharyngeal tumorigenesis.
Figure 6. Targeted E6 and E7 Expression Induces Hyperplasia and Dysplasia with Reduced Off-Target Effects.
(A) General strategy for generating conditional and inducible iKH mice by crossing the KRT14-CreERtam (iK) strain to our knockin HPV16 strain (H).
(B) Representative photographs of iKH animals at 8–10 months.
(C) Comparison of oral volume fold change between LH, KH, and iKH and control littermate mice 42 weeks post-tamoxifen injection reveals that restricting expression to the lingual and oropharyngeal epithelia significantly reduces off-target effects on external oral epithelial volume (n = 3, *p < 0.01).
(D) Quantitative real-time PCR analysis of E6 and E7 transcript copy numbers from dissected whole tongue of iKH and control littermate mice calculated using respective standard curves (n = 3, **p < 0.001, ***p < 0.0001).
(E) Body weight remains unchanged with lingual TAM administration. Comparison of male versus female body weights over the course of the study reveals no significant acute or long-term impact of i.l. TAM delivery either during or following administration, respectively (n = 3–8, mean ± SEM). Yellow shading highlights tamoxifen injection time frame. Blue shading highlights daily animal monitoring performed immediately post-injection followed thereafter by periodic weight and health assessment.
(F) Histologic analysis of epithelial tissues. Representative sagittal sections from adult tongue and cutaneous epithelia were formalin fixed, embedded, and H&E stained. Black arrowheads denote suprabasal mitoses. Magnification, 400×; bar, 50 µm.
(G) Comparison of cutaneous epithelial thickness fold change between LH, KH, and iKH and control littermate mice reveals that restricting expression to the lingual epithelia significantly reduces general cutaneous epithelial manifestations (n = 3, mean ± SEM; **p < 0.001, *p < 0.01).
(H) Increased expression of a E7/E2F1 target within iKH tongues. IF analysis of representative sections from adult iKH tongues stained for the HPV16 biomarker MCM7. White arrowheads denote suprabasal events. Magnification, 400×; bar, 50 µm.
(I) Targeted expression of E6 and E7 together with 4NQO treatment drives oropharyngeal squamous cell carcinoma development. Schematic overview of the animal experiment (top) and representative images of the gross morphology for tumors (yellow arrow) that develop in the iKHP mice provided 4NQO (20 µg/ml) in the drinking water (bottom). Dotted lines delineate the tumor boundary.
(J) Histologic and immunohistochemical analysis of immune cell infiltration. Representative sections from formalin-fixed, embedded, and H&E-stained iKHP tongues (left) and reveal morphologic and histologic abnormalities consistent with oropharyngeal squamous cell carcinoma. Inset highlights a region with an invasive epithelial island (yellow arrowhead). IHC staining of infiltrating Cd8a+ cytotoxic T cells (right). Magnification, 200×; bar, 200 µm.
Data presented in (C), (D), and (G) are shown as mean ± SEM of a representative experiment (n = 3 experiments). Two-way ANOVA with Bonferroni’s post hoc tests were used to determine significance. Related to Figure S7.
Autochthonous HPV16 E7iresE6 and PIK3CAE545K Expression Induces Oropharyngeal Carcinoma
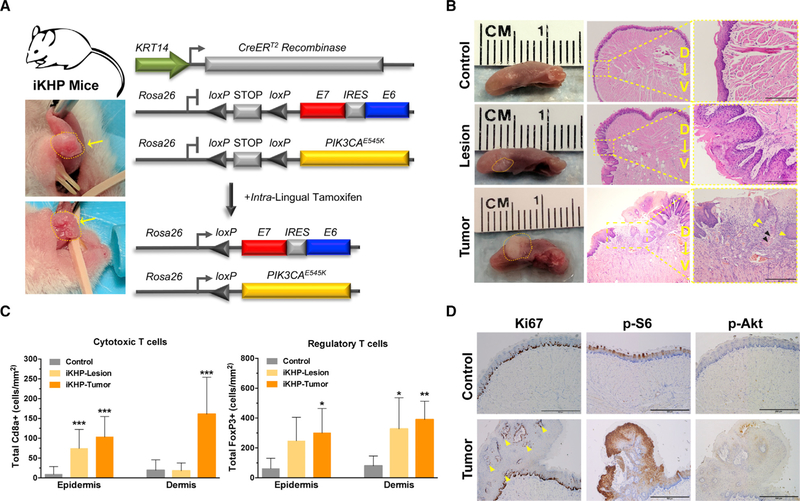
HPV(+) HNSCC in humans is characterized by low relative mutational burden, suggesting that few genomic alterations are required in combination with HPV16 E6 and E7 oncogene expression to drive tumorigenesis (Leemans et al., 2018; Cancer Genome Atlas Network, 2015). Indeed, integrated bioinformatics analyses revealed that of the downstream pathways activated in HNSCC, PIK3CA alterations are by far among the dominant driver events, where nearly 40% of all HPV(+) HNSCC cases harbor gain-of-function PIK3CA mutations at two key hotspots within the helical domain, namely, E542K and E545K (Hayes et al., 2015; Ng et al., 2018).
To recapitulate this HNSCC PIK3CA alteration, we generated a GEMM that combines Rosa26-LSL-E7iresE6 and mutant PIK3CA (Rosa26-LSL-PIK3CAE545K) (Meyer et al., 2013) under the control of KRT14-CreERtam. Offspring of these crosses (iKHP mice) were born with normal Mendelian distribution, but excitingly, restricting post-natal activation of these transgenes by using our i.l. tamoxifen delivery method induced the rapid development of oropharyngeal tumors (Figure 7A). Interestingly, macroscopic examination revealed that multifocal premalignant lesions develop with 100% penetrance and moderate burden (2–5 lesions/tongue) during a short (4–6 week) time period post-tamoxifen and that, of these lesions, oropharyngeal tumors develop with ~40% penetrance (1–2 tumors/tongue) starting as early as 6–8 weeks post-tamoxifen (Figure 7B; Table S5). These tumors primarily developed within the posterior two-thirds of the mouse tongue that represent the murine oropharynx equivalent at the dorsallateral borders and the base-of-tongue, although occasional ventral tumors also developed. Histologic and immunohistochemical analysis confirmed the presence of epithelial hyperplasia and moderate-to-severe dysplasia in the pre-malignant lesions and that some of these lesions progress to oropharyngeal squamous cell carcinoma (Figures 7B and 7C). Moreover, these analyses confirmed that although the majority of these oral tumors are moderately differentiated carcinoma in situ that display a largely exophytic, papillary appearance (Figure S7B), some are ulcerative and display invasive features with basement membrane disruption and muscle invasion accompanied by poorly differentiated areas characteristic of HPV(+) HNSCC (Figure 7B).
Figure 7. Autochthonous HPV16 E6 and E7 Coupled with PIK3CAE545K Expression Induces Preneoplastic Lesions That Rapidly Progress to Squamous Cell Carcinoma.
(A) General strategy for generating conditional and inducible iKHP mice by crossing the iKH strain to an inducible PIK3CAE545K strain (P). Representative photographs of independent tumors from iKHP animals at 8 weeks.
(B) Mutant PIK3CA cooperates with HPV16 E6 and E7 oncogenes to drive tumorigenesis. Representative images of the gross morphology for control and iKHP tongues at various times post-tamoxifen (left). Representative H&E-stained sections of control tongues compared to iKHP tongues harboring early, pre-malignant lesions (4–6 weeks) or tumors (6–8 weeks; middle). Representative invasive squamous cancer displaying both papillary, exophytic growth, and ulcerative lesion with basement membrane disruption, muscle invasion, and poorly differentiated areas. Tumor inset highlights invasive features (right, 2003) with mitotic figures, nuclear pleomorphism, and hyperchromatism (yellow arrowheads) and keratin pearls (black arrowheads). Magnification (left), 40×; bar, 200 µm.
(C) Quantification of epidermal versus dermal Cd8a+ cytotoxic T cell infiltration (left) or FoxP3+ regulatory T cell infiltration (right) in control versus pre-malignant lesions or frank tumors (n = 3, mean ± SEM; ***p < 0.0001, **p < 0.001, *p < 0.01).
(D) IHC staining of Ki67 demonstrating increased suprabasal proliferation of tumors (left) accompanied by marked increases in p-S6Ser235/236 (middle) staining but not pAKTSer473 (right). Magnification, 200×; bar, 200 µm.
Data presented in (C) is shown as mean ± SEM. Student’s t test (2 tailed) compared to control was used to determine significance. Related to Figure S7.
Similar to the oropharyngeal tumors that arise in the iKH mice with combined chemical carcinogenesis (Figures 6I and 6J), iKHP oropharyngeal tumors displayed significant leukocyte infiltration (Figure 7C). Notably, this immune cell recruitment is an early feature in disease progression, as these infiltrating cells are also present in pre-malignant lesions (Figure S7D). Previous work demonstrated that this enhanced leukocyte infiltration is associated with PIK3CA activation and that HPV(+) HNSCCs harboring PIK3CA mutations are associated with activation of mTOR but not of AKT, suggesting that E6 and E7 expression inhibits the ability of wild-type and/or mutant PIK3CA to activate AKT (Du et al., 2016). To investigate these mechanisms, we next examined S6 and Akt activation by immunohistochemistry and compared to control littermate tongues. We confirmed robust S6 phosphorylation at Ser235/236 in the iKHP oropharyngeal tumors but did not detect significant differences in AKT phosphorylation at Ser473 between control oral tissues and GEMM tumors (Figure 7D). Furthermore, similar to previous observations (Aguilar-Martinez et al., 2015; Rong et al., 2019; Sewell et al., 2014), we also found that these tumors exhibit low-level ERK1/2 activation (Thr202/Tyr204 phosphorylation) despite robust suprabasal total ERK1/2 expression (Figure S7E). Collectively, these data indicate that mTOR activation contributes to tumor progression in our HNSCC GEMM and that mutant PIK3CA in combination with balanced expression of HPV E6 and E7 is sufficient to drive oral tumorigenesis.
DISCUSSION
HPV is the most common sexually transmitted infection in the United States (Satterwhite et al., 2013), with over 20 million active cases and an estimated 6 million new cases each year according to the Centers for Disease Control and Prevention. This rising HPV epidemic poses significant health risks due to the causal role that HPV plays in the development of several cancers, including a distinct anatomic subset of head and neck cancers (OPSCCs) (Ha and Califano, 2016; Moody and Laimins, 2010; Münger et al., 2004). In fact, there is a rising incidence of HPV(+) HNSCCs linked to changes in sexual behaviors that promote viral transmission and oral infection (Chaturvedi et al., 2011; Gillison et al., 2008; Stein et al., 2014), but the cooperating genetic insults that drive development and progression of this cancer remain poorly understood.
Several in vitro and in vivo model systems exist for investigating HPV-associated pathogenesis (Doorbar, 2016; Lambert, 2016; Spiotto et al., 2013), but the currently available GEMMs each have a unique set of limitations that must be considered when designing studies (Table S4). These HPV GEMM models fall into two broad categories, namely, those that display constitutive versus those with inducible HPV16 E6 and/or E7 oncogene expression. Constitutive GEMMs for in vivo study include the αA-HPV16-E6/E7 (Griep et al., 1993; Lambert et al., 1993), K14-HPV16 (Arbeit et al., 1994), K14HPV16E6 (Song et al., 1999), and K14HPV16E7 (Herber et al., 1996) transgenic mouse models, where HPV16 genes are expressed by the αA crystallin or keratin 14 (K14) promoters. Studies using aA-HPV16-E6/E7 transgenic mice revealed that the αA crystallin promoter successfully directs E6/E7 expression to the mouse ocular lens, but this model is confounded by non-specific expression within cutaneous epithelia, limiting the ability to model HPV-induced oropharyngeal, cervical, or anogenital cancers (Griep et al., 1993). Consequently, subsequent studies used a basal epithelial-cell-specific keratin promoter (K14) to direct oncogene expression to cutaneous and mucosal epithelia, but these models have low tumor penetrance and the K14 promoter is activated early in embryogenesis by E12.5 such that constitutive in utero oncogene expression impacts normal epithelial development and viral antigens induce immune tolerance, minimizing usefulness as an immunocompetent model (Doan et al., 1999; Melero et al., 1997).
To generate systems that model the onset of HPV infection in adult humans, two inducible transgenic models were recently generated that enable conditional basal epithelial-cell-specific and tamoxifen-(Zhong et al., 2014) or doxycycline-(Callejas-Valera et al., 2016) inducible expression (K14-CreERTam or K5-rtTA, respectively). However, both models are transgenic (and thus subject to founder effects linked to sites of transgene integration) and require high doses of tamoxifen (100 mg; 20 mg/ml for 5 days) or doxycycline (6 g/kg chow). Moreover, E6 and E7 promote cancer by distinct mechanisms that disrupt cellular proliferation (E7/E2F1) and apoptotic (E6/TP53) programs while also promoting genomic instability and immune cell invasion (Moody and Laimins, 2010; Münger et al., 2004; Rautava and Syrjänen, 2012). Unfortunately, both models use the E6/E7 ORF amplified from the HPV viral genome and lack proper control of E7 expression, which is dependent on several variables, including host factors that regulate cellular differentiation, EGFR-ERK1/2 signaling, and, most importantly, mRNA splicing (Graham and Faizo, 2017; McFarlane et al., 2015; Rosenberger et al., 2010; Wu et al., 2017). Due to a limited number of nucleotides located between the E6 and E7 ORFs, splicing of the polycistronic E6 mRNA transcript into the E6*I variant is necessary to relieve spacing constraints imposed on the ribosome and enhance E7 translation efficiency (Ajiro et al., 2012; Tang et al., 2006). Notably, E7 expression levels are either not reported (Zhong et al., 2014) or ratios appear to be lower than E6 based on available western blot data (Callejas-Valera et al., 2016), suggesting that these models may suffer from inadequate E7 expression that do not faithfully recapitulate the combined effects of E6 and E7 oncogene actions on TP53 and RB, respectively, in promoting HPV(+) oropharyngeal tumorigenesis. In fact, studies have identified HPV16 E7 as the dominant oncogene with respect to promoting cancer development (Halbert et al., 1991; Riley et al., 2003; Strati and Lambert, 2007), and repression of E7 provokes regression of high-grade cervical dysplasia and cervical tumors irrespective of E6 expression (Jabbar et al., 2009, 2012). Importantly, mice that express both the E6 and E7 transgenes display an increased incidence of invasive carcinomas (Jabbar et al., 2010), further supporting the need for GEMMs that accurately reflect the stoichiometry driving this pathobiology.
We developed a knockin GEMM of HPV(+) HNSCC whose autochthonous disease progression reflects the histopathological and molecular features observed in human HPV(+) OPSCC Our approach (Rosa26-loxP-STOP-loxP-E7iresE6; ‘‘H’’ mice) spatiotemporally restricts expression of the high-risk HPV16 E6 and E7 oncogenes to the oropharyngeal basal epithelium in post-natal immunocompetent mice. This approach recapitulates the in vivo E6/E7 expression levels and stoichiometric ratios seen in human HPV(+) HNSCC tumors and HPV(+) HNSCC cell lines. Although some challenges still exist in modeling human biology in a rodent, this is an unfortunate limitation of many animal models. We confirmed that autochthonous mosaic transgene expression is achievable within relevant oropharyngeal sites of HPV infection by using an i.l. targeting methodology coupled with a second knockin GEMM we generated using our recently described bioluminescent-fluorescent LumiFluor optical reporter tool (Rosa26-loxP-STOP-loxP-LumiFluor). Therefore, this method also provides a tractable approach to modeling field cancerization in GEMMs.
Two recent genomics studies reported that the alternatively spliced E6*I transcript is the dominant variant expressed in HPV(+) tumors and that full-length E6 transcripts are present at substantially lower levels (Gillison et al., 2018; Nulton et al., 2017). Consequently, these observations further limit the utility of current inducible HPV GEMMs because full-length E6 transcripts rather than E6*I predominate in those models, so the impacts of E7 are minimal. In stark contrast, we discovered that full-length E6 and E6*I transcripts are expressed to near equivalent levels in most human primary HPV(+) HNSCCs. The observed difference in our results can be accounted for by the bioinformatics approaches we used. Specifically, there is an apparent decrease in overall reads mapping to the alternatively spliced region missing from the E6*I transcript when viewing read pileup data (nt 226 – 409), but reads mapping 5′ or 3′ to this region actually represent reads that can arise from shared sequences common to both full-length E6 and spliced E6*I transcripts. Therefore, we addressed this caveat by only considering junction-spanning reads unique to each splice variant. Furthermore, relative E6:E7 ratios correlate with distinct molecular subtypes and available HPV(+) HNSCC cell lines express increased E7 levels marked by increased E7/E2F1-regulated target gene expression, such as the canonical HPV biomarker MCM7 (Strati et al., 2006). This feature may be due to the loss of selective pressures required to maintain E6 expression following TP53 deletion or mutation that occurred during the establishment of these cell lines. Thus, currently available HPV(+) cell lines may best model only a small subset of HPV(+) HNSCC cases where high E7 expression is coupled with alternative mechanisms of TP53 pathway inhibition.
Our HPV16 GEMM allows one to define the genetic events that drive multi-stage tumorigenesis. Interestingly, post-natal induction of these HPV oncogenes within lingual epithelia of our GEMM circumvents the immune tolerance phenotypes that plague constitutive model systems (Doan et al., 1999; Melero et al., 1997). This is reflected by the robust leukocyte infiltration we observe in early pre-malignant lesions and tumors that develop in our GEMM, suggesting that this model preserves the immune system’s ability to respond to exogenous viral antigen exposure. These findings have important applications and support the use of our HPV16 GEMM as an immunocompetent preclinical platform for investigating mechanisms of immunosuppression in HPV(+) HNSCC. A limitation of our current HPV GEMM mice is that oropharyngeal tumors that develop are primarily carcinoma in situ with only occasional poorly differentiated invasive features, suggesting that additional genetic alteration(s) are required for their progression. However, the spontaneous and incomplete conversion of all pre-malignant lesions to HNSCC also presents an intriguing opportunity to evaluate the utility of this model for investigating those molecular events responsible for the progression of only a subset of lesions to malignancy. Although previous models of oral tumorigenesis expressed CCND1, mutant Kras, Tp53, Tgfbr1, or Notch1 (Spiotto et al., 2013), we specifically chose to investigate the contribution of mutant PIK3CA in promoting the development of HNSCCs given accumulating genomic sequencing evidence pointing to a potential role for PIK3CA amplification and/or mutation as a key driver event (Gillison et al., 2018; Hayes et al., 2015; Leemans et al., 2018; Cancer Genome Atlas Network, 2015). Aberrant PI3K signaling enhances tumorigenic potential by increasing cell proliferation and survival and promoting migration, invasion, metabolism, angiogenesis, as well as resistance to chemotherapy (Hafsi et al., 2012; Wong et al., 2010; Yuan and Cantley, 2008). Most gain-of-function PIK3CA mutations occur in either the kinase (H1047R) or helical domains (E542K and E545K) of p110a and create a constitutively active enzyme (Samuels et al., 2004). But unlike breast carcinoma and HPV(—) HNSCC, nearly all HPV(+) HNSCCs selectively accumulate mutations within the helical domain, consistent with mRNA editing induced by APOBEC-mediated cytosine deaminase mutagenesis (Hayes et al., 2015; Ng et al., 2018). Indeed, deregulation of pathways that converge on mTOR signaling are characteristic of human HPV(+) HNSCC, and activation of this pathway also occurs in mouse models of HPV-associated cervical and oropharyngeal cancers (Callejas-Valera et al., 2016). Thus, our findings demonstrating selective activation of mTOR rather than AKT signaling are aligned with observations made in both human HPV(+) HNSCC and mouse models of these cancers. Future studies aimed at defining the genetic events that cooperate with E6/E7;PIK3CAE545K to promote invasion and migration will be important to understand the mechanisms that underlie HPV(+) cases with poor prognosis.
Collectively, we demonstrate that significant similarities exist between human HPV(+) HNSCC and our HPV GEMM, establishing this model system as a powerful platform for gaining new insights into the underlying molecular mechanisms governing HPV(+) HNSCC development and progression and investigating the efficacy of currently available and next-generation therapies.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Antonio L. Amelio (antonio_amelio@unc.edu). The plasmids, mouse lines, and cell lines generated in this study are available upon request via a material transfer agreement (MTA).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Models
All animal studies were reviewed and approved by The University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee. The Rosa26-LSL-E7iresE6 mice were generated by knocking-in a polycistronic cassette containing the high risk HPV16 E7 and E6 cDNAs separated by an IRES element into a Rosa26-loxP-Stop-loxP cassette, using the pBigT and pRosa26-PA vectors, as previously described (Srinivas et al., 2001). Briefly, the E7 cDNA was cloned into pBigT via XhoI and NotI while the E6 cDNA was cloned into the MIGR1 plasmid downstream of the IRES element via NcoI and SalI. An IRES-E6 amplicon containing NotI and SacI sites was then subcloned into the pBigT-E7 construct to create pBigT-E7iresE6 prior to shuttling into the Rosa26-PA destination vector via Pac1 and AscI. Similarly, Rosa26-LSL-GpNLuc mice were generated by knocking-in the GpNLuc (Schaub et al., 2015) LumiFluor optical reporter cDNA into a Rosa26-loxP-Stop-loxP cassette by first subcloning GpNLuc into pBigT prior to shuttling into the Rosa26-PA destination vector. MEFs from founder lines were screened by transducing cells with Adeno-Cre virus (MOI = 20) and analyzing the recombined allele from the Rosa26-LSL-GpNLuc mice by measuring bioluminescent light output or by PCR and western blot for the Rosa26-LSL-E7iresE6 mice. The L2-Cre (Stairs et al., 2011) (a gift from Anil Rustgi, University of Pennsylvania, Philadelphia, PA), KRT14-Cre (a gift from Elaine Fuchs, The Rockefeller University, New York City, NY), and KRT14-Cretam (The Jackson Laboratory, Stock #: 005107) Cre recombinase strains were crossed to the Rosa26-LSL-E7iresE6 mice to generate the LH, KH, or iKH models, respectively. The iKH mice were crossed to Rosa26-LSL-PIK3CAE545K (a gift from Mohamed Bentires-Alj, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) (Meyer et al., 2013) to generate the oropharyngeal cancer GEMM (iKHP). The KRT14-Cretam mouse was crossed with Rosa26-LSL-GpNLuc to generate the iLumiFluor reporter model. All animals were backcrossed to C57BL/6J or albino B6(Cg)-Tyrc−2J/J mice (The Jackson Laboratory, Stock #: 000664 or Stock #: 000058) for seven to ten generations.
To induce Cre-mediated transgene recombination in the tongues of iLumiFluor, iKH, or iKHP mice, tamoxifen was administered to the lingual submucosa by direct injection. Optical reporter signal or tumor development were regularly monitored by IVIS BLI imaging and/or oral examination, respectively. Male and female mice were used for experiments, and randomly assigned to experimental groups. Mice were sacrificed for the humane endpoints as follows. For the autochthonous mouse models, mice were sacrificed for weight loss more than 20% of the initial animal weight or tumor size volume of > 40 mm3 as evaluated by caliper and/or BLI measure. In our studies, all mice were sacrificed because of tumor size. The endpoint for allograft models was tumor volume > 500 mm3 or weight loss greater than 20% body weight.
Cell lines
Mouse embryonic fibroblasts (MEFs) were prepared as previously described and cultured in DMEM (GIBCO, cat. #: 11965–092) supplemented with 10% FBS (Atlanta Biologicals, cat. #: S11550), 1x penicillin, streptomycin, glutamine (1xPSG - GIBCO, cat. #: 10378016,) 1x GlutaMAX (GIBCO, cat. #: 35050–061), and 0.1mM β-Mercaptoethanol (Fisher Chemicals, cat. #: 034462). ‘Normal’ oral keratinocyte (NOK) cell lines OKF4-TERT1, OKF4-E6/E7, OKF6-TERT1, and OKF6-E6/E7 (a gift from Jim Rheinwald and Matthew Ramsey, Harvard University, Boston, MA) and the GMSM-K (Gilchrist et al., 2000) (a gift from Valerie Murrah, UNC-Chapel Hill, Chapel Hill, NC) were grown in keratinocyte serum free media (KSFM) from (GIBCO, cat. #: 17004–042). The OKF6/OKF4 lines were grown in KSFM media supplemented with 25 µg of bovine pituitary gland extract (BPE), 0.2 ng/ml of EGF, 300mM of CaCl2, and 1x of PSG (Dickson et al., 2000; Schön and Rheinwald, 1996). The GMSMK cells were cultured in KSFM supplemented with 50mg/ml of BPE, 50ng/ml of EGF, and 1x PSG. The HPV-negative cell lines UM-SCC-74A, UM-SCC-5, UM-SCC-11A, and UM-SCC-14A (a gift from Thomas Carey, University of Michigan, Ann Arbor, MI) were grown in DMEM (GIBCO, cat. #: 11965–092) supplemented with 10% heat-inactivated FBS, 1x PSG, 1x GlutaMAX, and 1x MEM non-essential amino acids (GIBCO, cat. #: 11140050). The additional HPV-negative cell lines SCC-15 and SCC-25 (a gift from Ben Major, UNC-Chapel Hill, Chapel Hill, NC) were grown according to ATCC cell culture conditions. The HPV-positive cells UM-SCC-47, 93-VU-147T, UD-SCC-2, and UPCI:SCC090 (a gift from Randall Kimple, University of Wisconsin, Madison, WI) were cultured in DMEM media supplemented with 10% FBS, 1x PSG, and 1x GlutaMAX.
METHOD DETAILS
Bioinformatics
Reads from TCGA HNSC RNA-seq were aligned to the human genome using STAR v2.4.2a (Dobin et al., 2013) allowing for 5 mismatches and 1080 multi-maps. Reads that did not align to the human genome were aligned to a curated vertebrate virus reference downloaded from GenBank December 2015, with the addition of HPV16 from the PaVE database using STAR v2.4.2a allowing for 4 mismatches and 52 multi-maps. Unspliced and spliced HPV16 early gene transcripts were quantified by counting reads that included the splice junction (nt 226^409) or spanning reads (nt 226/227 and 408/409) and calculating the mean coverage for each sample. Reads were counted as splicing when their cigar string in the SAM file included the splice junction, while reads were counted as spanning when they started or ended within 50 bp of the splice junction but had a cigar string that indicated that they went into the intron rather than being spliced. We calculated average coverage by taking each read that was more than 50 bp past the end of the splice and parsing its cigar string to determine which genomic positions it covers, then calculating the mean of the coverage of that region. Differentially expressed genes were determined using DESeq2 (Love et al., 2014) and spliced E6*I ratio as the predictor. Gene expression modules were screened for association with E6*I ratios using the Wald statistic from DESeq2. Genes within each module were stratified by the sign of the statistic and scores calculated for each stratum by summing the statistic for each gene in the module and dividing that sum by the square root of the sum of the statistic of the genes in the module. Thus, each module is represented by two statistics representing the strength of increasing and decreasing gene expression. Permutation tests were performed to characterize the false discovery rate for thresholds of the module scores by shuffling the E6*I ratios and repeating the gene level DESeq2 statistic. This test yielded scores more extreme than 12 or —12 approximately 5 times each out of 100, thus a pathway score more extreme than 12 or —12 was assigned an empirical q-value of 0.05.
Tamoxifen and 4NQO administration
To conditionally induce GpNLuc or E7iresE6 expression, iLumiFluor or iKH and iKHP animals were intra-lingually injected using a 27-gauge needle with 0.5mg of tamoxifen (Sigma-Aldrich, cat. #: T5648) dissolved in 30µl corn oil (MP Biomedical, cat. #: 901414) either two and/or three times over a five-day period, respectively.
4NQO (4-Nitroquinoline 1-oxide; Sigma, cat. #: N8141) was dissolved in propylene glycol (Sigma, cat. #: 398039) at a concentration of 5 mg/mL and then further diluted to a final concentration of 10 or 20 µg/ml in sterile ddH2O. Following intra-lingual tamoxifen administration, animals were allowed a one-week recovery period prior to being administered 4NQO treated water for 8 weeks as previously described (Vitale-Cross et al., 2009). The 4NQO treated water was changed weekly.
Histological analysis and immunohistochemistry
Tissue harvesting and histological analysis were performed as previously described with minor modification (Herber et al., 1996). Briefly, all animals showing obvious tumors or other signs of distress were euthanized and subjected to full necropsy. For histological analysis, all tissues including whole head and/or tumors from LH, KH, iKH, and iKHP animals were fixed in 10% neutral buffered formalin for approximately 108 hr at room temperature. Following fixation, forehead and left cheek skin and tongues were extracted, and processed on an ASP6025 automated tissue processor (Leica Biosystems), and embedded in paraffin wax. Blocks were sectioned at 4–6 µm, mounted on glass slides, and FFPE tissue sections were deparaffinized prior to staining. Hematoxylin and eosin (H&E) staining was performed using pre-mixed hematoxylin, clarifier, bluing reagent, and eosin (Richard Allan Scientific). Immunohistochemistry was performed on the Discovery Ultra (Ventana Medical Systems) using manufacturers reagents on 4µm sections. Briefly, anti-Mouse CD8a (eBioscience, cat. #: 14–0808) was prepared using Discovery PSS Diluent (cat. #: 760–212). Antigen retrieval was performed using Ventana’s CC1 (pH 8.5) for 64 min at 90°C. The slides were given a hydrogen peroxide block for 8 min at room temperature and then incubated in the primary antibody diluent (1:100) for 1 hr at room temperature, followed by anti-Rat HRP secondary antibody (Ventana Omap OmniMap, cat. #: 760–4457) for 32 min at room temperature. The slides were then treated with DAB and counterstained with Hematoxylin II for 12 min and then Bluing Reagent for 4 min.
Alternatively, the anti-Mouse/Rat Foxp3 (eBioscience, cat. #: 14–5773) was prepared using Discovery Ab Diluent (Ventana Omap OmniMap, cat. #: 760–108). Antigen retrieval was performed using Ventana’s CC1 (pH 8.5), for 24 min at 100°C and then blocked with a protein block for 1 hr at room temperature. The slides were given a hydrogen peroxide block for 8 min at room temperature and then incubated in the primary antibody diluent (1:50) for 2 hr at room temperature, followed by the anti-Rat HRP secondary antibody for 32 min at room temperature. The slides were treated with Discovery Purple for 100 min. For each genotype, ≥ 3 animals from any given time point were examined and multiple similar grade tumors scored.
Immunofluorescence
Back skin or oral tissues (tongue) from p45 animals were fixed in 4% Paraformaldehyde for 0.5–1 hr followed by PBS washes. Tissues were incubated in 15% and 30% sucrose gradient at 4°C prior to embedding in O.C.T compound (Tissue Tek, cat. #: 4583) without prior fixation. Tissues were sectioned to 8µm on a Leica CM1950 cryostat using DB80LX blades (cat. #: 14035843496), mounted on SuperFrost Plus glass slides (Fisher Scientific, cat. #: 12–550-15), and dried 10 min at 37°C. Fresh frozen tissue from p0 mice and fixed p45 skin sections were fixed for 5 min with 4% PFA, washed with PBS, and blocked in a gelatin block (5% normal donkey serum (Jackson Immunoresearch, cat. #: 017–000-121), 3% BSA, 8% gelatin (Sigma cat. #: G7765), and 0.05% Triton X-100 in PBS) for 1 hr. Sections were stained with primary antibodies diluted in gelatin block and incubated overnight at 4°C, washed three times with PBS, incubated 2 hr with secondary antibodies at room temperature, counterstained with DAPI for 5 min at room temperature, and mounted in ProLong Gold (Invitrogen, cat. #: P36930). Tissues were stained with primary and secondary antibodies (Table S6) and images acquired using LAS AF software on a Leica TCS SPE-II 4 laser confocal system on a DM5500 microscope with ACS Apochromat 20 3 /0.60 multi-immersion, ACS Apochromat 40 3 /1.15 oil, and ACS Apochromat 63 3 /1.30 oil objectives.
In vivo BLI
Bioluminescent-fluorescent BRET signal was measured non-invasively as previously described (Schaub et al., 2015) with minor modification. Briefly, animals were injected tail-vein with 250 µM (1:20 dilution, ~500 µg/kg) Nano-Glo Luciferase Assay Substrate (Promega, cat. #: N1138) in sterile PBS. Isoflurane-anesthetized animals were then imaged using an IVIS Kinetic (PerkinElmer) 5 min after injection. Images were captured with open filter and acquisition times of 60 s or less at the indicated settings. Data were analyzed using the Living Image software.
Plasmids
The pBigT and pRosa26-PA (Srinivas et al., 2001) constructs (Addgene plasmid #21270 and #21271, respectively) were gifts from Frank Costantini, MIGR1 (Pear et al., 1998) (Addgene plasmid #27490) was a gift from Warren Pear, and the pRetroX-Tight-MCS_PGK-GpNLuc construct (Addgene plasmid #70185) containing the LumiFluor optical reporter was previously described (Schaub et al., 2015).
PCR and qPCR
Recombination was assessed by lysing cells (100 mM NaCl, 10 mM Tris pH 8, 25mM EDTA pH 8, 0.5% SDS, and 50 µg/ml Proteinase K) and extracting genomic DNA using phenol-chloroform and ethanol precipitation. Briefly, MEFs from founders and established lines were tested by transducing cells with Adeno-Cre virus (MOI = 20) and analyzing the recombined allele from the Rosa26-LSL-E7iresE6 mice by PCR (PrimeSTAR GXL DNA polymerase - Takara Bio USA, cat. #: R050A) using pBigT-E7iresE6 as a control with the following primers (Table S7) and cycling conditions: 95 °C for 3 min followed by 98 °C for 10 s, 55 °C for 15 s and 68 °C for 40 s for 34 cycles and 5 min for 68°C.
For cell lines, gene expression was measured by extracting RNA using a Nucleospin RNA kit (Machery-Nagel, cat #: 740955) as per the manufacturer’s recommendations. cDNA was made from 0.65 µg (HNSCC and NOKs) or 1 µg (MEFs) of RNA using iScript cDNA synthesis kit (Bio-Rad, cat. #: 170–8890). For tissues, gene expression was measured by extracting RNA from tissues snap frozen in liquid nitrogen and homogenized with liquid nitrogen using mortar and pestle. RNA was then prepared using NucleoZOL (Macherey-Nagel, cat. #: 740404.200) in accordance with the manufacturer’s instructions. cDNA was made from 0.5–1 µg of RNA using SuperScript IV Reverse Transcriptase (Invitrogen, cat. #: 18090050) with dNTPs (NEB, cat. #: N0446S), RNase inhibitor (Applied Biosystems, cat. #: N808–0119), and 50 µM oligo d(T)20 primers (Invitrogen, cat. #: 100023441). HPV16 E6 and E7 copy number was determined by establishing standard curves with 100 to 1×106 copies of the pBigT-E7iresE6 construct. Relative gene expression of E6, E7, E6/TP53, and E7/E2F1 targets was determined using the 2∆∆Ct method and normalized using human and mouse RPL23. Expression in HNSCC cell lines relative to NOKs was calculated using the ∆CT of OKF4-TERT1. qPCR was performed using FastStart Universal SYBR Green Master (Rox) Mix (Roche, cat. #: 04913850001) with 1/20 (tissue) or 1/40 (cells) volume of the cDNA iScript reaction, and 0.25 µM of primers (Table S7).
For basal keratinocytes, tissues were treated with Dispase overnight to enable separation of the epidermis from the dermis. The epidermal layer was then further processed with Typsin and passed through a 70 µm cell strainer, snap frozen in liquid nitrogen, and RNA was then isolated using QIAGEN RNeasy (QIAGEN, cat. #: 74104) in accordance with the manufacturer’s instructions. cDNA was made from 250 ng of RNA using iScript (Biorad, cat#1708891) Reverse Transcriptase, diluted 1:5 in ddH2O, and qPCR performed using iTaq Universal SYBR green supermix (BioRad, cat. #: 1725121).
Western blotting
Whole cell lysates were prepared in buffer containing 250 mM NaCl, 50 mM Tris (pH 7.4), 50 mM NaF, 0.1 mM NaVO4, 5 mM EDTA, and 0.1% Triton X-100 supplemented with protease inhibitors (cOmplete, EDTA-free Protease Inhibitor Cocktail - Roche, cat. #: 04693132001) and phosphatase inhibitors (phosphoSTOP, - Roche, cat. #: 10917400). Lysates (30–50 µg) were loaded onto mini-16% tris-glycine polyacrylamide gels and proteins separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to a 0.2 µm nitrocellulose membrane (GE Healthcare Life Sciences, cat. #: 10600011) using a Bio-Rad Transblot Turbo System set at 1.0 Amps (constant), 25 V, and run for 22 min. Membranes were blocked for 1 hr at room temperature in TBS-T + 5% milk and then incubated over night at 4°C with primary antibodies (Table S6): HPV16 E6 (1:1000–1:2000 - Gene Tex, cat. #: GTX132686) or HPV16 E7 (1:1000 - Gene Tex, cat. #: GTX133411) diluted in TBS-T + 3% milk. Following primary antibody incubation, membranes were washed and probed with Horseradish peroxidase (HRP)-conjugated goat anti-mouse (1:2500 - Thermo Fisher, cat. #: 31432) or donkey anti-rabbit (1:2500 - Thermo Fisher, cat. #: 31458) secondary antibodies for 1–2 hr at room temperature. Blots were imaged using Clarity ECL (Bio-Rad, cat. #: 170–5060) and ImageQuant LHS4000 (GE). Data are normalized using rabbit anti-Gapdh for MEFs (1:2000 - Santa Cruz, cat. #: sc-25778) or mouse anti-b-actin (1:5000 - Sigma Life Sciences, cat. #: A3854). Densitometry analysis was performed using ImageQuant TL software (GE).
QUANTIFICATION AND STATISTICAL ANALYSIS
Each in vivo and in vitro experiment was performed in triplicate and repeated at least three times. All statistical tests were executed using GraphPad Prism software or the statistical software R (version 3.1.2). Differences between variables were assessed by 2-tailed Student’s t test or 2-way ANOVA with Bonferroni’s post hoc tests, where appropriate. Data are expressed as mean ± SEM P values less than 0.05 were considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-HPV16 E6 (1:1000) | Gene Tex | Cat# GTX132686 |
| Rabbit anti-HPV16 E7 (1:1000) | Gene Tex | Cat# GTX133411 |
| Rabbit anti-GAPDH (1:2000) | Santa Cruz | Cat# sc25778 |
| Mouse anti-Actin (1:5000) | Sigma | Cat# A3854 |
| Guinea pig anti-Cytokeratin 14 (1:500) | Acris | Cat# BP5009 |
| Chicken anti-Cytokeratin 14 (1:1000) | Biolegend | Cat# 906001 |
| Rabbit anti-Cytokeratin 4 (1:1000) | Proteintech | Cat# 16572–1-AP |
| Rabbit anti-Cytokeratin 10 (1:500) | Covance | Cat# 905401 |
| Rat anti-Integrin B4 clone 346–11a (1:1000) | BD Biosciences | Cat# 553745 |
| Rabbit anti-Cyclin E (1:1000) | Abcam | Cat# ab52189 |
| Goat anti-RRM2 (1:50) | Santa Cruz | Cat# Sc10814 |
| Rabbit anti-Involucrin (1:500) | Covance | Cat# 924401 (was Cat# PRB-140C) |
| Rat anti-Phospho-Histone H3 (1:1000) | Abcam | Cat# a10543 |
| Rabbit anti-MCM7 (1:500) | Santa Cruz | Cat# sc22782 |
| Chicken anti-GFP (1:2000) | Abcam | Cat# ab13970 |
| Donkey Anti-Rabbit HRP (1:2500) | Thermo Fisher | Cat# 31458 |
| Goat Anti-Mouse (1:2500) | Thermo Fisher | Cat# 31432 |
| Rhodamine Red™-X (RRX) AffiniPure Donkey Anti-Rabbit IgG (H+L) (1:500) | Jackson ImmunoResearch Labs | Cat# 711–295-152 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) (1:1000) | Jackson ImmunoResearch Labs | Cat# 706–545-148 |
| Cy™5 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 706–175-148 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) (1:1000) | Jackson ImmunoResearch Labs | Cat# 703–545-155 |
| Donkey Anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor® 488 (1:1000) | Invitrogen | Cat# A-21208 |
| Rhodamine Red™-X (RRX) AffiniPure Donkey Anti-Rat IgG (H+L) (1:500) | Jackson ImmunoResearch Labs | Cat# 712–295-150 |
| Cy™5 AffiniPure Donkey Anti-Goat IgG (H+L) (1:400) | Jackson ImmunoResearch Labs | Cat# 705–175-147 |
| Rabbit anti-wide spectrum Cytokeratin (1:300) | Abcam | Cat# ab9377; RRID: AB_307222 |
| Rat anti-mouse Cd8a | eBioscience, | Cat# 14–0808 |
| Rat anti-mouse Foxp3 | eBioscience | Cat# 14–5773 |
| Rat Anti-Ly6g | BioLegend | Cat# 127601 |
| Rabbit mAb anti-p44/42 MAPK (Total Erk1/2) | Cell Signaling | Cat# 4695 |
| Rabbit mAb anti-Phospho-p44/42 MAPK (Thr202/Tyr204) (p-Erk1/2) | Cell Signaling | Cat# 4370 |
| Anti-Rat HRP conjugated secondary antibody | Ventana Omap OmniMap | Cat# 760–4457 |
| Anti-Rabbit HRP conjugated secondary antibody | Ventana Omap OmniMap | Cat# 760–4311, |
| Bacterial and Virus Strains | ||
| Adeno-GFP with CMV promoter (20 MOI) | Gift from Marc Montminy (Salk Institute) | N/A |
| Adeno-Cre with CMV promoter (20 MOI) | Gift from Marc Montminy (Salk Institute) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648; CAS: 10540–29-1 |
| Corn oil | MP Biomedical | Cat# 901414; CAS: 8001–30-7 |
| 4-Nitroquinoline 1-oxide | Sigma | Cat# N8141; 56–57-5 |
| Propylene Glycol | Sigma | Cat# 398039; 57–55-6 |
| Critical Commercial Assays | ||
| NucleoZOL | Macherey-Nagel | Cat# 740404.200 |
| Nucleospin RNA kit | Macherey-Nagel | Cat# 740955 |
| SuperScript IV | Invitrogen | Cat# 18090050 |
| dNTPs | NEB | Cat# N0446s |
| RNase inhibitor | Applied Biosystems | Cat# N808–0119 |
| 50µM oligo d(T)20 primers | Invitrogen | Cat# 100023441 |
| FastStart Universal SYBR Green Master (ROX) Mix | Roche | Cat# 04913850001 |
| Nano-Glo Luciferase Assay Substrate | Promega | Cat# N1120 |
| Deposited Data | ||
| PaVE Database | Van Doorslaer et al., 2017 | https://pave.niaid.nih.gov/ |
| TCGA Head and Neck Squamous Cell Carcinoma gene expression data | Genomic Data Commons | https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga |
| Experimental Models: Cell Lines | ||
| Mouse: Mouse Embryonic Fibroblast cells | This paper | MEF-H (HPV16_E7_ires_E6) |
| Human: OKF4-TERT1 cell line | Gift from Jim Rheinwald and Matthew Ramsey (Harvard University, Boston, MA) | RRID: CVCL_L227 |
| Human: OKF4-E6/E7 cell line | Gift from Jim Rheinwald and Matthew Ramsey (Harvard University, Boston, MA) | RRID: CVCL_L226 |
| Human: OKF6-TERT1 cell line | Gift from Jim Rheinwald and Matthew Ramsey (Harvard University, Boston, MA) | RRID: CVCL_L224 |
| Human: OKF6-E6/E7 cell line | Gift from Jim Rheinwald and Matthew Ramsey (Harvard University, Boston, MA) | RRID: CVCL_L223 |
| Human: GMSM-K cell line | Gift from Valerie Murrah (UNC-Chapel Hill, NC) | RRID: CVCL_6A82 |
| Human: UM-SCC-74A HPV-negative HNSCC cell line | Gift from Thomas Carey (University of Michigan (Ann Arbor, MI) | RRID: CVCL_7779 |
| Human: UM-SCC-5 HPV-negative HNSCC cell line | Gift from Thomas Carey (University of Michigan (Ann Arbor, MI) | RRID: CVCL_7762 |
| Human: UM-SCC-11A HPV-negative HNSCC cell line | Gift from Thomas Carey (University of Michigan (Ann Arbor, MI) | RRID: CVCL_7715 |
| Human: UM-SCC-14A HPV-negative HNSCC cell line | Gift from Thomas Carey (University of Michigan (Ann Arbor, MI) | RRID: CVCL_7719 |
| Human: SCC15 HPV-negative HNSCC cell line | Gift from Ben Major (UNC-Chapel Hill, NC) | RRID: CVCL_1681 |
| Human: SCC25 HPV-negative HNSCC cell line | Gift from Ben Major, (UNC-Chapel Hill, NC) | RRID: CVCL_1682 |
| Human: UM-SCC-47 HPV-positive HNSCC cell line | A gift from Randall Kimple (University of Wisconsin, Madison, WI) | RRID: CVCL_7759 |
| Human: 93-VU147T HPV-positive HNSCC cell line | A gift from Randall Kimple (University of Wisconsin, Madison, WI) | VU-SCC-147; RRID:CVCL_L895 |
| Human: UD-SCC-2 HPV-positive HNSCC cell line | A gift from Randall Kimple (University of Wisconsin, Madison, WI) | RRID: CVCL_E325 |
| Human: UPCI:SCC090 HPV-positive HNSCC cell line | A gift from Randall Kimple (University of Wisconsin, Madison, WI) | RRID: CVCL_7794 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Rosa26-LSL-E7iresE6: Albino B6(Cg)-Tyrc−2J/J Male and Female mice were used for experiments, and randomly assigned to experimental groups. | This paper | N/A |
| Mouse: Rosa26-LSL-GpNLuc: Albino B6(Cg)-Tyrc−2J/J Male and Female mice were used for experiments, and randomly assigned to experimental groups. | This paper | N/A |
| Mouse: Albino B6(Cg)-Tyrc−2J/J Male and Female mice used for breeding. | Jackson Laboratory | Stock# 000058 |
| Mouse: C57BL/6J Male and Female mice used for breeding. | Jackson Laboratory | Stock# 000664 |
| Mouse: Rosa26-LSL-PIK3CAE545K: Albino B6(Cg)-Tyrc−2J/J Male and Female mice were used for experiments, and randomly assigned to experimental groups. | Meyer et al., 2013; a gift from Mohammed Bentires-Alj (University of Basel, Basel, Switzerland) and backcrossed in this paper | N/A |
| Mouse: L2-Cre: Albino B6(Cg)-Tyrc−2J/J Male and Female mice were used for experiments, and randomly assigned to experimental groups. | Stairs et al., 2011; a gift from Anil Rustgi (University of Pennsylvania, Philadelphia, PA) and backcrossed in this paper | N/A |
| Mouse: KRT14-Cre: C57BL/6J Male and Female mice were used for experiments, and randomly assigned to experimental groups. | A gift from Elaine Fuchs (The Rockefeller University, New York City, NY) | N/A |
| Mouse: KRT14-Cretam: Albino B6(Cg)-Tyrc−2J/J Male and Female mice were used for experiments, and randomly assigned to experimental groups. | Jackson Laboratory and backcrossed in this paper | Stock# 005107 |
| Oligonucleotides | ||
| Primers for gene expression analysis, see Table S7 | This paper | N/A |
| Primers for Genotyping animal models, see Table S7 | This Paper | N/A |
| Primers used to verify DNA Recombination, see Table S7 | Forward Primer: from this paper Reverse Primer: Zhong et al., 2014 | N/A |
| Recombinant DNA | ||
| pBigT | Addgene; Srinivas et al., 2001 | Plasmid# 21270 |
| pRosa26-PA | Addgene; Srinivas et al., 2001 | Plasmid# 21271 |
| MIGR1 | Addgene; Pear et. al., 1998 | Plasmid# 27490 |
| pRetroX-Tight_MCS_PGK-GpNLuc | Addgene; Schaub et al., 2015 | Plasmid# 70185 |
| Software and Algorithms | ||
| Gene Set Enrichment Analysis (GSEA) | Mootha et al., 2003; Subramanian et al., 2005 | http://software.broadinstitute.org/gsea/index.jsp |
| DESeq2 | Love et al., 2014 | N/A |
| STAR v2.4.2a | Dobin et al., 2013 | N/A |
| R (version 3.1.2) | https://www.r-project.org/ | N/A |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| GraphPad Prism 6 | N/A | https://www.graphpad.com |
| LAS AF Software | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
Highlights.
Creation of a knockin allele for balanced HPV16 E6 and E7 oncogene expression
Intra-lingual tamoxifen delivery enables mosaic oropharyngeal transgene activation
E6/E7 and PIK3CA activation leads to oropharyngeal squamous cell carcinoma
Post-natal transgene activation maintains robust immune cell infiltration
ACKNOWLEDGMENTS
We thank members of the Amelio lab, including Kotaro Sato, Adele Musicant, and Ryan Murphy, for technical support and helpful discussions as well as David Marron in the Lineberger Bioinformatics Core and Charlene Santos in the Animal Studies Core. We also thank Barbara Savoldo, Jennifer Webster-Cyriaque, Valerie Murrah, Cary Moody, Wendell Yarbrough, and Norman Sharpless for helpful comments, suggestions, and scientific review during the preparation of this manuscript. This work was supported by NIH/NIDCR T90-DE021986 and NIH/NCI T32-CA211056 (to M.B.C.); NIH/NIDCR K08-DE026537 (to K.M.B.); NIH/NIDCR R21-DE025725 (to S.E.W. and A.L.A.); the Cortner-Couch Chair for Cancer Research from the University of South Florida School of Medicine (to J.L.C.); Dental Foundation of North Carolina (to A.L.A.); University Cancer Research Fund (UCRF; to A.L.A.); UNC Lineberger Tier 3 Developmental Award (to A.L.A.); and NIH/NCI Howard Temin Pathway to Independence Award in Cancer Research R00-CA157954 (to A.L.A.). This work was also supported, in part, by NIH/NCI Comprehensive Cancer Center grants P30-CA076292, awarded to the Moffitt Comprehensive Cancer Center, and P30-CA016806 to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.005.
DECLARATION OF INTERESTS
The authors declare no competing interests. Received: May 9, 2019
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
REFERENCES
- Aguilar-Martinez E, Morrisroe C, and Sharrocks AD. (2015). The ubiquitin ligase UBE3A dampens ERK pathway signalling in HPV E6 transformed HeLa cells. PLoS One 10, e0119366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajiro M, Jia R, Zhang L, Liu X, and Zheng Z-M. (2012). Intron definition and a branch site adenosine at nt 385 control RNA splicing of HPV16 E6*I and E7 expression. PLoS One 7, e46412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeit JM, Münger K, Howley PM, and Hanahan D. (1994). Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol 68, 4358–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H-U, Burk RD, Chen Z, Van Doorslaer K, zur Hausen H, and de Villiers E-M. (2010). Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401, 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzer GC, Smith MA, Harris SL, and Kimple RJ. (2014). Review of the clinical and biologic aspects of human papillomavirus-positive squamous cell carcinomas of the head and neck. Int. J. Radiat. Oncol. Biol. Phys 88, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, et al. (1996). Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 56, 2488–2492. [PubMed] [Google Scholar]
- Callejas-Valera JL, Iglesias-Bartolome R, Amornphimoltham P, Palacios-Garcia J, Martin D, Califano JA, Molinolo AA, and Gutkind JS. (2016). mTOR inhibition prevents rapid-onset of carcinogen-induced malignancies in a novel inducible HPV-16 E6/E7 mouse model. Carcinogenesis 37, 1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraresi L, Tripodi SA, Mulder LC, Bertini S, Nuti S, Schuerfeld K, Cintorino M, Bensi G, Rossini M, and Mora M. (2001). Thymic hyperplasia and lung carcinomas in a line of mice transgenic for keratin 5-driven HPV16 E6/ E7 oncogenes. Oncogene 20, 8148–8153. [DOI] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, et al. (2011). Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J. Clin. Oncol 29, 4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Cancer Collaboration; Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al. (2017). Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 3, 524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza G, Westra WH, Wang SJ, van Zante A, Wentz A, Kluz N, Rettig E, Ryan WR, Ha PK, Kang H, et al. (2016). Differences in the Prevalence of Human Papillomavirus (HPV) in Head and Neck Squamous Cell Cancers by Sex, Race, Anatomic Tumor Site, and HPV Detection Method. JAMA Oncol 3, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De A, Jasani A, Arora R, and Gambhir SS. (2013). Evolution of BRET Biosensors from Live Cell to Tissue-Scale In vivo Imaging. Front. Endocrinol. (Lausanne) 4, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, and Rheinwald JG. (2000). Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. In Mol. Cell Biol 20, 1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Herd K, Street M, Bryson G, Fernando G, Lambert P, and Tindle R. (1999). Human papillomavirus type 16 E7 oncoprotein expressed in peripheral epithelium tolerizes E7-directed cytotoxic T-lymphocyte precursors restricted through human (and mouse) major histocompatibility complex class I alleles. J. Virol 73, 6166–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J. (2016). Model systems of human papillomavirus-associated disease. J. Pathol 238, 166–179. [DOI] [PubMed] [Google Scholar]
- Du L, Chen X, Cao Y, Lu L, Zhang F, Bornstein S, Li Y, Owens P, Malkoski S, Said S, et al. (2016). Overexpression of PIK3CA in murine head and neck epithelium drives tumor invasion and metastasis through PDK1 and enhanced TGFb signaling. Oncogene 35, 4641–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabani P, Lin AJ, Barnes J, Oppelt P, Adkins DR, Rich JT, Zevallos JP, Daly MD, Gay HA, and Thorstad WL. (2019). Radiation therapy dose de-escalation compared to standard dose radiation therapy in definitive treatment of HPV-positive oropharyngeal squamous cell carcinoma. Radiother. Oncol 134, 81–88. [DOI] [PubMed] [Google Scholar]
- Gilchrist EP, Moyer MP, Shillitoe EJ, Clare N, and Murrah VA. (2000). Establishment of a human polyclonal oral epithelial cell line. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod 90, 340–347. [DOI] [PubMed] [Google Scholar]
- Gillison ML, D’Souza G, Westra W, Sugar E, Xiao W, Begum S, and Viscidi R. (2008). Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J. Natl. Cancer Inst 100, 407–420. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Akagi K, Xiao W, Jiang B, Pickard RKL, Li J, Swanson BJ, Agrawal AD, Zucker M, Stache-Crain B, et al. (2018). Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res 29, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SV, and Faizo AAA. (2017). Control of human papillomavirus gene expression by alternative splicing. Virus Res. 231, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep AE, Herber R, Jeon S, Lohse JK, Dubielzig RR, and Lambert PF. (1993). Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J. Virol 67, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha PK, and Califano JA. (2016). The Role of Human Papillomavirus in Oral Carcinogenesis. Critical Rev. Oral Biol. Med 15, 188–196. [DOI] [PubMed] [Google Scholar]
- Hafsi S, Pezzino FM, Candido S, Ligresti G, Spandidos DA, Soua Z, McCubrey JA, Travali S, and Libra M. (2012). Gene alterations in the PI3K/PTEN/AKT pathway as a mechanism of drug-resistance (review). Int. J. Oncol 40, 639–644. [DOI] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, and Galloway DA. (1991). The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol 65, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes DN, Van Waes C, and Seiwert TY. (2015). Genetic Landscape of Human Papillomavirus-Associated Head and Neck Cancer and Comparison to Tobacco-Related Tumors. J. Clin. Oncol 33, 3227–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herber R, Liem A, Pitot H, and Lambert PF. (1996). Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol 70, 1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar SF, Abrams L, Glick A, and Lambert PF. (2009). Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res 69, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar S, Strati K, Shin MK, Pitot HC, and Lambert PF. (2010). Human papillomavirus type 16 E6 and E7 oncoproteins act synergistically to cause head and neck cancer in mice. Virology 407, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbar SF, Park S, Schweizer J, Berard-Bergery M, Pitot HC, Lee D, and Lambert PF. (2012). Cervical cancers require the continuous expression of the human papillomavirus type 16 E7 oncoprotein even in the presence of the viral E6 oncoprotein. Cancer Res 72, 4008–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert PF. (2016). Transgenic Mouse Models of Tumor Virus Action. Annu. Rev. Virol 3, 473–489. [DOI] [PubMed] [Google Scholar]
- Lambert PF, Pan H, Pitot HC, Liem A, Jackson M, and Griep AE. (1993). Epidermal cancer associated with expression of human papillomavirus type 16 E6 and E7 oncogenes in the skin of transgenic mice. Proc. Natl. Acad. Sci. USA 90, 5583–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans CR, Snijders PJF, and Brakenhoff RH. (2018). The molecular landscape of head and neck cancer. Nat. Rev. Cancer 18, 269–282. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, Şenbabaoğlu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, Lee K-W, Ganly I, Hakimi AA, Chan TA, and Morris LG. (2016). The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 1, e89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane M, MacDonald AI, Stevenson A, and Graham SV. (2015). Human Papillomavirus 16 Oncoprotein Expression Is Controlled by the Cellular Splicing Factor SRSF2 (SC35). J. Virol 89, 5276–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero I, Singhal MC, McGowan P, Haugen HS, Blake J, Hellstrom KE, Yang G, Clegg CH, and Chen L. (1997). Immunological ignorance of an E7-encoded cytolytic T-lymphocyte epitope in transgenic mice expressing the E7 and E6 oncogenes of human papillomavirus type 16. J. Virol 71, 3998–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DS, Koren S, Leroy C, Brinkhaus H, Müller U, Klebba I, Müller M, Cardiff RD, and Bentires-Alj M. (2013). Expression of PIK3CA mutant E545K in the mammary gland induces heterogeneous tumors but is less potent than mutant H1047R. Oncogenesis 2, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael S, Lambert PF, and Strati K. (2013). The HPV16 oncogenes cause aberrant stem cell mobilization. Virology 443, 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, and Laimins LA. (2010). Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10, 550–560. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273. [DOI] [PubMed] [Google Scholar]
- Münger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, and Huh K. (2004). Mechanisms of Human Papillomavirus-Induced Oncogenesis. J. Virol 78, 11451–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Wang TC, Zukerberg L, Odze R, Togawa K, May GH, Wilson J, and Rustgi AK. (1997). The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach. Oncogene 14, 1185–1190. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PK-S, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, Sengupta S, Wang Z, Bhavana VH, Tran R, et al. (2018). Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer Cell 33, 450–462.e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nulton TJ, Olex AL, Dozmorov M, Morgan IM, and Windle B. (2017). Analysis of The Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget 8, 17684–17699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz OG, Harada H, Suliman Y, Rhoades B, Sharpless NE, Kent R, Kopelovich L, Nakagawa H, and Rustgi AK. (2002). A mouse model of human oral-esophageal cancer. J. Clin. Invest 110, 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuszak-Lewandoska D, Bartosińska-Dyc A, Migdalska-Sęk M, Czarnecka KH, Nawrot E, Domańska D, Szyłło K, and Brzeziańska E. (2014). HPV16 E6*II gene expression in intraepithelial cervical lesions as an indicator of neoplastic grade: a pilot study. Med. Oncol 31, 842. [DOI] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, et al. (1998). Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92, 3780–3792. [PubMed] [Google Scholar]
- Rautava J, and Syrjänen S. (2012). Biology of Human Papillomavirus Infections in Head and Neck Carcinogenesis. Head Neck Pathol 6, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley RR, Duensing S, Brake T, Münger K, Lambert PF, and Arbeit JM. (2003). Dissection of Human Papillomavirus E6 and E7 Function in Transgenic Mouse Models of Cervical Carcinogenesis. Cancer Res 63, 4862–4871. [PubMed] [Google Scholar]
- Rong C, Muller M, Flechtenmacher C, Holzinger D, Dyckhoff G, Bulut OC, Horn D, Plinkert P, Hess J, and Affolter A. (2019). Differential Activation of ERK Signaling in HPV-Related Oropharyngeal Squamous Cell Carcinoma. Cancers (Basel) 11, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger S, De-Castro Arce J, Langbein L, Steenbergen RDM, and Rösl F. (2010). Alternative splicing of human papillomavirus type-16 E6/E6* early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc. Natl. Acad. Sci. USA 107, 7006–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. (2004). High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554–554. [DOI] [PubMed] [Google Scholar]
- Satterwhite CL, Torrone E, Meites E, Dunne EF, Mahajan R, Ocfemia MCB, Su J, Xu F, and Weinstock H. (2013). Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm. Dis 40, 187–193. [DOI] [PubMed] [Google Scholar]
- Schaub FX, Reza MS, Flaveny CA, Li W, Musicant AM, Hoxha S, Guo M, Cleveland JL, and Amelio AL. (2015). Fluorophore-NanoLuc BRET Reporters Enable Sensitive In Vivo Optical Imaging and Flow Cytometry for Monitoring Tumorigenesis. Cancer Res. 75, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön M, and Rheinwald JG. (1996). A limited role for retinoic acid and retinoic acid receptors RAR alpha and RAR beta in regulating keratin 19 expression and keratinization in oral and epidermal keratinocytes. J. Invest. Dermatol 107, 428–438. [DOI] [PubMed] [Google Scholar]
- Sewell A, Brown B, Biktasova A, Mills GB, Lu Y, Tyson DR, Issaeva N, and Yarbrough WG. (2014). Reverse-phase protein array profiling of oropharyngeal cancer and significance of PIK3CA mutations in HPV-associated head and neck cancer. Clin. Cancer Res 20, 2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter DP, Southwick HW, and Smejkal W. (1953). Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 6, 963–968. [DOI] [PubMed] [Google Scholar]
- Song S, Pitot HC, and Lambert PF. (1999). The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J. Virol 73, 5887–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiotto MT, Pytynia M, Liu G-FF, Ranck MC, and Widau R. (2013). Animal models to study the mutational landscape for oral cavity and oropharyngeal cancers. J. Oral Maxillofac. Res 4, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F. (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs DB, Bayne LJ, Rhoades B, Vega ME, Waldron TJ, Kalabis J, Klein-Szanto A, Lee J-S, Katz JP, Diehl JA, et al. (2011). Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell 19, 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein AP, Saha S, Yu M, Kimple RJ, and Lambert PF. (2014). Prevalence of human papillomavirus in oropharyngeal squamous cell carcinoma in the United States across time. Chem. Res. Toxicol 27, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati K, and Lambert PF. (2007). Role of Rb-dependent and Rb-independent functions of papillomavirus E7 oncogene in head and neck cancer. Cancer Res 67, 11585–11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati K, Pitot HC, and Lambert PF. (2006). Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc. Natl. Acad. Sci. USA 103, 14152–14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Amelio I, Guerrero Urbano T, and Tavassoli M. (2014). Clinical update on cancer: molecular oncology of head and neck cancer. Cell Death Dis 5, e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Tao M, McCoy JP Jr., and Zheng Z-M. (2006). The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol 80, 4249–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuting PM, and Dintzis SM. (2012). Comparative Anatomy and Histology (Academic Press; ). [Google Scholar]
- Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D, Sun Q, Kaur R, Huyen Y, and McBride AA. (2017). The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res 45, D499–D506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale-Cross L, Czerninski R, Amornphimoltham P, Patel V, Molinolo AA, and Gutkind JS. (2009). Chemical carcinogenesis models for evaluating molecular-targeted prevention and treatment of oral cancer. Cancer Prev. Res. (Phila.) 2, 419–422. [DOI] [PubMed] [Google Scholar]
- Wong K-K, Engelman JA, and Cantley LC. (2010). Targeting the PI3K signaling pathway in cancer. Curr. Opin. Genet. Dev 20, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Kajitani N, and Schwartz S. (2017). Splicing and Polyadenylation of Human Papillomavirus Type 16 mRNAs. Int. J. Mol. Sci 18, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, and Cantley LC. (2008). PI3K pathway alterations in cancer: variations on a theme. Oncogene 27, 5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Pytynia M, Pelizzari C, and Spiotto M. (2014). Bioluminescent imaging of HPV-positive oral tumor growth and its response to image-guided radiotherapy. Cancer Res. 74, 2073–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.