Abstract
Purpose:
Several Monte Carlo transport codes are available for medical physics users. To ensure confidence in the accuracy of the codes, they must be continually cross-validated. This study provides comparisons between MC2 and Tool for Particle Simulation (TOPAS) simulations, that is, between medical physics applications for Monte Carlo N-Particle Transport Code (MCNPX) and Geant4.
Materials and Methods:
Monte Carlo simulations were repeated with 2 wrapper codes: TOPAS (based on Geant4) and MC2 (based on MCNPX). Simulations increased in geometrical complexity from a monoenergetic beam incident on a water phantom, to a monoenergetic beam incident on a water phantom with a bone or tissue slab at various depths, to a spread-out Bragg peak incident on a voxelized computed tomography (CT) geometry. The CT geometry cases consisted of head and neck tissue and lung tissue. The results of the simulations were compared with one another through dose or energy deposition profiles, r90 calculations, and γ-analyses.
Results:
Both codes gave very similar results with monoenergetic beams incident on a water phantom. Systematic differences were observed between MC2 and TOPAS simulations when using a lung or bone slab in a water phantom, particularly in the r90 values, where TOPAS consistently calculated r90 to be deeper by about 0.4%. When comparing the performance of the 2 codes in a CT geometry, the results were still very similar, exemplified by a 3-dimensional γ-analysis pass rate > 95% at the 2%–2-mm criterion for tissues from both head and neck and lung.
Conclusion:
Differences between TOPAS and MC2 were minor and were not considered clinically relevant.
Keywords: Monte Carlo simulations, intercomparison
Introduction
Radiation therapy with charged ions, such as proton therapy, presents a promising alternative to conventional radiotherapy methods, such as 3-dimensional–conformal x-ray therapy or even the more-advanced intensity-modulated radiotherapy. In the United States, > 24 proton centers are operating today, and more are under construction or in the planning phase [1]. Although the dosimetric advantage of proton therapy has been investigated in numerous publications [2–5], the expected improvements in clinical outcomes have not yet been fully realized, which is, in part, attributed to the extremely complex behavior of the ions in therapeutic applications. Proton therapy presents many challenges to medical physicists, including uncertainties in radiobiological effectiveness, inaccuracies in the calculation of proton tracks because of multiple Coulomb scattering along heterogeneities in the ions' paths, and the resulting overall uncertainty caused by interfractional and intrafractional motion of the patients [6, 7]. One of the dominant problems is the uncertainty in the physical dose distribution in patients. Although measurements can easily be performed in water and simple phantoms of various materials, the dose distribution in a patient relies mostly on predictive calculations. Contemporary treatment planning systems rely on analytic or semiempirical dose-prediction algorithms, and only recently, some commercial products have begun to provide Monte Carlo (MC) simulations, which propagate the path of protons track by track, based on first principles. The MC dose predictions are typically considered the “best estimate” or “gold standard,” especially in highly complex and heterogeneous environments, such as a human body. However, these codes require considerable expertise in MC techniques and programming and enormous amounts of central processing unit time. To facilitate MC simulations of clinical dose distributions, a variety of adaptations of general purpose MC systems have been developed, including MC2 (based on MCNPX) and Geant4 Application for Emission Tomography (GATE) and Tool for Particle Simulation (TOPAS) (both based on Geant4) [8–13]. Other codes, such as Fluktuierende Kaskade (FLUKA), have been developed with user interfaces that enable the quick implementation of digital imaging and communications in medicine (DICOM) computed tomography (CT) images into simulations [14]. The application of these wrapper-codes is substantially simpler than the application of the underlying general-purpose MC systems. Nevertheless, they require a thorough configuration, and a detailed validation process has to be performed to provide accurate results.
In this study, we compared the performance of 2 of these adapted codes: MC2, implemented at the University of Texas MD Anderson Cancer Center (Houston, Texas), and TOPAS, installed at Massachusetts General Hospital (Boston, Massachusetts) and at MD Anderson. Comparisons were performed on multiple phantoms as well as 2 patient CTs. Variations in dose calculations when using different Monte Carlo systems must be expected and evaluated. Comparing computational results between 2 or more codes has a well-established precedent [15–18]. MC simulations are being increasingly adopted for both clinical and research use in proton therapy. The results of the phantom simulations were compared to estimate systematic differences in the MC systems. Results of the patient treatment simulation were assessed with γ-analyses [19], and their potential clinical impacts were estimated.
Materials and Methods
Monte Carlo Codes
TOPAS
TOPAS [9] is a user-friendly wrapper that sits on top of the MC code Geant4, allowing the user to quickly and easily create their own simulations without the need to write or compile code. It is a multipurpose program that includes the entire Geant4 functionality. All simulations were performed with TOPAS version 3.0.p1 (TOPAS MC Inc) using the default physics list recommended for use in proton therapy. This physics list has models that work for protons and secondary particles [20]. TOPAS fully supports multithreading, and each simulation was set to use 24 threads on a computer cluster. Each node on the cluster is an HP BL465c G7 blade (Hewlett Packard, Palo Alto, California) with 2, 12-core AMD (Santa Clara, California) 6174 processors, for 24 cores per node. Each node has a minimum of 64 GB memory. The cluster's operating system is Linux 6.5 (Red Hat Enterprise, Raleigh, North Carolina) with a queuing interface consisting of a combination of PBS/Torque and Moab (Adaptive Computing, Naples, Florida). TOPAS simulations at Massachusetts General Hospital were performed on a large research computing cluster, splitting jobs into up to 30 parallel jobs, each using 2 to 4 threads to improve calculation speed. TOPAS converts the Hounsfield unit (HU) from a DICOM CT file to stopping power via a CT calibration curve that the user must designate in the simulation. For our simulations, we used the CT calibration curve used by the Proton Therapy Center at Houston (PTCH, Houston, Texas). For each HU encountered, TOPAS generates unique stopping power based on the provided HU-stopping power curve.
MC2
MC2 is a wrapper for Monte Carlo N-Particle Transport Code (MCNPX), a general-purpose MC radiation-transport code developed at Los Alamos National Laboratories (Los Alamos, New Mexico) [11]. MCNPX was adapted and applied at MD Anderson for research purposes as early as 2003, and in 2011, MC2 was developed at the MD Anderson PTCH to facilitate easier processing of proton treatment plans. MC2 is equipped with a graphical user interface, limiting the user's choice of parameters, so that an established, calibrated, and validated performance of the underlying MCNPX code produces predictions of the highest possible accuracy. MC2 uses MCNPX, with added features enabling the user to import treatment plans created by a commercial treatment planning system and, then, runs an MCNPX simulation of that plan based on a validated model of the Hitachi (Tokyo, Japan) proton machine at PTCH. Details of the physics parameters of the MCNPX code are described by Titt et al [21] and in references therein. All MC2 simulations were performed on a Linux cluster using 64 core nodes with 256-GB memory. The operating system installed on this cluster is RHEL 6 (Red Hat Enterprise) with a Torque queuing system. To handle the conversion from HU to stopping power, MC2 uses a lookup table to assign all HU numbers within a certain range to a material with a certain density. For instance, MC2 has multiple “bone” stopping powers that correspond to ranges of CT numbers. The stopping powers for these materials are then calculated based on their elemental composition.
Tallies
Most results in this study are reported with units of MeV cm−3 h−1, with h being the number of source-particle histories. Use of these units allows comparison between MC2 and TOPAS, independent of the material being irradiated. Conversion to dose is straightforward: simply divide the energy deposited by the mass density of the material to get the absorbed dose per proton. Thus, the terms dose and energy deposition are used interchangeably throughout this work. The dimensions and volumes of the tallies in MC2 and TOPAS were identical and are described in “Water Phantom,” “Bone and Lung Heterogeneities,” and “Patient CT Geometry sections. The low-range cutoff in TOPAS was set to 0.05 mm, and the step size was set to 1 mm. MC2 tracked all particles down to a kinetic energy of 1 keV. Further details on the physics settings used by MC2 are described in Titt et al [8].
Water Phantom
A 50 × 50 × 50 cm3 water phantom was placed at the center of the simulation geometry with the proton beam source located 5 cm from the face of the phantom. The proton beam-source geometry was a planar circle with a radius of 5 mm. Protons were emitted in a monoenergetic beam aimed at the water phantom at 3 energies—90, 150, and 200 MeV—corresponding to ranges in water (continuous slowing-down approximation ranges) of 6.4, 15.8, and 26.0 cm [22], respectively. The proton beam was emitted in the −Z direction, and 100 million proton histories were simulated in both TOPAS and MC2 to keep statistical uncertainties < 1% (1 σ) in locations with energy depositions > 10% of the maximum. Voxels scored energy deposited in the in-plane and cross-plane directions and had dimensions of 1 × 0.5 × 1-mm3 (in-plane) and 0.5 × 1 × 1-mm3 (cross-plane). The integrated depth dose (IDD) was scored in 3 cylindrical geometries with radii of 4.08 cm, 10 cm, and 25 cm. All IDD scorers had a resolution of 0.1 mm in the beam-depth direction.
Half-profiles for each simulation were taken at depths of 40 mm (90 MeV), 90 mm (150 MeV), and 130 mm (200 MeV). r90, or the depth at which the dose is 90% of the maximum and distal to the Bragg peak, was determined for each code and compared. The differences between depth profiles scored with the large-radius (25 cm) scorer and the small-radius (4.08 cm) scorer were assessed to understand possible differences between the codes' scattering algorithms. Variations in large-angle scattering lead to differences in the depth profiles, when comparing tallies with greatly different radii. An r = 4.08 cm scorer was chosen to mimic the Bragg peak ionization chamber (PTW, Freiburg, Germany), whereas the r = 25 cm scorer is able to capture energy deposited far from the central axis of the beam. Two-dimensional energy-deposition distributions in-plane and cross-plane for the MC2 and TOPAS simulations were compared by γ-analysis.
Bone and Lung Heterogeneities
Several geometries were created to compare performance between each code. All beams had a planar circular source with radius of 5 mm and were placed 0.5 mm from the surface of a water phantom, on its central axis. The water phantom had dimensions of 20 × 20 × 30 cm3. Lung and bone inserts were created with dimensions of 10 × 10 × 1 cm3. The bone material was “G4_BONE_COMPACT_ICRU” found in TOPAS/Geant4 and had a density of 1.85 g cm−3. Lung material was also the default TOPAS/Geant4 lung material, “G4_LUNG_ICRP,” except the density was set to 0.25 g cm−3 to simulate inflated lung. The lung and bone material compositions in the MC2 simulations are given in Table S1; the densities and compositions exactly matched those used in TOPAS.
Lung and bone inserts were placed at a variety of depths, with 2 different proton energies (100 and 200 MeV). Table S2 summarizes the geometries of each simulation. Energy deposition was scored in-plane and cross-plane in voxels with dimensions of 0.5 × 1 × 0.5 mm3 in the in-plane and 1 × 0.5 × 0.5 mm3 in the cross-plane. Profiles for each simulation were compared to each other. The IDDs were scored in cylinders with a radius of 10 cm and a resolution in the Z-direction of 0.1 mm. The r90 was calculated for the TOPAS and MC2 simulations and compared.
Patient Computed Tomography Geometry
The CT of the head and neck of a patient with an oropharynx tumor, and the CT of a patient with a tumor in the right lung were chosen for simulation because of the complex heterogeneities or dose-calculation difficulties present in such cases. The complexity of CT geometries was chosen to maximize any differences between the different MC systems. The beam was designed to generate a spread-out Bragg peak (SOBP) with a range of 14 cm in water and a modulation width of 5 cm. The modulation width and range for the SOBP were chosen because they are the beam parameters that most closely matched real treatment plans created for those patients. We chose to simulate a single treatment-like field, rather than multiple fields comprising many spots, to highlight the dose-calculation differences between the 2 codes. Differences between the 2 codes would likely be blurred across multiple fields. In addition, a comparison of full-treatment plans among MC codes and treatment planning software has been previously published [23]. To create the SOBP in TOPAS, a parameterized beam was configured to give the desired modulation width and range. A phase space was generated for MC2 based on the TOPAS parameters. Both the TOPAS and MC2 beam were compared with each other in a water phantom (no heterogeneities) to ensure agreement before simulating the patient CT irradiation. The beam had a radius of 2.5 cm and irradiated the patient from the posterior, with the central axis aligned to the treatment plan's isocenter. For all simulations, 1 × 108 proton histories were simulated. The CT had a voxel size of 0.5 × 0.5 × 2.5 mm3 (2.5 mm slice thickness), and energy deposited was scored in those voxels. To convert from HU to stopping power, the CT conversion table used at PTCH was implemented in both codes.
Results
The results of the water-phantom simulations, as well as the bone and lung slab simulations, are presented in further detail in supplemental tables and figures. Supplemental figures and tables are denoted with the prefix S before their number.
Simple Geometry, TOPAS-MC2 Comparisons (Water Phantom)
The IDDs from each code are shown in Figure S1. To compare, the depth of the 90% dose fall-off distal to the Bragg peak was determined. For the 90-MeV beams, MC2 had r90 = 6.37 cm and TOPAS r90 = 6.40 cm. For the 150-MeV beams, MC2 had r90 = 15.71 cm and TOPAS r90 = 5.78 cm. For the 200-MeV beams, MC2 had r90 = 25.87 cm and TOPAS r90 = 25.96 cm. Lateral energy-deposition profiles were taken at a variety of depths and are presented in Figure S2. The ratio of the depth profile scored by the 25-cm radius scorer to that scored by the 4.08-cm radius scorer for each code as a function of depth is plotted in Figure S3. Differences in the Dr25/Dr4.08 ratio between the 2 codes indicate differences in the way large-angle proton scattering is modeled between TOPAS and MC2. Two-dimensional γ-analyses were performed for dose distributions in the in-plane and cross-plane directions in MC2 and TOPAS, as shown in Table S1. Two passing criteria were tested: 2%–2-mm and 1%–1 mm. γ-Analysis was performed only between voxels that had ≥ 10% of the maximum dose from the reference-dose distribution, which was defined as the TOPAS dose.
Heterogeneous Phantom Geometry, TOPAS-MC2 Comparisons
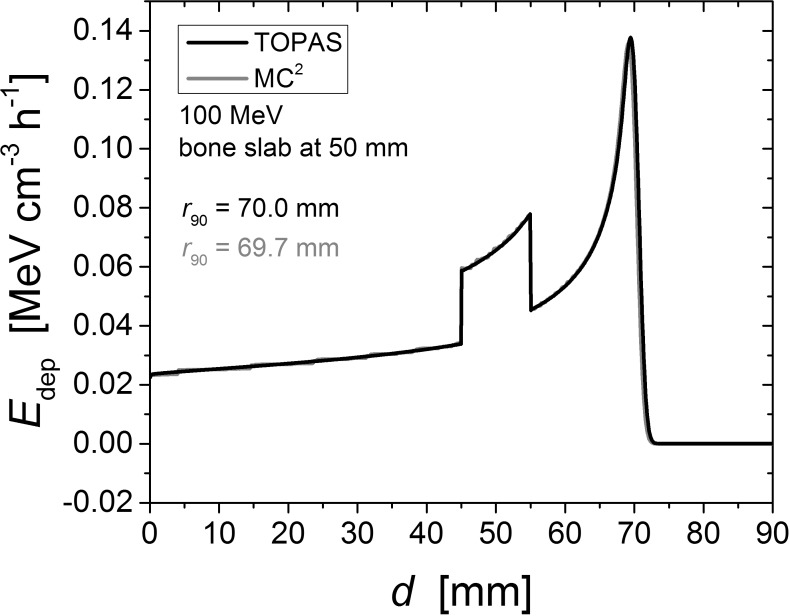
Results of the simulations detailed in the Bone and Lung Heterogeneities section are shown in Figure 1 and Table S3. The material compositions for each simulation are given in Table S2, and a summary of the geometries for each simulation are provided in Table S3. Figure 1 shows an example of depth–dose curve comparison between TOPAS and MC2, with the 100 MeV incident beam on a bone slab centered at a depth of 50 mm. All r90 depths calculated by MC2 and TOPAS agreed within 0.5%. Table S4 shows the differences in r90 between the MC2 and TOPAS bone and lung slab simulations.
Figure 1.
Depth-dose profile of the 100-MeV beam with a 10-mm-thick bone insert centered at a depth of 50 mm. This plot is representative of all TOPAS-MC2 comparisons with the lung or bone slab geometry. All r90 depths agreed to within < 1 mm.
Simulations in a Computed Tomography Geometry
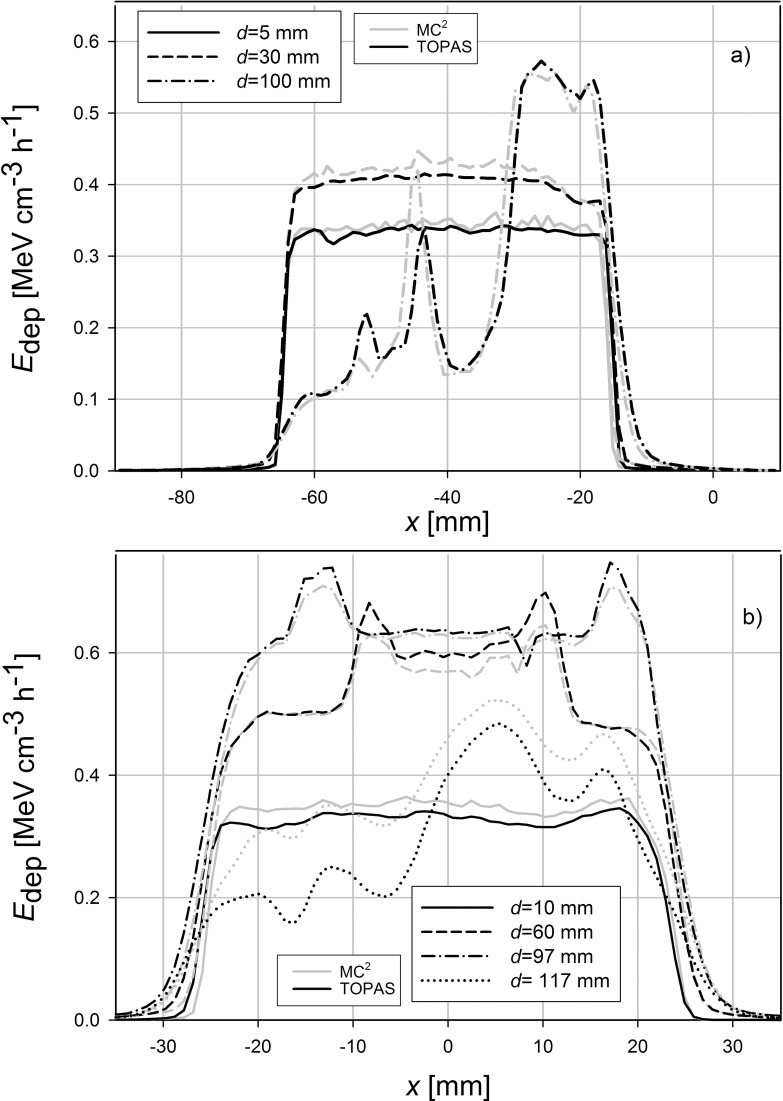
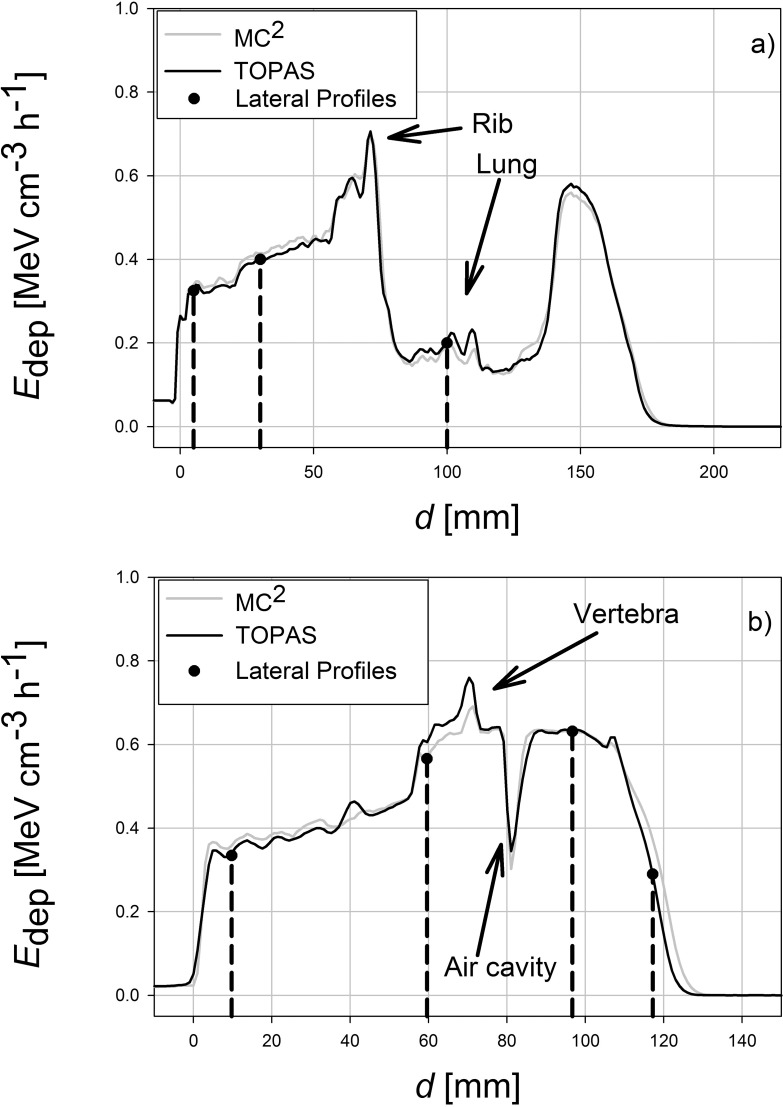
The planning CTs for the patients with a head and neck tumor and a lung tumor was imported into each MC system and each system simulated an irradiation with an SOBP. Profiles of energy deposited were obtained at 4 different depths as indicated in Figure 2. These profiles are shown in Figure 3. Table 1 shows the 2-dimensional γ-analysis results for the TOPAS-MC2 simulations. For these γ-tests, the slice containing the treatment plan's isocenter, along with slices 10 mm superior and 10 mm inferior to the isocenter, were analyzed. In addition, the simulations were analyzed using a 3-dimensional γ-test, and the results are shown in Table 2. Figure 4 shows the depth–dose curve along the beam path averaged > 9 voxels at each depth, centered on the beam's central axis.
Figure 2.
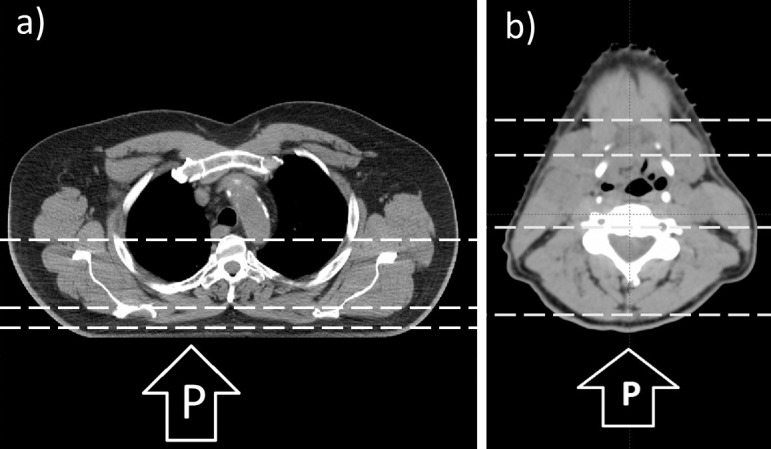
Planning CTs of a patients with a lung tumor in the right lung (a), an oropharynx tumor (b); the slice shown contains the treatment plan isocenter. This slice also corresponds to the plots in Figures 3 and 4. The proton beam in both cases is in the posterior-anterior direction, as shown. The horizontal dashed lines indicate the depths at which lateral profiles were obtained (see Figures 3 and 4). The depths chosen correspond to the beam entrance, proximal SOBP, distal SOBP, and, in the CT of the patient with head and neck, the distal fall-off region. Abbreviations: CT, computed tomography; P, proton beam; SOBP, spread-out Bragg peak.
Figure 3.
Lateral profiles (in the x-direction in the MC geometry) of Edep, for each code at different depths. (a) Three profiles taken in the patient with a lung tumor. (b) Four profiles taken in the patient with head and neck cancer. The profiles were taken in the isocenter slice, which is shown in Figure 2. The location of each profile on the depth dose curve is shown in Figure 4, and their locations correspond to the beam entrance, proximal SOBP, distal SOBP, and, in the head and neck case, the distal fall-off region. Abbreviations: Edep, energy deposited; MC, Monte Carlo; SOBP, spread-out Bragg peak.
Table 1.
Two-dimensional γ-analysis results of in-patient simulations. Pass rates are given as percentages. Only voxels that received > 10% of the maximum dose are included in the analysis.
|
Slice location |
Site being compared |
Pass rate |
|
|
2%–2 mm |
1%–1 mm |
||
| Isocenter | Lung | 93.2 | 73.5 |
| H&N | 94.0 | 63.0 | |
| 1-cm superior of isocenter | Lung | 94.4 | 72.8 |
| H&N | 90.2 | 60.9 | |
| 1-cm inferior of isocenter | Lung | 93.0 | 73.5 |
| H&N | 93.9 | 65.5 | |
Abbreviation: H&N, computed tomography of patient with an oropharynx tumor.
Table 2.
Three-dimensional γ-analysis results of in-patient simulations. Pass rates are given as percentages. Only voxels that received > 10% of the maximum dose are included in the analysis.
|
Site being compared |
Pass rate |
|
|
2%–2 mm |
1%–1 mm |
|
| Lung | 97.9 | 91.7 |
| H&N | 96.4 | 67.2 |
Abbreviation: H&N, computed tomography of patient with an oropharynx tumor.
Figure 4.
The Edep, along the beam's central axis, as a function of d, centered on the slice containing the treatment plan's isocenter. For smoother curves and better statistics, the energy deposited into the central region was defined as the Edep into a 9-voxel region at each depth: the voxel on the beam's central axis, the adjacent 2 voxels on either side (left and right), and those same voxels in the slices that are superior and inferior to the isocenter slice. Arrows indicate anatomic features in each patient that can be identified in Figure 2. Abbreviations: d, depth; Edep, energy deposited.
Discussion
Simple Geometry: MC2-TOPAS Comparisons (Water Phantom)
To evaluate differences between MC2 and TOPAS, we performed a series of simulations, starting with simple water phantoms and advancing to heterogeneous phantoms containing lung- and bone-equivalent slabs. Finally, we simulated 1 field of a treatment scenario for a patient with a head and neck tumor in the oropharynx and 1 for a patient with a tumor in the right lung by importing the planning CTs into the MC simulation and irradiating it with an SOBP. Our choice of a patient with a head and neck tumor was to use a patient geometry with a maximum amount of target heterogeneity, which, we hypothesized, would maximize the differences in dose distributions between the 2 codes. Dose calculation in the lung is a well-known challenge for dose-calculation algorithms, particularly in proton therapy [24, 25]. The MC simulations have also revealed differences in proton ranges compared with experimental values that can be significant [26, 27]. Thus, it is important to compare MC dose distributions in patients with both head and neck and lung cancer. The primary reason to compare calculated dose distributions on the same patient CT is that the CT geometry is independent of both MC2 and TOPAS. We used the same CT number-to-stopping power ratio curve in both codes, which converts the DICOM files of patient CT images into MC-compatible geometries.
As seen in Figure S1, the Bragg curves simulated in each code agreed well. The depths of r90 at each energy differed by less than 1 mm. The MC2 and TOPAS half-profiles agree well with each other in both the high-dose region, close to the beam's central axis, and in the low-dose tails, far from the beam's central axis, as shown in Figure S2. Each beam was symmetric in-plane and cross-plane.
Figure S3 demonstrates the effect of scorer radius on a simulation result. That each curve is greater than unity at all depths indicates that a larger-radius scorer will have more secondary particles deposit energy inside it than a smaller-radius scorer. The difference between the different-sized scorers is small for low-energy beams but can be as high as several percent in higher-energy beams (eg, 200 MeV). The results indicate that there are either small, but noticeable, differences between MC2 and TOPAS in large-angle, particle-scattering algorithms; or differences in the underlying cross-sectional data. However, because of the small doses at those large distances from the central axis, those differences were not significant in this study. However, in treatment plans with large numbers of spots, those small dose differences in the halo of each individual spot might contribute to differences in dose calculations between these 2 codes.
Small differences in beam ranges, on the order of 1 mm or less, were observed in Figure 1 and Table S4. We hypothesized the cause in those different ranges for a monoenergetic proton source was due to the slightly different transport parameters in each code. We note that potential differences in predicted range in water between 2 MC codes do not have a clinical effect because each MC system would be calibrated to the measured proton beam range by tuning the proton source parameters, such as mean energy, energy distribution, and spatial distribution. Thus, to match the measured beam ranges, each code would have slightly different beam source characteristics yet both codes would accurately simulate the proton beam in water.
Heterogeneous Geometry: MC2-TOPAS Comparisons
As shown in Table S3, simulation results for MC2 and TOPAS with a bone or lung slab insert showed good agreement. For 100-MeV beams, the r90 depths for each code agreed within 0.5 mm. For 200-MeV beams, the r90 depths varied on the order of 1 mm. These results indicate that, in simulations involving simple heterogeneities, MC2 and TOPAS give very similar results.
Simulations in Computed Tomography Geometries
This study shows that even in heterogeneous targets, such as a patient CT, MC2 and TOPAS results show very similar dose distributions. Locally, the discrepancy can be large in regions of high gradients (eg, at a depth of 117 mm in Figure 3b). In 3 dimensions, the rate of agreement for both codes in both the head and neck and lung CTs was > 95%, even using the relatively strict γ-criterion of 2%–2 mm. The results do highlight a consistent difference between MC2 and TOPAS in how each code models energy deposition in dense bone. TOPAS tends to calculate larger energy deposition in bone. Evidence of this is seen in Figure 4b in a region centered at d = 63 mm. That location corresponds to the vertebral body, which is visible in Figure 2b and suggests a systematic difference between the 2 codes.
TOPAS provides a default CT number-to-stopping power conversion that follows a method outlined by Schneider et al [28]. We performed the patient CT simulations using that conversion and repeated the simulations with the CT-number conversion table unique to PTCH. There were noteworthy differences in the γ-passing rates when comparing the dose distributions calculated by each code with different CT number-to-stopping power curves. When the same curve was used in both TOPAS and MC2, the γ-passing rates increased and the dose distributions agreed more closely. We present γ-analyses for only the simulations using the same CT calibration curve (which is what is used for patient treatments at the PTCH) in Tables 1 and 2. We would like to highlight the importance of using the same CT number to calibration curve in comparisons among MC codes.
Despite using an identical CT number-to-stopping power conversion curves, there were still systematic differences between the codes when simulating a patient-irradiation field. We attribute that to differences in the way MC2 and TOPAS handle converting an HU number to a material and, thus, to stopping power. Both codes use slightly different methods for that conversion, as outlined in the TOPAS and MC2 sections. We believe the slight differences in stopping powers calculated by MC2 and TOPAS is the reason why, in CT geometry, there are notable differences that are most pronounced at the end of the beam range, as seen in the lateral profile in the distal fall-off region in the CT of the patient with head and neck cancer in Figure 3b and the energy deposition curves in Figure 4. These findings are at odds with the bone slab results shown in Table S4, in which TOPAS typically calculated greater ranges for the beam than MC2 did. The reason for that discrepancy, we hypothesized, was due to systematic differences in the way CT numbers are converted to stopping powers in each respective code. In both patient CT cases, MC2 calculated a greater proton range than TOPAS did. In the γ-analyses, the regions in which the most failures occurred were in the distal–fall-off and high-gradient regions, where large differences in the beam range occurred because of the different stopping powers used.
Conclusions
In summary, a comparison of simulation results between MC2 and TOPAS of several geometries was performed to assess differences between the codes. In simple geometries, such as a water phantom and a single heterogeneity in a water phantom, the results between TOPAS and MC2 agree well. For irradiation of a water phantom, r90 calculated by MC2 and TOPAS agree to within 1 mm. The γ-analysis also shows a high degree of agreement as evidenced by the high rate of pixels passing the γ-analysis, even with criteria as strict as 1% and 1 mm. When simulating a bone or lung slab in a water phantom, both MC2 and TOPAS r90 depths agreed to within 0.5%. When the 2 codes were used to simulate the same beam in the same CT geometry, the results showed noticeable differences in some locations (in bone), as shown by γ-analysis results comparing the energy-deposition distribution inside the CT geometry. An earlier study comparing the 2 underlying codes of MC2 and TOPAS and MCNPX and Geant4, respectively, showed a similar level of agreement in energy deposition inside patient CT geometry [21]. In the aforementioned study, MCNPX and Geant4 simulated the irradiation of a simple phantom, half-beam blocks of bone and lung, and a patient CT. Titt et al [21] found that, in a γ-analysis on the dose distributions calculated by MCNPX and Geant4, there was a mean passing rate of about 75% with 1%–1 mm criteria and a 95% passing rate with 2%–2 mm criteria. This work, using MC2 and TOPAS, confirms and expands upon those earlier results. Typically, the standard for an intensity-modulated radiotherapy plan to pass quality assurance testing is a 95% passing rate with 3%–3 mm tolerance. The 3, 2-dimensional slices selected in Table 1 show that TOPAS and MC2 agree to within that standard. Both codes have been found to agree well, similar to the results previously reported [15–18]. We conclude that the different results calculated by each code are not clinically significant in the geometries we simulated. This work increases our confidence in the use of MC simulations in ongoing proton therapy research across multiple institutions.
Supplementary Material
ADDITIONAL INFORMATION AND DECLARATIONS
Conflicts of Interest: The authors have no conflicts of interest to disclose.
Acknowledgments: The authors would like to thank Kathryn Hale at the Department of Scientific Publications at MD Anderson Cancer Center for her help in preparing this manuscript.
Funding: This work was supported in part by the NIH/NCI grant 2U19CA021239.
Ethical Approval: All patient data has been collected under internal review board–approved protocols.
References
- 1.Particle Therapy Co-Operative Group. Particle therapy facilities in operation. 2018 https://ptcog.ch/index.php/facilities-in-operation Published. Accessed October 26, 2018.
- 2.Harrabi SB, Bougatf N, Mohr A, Haberer T, Herfarth K, Combs SE, Debus J, Adeberg S. Dosimetric advantages of proton therapy over conventional radiotherapy with photons in young patients and adults with low-grade glioma. Strahlenther Onkol. 2016;192:759–69. doi: 10.1007/s00066-016-1005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Schoot AJAJ, de Boer P, Crama KF, Visser J, Stalpers LJA, Rasch CRN, Bel A. Dosimetric advantages of proton therapy compared with photon therapy using an adaptive strategy in cervical cancer. Acta Oncol. 2016;55:892–9. doi: 10.3109/0284186X.2016.1139179. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Howell RM, Taddei PJ, Giebeler A, Mahajan A, Newhauser WD. A comparative study on the risks of radiogenic second cancers and cardiac mortality in a set of pediatric medulloblastoma patients treated with photon or proton craniospinal irradiation. Radiother Oncol. 2014;113:84–8. doi: 10.1016/j.radonc.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandula S, Zhu X, Garden AS, Gillin M, Rosenthal DI, Ang KK, Mohan R, Amin MV, Garcia JA, Wu R, Sahoo N, Frank SJ. Spot-scanning beam proton therapy vs intensity-modulated radiation therapy for ipsilateral head and neck malignancies: a treatment planning comparison. Med Dosim. 2013;38:390–4. doi: 10.1016/j.meddos.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Knopf AC, Lomax A. In vivo proton range verification: a review. Phys Med Biol. 2013;58:R131–60. doi: 10.1088/0031-9155/58/15/R131. [DOI] [PubMed] [Google Scholar]
- 7.Schuemann J, Dowdell S, Grassberger C, Min CH, Paganetti H. Site-specific range uncertainties caused by dose calculation algorithms for proton therapy. Phys Med Biol. 2014;59:4007–31. doi: 10.1088/0031-9155/59/15/4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Titt U, Liu A, Mirkovic D. SU-E-T-523: runtime optimization for the automatic monte carlo dose computation system MC2 [abstract] Med Phys. 2013;40:326. [Google Scholar]
- 9.Perl J, Shin J, Schümann J, Faddegon B, Paganetti H. TOPAS: An innovative proton Monte Carlo platform for research and clinical applications. Med Phys. 2012;39:6818–37. doi: 10.1118/1.4758060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agostinelli S, Allison J, Amako K, Apostolakis J, Araujo H, Arce P, Asai M, Axen D, Banerjee S, Barrand G, Behner F, Bellagamba L, Boudreau J, Broglia L, Brunengo A, Burkhardt H, Chauvie S, Chuma J, Chytracek R, Cooperman G, Cosmo G, Degtyarenko P, Dell'Acqua A, Depaola G, Dietrich D, Enami R, Feliciello A, Ferguson C, Fesefeldt H, Folger G, Foppiano F, Forti A, Garelli S, Giani S, Giannitrapani R, Gibin D, Gómez Cadenas JJ, González I, Gracia Abril G, Greeniaus G, Greiner W, Grichine V, Grossheim A, Guatelli S, Gumplinger P, Hamatsu R, Hashimoto K, Hasui H, Heikkinen A, Howard A, Ivanchenko V, Johnson A, Jones FW, Kallenbach J, Kanaya N, Kawabata M, Kawabata Y, Kawaguti M, Kelner S, Kent P, Kimura A, Kodama T, Kokoulin R, Kossov M, Kurashige H, Lamanna E, Lampén T, Lara V, Lefebure V, Lei F, Liendl M, Lockman W, Longo F, Magni S, Maire M, Medernach E, Minamimoto K, Mora de Freitas P, Morita Y, Murakami K, Nagamatu M, Nartallo R, Nieminen P, Nishimura T, Ohtsubo K, Okamura M, O'Neale S, Oohata Y, Paech K, Pfeiffer A, Pia MG, Ranjard F, Rybin A, Sadilov S, Di Salvo E, Santin G, Sasaki T, Savvas N, Sawada Y, Scherer S, Sei S, Sirotenko V, Smith D, Starkov N, Stoecker H, Sulkimo J, Takahata M, Tanaka S, Tcherniaev E, Safai Tehrani E, Tropeano M, Truscott P, Uno H, Urban L, Urban P, Verderi M, Walkden A, Wander W, Weber H, Wellisch JP, Wenaus T, Williams DC, Wright D, Yamada T, Yoshida H, Zschiesche D. Geant4—a simulation toolkit. Nucl Instrum Methods Phys Res A. 2003;506:250–303. [Google Scholar]
- 11.McKinney GW, Durkee JW, Hendricks JS, James MR, Pelowitz DB, Waters LS, Gallmeier FX. MCNPX version 2.5.0—new features demonstrated. In: Monte Carlo 2005 Topical Meeting: The Monte Carlo Method: Versatility Unbounded in a Dynamic Computing World. Chattanooga, TN: American Nuclear Society; 2005:1–14. https://mcnp.lanl.gov/pdf_files/la-ur-04-8695.pdf Accessed December 21, 2017.
- 12.Jan S, Benoit D, Becheva E, Carlier T, Cassol F, Descourt P, Frisson T, Grevillot L, Guigues L, Maigne L, Morel C, Perrot Y, Rehfeld N, Sarrut D, Schaart DR, Stute S, Pietrzyk U, Visvikis D, Zahra N, Buvat I. GATE. V6: a major enhancement of the GATE simulation platform enabling modelling of CT and radiotherapy. Phys Med Biol. 2011;56:881–901. doi: 10.1088/0031-9155/56/4/001. [DOI] [PubMed] [Google Scholar]
- 13.Sarrut D, Bardiès M, Boussion N, Freud N, Jan S, Létang J-M, Loudos G, Maigne L, Marcatili S, Mauxion T, Papadimitroulas P, Perrot Y, Pietrzyk U, Robert C, Schaart DR, Visvikis D, Buvat I. A review of the use and potential of the GATE Monte Carlo simulation code for radiation therapy and dosimetry applications. Med Phys. 2014;41:064301. doi: 10.1118/1.4871617. [DOI] [PubMed] [Google Scholar]
- 14.Battistoni G, Bauer J, Boehlen TT, Cerutti F, Chin MPW, Dos Santos Augusto R, Ferrari A, Ortega PG, Kozłowska W, Magro G, Mairani A, Parodi K, Sala PR, Schoofs P, Tessonnier T, Vlachoudis V. The FLUKA code: an accurate simulation tool for particle therapy. Front Oncol. 2016;6:116. doi: 10.3389/fonc.2016.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bednarz B, Chen G, Paganetti H, Han B, Ding A, Xu XG. Comparison of particle-tracking features in GEANT4 and MCNPX codes for applications in mapping of proton range uncertainty. Nucl Technol. 2011;175:2–5. doi: 10.13182/nt11-a12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimstrand P, Tilly N, Ahnesjö A, Traneus E. Experimental test of Monte Carlo proton transport at grazing incidence in GEANT4, FLUKA and MCNPX. Phys Med Biol. 2008;53:1115–29. doi: 10.1088/0031-9155/53/4/020. [DOI] [PubMed] [Google Scholar]
- 17.Sadoughi HR, Nasseri S, Momennezhad M, Sadeghi HR, Bahreyni-Toosi MH. A comparison between GATE and MCNPX Monte Carlo codes in simulation of medical linear accelerator. J Med Signals Sens. 2014;4:10–7. [PMC free article] [PubMed] [Google Scholar]
- 18.Randeniya SD, Taddei PJ, Newhauser WD, Yepes P. Intercomparision of Monte Carlo radiation transport codes MCNPX, GEANT4, and FLUKA for simulating proton radiotherapy of the eye. Nucl Technol. 2009;168:810–4. doi: 10.13182/nt09-a9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low DA, Dempsey JF. Evaluation of the gamma dose distribution comparison method. Med Phys. 2003;30:2455–64. doi: 10.1118/1.1598711. [DOI] [PubMed] [Google Scholar]
- 20.Testa M, Schumann J, Lu HM, Shin J, Faddegon B, Perl J, Paganetti H. Experimental validation of the TOPAS Monte Carlo system for passive scattering proton therapy. Med Phys. 2013;40:121719. doi: 10.1118/1.4828781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titt U, Bednarz B, Paganetti H. Comparison of MCNPX and Geant4 proton energy deposition predictions for clinical use. Phys Med Biol. 2012;57:6381–93. doi: 10.1088/0031-9155/57/20/6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.n.a. ICRU reports. J ICRU. 1973;(os12):26–9. [Google Scholar]
- 23.Schuemann J, Giantsoudi D, Grassberger C, Moteabbed M, Min CH, Paganetti H. Assessing the clinical impact of approximations in analytical dose calculations for proton therapy. Int J Radiat Oncol Biol Phys. 2015;92:1157–64. doi: 10.1016/j.ijrobp.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor PA, Kry SF, Followill DS. Pencil beam algorithms are unsuitable for proton dose calculations in lung. Int J Radiat Oncol Biol Phys. 2017;99:750–6. doi: 10.1016/j.ijrobp.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grassberger C, Daartz J, Dowdell S, Ruggieri T, Sharp G, Paganetti H. Quantification of proton dose calculation accuracy in the lung. Int J Radiat Oncol Biol Phys. 2014;89:424–30. doi: 10.1016/j.ijrobp.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Titt U, Sell M, Unkelbach J, Bangert M, Mirkovic D, Oelfke U, Mohan R. Degradation of proton depth dose distributions attributable to microstructures in lung-equivalent material. Med Phys. 2016;42:6425–32. doi: 10.1118/1.4932625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.España S, Paganetti H. Uncertainties in planned dose due to the limited voxel size of the planning CT when treating lung tumors with proton therapy. Phys Med Biol. 2011;56:3843–56. doi: 10.1088/0031-9155/56/13/007. [DOI] [PubMed] [Google Scholar]
- 28.Schneider W, Bortfeld T, Schlegel W. Correlation between CT numbers and tissue parameters needed for Monte Carlo simulations of clinical dose distributions. Phys Med Biol. 2000;45:459–78. doi: 10.1088/0031-9155/45/2/314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.