Unraveling the full genetic basis of cardiometabolic diseases such as impaired glucose metabolism, diabetes, obesity, dyslipidemias, hypertension – that are among the key causes of stroke and coronary artery disease (CAD) - is proving to be one of the most profoundly complicated tasks facing contemporary biomedical research. In this issue of JACC, the Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators present the latest in a series of landmark studies that have added piecemeal to our understanding of the genetic basis of CAD. The backdrop to this study was another recently published undertaking, where this well-known consortium assembled a large DNA sequence dataset from 72,868 CAD patients and 120,770 controls to explore and identify rare coding sequence variants associated with CAD (1). Notably, the consortium was successful in identifying low-frequency variants and several novel loss-of-function mutations in the genes LPL, SVEP1 and ANGPTL4, the latter being associated with altered triglyceride levels (1). In the current study in JACC, the consortium leveraged the same dataset to look for novel common variants associated with CAD. As a point of distinction, in the former study the consortium used analytic techniques to specifically study and identify variants with a minor allele frequency of < 5% (‘rare variants’); for the current study the focus was variants with minor allele frequency > 5% (‘common variants’). Subdividing their dataset into discovery and replication cohorts, as well as replicating previously known CAD risk loci, the consortium identified and validated six new variants associated with CAD at genome-wide significance, being respectively associated with the genes KCNJ13-GIGYF2, C2, MRVI1-CTR9, LRP1, SCARB1 and CETP. As has been a consistent trend with prior studies, some of these new loci are associated with genes that might plausibly be related to CAD based on prior knowledge: LRP1 (low density lipoprotein receptor related protein-1), SCARB1 (which encodes SR-B1, a receptor for HDL cholesterol) and CETP (cholesterol ester transfer protein). On the other hand and again consistent with prior studies, the genes associated with the other new loci do not have immediately apparent links to known major biological aspects of CAD, although, the variant associated with C2 (which encodes complement C2 protein) introduces the possibility that the compliment system may have a more prominent role in atherosclerosis and CAD than currently appreciated. Previously, there were 56 validated CAD risk loci (2,3), which when added to the 6 new novel variants gave a total of 62 validated CAD risk loci. Of note, there are at least another 100 identified loci that are potentially associated with CAD but which are yet to be validated (2–4). Furthermore, in another study that appeared very recently, 17 additional novel CAD risk loci were identified (5).
The consortium then evaluated potential associations between the 62 validated CAD risk loci and both traditional CAD risk factors (lipid traits, blood pressure, body mass index, diabetes and smoking) and a wide range of other diseases and traits - including diseases/traits as diverse as coronary artery calcification, stroke, lupus and autism. These analyses were motivated by mounting evidence suggesting that single nucleotide polymorphisms (SNPs), such as the authors identified here, exhibit substantial overlap across common complex disorders. Adding support to this paradigm, they found that 24/62 loci (38.7%) showed statistical association with traditional cardiovascular risk factors (most commonly with lipid traits), and with some loci showing multiple associations. Furthermore, almost half of the 62 SNPs (29/62; 47%) were associated with other diseases/traits.
This paper is an important undertaking and a notable advance in our knowledge of CAD risk SNPs and the heritability of common complex disorders. Before considering the advances this study brings, let us first address its limitations. The study was performed by compiling 20 individual studies for the discovery cohort and a further 8 studies for validation. While every effort was made to harmonize these datasets, it is inevitable that biases and inconsistencies, both known and unknown, were present across these studies. For example, among the 28 studies the definition of a CAD ‘case’ included such diverse designations as: physician-assigned ICD codes, coronary stenosis ≥ 50%, abnormal stress test (that may include false positives), and fatal myocardial infarction (MI). This inhomogeneity in the definition of a CAD ‘case’ adds uncertainty to this study, particularly because the biology of acute MI (plaque rupture and thrombosis) is not the same as the biology of stable CAD. Furthermore, the inconsistent definition of CAD cases also affects the definition of the control group, where a significant but unknown portion may have had subclinical CAD (6). As further limitations, the populations studied were overwhelmingly Caucasian Europeans, and the generalizability of these SNPs to other ethnicities remains to be proven. In addition, by nature of the retrospective compilation of existing datasets, this study used a now superseded Illumina platform from circa 2011 that looked for variance at ~30,000 loci. Contemporary beadchips cover dramatically more loci, while DNA sequencing (that can disclose novel SNPs) is rapidly becoming the gold standard for gene discovery studies. In time, as these more powerful methods are used to interrogate large cohorts, we can expect to see the list of CAD risk loci to further grow.
Scientifically, what does this study add? Foremost, this study discloses the profound pleiotropy that exists not just among CAD risk SNPs, but across common complex disorders and traits in general. To be clear, what is meant by pleiotropy is that a single risk locus is associated with multiple different diseases and traits. Perhaps it might have been anticipated that a proportion of CAD risk SNPs would also show association with cardiovascular risk factors (because certain SNPs may promote CAD by affecting the risk factor itself – for example by raising LDL cholesterol). Indeed, this was the case for 24/62 loci. However, what is particularly revealing about the biology of CAD and other complex disorders is that almost half of the CAD risk SNPs (29/62) were also associated with other diseases or traits. What this tells us about the biology of CAD, common complex disorders and the human genome is that an extremely complex balance exists among the heritability of different disease states. Indeed, not only can a single SNP be associated with several different diseases, but adding even greater complexity and by entirely unknown mechanisms, the risk association can be in a different direction for different diseases. For example, at the SNP rs9349379, which is associated with the gene PHACTR1, either an adenine [A] or guanine [G] may be present. Remarkably, regardless of which nucleotides are present (AA, AG, or GG), a disease association exists. Thus, an [A] at rs9349379 is associated with increased risk of cervical artery dissection (7) migraine headache (8) and fibromuscular dysplasia (9). Conversely, the [G] allele at rs9349379 is associated with CAD (10–12), coronary artery calcification (13) and MI (10,12,13). Therefore, the risk of having CAD or other diseases is related, in certain cases both directly and reciprocally, with the risk of having other diseases.
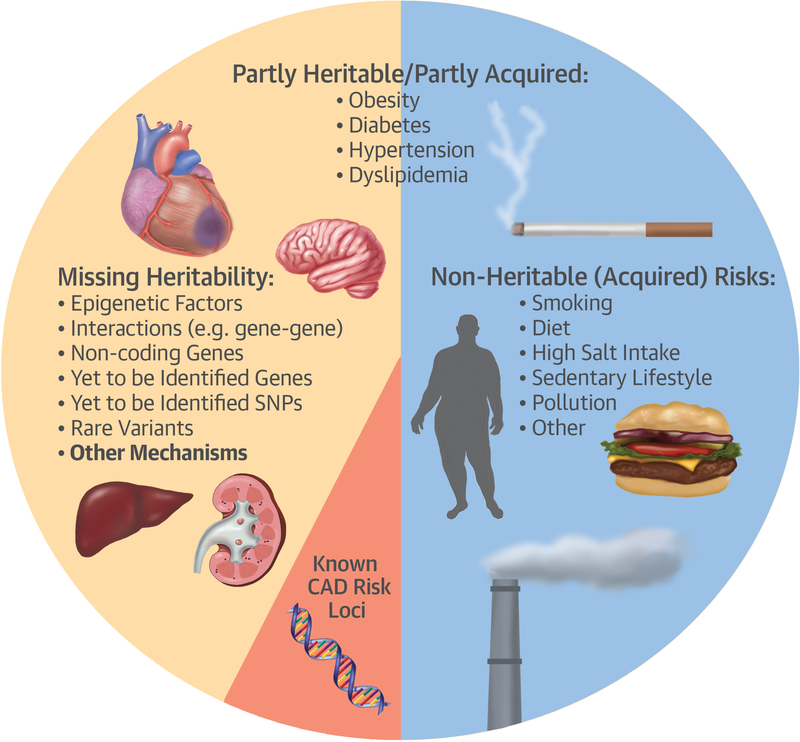
Seminal studies from the 1990s suggested that heritability accounts for ~50% of the likelihood of developing CAD (14), with the remaining ~50% of risk being likely attributable to environmental and lifestyle-related factors such as smoking, sedentary lifestyle, obesity, salt intake, diet and other factors (Figure). At present, one of the great mysteries of common complex diseases is that of ‘missing heritability.’ That is, if we take all 62 validated CAD risk loci and even if we include the >100 non-validated loci, these directly account for only ~15% of the heritable likelihood of having clinically manifest CAD. On the one hand, the addition of 6 new SNPs from the current study to the already known or suspected CAD risk loci does little to increase our understanding of ‘missing heritability.’ However, the truly fascinating aspect of the current paper is perhaps not the 6 new loci, but the profound pleiotropy that was shown among complex diseases. Along with other recent studies like STARNET (11), these papers have opened up the possibility that heritable risk for CAD and common complex diseases may be partially attributable to interactions among diseases and traits. Some of these interactions are logical (e.g. LDL cholesterol levels and CAD), but many are less intuitive. An example of this is inflammatory diseases like Rheumatoid arthritis and lupus. These diseases are strongly represented among those showing pleiotropy with CAD risk SNPs (see Table 3 in current JACC article). While it has been assumed that the association of inflammatory diseases with CAD was via generalized inflammatory activation, the current manuscript suggests that their association with CAD may be more formally enshrined in discrete pathways of action related to individual risk SNPs. Therefore, we can surmise by saying that a broader picture is emerging, where part of the missing heritability of CAD and other common complex disorders may be attributable to interactions among genes, proteins, tissues (15), and diseases (11) – all of which have an added ‘wildcard’ element which is the influence of environmental factors on these interactions (e.g. smoking, diet, exercise, pollution).
Figure – Some of the currently suspected and known factors responsible for CAD.
The basis of ‘missing heritability’ remains a topic of intense ongoing speculation. The factors depicted here are merely meant to illustrate specific aspects that appear to be of potential relevance, rather than being in any way a definitive or exhaustive list of all factors that cause CAD.
Achieving a definitive understanding of the interactions that underlie common complex disorders and the added role of environmental influences is a task of daunting size and complexity. However, the task is an especially compelling one that is directly related to the vast burden of morbidity and mortality suffered by humankind. It may be years before we acquire even a basic knowledge of the full heritability and biology of CAD and common complex disorders including epigenetics, pleiotropy and a multitude of other factors, but without question, this will be a truly fascinating scientific journey to the core of the human condition.
Acknowledgements:
Jason Kovacic acknowledges research support from National Institutes of Health (K08HL111330, R01HL130423), the American Heart Association (14SFRN20490315; 14SFRN20840000) and The Leducq Foundation (Transatlantic Network of Excellence Award).
Footnotes
Disclosures: None
References
- 1.Stitziel NO, Stirrups KE, Masca NG et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med 2016;374:1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium CAD. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium CAD, Deloukas P, Kanoni S et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schunkert H, Konig IR, Kathiresan S et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iotchkova V, Huang J, Morris JA et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat Genet 2016;48:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorkegren JL, Kovacic JC, Dudley JT, Schadt EE. Genome-wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J Am Coll Cardiol 2015;65:830–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debette S, Kamatani Y, Metso TM et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet 2015;47:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anttila V, Winsvold BS, Gormley P et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 2013;45:912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiando SR, Tucker NR, Castro-Vega LJ et al. PHACTR1 Is a Genetic Susceptibility Locus for Fibromuscular Dysplasia Supporting Its Complex Genetic Pattern of Inheritance. PLoS Genet 2016;12:e1006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikpay M, Goel A, Won HH et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzen O, Ermel R, Cohain A et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 2016;353:827–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaudoin M, Gupta RM, Won HH et al. Myocardial Infarction-Associated SNP at 6p24 Interferes With MEF2 Binding and Associates With PHACTR1 Expression Levels in Human Coronary Arteries. Arterioscler Thromb Vasc Biol 2015;35:1472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Donnell CJ, Kavousi M, Smith AV et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 2011;124:2855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 1994;330:1041–1046. [DOI] [PubMed] [Google Scholar]
- 15.Talukdar HA, Foroughi Asl H, Jain RK et al. Cross-Tissue Regulatory Gene Networks in Coronary Artery Disease. Cell Syst 2016;2:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]