Abstract
Purpose:
To develop a clinical infrastructure that allows for routine Monte Carlo dose calculation verification of spot scanning proton treatment plans and includes a simple biological model to aid in normal tissue protection.
Materials and Methods:
A graphical processing unit accelerated Monte Carlo dose engine was used as the calculation engine for dose verification on spot scanning proton plans. An infrastructure was built around this engine that allows for seamless exporting of treatment plans from the treatment planning system and importing of dose distribution from the Monte Carlo calculation via DICOM (digital imaging and communications in medicine). An easy-to-use Web-based interface was developed so that the application could be run from any computer. In addition to the standard relative biological effectiveness = 1.1 for proton therapy, a simple linear equation dependent on dose-weighted linear energy transfer was included. This was used to help detect possible high biological dose in critical structures.
Results:
More than 270 patients were treated at our proton center in the first year of operation. Because most plans underwent multiple iterations before final approval, more than 1000 plans have been run through the system from multiple users with minimal downtime. The average time from plan export to importing of the Monte Carlo doses was less than 15 minutes. Treatment plans have been modified based on the nominal Monte Carlo dose or the biological dose.
Conclusion:
Monte Carlo dose calculation verification of spot scanning proton treatment plans is feasible in a clinical environment. The 3-dimensional dose verification, particularly near heterogeneities, has resulted in plan modifications. The biological dose data provides actionable feedback for end of range effects, especially in pediatric patients.
Keywords: proton therapy, dose verification, Monte Carlo
Introduction
Proton beam therapy (PBT) is emerging as a promising form of external beam radiation therapy for cancer due to its superior ability to spare healthy tissue while treating the clinical target to the required tumoricidal dose. At the present time, PBT can be delivered in 3 modes: passive scattering, uniform scanning, and pencil beam scanning (PBS). Of these 3 delivery modes, PBS is the most advanced as it allows for intensity modulated PBT (IMPT).
Mayo Clinic is establishing a PBT program at 2 of its sites, one in Rochester, Minnesota, and the other in Phoenix, Arizona. The PBT facilities (Hitachi Ltd, Tokyo, Japan) at both sites are identical in all meaningful aspects; each features a synchrotron capable of producing proton beams with energies ranging from 71.3 MeV to 228.8 MeV, 4 gantry treatment rooms, and 1 fixed beam treatment room. All treatment rooms are equipped with a PBS nozzle for the delivery of IMPT. Treatment planning is carried out using the PBT treatment-planning software (TPS) Eclipse, version 13.5 (Varian Medical Systems, Palo Alto, California).
As part of a comprehensive quality assurance (QA) program, we identified a need for an independent secondary verification of the dose calculation carried out in the Eclipse TPS. The practice of performing an independent secondary dose calculation, which is well established in photon external beam therapy [1] is slowly being adopted in PBT. Secondary dose calculations for passive scattering PBT [2–4], uniform scanning [5, 6], and PBS [7, 8] have recently been reported. The requirements to use the second check at both centers, provide superior accuracy to the commercial dose calculation, and run quickly enough to be deployable in the clinic for every patient prompted us to implement a graphical processing unit (GPU) based fast Monte Carlo (MC) dose calculation engine [9].
In addition to a more accurate dose calculation, the MC calculation allows the inclusion of a relative biological effectiveness (RBE) model based on the dose-weighted linear energy transfer (LETd) of the protons [10]. This was deemed valuable given the uncertainty [11, 12] of the proton RBE as a function of linear energy transfer (LET), the complexity of IMPT plans, and the recent reports of necrosis following PBT [13, 14].
The purpose of this study is to report on a novel, clinically practical, and innovative design that allows the sharing of computational resources over large geographical distances utilizing a fast GPU-based MC algorithm for routine secondary proton dose calculation and RBE information for clinical use in a PBS system.
Materials and Methods
Graphical Processing Unit Accelerated Monte Carlo Dose Calculation
The functional details and benchmarking of the GPU accelerated MC dose calculation are described elsewhere [9, 15]. In short, the Compute Unified Device Architecture framework, which is a parallel computing platform and application programming interface, was used to implement GPU kernels for the following tasks: (1) simulation of beam spots from our various scanning nozzle configurations; (2) proton propagation through the simulated patient based on computed tomography (CT) image data; (3) modeling of the intranuclear cascade stage of nonelastic interactions when they occur; (4) simulation of nuclear evaporation; and (5) statistical error estimates on the dose.
Validation of the GPU MC calculation consisted of secondary particle yield calculations in proton collisions with therapeutically relevant nuclei, dose calculations in homogeneous phantoms, and recalculations of complex head and neck treatment plans from a commercially available TPS, all of which were compared with Geant 4.9.6p2/TOPAS [16, 17].
Commissioning of the GPU MC code consisted of matching the lateral spot shape, depth and width of the distal edge, and number of protons per monitor unit for each of the 97 pencil beam energies, which ranged from 70 to 232 MeV, available for treatment to the measured values. The conversion of CT Hounsfield units to material and density was also commissioned following the formalism in Zacharatou et al [18], including material overrides for implanted device, i.e. titanium or stainless steel implants, details given in Zhao et al [5] Extensive comparisons to measurements were conducted to validate the commissioning of the GPU MC.
Simplified Biological Model
Many different proton RBE models have been proposed [12, 19–21]. These models are usually a function of LETd, the α and β parameters from the linear quadratic formula, and the physical dose (PD). In the system described here, in addition to the conventional 1.1 constant, a simple linear model that is independent of α and β is used.
During the MC simulation, LETd [10] is calculated along with the PD distribution. For each voxel of the CT data set the biological dose (BD) was computed from PD and LETd. In addition to the conventional 1.1*PD dose file, a dose file with an LETd weighted BD is created based on the following equation:
where LETd is in units of keV/μm, PD has units of Gy, the constant 0.08 has units of μm/keV, and the constant 0.88 is unitless. It is important to note here that we are not claiming that the RBE of protons follows this equation; rather, the implementation of this equation aids in detection of possible at-risk areas. Fractionation is explicitly excluded from this formula. This model agrees well with other published models for α/β values of 2.0 + 1.0 and a dose of 2 Gy per fraction. The constant values were chosen to give an RBE of 1.5 for an LETd of 6.0 keV/μm and 1.1 for an LETd of 1.5.
System Configuration
The requirements of the application presented several unique challenges. Because this application was to be used clinically, it needed to be built using institutionally approved tools to ensure reliability and maintainability, so the user interface and scheduler services were developed using common Windows technologies including C# programming language, .NET framework, Microsoft SQL Server databases, and Visual Studio development environment. The system also had to interoperate with a TPS. This interoperability was achieved through the use of a standard medical imaging format and communications protocol, namely DICOM (digital imaging and communications in medicine). The dose simulation engines, for performance reasons, were developed to be executed on Linux-based servers. To seamlessly interact with Linux executables from a Windows application, Python-based Web services were employed on the Linux side, allowing simple Web requests to be made from the Windows servers. A file share was created, allowing data to be accessed from all servers, regardless of the operating system. These last 2 were critical to the successful integration of a system with some key components developed and deployed for Windows and others that must be run under Linux. For improved usability, the user interface is Web based so users can run anywhere, provided the user is within the institutional firewall. Automatic authorization is provided using an Active Directory service.
Digital Imaging and Communications in Medicine Handling
The MC dose verification system, called 2nd Check, begins with a DICOM export from the TPS to the 2nd Check DICOM listener. The export filter is set up to push all chosen DICOM files to the listener running as a service on a Windows server. To successfully complete the dose verification, the user must export the desired radiation therapy ION plan along with the associated radiation therapy dose, radiation therapy structure set, and all CT images associated with the plan .
To prepare the dose files for import back into the TPS, a windows service called the Import Preparer makes a copy of the original radiation therapy plan file for each newly generated dose file. New unique identifiers are created and properly matched for each of the new radiation therapy plan and radiation therapy dose files, which is needed for successful import back into the TPS. These files are then all copied over to an outgoing directory that is already preset within Eclipse, greatly simplifying the import process.
User Experience and Data Management
One of the key design considerations was to keep the process for users as smooth and intuitive as possible. The user experience should consist of signaling to the system that they are ready to run the 2nd Check and clearly see the status of the process. Another requirement was that inputs would need to be made from across the institutional network on a variety of devices. The last requirement was to build a system with a high degree of fault tolerance and segment independence.
The functionality of all pieces of this system is based on virtual concepts created in a relational database. The primary virtual concept is the queue. All activity the user expects to occur automatically happens in a virtual queue.
To meet the requirement for the user experience, it was decided to build a Web application to accommodate the run anywhere philosophy. The contents of the website were chosen for simplicity and ease of navigation. When the user enters the website, the first page is simply the instruction and any bulletins with simple tabs across the top. The tabs take users to where they need to be, either looking for patients or viewing the current state of the queue. Patient navigation occurs by searching for either patient ID or the patient name. Once the patient is found, the user can drill down to the plans.
At this level, users will see the actions available for their plans or its current state in the QA system. The queue is simply the view of all plans currently moving through all steps in relation to their current state and the time they entered the queue.
The plans remain in the system until they are removed. Two processes exist to remove a plan from the system; deleting the patient or importing all of the patients plans back into the TPS.
Clinical Workflow
After the treatment plan is exported and results are brought back into the TPS, as detailed in the sections “DICOM Handling” to :User Experience and Data Management,” the clinical evaluation is performed. The first step in the evaluation is performed by the medical physicist and consists of comparing the physical dose calculated by the TPS and the MC system (both include the conventional 1.1 biological factor). Line profiles in each CT slice, particularly in heterogeneous areas, are examined, dose-volume histogram statistics are evaluated, and dose output is examined. If major issues are detected, such as dose differences >5%, the root cause is explored, and typically, a new planning approach is devised. Minor issues are noted and brought to the treating physician's attention during their review. The medical physicist then evaluates the BD distribution. This step primarily includes evaluating sensitive critical structures with the key feature of interest being a comparison of the enhanced target BD compared with enhanced critical structure dose. As a note, independent of this system, the robustness of the plan with respect to stopping power and position uncertainties are also evaluated at this stage; this can be done in the TPS or via the MC system. In routine practice, the TPS is generally used. If only minor issues are found (ie, sporadic dose differences <2%), the plan is prepared for the physician review with the minor issues noted verbally or in writing. The physician reviews the TPS dose distribution along with the MC and BD distributions. If major discrepancies between the MC and TPS dose (ie, systematic differences >2%) or concerns based on the BD exist that cannot be easily remedied by plan modification; the physician is contacted for clinical guidance.
Results
Calculation Times
The calculation time is broken up into the following 5 components: (1) DICOM export from the TPS, (2) submission of the plan to the GPU queue, (3) MC conversion and calculation, (4) conversion of the dose files (PD, BD, LETd) to DICOM, and (5) DICOM import to the TPS.
The DICOM export is typically around 1 minute but depends primarily on the size of the CT data set. The plan is generally in the queue for less than 30 seconds. The system is designed to handle several submissions simultaneously, but occasionally all servers may be occupied and queue time can be up to 10 minutes. The MC calculation time can range from 3 minutes for a 2-field plan with a clinical target volume <200 cm3 to 9 minutes for a multiple field plans with a clinical target volume >1000 cm3. Currently, 108 protons are simulated for each plan, divided among all the proton spots according to their relative weight. Conversion of the MC dose cubes to DICOM generally takes 2 minutes; this time also includes archiving and transferring the files to the import directory. Importing the DICOM dose cubes into the TPS takes 2 minutes. The entire process takes anywhere from 7 to 18 minutes with about half of the time occupied by the MC calculation. This meets our initial clinical specification of <15 minutes for the majority of the plans.
Clinical Results
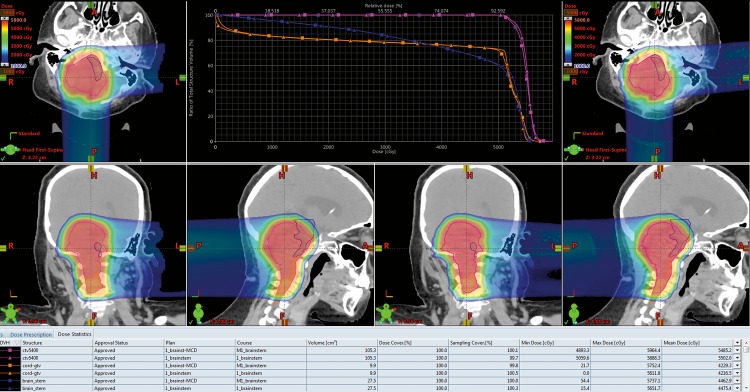
Figure 1 represents a case where the dose calculation from the TPS and the 2nd Check system agree quite well.
Figure 1.
The dose colorwash and dose-volume histogram (DVH) plots for the treatment planning system (TPS) and Monte Carlo (MC) dose calculation are displayed. The images on the left are from the TPS calculation, and on the right is the MC. The magenta DVH is the target; the other 2 are serial-type critical structures. The triangle markers are for the TPS DVH; the square is for the MC. For this patient, there were no meaningful differences in the 2 calculations, so no intervention was required.
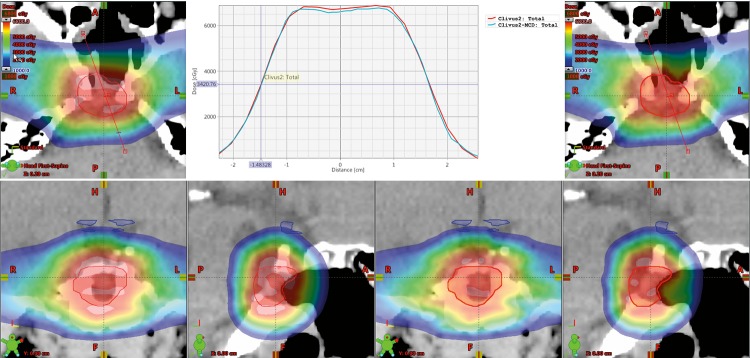
Figure 2 shows a case where the TPS and MC dose disagree slightly. The MC dose distribution is 2.2% lower than that from the TPS. This difference is attributed to the small target size (∼ 2 cm in diameter) and the complex heterogeneous surroundings. This plan would be suitable for renormalization based on the MC dose calculation.
Figure 2.
The dose colorwash of the treatment planning system (TPS) generated distribution (left) and the Monte Carlo generated dose distribution (right) for a central brain tumor with a clinical target volume (CTV) (red) that is approximately 2 cm in diameter. The graph in the center displays the dose profile through the center of the CTV, with red representing the TPS calculation. Note that the Monte Carlo dose distribution is 2.2% lower than that from the TPS in the center region of the CTV. This difference is attributed to the small target size and the complex heterogeneous surroundings. This plan would be suitable for renormalization.
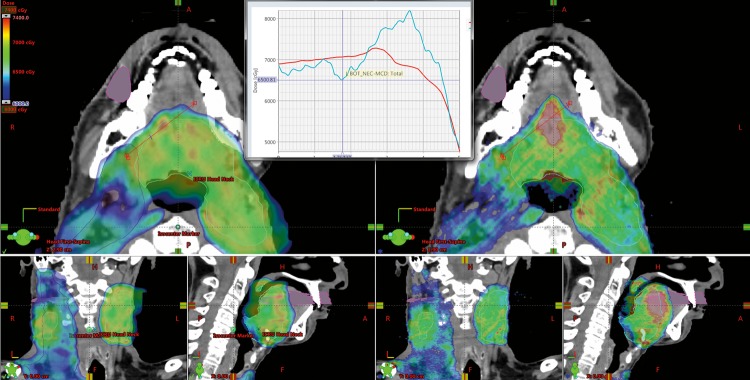
Figure 3 is an example of a large disagreement between the TPS and MC dose. This error in the TPS calculation would not be picked up by a patient-specific measurement on a flat water phantom as in that case both calculations agreed to within 2% in dose. Upon investigation it was determined that the beam was traveling parallel to a large bone/air interface, which resulted in the discrepancy.
Figure 3.
The dose colorwash of the treatment planning system generated distribution (left) and the Monte Carlo generated dose distribution (right) for a head and neck treatment. There exists a notable region where the Monte Carlo calculation results in an approximately 15% increased dose. Upon investigation it was determined that the beam was traveling parallel to a large bone/air interface near the trachea and vertebral bodies, resulting in the discrepancy, which is not located immediately adjacent to those interfaces.
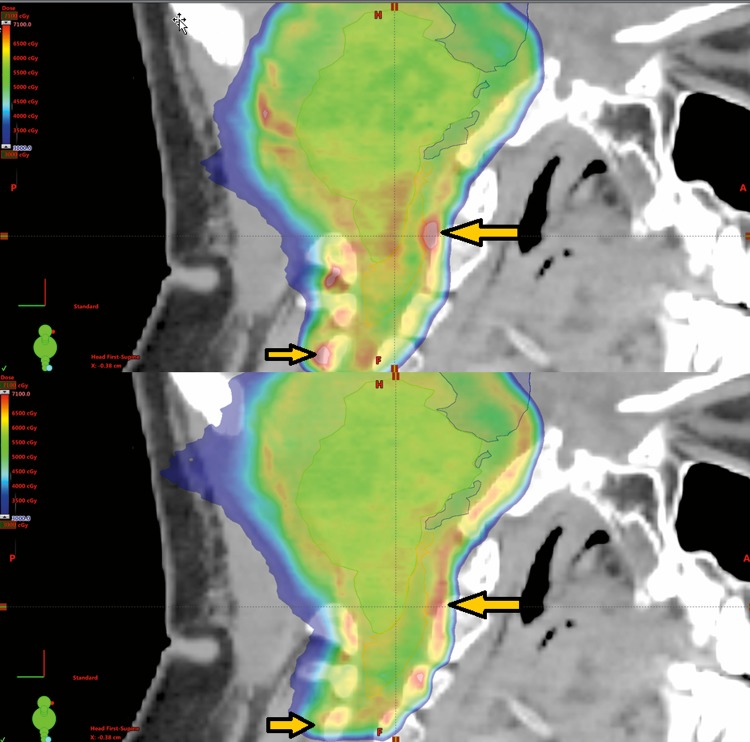
Shown in Figure 4 is the dose colorwash for the LETd weighted biological plan. This is an example of a plan modification based on the BD. The top image is the initial plan and the bottom image is after mitigation for high BD. In the region of the biological hot spots (arrows), the physical dose of the initial plan is 2% higher than the modified plan; however, the BD is approximately 10% higher. A slight margin and gantry angle change are the main differences in the plan; the optimization objectives are similar.
Figure 4.
The dose colorwash for the dose-weighted linear energy transfer weighted biological dose calculation for the initial plan (top) and after modification (bottom). The modification was done to reduce the high biological dose indicated by the yellow arrows. In the region of the biological hot spots, the physical dose of the initial plan is 2% higher than the modified plan, but the biological dose is ∼10% different. Small modifications of margin and gantry angle resulted in an improved biologic dose profile in critical locations such as the brainstem and spinal cord.
More than 270 patients were treated at our proton center in the first year of operation, all of whom used this system. Because most plans underwent multiple iterations before final approval, more than 1000 plans have been run through the system from multiple users with minimal downtime. The average time from plan export to importing of the MC doses is less than 15 minutes. Modifications of the treatment plans have been made based on either the nominal MC dose or the BD. An exact quantification of the percent of plans that have been modified based on this system is difficult because both the MC dose and BD are included in the instructions to the dosimetrists. One example of this instruction is for breast treatments. For those treatments, there is a constraint on the brachial plexus of less than 60 Gy as represented in the BD.
Discussion
In this report, we demonstrate that a GPU-based MC dose second-check system is feasible and has been implemented for routine use on every patient treated with spot scanned protons in an enterprise-wide infrastructure allowing fast, accurate, and simple dose second checks to be integrated into the patient care path. Early work by Kooy et al [2] demonstrated the need for and value of a calculative infrastructure that was independent of the planning and delivery system. Based on scattered PBT, the system took into account a limited number of routinely used treatment components to allow for an independent quality check. Sahoo et al [3], also working with scattered beams, demonstrated that an monitor unit-check algorithm similar to that in photon beam therapy was possible if each physical characteristic and variable was taken into account. In both cases, an independent and accurate second check was created. More recently, Mackin [7] reported on the use of an analytical dose second-check algorithm to perform patient-specific spot scanning QA with intent to reduce or eliminate time-consuming measurements. The current work extends beyond each of these in that it is dedicated to spot scanning, creates a highly accurate and very efficient GPU-based MC second-check system, is integrated into a robust and enterprise-wide interface, calculates and compares physical dose with the commercial TPS, and provides key insight into LET and simple BD distributions.
Recent reports of brainstem injury in patients undergoing proton therapy have highlighted the importance of understanding the biologic dose and end of range effects, especially in pediatric patients [8, 9]. Although RBE is dependent on multiple factors, understanding differences in LET along the proton path, particularly at the Bragg peak is important because it is an actionable parameter, especially in spot-scanning systems. The BD 2nd Check process described herein provides visual feedback of areas of relative intensification of biologic dose. By visually inspecting these BD maps for clinical treatment plans, clinicians can easily see the effects of beam arrangements, optimization parameters, and planning margins that may result in regions of biological intensification. However, because the calculation is based on a simple model, the absolute BDs are currently unknown. Further refinements in understanding proton LET from cell models, as well as patient data, can be incorporated in future models.
In the initial implementation now in clinical use, end-to-end second check per patient has met required times of less than ∼15 minutes, running from anywhere on the network, and by any physics or dosimetry staff member. Current limitations of the system currently are the queue/scheduler, which can result in longer wait times if multiple plans are in queue. In addition, a fixed number of protons are currently used in the simulation, but we anticipate that improvement in calculation times can be achieved with a statistical error–based stopping criterion. Finally, this system does not replace patient-specific machine-based measurement since machine performance cannot be predicted by the software; however, with the use of this system to verity 3-dimensional dose calculation, patient-specific measurement at multiple depths would no longer be necessary.
Conclusion
MC dose calculation verification of spot scanning proton treatment plans is feasible and practical in the clinical environment. The 3-dimensional dose verification, particularly near heterogeneities, has resulted in plan modifications. The BD data provide actionable feedback for end-of-range effects, especially in pediatric cases.
ADDITIONAL INFORMATION AND DECLARATIONS
Conflicts of Interest: The authors have no conflicts of interest to disclose.
References
- 1.Stern RL, Heaton R, Fraser MW, Goddu SM, Kirby TH, Lam KL, Molineu A, Zhu TC, Group AT Verification of monitor unit calculations for non-IMRT clinical radiotherapy: report of AAPM Task Group 114. Med Phys. 2011;38:504–30. doi: 10.1118/1.3521473. [DOI] [PubMed] [Google Scholar]
- 2.Kooy HM, Rosenthal SJ, Engelsman M, Mazal A, Slopsema RL, Paganetti H, Flanz JB. The prediction of output factors for spread-out proton Bragg peak fields in clinical practice. Phys Med Biol. 2005;50:5847–56. doi: 10.1088/0031-9155/50/24/006. [DOI] [PubMed] [Google Scholar]
- 3.Sahoo N, Zhu XR, Arjomandy B, Ciangaru G, Lii M, Amos R, Wu R, Gillin MT. A procedure for calculation of monitor units for passively scattered proton radiotherapy beams. Med Phys. 2008;35:5088–97. doi: 10.1118/1.2992055. [DOI] [PubMed] [Google Scholar]
- 4.Hsi WC, Schreuder AN, Moyers MF, Allgower CE, Farr JB, Mascia AE. Range and modulation dependencies for proton beam dose per monitor unit calculations. Med Phys. 2009;36:634–41. doi: 10.1118/1.3056466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Q, Wu H, Cheng CW, Das IJ. Dose monitoring and output correction for the effects of scanning field changes with uniform scanning proton beam. Med Phys. 2011;38:4655–61. doi: 10.1118/1.3609417. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y, Ramirez E, Mascia A, Ding X, Okoth B, Zeidan O, Hsi W, Harris B, Schreuder AN, Keole S. Commissioning of output factors for uniform scanning proton beams. Med Phys. 2011;38:2299–306. doi: 10.1118/1.3569581. [DOI] [PubMed] [Google Scholar]
- 7.Mackin D, Li Y, Taylor MB, Kerr M, Holmes C, Sahoo N, Poenisch F, Li H, Lii J, Amos R, Wu R, Suzuki K, Gillin MT, Zhu XR, Zhang X. Improving spot-scanning proton therapy patient specific quality assurance with HPlusQA, a second-check dose calculation engine. Med Phys. 2013;40:121708. doi: 10.1118/1.4828775. [DOI] [PubMed] [Google Scholar]
- 8.Meier G, Besson R, Nanz A, Safai S, Lomax AJ. Independent dose calculations for commissioning, quality assurance and dose reconstruction of PBS proton therapy. Phys Med Biol. 2015;60:2819–36. doi: 10.1088/0031-9155/60/7/2819. [DOI] [PubMed] [Google Scholar]
- 9.Wan Chan Tseung H, Ma J, Beltran C. A fast GPU-based Monte Carlo simulation of proton transport with detailed modeling of nonelastic interactions. Med Phys. 2015;42:2967–78. doi: 10.1118/1.4921046. [DOI] [PubMed] [Google Scholar]
- 10.Wan Chan Tseung HS, Ma J, Kreofsky CR, Ma DJ, Beltran C. Clinically applicable Monte Carlo-based biological dose optimization for the treatment of head and neck cancers with spot-scanning proton therapy. Int J Radiat Oncol Biol Phys. 2016;95:1535–43. doi: 10.1016/j.ijrobp.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 11.Paganetti H, Niemierko A, Ancukiewicz M, Gerweck LE, Goitein M, Loeffler JS, Suit HD. Relative biological effectiveness (RBE) values for proton beam therapy. Int J Radiat Oncol Biol Phys. 2002;53:407–21. doi: 10.1016/s0360-3016(02)02754-2. [DOI] [PubMed] [Google Scholar]
- 12.Carabe A, Moteabbed M, Depauw N, Schuemann J, Paganetti H. Range uncertainty in proton therapy due to variable biological effectiveness. Phys Med Biol. 2012;57:1159–72. doi: 10.1088/0031-9155/57/5/1159. [DOI] [PubMed] [Google Scholar]
- 13.Kralik SF, Ho CY, Finke W, Buchsbaum JC, Haskins CP, Shih CS. Radiation necrosis in pediatric patients with brain tumors treated with proton radiotherapy. AJNR Am J Neuroradiol. 2015;36:1572–8. doi: 10.3174/ajnr.A4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther JR, Sato M, Chintagumpala M, Ketonen L, Jones JY, Allen PK, Paulino AC, Okcu MF, Su JM, Weinberg J, Boehling NS, Khatua S, Adesina A, Dauser R, Whitehead WE, Mahajan A. Imaging changes in pediatric intracranial ependymoma patients treated with proton beam radiation therapy compared to intensity modulated radiation therapy. Int J Radiat Oncol Biol Phys. 2015;93:54–63. doi: 10.1016/j.ijrobp.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Wan Chan Tseung HS, Beltran C. A graphics processor-based intranuclear cascade and evaporation simulation. Comput Phys Commun. 2014;185:2029–33. [Google Scholar]
- 16.Agostinelli S, Allison J, Amako K, Apostolakis J, Araujo H, Arce P, Asai M, Axen D, Banerjee S, Barrand G. Geant4—a simulation toolkit. Nucl Instrum Methods Phys Res A. 2003;506:250–303. [Google Scholar]
- 17.Perl J, Shin J, Schumann J, Faddegon B, Paganetti H. TOPAS: an innovative proton Monte Carlo platform for research and clinical applications. Med Phys. 2012;39:6818–37. doi: 10.1118/1.4758060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharatou Jarlskog C, Lee C, Bolch WE, Xu XG, Paganetti H , , , , . Assessment of organ-specific neutron equivalent doses in proton therapy using computational whole-body age-dependent voxel phantoms. Phys Med Biol. 2008;53:693–717. doi: 10.1088/0031-9155/53/3/012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhary P, Marshall TI, Perozziello FM, Manti L, Currell FJ, Hanton F, McMahon SJ, Kavanagh JN, Cirrone GA, Romano F, Prise KM, Schettino G. Relative biological effectiveness variation along monoenergetic and modulated Bragg peaks of a 62-MeV therapeutic proton beam: a preclinical assessment. Int J Radiat Oncol Biol Phys. 2014;90:27–35. doi: 10.1016/j.ijrobp.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Wedenberg M, Lind BK, Hardemark B. A model for the relative biological effectiveness of protons: the tissue specific parameter alpha/beta of photons is a predictor for the sensitivity to LET changes. Acta Oncol. 2013;52:580–8. doi: 10.3109/0284186X.2012.705892. [DOI] [PubMed] [Google Scholar]
- 21.Paganetti H. Significance and implementation of RBE variations in proton beam therapy. Technol Cancer Res Treat. 2003;2:413–26. doi: 10.1177/153303460300200506. [DOI] [PubMed] [Google Scholar]