Abstract
The objective of this study was to characterize the sensitivity of voxel‐based morphometry (VBM) results to choice field strength. We chose to investigate the two most widespread acquisition sequences for VBM, FLASH and MP‐RAGE, at 1.5 and 3 T. We first evaluated image quality of the four acquisition protocols in terms of SNR and image uniformity. We then performed a VBM study on eight subjects scanned twice using the four protocols to evaluate differences in grey matter (GM) density and corresponding scan‐rescan variability, and a power analysis for each protocol in the context a longitudinal and cross‐sectional VBM study. As expected, the SNR increased significantly at 3 T for both FLASH and MP‐RAGE. Image non‐uniformity increased as well, in particular for MP‐RAGE. The differences in CNR and contrast non‐uniformity cause regional biases between protocols in the VBM results, in particular between sequences at 3 T. The power analysis results show an overall decrease in the number of subjects required in a longitudinal study to detect a difference in GM density at 3 T for MP‐RAGE, but an increase for FLASH. The number of subjects required in a cross‐sectional VBM study is higher at 3 T for both sequences. Our results show that each protocol has a distinct regional sensitivity pattern to morphometric change, which goes against the classical view of VBM as an unbiased whole brain analysis technique, complicates the combination of data within a VBM study and the direct comparison of VBM studies based on different protocols. Hum Brain Mapp, 2010. © 2009 Wiley‐Liss, Inc.
Keywords: magnetic resonance imaging, neuroimaging, morphology, voxel‐based morphometry, field strength comparison
INTRODUCTION
Although high‐field (≥3 Tesla) magnetic resonance imaging (MRI) is attractive due to the increase in sensitivity, technical issues originally limited its use in automated whole‐brain morphometric analysis techniques such as voxel‐based morphometry. The gain in equilibrium magnetization allows for more flexibility in the acquisition process. The SNR can be traded off to vary contrast characteristics and/or increase spatial resolution, for instance. However, high field imaging also brings new technical challenges including an increase in transmission field inhomogeneity and modified relaxation properties.
Voxel‐based morphometry (VBM) [Ashburner and Friston2000; Wright et al.,1995] is a whole brain analysis technique that detects local differences in tissue composition across subjects once gross anatomical differences have been accounted for by linear or non‐linear registration to a model. The images are pre‐processed to reduce acquisition artifacts and improve sensitivity to biological differences. The stability of the morphometric analysis process, over time and, if applicable, across sites, is crucial in such studies to enhance the statistical power of results and reduce the number of subjects required. The accuracy of the morphometric results is equally important, as there is questionable value in using a consistent measure of morphometric difference that is not related to true anatomical variation.
The accuracy and precision of VBM relies on the quality of the input images and the image processing algorithms used. An important feature of VBM analysis is that it is not biased to a particular region or structure of the brain. Likewise, the anatomical images should not be biased by regional variations in signal‐to‐noise (SNR) and contrast‐to‐noise (CNR), or by imaging artifacts. These regional biases, whether caused by transmission inhomogeneity, the contrast sensitivity of the acquisition protocol or natural biological variation in the MR properties of brain tissues, could lead to regional variations in the accuracy and precision of the VBM results. Traditionally, VBM has been performed at 1.5 Tesla (T). However, the prevalence of 3 T scanners, which provide an approximate two‐fold increase in intrinsic SNR over 1.5 T, have resulted in an increased interest in and use of 3 T for VBM studies. The increase in transmission non‐uniformity and T1 relaxation times at 3 T may affect accuracy and/or deteriorate local reproducibility; thus counteracting the potential gains from an increase in SNR.
There have been very few studies examining the impact of MRI acquisition protocols at different field strengths on morphometric analysis methods [Briellmann et al.,2001; Scorzin et al.,2008; Stankiewicz et al.,2009; Vidal et al.,2008]; none to the best of our knowledge from the specific perspective of VBM. Han et al. studied the effect of field strength on the reliability of cortical thickness measurements [Han et al.,2006]. Dickerson et al. performed a similar reliability study on the detection of cortical thickness correlates of cognitive performance [Dickerson et al.,2008]. However, neither of these studies analyzed the scan‐rescan reproducibility of the morphometric analysis at both field strengths.
The purpose of this paper is to characterize the sensitivity of VBM (as it is commonly performed) to choice of field strength. We chose the two most widespread T1‐weighted sequences for VBM: FLASH (Fast Low Angle Shot) [Frahm et al.,1986; Haase1990] and MP‐RAGE (Magnetization Prepared Rapid Acquisition by Gradient Echo) [Bluml et al.,1996; Deichmann et al.,2000; Epstein et al.,1994; Mugler and Brookeman,1990,1991; Runge et al.,1991]. Our objective was not to optimize acquisition protocols; rather, we chose to compare the leading acquisition protocols at 3 T to published protocols for VBM at 1.5 T. We first evaluated these protocols in terms of basic image quality metrics, including SNR and image uniformity, with a phantom study. To make a fair comparison between sequences and field strengths, the scan time, field‐of‐view and resolution were matched. Next, we investigated the sensitivity of VBM results to field strength to assess whether there is an advantage in performing VBM at 3 T as opposed to 1.5 T, and to assess the extent to which existing studies at both field strengths can be compared to each other. We also investigated the sensitivity of VBM to choice of pulse sequence, FLASH or MP‐RAGE, to determine whether differences between sequences were consistent across field strengths. The data set acquired consists of eight healthy volunteers scanned twice at 1.5 T and 3 T using the two sequences. The data were analyzed using published image processing tools and a probabilistic neuro‐anatomical atlas, developed at the Montreal Neurological Institute. Finally, to place these results into the context of a VBM study, we performed a power analysis to compare the population sample size required to detect a predefined difference in GM density at 1.5 T and 3 T, for both FLASH and MP‐RAGE. We also performed a power analysis based on the variance across field‐strengths to simulate a multi‐center study where the data may be pooled from different field strengths.
METHODS
Phantom Design
A uniform elliptical cylinder phantom with similar dimensions, electrical properties and relaxation times as the human brain was designed to study the B1 transmission field and the resulting image non‐uniformity [Sled and Pike,1998]. The phantom solution consists of distilled H2O, 77 mM NaCl to adjust the conductivity to σ ≈ 0.6 S/m, and 103 μM MnCl2 to modify the relaxation time to 822 ± 44 ms at 1.5 T and 879 ± 27 ms at 3 T, estimated using DESPOT1 [Deoni et al.,2003] with B1 field correction. The permittivity of brain tissues at 64 MHz (εr ≈ 88) and 128 MHz (εr ≈ 70) is comparable with that of water (εr ≈ 80) [Chen et al.,1998].
The ΔB1 field map was computed from two slice‐selective magnetization‐prepared turbo spin echo (TSE) acquisitions [Sled and Pike,2000]. The preparation of the first acquisition consists of a non‐selective flip angle α and time delay τ/2 followed by a TSE readout with echo spacing τ. The acquisition is repeated with a magnetization preparation of 2α. The ΔB1 field map is computed using the following equation,
where αnom is the nominal flip angle, αreal is the real flip angle and y = I 2/I 1 is the ratio of signal intensities. An echo spacing of 11 ms and repetition time of 2 s was used. The flip angle αnom was 33° at 1.5 T and 20° at 3 T.
As shown in Figure 1, the elliptical phantom ΔB1 field map has a similar range and shape as that of a human brain at both field strengths.
Figure 1.
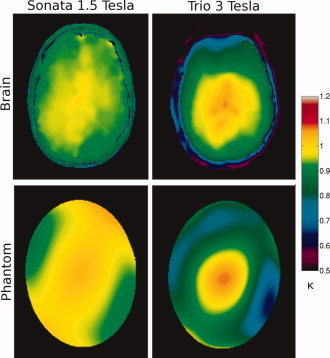
Transmission field inhomogeneity maps, where κ = B 1measured/B 1nominal, of a volunteer (top) and the elliptical uniformity phantom (bottom) at 1.5 T (left) and 3 T (right).
Subjects
Eight healthy young volunteers (mean age 28 years, SD 8 years; 1 male; all right handed) were scanned twice at both field strengths. Informed consent was obtained from all subjects who participated in this study, which was approved by the Research Ethics Board of the Montreal Neurological Institute and Hospital.
Acquisition Protocols
Images were acquired on a MAGNETOM Sonata 1.5 T and Trio 3 T MR imaging system with the Syngo 2004A VA25 software platform (Siemens Medical Systems, Erlangen, Germany). For the phantom uniformity study, the circularly polarized transmit/receive head coil was used to minimize the non‐uniformity of the reception field. For the human imaging study, 8‐channel receive‐only head coils were used to increase the SNR and CNR. The eight subjects were scanned twice using FLASH and MP‐RAGE at both field strengths, for a total of eight acquisitions per subject. Scans at the same field strength took place within a two‐week period; the scans at 1.5 T were scheduled about three months after those at 3 T. The sequence order was reversed between scanning sessions to minimize potential systematic bias such as subject motion. Acquisitions were sagittal with a field‐of‐view of 256 × 224 × 176 mm3 and 1 mm isotropic resolution.
1.5 Tesla
The acquisition protocols at 1.5 T are published standards for VBM studies. The FLASH acquisition protocol corresponds to the ICBM (International Consortium for Brain M) protocol [Mazziotta et al.,2001], with parameters α/TE/TR of 30°/8 ms/25 ms. The MP‐RAGE acquisition protocol corresponds to the ADNI (Alzheimer's Disease Neuroimaging Initiative) protocol [Mueller et al.,2005], with parameters TI/α/TE/TEcho Spacing/TR set to 1,000 ms/8°/3.8 ms/9.3 ms/2,400 ms, and partition oversampling of 20%. The bandwidths were matched at 180 Hz/pixel to minimize differences in geometric distortions. The scan times are equal to 16:27 and 8:59 (min:sec) for FLASH and MP‐RAGE, respectively. Even though the scan time of the FLASH ICBM protocol is considerably longer than the 1.5 T MP‐RAGE protocol and both 3 T protocols described below, we chose to not alter the protocol as it was used in many published VBM studies and serves as an important benchmark for comparison. Apart from the 1.5 T FLASH protocol, all scan times were matched to ∼9.5 min to provide a fair comparison.
3 Tesla
To reduce the scan time of the 3 T FLASH protocol to ∼9.5 min, the repetition time TR was set to 19 ms and elliptical k‐space coverage was implemented. The acquisition parameters α/TE were set to 18°/5.67 ms to maintain a maximal WM‐GM contrast. The MP‐RAGE protocol was adapted from the 3 T ADNI protocol, with TI/α/TE/TEcho Spacing/TR set to 960 ms/9°/4.19 ms/9.9 ms/2,420 ms. Minor modifications were made to the ADNI MP‐RAGE protocol in order to match the acquisition bandwidths and scan times. A comparison of the modified and original MP‐RAGE protocols for a human subject revealed changes in SNR and CNR efficiency of +36% and +2%. The bandwidths were matched at 150 Hz/pixel; and the scan times are 9:49 and 9:04 for FLASH and MP‐RAGE, respectively.
Image Processing
The images of the eight subjects were corrected for intensity non‐uniformity using N3 (Non‐parametric Non‐uniform intensity Normalization) [Sled et al.,1998], with smoothing distance of 200 mm, and linearly registered using ANIMAL (Automated Non‐linear Image Matching and Anatomical Labeling) [Collins et al.,1994,1995] to the ICBM 152 non‐linear symmetric brain template [Mazziotta et al.,2001]. This neuro‐anatomical atlas consists of 152 T1‐weighted whole brain volume images that have been independently non‐linearly registered to stereotaxic space and averaged. Finally, using BET (Brain Extraction Tool) [Smith,2002] from the FSL tools [Smith et al.,2004], with fractional intensity threshold of 0.5, the images were masked to remove all non‐brain voxels. N3 was applied a second time, to the skull‐stripped images in stereotaxic space at both field strengths. Because of the increase in image non‐uniformity at 3 T, the spline smoothing distance of N3 was reduced to 50 mm, as suggested by [Boyes et al.,2008]. Tissue maps were created in a two step process. First, discrete tissue maps were created from these pre‐processed images using INSECT (Intensity Normalized Stereotaxic Environment for Classification of Tissues) [Cocosco et al.,2003], where each image voxel was classified as white matter, grey matter, cerebral‐spinal fluid, or non‐brain. Second, the binary GM map is then corrected for partial volume effects [Tohka et al.,2004] to create a final volume that is continuously classified. The value at each voxel indicates the estimated percentage of GM.
Although the MNI image processing tools are widely used in morphometric studies, to ensure the widest applicability of our results, we also processed the data using the unified segmentation approach (with default settings) of SPM5 [Ashburner and Friston,2005], the latest version of Statistical Parametric Mapping, a Matlab software package for analyzing neuroimaging data (available from http://www.fil.ion.ucl.ac.uk/spm).
Statistical Analysis
We performed a two‐tailed paired‐sample t‐test to look at voxel‐wise differences in GM tissue density between the FLASH and MP‐RAGE images at both field strengths, as well as between field strengths for each sequence. Prior to statistical analysis, the partial volume corrected GM tissue maps were smoothed by an 8‐mm isotropic full‐width at half the maximum Gaussian kernel. The differences at a level of significance of α = 0.05, for both local maxima and clusters, were corrected for multiple dependent comparisons using the most limiting of Bonferroni correction, Gaussian random field theory and discrete local maxima analyzes [Worsley et al.,1996]. We also evaluated differences in GM tissue density variability, where the GM variability maps were estimated as the absolute difference between the scan and rescan GM percentage maps. As with the GM tissue maps, the GM variability maps were smoothed prior to statistical analysis.
The population sample size required to detect a predefined change in GM density is a function of the total variance in GM density within the populations. For longitudinal VBM, the sample variance was estimated as the variance of the eight difference images between scan‐rescan blurred GM maps for each acquisition protocol. This variance includes measurement error and the biological variance that may occur over time and which may be of interested, such as GM atrophy. For cross‐sectional VBM, the sample variance was estimated as the variance of the eight blurred GM maps for each acquisition protocol, which includes the measurement error as well as the biological variability in GM morphology within the population.
We performed a voxel‐wise power analysis to determine how many subjects N would be required to detect an absolute difference in GM density of 0.05 within a voxel for a level of significance of α = 0.05 (one‐sided) and statistical power of 1‐β = 0.90 [Cohen,1988]. The power analysis was performed for the four acquisition protocols individually and in groups to simulate a multi‐center study where images acquired using different protocols are pooled together. The resulting N‐maps were masked by the average of all blurred GM maps, where GM represented at least 25% of the voxel volume, in order to include only those voxels where a change in GM density is likely to occur. Quartiles of the whole masked N‐maps were evaluated.
RESULTS
Image Quality
The elliptical uniformity phantom was scanned at both field strengths to evaluate the image intensity non‐uniformity of the four imaging protocols over the range of transmission field inhomogeneity expected in the human brain. Image uniformity was quantified by the standard deviation σ and signal range of the elliptical phantom images, which have been Normalized by the mean signal intensity. The uniformity results listed in Table I indicate that FLASH and MP‐RAGE image intensity non‐uniformity increases with field strength due to the increase in transmission field inhomogeneity. The standard deviation of the Normalized phantom images is higher for MP‐RAGE than for FLASH at both field strengths. The increase in image non‐uniformity from 1.5 to 3 T is also higher for this MP‐RAGE protocol, which is more sensitive than FLASH to variations in the RF transmission field. FLASH signal standard deviation increased by 92.9% in response to an increase of 123.8% in transmission field (B1) inhomogeneity from 1.5 to 3 T, whereas the MP‐RAGE signal standard deviation increased by 192.1%.
Table I.
Uniformity and signal‐to‐noise ratio (SNR) results from the elliptical uniformity phantom scanned at both field strengths, including the standard deviation σ and range of the ΔB1 map and Normalized FLASH and MP‐RAGE images
| Sequence | σ | Min | Max | SNR (/√time) |
|---|---|---|---|---|
| 1.5 Tesla | ||||
| ΔB1 map | 0.042 | 0.879 | 1.075 | — |
| FLASH | 0.070 | 0.635 | 1.248 | 35.27 (1.12) |
| MP‐RAGE | 0.076 | 0.606 | 1.349 | 19.34 (0.83) |
| 3.0 Tesla | ||||
| ΔB1 map | 0.094 | 0.537 | 1.107 | — |
| FLASH | 0.135 | 0.502 | 1.465 | 91.46 (3.77) |
| MP‐RAGE | 0.222 | 0.410 | 1.630 | 48.51 (2.08) |
The elliptical uniformity phantom was scanned using a CP transmit/receive head coil, which has a more uniform reception field than the phased array reception coil used for the human subjects, and a less uniform transmission field than the body coil. Because the transmission field inhomogeneity increase from 1.5 to 3 T is mainly due to the decrease in RF wavelength in comparison to the size of the human head, we don't think our choice of transmission coil will significantly affect our results.
Absolute SNR and SNR efficiency measurements, i.e. divided by the square root of the scan time, of the elliptical phantom images are also listed in Table I. The phantom was segmented from the background noise in the axial slice matching the 2D ΔB1 map, and the average signal intensity was used for the region of interest SNR measurements. Given that equilibrium magnetization is approximately proportional to the magnetic field strength, the SNR of FLASH and MP‐RAGE is significantly higher at 3 T than at 1.5 T. The sequence acquisition parameters and phantom T1 values are not equivalent at both field strengths, and thus the SNR is not exactly doubled in this case. The FLASH protocols produce a significantly higher SNR, and SNR efficiency, than the MP‐RAGE protocols at both field strengths. Recall that at 1.5 T, the FLASH scan time is approximately 15:00, much longer than the average scan time of the three other protocols of 9:30. MP‐RAGE is characterized by a higher CNR efficiency, particularly at the cortex [Tardif et al.,2009]. MP‐RAGE is thus often preferred over FLASH for studies that require tissue classification, particularly when the low SNR efficiency is compensated for by the use of high field strengths and multi‐channel receiver coils.
Four sample images of the same subject acquired with each protocol at 1.5 and 3 T are presented, along with their corresponding grey matter maps, in Figures 2 and 3. These images have been non‐uniformity corrected and registered to stereotaxic space. The improvement in WM‐GM contrast for MP‐RAGE at 3 T is clearly visible, particularly at the motor cortex in Figure 2 and the occipital cortex in Figure 3. A recent study of ours includes a more complete assessment of image quality of three T1‐weighted acquisition protocols at 3 T, as well as their impact on VBM results [Tardif et al.,2009].
Figure 2.
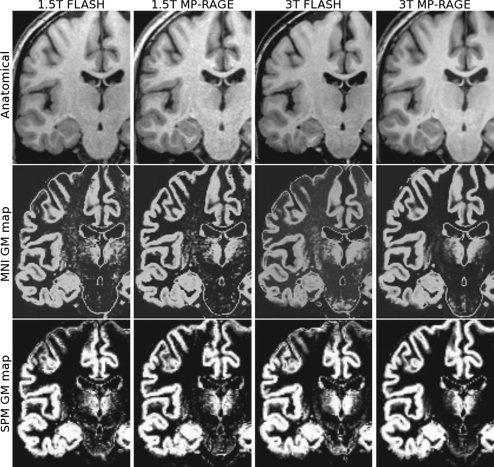
Coronal T1‐weighted images (top) and corresponding grey matter maps derived using the MNI tools (middle) and the SPM tools (bottom) for the four acquisition protocols. Differences in contrast and tissue classification are visible in the motor cortex.
Figure 3.
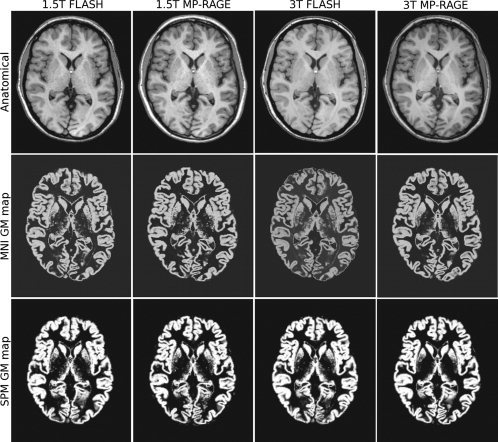
Axial T1‐weighted images (top) and corresponding grey matter maps derived using the MNI tools (middle) and the SPM tools (bottom) for the four acquisition protocols. Differences in contrast and tissue classification are visible in the occipital cortex.
Elliptical k‐space sampling was implemented in the 3 T FLASH protocol to reduce the scan time to more closely match that of the MP‐RAGE protocol, thus impacting the SNR and resolution by removing the contribution of high frequencies in the corners of k‐space. The absolute SNR is increased by ∼10%. The point spread function is widened most at an angle of 45° between the phase‐ and partition‐encode axes, where the full width at half the maximum will be increased by 0.16 mm.
Voxel‐Based Morphometry
GM probability maps, derived using the MNI and SPM image analysis tools, from a representative subject are shown below the corresponding T1‐weighted images in Figures 2 and 3. Differences in tissue classification between sequences and field strengths are clearly visible in the GM maps derived by both MNI and SPM tools, particularly at the motor cortex and occipital cortex. The VBM results related below are based on the MNI tools. However, SPM results are included in the tables, and the main differences between the two analysis tools are highlighted in the Results and Discussion sections.
Grey matter density
The VBM analysis of GM density between field strengths was performed for each sequence independently with the images from the first scanning session, i.e. for N = 8. As seen in Figure 4, FLASH produces clusters of higher GM density estimates at 1.5 T in the left occipital lobe, superior frontal gyrus, frontopolar gyrus, orbital frontal cortex, left precentral gyrus, left calcarine sulcus, inferior temporal gyrus, right lateral orbital gyrus, right middle occipital gyrus, right middle frontal gyrus and right cerebellum. FLASH produces clusters of higher GM density estimates at 3 T in the right cerebellum, right thalamus and left cingulate gyrus.
Figure 4.
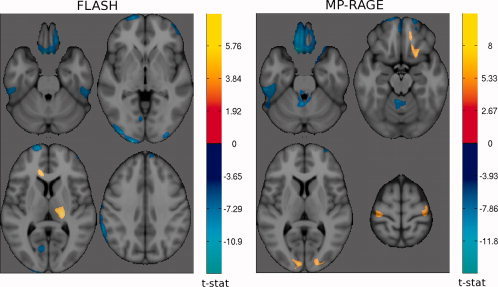
t‐maps of significant differences (α = 0.05) in grey matter density between field strengths for FLASH (left) and MP‐RAGE (right). The cool colormap indicates areas of higher GM density at 1.5 Tesla; and the hot colormap indicates higher GM density at 3 Tesla.
MP‐RAGE produces clusters of higher GM density at 1.5 T in the orbito‐frontal cortex as well, in the left inferior temporal gyrus, the medulla oblongata in the brainstem and the right superior frontal sulcus. MP‐RAGE produces clusters of higher GM density at 3 T in the striate cortex, superior frontal gyrus, right orbital sulcus, precentral gyrus, right medial orbital gyrus and left cerebellum.
We also performed a VBM analyze between FLASH and MP‐RAGE protocols at both field strengths. There is a distinct regional bias in GM density estimates between FLASH and MP‐RAGE, which is consistent across field strengths and more extensive at 3 T. As illustrated in Figures 5 and 6, MP‐RAGE images produce higher GM density estimates than FLASH at both field strengths towards the cortex, in particular the motor and somatosensory cortices, inferior temporal gyri, as well as the cerebellar cortex at both field strengths. At 3 T, MP‐RAGE also produces higher GM density estimates than FLASH in the left occipital lobe, the superior frontal gyri, the right middle frontal gyrus and the right caudate.
Figure 5.
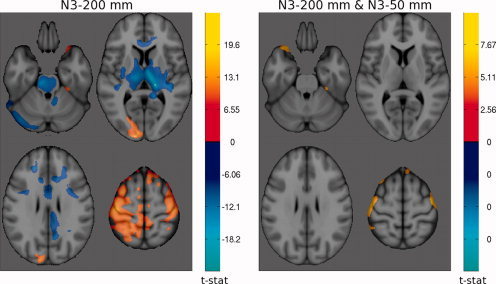
t‐maps of significant differences (α = 0.05) in grey matter density between FLASH and MP‐RAGE at 1.5 Tesla. The cool colormap indicates areas of higher GM density for FLASH; and the hot colormap indicates higher GM density for MP‐RAGE. Left: N3 was applied once with a spline distance of 200 mm. Right: N3 was applied a second time after brain extraction and spatial Normalization with spline distance 50 mm.
Figure 6.
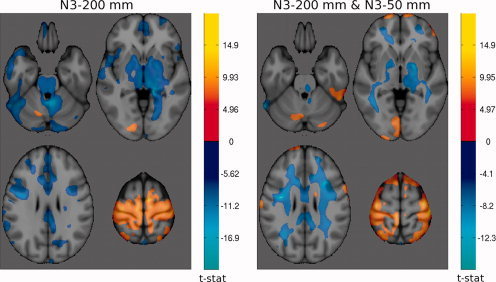
t‐maps of significant differences (α = 0.05) in grey matter density between FLASH and MP‐RAGE at 3 Tesla. The cool colormap indicates areas of higher GM density for FLASH; and the hot colormap indicates higher GM density for MP‐RAGE. Left: N3 was applied once with a spline distance of 200 mm. Right: N3 was applied a second time after brain extraction and spatial Normalization with spline distance 50 mm.
GM density estimates produced by FLASH, on the other hand, are higher in the deep grey matter structures. FLASH images produce higher GM density estimates than MP‐RAGE at both field strengths adjacent to the superior sagittal sinus. At 3 T, FLASH also produces higher GM density estimates than MP‐RAGE in the pons, thalamus, globus palidus, internal capsule, adjacent the optic radiations and along the WM‐GM boundary of the sulci in the frontal lobe. A significant difference in GM density estimates between FLASH and MP‐RAGE was observed in a total volume of 51 453 mm3 at 1.5 T in comparison to a total volume of 319 444 mm3 at 3 T.
As shown by the total volumes of significant differences in GM density listed in Tables II and III, the SPM tools are more sensitive than the MNI tools to differences in GM density between acquisition protocols. The focal anatomical areas of GM density difference are generally consistent across both analysis methods, although the spatial extent varies. The SPM tools produce a more widespread regional bias between field strengths than the MNI tools: for a total of 156 827 mm3 vs. 33 107 mm3 for FLASH, and 242 741 mm3 vs. 33 394 mm3 for MP‐RAGE. The SPM tools also produce a larger regional bias than the MNI tools between FLASH and MP‐RAGE protocols, both at 1.5 T (175 722 mm3 vs. 51 453 mm3) and at 3 T (585 547 mm3 vs. 319 444 mm3).
Table II.
Total volumes (mm3) of significant difference (α = 0.05) in grey matter density and corresponding variability between field strengths for both FLASH and MP‐RAGE sequences
| Sequence | MNI tools | SPM tools | ||||
|---|---|---|---|---|---|---|
| 1.5 T < 3 T | 3 T < 1.5 T | Total | 1.5 T < 3 T | 3 T < 1.5 T | Total | |
| Grey matter density | ||||||
| FLASH | 5,865 | 27,242 | 33,107 | 81,625 | 75,202 | 156,827 |
| MP‐RAGE | 9,586 | 23,808 | 33,394 | 187,310 | 55,431 | 242,741 |
| Grey matter density variability | ||||||
| FLASH | 7,812 | 2,958 | 10,770 | 0 | 0 | 0 |
| MP‐RAGE | 0 | 6,456 | 6,456 | 0 | 719 | 719 |
Table III.
Total volumes (mm3) of significant difference (α = 0.05) in grey matter density variability between FLASH and MP‐RAGE at both field strengths (1.5 and 3 Tesla)
| Field strength | MNI tools | SPM tools | ||||
|---|---|---|---|---|---|---|
| FLASH < MP‐RAGE | MP‐RAGE < FLASH | Total | FLASH < MP‐RAGE | MP‐RAGE < FLASH | Total | |
| Grey matter density | ||||||
| 1.5 Tesla | 50,899 | 554 | 51,453 | 175,722 | 0 | 175,722 |
| 3.0 Tesla | 131,363 | 188,081 | 319,444 | 312,317 | 273,230 | 585,547 |
| Grey matter density variability | ||||||
| 1.5 Tesla | 0 | 3,855 | 3,855 | 1,540 | 573 | 2,113 |
| 3.0 Tesla | 0 | 0 | 0 | 0 | 9,478 | 9,478 |
VBM analysis was used to detect local differences in estimated tissue composition between acquisition protocols. This study does not include a gold standard reference to measure the true accuracy of each protocol. However, as illustrated in Figures 2 and 3, it is visibly clear that the accuracy of segmentation of several anatomical structures improves at 3 T, in particular for the 3 T MP‐RAGE protocol. These areas, which are typically difficult to segment due to low T1 contrast and/or high gyrification, include the motor and somato‐sensory cortices, the occipital cortex, the cerebellar cortex, and the putamen.
Originally, we performed non‐uniformity correction only once, prior to spatial registration, with a smoothing distance of 200 mm. Due to the increase in RF field inhomogeneity and resulting image non‐uniformity at 3 T, we decided to perform non‐uniformity correction a second time at both field strengths, to the skull‐stripped images in stereotaxic space with a shorter smoothing distance of 50 mm. The comparison of the two t‐maps of GM density differences between FLASH and MP‐RAGE protocols at 1.5 T in Figure 5 show the reduction in regional differences due to improved non‐uniformity correction. At 3 T, although the total spatial extent of the differences is similar, the structure of the regional bias has changed as seen in Figure 6.
Grey matter density variability
Absolute scan‐rescan differences between GM maps were analyzed to investigate regional differences in GM density variability. The total volumes of significant differences between field strengths are listed in Table II. FLASH produces clusters of higher GM density variability estimates at 3 T, compared to 1.5 T, in the superior parietal lobules medial to the longitudinal fissure, the angular gyri, and the left precentral gyrus. FLASH produces clusters of higher GM density variability estimates at 1.5 T, compared with 3 T, in the right thalamus and putamen. MP‐RAGE, on the other hand, produces clusters of higher variability estimates at 1.5 T, compared with 3 T, in the left posterior end of the thalamus, the cerebral peduncle, the putamens, and the left cingulated gyrus. MP‐RAGE does not produce areas of higher GM density variability estimates at 3 T.
The total volumes of significant differences between sequences are listed in Table III. MP‐RAGE did not produce any regions of significantly higher GM density variability estimates than FLASH at either field strength. FLASH, however, produces clusters of higher GM density variability than MP‐RAGE at 1.5 T in the left cerebellar cortex.
As for the GM density results, the t‐maps comparing GM density variability estimates derived using the SPM and MNI tools are very similar in structure. However, the volume and location of statistically significant clusters is not consistent for both sets of image analysis tools as seen in Tables II and III. In particular, SPM tools produce fewer differences in GM density variability between field strengths than the MNI tools.
Power analysis
Using the smoothed GM maps of the eight volunteers to estimate the measurement error, we performed a power analysis for each of the four acquisition protocols, i.e. for FLASH and MP‐RAGE at both 1.5 and 3 T. The N‐maps in Figure 7 represent the spatial distribution of the required sample size, i.e. number of subjects per group, to detect a longitudinal difference in GM density of 0.05 within a voxel for P < 0.05 and statistical power of 90%. The N‐maps are inversely proportional to the measurement error, which in this case is mainly due to methodology, i.e. image acquisition and processing techniques. In a typical cross‐sectional VBM study, the anatomical variance within each population, which is usually much higher, should also to be taken into consideration. The resulting required sample size is quite higher as seen in the N‐maps of Figure 8 and the power analysis quartiles in Table IV.
Figure 7.
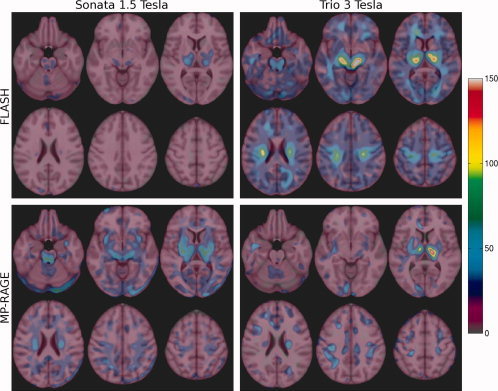
N‐maps for a longitudinal VBM study, where N represents the number of subjects per group required to detect a difference in GM density of 0.05 within a voxel with a level of significance of α = 0.05 and with power 1‐β = 0.90.
Figure 8.
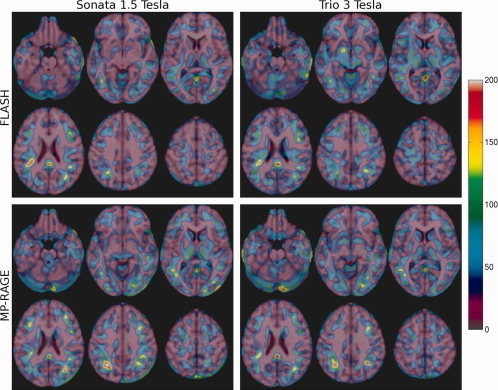
N‐maps for a cross‐sectional VBM study, where N represents the number of subjects per group required to detect a difference in GM density of 0.05 within a voxel with a level of significance of α = 0.05 and with power 1‐β = 0.90.
Table IV.
Quartiles of the number of subjects required to detect a difference of 0.05 in GM density in the brain in the context of a longitudinal and cross‐sectional VBM study using a single acquisition protocol and the MNI analysis tools
| Sequence | 1st quartile | Median | 3rd quartile |
|---|---|---|---|
| Longitudinal | |||
| FLASH 1.5 T | 6 | 8 | 11 |
| MP‐RAGE 1.5 T | 11 | 15 | 20 |
| FLASH 3 T | 16 | 21 | 27 |
| MP‐RAGE 3 T | 7 | 10 | 16 |
| Cross‐sectional | |||
| FLASH 1.5 T | 14 | 24 | 37 |
| MP‐RAGE 1.5 T | 18 | 28 | 45 |
| FLASH 3 T | 21 | 31 | 46 |
| MP‐RAGE 3 T | 19 | 30 | 46 |
Areas of relatively high measurement variance, seen on the longitudinal N‐maps, that are common to all four protocols include the motor and somato‐sensory cortices, occipital and temporal cortices (left in particular), cingulate gyrus, thalamus, basal ganglia, cerebellum and brainstem.
An interesting reversal in performance for FLASH and MP‐RAGE is observed across field strengths, as seen from the longitudinal VBM power analysis quartiles listed in Table IV. The number of subjects required to detect the above‐defined difference in GM density with FLASH is lower at 1.5 T than at 3 T. However, the opposite is true for MP‐RAGE: a lower number of subjects is required at 3 T. These trends were also observed in the SPM power analysis results. The required sample size is lowest for FLASH at 1.5 T (1st quartile/median/3rd quartile = 6/8/11 subjects) followed by MP‐RAGE at 3 T (7/10/16 subjects).
FLASH at 1.5 T requires the smallest sample size to detect a predefined GM difference in a cross‐sectional VBM study, as for longitudinal VBM. In contrast, MP‐RAGE requires more subjects at 3 T than at 1.5 T in a cross‐sectional study.
In multi‐center or long‐term imaging studies, data from different field strengths is often pooled together in the same VBM analysis. We investigated the impact of pooling data acquired with different acquisition protocols in both a longitudinal and cross‐sectional VBM study. The power analysis results in Table V show that the required sample size for a study that combines FLASH images acquired at both field strengths is intermediate to the size required at either field strength. The same is observed for MP‐RAGE. On the basis of the results of Table IV, another option could be to use the sequence with the lowest variance at the respective field strength, i.e. FLASH at 1.5 T and MP‐RAGE at 3 T. However, the required sample size is higher than for each protocol individually.
Table V.
Quartiles of the number of subjects required to detect a difference of 0.05 in GM density in the brain in the context of a multi‐center longitudinal and cross‐sectional VBM study, where images acquired with different protocols are pooled together and analyzed with the MNI tools
| Sequence | 1st quartile | Median | 3rd quartile |
|---|---|---|---|
| Longitudinal | |||
| FLASH 1.5 T – FLASH 3 T | 10 | 13 | 17 |
| MP‐RAGE 1.5 T – MP‐RAGE 3 T | 8 | 12 | 19 |
| FLASH 1.5 T – MP‐RAGE 3 T | 9 | 12 | 18 |
| Cross‐sectional | |||
| FLASH 1.5 T – FLASH 3 T | 17 | 27 | 41 |
| MP‐RAGE 1.5 T – MP‐RAGE 3 T | 18 | 29 | 47 |
| FLASH 1.5 T – MP‐RAGE 3 T | 19 | 31 | 49 |
DISCUSSION
The main objective when designing an acquisition protocol for VBM is to maximise sensitivity to subtle but real change in morphology. The sensitivity of VBM depends on the input image characteristics and their impact on each step of the image processing pipeline, including image registration to an anatomical model, brain extraction, intensity non‐uniformity correction, spatial blurring and tissue classification. The purpose of this study was to evaluate the regional differences in VBM results between widely used acquisition protocols, as well as the reproducibility of results in order to assess the impact of changing field strength from 1.5 to 3 T for VBM studies.
The same eight subjects were scanned twice with four different protocols: FLASH and MP‐RAGE at both 1.5 T and 3 T. While slight variations in brain volumes may be possible due to differing hydration levels or diurnal period, we attempted to scan subjects at the same time of the day, and the maximum amount of time separating two scans of the same healthy young subject is three months. Since VBM highlights subtle yet consistent differences in morphometry, we assume that the differences output by the VBM analysis reflect the differences in the tissue classification results caused by the image characteristics of the different acquisition protocols and are typical of those found in many published VBM studies.
The images were processed using a collection of algorithms described in the Methods section and collectively referred to as the MNI tools, as well as with the latest statistical parametric mapping release (SPM5). A recent study showed that the choice of pre‐processing methods, in particular skull‐stripping and RF bias correction, has a major impact on VBM results [Acosta‐Cabronero et al.,2008]. Therefore, we expected some discrepancies between the VBM results derived using the MNI and SPM tools. Yet, the main significant differences revealed between the four acquisition protocols are consistent across both image processing methods, and thus strengthen our conclusions regarding the impact of moving from 1.5 to 3 T for VBM.
Grey Matter Density
According to the image quality and VBM results shown above, there are advantages and disadvantages of moving to 3 T for VBM. Imaging at 3 T has the advantage of an increase in baseline magnetization, which leads to about a two‐fold increase in SNR. A uniform increase in SNR and CNR is expected to improve the accuracy and reproducibility of tissue classification results, thus hopefully improving the sensitivity of VBM to true morphometric change. For instance, the gain in CNR in the motor cortex for MP‐RAGE at 3 T leads to a, visually, more accurate segmentation of the WM‐GM tissue boundary as shown in Figure 2.
Unfortunately, RF transmission field inhomogeneity also increases at 3 T compared to 1.5 T, enhancing signal and contrast non‐uniformity in the resulting images. The INSECT tissue classification algorithm [Cocosco et al.,2003], of the MNI tools, is based on a threshold operation applied to the whole brain, and is thus sensitive to local variations in signal intensity and contrast. The image is first registered to an atlas with a set of predefined tag points with a high probability of belonging to an assigned tissue class. These tag points initialise the classification, but are then purged to create a customised set of tag points for the image. The final classification, based on the customised tag points, is then performed by an artificial neural network, but is still intensity‐based.
Segmentation using the MNI tools consists of several sequential processing steps. SPM image segmentation, on the other hand, combines tissue classification, image registration and non‐uniformity correction into a single framework [Ashburner and Friston,2005]. The images need to be registered to a tissue probability map, the atlas. The latter represents the prior probability of the tissue classes at each voxel. Using the Bayes rule, the final tissue probability maps are derived from these priors in combination with tissue probabilities derived from the image voxel intensities. Since the classification requires an initial registration to the atlas and, conversely, the registration to the atlas requires an initial classification, these steps are integrated into a unified segmentation approach. SPM also includes non‐linear warping to the atlas, which improves the accuracy of the spatial Normalization and potentially increases the sensitivity to small differences in tissue density.
The inhomogeneity of the reception coil's sensitivity profile causes SNR variations across the image, where the signal is strongest near the coil. The signal intensity variations, which are independent of the tissue properties, can be corrected by a multiplicative field that is estimated from the coil's sensitivity profile during image reconstruction.
The inhomogeneity of the RF transmission field cause both signal and contrast non‐uniformity that depend on the tissue type. The signal non‐uniformity may be compensated for, not corrected, by a smoothly varying field estimated using N3, for instance, with settings optimized for the protocol at hand. Uncorrected signal non‐uniformity will cause stronger regional biases in the tissue classification results, which may reduce the sensitivity of VBM to true morphometric change in some areas. This would go against the classical view of VBM as a whole brain analysis. The contrast non‐uniformity will remain post N3 correction. Discrete tissue classification of large homogeneous structures may not be compromised. However, when the tissue classification algorithm takes partial volume effects into account in order to create a continuously classified volume based on signal intensities, the regional bias due to contrast non‐uniformity will become apparent. The final GM probability associated with a voxel will be a function of CNR, partial volume effects, as well as contrast non‐uniformity due to spatially varying grey matter T1 values and RF transmission field inhomogeneity.
Contrast non‐uniformity causes regional differences in GM density between field strengths and between sequences, particularly at 3 T. The regional differences are more extensive between sequences at 3 T than between field strengths because FLASH and MP‐RAGE each have different contrast non‐uniformity patterns due to their different signal intensity dependence on B1 and T1. Non‐uniformity at 3 T leads to higher contrast towards the centre of the brain for the FLASH protocol in this study, whereas it leads to higher contrast towards the cortex for the MP‐RAGE protocol considered here, given constant T1 times for WM and GM [Tardif et al.,2009].
The 3 T ADNI MP‐RAGE protocol was designed for optimal contrast at the cortex. Although this study does not include a ground truth, it is visually clear from the tissue classification maps that accuracy is improved in several cortical areas. The deep GM‐WM CNR also increases at 3 T; but more importantly, the deep GM‐WM contrast is much lower than cortical GM‐WM contrast at 3 T. This is due to the different T1 times for deep and cortical GM, but also due to the local B1 field. This contrast non‐uniformity at 3 T shifts the tissue boundaries and lowers the GM probability of deep structures, such as the thalamus, for MP‐RAGE in comparison to FLASH. The MP‐RAGE inversion time could be shortened to improve contrast between subcortical structures and minimize differences with FLASH; but contrast non‐uniformity will still remain and affect the GM density distribution.
A recent study showed that using BET in combination with N3 outperforms SPM5 (with default settings) with respect to non‐uniformity correction [Acosta‐Cabronero et al.,2008]. This study suggests that residual non‐uniformity could cause small errors in spatial registration to the atlas, and result in an apparent increase in sensitivity to change in certain areas of the brain. The latter is a plausible explanation for the stronger regional differences estimated with SPM than with MNI tools between field strengths and between sequences at 3 T, particularly for MP‐RAGE since it is less uniform than FLASH. Another possible cause is the use of non‐linear registration in SPM, which will improve the precision of spatial Normalization and thus enhance the sensitivity to tissue density differences between the protocols.
Grey Matter Density Variability and Power Analysis
The regional differences in GM density variability between the four protocols are not very extended, affecting mainly the thalamus and the brainstem where the CNR is low relative to other GM structures, as well as in the cerebellum due to low CNR and the fine cerebellar cortex gyrification. Tissue classification in these areas is thus more sensitive to CNR and to image non‐uniformity, and is more likely to vary as a function of the image acquisition and processing protocols.
An increase in CNR may in some cases cause a spatial shift in the tissue classification variability, since variability usually increases about tissue boundaries. For example, some areas that were consistently misclassified at 1.5 T due to a low CNR, such as the motor and somatosensory cortices, are visually more accurately classified at 3 T. As a result, the region of higher classification variability shifted inward, towards the true WM‐GM tissue boundary. Although the mean variance over the entire motor cortex may not change, due to the increase in accuracy the method may be more sensitive to true anatomical variation in this area.
The GM variability results between field strengths are consistent with the power analysis results for a longitudinal VBM study. For FLASH, more widespread clusters of higher GM density variability were revealed at 3 T. The power analysis results show that the number of subjects required to detect a predefined difference in GM density more than doubles at 3 T for FLASH. On the other hand, MP‐RAGE produces clusters of higher variability at 1.5 T only, and the number of subjects required decreases at 3 T. At 1.5 T, MP‐RAGE has the disadvantage of a lower SNR and CNR, which increases the variance of the tissue classification results. At 3 T however, MP‐RAGE is more efficient than FLASH at producing good T1‐weighted contrast. Unfortunately, the power analysis results show that the variance of the tissue classification results for a sequence does not always decrease with field strength.
The FLASH ICBM protocol at 1.5 T produces the lowest measurement variance in GM density in a longitudinal and cross‐sectional study. This may be because of better registration to the ICBM atlas, due to the exact match in acquisition protocol. The spatial structure of the differences in the longitudinal power analysis, and the VBM results between the FLASH ICBM protocol at 1.5 T and the other protocols investigated, suggest that this is not the main factor contributing to the superior tissue classification reproducibility of the FLASH ICBM protocol. However, precise registration to the atlas could lower the apparent anatomical variance between subjects in a population and decrease the number of subjects required in a cross‐sectional VBM study.
The 3 T MP‐RAGE protocol requires fewer subjects than the 1.5 T MP‐RAGE protocol in a longitudinal VBM study but more in a cross‐sectional study, suggesting that there is a higher anatomical variance between images acquired with MP‐RAGE at 3 T than at 1.5 T. This may either be due to imperfect registration to the atlas, or more likely higher sensitivity to small differences in morphology.
Due to the regional differences in tissue classification between acquisition protocols, combining data acquired with different protocols within the same VBM study is generally not recommended, as it will add an additional source of variance to the study. However, in multi‐center studies this is often the case due to different scanner field strengths. The power analysis results in Table IV and Table V indicate that the additional methodological variance from combining data acquired at different field strengths, but with the same sequence, does not penalise the power of the study. In a multi‐center study where data is acquired at both field strengths, one could be tempted to use the protocol that requires the least number of subjects for the respective field strength. Thus the study would combine FLASH images acquired at 1.5 T with MP‐RAGE images acquired at 3 T. However, the power analysis results in Table V show that this would in fact increase the number of subjects required to detect a change in morphology due to the strong regional differences in GM density estimates between the sequences. If different acquisition protocols are used in a study, it is important to not mistake interactions with the field strength/sequence variable as a true morphometric difference.
The power analysis results presented here are in close agreement with those presented for a similar effect size in cortical thickness measurements by Han et al. [Han et al.,2006]. Small sample sizes close to these minima are sometimes seen in the literature; for instance, in a longitudinal VBM study on children and adolescents with obsessive‐compulsive disorder before and after treatment, 15 patients and 15 healthy controls were scanned [Lázaro et al.,2009]. Typical sample sizes in cross‐sectional studies range from 20 to 40 subjects per group. Larger sample sizes are also seen due to the increased anatomical variability of the disease population or to enhance the statistical power of the study, such as the multi‐site collaborative study on schizophrenia including 237 patients and 266 controls [Segall et al.,2009]. The sample sizes reported here can be used as guidelines in designing a VBM study. For better estimates, a power analysis can be performed on initial VBM results to take into account the exact image acquisition protocol, image analysis pipeline (including the choice of atlas, non‐uniformity correction, spatial registration, segmentation and blurring), the anatomical variance of the populations under study, as well as the effect size.
The regional differences in tissue classification and corresponding variability cause regional biases in sensitivity to change, thus complicating the direct comparison of VBM studies performed at different field strengths. In addition, this study also showed that the differences in GM density estimates between the different sequences acquired at the same field strength are stronger and broader than those using the same sequence at different field strengths. Caution should thus be taken when comparing VBM studies based on different acquisition sequences and sequence parameters as well.
CONCLUSION
This study was carried out to characterize the sensitivity of VBM to choice of field strength. Our results are only strictly applicable to the FLASH and MP‐RAGE protocols chosen in this study, and to the MNI and SPM analysis packages. However, since the implementation and acquisition parameters of these sequences are relatively stable across imaging studies, we believe that these results are representative of the typical performance of these sequences at both field strengths.
The increase in SNR and CNR at 3 T has the potential to improve the accuracy and reproducibility of the tissue classification results, thus hopefully improving the sensitivity of VBM to true morphometric change. These potential benefits may be hampered by the increase in transmission field inhomogeneity at 3 T if not adequately compensated for by image processing techniques. Although regional improvements in accuracy were visually evident, a quantitative assessment of tissue classification accuracy, a complex task, was not within the scope of this study. However, the VBM study of GM density revealed extensive regional differences between sequences and field strengths. These differences, minimized by proper VBM pre‐processing, are due to differences in CNR and contrast non‐uniformity caused by the protocol's signal dependence on B1 and T1.
The power analysis indicates that, averaged over a whole brain GM mask, the 1.5 T FLASH protocol requires the least subjects of the four protocols to detect a difference in GM density in a longitudinal VBM study, closely followed by the 3 T MP‐RAGE protocol. The 1.5 T FLASH protocol may be advantaged due to the match in image characteristics with the ICBM atlas. In the context of a cross‐sectional VBM study, the number of subjects required is higher at 3 T than at 1.5 T.
Unlike its classical application suggests, VBM based on a single T1‐weighted contrast is characterized by a distinct regional sensitivity to morphometric change, complicating the combination of data within a VBM study and the direct comparison of VBM studies based on different image acquisition protocols.
REFERENCES
- Acosta‐Cabronero J, Williams GB, Pereira JMS, Pengas G, Nestor J ( 2008): The impact of skull‐stripping and radio‐frequency bias correction on grey‐matter segmentation for voxel‐based morphometry. NeuroImage 39: 1654–1665. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ ( 2000): Voxel based morphometry—The methods. NeuroImage 11: 805–821. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ ( 2005): Unified segmentation. NeuroImage 26: 839–851. [DOI] [PubMed] [Google Scholar]
- Bluml S, Schad LR, Scharf J, Wenz F, Knopp MV, Lorenz WJ ( 1996): A comparison of magnetization prepared 3D gradient‐echo (MP‐RAGE) sequences for imaging of intracranial lesions. Magnet Resonance Imaging 14: 329–335. [DOI] [PubMed] [Google Scholar]
- Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DLG, Bernstein MA, Thompson PM, Weiner MW, Schuff N, Alexander GE, Killiany RJ, DeCarli C, Jack CR, Fox NC, Study FTA ( 2008): Intensity non‐uniformity correction using N3 on 3‐T scanners with multichannel phased array coils. NeuroImage 39: 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briellmann RS, Syngeniotis A, Jackson GD ( 2001): Comparison of hippocampal volumetry at 1.5 Tesla and 3 Tesla. Epilepsia 42: 1021–1024. [DOI] [PubMed] [Google Scholar]
- Chen J, Feng Z, Jin J‐M ( 1998): Numerical simulation of SAR and B1‐field inhomogeneity of shielded RF coils loaded with the human head. IEEE Trans Biomed Eng 45: 650–659. [DOI] [PubMed] [Google Scholar]
- Cocosco CA, Zijdenbos AP, Evans AC ( 2003): A fully automatic and robust brain MRI tissue classification method. Med Image Anal 7: 513–527. [DOI] [PubMed] [Google Scholar]
- Cohen JA ( 1988): Statistical Power Analysis for the Behavioral Sciences. Hillsdale, NJ: L. Erlbaum Associates. [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC ( 1994): Automatic 3D intersubject registration of MR volumetric data in standardized tailarach space. J Comput Assisted Tomography 18192–205. [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC ( 1995): Automatic 3D model‐based neuroanatomical segmentation. Hum Brain Mapp 3: 190–208. [Google Scholar]
- Deichmann R, Good CD, Josephs O, Ashburner J, Turner R ( 2000): Optimization of 3D MP‐RAGE sequences for structural brain imaging. NeuroImage 12: 112–127. [DOI] [PubMed] [Google Scholar]
- Deoni SC, Rutt BK, Peters TM ( 2003): Rapid combined T1 and T2 m using gradient recalled acquisition in the steady state. Magnet Resonance Med 49: 515–526. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Fenstermacher E, Salat DH, Wolk DA, Maguire RP, Desikan R, Pacheco J, Quinn BT, van der Kouwe A, Greve DN, Blacker D, Albert MS, Killiany RJ, Fischl B ( 2008): Detection of cortical thickness correlates of cognitive performance: Reliability across MRI scan sessions, scanners, and field strengths. NeuroImage 39: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein FH, Mugler JP III, Brookeman JR ( 1994): Optimization of parameter values for complex pulse sequences by simulated annealing: Application to 3D MP‐RAGE imaging of the brain. Magnet Resonance Med 31: 164–177. [DOI] [PubMed] [Google Scholar]
- Frahm J, Haase A, Matthaei D ( 1986): Rapid three‐dimensional NMR imaging using the FLASH technique. J Comput Assisted Tomography 10: 363–368. [DOI] [PubMed] [Google Scholar]
- Haase A ( 1990): Snapshot FLASH MRI: Applications to T1, T2, and chemical‐shift imaging. Magnet Resonance Med 13: 77–89. [DOI] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Pacheco J, Albert M, Killiany R, Maguire P, Rosas HD, Makris N, Dale A, Dickerson B, Fischl B ( 2006): Reliability of MRI‐derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. NeuroImage 32: 180–194. [DOI] [PubMed] [Google Scholar]
- Lázaro L, Bargalló N, Castro‐Fornieles J, Falcón C, Andrés S, Calvo R, Junqué C ( 2009): Brain changes in children and adolescents with obsessive‐compulsive disorder before and after treatment: A voxel‐based morphometric MRI study. Psychiatric Res 172: 140–146. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero‐Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B ( 2001): A probabilistic atlas and reference system for the human brain: International Consortium for Brain M (ICBM). Philosophical Trans Royal Soc Lond, Series B: Biol Sci 356: 1293–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L ( 2005): The Alzheimer's disease neuroimaging initiative. Neuroimaging Clinics N Am 15: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler JP III, Brookeman JR ( 1990): Three‐dimensional magnetization‐prepared rapid gradient‐echo imaging (3D MP‐RAGE). Magnet Resonance Med 15: 152–157. [DOI] [PubMed] [Google Scholar]
- Mugler JP III, Brookeman JR ( 1991): Rapid three‐dimensional T1‐weighted MR imaging with the MP‐RAGE sequence. J Magnet Resonance Imaging 1: 561–567. [DOI] [PubMed] [Google Scholar]
- Runge VM, Kirsch JE, Thomas GS, Mugler JP III ( 1991): Clinical comparison of three‐dimensional MP‐RAGE and FLASH techniques for MR imaging of the head. J Magnet Resonance Imaging 1: 493–500. [DOI] [PubMed] [Google Scholar]
- Scorzin JE, Kaaden S, Quesada CM, Müller CA, Fimmers R, Urbach H, Schramm J ( 2008): Volume determination of amygdala and hippocampus at 1.5 and 3.0 T MRI in temporal lobe epilepsy. Epilepsy Res 82: 29–37. [DOI] [PubMed] [Google Scholar]
- Segall JM, Turner JA, van Erp T, White T, Bockholt HJ, Gollub RL, Ho BC, Magnotta V, Jung RE, McCarley RW, Schulz SC, Lauriello J, Clark VP, Voyvodic JT, Diaz MT, Calhoun VD ( 2009): Voxel‐based morphometru multisite collaborative study on schizophrenia. Schizophrenia Bull 35: 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Pike GB ( 1998): Standing‐wave and RF penetration artifacts caused by elliptic geometry: An electrodynamic analysis of MRI. IEEE Trans Med Imaging 17: 653–662. [DOI] [PubMed] [Google Scholar]
- Sled JG, Pike GB ( 2000): Correction for B1 and B0 variations in quantitative T2 measurements using MRI. Magnet Resonance Med 43: 589–593. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC ( 1998): A non‐parametric method for automatic correction of intensity non‐uniformity in MRI data. IEEE Trans Med Imaging 17: 87–97. [DOI] [PubMed] [Google Scholar]
- Smith SM ( 2002): Fast robust automated brain extraction. Hum Brain Mapp 17: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen‐Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy R, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM ( 2004): Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23( Suppl 1): 208–219. [DOI] [PubMed] [Google Scholar]
- Stankiewicz JM, Neema M, Alsop DC, Healy BC, Arora A, Buckle GJ, Chitnis T, Guttmann CR, Hackney D, Bakshi R. ( 2009): Spinal cord lesions and clinical status in multiple sclerosis: A 1.5 T and 3 T MRI study. J Neurol Sci. 279, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif CL, Collins DL, Pike GB ( 2009): Sensitivity of voxel‐based morphometry analysis to choice of imaging protocol at 3 Tesla. NeuroImage 44: 827–838. [DOI] [PubMed] [Google Scholar]
- Tohka J, Zijdenbos A, Evans A ( 2004): Fast and robust parameter estimation for statistical partial volume models in brain MRI. NeuroImage 23: 84–97. [DOI] [PubMed] [Google Scholar]
- Vidal A, Bureau Y, Wade T, Spence JD, Rutt BK, Fenster A, Parraga G ( 2008): Scan‐rescan and intra‐observer variability of magnetic resonance imaging of carotid atherosclerosis at 1.5 T and 3.0 T. Phys Med Biol 53: 6821–6835. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC ( 1996): A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp 4: 58–73. [DOI] [PubMed] [Google Scholar]
- Wright IC, McGuire PK, Poline J‐B, Travere JM, Murray RM, Frith CD, Frackowiak RSJ, Friston KJ ( 1995): A voxel‐based method for the statistical analysis of grey and white matter density applied to schizophrenia. NeuroImage 2: 244–252. [DOI] [PubMed] [Google Scholar]


