Abstract
As a result of exome-based sequencing work performed by the DDD study, de novo variants in CNOT3 have emerged as a newly recognised cause of a developmental disorder. This paper describes molecular and clinical details of 16 probands with developmental disorders and de novo CNOT3 variants. It is the first such description of the developmental phenotype associated with CNOT3 variants. Eight of these cases were discovered as part of the DDD study, while the other eight were found as a result of large-scale sequencing work performed by other groups. A highly specific phenotype was not recognised in these 16 cases. The most consistent phenotypic features seen in subjects with de novo variants in CNOT3 were hypotonia, relatively small stature, developmental delay, behavioural problems and intellectual disability. There is no easily recognisable facial phenotype, but some common dysmorphic features such as anteverted nares, thin upper lip and low set eyebrows were shared among some of the probands. Haploinsufficiency appears to be the most likely mechanism of action, with eight cases found to have protein-truncating variants. Of the other eight cases (all missense variants), three share an amino acid substitution at the same position which may therefore represent an important functional domain.
Subject terms: Disease genetics, Next-generation sequencing
Introduction
The Deciphering Developmental Disorders (DDD) study used family exome studies to identify de novo variants in children with severe developmental disorders [1–3]. Initial results from this study identified seven patients with de novo variants in CNOT3. CNOT3 is therefore a gene significantly enriched for de novo variants in the DDD cohort; this provides compelling evidence for CNOT3 being a new developmental disorder-associated gene [4].
Since the initial seven cases discovered by the DDD study, additional patients with developmental disorders and CNOT3 de novo variants have been identified (some as a result of further analyses by the DDD study and others as a result of large-scale sequencing work performed by other groups). Clinical and molecular descriptions of 16 cases are described in this paper, representing the first known developmental disorders to be associated with variants in this gene.
CNOT3 comprises part of the CCR4-NOT protein complex, which is a global regulator of RNA polymerase II transcription [5, 6]. It has previously been identified as a potential tumour suppressor gene in T-cell acute lymphoblastic leukaemias, with loss of function variants in CNOT3 being observed in these tumours [7]. Prior to the DDD study it had not been firmly linked to developmental disorders.
Materials and methods
DDD cases
The DDD study recruited over 13,000 individuals with a severe undiagnosed developmental disorder, via Clinical Genetics centres in the UK and the Republic of Ireland. Copy number analysis and exome based sequencing were performed. Results were published in 2017 from 4293 trios (i.e. 4293 single affected children with unaffected parents) [4]. Data were filtered to look specifically for de novo variants in the coding or splicing sequences of the probands. After excluding probands with likely pathogenic variants in other developmental disorder-associated genes and combining the statistical analysis with results from 3287 individuals recruited to 13 other exome or genome sequencing studies, 14 new developmental disorder-associated genes were initially identified. CNOT3 was one of these genes; initial analyses by the DDD study identified seven patients with de novo variants in CNOT3 [4].
Subsequent exome-based analysis by the DDD study has identified at least a further 16 subjects who have CNOT3 variants; two of these are known to have occurred de novo (making a total of nine de novo DDD cases, eight of which are included in this series). Four out of sixteen (all missense or in-frame deletions) are inherited from a parent and so are presumed not to be the cause of the child’s phenotype. Inheritance data are not currently available for the remaining ten DDD variants (one frameshift; nine missense or in-frame deletions).
Non-DDD cases
Other cases of de novo CNOT3 variants in probands with developmental disorders have since emerged from large-scale sequencing work performed by other groups. Phenotypic and molecular data on eight such patients are included in this case series. Cases 3 and 10 (see Table 1) underwent diagnostic exome sequencing in Utrecht because of a developmental disorder. Case 4 was tested for 217 genes of interest as part of a study which recruited probands with intellectual disability (ID) of unknown aetiology [8]. Cases 11, 13 and 15 had exome sequencing in Dijon as part of a whole exome sequencing study involving probands with severe ID or epileptic encephalopathy [9]. Cases 12 and 14 had exome sequencing in Leiden.
Table 1.
Molecular and phenotypic summary of 16 probands with de novo variants in CNOT3
| Case | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | c.58G > C | c.142C > G | c.355A > G | c.439G > A | c.562C > T | c.563G > A | c.563G > A | c.2090G > A | c.732dup | c.821_825dup | c.1127_1145del | c.1473_1474del | c.1538_1541del | c.1866G > A | c.1926_1927del | c.2080C > T |
| Effect | p.(Glu20Gln) | p.(Leu48Val) | p.(Lys119Glu) | p.(Glu147Lys) | p.(Arg188Cys) | p.(Arg188His) | p.(Arg188His) | p.(Arg697Gln) | p.(Ser245Glnfs*8) | p.(Asn276Glnfs*61) | p.(Ala376Glyfs*61) | p(Gly493Thrfs*21) | p.(Ser513Metfs*27) | p.(Trp622Ter) | p.(Cys643Serfs*39) | p.Gln694Ter |
| Type of variant | Missense | Missense | Missense | Missense | Missense | Missense | Missense | Missense | Truncating | Truncating | Truncating | Truncating | Truncating | Truncating | Truncating | Truncating |
| Open access DECIPHER ID | 261122 | 276825 | 262450 | 259521 | 268198 | 263217 | 327692 | 357220 | 257816 | |||||||
| Age at diagnosis (years) | 8 | 5 | 17 | 12 | 12 | 21 | 6 | 8 | 12 | 15 | 18 | 18 | 7 | 55 | 4 | 19 |
| Gender | F | M | M | M | M | F | F | F | M | M | F | M | M | M | M | F |
| Ethnicity | Caucasian | White British | Dutch Caucasian | French Caucasian | ? | White British | White British | White British | Caucasian | Dutch | French Caucasian | Caucasian | French Caucasian | Dutch Caucasian | Caucasian | Caucasian |
| Birth weight | 3487 g | 2211 g | 2970 g | 1900g | 2100 g | 2400 g | ? | 3005 g | 3856 g | 3120 g | 2820 g | 2000g | 2870 g | 3500 g | 3220 g | 2870 g |
| Gestation (weeks) | 40 + 6 days | 39 | 39 | 32 | 38 | 40 | 40 + 6 days | 37 | 38 | 40 | 37 | 36 | 37 + 2 days | 40 | 40 | 38 |
| Peri/postnatal issues | Hip dysplasia requiring harness | Fetal tachycardia, cord round neck | CS (breech) | CS | Remained in hospital for 1 week requiring incubation, oxgyen and antibiotics | Brachycephaly, floppy, readmitted day 10 with seizures | Induced because of pre-eclampsia, cyanotic episode 2 h after birth | Oblique lie, elective CS | None | None | None | None | Feeding difficulties | None | None | None |
| Walked independently (age - in months) | 27 | 35 | 36 | 30 | ? | 22 | 21 | 26 | Cruising furniture at 30 months | 15 | 17 | 24 | 35 | 24 | 55 | 14 |
| Motor development | Fine motor skills remain delayed | ? | Mild–moderate fine and gross motor delay | Fine and gross motor delay | Able to walk but uses wheelchair for longer distances | ? | Delay in gross and fine motor skills | Fine and gross motor delay | Comparatively good fine motor skills | Fine motor development normal, some delay of gross motor skills but now able to cycle | Delayed fine motor skills, can run with difficulty and slowly climb stairs | Fine and gross motor delay | Fine and gross motor delay | Fine and gross motor delay, able to cycle at 10 years old | Fine and gross motor delay | ? |
| Abnormailty of tone | Hypotonia (resolved) | Hypotonia | Hypotonia | No | No | Early hypotonia | Hypotonia | Hypotonia | Borderline hypotonia | ? | Hypotonia | ? | Hypotonia | ? | Hypotonia | ? |
| Speech and language development | 50 makaton signs only at 3 years; some speech, good understanding at 6.5 years | First words after 2 years; at 4 years mainly repeats words | Nasal speech | No language | No words | No words | 3 single words by 21 months, now talks in sentences although words can be indistinct | Delayed | No expressive language skills, hoarse voice | Delayed, nasal speech | First words aged 17 months, diffculty in communicating with others but does form some complete sentences | Delayed, talks in full sentences now | Delayed | First words aged 3 years, has developed speech, verbal dyspraxia | No language | Speech from 5 to 6 years, now short phrases and echolalia |
| Head circumference centile | 5.5 cm < 0.4th centile | 75 | 25–50 | 25–50 | 75 | 75–91 | 50–75 | 1.5 cm < 0.4th | 75–91 | 25–50 | 9–25 | 25–50 | 25–50 | 25 | 50 | 50–75 |
| Height centile | 25 | 0.4–2 | 25 | 0.4–2 | 2–9 | 0.4–2 | 9–25 | 50 | 2 | 9–25 | 50–75 | 9 | 9–25 | (Adult height 173 cm) | 0.4–2 | 0.4–2 |
| Weight centile | 2–9 | 9–25 | 25–50 | 0.4 | 75 | 2–9 | 25–50 | 25 | 0.4–2 | 9–25 | 91 | 75–91 | 50 | (Adult weight 72 kg) | 0.4–2 | ? |
| Learning and behaviour | 1:1 full time help in mainstream school | Moderate LD; attends mainstream school and special needs nursery aged 4; reduced eye contact and sensory processing disorder | IQ 52, hand flapping, pervasive developmental disorder not otherwise specified | Severe LD, autism, behavioural steriotypies | Severe autism | Profound LD | Requires extra support at mainstream school | Attends mainstream school with additional support | Marked autism | Autistic spectrum disorder, learning difficulty | Anxiety, aggression, unable to read or write, special needs school | Autism, attends special needs school | Autism | Unable to read or write, learning level of a 6 year old, works in a social empoyment facility | Autistic spectrum disorder, learning diffculty | Severe LD, sensory issues and rocking movements |
| Hearing and vision | Glue ear | Conductive hearing loss | Hypermetropia (+5 dioptres) | Nystagmus | Strabismus | Normal | Moderate− severe left-sided SNHL; right conductive hearing loss; intermittent right strabismus | ? | Normal | Strabismus | Esotropia | ? | Normal | Strabismus, optic nerve atrophy | Normal | Strabismus, visual processing disorder, bilateral discrete raised lesions superior to optic discs?calcium deposits secondary to retinal haemorrhage |
| Dysmorphic features | Large central incisors, downturned mouth, flat broad nose, 2 right ear pits | Prominent forehead, mild cranial asymmetry, hypotelorism, deep-set eyes, right helix squared off and dimple on lobe, short nose with depressed bridge, upturned nares | Prominent occiput and foreheed, low set ears, prominent nasal tip, mild prognathism, short philtrum, tapering fingers, bilateral 5th finger clinodactyly, flat feet | Malar hypoplasia, prognathism, doliocephaly | Brachycephaly, flat midface, hypertelorism, upslanting palpebral fissures, two crowns over posterior parietal area with midline join of the hairgrowth, tapering fingers | Prominent forehead, deep set eyes, small hands and feet | Left plagiocephaly, low post hairline, upper back and arms hirsute, prominent lower jaw | Plagiocephaly and brachycephaly, small ears with short upturned lobes, short philtrum, dimples over shoulders, bilateral talipes calcaneo-valgus | Prominent forehead, flat nasal bridge, macroglossia, short tapering fingers, small feet | Small triangular skull, coarse hair, narrow nasal bridge, long columella, short philtrum, prominent eyebrows | Slightly descending columella, tubular nose, narrow palate, misaligned teeth, upslanting palpebral fissures, malar hypoplasia, 5th finger clinodactyly, short hands | Low frontal hairline, simple helices with fleshy earlobes | Small chin, malar hypoplasia, dolicocephaly | Deep set eyes, slight hypotelorism | Prominent forehead | Straight, heavy eyebrows, deep set eyes, upslanting palpebral fissures, short philtrum, full lower lip, full slightly overhanging nasal tip, long fingers |
| MRI brain | Myelination delayed at 2 years, resolved by 3 years; slim corpus callosum | Aged 11 months mild prominence of ventricular system with normal brain parenchyma | Normal 3.5 years | Thin corpus callosum (4 years) | Small hippocampus and amygdala aged 3 | Global cerebral and cerebellar atrophy aged 18 years, with mega cisterna magna and thickened corpus callosum | Not done | Not done (did have normal CT brain) | Normal aged 2 years | Normal aged 6 years | Occipital atrophy 3.5 years | ? | Normal | Normal | Possible partial agenesis of the corpus callosum | Not done |
| Heart and kidneys | Normal echo and renal USS | Small left kidney (VUR) | ? | Pyelocalyceal hypotonia | Perimembranous VSD | ? | Normal renal USS | Normal renal USS | Trivial aortic regurgitation aged 5 years; normal renal USS | Normal echo aged 5 years | Normal echo | ? | Normal renal USS; abnormal right pulmonary venous return | ? | Normal echo and renal USS aged 3 years | ? |
| Other features | Some excess hair over cheek bones, marked microcephaly | Elbow dimples, deep plantar creases, right lower leg capillary naevus, reduced pain sensation | Scoliosis, persistent primary dentition (at 12 years), frequent infections, seizures from 4 years | Paroxysmal staring events with focal abnormalities on EEG, delayed bone age, wormian bones, delayed dentition, hypermobility, persistent fontanelle | Seizures started 10 weeks of age, frontal lobe epilepsy from around age 3; stable thoracolumbar scoliosis | Borderline biotinase level (replacement started december 2015), high-pitched scream as an infant, torticollis noted at 6 weeks | Tongue tie, blind ending sacral dimple | Delayed bone age, lower thoracic kyphosis, possible small genitalia | Pes planovalgus | Varus foot deformity | Soft skin, hypermobile joints, penile hypopigmentation, inguinal hyperpigmentation, temporal lobe epilepsy diagnosed aged 13 years, melatonin for sleep | Tremor left hand, prone to respiratory tract infections, high cholesterol, late puberty | Positional plagiocephaly | Scoliosis, hairy, episodes of hyperventilation |
RefSeq NM_014516.3
All CNOT3 variants are described using the reference sequence NM_014516.3.
Results
Molecular and clinical descriptions for the 16 patients with de novo variants in CNOT3 are shown in Table 1. Cases 1–8 are missense variants; cases 9–16 are protein-truncating variants. A diagram adapted from Boland et al. [5] shows the distribution of the de novo variants found in CNOT3 (Fig. 1). The de novo missense variants all result in amino acid substitutions at highly conserved positions (details shown in Table 2, adapted from data taken from Alamut [10]).
Fig. 1.
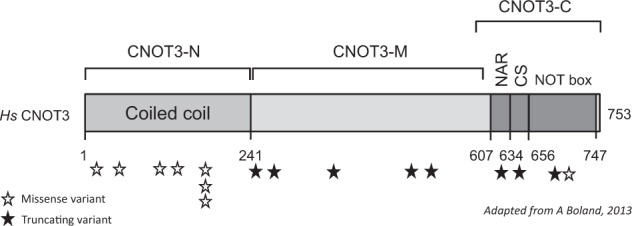
Distribution of variants in CNOT3
Table 2.
Aligned amino acid sequences at each missense variant position
| Case | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Position | 20 | 48 | 119 | 147 | 188 | 188 | 188 | 697 |
| New amino acid | Q | V | E | K | C | H | H | Q |
| Orthologs | ||||||||
| Human | E | L | K | E | R | R | R | R |
| Chimp | E | L | K | E | R | R | R | R |
| Olive baboon | E | L | K | E | R | R | R | R |
| Rat | E | L | K | E | R | R | R | R |
| Mouse | E | L | K | E | R | R | R | R |
| Dog | E | L | K | E | R | R | R | R |
| Frog | E | L | K | E | R | R | R | R |
| Tetraodon | E | L | K | E | R | R | R | R |
| Zebrafish | E | L | K | E | R | R | R | R |
| Fruitfly | E | L | R | E | R | R | R | R |
| C. elegans | E | L | K | E | R | R | R | R |
| Baker’s yeast | E | L | R | D | R | R | R | K |
Development, learning and behaviour
All 16 probands have developmental delay and learning difficulties; these vary in severity. Only three children were known to be walking by 18 months of age although at least 15/16 did acquire the ability to walk independently. The median age of walking independently was 25 months (range 14–55 months; cases 5 and 9 excluded due to insufficient data). In terms of speech development, 5/16 probands have no speech at all. The others range from repeating words to talking in full sentences. Four of sixteen are known to attend a form of mainstream education, all four require additional learning support at school. Eight of sixteen have behavioural problems ranging from pervasive developmental disorder to autism.
Growth
There is a tendency towards short stature with 13/16 cases having heights ≤25th centile; indeed 6/16 have heights ≤2nd centile. Case 1 has a severe primary microcephaly for which an alternative explanation is felt to be highly likely. Case 8 has a less pronounced microcephaly, but otherwise the trend is to have a relatively well-preserved head circumference compared to height (thirteen cases had head circumferences ≥25th centile, including seven ≥50th centile).
Other features
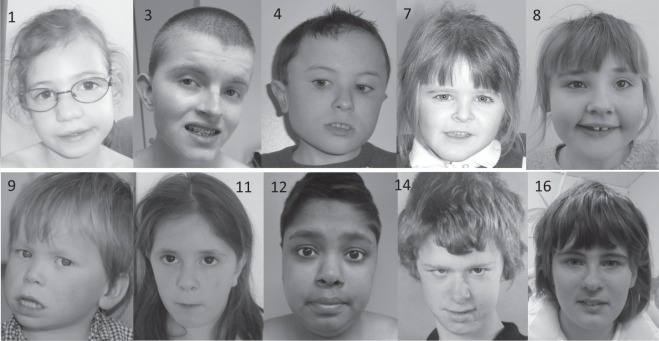
Although some children share common facial features, a consistent or recognisable facial gestalt was not felt to be present (see Table 1 and photographs in Fig. 2). Twelve of sixteen cases had had an MRI brain scan; 5/12 were said to be normal, the other seven cases had various reported findings including partial agenesis of the corpus callosum (3/7) and parenchymal atrophy (2/7). Hypotonia appears to be common, affecting 10/16 cases.
Fig. 2.
Clinical photographs of ten probands with de novo CNOT3 variants (case numbers shown, corresponding to Table 1)
Clinical photographs are available for 10 subjects (shown in Fig. 2). There is no easily recognisable facial phenotype, but some common dysmorphic features such as anteverted nares, thin upper lip and low set eyebrows were shared among some of the probands.
Discussion
The initial seven de novo variants in CNOT3 identified by the DDD study provide firm statistical evidence for this being a developmental disorder-associated gene [4]. Haploinsufficiency would appear to be the likely mechanism of action given the eight cases with protein-truncating variants; data from ExAC and Decipher support the hypothesis that CNOT3 is highly intolerant of loss of function variation [11, 12]. There are a total of 66 patients in the Decipher [12] database with copy number or sequence variation in CNOT3. Of these, 30 are copy number gains; there are no copy number losses. According to the ExAC database, CNOT3 is highly intolerant of loss of function variants (expected number of LOF variants 28.9; observed number 0; pLI 1.00). CNOT3 is also significantly less tolerant of missense variation than expected (expected number of missense variants 292.1; observed number 156; z = 3.89) [11].
There are a total of 36 sequence variants in CNOT3 listed on Decipher [12]. Fourteen of these are de novo variants (9 of which are included in this case series).
The most consistent phenotypic features seen in subjects with de novo variants in CNOT3 in this case series are hypotonia, relatively small stature, developmental delay, behavioural problems and ID. The de novo variants identified in CNOT3 do not appear to result in an easily recognised or highly specific phenotype. This is not surprising; it can be hypothesised that the majority of hitherto undiscovered developmental disorder-associated genes will not produce an easily recognised phenotype which is one major reason why they eluded discovery prior to the advent of exome-based sequencing.
Phenotypic comparison was made between the eight missense variants and the eight truncating variants, primarily to ascertain whether the cases with truncating variants might tend towards a more severe phenotype. It is interesting to note that all four probands who attend a form of mainstream education have missense variants (cases 1, 2, 7 and 8), lending some weight to this hypothesis.
Using the ACMG guidelines on variant interpretation [13], loss of function variants in CNOT3 are more likely to be definitively classified (as pathogenic) than missense variants. Inheritance data will be helpful in further interrogating missense variants, but this will not always be available. Showing that a variant has occurred de novo is less powerful evidence in favour of pathogenicity when a highly specific phenotype is not present, such as appears to be the case here. In addition, given the apparent phenotypic spectrum resulting from CNOT3 variants, there may be families where a pathogenic variant has been inherited from a mildly affected parent.
Three cases have missense variants affecting the same amino acid position (cases 5, 6 and 7). These probands did not appear to have a very similar phenotype; cases 5 and 6 having severe developmental delay and case 7 being one of the most mildly affected probands. There are two further cases on Decipher [12] with de novo missense variants at this position. It is hypothesised that this amino acid position represents an important functional domain.
Missense variation is tolerated in some residues of CNOT3 as evidenced by ExAC and at least some of the inherited variants reported in Decipher. Functional work to aid more definitive classification of missense variation within CNOT3 is needed. Additional cases to better delineate the phenotype associated with CNOT3 variants would be welcomed and will likely arise as a result of further whole exome-based sequencing studies in probands with developmental disorders.
Acknowledgements
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of Wellcome or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. This study makes use of data generated by the DECIPHER community. A full list of centres who contributed to the generation of the data are available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deciphering Developmental Disorders Study Largescale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firth HV, Wright CF. The Deciphering Developmental Disorders (DDD) study. Dev Med Child Neurol. 2011;53:702–3. doi: 10.1111/j.1469-8749.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 3.Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–14. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland A, Chen Y, Raisch T, Jonas S, Kuzuoğlu-Öztürk D, Wohlbold L, et al. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat Struct Mol Biol. 2013;20:1289–97. doi: 10.1038/nsmb.2681. [DOI] [PubMed] [Google Scholar]
- 6.Online Mendelian Inheritance in Man, OMIM®. MIM number: 604910: last edited 04/09/2013. Johns Hopkins University, Baltimore, MD. https://www.omim.org/entry/604910. Accessed 01 Jun 2018.
- 7.De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, et al. 2013. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–90. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redin C, Gerard B, Lauer J, Herenger Y, Muller J, Quartier A, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J Med Genet. 2014;51:724–36. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thevenon J, Duffourd Y, Masurel-Paulet A, Lefebvre M, Feillet F, El Chehadeh-Djebbar S, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first line diagnostic test. Clin Genet. 2016;89:700–7. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 10.Alamut Visual. http://www.interactive-biosoftware.com/alamut-visual/. Accessed 26 Apr 2018.
- 11.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]