Abstract
Malnutrition is a severe non-communicable disease, which is prevalent in children from low-income countries. Recently, a number of metagenomics studies have illustrated associations between the altered gut microbiota and child malnutrition. However, these studies did not examine metabolic functions and interactions between individual species in the gut microbiota during health and malnutrition. Here, we applied genome-scale metabolic modeling to model the gut microbial species, which were selected from healthy and malnourished children from three countries. Our analysis showed reduced metabolite production capabilities in children from two low-income countries compared with a high-income country. Additionally, the models were also used to predict the community-level metabolic potentials of gut microbes and the patterns of pairwise interactions among species. Hereby we found that due to bacterial interactions there may be reduced production of certain amino acids in malnourished children compared with healthy children from the same communities. To gain insight into alterations in the metabolism of malnourished (stunted) children, we also performed targeted plasma metabolic profiling in the first 2 years of life of 25 healthy and 25 stunted children. Plasma metabolic profiling further revealed that stunted children had reduced plasma levels of essential amino acids compared to healthy controls. Our analyses provide a framework for future efforts towards further characterization of gut microbial metabolic capabilities and their contribution to malnutrition.
Keywords: child malnutrition, gut microbiota, genome-scale metabolic modeling, bacterial interactions, metabolic capabilities, plasma metabolic profiling
INTRODUCTION
Malnutrition is a severe non-communicable disease in low- and middle-income countries, which includes two forms, namely undernutrition (stunting and wasting) and overnutrition (obesity). According to a recent World Health Organization (WHO) report, stunting and wasting have affected 160 million and 50 million children, respectively worldwide (Chan, 2017). Child stunting alone affected 22.6% children under the age of 5 years globally in 2016 (UNICEF et al., 2017). WHO recommends breastfeeding until 6 months of age for healthy growth of infants. Moreover, in the case of malnutrition, peanut butter and dried skimmed milk based ready-to-use-therapeutic food (RUTF) is recommended (Briend et al., 2015). Although significant progress has been made over the past few decades in reducing the prevalence of stunting, interventions applied to treat child stunting have not always been successful (Bhutta et al., 2008; Ramakrishnan et al., 2009). Recently, several studies have been conducted to understand the effect of nutrients intervention, such as lipid-based nutrient supplements or RUTFs, micronutrients such as vitamin A, zinc, iron individually or in multiple combinations, on stunted children (Ashorn et al., 2015a; Mayo-Wilson et al., 2014; Ramakrishnan et al., 2009). Results of these studies suggested that interventions were unable to reverse the linear growth deficit of stunting.
Apart from dietary nutrients, the human gut microbiota has been found to be associated with malnutrition (Blanton et al., 2016; Smith et al., 2013; Subramanian et al., 2014). The human gut harbors trillions of microbial cells, which collectively act like a bioreactor for fermenting dietary macronutrients to health-promoting metabolites, for instance, short chain fatty acids (SCFAs) (De Vadder et al., 2014; Louis and Flint, 2017), amino acids (AAs) (Husted et al., 2017; Metges, 2000; Shoaie et al., 2015) and vitamins (Degnan et al., 2014; Gominak, 2016; Östman et al., 2001). Moreover, previous studies demonstrated the interaction of gut microbiota with bile acid metabolism, which may have significant impact on host metabolism (Wahlström et al., 2016).
Recently, few studies based on comparative metagenomic analysis addressed the questions related to variations in taxonomic composition of gut microbiota between health and malnutrition. These studies demonstrated the relative taxonomic abundance of gut microbial species using ribosomal 16S gene sequencing (Subramanian et al., 2014) or shotgun metagenomic sequencing (Blanton et al., 2016; Smith et al., 2013). However, metabolic differences and underlying mechanisms of the functional variations in gut microbiota in malnutrition are poorly known due to the limitations of available systems biology tools (Faust and Raes, 2012). Hence, there is a clear requirement of developing novel computational approaches to investigate the uncharacterized mechanisms connected with gut microbial ecosystem and its associations with diseases. Therefore, we propose a constraint-based modeling framework using genome-scale metabolic models (GEMs), to examine the metabolic variations in health and malnutrition. Previously, GEMs have been successfully used to study the metabolism of human and microbial species (Agren et al., 2012; Kim et al., 2017). Here, we reconstructed GEMs of the most abundant gut bacterial species from healthy and malnourished children groups. Although genome-scale metabolic models have been proven to be useful tools to investigate a target organism’s metabolic capabilities, the majority of high-quality GEMs have been biased towards well-studied species such as E. coli (Feist et al., 2007; Reed and Palsson, 2003), B. subtilis (Henry et al., 2009), S. cerevisiae (Duarte et al., 2004), and H. sapiens (Duarte et al., 2007; Thiele et al., 2013). The reason for this tendency is the vast availability of data for these organisms, which is required both for manual curation and evaluation of the model predictions. However, when it comes to the members of the human gut microbiota repertoire, most of these strains are yet to be cultured to be better studied and for their biosynthetic capabilities to be further characterized. There have been some recent efforts towards this direction. For instance, a valuable research has been conducted in which 96 phylogenetically diverse strains from the human gut symbionts were cultured in rich and also defined media to gain insight into their growth rates and nutritional dependencies (Tramontano et al., 2018). Another work has been focused on reconstruction of a collection gut microbiota strains (Magnúsdóttir et al., 2016), however, there seems to been little accordance between these two, suggesting that there is still need for more characterization of these under-studied strains. From analysis of these models we found that due to automatic reconstruction the models are highly similar and they are therefore not suitable for looking into metabolic alterations (Babaei et al., 2018). To overcome this we here combined automatic reconstruction with detailed manual curation of the draft GEMs. In this process we first evaluated simulation performance of the GEMs on two media (human breast milk (HBM) and RUTF) to predict the microbial growth and secretion of small molecules such as SCFAs and AAs. Next, gut microbial GEMs allowed predicting the community-level metabolic potentials of gut microbes and examining patterns of pairwise competition/synergism of key species. Collectively, our modeling framework presents a potential route to model the gut microbial species and examine the role of individual microbes in terms of growth and synthesis of vital metabolites in health and diseases, and hereby allowed us to gain new functional insight into the gut microbiota associated with malnutrition. Furthermore, the modeling framework can be used to explore the metabolic crosstalk between different gut microbial species and how these metabolic interactions integrate with the host, which will enable design of effective strategies for modulating the gut microbiome for health benefits. Causal mechanisms of malnutrition are also still poorly known, and there is a clear need to use application of advances in omics-based approaches to examine host’s mechanistic pathways associated with malnutrition (Huey and Mehta, 2016). To begin to address this issue, we followed a birth cohort of children in Bangladesh up to the age of 2 and performed targeted plasma metabolic profiling on 25 children assessed to be stunted with 25 healthy controls. The main goal of our study was to assess any differences in circulating levels of 50 metabolites that may be influenced by the microbiota between the healthy and stunted groups. Results demonstrated that stunted children had reduced levels of essential amino acids as well as lower ratio of tryptophan to other neutral amino acids compared to the healthy group. Overall, ten plasma metabolites were found to be correlated with Height-for-Age Z score (HAZ) of children.
MATERIALS AND METHODS
Analysis of the gut microbial relative abundance data
Relative taxonomic abundance data of gut bacteria were obtained from three published studies based on children groups from three different countries (Malawi, Bangladesh, and Sweden) (Bäckhed et al., 2015; Smith et al., 2013; Subramanian et al., 2014). Only species-level relative abundances were used for selecting the 20 most abundant bacterial species from gut microbiota of healthy and malnourished children in order to reconstruct GEMs. This assumption was made to simplify modeling framework in order to understand the metabolic capabilities of the most dominant part of child gut microbiome in health and malnutrition. A total of 68 species were selected. Relative taxonomic abundances of selected species can be seen in Table S1 and Figure S2.
Firmicutes/Bacteroidetes ratio was calculated for healthy and malnourished children using relative taxonomic abundance data from four published studies (Bäckhed et al., 2015; Blanton et al., 2016; Smith et al., 2013; Subramanian et al., 2014). Among the four datasets two were from Malawian children (Blanton et al., 2016; Smith et al., 2013) and the remaining two were from Bangladeshi (Subramanian et al., 2014) and Swedish children (Bäckhed et al., 2015). Relative taxonomic abundance data of Swedish children were available only for healthy children. Further species abundance datasets were used to determine the Shannon diversity index or Shannon-Wiener index of the gut microbiota for each children group. Shannon-Wiener index is defined as following (Hurlbert, 1971):
Where, Pi is the relative abundance of species i. S is the number of species in microbial community. diversity function of package vegan was deployed to obtain this index in R version 3.3.0.
Reconstructions of the genome-scale metabolic models
Due to the partial sequencing and annotation or no availability in public databases of genome of ten species, draft GEMs were reconstructed for 58 species (Table S2). These models are represented by a stoichiometric matrix (S), which incorporates metabolic conversion and mass balance information of biochemical reactions (enzymatic and transport) in metabolic network of organism and growth is determined from biomass reaction, which is connected with other biochemical reactions in the network. Annotated genomes based on Rapid Annotation using Subsystem Technology (RAST) sever (Aziz et al., 2008) were used for reconstructing the draft GEMs at KBase platform (The U.S. Department of Energy Systems Biology Knowledgebase, https://kbase.us/). KBase platform uses the modelSEED pipeline (Henry et al., 2010) for generating GEMs.
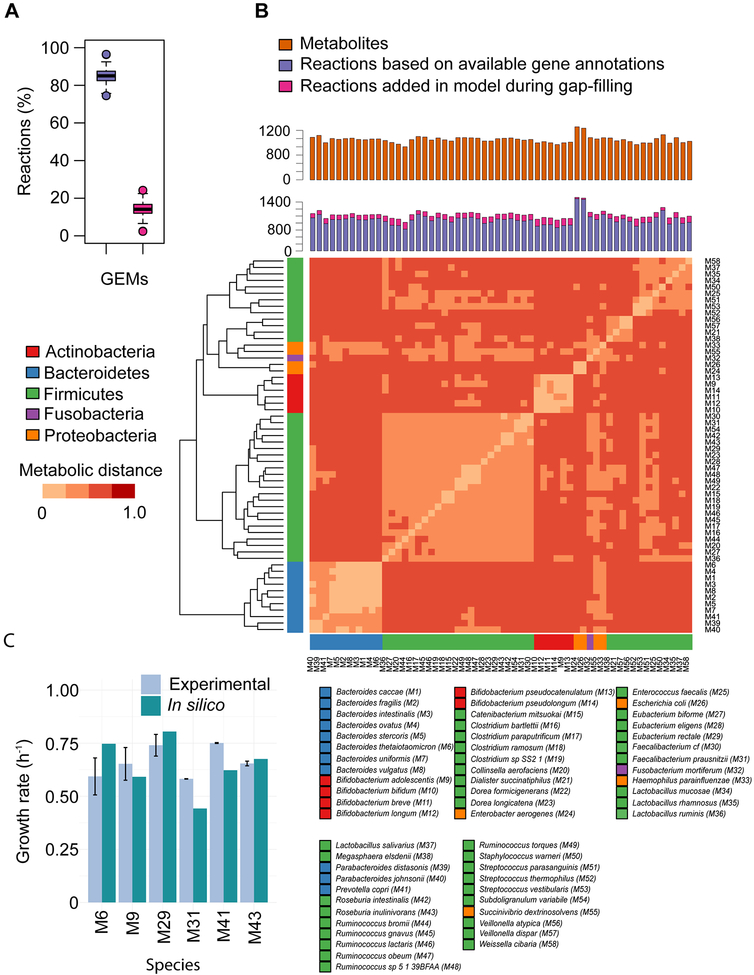
To fill gaps in the GEMs we have used the KBase/modelSEED pipeline to automatically add a minimal set of reactions to the draft networks in order to produce biomass (Henry et al., 2010). Updated KBase/modelSEED pipeline uses a previously published method for gap-filling (Latendresse, 2014). GEMs, which contained at least 75% reactions based on available gene annotations, were considered for further analysis to lessen the possible influence of automatically gap-filled reactions. For details of the steps followed in curation of the GEMs see Supplemental Text. All 58 GEMs are available on GitHub (https://github.com/SysBioChalmers/ChildrenGutMicrobialGEMs). In the next step, we compared the GEMs to determine the metabolic dissimilarities from each other. Models dissimilarity was examined in terms of metabolic distances between models based on their list of reactions. Metabolic distance was calculated using Jaccard coefficient, as metabolic distance = 1 – Jaccard coefficient (Levandowsky, 1971) (Figure 2B and Table S4). If A is the set of reactions of model a and B is the set of reactions of model b, then metabolic distance between a and b is defined as 1 – |A∩B| /|A∪B|.
Figure 2: Features of GEMs.
(A) Proportion of reactions associated with gene annotations (violet bar) and added during gap filling (pink bar). (B) Number of metabolites and reactions in GEMs of gut bacteria, and metabolic distances between models based on shared and unique reactions, which was calculated using the Jaccard coefficient (Experimental Procedure). Models representing the same phylum share more reactions than models from taxonomically distinct species. heatmap.2 function of gplots package, and dist and hclust functions of R were used for generating heatmap and dendrogram, respectively, in R version 3.3.0. (C) Validation of GEMs. Growth rate predictions of the GEMs of six gut bacterial species were validated with experimental observations on YCFA medium with different glucose uptake rates (mmol gDW-1 h-1) under anaerobic growth conditions (Table 1). Species involved in this analysis were Bacteroides thetaiotaomicron (M6), Bifidobacterium adolescentis (M9), Eubacterium rectale (M29), Faecalibacterium prausnitzii (M31), Prevotella copri (M41), and Roseburia inulinivorans (M43).
Model simulations
All Flux Balance Analysis (FBA) simulations were performed in MATLAB R2015b and R2016b (The MathWorks, Inc., Natick, MA) using the COBRA Toolbox 2.0.5 (https://opencobra.github.io/) and linear programming solver Gurobi 6.5.2 with academic license (GUROBI OPTIMIZATION, http://www.gurobi.com/index). GEMs were employed to predict the in silico optimal growth rates on complete media, human breast milk, and ready-to-use therapeutic food. Complete media is an in silico growth media, which does not have a fixed composition. Instead, complete media allows models to grow on all metabolites, for which transport reactions are available in the model. The other two media are the common diets for healthy and malnourished children and the composition of these were based on previously published studies (Bahwere et al., 2016; Nilsson et al., 2017; Peng et al., 2007; Yamawaki et al., 2005) (Table S6). We have also quantified fluxes of several metabolites (short chain fatty acids and amino acids). Models were constrained with maximum biomass production during quantification of secretion of metabolites. Using fluxes of these metabolites and relative abundances of species, the metabolic potential of gut microbiota in different species, communities and conditions were determined in terms of Estimated Maximal Metabolic Potential (EMPP). EMPP = Ai × vj denotes the estimated maximal metabolic potential of metabolite j in species i. A and v represent the relative abundance of a species and flux of a secreted metabolite, respectively.
Reconstruction of community metabolic models for gut microbes
Community Metabolic Models (CMMs) were reconstructed using single species GEMs, following a previously published method for generating mixed-bag models for microbial communities (Henry et al., 2016). For this purpose, five gut microbial communities based on five different children groups, namely Malawian children (healthy), Bangladeshi children (healthy), Swedish children (healthy), Malawian children (malnourished), and Bangladeshi children (malnourished) were selected (Table S5).
Pairwise interactions among species
Pairwise simulations were performed by formulating a linear programming in which the media metabolites were set to be the only available resources for the models. If a metabolite was required by both of the models, a constraint would imply that the sum of its associated consumption flux would be less or equal to the available amount in the media. Any produced metabolites by the community members that was used by the other member were under a constraint implying that the consumed flux is less or equal to the produced flux. The objective function for pairwise growth simulation and potential metabolite production were sum of biomass reactions and the sum of exchange reactions producing the metabolite of interest, respectively. For identifying the type of effect on growth, the growth rate in pairwise simulations were subtracted from the growth rate in single species optimization and depending on the change in the growth rates the type of effect was classified.
Validation of models
GEMs of Bacteroides thetaiotaomicron (M6), Bifidobacterium adolescentis (M9), Eubacterium rectale (M29), Faecalibacterium prausnitzii (M31), Prevotella copri (M41), and Roseburia inulinivorans (M43) were employed to predict growth rates and were validated with in vitro experimental data based on mono-cultures. In experimental setup, Bifidobacterium adolescentis L2–32, Eubacterium rectale A1–86 (DSM 17629), Faecalibacterium prausnitzii A2–165 (DSM 17677), and Roseburia inulinivorans A2–194 (DSM 16841) were provided by Dr. Karen Scott, The Rowett Institute of Nutrition and Health, Aberdeen. Bacteroides thetaiotaomicron ATCC 29148 and Prevotella copri DSM 18205 were obtained from American Type Culture Collection (ATCC) and Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), respectively. Originally, above all strains have been isolated from their human faecal habitat. B. adolescentis, B. thetaiotaomicron, E. rectale, F. prausnitzii, and R. inulinivorans were maintained in Hungate like tube (Ochs Laborbedarf, Germany) in YCFA medium (Supplemental Experimental Procedures) under an anaerobic environment using oxygen-free CO2 (Ze et al., 2012). However, P. copri was maintained in PYG medium (Media 104 in the DSMZ catalog).
An anaerobic chamber (Coy Lab Products, Grass Lake, MI, USA) was used to maintain oxygen-free environment for sub-culturing each bacterium on an agar plate containing YCFA medium. For inoculum preparation, a single colony was transferred to liquid YCFA medium (7.5 ml in each Hungate tube) and incubated for 12–15 h. Further, 100 ml serum bottles (Ochs Laborbedarf, Germany) with 50 ml working volume of autoclaved YCFA medium were used to perform fermentation experiments under anaerobic environment that maintained by oxygen free CO2. Each bottle was inoculated with 2% (v/v) inoculum and incubated for a maximum 30 h. All experiments were performed in triplicate.
For simulating the GEMs of above mentioned species, all components of YCFA medium were separated in individual metabolites (Supplemental Experimental Procedure and Table S7) that were used as input for GEMs. Glucose uptake rates (mmol gDW−1 h−1) were constrained as measured by experimental observations (Table 1). The uptake rates of remaining medium metabolites were kept unconstrained (Table S7). Biomass production was set as an objective function for simulating these GEMs.
Table 1:
Comparison between predicted growth rates and experimental growth rates (n=3)
| Bacterial species | Glucose uptake rate (mmol gDW−1 h−1) | Experimental growth rate (h−1) | In silico growth rate (h−1) | |
|---|---|---|---|---|
| Mean | SD | |||
| Bacteroides thetaiotaomicron (M6) | 2.848 | 0.593 | 0.747 | 0.087 |
| Bifidobacterium adolescentis (M9) | 7.085 | 0.653 | 0.592 | 0.077 |
| Eubacterium rectale (M29) | 5.601 | 0.740 | 0.805 | 0.051 |
| Faecalibacterium prausnitzii (M31) | 1.652 | 0.582 | 0.442 | 0.001 |
| Prevotella copri (M41) | 5.204 | 0.751 | 0.622 | 0.002 |
| Roseburia inulinivorans (M43) | 1.598 | 0.654 | 0.676 | 0.011 |
Experimental design for plasma metabolic profiling analysis
Subjects for this study were part of a longitudinal birth cohort study (PROVIDE) conducted at International Centre for Diarrheal Disease Research, Dhaka, Bangladesh (icddr,b) (Kirkpatrick et al., 2015). This study followed a cohort of 700 infants in an urban slum in Dhaka, Bangladesh over the first 2 years of life to understand factors associated with vaccine efficacy, enteric dysfunction and stunting. Plasma samples were collected from 25 subjects with the greatest growth deficit (HAZ<−2) at 2 years of age and from 25 control subjects were used for targeted metabolite analysis to identify discriminant biomarkers associated with stunting (Table 2).
Table 2:
Characteristics of healthy and *stunted children
| Items | Details |
|---|---|
| Healthy (non-stunted) children (% female) | 25 (48) |
| Stunted children (% female) | 25 (32) |
| Age (weeks) | 40–104 |
| **HAZ, healthy children, mean (SD) | 0.22 (0.64) |
| HAZ, stunted children, mean (SD) | −2.86 (0.76) |
| ***WAZ, healthy children, mean (SD) | 0.35 (1.13) |
| WAZ, stunted children, mean (SD) | −2.42 (0.62) |
Child stunting, typically assessed using the anthropometric measurement Height-for-Age Z score (HAZ score), results not just in short stature but also in long term cognitive deficit (de Onis and Branca, 2016). If HAZ scores of children are below three standard deviations (−3 s.d.) from the median of World Health Organization (WHO) references growth standards, these children are classified as having severe stunting, whereas, children with HAZ scores between −3 and −2 s.d. are defined as moderate stunted children (Reyes et al., 2004). The period up to age 2 is thought be critical for the establishment of growth deficit -associated processes that define stunting (Victora et al., 2010).
Height-for-age Z-score
Weight-for-age Z-score
Metabolite profiling and quantitative analysis
Plasma samples stored at −80°C were shipped on dry ice to University of Virginia for analysis. Amino acid levels were assayed using the EZ:faast™ Amino acids kit from Phenomenex (Torrance, CA) following manufacturer’s instructions. A method for derivatization of carboxylates (Han et al., 2013) and short-chain fatty acids (Han et al., 2015) by 3-nitrophenylhydrazine (3-NPH) for analysis by UPLC-MS/MS was adapted to quantify these molecules from plasma. 25 μL of plasma was mixed with 10 μL of internal standards comprising 10mM acetic acid-D4, 5mM sodium butyrate-D5, 1mM propionic acid -D5 (all from Cambridge Isotope Labs, Andover, MA, USA) and 1 μg/mL pyruvate and methanol was added to 80%. The mix was incubated on ice for 10 minutes, followed by centrifugation to remove the precipitate. 100 mL of the supernatant was combined in a mixture containing 25 μL of 150 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) in 75% methanol, 25 mL of 7.5% pyridine in 75% methanol and 50 mL of 200 mM 3-NPH in 75% methanol, and reacted sequentially at 4°C for 30 minutes, 40°C for 30 minutes and 50°C for 30 minutes to facilitate optimal derivatization of different compounds. The sample was diluted 1:1 with water for analysis by UPLC-MS/MS. Standard stocks of individual metabolites were serially diluted and analyzed similarly for generation of standard curves. Samples were separated on a 1.0 × 10cm Waters BEH column with an I-class Acquity using water:formic acid (100:0.01, v/v) and acetonitrile:formic acid (100:0.01, v/v) as the mobile phases for gradient elution.. The column flow rate was 0.1 mL/min; the column temperature was 40 °C, and the autosampler was kept at 5 °C. Eluate was analyzed on a Waters TQ-S mass spectrometer using electrospray ionization in negative ion mode using optimized MRM parameters and the resultant data was process using TargetLynx 4.1.
Statistical analysis
The data adjusted by age and gender are expressed as medians and interquartile range (upper, lower) (Buffie et al., 2015) (Table S9 and S10). All statistical comparisons between healthy and stunted children data were made using Mann-Whitney U and Wilcoxon rank-sum test. When multiple hypotheses were considered simultaneously, P values were adjusted to control the false discovery rate using a previously published method (Benjamini and Hochberg, 1995). Correlation between concentrations of plasma amino acids and HAZ score of children groups were examined by determining the Spearman’s correlation coefficients (Wu et al., 2011) (Table S11). The adjusted P value cut-off of 0.05 was considered to define the statistically significant correlation.
All of the dendrograms were obtained by hierarchical clustering of distances (or dissimilarities) measures between rows or columns in the heatmaps. Distance measures were obtained using Euclidean method as following (Deza and Deza, 2014):
where, x and y represent the vectors of a pair of rows or columns i, which depends on distances calculated for rows or columns. n represents the number of rows or columns in the heatmaps. heatmap.2 function of gplots package, and dist and hclust functions of R were used for generating heatmaps and dendrograms, respectively, in R version 3.3.0.
RESULTS
Representative microbial species in healthy and malnourished children
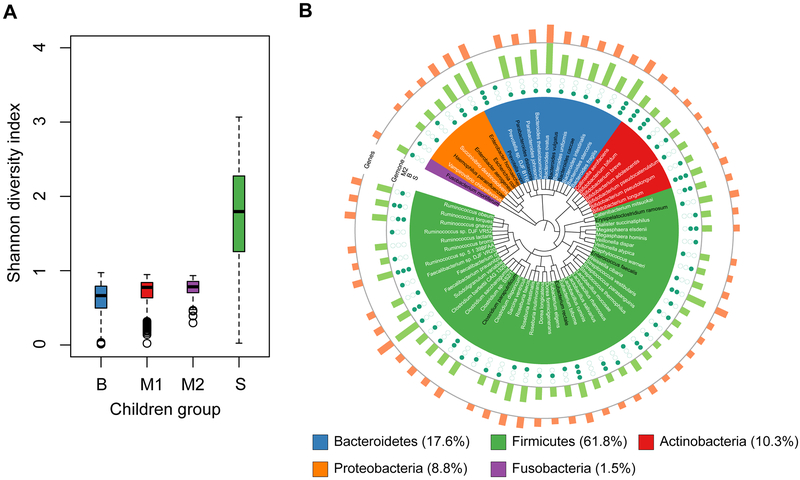
Relative taxonomic abundances of species in the gut microbiota of four groups of healthy and malnourished children from three different countries (Malawi, Bangladesh, and Sweden) were used to calculate the Shannon diversity index as low diversity has been associated with a dysfunctional gut microbiota (Menni et al., 2017). Two datasets were from Malawian children (Blanton et al., 2016; Smith et al., 2013), one from Bangladeshi (Subramanian et al., 2014) and one from Swedish children (Bäckhed et al., 2015). The relative taxonomic abundance data for Swedish children were available only for healthy children. To investigate the gut microbial diversity of healthy and malnourished children, we compared the diversity index of the gut microbiota between children having more (Malawian and Bangladeshi children) and less (Swedish children) prevalence of malnutrition. The diversity index data showed that the Malawian and Bangladeshi children have a significantly lower diversity index than the Swedish children (P < 0.01 for all comparisons, Mann-Whitney U test; Figure 1A). Similarly we calculated the Firmicutes to Bacteroidetes ratio (F/B ratio) as this ratio has previously been reported to be associated with several disease states, namely Inflammatory Bowel Disease (Sokol et al., 2009) and obesity (Turnbaugh et al., 2006). The F/B ratio was found to be significantly lower in Swedish children than the children from the other two countries (P < 0.01 for all comparisons, Mann-Whitney U test; Figure S1).
Figure 1: Gut Microbes in Different Healthy and Malnourished Children Groups.
(A) Shannon diversity index of the total gut microbiota of four groups of healthy and malnourished children from three countries (Bangladesh (B), Malawi (M1, M2) and Sweden (S)). These groups include both healthy and malnourished children, except for Swedish group, which doesn’t include malnourished children. Species-level relative abundance data was deployed to calculate the Shannon diversity index (Experimental Procedure) using diversity function of package vegan in R version 3.3.0. Mann-Whitney U test (two-sided, α = 0.01) was used to compare the Shannon diversity index between different children groups. (B) Taxonomic relationship among selected 20 most abundant bacteria each from gut microbiota of M2, B, and S children groups. Total number of selected bacterial species is 68. Bacterial species’ names colored in black are pathogens or conditionally-pathogens. Green solid points and circles denote the presence and absence, respectively, of each species in corresponding children group. In outer two layers, Green and orange bars represent the genome size and number of genes, respectively. This phylogenetic tree was generated using phyloT (https://phylot.biobyte.de/index.cgi) and visualized with iTOL v3 (Letunic and Bork, 2016) based on NCBI taxonomy.
In order to gain insight into the metabolic functions of the gut microbiome in the three cohorts of children we reconstructed GEMs of gut bacterial species. For this we selected the top 20 most abundant bacterial species using species-level relative taxonomic abundances from the gut microbiota of children from different time points and different cohorts of children in three countries (a total of 11 sample groups) (Table S1 and Figure S2). The focus was to investigate if there are differences in metabolic capabilities of the most abundant part of the gut microbiota of healthy and malnourished children. A total of 68 species, representing five phyla, viz. Firmicutes (61.8%), Bacteroidetes (17.6%), Actinobacteria (10.3%), Proteobacteria (8.8%), and Fusobacteria (1.5%), were selected after considering species abundances data from all children groups and time points (Figure 1B). There were 7 species common among all children groups, whereas 18, 9, and 22 species were unique in Malawian, Bangladeshi, and Swedish children, respectively (Figure S3).
GEMs reconstruction and their features
Due to only partial sequencing and annotation or no availability in public databases of genome sequences GEMs could not be reconstructed for 10 of the 68 species. These 10 species were among the least abundant species in the group of the 68 species (Figure S3). Draft GEMs were reconstructed for the remaining 58 species (Table S2). The ModelSEED pipeline (Henry et al., 2010) on the KBase platform (The U.S. Department of Energy Systems Biology Knowledgebase, https://kbase.us/) was used to reconstruct the GEMs. Available annotated genomes of the selected species were downloaded from KBase platform. These bacterial genomes have been annotated using the Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al., 2008). Further annotated bacterial genomes were used to reconstruct the draft GEMs. The draft models consisted of a network of reactions with Gene-Protein-Reaction (GPR) relationships, and a biomass reaction, which represents the growth of microbes. Due to lack of the elemental analysis data of biomass of gut microbes only two different kinds of biomass reactions were included in the models, one for Gram-positive and one for Gram-negative taxa. An automatic pipeline was used to fill the gaps in the draft models under specific growth condition of human breast milk (HBM) as media (Materials and Methods) (Latendresse, 2014). Hereby different sets of reactions were incorporated into each model, which is necessary due to non-perfect genome annotations and variations in growth requirements for different organisms. Growth, i.e. biomass formation, was considered as an objective function during the gap-filling. An overview of key parameters for all 58 gut bacterial GEMs (M1–M58,) is summarized in Figure 2. In the next step, we manually curated the gap-filled models in terms of enabling anaerobic growth, removal of unused gap-filled reactions, enabling key metabolic tasks, such as synthesis of SCFAs and AAs, corrections in reaction directionalities, and improving model annotation (Supplemental Text). GEMs that contained more than 75% of their reactions based on available gene annotations were further analyzed to reduce the possible influence of over-fitting in the automatic gap-filling process (Figure 2A). Collectively, the models incorporate a total number of reactions and metabolites of 2160 (including exchange reactions) and 1557, respectively. Moreover, these 58 GEMs contain genes, reactions, and metabolites in the range of 1645 to 6680, 818 to 1547, and 805 to 1248, respectively (Figure 2B, Table S2 and S3).
A metabolic dissimilarity test was performed between the models to elucidate how these models are metabolically distinct from each other (Materials and Methods). The metabolic dissimilarity was examined in terms of metabolic distances, which was determined using the Jaccard coefficient considering shared and unique reactions between the models. Results of this analysis inferred that models representing the same phyla contained more reactions in common than models representing different phyla (Figure 2B). Moreover, models shared only 41% of the reactions and as expected, models of Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria incorporated 20, 50, 163, and 132 unique reactions, respectively, which showed distinctiveness between the models based on genomic differences of the bacterial species (Figure S4).
Validation of GEMs
In the next step, the accuracy of the reconstructed GEMs was examined by predicting growth rates and comparing these with experimentally generated data for six gut bacterial species grown on YCFA medium under anaerobic conditions. Species involved in this analysis are Bacteroides thetaiotaomicron (M6), Bifidobacterium adolescentis (M9), Eubacterium rectale (M29), Faecalibacterium prausnitzii (M31), Prevotella copri (M41), and Roseburia inulinivorans (M43) (Materials and Methods). GEMs were simulated using components of the YCFA medium (Table S7) with different glucose uptake rates (mmol gDW−1 h−1) as demonstrated by experimental observations (Table 1). As shown in Figure 2C and Table 1, GEM predictions agreed well with experimental results.
Estimation of growth and metabolic capabilities of gut bacterial species
HBM and RUTF (Table S6) provide vital dietary requirements for children during their early age. Using these two diets, gut bacterial GEMs were simulated for predicting the growth and secretion rate of metabolites, such as short chain fatty acids (SCFAs) and amino acids (AAs). Another growth media, which is called complete media (Benedict et al., 2014), was also used to examine the growth and metabolic capabilities of gut microbial species. Complete media is an in silico growth media, which does not contain fixed components. Instead, complete media allows the models to simulate growth on all metabolites for which transport reactions are available in the model (Supplemental Text).
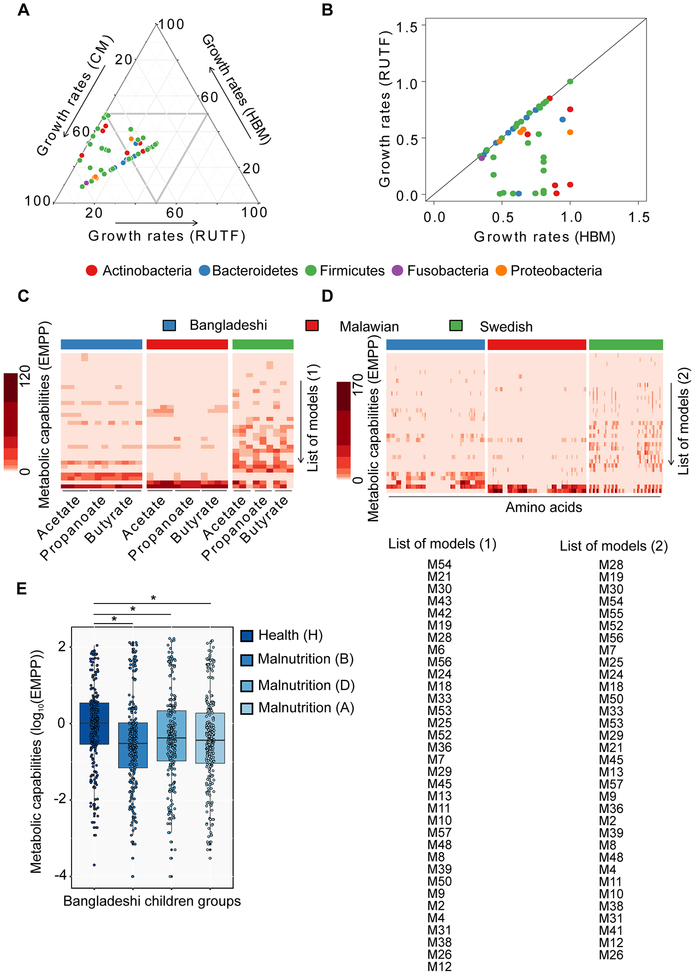
Model predictions suggested large variations in growth rates of the gut microbes on the three different media (Figure 3A). In comparison, 33 models predicted faster growth on CM than on the other two media (HBM and RUTF) (P < 0.01 for all comparisons, Mann-Whitney U test). This is to be expected because CM contains all of the compounds that are required for growth of each bacterium. This shows good quality of the models in terms of making biologically meaningful predictions. The remaining 25 models predicted similar growth rates (h−1) on all three media. Next, we compared predicted growth rates (h−1) of bacteria on HBM and RUTF (Figure 3B). The results showed that all of the models either predicted faster growth on HBM (P < 0.01 for all comparisons, Mann-Whitney U test) or similar growth rates on HBM and RUTF. Actinobacteria, Proteobacteria, and Fusobacteria were predicted to grow faster on HBM except Bifidobacterium breve (Actinobacteria), whereas Firmicutes and Bacteroidetes grew either with similar growth rates on both media or faster on HBM. This clearly shows that RUTF lacks certain nutrients compared to HBM in terms of enabling fast growth of gut symbionts. Thus, the unavailability of human breast milk and compensation by RUTF may have an effect on shaping the composition of the gut microbiota of malnourished children during their early life. This could be because of the nutritional differences between HBM (Dewey et al., 1995; Gale et al., 2012; Lanting et al., 1994) and RUTF, and this may explain why human breast milk is vital for shaping the gut microbiota in infants (Subramanian et al., 2015).
Figure 3: In silico predictions of growth rates of gut bacteria and secretion of health-promoting small molecules.
(A) Triplot illustrates comparative representation of predicted growth rates (h−1) on three media/diets, (i.e. complete media (CM), human breast milk (HBM), and ready-to-use therapeutic food (RUTF)). R packages ggtern and ggplot were used to plot the triplot. (B) Absolute values of the growth rates (h−1) on two media (human breast milk (HBM) and ready-to-use therapeutic food (RUTF)) (C) Estimated maximal production potential (EMPP) represents metabolic capabilities of the gut microbiota in terms of secretion of short chain fatty acids (SCFAs; left to right for each children group, acetate, propanoate, and butyrate) and (D) amino acids (AAs; left to right for each children group, L-Glutamate, Glycine, L-Alanine, L-Lysine, L-Aspartate, L-Arginine, L-Glutamine, L-Serine, L-Methionine, L-Tryptophan, L-Phenylalanine, L-Tyrosine, L-Cysteine, L-Leucine, L-Histidine, L-Proline, L-Valine, L-Threonine, L-Isoleucine, and L-Asparagine)) using GEMs of gut bacteria from Malawian, Bangladeshi, and Swedish children. heatmap.2 function of gplots package, and dist and hclust functions of R were used for generating heatmaps and dendrograms, respectively, in R version 3.3.0. (E) Box plots represent comparative metabolic capabilities of gut bacteria between healthy and three stages of malnourished children (*P < 0.01 for all comparisons, Mann-Whitney U test). Metabolic capabilities of gut bacteria have been illustrated in the form of EMPP collectively in terms of SCFAs and AAs secretion. For the details of different stages of health and malnutrition of Bangladeshi children, Tables S1 and Figure S2 can be referred. List of models (1) and List of models (2) represent the labels for y-axis in panel (C) and (D), respectively. Abbreviations of models are consistent with Figure 2.
Previous studies suggest that the human gut harbors trillions of microbes and both host and microbes maintain a homeostasis in terms of exchange of metabolites for health benefits along with other mutual relationships (Schroeder and Bäckhed, 2016). SCFAs and branched chained fatty acids (BCFAs) are some of the most vital gut microbiota-derived metabolites, for instance, acetate, propanoate, butyrate, and isobutyrate etc. (De Vadder et al., 2014; Louis and Flint, 2017), which significantly contribute to the daily energy requirement, in particular for proper development of the gastrointestinal system (Bergman, 1990). Moreover, gut microbes also produce amino acids, importantly, essential amino acids, like tryptophan and phenylalanine, and derivatives thereof such as kynurenine that serves an important role in regulating the immune responses (Husted et al., 2017; Metges, 2000; Nguyen et al., 2010; Shoaie et al., 2015). We therefore used the GEMs to predict the maximal production capacity of three SCFAs (acetate, propanoate, and butyrate) and 20 AAs (L-Glutamate, Glycine, L-Alanine, L-Lysine, L-Aspartate, L-Arginine, L-Glutamine, L-Serine, L-Methionine, L-Tryptophan, L-Phenylalanine, L-Tyrosine, L-Cysteine, L-Leucine, L-Histidine, L-Proline, L-Valine, L-Threonine, L-Isoleucine, and L-Asparagine) for the selected 58 bacterial species. The metabolic capabilities of the individual gut microbes were estimated in terms of Estimated Maximal Production Potential (EMPP) of a species for secreting SCFAs and AAs (Figure 3C and 3D). EMPP was calculated by multiplying the flux of metabolite production and the abundance of the corresponding species (Materials and Methods). This analysis showed a much larger metabolic diversity of the gut microbiota in Swedish children in terms of SCFAs and AAs secretions, compared with the Malawian and Bangladeshi children, who have much lower EMPP for most of the metabolites evaluated by our analysis. This difference in metabolic diversity was noticed irrespective of health and disease states of the Malawian children groups, i.e. there were no significant differences between metabolic capabilities of healthy and malnourished children groups from Malawi. However, metabolic capabilities of the gut microbiota in Bangladeshi children were significantly different between healthy and malnourished children (P < 0.01 for all comparisons, Mann-Whitney U test; Figure 3E). These results indicated that there were gut microbiota-driven metabolic variations between children groups, which is likely driven by environmental factors, e.g. dietary differences in Sweden versus Malawi and Bangladesh, but also that the reduced metabolic diversity in the Malawian and Bangladeshi children may increase the risk of malnutrition. This is consistent with an overall lower microbial diversity in these children groups. More importantly, to explore any possible associations between significantly lower metabolic capabilities of gut microbiota in malnourished Bangladeshi children than healthy group with plasma biochemistry, plasma metabolic profiling was performed using blood samples from this children group (see later).
Metabolic potential of communities of gut bacterial species in health and malnutrition
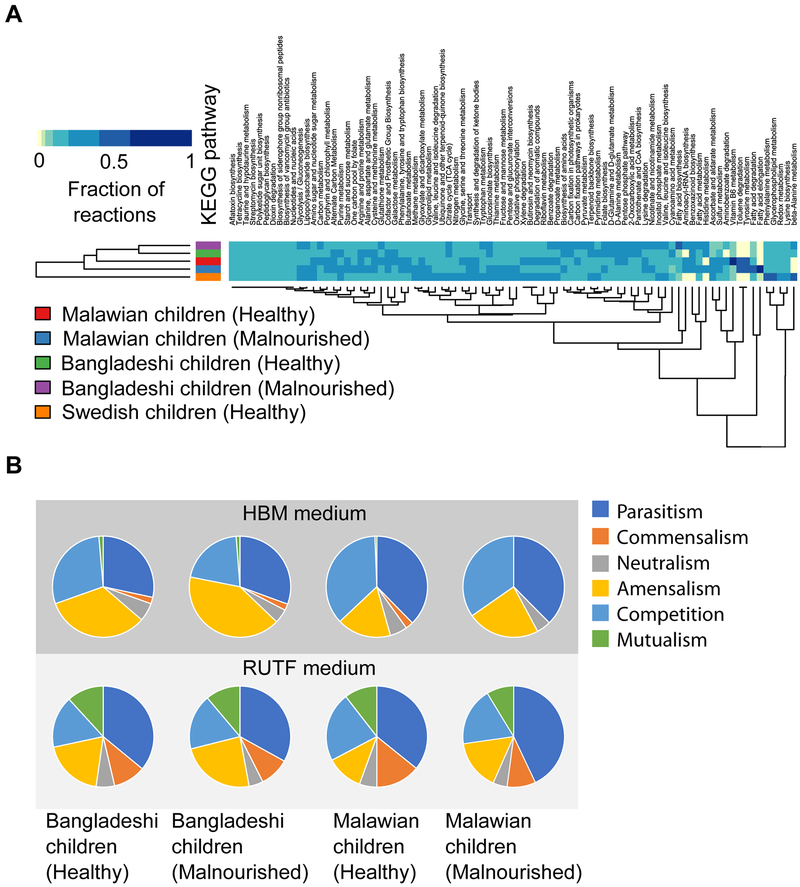
Using single species GEMs, Community Metabolic Models (CMMs) were reconstructed, following a previously published method (Henry et al., 2016). This method allows single species GEMs to merge into one model with a single intracellular compartment, which is also called mixed-bag network modeling. The objective of this analysis was to estimate the functional enrichment in gut microbial communities during health and malnutrition. Here, functional enrichment was defined as the number of active reactions representing particular metabolic pathways in CMM of a microbial community, when CMMs were simulated on HBM. For this analysis, five gut microbial communities based on five different children groups, namely Malawian children (healthy), Malawian children (malnourished), Bangladeshi children (healthy), Bangladeshi children (malnourished), and Swedish children (healthy) were considered to reconstruct the CMMs (Table S5). The total number of active reactions in the CMMs were 752, 755, 785, 815, and 832, respectively. Functional categories were assigned to the reactions using KEGG (Kanehisa et al., 2004) and BIGG (Schellenberger et al., 2010) databases. Some metabolic functional categories, for instance, phenylalanine metabolism, glycerophospholipid metabolism, redox metabolism, lysine biosynthesis, and beta-alanine metabolism were dominated by reactions in Swedish children compared with the other children groups (Figure 4A). In contrast, there was no pattern found in the case of other metabolic functions. The highest number of reactions lie in amino acid metabolism functional category covering 19% to 23% of total number of reactions in CMMs.
Figure 4: Community-level analyses of the gut microbiota during health and malnutrition.
(A) Functional enrichment in gut microbial communities during health and malnutrition. Here, we defined the functional enrichment in terms of number of active reactions representing particular functional categories in CMMs while simulating on HBM. The heatmap represents the fraction of reactions of each pathway present in each CMM. Table S5 can be referred for the composition of each microbial community. Functional enrichment was determined using KEGG and BIGG databases. heatmap.2 function of gplots package, and dist and hclust functions of R were used for generating heatmap and dendrogram, respectively, in R version 3.3.0. (Experimental Procedure). (B) Pairwise growth simulations. These pie charts represent the possible effects of pairwise growth in gut microbial communities based on Bangladeshi and Malawian healthy and malnourished children on (A) HBM and (B) RUTF medium. The effects of pairwise growth was determined in terms of the number of pairs of species involved in each kind of species-species interactions, namely competition, parasitism, commensalism, mutualism, amensalism, and neutralism.
Pairwise interactions between species from different microbial communities in health and malnutrition
Competition and interaction
In order to elucidate the potential effect of bacteria on each other in terms of growth, we analyzed the extent of competition and cooperation existing between the species in each of the four aforementioned microbial communities based on four children groups, namely Malawian children (healthy), Malawian children (malnourished), Bangladeshi children (healthy), and Bangladeshi children (malnourished), in a pairwise manner (Table S5). We inspected the consumed and the produced metabolites for each of the bacterial models while simulating growth on HBM and RUTF (Table S6). We then identified the potential metabolites that could be regarded as a source of competition, in a sense that they are required by both of the models for growth optimization.
However, this could include metabolites that do not affect the growth rate and we therefore analyzed the reduced costs of the exchange reactions associated with the metabolites that each pair of bacteria have in common as their consumed metabolites while maximizing their growth. We performed the same analysis for the metabolites that could be considered as a means of cooperation between the bacterial species, meaning when the metabolite is produced by one and consumed by the other. According to our results, the bacteria are mostly competing for resources that they need for growth and there are less interactions in terms of cooperation, i.e. one species needs a metabolite for growth that is produced by another species (Figure S5). There are, however, also cases that they cooperate, but such cases are fewer than competition, which is in line with the fact that competition has been reported to be a more stabilizing force for a microbial community (Coyte et al., 2015).
Effect on growth rate
Next, we checked how these interactions, either competition or cooperation, would influence the growth rate of bacteria when they grow together. In order to do so, we simulated co-growth of every possible pairwise combination of the bacterial models on HBM and RUTF media in the four aforementioned microbial communities. In pairwise growth simulations, the media metabolites were shared between the two models and the metabolites that were produced by either of them, was added to the available media resources. A linear programming (LP) was formulated applying these constraints concerning the exchanged metabolites (Materials and Methods). The objective function for this LP was the sum of biomass reactions of the two models. When the models were optimized in this manner, each would reach a specific growth rate that might be different from its growth rate when it was optimized for growth as a single model. Pairwise growth rate might be higher or lower than the single-species growth rate, or it might not be affected. Depending on the effect that the bacteria could have on each other in terms of growth rates, there are six possible outcomes: 1) competition, 2) parasitism, 3) commensalism, 4) mutualism, 5) amensalism, and 6) neutralism (Heinken and Thiele, 2015). Our results show that on HBM media, the most dominant effects that the microbes have on the growth rate of each other are parasitism, amensalism and competition (Figure 4B). There are fewer cases of amensalism in healthy communities compared to the malnourished ones, while parasitism is quite similar between the two communities in both Bangladeshi and Malawian cases. Competition, commensalism, neutralism and mutualism is marginally higher in healthy communities compared with malnourished communities on HBM media. The results on RUTF showed that the most frequent effects are parasitism, followed by competition and amensalism similar to the HBM media. However, on RUTF we observed more commensalism and mutualism due to the fact that RUTF contains a different composition compared to HBM Table S6, but it is contributing to mutualism, which is generally not considered a strong force in community stabilization.
Metabolite production
Lastly, we investigated the effect on microbial co-growth in terms of metabolite production. To this end, the same procedure for pairwise growth simulation was used as explained above regarding exchanged metabolites. In single-species metabolite production simulations (Figure 3C and 3D), each model was first optimized for growth, and subsequently for metabolite (SFCAs or AAs) production while setting the optimum achieved growth rate as the lower bound for the biomass reaction (Materials and Methods). In pairwise growth simulations, for every possible pairwise combination of bacteria, first an LP was solved with the objective function being the sum of biomass reactions to get the growth rates of the microbes while growing together. Subsequently these rates were used as the lower bound of the biomass reactions and a demand reaction was added to each model for production of the metabolite of interest. Finally, by comparing the rates of the produced metabolite in single versus pairwise manner for each bacteria, we investigated how the microbes influence each other’s potential for metabolite productions that are afterwards metabolized by the host. We performed these analyses for all four communities on HBM and RUTF and the results are shown in Table S8. Regarding SCFAs, we noted that the bacteria affect each other in dissimilar ways. In case of acetate, in almost all communities on both HBM and RUTF media, there is no change of production when comparing pairwise with single microbe simulations. Contrarily, we observed that bacteria influence the rate of propionate and butyrate production notably, however in different ways. For butyrate, the rate of production is increased in pairwise simulation compared to single species analysis and this increase is higher in healthy communities in both Bangladeshi and Malawian children, which is consistent with the beneficial roles associated to butyrate. Butyrate can serve as an energy source for intestinal epithelial cells and might prevent colonic carcinogenesis (Beyer-Sehlmeyer et al., 2003; Velázquez et al., 1996). Besides, this increase in butyrate production is marginally higher in RUTF in Malawian groups, but not in Bangladeshi, which reflects the compositional effects of microbial communities. In case of propionate production, the results showed that the bacterial species tend to produce less propionate when simulated in pairwise manner in all communities, however, the amount of decrease is more noticeable in Bangladeshi communities while there are more cases of increase of propionate production in Malawian children. In regard to amino acid production potential, we observed a decrease in the potential production for five out of the nine essential amino acids, i.e. lysine, tryptophan, histidine, valine and isoleucine, as well as for the conditionally essential amino acid arginine, for pairwise simulations. Comparing Malawian healthy and malnourished communities, there is a decreased production potential for three out of the nine essential amino acids, i.e. phenylalanine, leucine and methionine, for two conditionally essential amino acids, i.e. tyrosine and proline, and for the non-essential amino acid alanine. For Bangladeshi groups where two essential amino acids, i.e. valine and isoleucine are less produced in malnourished children and the others are either not changed or altered slightly.
Plasma metabolic profiling in health and malnutrition (stunting)
Our GEMs-based analyses suggested that there were significantly lower metabolic capabilities of gut microbiota in malnourished children group from Bangladesh compared to the healthy group. To explore any possible associations between variations in gut microbiota with plasma biochemistry of these children, we therefore performed plasma metabolic profiling of a group of healthy and stunted children from Bangladesh (from the PROVIDE cohort (Naylor et al., 2015; Subramanian et al., 2014), which is the same cohort used for the gut microbiota study analyzed above). Each group included 25 children and the characteristics of total 50 children are shown in Table 2. Healthy and stunted children groups included female subjects of 48 % and 32 %, respectively. The design of our study is described in Materials and Methods, and depicted in Figure S6. Here, we quantitated a total of 50 plasma metabolites that included amino acids, short chain and branched chain fatty acids, TCA cycle intermediates, and sugars up to the age of 2 years (at which time the growth status of the children was assessed using the HAZ score, with HAZ < 2 indicative of stunting). Age- and gender-adjusted concentrations of these metabolites were compared between the children groups with and without stunting using Wilcoxon rank-sum test (Materials and Methods). A summary of these comparisons is depicted in Table S9 and S10.
Ratio of Tryptophan to Neutral Amino Acids
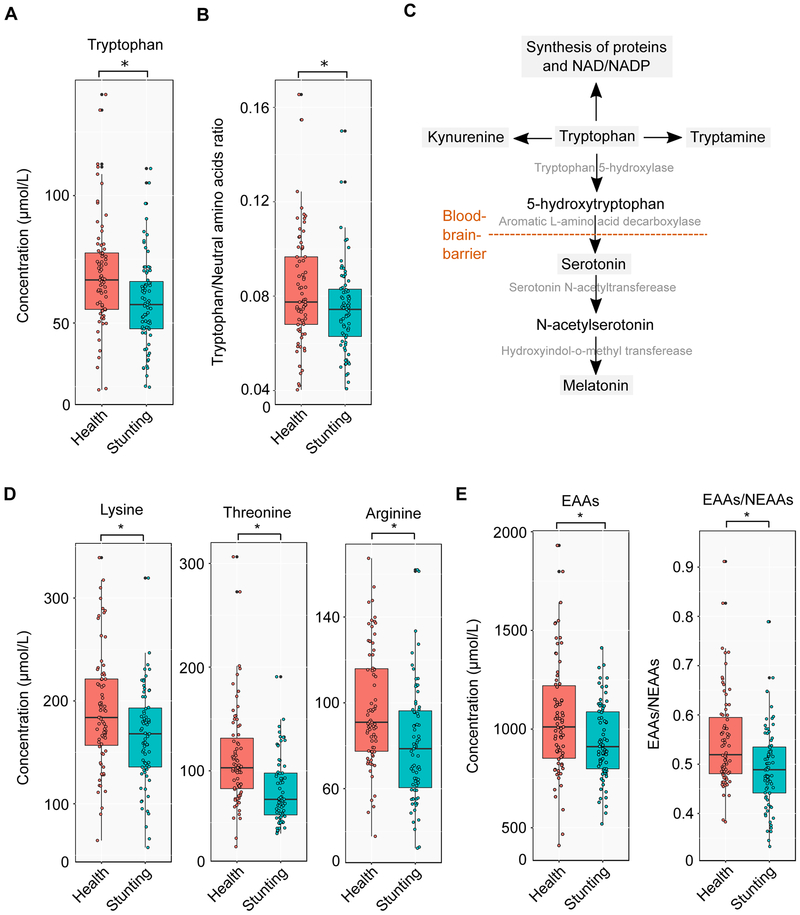
A significantly lower level of Tryptophan (adjusted P < 0.05, Wilcoxon rank-sum test) was recorded in plasma of stunted children compared to healthy control group (Figure 5A and Table S9). Previous studies suggested that a ratio of tryptophan to the sum of other neutral amino acids (T/NAAs), for example histidine, isoleucine, leucine, methionine, phenylalanine, threonine, tyrosine, and valine, was associated with maturation and functioning of brain, in particular at the early age of life (Heine et al., 1999; Richard et al., 2009). Therefore, we examined T/NAAs ratio between heathy and stunted children groups, and stunted children had significantly reduced level of T/NAAs ratio compared to healthy children (adjusted P < 0.05, Wilcoxon rank-sum test) (Figure 5B).
Figure 5: Distinct profiles of essential and conditionally-essential amino acids in healthy and stunted children.
(A) Plasma concentrations of tryptophan in health (n=25) and stunting (n=25). (B) Ratio of tryptophan to other neutral amino acids such as histidine, isoleucine, leucine, methionine, phenylalanine, threonine, tyrosine, and valine in health and stunting (n=25). (C) Some important pathways associated with tryptophan such as synthesis of protein, tryptamine, kynurenine, serotonin, and melatonin. (D) Plasma concentrations of lysine, threonine, and arginine in health (n=25) and stunting (n=25). (E) Overall concentrations of nine essential amino acids (lysine, histidine, threonine, methionine, tryptophan, isoleucine, leucine, phenylalanine, and valine) in health (n=25) and stunting (n=25) and ratio of total concentration of essential to non-essential amino acids in health and stunting.
*adjusted P < 0.05 (statistical significance). All statistical comparisons between healthy and stunted children data were performed using Wilcoxon rank-sum test. We adjusted P values based on multiple testing of all metabolites using a previously published method.
Profiles of Essential and Non-essential amino acids
The importance of protein quality in the mother’s diet for improved birth weight and reduced risk of small gestational size is well recognized (Bhutta et al., 2013). In a recent cross-sectional study with children between 1 and 5 years of age in Malawi, stunted children were found to have low levels of essential amino acids in plasma (Semba et al., 2016). We therefore measured all nine essential amino acids (lysine, histidine, threonine, methionine, tryptophan, isoleucine, leucine, phenylalanine, and valine), together with seven conditionally-essential amino acids (arginine, glutamine, glycine, proline, serine, tyrosine, and asparagine) and sixteen non-essential amino acids in plasma samples from both groups at several different time points (Experimental Procedure; Table S9). At individual-level, concentration of two essential amino acids, namely lysine and threonine and one conditionally-essential amino acid, namely arginine, were found to be significantly lower in stunted children compared to healthy group (Figure 5D). Moreover, the reduced level of metabolites in stunted children were consistent in the case of overall concentration of essential amino acids (EAAs) and ratio of overall concentration of essential to non-essential amino acids (EAAs/NEAAs) (Figure 5E). It was also noticed that two non-essential amino acids (hydroxyproline and α-aminobutyric acid; Figure S8A and B) had a reduced concentration in plasma of stunted children.
Metabolites Elevated in Plasma of Stunted Children
One non-essential amino acid (cystathionine), two short-chain fatty acids (propionate and butyrate), and two TCA cycle intermediates (Pyruvate and malate) were found to be significantly higher in the plasma of stunted children compared to the healthy group (Figure S9, Table S9 and S10). However, there was no significant difference noticed in plasma sugars (fructose, mannose, and glucose) level between health and stunting (Table S10).
Correlations between plasma metabolites and HAZ score of children
Spearman’s correlation coefficient was determined to examine the correlation between plasma metabolites and HAZ score of children groups (Experimental Procedure). The outcomes of this analysis are summarized in Table S11. This analysis suggested that six essential (lysine, isoleucine, leucine, valine, tryptophan, and threonine), one conditionally-essential (arginine), and one non-essential (α-aminobutyric acid) amino acids were positively correlated with HAZ score (adjusted P < 0.05, Wilcoxon rank-sum test; Figure 6). The significant negative associations were noted between two other metabolites (pyruvate and butyrate) and HAZ score (adjusted P < 0.05, Wilcoxon rank-sum test; Figure 6).
Figure 6: Correlations between plasma metabolites and HAZ score of children.
Volcano plot represent the Spearman’s correlation coefficients versus minus logarithm of adjusted P values between plasma amino acids and HAZ score of children. Red dots denote plasma metabolites that significantly correlated with HAZ score of children (adjusted P < 0.05, Wilcoxon rank-sum test). Plot shows that eight metabolites (lysine, isoleucine, leucine, valine, tryptophan, and threonine, arginine, and α-aminobutyric acid) are positively and two metabolites (pyruvate and butyrate) negatively correlated with HAZ score of children.
DISCUSSION
Metabolic functions and interactions between individual species in the gut microbiota during health and malnutrition
The main goal of applying computational approaches in gut microbiome studies is to understand the underlying mechanisms of alterations in the gut microbiota during different health and disease states. A detailed mechanistic understanding of the gut microbial ecosystem can help to modify the microbiome towards health benefits. Here, we used constraint-based modeling to estimate the metabolic capabilities of the gut microbial species in health and disease. Moreover, we performed data mining of gut microbial species abundance data from children groups of low- and high-income countries.
Interestingly, the poor state of the gut microbiota of the Malawian and Bangladeshi children is also clearly, illustrated by much lower Shannon diversity index for these children, something that has been ascribed to increased health risk for over-weight and obese subjects (Menni et al., 2017). Species-level abundances demonstrated a higher F/B ratio in Malawian and Bangladeshi children than Swedish children. The F/B ratio was not found to be directly associated with malnutrition, but this ratio was found to be higher in children groups having more prevalence of malnutrition. The difference in F/B ratio between the children groups was mainly due to the variations in abundance of Bacteroidetes. Here, it should be noted that like Bifidobacterium, several species of Bacteroidetes have been shown to play an important role in consuming undigested human milk oligosaccharides (HMOs) (Marcobal et al., 2011).
Our modeling allowed us to quantify the metabolic potential of gut microbial species in health and malnutrition. However, due to knowledge gaps in the understanding of gut microbial ecology, this framework is based on some simplifying assumptions. First, all constraint-based models in this study were reconstructed using fixed annotated genomes from publicly available databases and gene copy numbers in metagenomic datasets were not considered. More precise attempts towards annotation of strain-specific genomes and multi-omics data could improve this modeling framework for more accurate predictions. Second, to reduce the complexity in model reconstruction and analysis, only the 20 most abundant bacteria were selected to represent the microbial community from all children groups in health and malnutrition. This problem can be overcome, partially, in future studies by extending this framework to whole microbial communities for different metagenomic datasets. Third, our modeling framework is limited for analyzing cross-feeding between more than two species. So, the presented metabolic variations in microbial communities are based on independent single species. It is believed that there could be entirely different scenarios of metabolic potentials if cross-feeding was considered between several species.
However, despite these shortcomings our modeling efforts clearly provided new insights into the function of the gut microbiota of children in three different countries, and two key lessons from our study are: 1) The higher species diversity of the gut microbiota of Swedish children compared with Malawian and Bangladeshi children directly translates to a reduced metabolite production capabilities in the latter group of children. This can have an important impact on the development of the epithelial cells of the gut, and hereby lead to poor development of this vital organ. However, it may also have a direct impact on the absorption of certain essential amino acids, which may in particular be important if the children have restricted provision of these amino acids from their food. 2) Each bacterium requires a specific set of metabolites to grow and produces some metabolites as metabolic by-products. When two bacterial species need the same metabolites for growth, they will compete and this competition might have an effect on their growth rates. However, a metabolite may be required by both while not having a significant effect, or any effect at all on the growth rate such as water or ions that are available in excess in the environment. On the other hand, a metabolic by-product of one bacterium might be consumed by another one and in turn influence the growth of one or both when growing in proximity to each other. From our analysis of metabolic interactions between species we found clear differences in output between gut microbiota from malnourished and healthy children in both the Malawian and the Bangladeshi cohorts. Finally, our modeling showed reduced capacity for the gut microbiome to produce several essential amino acids in stunted children, which was supported by our metabolome analysis.
Alterations in metabolism of stunted children
Here we present the plasma metabolomics data from malnourished children and our analysis showed that tryptophan was one of the amino acids found to be at significantly lower levels in the malnourished children. Apart from its role in protein and synthesis (Sainio et al., 1996), tryptophan is also a key precursor for the pathways associated with synthesis of serotonin, melatonin, kynurenine, and tryptamine (Figure 5C). For synthesis of serotonin and melatonin, tryptophan is transported across the blood-brain-barrier into the brain. During this process, tryptophan competes with other NAAs (for example histidine, isoleucine, leucine, methionine, phenylalanine, threonine, tyrosine, and valine) for the blood-brain-barrier transporter (Fernstrom, 1983; Fernstrom and Wurtman, 1972). Therefore, the T/NAAs ratio is a crucial factor and changing this ratio can cause significant differences in availability of tryptophan for the synthesis of serotonin and melatonin in brain (Heine et al., 1999). The lower T/NAAs ratio found here in stunted children group can be associated with adverse effect on brain maturation and function. Similarly, decreased plasma tryptophan concentration can affect the synthesis of kynurenine and tryptamine. Kynurenine is precursor of kynurenic and quinolinic acid that affect other neurotransmitters, for example glutamate (Moroni, 1999). Through the kynurenine and quinolinic acid pathway, tryptophan can also act as precursor for niacin (or nicotinic acid and nicotinamide or water-soluble vitamin B3) synthesis, which is essential for NAD+ and NADP+ synthesis (Moffett and Namboodiri, 2003). Tryptamine is an important neuromodulator of serotonin that plays an important role in controlling the functions of serotonin (Jones, 1982). Additionally, several recent studies have shown that the gut microbiota metabolism impacts the level of plasma metabolites (Fujisaka et al., 2018; Krishnan et al., 2018; Org et al., 2017; Ottosson et al., 2018; Wikoff et al., 2009; Yano et al., 2015).
Apart from tryptophan, the concentration of several other essential amino acids, e.g. lysine and threonine were significantly lower in stunted children (Figure 5D). Lysine is an important precursor for the protein synthesis (Tomé and Bos, 2007) as well as the biosynthesis of carnitine that plays a very vital role in β-oxidation (Tanphaichitr and Broquist, 1973). Lysine also acts as a regulator for nitric oxide (NO) synthesis pathway from arginine (Wu, 2009). A WHO report suggested that there is a higher requirement of lysine in infants than adults (WHO/FAO/UNU Expert Consultation, 2007). Threonine plays a role in synthesis of the mucin proteins, which are important for gut immunity and necessary for maintaining intestinal integrity and function (Wu, 2009). In our study, the level of another amino acid, arginine, which is a conditionally-essential amino acids, was found to be lower in plasma of stunted children compared to healthy group (Figure 5D). Arginine is a vital precursor for synthesis of not only proteins but also of NO, proline, ornithine, glutamate, citrulline, creatine, and agmatine (Wu and Morris Jr., 1998). Arginine directly or through its products is involved in several important functions, for example, in tissue repairing, wound healing, blood flow modulating, mitochondrial respiration, and ammonia detoxification (Figure S7) (Wu, 2009). In the study, it has been noticed that stunted children had significantly lower overall concentration of plasma essential amino acids (lysine, histidine, threonine, methionine, tryptophan, isoleucine, leucine, phenylalanine, and valine) as well as lower ratio of essential to non-essential amino acids than healthy children (Figure 5E). Mammals do not possess the enzymatic machinery for synthesis of essential amino acids that shows mammals’ dependency on dietary intake and gut microbiome for availability of these amino acids (Moffett and Namboodiri, 2003). The lower concentrations of plasma essential amino acids in stunted children may be because of possible lack of proper proportion of amino acids in diet or/and inability of gut to absorb the nutrients. Moreover, ratio of essential to non-essential amino acids has been proposed as an indicator of protein nutrition previously (Arroyave, 1970; Whitehead, 1964).
We also determined the correlation between plasma metabolites and HAZ score of children. For this purpose, we calculated Spearman’s correlation coefficients and results demonstrated positive coefficients for eight metabolites (lysine, isoleucine, leucine, valine, tryptophan, and threonine, arginine, and α-aminobutyric acid) and negative coefficients for two metabolites (pyruvate and butyrate).
The findings of this study demonstrate alterations in plasma biochemistry in health and stunting. Our results mainly highlight a significantly (i) reduced level of essential amino acids, (ii) reduced level of arginine (conditionally-essential amino acid), and (iii) reduced ratio of essential to non-essential and ratio of tryptophan to other neutral amino acids, in stunted children compared to healthy group. Inadequate proportion of plasma amino acids can be one of the possible cues for inability of micronutrients supplementation for enhancing child growth (Ashorn et al., 2015b; Garza, 2015; Mayo-Wilson et al., 2014; Ramakrishnan et al., 2009). These reduced levels of metabolites, in particular, essential amino acids in the plasma samples of malnourished children were consistent with lower predicted metabolic capabilities of the gut microbiota using GEM simulations. This suggested that there may be potential associations between functional capabilities of gut microbiota and plasma metabolite levels in malnutrition. In order to test this hypothesis, more future research efforts are required to uncover the mechanisms underlying how the gut microbiota as well as other cues (since the altered levels of plasma metabolites may not be the only reflection of gut microbiome) play a role in altering the host metabolism in disease states.
Besides these important biological findings, we believe that our proposed modeling framework can be also used to evaluate other scenarios in particular as the set of 58 high quality GEMs for gut bacteria represents a valuable resource for further studies of metabolic diseases associated with an altered gut microbiota.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge financial support from Bill & Melinda Gates Foundation, the Novo Nordisk Foundation, Vetenskapsrådet, Knut and Alice Wallenberg Foundation, MetaCardis, FORMAS, and the Swedish Foundation for Strategic Research. We thank Dr Karen Scott, Dr Sylvia Duncan and Jenny Martin from Rowett Institute in Nutrition and Health, Aberdeen, UK, for providing Bifidobacterium adolescentis L2-32, Eubacterium rectale A1-86 (DSM 17629), Faecalibacterium prausnitzii A2-165 (DSM 17677) and Roseburia inulinivorans A2-194 (DSM 16841).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agren R, Bordel S, Mardinoglu A, Pornputtapong N, Nookaew I, Nielsen J, 2012. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput. Biol 8, e1002518 10.1371/journal.pcbi.1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyave G, 1970. Comparative sensitivity of specific amino acid ratios versus “essential to nonessential” amino acid ratio. Am. J. Clin. Nutr 23, 703–6. [DOI] [PubMed] [Google Scholar]
- Ashorn P, Alho L, Ashorn U, Cheung YB, Dewey KG, Gondwe A, Harjunmaa U, Lartey A, Phiri N, Phiri TE, Vosti SA, Zeilani M, Maleta K, 2015a. Supplementation of Maternal Diets during Pregnancy and for 6 Months Postpartum and Infant Diets Thereafter with Small-Quantity Lipid-Based Nutrient Supplements Does Not Promote Child Growth by 18 Months of Age in Rural Malawi: A Randomized Controlled Tria. J. Nutr 145, 1345–1353. 10.3945/jn.114.207225 [DOI] [PubMed] [Google Scholar]
- Ashorn P, Alho L, Ashorn U, Cheung YB, Dewey KG, Gondwe A, Harjunmaa U, Lartey A, Phiri N, Phiri TE, Vosti SA, Zeilani M, Maleta K, 2015b. Supplementation of Maternal Diets during Pregnancy and for 6 Months Postpartum and Infant Diets Thereafter with Small-Quantity Lipid-Based Nutrient Supplements Does Not Promote Child Growth by 18 Months of Age in Rural Malawi: A Randomized Controlled Tria. J. Nutr 145, 1345–1353. 10.3945/jn.114.207225 [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O, Meyer F, Goesmann A, McHardy A, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Bryson K, Loux V, Bossy R, Nicolas P, Chaillou S, Guchte M van de, Penaud S, Maguin E, Hoebeke M, Bessieres P, Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Medigue C, Overbeek R, Begley T, Butler R, Choudhuri J, Chuang H, Cohoon M, Crecy-Lagard V. de, Diaz N, Disz T, Edwards R, Tatusov R, Natale D, Garkavtsev I, Tatusova T, Shankavaram U, Rao B, Kiryutin B, Galperin M, Fedorova N, Koonin E, Schneider M, Tognolli M, Bairoch A, Kanehisa M, Goto S, Haft D, Loftus B, Richardson D, Yang F, Eisen J, Paulsen I, White O, Overbeek R, Bartels D, Vonstein V, Meyer F, Wu C, Shivakumar S, Lowe T, Eddy S, Delcher A, Harmon D, Kasif S, White O, Salzberg S, DeJongh M, Formsma K, Boillot P, Gould J, Rycenga M, Best A, Becker S, Palsson B, 2008. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9, 75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaei P, Shoaie S, Ji B, Nielsen J, 2018. Challenges in modelling the human gut microbiome. Nat. Biotechnol. In Press. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J, 2015. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 17, 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Bahwere P, Balaluka B, Wells JCK, Mbiribindi CN, Sadler K, Akomo P, Dramaix-Wilmet M, Collins S, 2016. Cereals and pulse-based ready-to-use therapeutic food as an alternative to the standard milk- and peanut paste-based formulation for treating severe acute malnutrition: A noninferiority, individually randomized controlled efficacy clinical trial. Am. J. Clin. Nutr 103, 1145–1161. 10.3945/ajcn.115.119537 [DOI] [PubMed] [Google Scholar]
- Benedict MN, Mundy MB, Henry CS, Chia N, Price ND, 2014. Likelihood-Based Gene Annotations for Gap Filling and Quality Assessment in Genome-Scale Metabolic Models. PLoS Comput. Biol 10, e1003882 10.1371/journal.pcbi.1003882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the False Discovery Rate : A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc 57, 289–300. [Google Scholar]
- Bergman EN, 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev 70, 567–90. [DOI] [PubMed] [Google Scholar]
- Beyer-Sehlmeyer G, Glei M, Hartmann E, Hughes R, Persin C, Böhm V, Rowland I, Schubert R, Jahreis G, Pool-Zobel BL, 2003. Butyrate is only one of several growth inhibitors produced during gut flora-mediated fermentation of dietary fibre sources. Br. J. Nutr 90, 1057–1070. 10.1079/BJN20031003 [DOI] [PubMed] [Google Scholar]
- Bhutta ZA, Ahmed T, Black RE, Cousens S, Dewey K, Giugliani E, Haider BA, Kirkwood B, Morris SS, Sachdev H, Shekar M, 2008. What works? Interventions for maternal and child undernutrition and survival. Lancet 371, 417–440. 10.1016/S0140-6736(07)61693-6 [DOI] [PubMed] [Google Scholar]
- Bhutta ZA, Das JK, Rizvi A, Gaffey MF, Walker N, Horton S, Webb P, Lartey A, Black RE, 2013. Evidence-based interventions for improvement of maternal and child nutrition: What can be done and at what cost? Lancet 382, 452–477. 10.1016/S0140-6736(13)60996-4 [DOI] [PubMed] [Google Scholar]
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan Y. -m., Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, Gordon JI, 2016. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science (80-. ). 351, aad3311–aad3311. 10.1126/science.aad3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briend A, Akomo P, Bahwere P, Pee S. De, Dibari F, Golden MH, Manary M, Ryan K, 2015. Developing food supplements for moderately malnourished children : Lessons learned from ready-to-use therapeutic foods. Food Nutr. Bull 36, S53–S58. 10.1177/15648265150361S109 [DOI] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG, 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–8. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M, 2017. Ten Years in Public Health 2007–2017. Geneva. [Google Scholar]
- Coyte KZ, Schluter J, Foster KR, 2015. The ecology of the microbiome: Networks, competition, and stability. Science (80-. ). 350, 663–666. 10.1126/science.aad2602 [DOI] [PubMed] [Google Scholar]
- de Onis M, Branca F, 2016. Childhood stunting: A global perspective. Matern. Child Nutr. 12, 12–26. 10.1111/mcn.12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G, 2014. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96. 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Degnan PH, Taga ME, Goodman AL, 2014. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778. 10.1016/j.cmet.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey KG, Heinig MJ, Nommsen-Rivers LA, 1995. Differences in morbidity between breast-fed and formula-fed infants. J. Pediatr 126, 696–702. 10.1016/S0022-3476(95)70395-0 [DOI] [PubMed] [Google Scholar]
- Deza MM, Deza E, 2014. Encyclopedia of Distances. Springer Berlin Heidelberg, Berlin, Heidelberg: 10.1007/978-3-662-44342-2 [DOI] [Google Scholar]
- Duarte NC, Becker S. a, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ, 2007. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. U. S. A 104, 1777–82. 10.1073/pnas.0610772104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Herrgård MJ, Palsson BØ, 2004. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 14, 1298–309. 10.1101/gr.2250904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Raes J, 2012. Microbial interactions: from networks to models. Nat. Rev. Microbiol 10, 538–550. 10.1038/nrmicro2832 [DOI] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson B, 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol 3, 1–18. 10.1038/msb4100155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernstrom JD, 1983. Role of precursor availability in control of monoamine biosynthesis in brain. Physiol. Rev 63, 484–546. [DOI] [PubMed] [Google Scholar]
- Fernstrom JD, Wurtman RJ, 1972. Brain serotonin content: physiological regulation by plasma neutral amino acids. Science (80-. ). 178, 414–416. 10.1126/science.178.4059.414 [DOI] [PubMed] [Google Scholar]
- Fujisaka S, Avila-Pacheco J, Soto M, Kostic A, Dreyfuss JM, Pan H, Ussar S, Altindis E, Li N, Bry L, Clish CB, Kahn CR, 2018. Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites. Cell Rep. 22, 2809–2817. 10.1016/j.celrep.2018.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale C, Logan KM, Santhakumaran S, Parkinson JRC, Hyde M, Modi N, 2012. Effect of Breast Versus Formula Feeding on Infant Body Composition: a Systematic Review and Meta-Analysis. Am. J. Clin. Nutr 95, 656–669. 10.3945/ajcn.111.027284.INTRODUCTION [DOI] [PubMed] [Google Scholar]
- Garza C, 2015. Commentary: Please sir, I want some more (and something else). Int. J. Epidemiol 44, 1876–1878. 10.1111/mcn.12049. [DOI] [PubMed] [Google Scholar]
- Gominak SC, 2016. Vitamin D deficiency changes the intestinal microbiome reducing B vitamin production in the gut. The resulting lack of pantothenic acid adversely affects the immune system, producing a “pro-inflammatory” state associated with atherosclerosis and autoimmun. Med. Hypotheses 94, 103–107. 10.1016/j.mehy.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Han J, Gagnon S, Eckle T, Borchers CH, 2013. Metabolomic Analysis of Key Central Carbon Metabolism Carboxylic Acids as Their 3-Nitrophenylhydrazones by UPLC/ESI-MS. Electrophoresis 34, 2891–2900. 10.1002/elps.201200601.Metabolomic [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lin K, Sequeira C, Borchers CH, 2015. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 854, 86–94. 10.1016/j.aca.2014.11.015 [DOI] [PubMed] [Google Scholar]
- Heine W, Radke M, Wutzke KD, 1999. The Significance of Tryptophan in Human-Nutrition, in: Tryptophan, Serotonin, and Melatonin. Advances in Experimental Medicine and Biology. pp. 705–710. [Google Scholar]
- Heinken A, Thiele I, 2015. Anoxic conditions promote species-specific mutualism between gut microbes In Silico. Appl. Environ. Microbiol 81, 4049–4061. 10.1128/AEM.00101-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CS, Bernstein HC, Weisenhorn P, Taylor RC, Lee JY, Zucker J, Song HS, 2016. Microbial Community Metabolic Modeling: A Community Data-Driven Network Reconstruction. J. Cell. Physiol 231, 2339–2345. 10.1002/jcp.25428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL, 2010. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol 28, 977–982. 10.1038/nbt.1672 [DOI] [PubMed] [Google Scholar]
- Henry CS, Zinner JF, Cohoon MP, Stevens RL, 2009. iBsu1103: A new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol. 10, 1–15. 10.1186/gb-2009-10-6-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey SL, Mehta S, 2016. Stunting: The Need for Application of Advances in Technology to Understand a Complex Health Problem. EBioMedicine 6, 26–27. 10.1016/j.ebiom.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert SH, 1971. The nonconcept of species diversity: A critique and alternative parameters. Ecology 52, 577–586. 10.15063/rigaku.KJ00008826560 [DOI] [PubMed] [Google Scholar]
- Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW, 2017. GPCR-Mediated Signaling of Metabolites. Cell Metab. 25, 777–796. 10.1016/j.cmet.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Jones RSG, 1982. Tryptamine: a neuromodulator or neurotransmitter in mammalian brain? Prog. Neurobiol 19, 117–139. 10.1016/0301-0082(82)90023-5 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M, 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–80. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Kim HU, Lee SY, 2017. Current state and applications of microbial genome-scale metabolic models. Curr. Opin. Syst. Biol 2, 9–17. 10.1016/j.coisb.2017.03.001 [DOI] [Google Scholar]
- Kirkpatrick BD, Colgate ER, Mychaleckyj JC, Haque R, Dickson DM, Carmolli MP, Nayak U, Taniuchi M, Naylor C, Qadri F, Ma JZ, Alam M, Walsh MC, Diehl SA, Petri WA, 2015. The “Performance of Rotavirus and Oral Polio Vaccines in Developing Countries” (PROVIDE) study: Description of methods of an interventional study designed to explore complex biologic problems. Am. J. Trop. Med. Hyg 92, 744–751. 10.4269/ajtmh.14-0518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K, 2018. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages . Cell Rep. 23, 1099–1111. 10.1016/j.celrep.2018.03.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanting CI, Huisman M, Boersma ER, Touwen BCL, Fidler V, 1994. Neurological differences between 9-year-old children fed breast-milk or formula-milk as babies. Lancet 344, 1319–1322. 10.1016/S0140-6736(94)90692-0 [DOI] [PubMed] [Google Scholar]
- Latendresse M, 2014. Efficiently gap-filling reaction networks 15, 225 10.1186/1471-2105-15-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P, 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levandowsky M, 1971. Distance between Sets. Nature 234, 34–35. 10.1038/239174c0 [DOI] [Google Scholar]
- Louis P, Flint HJ, 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol 19, 29–41. 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, Greenhalgh K, Jäger C, Baginska J, Wilmes P, Fleming RMT, Thiele I, 2016. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol 35, 81–89. 10.1038/nbt.3703 [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL, 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10, 507–514. 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo-Wilson E, Junior J, Imdad A, Dean S, Chan X, Chan E, Jaswal A, Bhutta Z, 2014. Zinc supplementation for preventing mortality, morbidity, and growth failure in children aged 6 months to 12 years of age. Cochrane Collab. 1–442. 10.1002/14651858.CD009384.pub2.www.cochranelibrary.com [DOI] [PubMed] [Google Scholar]
- Menni C, Jackson MA, Pallister T, Steves CJ, Spector TD, Valdes AM, 2017. Gut microbiome diversity and high-fibre intake are related to lower long-term weight gain. Int. J. Obes 41, 1099–1105. 10.1038/ijo.2017.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metges CC, 2000. Contribution of microbial amino acids to amino acid homeostasis of the host. J. Nutr 130, 1857S–64S. [DOI] [PubMed] [Google Scholar]
- Moffett JR, Namboodiri MA, 2003. Tryptophan and the immune response. Immunol. Cell Biol 81, 247–265. 10.1046/j.1440-1711.2003.t01-1-01177.x [DOI] [PubMed] [Google Scholar]
- Moroni F, 1999. Tryptophan metabolism and brain function: Focus on kynurenine and other indole metabolites. Eur. J. Pharmacol 375, 87–100. 10.1016/S0014-2999(99)00196-X [DOI] [PubMed] [Google Scholar]
- Naylor C, Lu M, Haque R, Mondal D, Buonomo E, Nayak U, Mychaleckyj JC, Kirkpatrick B, Colgate R, Carmolli M, Dickson D, van der Klis F, Weldon W, Steven Oberste M, Ma JZ, Petri WA, The PROVIDE study teams, 2015. Environmental Enteropathy, Oral Vaccine Failure and Growth Faltering in Infants in Bangladesh . EBioMedicine 2, 1759–1766. 10.1016/j.ebiom.2015.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T, 2010. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc. Natl. Acad. Sci 107, 19961–19966. 10.1073/pnas.1014465107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A, Mardinoglu A, Nielsen J, 2017. Predicting growth of the healthy infant using a genome scale metabolic model. npj Syst. Biol. Appl 3, 3 10.1038/s41540-017-0004-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, Soininen P, Wang Z, Ala-Korpela M, Hazen SL, Laakso M, Lusis AJ, 2017. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 18, 1–14. 10.1186/s13059-017-1194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östman EM, Liljeberg Elmståhl HGM, Björck IME, 2001. Inconsistency between glycemic and insulinemic responses to regular and fermented milk products. Am. J. Clin. Nutr 74, 96–100. 10.1038/nature10213 [DOI] [PubMed] [Google Scholar]
- Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, Melander O, Orho-Melander M, 2018. Connection between BMI-Related Plasma Metabolite Profile and Gut Microbiota. J. Clin. Endocrinol. Metab 103, 1491–1501. 10.1210/jc.2017-02114 [DOI] [PubMed] [Google Scholar]
- Peng YM, Zhang TY, Wang Q, Zetterström R, Strandvik B, 2007. Fatty acid composition in breast milk and serum phospholipids of healthy term Chinese infants during first 6 weeks of life. Acta Paediatr. Int. J. Paediatr 96, 1640–1645. 10.1111/j.1651-2227.2007.00482.x [DOI] [PubMed] [Google Scholar]
- Ramakrishnan U, Nguyen P, Martorell R, 2009. Effects of micronutrients on growth of children under 5 years of age. Am. J. Clin. Nutr 89, 191–203. 10.3945/ajcn.2008.26862.Am [DOI] [PubMed] [Google Scholar]
- Reed JL, Palsson BØ, 2003. Thirteen Years of Building Constraint-Based In Silico Models of Escherichia coli MINIREVIEW Thirteen Years of Building Constraint-Based In Silico Models of Escherichia coli. Society 185, 2692–2699. 10.1128/JB.185.9.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes H, Pérez-Cuevas R, Sandoval A, Castillo R, Santos JI, Doubova SV, Gutiérrez G, 2004. The family as a determinant of stunting in children living in conditions of extreme poverty: A case-control study. BMC Public Health 4, 1–10. 10.1186/1471-2458-4-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DM, Dawes MA, Mathias CW, Acheson A, Hill-kapturczak N, Dougherty DM, 2009. L -Tryptophan : basic metabolic functions , behavioral research and therapeutic indications. Int. J. Tryptophan Res 2, 45–60. 10.2964/jsik.kuni0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainio EL, Pulkki K, Young SN, 1996. L-Tryptophan: Biochemical, nutritional and pharmacological aspects. Amino Acids 10, 21–47. 10.1007/BF00806091 [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Park JO, Conrad TM, Palsson BØ, 2010. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 11, 213 10.1186/1471-2105-11-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BO, Bäckhed F, 2016. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med 22, 1079–1089. 10.1038/nm.4185 [DOI] [PubMed] [Google Scholar]
- Semba RD, Shardell M, Sakr Ashour FA, Moaddel R, Trehan I, Maleta KM, Ordiz MI, Kraemer K, Khadeer MA, Ferrucci L, Manary MJ, 2016. Child Stunting is Associated with Low Circulating Essential Amino Acids. EBioMedicine 6, 246–252. 10.1016/j.ebiom.2016.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaie S, Ghaffari P, Kovatcheva-Datchary P, Mardinoglu A, Sen P, Pujos-Guillot E, de Wouters T, Juste C, Rizkalla S, Chilloux J, Hoyles L, Nicholson JK, Dore J, Dumas ME, Clement K, Bäckhed F, Nielsen J, 2015. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 22, 320–331. 10.1016/j.cmet.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau a. L., Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI, 2013. Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. Science (80-. ). 339, 548–554. 10.1126/science.1229000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Doraé J, 2009. Low counts of faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis 15, 1183–1189. 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Blanton LV, Frese SA, Charbonneau M, Mills DA, Gordon JI, 2015. Cultivating Healthy Growth and Nutrition through the Gut Microbiota. Cell 161, 36–48. 10.1016/j.cell.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam M. a., Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province M. a., Petri W. a. Jr, Ahmed T, Gordon JI, 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421. 10.1038/nature13421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanphaichitr V, Broquist HP, 1973. Role of Lysine and N-Trimethyllysine in Carnitine Biosynthesis. J. Biol. Chem 248, 2176–2181. [PubMed] [Google Scholar]
- Thiele I, Swainston N, Fleming RMT, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, Thorleifsson SG, Agren R, Bölling C, Bordel S, Chavali AK, Dobson P, Dunn WB, Endler L, Hala D, Hucka M, Hull D, Jameson D, Jamshidi N, Jonsson JJ, Juty N, Keating S, Nookaew I, Le Novère N, Malys N, Mazein A, Papin JA, Price ND, Selkov E, Sigurdsson MI, Simeonidis E, Sonnenschein N, Smallbone K, Sorokin A, Van Beek JHGM, Weichart D, Goryanin I, Nielsen J, Westerhoff HV, Kell DB, Mendes P, Palsson BO, 2013. A community-driven global reconstruction of human metabolism. Nat. Biotechnol 31, 419–425. 10.1038/nbt.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomé D, Bos C, 2007. Lysine requirement through the human life cycle. J. Nutr 137, 1642S–1645S. https://doi.org/137/6/1642S [pii] [DOI] [PubMed] [Google Scholar]
- Tramontano M, Andrejev S, Pruteanu M, Klünemann M, Kuhn M, Galardini M, Jouhten P, Zelezniak A, Zeller G, Bork P, Typas A, Patil KR, 2018. Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol 3, 1–9. 10.1038/s41564-018-0123-9 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald M. a, Magrini V, Mardis ER, Gordon JI, 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- UNICEF, WHO, World Bank Group, 2017. Levels and Trends in Child Malnutrition. 10.1016/S0266-6138(96)90067-4 [DOI] [Google Scholar]
- Velázquez OC, Lederer HM, Rombeau JL, 1996. Butyrate and the Colonocyte 4, 123–134. 10.1007/978-1-4615-5967-2_14 [DOI] [PubMed] [Google Scholar]
- Victora CG, de Onis M, Hallal PC, Blossner M, Shrimpton R, 2010. Worldwide Timing of Growth Faltering: Revisiting Implications for Interventions. Pediatrics 125, e473–e480. 10.1542/peds.2009-1519 [DOI] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall HU, Bäckhed F, 2016. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 24, 41–50. 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Whitehead RG, 1964. Rapid determination of some plasma amino acids in subclinical kwashiorkor. Lancet 283, 250–252. 10.1016/S0140-6736(64)92353-0 [DOI] [PubMed] [Google Scholar]
- WHO/FAO/UNU Expert Consultation, 2007. Protein and amino acid requirements in human nutrition, World Health Organization technical report series. https://doi.org/ISBN9241209356 [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G, 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A 106, 3698–703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, 2009. Amino acids: Metabolism, functions, and nutrition. Amino Acids 37, 1–17. 10.1007/s00726-009-0269-0 [DOI] [PubMed] [Google Scholar]
- Wu G, Morris SM Jr., 1998. Arginine metabolism: nitric oxide and beyond. Biochem. J 336, 1–17. 10.1016/S0928-4680(98)80513-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Keilbaugh S.a, Bewtra M, Knights D, Walters W. a, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, 2011. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science (80-. ). 334, 105–109. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki N, Yamada M, Kan-no T, Kojima T, Kaneko T, Yonekubo A, 2005. Macronutrient, mineral and trace element composition of breast milk from Japanese women. J. Trace Elem. Med. Biol 19, 171–181. 10.1016/j.jtemb.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY, 2015. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 161, 264–276. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Duncan SH, Louis P, Flint HJ, 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543. 10.1038/ismej.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.