Abstract
Simultaneous electroencephalography and functional magnetic resonance imaging (EEG/fMRI) have been proposed to contribute to the definition of the epileptic seizure onset zone. Following interictal epileptiform discharges, one usually assumes a canonical hemodynamic response function (HRF), which has been derived from fMRI studies in healthy subjects. However, recent findings suggest that the hemodynamic properties of the epileptic brain are likely to differ significantly from physiological responses. Here, we propose a simple and robust approach that provides HRFs, defined as a limited set of gamma functions, optimized so as to elicit strong activations after standard model‐driven statistical analysis at the single subject level. The method is first validated on healthy subjects using experimental data acquired during motor, visual and memory encoding tasks. Second, interictal EEG/fMRI data measured in 10 patients suffering from epilepsy are analyzed. Results show dramatic changes of activation patterns, depending on whether physiological or pathological assumptions are made on the hemodynamics of the epileptic brain. Our study suggests that one cannot assume a priori that HRFs in epilepsy are similar to the canonical model. This may explain why a significant fraction of EEG/fMRI exams in epileptic patients are inconclusive after standard data processing. The heterogeneous perfusion in epileptic regions indicates that the properties of brain vasculature in epilepsy deserve careful attention. Hum Brain Mapp, 2010. © 2010 Wiley‐Liss, Inc.
Keywords: functional MRI, electroencephalography, epilepsy, modelling, hemodynamics
INTRODUCTION
Simultaneous recordings in electroencephalography (EEG) and functional magnetic resonance imaging (fMRI) using the Blood Oxygenation Level Dependent (BOLD) contrast [Kwong et al., 1992; Ogawa et al., 1992] in patients suffering from epilepsy allows to identify the brain regions showing a correlation between interictal epileptiform discharges (IEDs) and hemodynamic signals [Al‐Asmi et al., 2003; Baudewig et al., 2001; Gotman et al., 2004; Hamandi et al., 2004; Jager et al., 2002; Krakow et al., 1999; Lazeyras et al., 2000; Patel et al., 1999; Salek‐Haddadi et al., 2006; Seeck et al., 1998; Warach et al., 1996]. Establishing this correlation may be important for accurate identification of the epileptogenic zone during presurgical evaluation [Rosenow and Luders, 2001].
Standard EEG/fMRI analyses in epilepsy assume that significant variations of BOLD signals are triggered by interictal EEG activity [Salek‐Haddadi et al., 2003a]. From a modelling point of view, paroxystic events in the EEG are equivalent to the stimulus function used in cognitive studies. Under the linear assumption, a prediction of the “epileptic BOLD activity” is thus obtained by convolving the time series of EEG events with a hemodynamic response function (HRF). Data from large cohorts of patients [Aghakhani et al., 2006; Salek‐Haddadi et al., 2006] have been analyzed in such a way, assuming a HRF very similar to the one used in healthy subjects [Glover, 1999]. Recent reviews indicate that a large fraction (about 30%) of the EEG/fMRI examinations show inconclusive results (no activations or activations discordant with the clinical description of the patient) [Aghakhani et al., 2006; Salek‐Haddadi et al., 2006].
While a canonical HRF is commonly used for data analyses, studies in healthy volunteers have shown a significant variability of the HRF among brain regions, across subjects and over peristimulus time [Aguirre et al., 1998; Buckner, 1998; Handwerker et al., 2004; Menz et al., 2006; Miezin et al., 2000; Neumann et al., 2003]. A plausible source of the relative lack of sensitivity of EEG/fMRI for IEDs is therefore the poor accuracy of the canonical HRF in epileptic regions. Indeed, several studies in patients have shown that the hemodynamic responses are highly variable and slower for negative than for positive BOLD signals [Bagshaw et al., 2004; Benar et al., 2002]. Also, long‐lasting hemodynamic changes have been shown to occur several minutes prior to seizures [Baumgartner et al., 1998; Federico et al., 2005; Makiranta et al., 2005; Weinand et al., 1997]. In this context, HRFs modeling early BOLD changes occurring before IEDs are non‐causal because they precede IEDs, used as input to the linear system [Hawco et al., 2007; Moeller et al., 2008].
Several approaches have been proposed to estimate the HRFs locally [Bagshaw et al., 2004; Benar et al., 2002; Buckner, 1998; Goutte et al., 2000; Josephs et al., 1997; Kang et al., 2003; Kershaw et al., 1999; Lu et al., 2006; Marrelec et al., 2003]. To be applicable routinely in the clinic, the methods for estimating HRFs in EEG/fMRI need (i) to be fully automatic, (ii) to provide relevant information in a limited number of images and (iii) to require a limited computational time. Most of the approaches listed above, though very interesting, present some partial limitations to fulfill completely the criteria required for optimal analysis of EEG/fMRI data in epilepsy departments.
The aim of this study was to improve significantly the sensitivity of EEG/fMRI exams by using an exploratory method based on patient specific HRFs. The HRFs are selected from a set of basis functions derived from a classical parametric model. Parameters are identified so as to maximize the number of activated and deactivated voxels. Using predefined basis functions ensures fast and robust estimation of HRFs. In the following, we first validate the method using experimental data acquired during motor, visual and memory encoding fMRI paradigms in healthy volunteers. We thereby demonstrate that it is unlikely that spurious, widely distributed, activations are created because of multiple parametric tests. Second, we show, in 10 epileptic patients, the important increase in detection sensitivity by comparing the activation patterns obtained with this approach and with standard statistical analyses. Interpretation issues are finally discussed.
MATERIALS AND METHODS
All experiments were approved by the ethics committee of the Grenoble University Hospital. All patients and healthy subjects gave their written informed consent.
Healthy Volunteers and Patients
Healthy volunteers
Thirtheen right‐handed healthy volunteers (five males; mean age, 24.3 years; range, 20–30 years) performed at least one task. Seven subjects were included in the motor and visual tasks and 10 subjects were used for the scene and face memory encoding tasks.
Patients
We studied 10 patients suffering from focal epilepsy (n = 6) and from idiopathic generalized epilepsy (n = 4). Patients were included if they exhibited frequent interictal spiking activity on EEG recordings during the EEG/fMRI exam (more than 10 interictal spikes or more than three generalized spike‐and‐wave discharges during the 30 min recording) and whatever the results of the classical EEG/fMRI analysis. Patients' clinical details are given in Table I.
Table 1.
Patients' clinical description
| Patient | Age/age of onset/sex | Epilepsy diagnosis | Seizure description | EEG | MRI/PET | Intracranial EEG | Treatment |
|---|---|---|---|---|---|---|---|
| 1 | 18/6/F | FLE | Frequent brief seizures with polypnea, head version to the right and contraversive eye deviation, followed by agitation and verbal automatisms | Delta slowing activity involving the right temporal anterior and basal lobe, and spiking activity located in right frontopolar and/or frontobasal regions | Normal brain MRI, FDG‐PET right frontobasal hypometabolism | Wide right frontal lobe involvement during ictal recordings | Carmazepine, 600 mg |
| 2 | 58/20/M | IGE | Rare tonic‐clonic seizures. Neither absence nor myoclonic seizures reported | 3 Hz spike‐and‐wave discharges without associated clinical symptoms | Normal brain MRI | ND | Phenobarbital 70 mg, Lamotrigine 100 mg |
| 3 | 24/18/F | IGE/JAE | Very frequent absence and rare tonic‐clonic seizures | 3Hz generalized spike‐and‐waves discharges accompanied by chewing or slight limb movements | Normal brain MRI | ND | Levetiracetam 1,000 mg, Lamotrigine 200 mg, Valproate 500 mg |
| 4 | 14/6/F | IGE | Myoclonic abscences | 3 Hz spike‐and‐wave discharges | Normal brain MRI | ND | Lamotrigine, Valproate |
| 5 | 44/36/F | Symptomatic partial epilepsy | Focal motor seizures | 3 Hz spike‐and‐wave discharges | Right perisylvian gliosis | ND | Clonazepam |
| 6 | 33/2/M | TLE | Sudden loss of contact, perseverative automatisms | Right frontotemporal interictal epileptiform discharges | Right periventricular heterotopia | ND | Lamictal 200 mg, Keppra 1000 mg |
| 7 | 21/11/M | FLE | Epilepsia partialis continua involving right hemibody | Left centro‐frontal interictal epileptiform discharges | Focal cortical atrophy | Left primary sensorimotor cortex | |
| 8 | 12/11/M | FLE | Initial loss of contact, rapid asymmetric secondary tonic‐clonic generalization | Theta rhythm in frontal lobe | Frontal dysembryoplasic neuroepithelial tumor | ND | Levetiracetam 2,000 mg + carbamazepine 400 mg |
| 9 | 15/M | Frontotemporal LE | Gamma rhythm in left frontotemporal. | Left frontoprecentral dysplasia | Left insulo‐opercular onset zone | Valproate | |
| 10 | 49/7/M | TLE | Epigastric aura, loss of contact and gestural and verbal automatism | Right frontotemporal interictal epileptiform discharges | Right periventricular heterotopia | ND | Carbamazepoine 800 mg, topiramate 200 mg, Phenobarbital 100 mg, clobazzam 20 mg |
F, female; M, male; FDG PET, fluorodeoxyglucose positron emission tomography; FLE, frontal lobe epilepsy; IGE idiopathic generalized epilepsy; JAE, juvenile absence epilepsy, TLE, temporal lobe epilepsy; ND, not done
Data Acquisition
Functional MRI was acquired either at 1.5 Tesla (1.5T Achieva, Philips Medical Systems, Best, The Netherlands) or at 3 Tesla (3T Bruker Biospin, Bruker Medizintechnik GmbH, Ettlingen, Germany). Experiments with healthy subjects were all performed at 3T. Functional images were obtained using a single‐shot T2* gradient‐echo Echo Planar Imaging (GE‐EPI) sequence (1.5T: TR = 3 s, voxel size: 4 × 4×4 mm3, 32 adjacent slices; 3T: TR = 3 s, voxel size: 3 × 3 × 3.5 mm3, 41 adjacent slices). A high resolution T1‐weighted scan was used for anatomical localization (1.5T: 3D T1TFE sequence, TR = 7.66 ms, voxel size: 1 × 1 × 1 mm3; 3T: 3D MPRAGE sequence, TR = 10 ms, voxel size: 1.33 × 1.75 × 1.37 mm3).
EEG data were acquired using a MR compatible EEG amplifier (SD32, Micromed, Treviso, Italy) with 17 c‐shaped electrodes positioned according to the 10/20 system (O1 and O2 were not used for the sake of the subjects' comfort). An anti‐aliasing hardware low‐pass filter at 270 Hz was used and the EEG sampling rate was 1,024 Hz. TTL triggers were used for offline temporal co‐registration of EEG and MR recordings.
Statistical Analysis of fMRI Data Using Optimal HRFs
The general flow of fMRI data processing is summarized in Figure 1.
Figure 1.
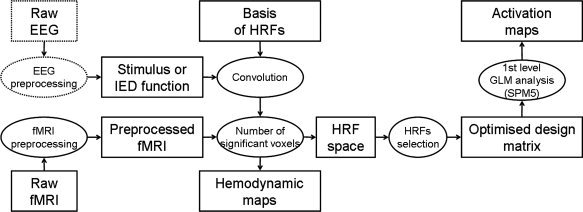
Flowchart of the HRF optimization procedure: after preprocessing of fMRI and EEG data, activation maps are obtained using a design matrix optimized so as to maximize the number of significant voxels. Hemodynamic parameters maps are produced during the optimization of the design matrix.
FMRI preprocessing
Standard fMRI preprocessing was performed using the SPM5 software (Wellcome Department of Imaging Neuroscience, University College London, UK, http://www.fil.ion.ucl.ac.uk/spm). Spatial preprocessing of functional images included: (i) slice‐timing correction, (ii) motion correction using rigid‐body coregistration, (iii) normalization to the MNI space and (iv) spatial smoothing with an isotropic Gaussian kernel of 6 mm full width at half‐maximum (FWHM). Finally, fMRI time series were whitened and serial correlations were modeled using an auto‐regressive filter of order 1. Low‐frequency drift was removed using a discrete cosine transform (DCT) basis set with a filter cut‐off period of 128 s.
HRF optimization
It is usually assumed that changes in the BOLD signal are induced by changes of synaptic activity that trigger a cascade of metabolic events [Friston et al., 2000]. In the general linear model (GLM), the BOLD signal is the output of a linear hemodynamic filter which receives a stimulus as input. The response of the hemodynamic filter is fully characterized by its impulse response function: the HRF. A typical HRF shows a time‐to‐peak of about 5 s, which is followed by an undershoot before going back to baseline [Kruger et al., 1996].
Several parameterizations of the HRF have been proposed: Poisson functions [Friston et al., 1994], Gaussian functions [Rajapakse et al., 1998], gamma functions [Boynton et al., 1996; Friston et al., 1998; Glover, 1999; Lange and Zeger, 1997], among others. In fact, the particular choice of the HRF model is here not critical. For simplicity, we used a reduction of the model presented in [Glover, 1999] and defined the “standard,” or “canonical,” HRF as a gamma function without undershoot:
| (1) |
where t is the time, a = 6 s, b = 0.9. The time‐to‐peak of the kernel h is equal to ab (5.4 s) and its FWHM is equal to 5.2 s. The standard HRF in Eq. (1) is used to construct a bivariate basis set of HRFs:
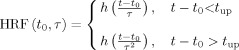 |
(2) |
where
 is the rise time. The two parameters (t
0, τ) of this basis set are the time of onset and the time scaling constant of the HRFs, respectively. The time of onset t
0 is the delay between the stimulus and the beginning of the hemodynamic response. A BOLD response preceding the stimulus corresponds to a negative value of t
0. The time‐to‐peak of the HRF relative to the stimulus is: t
peak = t
0 + t
up. The time scaling constant modifies the width of the HRF: if τ is greater than 1, the standard HRF is time‐dilated, otherwise it is time‐contracted. Following qualitative analysis of data, we applied different time scaling constants for the rise (τ) and for the fall (τ2) of the HRFs to allow a stronger dilation of the decaying phase of the HRF. Because τ and t
up are directly interconnected, in the following, we will refer to t
up only.
is the rise time. The two parameters (t
0, τ) of this basis set are the time of onset and the time scaling constant of the HRFs, respectively. The time of onset t
0 is the delay between the stimulus and the beginning of the hemodynamic response. A BOLD response preceding the stimulus corresponds to a negative value of t
0. The time‐to‐peak of the HRF relative to the stimulus is: t
peak = t
0 + t
up. The time scaling constant modifies the width of the HRF: if τ is greater than 1, the standard HRF is time‐dilated, otherwise it is time‐contracted. Following qualitative analysis of data, we applied different time scaling constants for the rise (τ) and for the fall (τ2) of the HRFs to allow a stronger dilation of the decaying phase of the HRF. Because τ and t
up are directly interconnected, in the following, we will refer to t
up only.
Parameters corresponding to the canonical HRF are t 0 = 0 s and t up = 5.4 s. We varied concomitantly t 0 from −15 s to 15 s and t up from 2 s to 15 s, so as to cover most of the physiological range of the parameter space of HRFs (see Fig. 2). The BOLD signals predicted in response to a stimulus or to epileptic events were obtained by convolving the HRFs with the stimulus function extracted from the experimental paradigm or from the EEG (see below), respectively.
Figure 2.
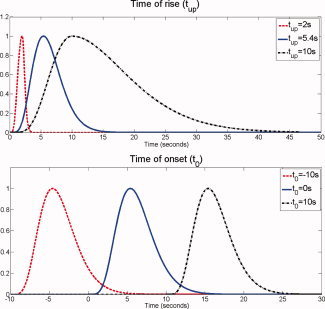
Construction of the basis of HRFs. Top: Effect of the time scaling factor on the standard HRF. Bottom: Effect of the time of onset on the standard HRF. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To identify optimal HRFs for each dataset, we computed the cross‐correlation coefficient r, and its associated p‐value testing the null hypothesis of no correlation, between the BOLD signal predicted using each HRF and the preprocessed time series from each brain voxel. When considering such a large set of basis functions, multiple comparisons have to be taken into consideration carefully. First, the false discovery rate (FDR) [Genovese et al., 2002] was used as a correction for multiple comparisons over brain voxels. Second, using principal component analysis, we measured that 98% of the variance of the set of basis functions is explained with only nine principal components. Therefore, a P value of 0.005 for the optimized analysis is approximately equivalent to a P value of 0.05 for a canonical analysis. Following these considerations, a voxel was considered in this study as significantly activated if the FDR issued from the observed P values is lower than 0.005. As a summary statistic, the number of activated (r > 0, FDR‐corrected P < 0.005) and deactivated (r < 0, FDR‐corrected P < 0.005) voxels was reported in the HRF space as a function of the parameters t 0 and t up. To limit MR spatial high‐frequency noise effects and because approximately 6 cm3 of cortex need to be active to produce a scalp potential [Ebersole, 1997], clusters with less than 20 activated voxels (equivalent to 6 cm3) were not considered when computing the total number of activated voxels. Because HRFs with similar parameters are correlated, the summary statistic in the HRF space is a smooth map (see Results section). Each local maximum of this map can be thought of as representing a hemodynamic mode of brain responses. These local maxima were automatically detected and corresponding HRFs were selected to construct the design matrix used to get optimal activation maps (see next step).
Activation maps were obtained from standard GLM analyses as performed in SPM5, using either the standard HRF [Eq. (1)] (“classical analysis”), or the HRFs corresponding to each local maximum detected in the HRF space (“optimized analysis”). Activation and deactivation maps of the different hemodynamic modes were obtained using T‐contrasts. To capture all regions in a single activation map, optimal HRFs were also put together in a large design matrix1 and an F‐contrast was applied to the parameter estimates. For EEG/fMRI data, all statistical analyses were performed using the alpha power as a confound variable [Tyvaert et al., 2008].
For voxels found significantly activated (P < 0.005, FDR‐corrected) in the map following F‐contrast analysis, hemodynamic parameter maps (onset time and rise time) were generated by reporting, for each of these voxels, the optimal parameters identified in the HRF space. These hemodynamic parameter maps allow easy visualization of the hemodynamics of brain activation.
Experimental Data and EEG Processing
Paradigms used in healthy subjects consisted of six alternations of activation and baseline blocks of 24 s for visual and motor tasks, and of eight alternations of activation and baseline blocks of 36 s for memory encoding tasks. Assuming a canonical HRF, the design efficiency was estimated to 95 for visual and motor tasks and to 191 for memory encoding tasks [Friston et al., 1999].
During the motor task, subjects were instructed to move sequentially each finger of their right hand to the thumb during the activation blocks and to stay at rest during the baseline blocks. Instructions (“action” or “rest”) were projected onto a screen viewed by the subject through a mirror attached to the head coil. During the visual paradigm, subjects were shown a circular checkerboard flickering at 8 Hz during the activation blocks and a fixation point during the baseline blocks. Subjects were instructed to maintain their gaze to the centre of the screen. Memory encoding tasks were based on [Golby et al., 2002]. Two types of visual stimuli (scenes or faces) were presented in separate sessions. During the control condition, stimuli were always the same two images whereas in the experimental condition novel images were presented. To maintain a high level of attention, subjects were instructed to indicate using button press if the scenes were indoor or outdoor or if the faces were male or female.
In patients suffering from epilepsy, EEG and fMRI data were simultaneously recorded during 30 min. Patients were asked to lie still and to keep awake, with their eyes closed. Imaging and cardiac artefacts in the EEG were removed offline using algorithms described in [Grouiller et al., 2007; Niazy et al., 2005]. EEG power in the frequency band of epileptic discharges (e.g. between 2.5 and 3 Hz averaged on all electrodes for spike‐and‐wave discharges in idiopathic generalized epilepsy) or timing of epileptic interictal spikes were used as a regressor of interest to be convolved with the HRF (see Fig. 3). The power in the alpha band (8–12 Hz) was also extracted and used as a confound (after convolution with the HRF) for fMRI statistical analyses [Tyvaert et al., 2008]. For each recording, design efficiency was estimated to check the specificity of the regressor and to evaluate the risk of fitting noise as compared to studies in healthy subjects.
Figure 3.
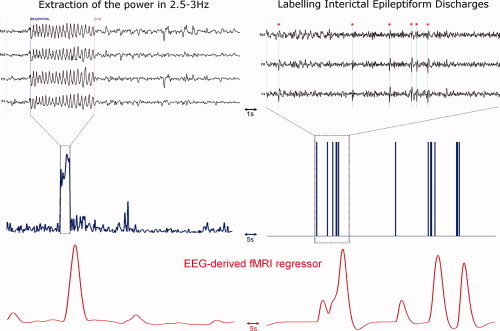
Construction of the fMRI regressors. Left: (top) EEG of an epileptic patient suffering from idiopathic generalized epilepsy, (middle) Power in 2.5–3 Hz band extracted from the EEG, (bottom) fMRI regressor after convolution of the power by the HRF. Right: (top) Interictal EEG of an epileptic patient suffering from right frontal lobe epilepsy. Spikes are indicated with a red star, (middle) manual labeling of the interictal spikes, (bottom) fMRI regressor after convolution of the spikes by the HRF. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
RESULTS
Functional Mapping in Healthy Volunteers
For each task and for each subject, both classical and optimized GLM fMRI analyses were performed. Results are summarized in Table II. The optimal HRF estimated separately for the motor, visual and memory encoding tasks were very similar to the canonical HRF (t 0 = 0.1 ± 2.0 s, t up = 5.3 ± 1.8 s for motor task; t 0 = −0.1 ± 2.3 s, t up = 6.2 ± 1.1 s for visual task; t 0 = 0.5 ± 2.2 s, t up = 4.5 ± 1.6 s for scene encoding task and t 0 = 0.8 ± 1.7 s, t up = 4.5 ± 1.4 s for face encoding task). The number of activated voxels using the optimized approach was thus, on average, respectively, only 5.4%, 11.8%, 6.1% and 5.4% larger than when using the conventional HRF.
Table 2.
Results of activation sizes obtained from motor, visual and memory encoding tasks
| Subject | Number of activated voxels using classical analysis | Number of activated voxels using optimal HRF | Optimal HRF time of onset (t 0) | Optimal HRF time of rise (t up) |
|---|---|---|---|---|
| Motor task | ||||
| 1 | 2,751 | 3,075 | 1 s | 4 s |
| 2 | 2,706 | 2,819 | −1 s | 5 s |
| 8 | 698 | 698 | 0 s | 5.4 s |
| 10 | 1,387 | 1,448 | −2 s | 7 s |
| 11 | 14,917 | 14,917 | 0 s | 5.4 s |
| 12 | 4,594 | 5,494 | −1 s | 7 s |
| 13 | 2,387 | 2,592 | 4 s | 2 s |
| Group | 4,206 ± 4,879 | 4,435 ± 4,860 | 0.1 ± 2.0 s | 5.3 ± 1.8 s |
| Visual task | ||||
| 2 | 4,622 | 4,955 | −5 s | 8 s |
| 3 | 1,607 | 1,607 | 0 s | 5.4 s |
| 5 | 2,842 | 2,842 | 0 s | 5.4 s |
| 10 | 552 | 651 | 2 s | 5 s |
| 11 | 1,592 | 2,016 | 1 s | 7 s |
| 12 | 1,725 | 2,735 | 1 s | 7 s |
| 13 | 2,837 | 2,837 | 0 s | 5.4 s |
| Group | 2,254 ± 1,312 | 2,520 ± 1,339 | −0.1 ± 2.3 s | 6.2 ± 1.1 s |
| Scene encoding task | ||||
| 1 | 10,707 | 11,093 | −1 s | 5 s |
| 2 | 9,257 | 9,804 | 4 s | 2 s |
| 3 | 9,237 | 9,455 | −2 s | 6 s |
| 4 | 8,660 | 9,226 | 1 s | 6 s |
| 6 | 8,392 | 8,963 | 0 s | 4 s |
| 7 | 5,416 | 5,679 | 1 s | 4 s |
| 8 | 7,660 | 7,931 | −3 s | 7 s |
| 9 | 7,056 | 7,405 | 3 s | 3 s |
| 10 | 7,972 | 8,017 | 0 s | 5 s |
| 13 | 2,707 | 3,106 | 2 s | 3 s |
| Group | 7,606 ± 2,203 | 8,068 ± 2,286 | 0.5 ± 2.2 s | 4.5 ± 1.6 s |
| Face encoding task | ||||
| 1 | 3,356 | 4,227 | 0 s | 4 s |
| 2 | 4,675 | 4,802 | 3 s | 3 s |
| 3 | 4,626 | 4,856 | −2 s | 6 s |
| 4 | 3,212 | 3,452 | 3 s | 3 s |
| 6 | 3,177 | 3,355 | 1 s | 4 s |
| 7 | 3,362 | 3,409 | 2 s | 4 s |
| 8 | 4,194 | 4,347 | 0 s | 4 s |
| 9 | 3,108 | 3,160 | −1 s | 7 s |
| 10 | 4,594 | 4,633 | 0 s | 6 s |
| 13 | 3,990 | 4,097 | 2 s | 4 s |
| Group | 3,829 ± 655 | 4,034 ± 643 | 0.8 ± 1.7s | 4.5 ± 1.4s |
Number of activated voxels (P < 0.005, FDR‐corrected, extent threshold = 20 voxels) for classical analyses (t 0 = 0 s, t up = 5.4 s) and for optimal HRF analyses (using the HRF parameters specified for each subject). Mean and standard deviation of the number of activated voxels, using classical HRF and using subject‐specific HRFs, are also indicated for the group.
Figure 4 shows: (i) The mean fraction of activated voxels in the HRF parameter space (average across subjects). Optimal HRFs over the group were: motor: t 0 = 2 s, t up = 4 s; visual: t 0 = 1 s, t up = 6 s; scene encoding: t 0 = 0 s, t up = 5 s; face encoding: t 0 = 0 s, t up = 5 s. (ii) The activation maps at the group level using the optimal HRF for the group. As anticipated, group analysis using either standard or optimized HRFs for each volunteer shows activation of the controlateral motor cortex and of the ipsilateral cerebellum for the motor task, activation of occipital regions for the visual task and bilateral activation of hippocampi, parahippocampal cortices and occipital regions for memory encoding tasks.
Figure 4.
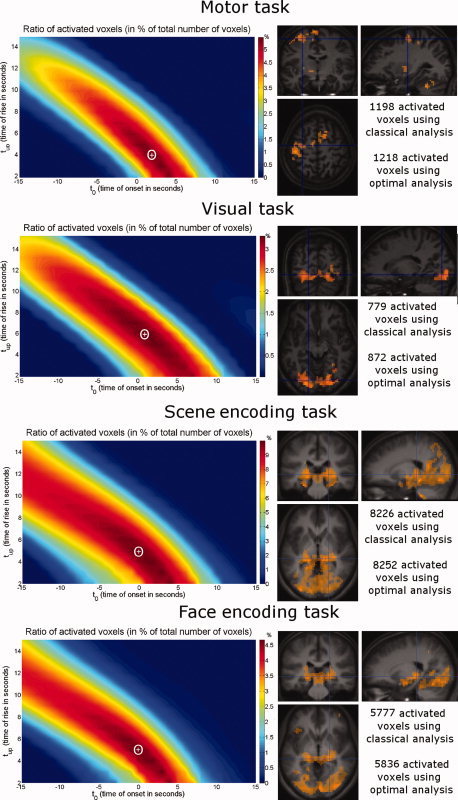
Functional tasks in healthy volunteers. From top to bottom: Motor task, visual task, scene encoding and face encoding. Left: mean ratio of activated voxels over all subjects. The white cross corresponds to the map local maximum whose coordinates give optimal HRF parameters. Right: group study activation (P < 0.005, uncorrected, extent threshold = 50 voxels) comparing classical analysis in yellow (t 0 = 0 s, t up = 5.4 s) and optimized analysis with HRF parameters corresponding to maximum mean ratio of activated voxels in red (Motor: t 0 = 2 s, t up = 4 s; Visual: t 0 = 1 s, t up = 6 s; Scene encoding: t 0 = 0 s, t up = 5 s; Face encoding: t 0 = 0 s, t up = 5 s). Orange areas, i.e. nearly all activated voxels, correspond to voxels activated with both analyses. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
These findings, together with results from preliminary simulations (not reported here), strongly suggest that the proposed method is unlikely to create large artefactual activations unrelated to the experimental context.
fMRI/EEG in Patients Suffering From Epilepsy
Results of fMRI/EEG recordings for all patients are summarized in Table III. For all 10 patients, activations or/and deactivations were partially concordant (i.e., at least one cluster was located in the same lobe as the epileptic focus for focal epilepsy, or widespread involvement is shown for idiopathic generalized epilepsy) with the optimal analysis. For the 3 patients without any (de)activation after classical analysis (patients 2, 9 and 10), optimized analysis produced nonetheless significant (de) activations. Schematically, one can classify the results into three groups: (i) activations well captured by the canonical model (patients 1, 7, 8); (ii) unimodal or multimodal hemodynamics dissimilar to canonical responses (patients 2, 3, 4, 5, 6); (iii) weak responses (similar to noise components?) (patients 9, 10). Interestingly, all patients showing rather standard hemodynamics suffered from frontal lobe epilepsy. Idiopathic generalized epilepsy patients were particularly prone to exhibit altered hemodynamics. Temporal lobe epilepsy patients showed less robust results. We report here detailed results for patients 1, 2 and 3.
Table 3.
Results of fMRI/EEG in epileptic patients using classical and optimal analysis
| Patient | EEG | fMRI regressor | Classical analysis | Optimal analysis |
|---|---|---|---|---|
| 1 | Delta slowing waves in right frontal and temporal electrodes | Power between 1.5 and 3 Hz averaged on T4, T6, F4, F8, Efficiency = 440.1 | Activation: 3,915 voxels—widespread to many cortical regions. Deactivation: 13,913 voxels—right frontotemporal cortex, cingulate gyrus, bilateral caudate nuclei | Activation (t 0 = 1s, t up = 4s): 4,850 voxels—widespread to many cortical regions. Deactivation (t 0 = 0 s, t up = 7 s): 15,262 voxels—right frontotemporal cortex, cingulate gyrus, bilateral caudate nuclei |
| 2 | Generalized spike‐and‐wave discharges (2.5‐3 Hz) | Power between 2.5 and 3 Hz averaged on all electrodes. Efficiency = 375.1 | No activation. No deactivation | Activation (t 0 = −5 s, t up = 5 s): 868 voxels. Deactivation (t 0 = −4 s, t up = 5 s): 34,611 voxels—prefrontal, parietal, occipital, insular cortices, cerebellum, caudate nuclei, and right hippocampus |
| 3 | Generalized spike‐and‐wave discharges (2.5‐3 Hz) | Power between 2.5 and 3 Hz averaged on all electrodes. Efficiency = 167.1 | Activation: 6,511 voxels—thalamus and occipital cortex. Deactivation: 390 voxels | Activation (t 0 = −8 s, t up = 8 s): 6,198 voxels—occipital and parietal lobes. Activation (t 0=−5 s, t up = 3 s): 8,280 voxels—occipital and parietal lobes. Activation (t 0 = 3 s, t up = 3 s): 15,750 voxels—thalamus, occipital and parietal lobes. Deactivation (t 0 = 0 s, t up = 2 s): 35,840 voxels—frontal and temporal lobes. Deactivation (t 0 = 4 s, t up = 9 s): 26,822 voxels—occipital and parietal lobes |
| 4 | Bilateral spike‐and‐wave discharges (2.6 Hz) | 4 runs of 2.6 Hz spike‐ and‐wave discharges (mean duration: 7.25 s). Efficiency = 310.1 | No activation. Deactivation: 565 voxels—cerebellum and cingulate cortex | Activation (t 0 = 10 s, t up = 2 s): 610 voxels—cingulate cortex. Deactivation (t 0 = 13 s, t up = 2 s): 29,474 voxels—thalamus, bilateral frontal, temporal superior and parietal lobes. Deactivation (t 0 = −2 s, t up = 10 s): 16,126 voxels—bilateral frontal, temporal superior and parietofrontal lobes. Deactivation (t 0 = 4 s, t up = 2 s): 2,609 voxels— thalamus, bilateral hippocampi, cerebellum and cingulate cortex |
| 5 | Bilateral spike‐and‐wave discharges (3 Hz) | 118 runs of 3 Hz spike‐and‐wave discharges (mean duration: 3.27 s). Efficiency = 555.6 | Activation: 1,125 voxels— cingulate cortex. Deactivation: 17,285 voxels—thalamus, cuneus, bilateral temporal and frontal lobes | Activation (t 0 = −7 s, t up = 5 s): 12,229 voxels—thalamus, bilateral temporal superior, frontal and parietal lobes. Activation (t 0 = 1 s, t up = 5 s): 1218 voxels—cingulate cortex. Deactivation (t 0 = −1 s, t up = 8 s): 23,145 voxels—thalamus, cuneus, bilateral temporal and frontal lobes |
| 6 | Spikes in right frontotemporal electrodes | 393 spikes in F8 and T4. Efficiency = 556.4 | Activation: 6,462 voxels—cuneus, bilateral sensorimotor cortex, occipital and right temporal posterior lobes. No deactivation | Activation (t 0 = −3 s, t up = 11 s): 23,771 voxels—cuneus, bilateral sensorimotor cortex, occipital and right temporal posterior lobes. No deactivation |
| 7 | Spikes in left frontoparietal electrodes | 219 spikes in F3, C3 and P3. Efficiency = 469.7 | Activation: 55 voxels—left frontal lobe. Deactivation: 7,382 voxels—cuneus, cingulate cortex, bilateral frontal lobes | Activation (t 0 = 0 s, t up = 5 s): 81 voxels—left frontal lobe. Deactivation (t 0 = −10 s, t up = 13 s): 19,966 voxels—cuneus, thalamus, cingulate cortex, bilateral frontal and insular lobes |
| 8 | Theta rhythm in frontal electrodes | Power between 3 and 6 Hz averaged on F3, F4, Fz. Efficiency = 596.1 | Activation: 6,056 voxels—frontal (around dysplasia) and occipital lobes. No deactivation | Activation (t 0 = −2 s, t up = 5 s): 9,104 voxels—frontal (around dysplasia) and occipital lobes, cuneus, bilateral insula and thalamus. No deactivation |
| 9 | Gamma rhythm in left frontal electrodes | Power between 36 and 38 Hz averaged on Fp1, F3 and F7. Efficiency = 552.0 | No activation. No deactivation | Activation (t 0= −15 s, t up = 5 s): 355 voxels—bilateral temporal lobes (predominant in left). Activation (t 0 = 6 s, t up = 15 s): 1,288 voxels—bilateral frontal lobes (predominant in left). Deactivation (t 0 = 1 s, t up = 11 s): 5,064 voxels— bilateral temporal lobes (much more predominant in left), left frontal lobe and cuneus |
| 10 | Spikes in right frontotemporal electrodes | 118 spikes on F8 and T4. Efficiency = 475.3 | No activation. No deactivation | Activation (t 0 = −11 s, t up = 8 s): 395 voxels—bilateral temporal lobes (predominant in right). Deactivation (t 0 = −1 s, t up = 13 s): 1864 voxels—bilateral temporal lobes (predominant in right) |
For each patient, EEG, fMRI regressor and its corresponding efficiency, number of (de)activated voxels and their localization using classical analysis and optimal analysis are indicated.
Patient 1 suffered from frontal lobe epilepsy. Using a standard HRF, small activation clusters were found in many cortical regions (Fig. 5A). On the contrary, for deactivation, large clusters were found in frontomesial, dorsolateral prefrontal, posterior cingular cortices, bilaterally in the superior temporal sulcus and in subcortical regions (particularly in the head of the caudate nucleus). Maps in the HRF space (Fig. 5B) led to estimating optimal HRFs very similar to the standard HRF (activation: t 0 = 1 s, t up = 4 s; deactivation: t 0 = 0 s, t up = 7 s). Consequently, activation patterns (Fig. 5C) using either the standard or the optimal HRFs were very similar, although clusters obtained with the optimal analysis were a bit larger than with the classical analysis (4,850/3,915 activated voxels, 15,262/13,913 deactivated voxels, respectively). Hemodynamic parameter maps (Fig. 5D) showed a certain degree of spatial variability.
Figure 5.
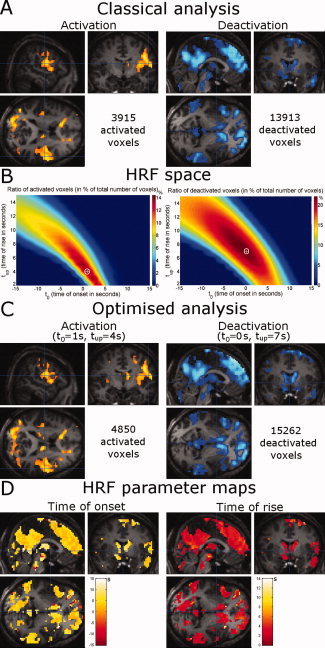
Patient 1: frontal lobe epilepsy. (A) Activation and deactivation patterns using classical HRF (P < 0.005, FDR‐corrected, extent threshold = 50 voxels). (B) Ratio of activated and deactivated voxels in the HRF space. (C) Activation and deactivation patterns using optimal HRF (P < 0.005, FDR‐corrected, extent threshold = 50 voxels). (D) HRF parameter maps (left: time of onset t 0; right: time of rise t up). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Patients 2 and 3 suffered from idiopathic generalized epilepsy. In Patient 2, neither activation nor deactivation was found after GLM analysis using the classical HRF (Fig. 6A). This examination would thus usually be classified as inconclusive. Exploration of the HRF space (Fig. 6B) revealed that the optimal HRFs occurred earlier (activation: t 0 = −5 s, t up = 5 s; deactivation: t 0 = −4 s, t up = 5 s) than the standard model. Reanalysing the data with the optimized HRF produced a small activation (868 activated voxels) and, more importantly, a widespread deactivation (34,611 deactivated voxels distributed mainly in prefrontal, parietal, occipital and insular cortices, in cerebellum, in caudate nucleus and in right hippocampus) (Fig. 6C). The data in HRF space (Fig. 6B) and the hemodynamic parameter maps (Fig. 6D) suggested quite homogenous behavior of the hemodynamics, in this patient.
Figure 6.
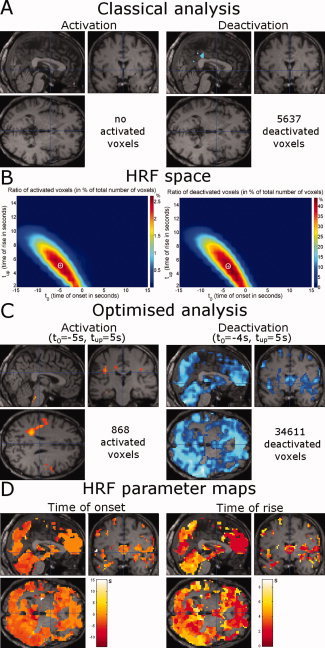
Patient 2: idiopathic generalized epilepsy. Same format as Figure 5. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In Patient 3, fMRI/EEG analysis using the standard HRF activated mainly the thalamus and the occipital cortex (6,511 voxels). Only a relatively small deactivation (390 voxels) was observed (Fig. 7A). HRF space maps of the fraction of activated voxels in the HRF space (Fig. 7B, left) indicated the benefit of using a HRF characterized by a late onset and fast changes (t 0 = 3 s, t up = 3 s). Doing so, a large activation (15,750 voxels) involving the occipital and parietal lobes was found (Fig. 6C). For the deactivation (Fig. 7B, right), multiple HRFs were identified in the HRF space, with fast (t 0 = 0s, t up = 2 s) and slow (t 0 = 4 s, t up = 9 s) components. Analyses using optimal HRFs (Fig. 7C) then revealed a widespread deactivation including frontal and temporal lobes for the fast response (35,840 voxels) and occipital and parietal lobes for the slow response (26,822 voxels). Hemodynamic parameter maps (Fig. 7D) showed that deactivation in frontal and parietal lobes occured prior to deactivation in the parietal and occipital lobes.
Figure 7.
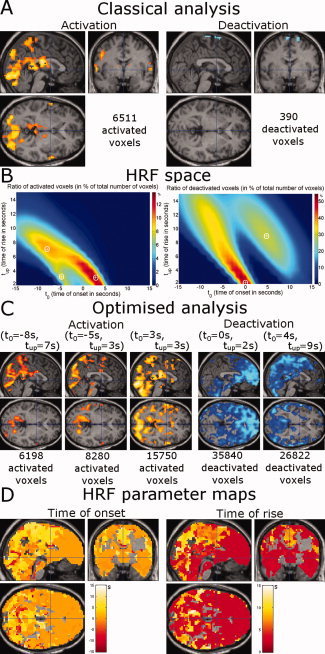
Patient 3: absence epilepsy. Same format as Figure 5. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 8 shows equivalent information obtained for remaining patients.
Figure 8.
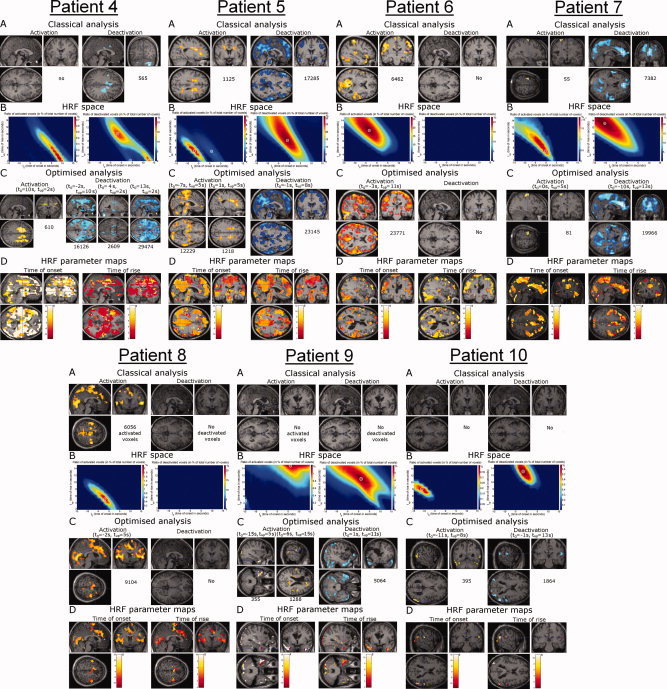
Patients 4–10. Same format as Figure 5. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
We proposed in this study an unsupervised parametric method that estimates the HRFs associated to IEDs, on an individual patient basis. The method can also be applied with standard neurocognitive paradigms in healthy subjects. Optimal HRFs maximized the number of (de)activated voxels following GLM analysis (e.g. as performed in SPM5). There was no particular neurocomputational motivation for this critical assumption since there is no greater truth in a large cluster of activated voxels than in a small cluster. Our perspective was rather pragmatic, with the goal to develop a technique which would capture all potential variability, in a restricted subspace, of epilepsy‐related hemodynamics. Because the same data are used to tune the model parameters to detect a signal of interest, this method should be considered as exploratory and should not replace more conventional model‐based statistical analyses.
Proposing this approach was motivated by the results from several studies [Baumgartner et al., 1998; Federico et al., 2005; Hawco et al., 2007; Makiranta et al., 2005; Moeller et al., 2008; Weinand et al., 1997] indicating that hemodynamics in the epileptic brain may behave differently from hemodynamics in response to exogenous stimuli. To deal with the potential variability of hemodynamics associated with epileptic discharges while keeping strong dynamical constraints (i.e. preserving the overall shape of a typical HRF), we adopted a deformable model of the HRF, within the GLM framework. A gamma function, with classical parameters that emulate physiological HRF [Glover, 1999], was used as a starting point. It was then deformed by time scaling and by time translation. We thus constructed a bi‐parametric HRF basis set that constituted a signal subspace into which fMRI signals were projected. Because causal relationships between the EEG and fMRI are not well established [Baumgartner et al., 1998; Federico et al., 2005; Hawco et al., 2007; Makiranta et al., 2005; Moeller et al., 2008; Weinand et al., 1997], we assumed non causal as well as causal HRFs (negative and positive values were assumed for the onset of HRFs). The unsupervised method for estimating relevant brain activations operates thus in two steps: (i) The HRFs that activate the largest brain regions are first identified; (ii) A GLM analysis is performed using the previously optimized HRFs. By construction, the method detects tiny activated regions in the case of significant hemodynamic correlation with larger regions. It does not if the HRFs in small regions (smaller than an adjustable threshold, set to 6 cm3 here) are very specific.
To validate the specificity of the proposed approach in healthy volunteers performing visual, motor and memory encoding tasks, we quantified spurious activation patterns that may occur because of the multiple parametric tests performed with the method. Activation maps obtained using either canonical or optimized HRFs showed the involvement of suspected regions, i.e. the primary visual and motor cortices and bilateral hippocampi. Because the optimized and canonical HRFs were very similar, activation maps were very much alike. The size of the activated clusters was still a bit larger under the optimized conditions, in agreement with the critical assumption of the method. These experiments confirm that our method is unlikely to produce widespread spurious activations.
For this methodological study, we selected 10 epileptic patients from the EEG/fMRI data available at Grenoble University Hospital. Here, we detailed the results from three of them who showed different hemodynamic behaviours (see Fig. 8 and Table III for remaining patients). In Patient 1—suffering from frontal lobe epilepsy—the estimated HRFs were close to the canonical HRF. The activation maps were thus very similar, even though cluster sizes increased after HRF optimization (see Fig. 5). The data from this patient suggest that the common hypothesis—hemodynamics in the epileptic brain is similar to that in the healthy brain—may be true in certain cases. Interestingly, this common hypothesis was verified in the three patients suffering from frontal lobe epilepsy, but not in others. In Patients 2 and 3 suffering from idiopathic generalized epilepsy, we found very little or even no deactivation patterns using the canonical HRF. This observation contradicts the results from other EEG/fMRI studies in idiopathic generalized epilepsy that have shown widespread cortical deactivations [Hamandi et al., 2006; Laufs et al., 2006; Salek‐Haddadi et al., 2003b]. However, massive brain deactivation was revealed using optimized HRFs in both patients. It appeared furthermore that hemodynamic modes were different in these two patients. Very much as was the case with the healthy subjects and Patient 1, Patient 2 revealed homogenous hemodynamic responses (i.e. only one hemodynamic mode). The optimized HRF preceded EEG activity, however. The hemodynamics in Patient 3 was more complex. Multimodal responses were observed whereby the frontal lobe was deactivated a few seconds prior to the occipital cortex. Similar findings obtained in the remaining patients indicated that hemodynamics in epilepsy is likely to vary a lot. However, because the number of patients included here was limited, we could not correlate significantly the hemodynamic properties with the type of epilepsy, although some trends emerged in particular for frontal lobe epilepsy (normal HRF) and idiopathic generalized epilepsy (abnormal HRF). Larger cohorts of patients are needed for future studies in that direction.
By imposing the shape of the HRFs, the method described here is much more constraining than the Fourier basis set procedure [Josephs et al., 1997] used in [Lemieux et al., 2008]. In the latter study, the authors concluded that (de)activations obtained using non‐canonical HRFs convolved with interictal epileptic discharges were almost suggestive of artefacts. Overall, our results do not support this interpretation because hemodynamics, as captured in the hemodynamic parameter maps, strongly differed between healthy subjects and epileptic patients. In addition, usual artefacts in fMRI such as high‐frequency noise, physiological noise (respiration, cardiac activity, large vessels) or residual motion are easily recognizable giving activations at the edges of the brain, near grey‐white matter interfaces, near ventricles and around main vessels. Denoising techniques, not applied in this study, could be useful to limit such spurious signals. To show up in the activation maps derived here, artefacts still need to be significantly correlated to the experimental paradigm or epileptic events, because the HRF space allows only a small departure from the canonical HRF despite multiple tests. Nonetheless, interpretation of the activation maps after optimization of the HRF requires careful attention to exclude such possible activations. In Figure 8 for instance, results obtained in Patients 9 and 10 might represent mostly noise, though activations lied mostly in the temporal lobes as expected. Fundamentally, the apparent smaller detection power for temporal lobe epilepsy patients might be explained by the difficulty to record on the scalp deep temporal activity [Nayak et al., 2004; Tao et al., 2005]. It is thus highly probable that the majority of electrical activity being actually correlated with measured hemodynamics is not adequately modeled in the regressor of interest.
The patients showed distinct hemodynamic parameter maps. Whether these maps can further be exploited, either to classify different types of epilepsy or to indicate potential markers of the physiopathology of neurovascular coupling, needs further investigation, all the more so that a possible influence of antiepileptic drugs on hemodynamics could not be excluded. In particular, lamotrigine, a neuronal voltage‐gated ion channel blocker and glutamate release inhibitor used in Patients 2, 3 and 4, has been showed in rodents to attenuate BOLD activation related to forepaws stimulation, but not basal signals [Kida et al., 2006]. Interpreting the hemodynamic maps is in fact difficult because little is known about altered brain hemodynamics in epilepsy. For instance, in a genetic rat model of absence epilepsy, we recently showed important differences between the hemodynamics of the cortical epileptic focus and the hemodynamics within the other areas of the network involved during spike‐and‐wave discharges [David et al., 2008]. Biophysical modeling of the fMRI time series suggested that processes of vasodilatation regulation by cerebral blood flow were a plausible source of such difference. Further studies using other neuroimaging or histological techniques are needed to better assess the validity of this first modeling attempt. More generally, it is hypothesized in the literature that the large metabolic demands following ictal and interictal events may render irrelevant the neurovascular coupling mechanisms that apply in the normal conditions [Schwartz, 2007]. In the extreme case of lesional epilepsy, such as temporal lobe epilepsy associated with hippocampal sclerosis, blood brain barrier disruption during seizures might even lead to neurovascular decoupling because of abnormal angiogenesis and vascular remodeling [Rigau et al., 2007].
Finally, the negative HRF onsets found in this work confirm results from other studies [Baumgartner et al., 1998; Federico et al., 2005; Hawco et al., 2007; Makiranta et al., 2005; Moeller et al., 2008; Weinand et al., 1997] and call for a cautious interpretation of the HRF in the context of epilepsy. In certain forms of epilepsy, the EEG might very well be a poor marker of underlying metabolic and hemodynamic activities. In particular, it is plausible that processes, such as glial activity [Takano et al., 2006; Tian et al., 2005], not measurable in the EEG but known to induce strong hemodynamic changes, cause precedence of the EEG over BOLD signals. In these conditions, interpreting hemodynamic maps derived from the EEG might well be an ill‐posed problem because of numerous hidden variables.
CONCLUSION
The estimation of epileptic networks in fMRI may fail when using the IEDs convolved with a canonical HRF. To minimize model misspecification in fMRI analyses, optimization of HRFs is thus required for at least two reasons: (i) the standard neurovascular model in which the EEG precedes fMRI signals is certainly not always valid; (ii) time constants of HRFs may differ significantly from those of the standard responses. Using a specific optimization procedure of the HRF, we significantly increased the size of networks whose activity was correlated with IEDs, whereas the size of functional networks in healthy subjects did not change much. This demonstrates the large variability of hemodynamics associated to IEDs, the reasons of which remain largely unknown. Further investigations are needed to improve our insights into the hemodynamics of the epileptic brain.
Footnotes
To increase the number of degrees of freedom, a singular value decomposition was applied to the design matrix. Only the first components explaining 90% of the variance were kept.
REFERENCES
- Aghakhani Y, Kobayashi E, Bagshaw AP, Hawco C, Benar CG, Dubeau F, Gotman J ( 2006): Cortical and thalamic fMRI responses in partial epilepsy with focal and bilateral synchronous spikes. Clin Neurophysiol 117: 177–91. [DOI] [PubMed] [Google Scholar]
- Aguirre GK, Zarahn E, D'Esposito M ( 1998): The variability of human, BOLD hemodynamic responses. Neuroimage 8: 360–369. [DOI] [PubMed] [Google Scholar]
- Al‐Asmi A, Benar CG, Gross DW, Khani YA, Andermann F, Pike B, Dubeau F, Gotman J ( 2003): fMRI activation in continuous and spike‐triggered EEG‐fMRI studies of epileptic spikes. Epilepsia 44: 1328–1339. [DOI] [PubMed] [Google Scholar]
- Bagshaw AP, Aghakhani Y, Benar CG, Kobayashi E, Hawco C, Dubeau F, Pike GB, Gotman J ( 2004): EEG‐fMRI of focal epileptic spikes: Analysis with multiple haemodynamic functions and comparison with gadolinium‐enhanced MR angiograms. Hum Brain Mapp 22: 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudewig J, Bittermann HJ, Paulus W, Frahm J ( 2001): Simultaneous EEG and functional MRI of epileptic activity: A case report. Clin Neurophysiol 112: 1196–1200. [DOI] [PubMed] [Google Scholar]
- Baumgartner C, Serles W, Leutmezer F, Pataraia E, Aull S, Czech T, Pietrzyk U, Relic A, Podreka I ( 1998): Preictal SPECT in temporal lobe epilepsy: Regional cerebral blood flow is increased prior to electroencephalography‐seizure onset. J Nucl Med 39: 978–982. [PubMed] [Google Scholar]
- Benar CG, Gross DW, Wang Y, Petre V, Pike B, Dubeau F, Gotman J ( 2002): The BOLD response to interictal epileptiform discharges. Neuroimage 17: 1182–1192. [DOI] [PubMed] [Google Scholar]
- Boynton GM, Engel SA, Glover GH, Heeger DJ ( 1996): Linear systems analysis of functional magnetic resonance imaging in human V1. J Neurosci 16: 4207–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL ( 1998): Event‐related fMRI and the hemodynamic response. Hum Brain Mapp 6: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O, Guillemain I, Saillet S, Reyt S, Deransart C, Segebarth C, Depaulis A ( 2008): Identifying neural drivers with functional MRI: An electrophysiological validation. PLoS Biol 6: 2683–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole JS ( 1997): Defining epileptogenic foci: Past, present, future. J Clin Neurophysiol 14: 470–483. [DOI] [PubMed] [Google Scholar]
- Federico P, Abbott DF, Briellmann RS, Harvey AS, Jackson GD ( 2005): Functional MRI of the pre‐ictal state. Brain 128: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Fletcher P, Josephs O, Holmes A, Rugg MD, Turner R ( 1998): Event‐related fMRI: Characterizing differential responses. Neuroimage 7: 30–40. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Jezzard P, Turner R ( 1994): Analysis of functional MRI time‐series. Hum Brain Mapp 1: 153–171. [Google Scholar]
- Friston KJ, Mechelli A, Turner R, Price CJ ( 2000): Nonlinear responses in fMRI: The Balloon model, Volterra kernels, and other hemodynamics. Neuroimage 12: 466–477. [DOI] [PubMed] [Google Scholar]
- sFriston KJ, Zarahn E, Josephs O, Henson RN, Dale AM ( 1999): Stochastic designs in event‐related fMRI. Neuroimage 10: 607–619. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T ( 2002): Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15: 870–878. [DOI] [PubMed] [Google Scholar]
- Glover GH ( 1999): Deconvolution of impulse response in event‐related BOLD fMRI. Neuroimage 9: 416–429. [DOI] [PubMed] [Google Scholar]
- Golby AJ, Poldrack RA, Illes J, Chen D, Desmond JE, Gabrieli JD ( 2002): Memory lateralization in medial temporal lobe epilepsy assessed by functional MRI. Epilepsia 43: 855–863. [DOI] [PubMed] [Google Scholar]
- Gotman J, Benar CG, Dubeau F ( 2004): Combining EEG and FMRI in epilepsy: methodological challenges and clinical results. J Clin Neurophysiol 21: 229–240. [DOI] [PubMed] [Google Scholar]
- Goutte C, Nielsen FA, Hansen LK ( 2000): Modeling the haemodynamic response in fMRI using smooth FIR filters. IEEE Trans Med Imaging 19: 1188–1201. [DOI] [PubMed] [Google Scholar]
- Grouiller F, Vercueil L, Krainik A, Segebarth C, Kahane P, David O ( 2007): A comparative study of different artefact removal algorithms for EEG signals acquired during functional MRI. Neuroimage 38: 124–137. [DOI] [PubMed] [Google Scholar]
- Hamandi K, Salek‐Haddadi A, Fish DR, Lemieux L ( 2004): EEG/functional MRI in epilepsy: The Queen Square Experience. J Clin Neurophysiol 21: 241–248. [DOI] [PubMed] [Google Scholar]
- Hamandi K, Salek‐Haddadi A, Laufs H, Liston A, Friston K, Fish DR, Duncan JS, Lemieux L. ( 2006): EEG‐fMRI of idiopathic and secondarily generalized epilepsies. Neuroimage 31: 1700–1710. [DOI] [PubMed] [Google Scholar]
- Handwerker DA, Ollinger JM, D'Esposito M ( 2004): Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage 21: 1639–1651. [DOI] [PubMed] [Google Scholar]
- Hawco CS, Bagshaw AP, Lu Y, Dubeau F, Gotman J ( 2007): BOLD changes occur prior to epileptic spikes seen on scalp EEG. Neuroimage 35: 1450–1458. [DOI] [PubMed] [Google Scholar]
- Jager L, Werhahn KJ, Hoffmann A, Berthold S, Scholz V, Weber J, Noachtar S, Reiser M ( 2002): Focal epileptiform activity in the brain: Detection with spike‐related functional MR imaging‐‐preliminary results. Radiology 223: 860–869. [DOI] [PubMed] [Google Scholar]
- Josephs O, Turner R, Friston KJ ( 1997): Event‐related fMRI. Hum Brain Mapp 5: 243–248. [DOI] [PubMed] [Google Scholar]
- Kang JK, Benar C, Al‐Asmi A, Khani YA, Pike GB, Dubeau F, Gotman J ( 2003): Using patient‐specific hemodynamic response functions in combined EEG‐fMRI studies in epilepsy. Neuroimage 20: 1162–1170. [DOI] [PubMed] [Google Scholar]
- Kershaw J, Ardekani BA, Kanno I ( 1999): Application of Bayesian inference to fMRI data analysis. IEEE Trans Med Imaging 18: 1138–1153. [DOI] [PubMed] [Google Scholar]
- Kida I, Smith AJ, Blumenfeld H, Behar KL, Hyder F ( 2006): Lamotrigine suppresses neurophysiological responses to somatosensory stimulation in the rodent. Neuroimage 29: 216–224. [DOI] [PubMed] [Google Scholar]
- Krakow K, Woermann FG, Symms MR, Allen PJ, Lemieux L, Barker GJ, Duncan JS, Fish DR ( 1999): EEG‐triggered functional MRI of interictal epileptiform activity in patients with partial seizures. Brain 122: 1679–1688. [DOI] [PubMed] [Google Scholar]
- Kruger G, Kleinschmidt A, Frahm J ( 1996): Dynamic MRI sensitized to cerebral blood oxygenation and flow during sustained activation of human visual cortex. Magn Reson Med 35: 797–800. [DOI] [PubMed] [Google Scholar]
- Kwong KK, Belliveau JW, Chesler DA, Goldberg IE, Weisskoff RM, Poncelet BP, Kennedy DN, Hoppel BE, Cohen MS, Turner R, Cheng H‐M, Brady TJ, Rosen BR ( 1992): Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci USA 89: 5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange N, Zeger SL ( 1997): Non‐linear Fourier time series analysis for human brain mapping by functional magnetic resonance imaging (with discussion). J Royal Stat Soc Appl Stat 46: 1–29. [Google Scholar]
- Laufs H, Lengler U, Hamandi K, Kleinschmidt A, Krakow K ( 2006): Linking generalized spike‐and‐wave discharges and resting state brain activity by using EEG/fMRI in a patient with absence seizures. Epilepsia 47: 444–448. [DOI] [PubMed] [Google Scholar]
- Lazeyras F, Blanke O, Perrig S, Zimine I, Golay X, Delavelle J, Michel CM, de Tribolet N, Villemure JG, Seeck M ( 2000): EEG‐triggered functional MRI in patients with pharmacoresistant epilepsy. J Magn Reson Imaging 12: 177–185. [DOI] [PubMed] [Google Scholar]
- Lemieux L, Laufs H, Carmichael D, Paul JS, Walker MC, Duncan JS ( 2008): Noncanonical spike‐related BOLD responses in focal epilepsy. Hum Brain Mapp 29: 329–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Bagshaw AP, Grova C, Kobayashi E, Dubeau F, Gotman J ( 2006): Using voxel‐specific hemodynamic response function in EEG‐fMRI data analysis. Neuroimage 32: 238–247. [DOI] [PubMed] [Google Scholar]
- Makiranta M, Ruohonen J, Suominen K, Niinimaki J, Sonkajarvi E, Kiviniemi V, Seppanen T, Alahuhta S, Jantti V, Tervonen O ( 2005): BOLD signal increase preceeds EEG spike activity‐‐a dynamic penicillin induced focal epilepsy in deep anesthesia. Neuroimage 27: 715–724. [DOI] [PubMed] [Google Scholar]
- Marrelec G, Benali H, Ciuciu P, Pelegrini‐Issac M, Poline JB ( 2003): Robust Bayesian estimation of the hemodynamic response function in event‐related BOLD fMRI using basic physiological information. Hum Brain Mapp 19: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz MM, Neumann J, Muller K, Zysset S ( 2006): Variability of the BOLD response over time: An examination of within‐session differences. Neuroimage 32: 1185–1194. [DOI] [PubMed] [Google Scholar]
- Miezin FM, Maccotta L, Ollinger JM, Petersen SE, Buckner RL ( 2000): Characterizing the hemodynamic response: Effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. Neuroimage 11: 735–759. [DOI] [PubMed] [Google Scholar]
- Moeller F, Siebner HR, Wolff S, Muhle H, Granert O, Jansen O, Stephani U, Siniatchkin M ( 2008): Simultaneous EEG‐fMRI in drug‐naive children with newly diagnosed absence epilepsy. Epilepsia 49: 1510–1519. [DOI] [PubMed] [Google Scholar]
- Nayak D, Valentin A, Alarcon G, Garcia Seoane JJ, Brunnhuber F, Juler J, Polkey CE, Binnie CD ( 2004): Characteristics of scalp electrical fields associated with deep medial temporal epileptiform discharges. Clin Neurophysiol 115: 1423–1435. [DOI] [PubMed] [Google Scholar]
- Neumann J, Lohmann G, Zysset S, von Cramon DY ( 2003): Within‐subject variability of BOLD response dynamics. Neuroimage 19: 784–796. [DOI] [PubMed] [Google Scholar]
- Niazy RK, Beckmann CF, Iannetti GD, Brady JM, Smith SM ( 2005): Removal of FMRI environment artifacts from EEG data using optimal basis sets. Neuroimage 28: 720–737. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K ( 1992): Intrinsic signal changes accompanying sensory stimulation: Functional brain mapping with magnetic resonance imaging. Proc Natl Acad Sci USA 89: 5951–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Blum A, Pearlman JD, Yousuf N, Ives JR, Saeteng S, Schomer DL, Edelman RR ( 1999): Echo‐planar functional MR imaging of epilepsy with concurrent EEG monitoring. AJNR Am J Neuroradiol 20: 1916–1919. [PMC free article] [PubMed] [Google Scholar]
- Rajapakse JC, Kruggel F, Maisog JM, von Cramon DY ( 1998): Modeling hemodynamic response for analysis of functional MRI time‐series. Hum Brain Mapp 6: 283–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigau V, Morin M, Rousset MC, de Bock F, Lebrun A, Coubes P, Picot MC, Baldy‐Moulinier M, Bockaert J, Crespel A, Lerner‐Natoli M ( 2007): Angiogenesis is associated with blood‐brain barrier permeability in temporal lobe epilepsy. Brain 130: 1942–1956. [DOI] [PubMed] [Google Scholar]
- Rosenow F, Luders H ( 2001): Presurgical evaluation of epilepsy. Brain 124: 1683–1700. [DOI] [PubMed] [Google Scholar]
- Salek‐Haddadi A, Diehl B, Hamandi K, Merschhemke M, Liston A, Friston K, Duncan JS, Fish DR, Lemieux L ( 2006): Hemodynamic correlates of epileptiform discharges: An EEG‐fMRI study of 63 patients with focal epilepsy. Brain Res 1088: 148–166. [DOI] [PubMed] [Google Scholar]
- Salek‐Haddadi A, Friston KJ, Lemieux L, Fish DR ( 2003a): Studying spontaneous EEG activity with fMRI. Brain Res Brain Res Rev 43: 110–133. [DOI] [PubMed] [Google Scholar]
- Salek‐Haddadi A, Lemieux L, Merschhemke M, Friston KJ, Duncan JS, Fish DR ( 2003b): Functional magnetic resonance imaging of human absence seizures. Ann Neurol 53: 663–667. [DOI] [PubMed] [Google Scholar]
- Schwartz TH ( 2007): Neurovascular coupling and epilepsy: Hemodynamic markers for localizing and predicting seizure onset. Epilepsy Curr 7: 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeck M, Lazeyras F, Michel CM, Blanke O, Gericke CA, Ives J, Delavelle J, Golay X, Haenggeli CA, de Tribolet N, Landis T ( 1998): Non‐invasive epileptic focus localization using EEG‐triggered functional MRI and electromagnetic tomography. Electroencephalogr Clin Neurophysiol 106: 508–512. [DOI] [PubMed] [Google Scholar]
- Takano T, Tian GF, Peng W, Lou N, Libionka W, Han X, Nedergaard M ( 2006): Astrocyte‐mediated control of cerebral blood flow. Nat Neurosci 9: 260–267. [DOI] [PubMed] [Google Scholar]
- Tao JX, Ray A, Hawes‐Ebersole S, Ebersole JS ( 2005): Intracranial EEG substrates of scalp EEG interictal spikes. Epilepsia 46: 669–676. [DOI] [PubMed] [Google Scholar]
- Tian GF, Azmi H, Takano T, Xu Q, Peng W, Lin J, Oberheim N, Lou N, Wang X, Zielke HR, Kang J, Nedergaard M ( 2005): An astrocytic basis of epilepsy. Nat Med 11: 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyvaert L, Levan P, Grova C, Dubeau F, Gotman J ( 2008): Effects of fluctuating physiological rhythms during prolonged EEG‐fMRI studies. Clin Neurophysiol 119: 2762–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warach S, Ives JR, Schlaug G, Patel MR, Darby DG, Thangaraj V, Edelman RR, Schomer DL ( 1996): EEG‐triggered echo‐planar functional MRI in epilepsy. Neurology 47: 89–93. [DOI] [PubMed] [Google Scholar]
- Weinand ME, Carter LP, el‐Saadany WF, Sioutos PJ, Labiner DM, Oommen KJ ( 1997): Cerebral blood flow and temporal lobe epileptogenicity. J Neurosurg 86: 226–232. [DOI] [PubMed] [Google Scholar]


