Abstract
The Open Material Transfer Agreement is a material-transfer agreement that enables broader sharing and use of biological materials by biotechnology practitioners working within the practical realities of technology transfer.
Supplementary information
The online version of this article (doi:10.1038/nbt.4263) contains supplementary material, which is available to authorized users.
Subject terms: Intellectual-property rights, Business
Material-transfer agreements (MTAs) underlie the legal frameworks within which biotechnology practitioners define the terms and conditions for sharing biomaterials ranging, for example, from plasmid DNA to patient samples. If MTAs are easy to use and well adapted to the needs of individual researchers, institutions, and broader communities, then more sharing, innovation, and translation can occur. However, the MTA frameworks currently in place were developed in the 1990s—before widespread adoption of the World Wide Web, genome sequencing, and gene synthesis—and are not always well adapted for contemporary research and translation practices or aligned with social objectives.
Here, we introduce a new MTA, the Open Material Transfer Agreement (OpenMTA), that relaxes restrictions on the redistribution and commercial use of biomaterials while maintaining aspects of standard MTAs that support widespread adoption (for example, incorporation into semiautomated administration systems). In developing the OpenMTA, our motivation was to realize a simple, standardized legal tool for sharing biological materials as broadly as possible without undue restrictions, while respecting the rights of creators and promoting safe practices and responsible research. Importantly, we wanted the tool to work within the practical realities of technology transfer and to be sufficiently flexible to accommodate the needs of many groups globally (for example, providing support for international transfers and compatibility with public and philanthropic funding policies).
Traditional MTAs
Currently, the most used MTA in biology and biotechnology is the Uniform Biological Material Transfer Agreement (UBMTA), which was developed and widely adopted in the 1990s (https://www.ott.nih.gov/resources/). The UBMTA represented a major step forward in providing a standard template intended to help decrease administrative transaction costs for material exchange among academic research institutions. Despite widespread adoption of the UBMTA, many institutions continue to insist on MTAs specific to their own institution1. In many cases, the effort required to implement MTAs has increased, because the complexity of included provisions requires protracted negotiation. Such transaction costs not only impede the distribution of materials within and beyond research communities but also place an unnecessary burden on technology transfer offices, thus leading some institutions to adopt a 'no MTA' policy2,3. Although eliminating the explicit execution of MTAs decreases the apparent administrative burden, the resulting reality is less than ideal, because MTAs also help with provenance tracking, recognition, and quality control. In addition, because some no-MTA policies state that transfers made without an explicit agreement default to the terms of the UBMTA, and because researchers may share materials informally without knowing that such terms apply by default4, the ability of everyone to legitimately use and further develop materials remains in limbo, at best.
Most widely used MTAs place two restrictions on material transfers, neither of which is often useful or desired (Table 1). First, MTAs typically disallow redistribution of materials (i.e., so-received materials cannot formally be shared with others). Second, any and all commercial uses of the so-received biomaterials are specifically prohibited. Although these two restrictions are appropriate for materials that require tight control of provenance for reasons of safety, security, or commercialization, such restrictions make little sense for most of the materials used widely throughout research (for example, basic samples, strains, or plasmids). Because the potential commercial value of most widely used materials is quite low, and MTAs are unlikely to ever be monitored and enforced1, blanket restrictions on redistribution and commercial use create unnecessary barriers and costs within research communities and to society at large.
Table 1.
Comparison of features and terms for standard MTAsa
| Features and terms of transfer | UBMTA (1995) | SLA (1999) | Science Commons (2005–2009) | OpenMTA (2018) |
|---|---|---|---|---|
| Similarities | ||||
| Use for research and teaching | Yes | Yes | Yes | Yes |
| Attribution | Yes | Yes | Yes | Yes |
| Compliance with laws and regulations | Yes | Yes | Yes | Yes |
| No warranty (for example, third-party rights) | Yes | Yes | Yes | Yes |
| Recipient assumes liability | Yes | Yes | Yes | Yes |
| Recipient indemnifies provider | No | No | No | No |
| Reach-through rights or restrictions | No | No | No | No |
| Fees for preparation and distribution (optional) | Yes | Yes | Yes | Yes |
| Fees for royalties | No | No | No | No |
| Provenance tracking | Yes | Yes | Yes | Yes |
| Alignment with policies of public and private funders of research | Yes | Yes | Yes | Yes |
| Differences | ||||
| Academia or nonprofit only | Yes | No | No | No |
| Ease of use internationally | No | Yes | Yes | Yes |
| Distribution of materials or derivatives | No | No | No | Yes |
| Use for commercial purposes | No | No | No | Yes |
aUBMTA, Uniform Biological Material Transfer Agreement (https://www.ott.nih.gov/resources/); SLA, NIH Simple Letter Agreement (https://www.ott.nih.gov/resources/); Science Commons MTA19.
For example, with the ongoing emergence of a second generation of biotechnology practitioners who are increasingly empowered by information-sharing networks and DNA sequencing and synthesis capacities—which together make genetic information and material interconvertible—both the 'no redistribution' and 'no commercial use' restrictions can be quite harmful. As one example, over the past 15 years, the international genetically engineered machines competition (http://igem.org/) has developed into a genetic engineering 'olympics' involving ∼6,000 students per year, in which students freely share biological materials under a get-and-give policy. Many of these students, having just prototyped a compelling biotechnology application, then wish to start a company to translate their innovation. Yet, the materials that they themselves developed via an open and collaborative community cannot be directly used in their subsequent commercial activities.
As a second specific example, Addgene has become a major public-benefit plasmid-sharing resource; it has shipped more than 950,000 plasmids to over 6,400 institutions in 93 countries since starting in 2004. Technology-transfer offices are enormously appreciative of the services that Addgene provides, which include electronic processing of MTAs. Yet, because the UBMTA was the default standard at the time Addgene's electronic MTA system was developed, commercial biotechnology researchers are unable to directly access most of the resources available via Addgene. This MTA-induced limitation arises even though many of the Addgene-hosted materials are not subject to any specific proprietary interest. Although well-funded commercial groups can gain access to such materials via ab initio DNA synthesis starting from sequences available on Addgene's website or other public databases, the companies that might benefit the most from such materials—the small start-up or midsized companies representing the lifeblood and future of biotechnology—are left out. Defaulting to restrictive terms that may not be uniformly necessary or appropriate is common with other repositories, including nonprofit culture collections, biobanks, gene banks, seed banks, and other plasmid repositories that have been established in recognition of their importance to various biotechnology communities.
Making research tools broadly available for use without restriction can accelerate the pace of research, improve the reproducibility of results, and save the time and costs that would otherwise be incurred by researchers recreating tools and materials in their own laboratories. Today, researchers in many branches of biotechnology including synthetic biology, genomics, and regenerative medicine are creating nucleic acid–based tools as well as other tools intended for widespread use and contribution to the public domain. Examples include molecular probes for drug discovery from the Structural Genomics Consortium, pluripotent stem cell lines from Boston University's Center for Regenerative Medicine, materials from the Montreal Neurological Institute and Hospital, functional genetic elements from the BIOFAB project, DNA part collections from OpenPlant, and the bionet.io and Free Genes projects from the BioBricks Foundation5,6,7,8,9,10. The desire of these researchers and organizations to share materials freely is rooted in the idea that, although there may be limited value in each of these tools individually, there is great value in their widespread use by researchers and others collectively. This idea is particularly relevant to synthetic biology, in which standards have been developed for the interoperability of modular DNA components or parts, and in which remixing of numerous parts is normal practice11,12. Nonetheless, accessing materials remains difficult and time consuming or impossible, thus leading to delays or lost opportunities.
Drafting the OpenMTA
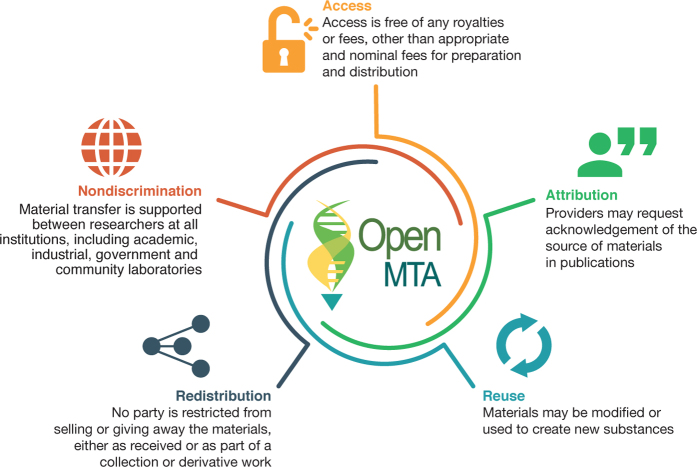
The OpenMTA was developed as a collaborative effort led by the BioBricks Foundation and the OpenPlant Synthetic Biology Research Centre. We began by convening a working group comprising researchers, technology-transfer professionals, social scientists, legal experts, and others interested in creating a legal framework that could improve sharing of biomaterials. This group, which met in person in June 2015 and continued with scheduled online meetings, developed the design goals for the OpenMTA, including access, attribution, reuse, redistribution, and nondiscrimination (Fig. 1). These five design goals were selected to be consistent with the Open Definition, which aims to “make precise the meaning of 'open' with respect to knowledge, promoting a robust commons in which anyone may participate, and interoperability is maximized” (https://opendefinition.org/). Additional design goals included issues of safety and, in particular, the sharing of biomaterials in an international context. For example, we wanted the agreement to be usable by institutions worldwide (i.e., to not be US- or UK-centric). After the design goals were in place, we drafted the legal text for the OpenMTA.
Figure 1. Design goals for the OpenMTA.
The design goals for the OpenMTA reflect the principles of 'openness' set out in the Open Definition (https://opendefinition.org/). These design goals, together with additional goals to ensure safety and enable sharing of biomaterials in an international context, were used to help guide the drafting of the legal text for the OpenMTA.
We used the text of the UBMTA as a starting point, removing any aspects that did not meet the design goals, and adding or refining text to strengthen aspects as needed. The resulting OpenMTA drafts went through several rounds of revision, beginning with review by the offices responsible for transfer of materials from OpenPlant, including Cambridge University, Cambridge Enterprise, and the John Innes Centre. Additional input was solicited by e-mail and by contacting the members of Praxis Unico and the Association for University Technology Managers. Revisions continued until the received comments reflected no new issues, at which point the OpenMTA text was officially posted online with digital-signature capability (http://openmta.org).
The OpenMTA has many features in common with other standard MTA templates, such as provisions on use, compliance, and liability, that help protect the provider and clarify the responsibility of the recipient (Table 1). Like all MTAs, the OpenMTA is a contract—a bargained-for exchange of consideration—wherein materials from the provider are given in exchange for a promise by the recipient to abide by the terms of the agreement, including attribution, reporting back, and payment of a fee for processing if requested. Although litigation of MTAs is exceedingly rare, a framework based on providing material in exchange for a promise meets the legal standards for adequate consideration1. As part of the promise, the OpenMTA includes a provision that requires recipients of the materials to “ensure compliance with all applicable laws, rules, and regulations.” This provision was included to provide flexibility and stability in use of the OpenMTA because, as is the case for all MTAs, the transfer of specific materials may be subject to laws, rules, and regulations that are context dependent, jurisdictional, and subject to change over time (for example, access and benefit-sharing obligations arising from the Convention for Biodiversity and Nagoya Protocol (https://www.cbd.int/abs/)). The OpenMTA also includes an optional 'catch-all' term specifying “information relevant to the status of the Material is provided in an attachment.” This optional term, suggested during the iterative review process as a means to capture unique or unforeseen circumstances, could be used to notify the recipient of any additional obligations that might apply to use of the material (for example, uses specified in patent claims). Finally, because the OpenMTA includes the same 'no warranty' provision as that provided in the UBMTA and other standard agreements, the recipients remain responsible for conducting their own due diligence for their use of the materials in their jurisdiction.
Differences between the OpenMTA and other standard templates arise via differences in design—specifically that researchers be allowed to use the materials for any lawful purpose, including commercial purposes, and may also redistribute the materials to others, subject to reporting back if requested by the providing institution. Reporting back was included as an optional term because technology transfer offices expressed different preferences. Some wanted reporting back as a means to measure the influence of research materials made freely available to others, whereas others did not want to track the materials beyond the first transfer.
Although the OpenMTA can be used as an integrated agreement, the online version of the OpenMTA is structured as a Master Agreement that can be approved at an institutional level. The online Master Agreement ensures consistency in the use of the OpenMTA (i.e., no altering of terms or editing), and also provides transparency for the individuals and institutions that have become signatories (i.e., signatories can be listed online for easy reference). After an institution becomes a signatory to the OpenMTA Master Agreement, transfers can be made with the use of an implementing letter. Doing so simplifies the material-transfer process by eliminating the need for review of terms and provides the documentation necessary for provenance tracking.
Importantly, we note that becoming a signatory to the OpenMTA Master Agreement provides an institution and its researchers the option of transferring materials under the terms of the OpenMTA but does not obligate its exclusive use. Institutions retain full discretion to handle the transfer of specific materials on a customized basis. Institutions also retain the discretion to designate authorized signatories for the implementing letter. In other words, use of the OpenMTA is not mandatory, even for signatory institutions, and institutional signatory authority is still required unless the institution decides otherwise. Additionally, because the OpenMTA does not include a 'viral' clause, institutions may accept incoming materials under the OpenMTA, use or modify so-received materials, and then subsequently redistribute the materials or derivatives under the same or different terms. This additional flexibility supports broad use of materials made available under the OpenMTA, even in cases in which more restrictive terms are best suited for redistribution.
As a standard template, the OpenMTA lends itself to automation that can further accelerate and simplify MTA processing while providing a less restrictive option for material transfer as appropriate. Within centralized repositories such as Addgene, for example, the OpenMTA should be able to be incorporated as an option alongside the UBMTA so that researchers and institutions can easily select the terms best suited for their needs. Additionally, the provenance-tracking capability inherent in Addgene's electronic MTA system13,14, with the necessary permissions, could provide increased transparency in the dissemination of research tools and help inform science policy. Stated differently, institutions may choose to use a centralized repository to distribute materials under the OpenMTA to be readily recognized for reaching the widest possible audience and enabling the greatest social good.
Incorporation of the OpenMTA within other electronic platforms, such as the MTAShare platform developed at Vanderbilt University (https://cttc.co/inventors/mtashare/) and the Transfer Agreement Dashboard hosted by the US National Institutes of Health15, could also enable less restrictive options for sharing biomaterials as appropriate. These platforms are designed for direct transfer of materials from one institution to another and provide an avenue for researcher-led sharing of materials. Although researcher-led sharing of materials may lack the quality control of centralized repositories, such an approach is practically essential for materials undergoing rapid iterative changes or to support broad collaborations and rapid scaling. Technology-transfer offices could still review and approve such transfers, and paperwork and individual negotiations could be replaced by electric communications and selection from a set of standard MTA templates. Such electronic platforms could also offer provenance tracking, so that researchers and their institutions would be better able to make informed choices about the materials they use in their research.
Use of the OpenMTA
We anticipate that the OpenMTA will be most useful for the transfer of biomaterials used in precompetitive research, such as plasmids, strains, and samples, whose quantities are not limited, owing to the easily replicated nature of the materials, and for which the value of individual materials is relatively low, owing to alternative sourcing options. We further anticipate that the OpenMTA may be well suited as a default institutional policy in contexts in which most materials are intended to be freely shared. We note that the OpenMTA is not suitable for all transfers, such as materials that are in limited quantity or subject to strict biosecurity regulations.
The OpenMTA does allow for recovery of costs associated with preparation and distribution of materials and therefore could also be used for the transfer of research reagents such as antibodies, cell lines, and fluorescent proteins for which patents have expired or were never sought. Indeed, the introduction of OpenMTA for sharing biological materials is particularly timely in that many patented materials are now entering the public domain as patents expire. As one example, a collection of patents on green fluorescent proteins initially aggregated and outlicensed by GE Healthcare Lifesciences16 have all expired (Table 2). Moreover, patent claims to nucleic acid sequences are now subject to heightened scrutiny and, if issued at all, are drawn much more narrowly than in the past17,18.
Table 2.
Patents on GFP initially aggregated and outlicensed by GE Healthcare
| Patent No. | Title | Legal status | Expiration date |
|---|---|---|---|
| US 5,491,084 | Uses of green fluorescent protein | Expired | 2013-09-10 |
| US 6,146,826 | Green fluorescent protein | Expired | 2017-11-14 |
|
US 5,625,048 US 5,777,079 US 6,319,669 US 6,066,476 |
Modified green fluorescent proteins Modified green fluorescent proteins Modified green fluorescent proteins Modified green fluorescent proteins |
Expired Expired Expired Expired |
2014-11-10 2014-11-10 2014-11-10 2014-11-10 |
|
US 5,804,387 US 6,090,919 |
FACS-optimized mutants of the green fluorescent protein FACS-optimized mutants of the green fluorescent protein |
Expired Expired |
2016-02-01 2017-01-31 |
| US 5,958,713 | Method of detecting biologically active substances by using GFP | Expired | 2015-01-31 |
| US 5,968,738 | Two-reporter FACS analysis of mammalian cells using GFP | Expired | 2015-12-06 |
| US 5,994,077 | Fluorescence-based isolation of differentially induced genes | Expired | 2015-01-31 |
|
US 6,054,321 US 6,077,707 US 6,124,128 US 6,403,374 |
Long wavelength engineered fluorescent proteins Long wavelength engineered fluorescent proteins Long wavelength engineered fluorescent proteins Long wavelength engineered fluorescent proteins |
Expired Expired Expired Expired |
2016-08-16 2016-08-16 2016-08-16 2016-08-16 |
|
US 6,172,188 US 6,818,443 US 7,314,915 |
Novel fluorescent proteins Novel fluorescent proteins Novel fluorescent proteins |
Expired Expired Expired |
2015-09-22 2015-09-22 2015-09-22 |
| CA 2232727 | Novel variants of green fluorescent protein, GFP | Expired | 2016-01-31 |
|
EP 0804457 EP 0815257 EP 0851874 |
Modified green fluorescent proteins A method of detecting biologically active substances Novel variants of green fluorescent protein, GFP |
Expired Expired Expired |
2014-11-10 2016-01-31 2016-01-31 |
|
JP 3283523 JP 3535177 |
Modified green fluorescent protein A new variant of a GFP is a green fluorescent protein |
Expired Expired |
2015-11-13 2016-01-31 |
We have received numerous inquiries about the suitability of the OpenMTA for the transfer of human-derived materials. Additional complexities including privacy, consent, and institutional-review-board approval must be addressed, and we are working with others to develop an OpenMTA for such materials. Extending these efforts to enable more open sharing of induced pluripotent stem cell lines, for example, in coordination with national and international registries, could vastly accelerate the development of useful biomedical applications.
Practically, the OpenMTA is already being used via the BioBricks Foundation's Free Genes project, wherein sequences requested by the synthetic biology research community are synthesized and made available without cost. The OpenPlant Synthetic Biology Research Centre plans to use the OpenMTA to distribute vectors that incorporate a common syntax with wide acceptance in the area of plant biotechnology. The online OpenMTA Master Agreement is already gaining traction, and it includes initial signatories from academic research institutions, companies, and community labs. We invite more signatories and welcome public comments on the OpenMTA Master Agreement, which is freely available via http://openmta.org and as Supplementary Data. Comments may be addressed to The OpenMTA Project, BioBricks Foundation, 77 Van Ness Avenue, Ste. 101-1626, San Francisco, California, 94102, USA, or sent by email to openmta@biobricks.org or to either corresponding author.
Supplementary information
Supplementary Data (PDF 130 kb)
Acknowledgements
This work was supported by grants from the Leona M. and Harry B. Helmsley Charitable Trust to the BioBricks Foundation, the joint Engineering and Physical Sciences Research Council/Biotechnological and Biological Sciences Research Council (BBSRC)-funded OpenPlant Synthetic Biology Research Centre (award BB/L014130/1), the BBSRC Institute Strategic Programme Grant 'Molecules from Nature' (BB/P012523/1), and the John Innes Foundation. We are especially grateful to the many people who gave freely of their time in providing insight and feedback on the manuscript, including the following individuals involved in the design, drafting, and review of the OpenMTA Master Agreement: Hal Abelson (Creative Commons and Free Software Foundation, and Electrical Engineering and Computer Science, MIT); Mohamed Abousalem (University of California at Santa Cruz); Sarah Ali-Khan (Centre for Intellectual Property Policy, McGill University); Mary Anderson (Norwich Bioscience Institutes); Anne Andrews (Human Subjects Protection Office, National Institute of Standards and Technology); Julia Barnes-Weiss (Global Health Innovation Alliance Accelerator, Research Triangle Park); Alan Bentley (Center for Technology Transfer & Commercialization, Vanderbilt University); Dominic Berry (London School of Economics); Mark Bloom (Technology Transfer and Research Integrity Officer, Sidra Medical and Research Center); Laura Bradford (Research Operations Office, University of Cambridge); Tania Bubela (Faculty of Health Sciences, Simon Fraser University); Sean Butler (Studies in Law, St Edmund's College); Diane Cabell (World Intellectual Property Organization); Jane Calvert (Science Technology and Innovation Studies, University of Edinburgh); Eggerton Campbell (Technology Transfer Office, US National Human Genome Research Institute); David Carr (Open Research, Wellcome Trust); Ruvani Chandrasekera (US Department of Health and Human Services); Lisa Chen (Stanford University); George Church (Harvard Medical School and MIT); Lionel Clarke (UK Synthetic Biology Leadership Council, Imperial College London and University of Manchester); Marie Connett (Global Development Technologies Portfolio, Global Good Fund); Case Cortese (California Institute of Technology); Anthony Cova (Addgene); Isha Datar (New Harvest); Brent Davidson (Division of International Health Security, US Department of Health and Human Services); Virginia de la Puenta (Lawrence Berkeley National Labs); Claire Driscoll (Technology Transfer Office, US National Human Genome Research Institute); Stephen Harsy (Contract & Research Support Program, University of Arizona); Wendy Harwood (John Innes Centre); Jodie Hecht (Technology Ventures Office, Beth Israel Deaconess Medical Center); Daphne Ioannidis (University of Cambridge); Mark Facciotti (UC Davis Genome Center); Fernan Federici (Synthetic Biology Lab, Pontificia Universidad Católica de Chile); Tony Ford (Open Forum Europe); Mario Gabiati (Counter Culture Labs); Richard Gold (Faculty of Law, McGill University); Henriette Fenger Grønfeldt (Technology Transfer Office, University of Copenhagen); Daniel Grushkin (Genspace); Alexandra Hall (Intellectual Property and Technology Transfer, University of Denver); Stephen Harsy (Contract & Research Support Program, University of Arizona); Nathan Hillson (Synthetic Biology Information, Joint Bioenergy Institute, US Department of Energy); Laura James (Doteveryone); Osmat Jefferson (Queensland University of Technology); Richard Jefferson (Queensland University of Technology, Canberra, Australia); Ellen Jorgensen (Biotech Without Borders); Maneshree Jugmohan-Naidu (Department of Science & Technology, Republic of South Africa); Vladimir Kaigorodov (Department of Molecular Biomedical Research, Ghent University); Robert Kiley (Wellcome Trust); Puneet Kishor (independent consultant; previously at Science Commons); Jamie Kitano (Stanford University); Tom Knight (Ginkgo Bioworks); Paul Labbett (Research Management & Innovation Directorate, King's College London); Meenakshi Lakshminarayanan (Center for Regenerative Medicine); John Liddicoat (University of Cambridge Faculty of Law); Kathy Liddell (University of Cambridge Faculty of Law); George Lomonossoff (John Innes Centre); Elizabeth Luna (University of Massachusetts Medical School); Mark Madsen (Counsel for Technology Transfer and Intellectual Property, United States Department of Commerce, National Institute of Standards and Technology); Eyal Maori (Tropic Biosciences); Ricardo Camilo Chávez Martínez (Scintia); Piers Millet (Woodrow Wilson International Center for Scholars); Chris Moran (Wellcome Trust, London); Sarah Munro (NIST Genome-Scale Measurements, National Institute of Standards and Technology); Richard Murray (Engineering and Applied Science, California Institute of Technology); Johnathan Napier (Rothamsted Research, Harpenden); Dianne Nicol (Centre for Law and Genetics, University of Tasmania); Jane Nielsen (Centre for Law and Genetics, University of Tasmania); Bárbara Ribbeck Neale (Pontificia Universidad Católica de Chile); Jayne Nicolson (Norwich Bioscience Institutes); Pamela Nunez (University of Cambridge); Anne Osbourn (John Innes Centre); Sally O'Neil (Industrial Contracts Office, Stanford University); Andreas Panagopoulos (Department of Economics, University of Crete); Geraint Parry (Cardiff University); Ben Pellegrini (Cambridge IP); James Headen Pfitzer (World Health Organization); Bernardo Pollack (University of Cambridge); Julia Powles (University of Cambridge); Scott Pownall (Open Science Network); John Puziss (Office of Cooperative Research, Yale University); Peter Ralph (Climate Change Cluster, University of Technology Sydney); Andrew Rens (Shuttleworth Foundation); Emily Rezendes (Office of University Counsel, Tufts University); Carolina Roa (International Center for Tropical Agriculture); Stephen J. Roberts (Technology Commercialization and Research Finance, Kent State University); Fiona Robertson (Department of Biochemistry, University of Zimbabwe); Rodrigo Sara (CGIAR System Organization); Jaysen Rajkomar (Office of Technology Licensing, Stanford University); David Rejeski (Science, Technology and Innovation Program, The Wilson Center); Mark Rohrbaugh (US National Institutes of Health); Marc Salit (Genome-Scale Measurements Group, National Institute of Standards and Technology); Richard Scheuermann (J. Craig Venter Institute); Joseph Secondine (New England BioLabs); Jacob Sherkow (Innovation Center for Law and Technology, New York Law School); Phyllis Leah Speser (King Mongkut's University of Technology Thonburi); Krishna Ravi Srinivas (Research and Information Systems for Developing Countries); Iain Thomas (Life Sciences, Cambridge Enterprise); Laurie Tzodikov (Office of Technology Licensing, Princeton University); Andrew Watson (Oregon Health & Science University); Julie Watson (Marshall, Gerstein & Borun LLP); Darius Whelan (University College Cork); John Wilbanks (Sage Bionetworks); Richard Wilder (Bill & Melinda Gates Foundation); Helen Young (Cambridge Enterprise); Justin Zook (Genome-in-a-Bottle Consortium, National Institute of Standards and Technology).
Competing interests
The authors declare no competing financial interests.
Contributor Information
Linda Kahl, Email: linda@biobricks.org.
Drew Endy, Email: endy@stanford.edu.
References
- 1.Bubela, T., Guebert, J. & Mishra, A. PLoS Biol.13, e1002060 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ku, K. & Henderson, J. Nat. Biotechnol.25, 721–724 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Stanford University et al. Material transfer agreements https://sites.stanford.edu/ico/sites/default/files/outgoingmtapolicy.pdf (2009).
- 4.Walsh, J.P., Cho, C. & Cohen, W.M. Science309, 2002–2003 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Temple, G. et al. Genome Res.19, 2324–2333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moran, B. CReM honored for sharing stem cell lines. BU Todayhttps://www.bumc.bu.edu/busm/2017/09/08/crem-honored-for-sharing-stem-cell-lines/ (2017).
- 7.Mutalik, V.K. et al. Nat. Methods10, 354–360 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Edwards A, et al. Sci Transl. Med.9, eaai9055 (2017).28566431 [Google Scholar]
- 9.Kahl, L. & Molloy, J. Towards an open material transfer agreement: OpenPlant IP working group report https://static1.squarespace.com/static/54a6bdb7e4b08424e69c93a1/t/5a81a054e4966bb7ff8a6885/1518444640740/OpenMTA+Report.pdf (2018).
- 10.BioBricks Foundation and Twist Bioscience. Twist Bioscience and the BioBricks Foundation announce partnership to provide 10,000 public-benefit genes to the synthetic biology community https://www.twistbioscience.com/company/press/twist-biobricks-foundation-partnership/ (2017).
- 11.Patron, N.J. et al. New Phytol.208, 13–19 (2015).26171760 [Google Scholar]
- 12.Zhang, M., McLaughlin, J.A., Wipat, A. & Myers, C.J. ACS Synth. Biol.6, 1150–1160 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Herscovitch, M., Perkins, E., Baltus, A. & Fan, M. Nat. Biotechnol.30, 316–317 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Kamens, J. Nucleic Acids Res.43, D1152–D1157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Institutes of Health. NIH eases tech transfer with online material transfer agreements https://www.nih.gov/news-events/news-releases/nih-eases-tech-transfer-online-material-transfer-agreements/ (2011).
- 16.G.E. Healthcare Life Sciences. GFP licensing statements http://cdn.gelifesciences.com/dmm3bwsv3/AssetStream.aspx?mediaformatid=10061&destinationid=10016&assetid=18861 (2013).
- 17.Jefferson, O.A., Köllhofer, D., Ehrich, T.H. & Jefferson, R.A. Nat. Biotechnol.33, 1033–1038 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Aboy, M., Liddicoat, J., Liddell, K., Jordan, M. & Crespo, C. Nat. Biotechnol.35, 820–825 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Nguyen, T. Innov. (Camb., Mass.)2, 137–143 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data (PDF 130 kb)