Abstract
Transcription and RNA decay are key determinants of gene expression; these processes are typically considered as the uncoupled beginning and end of the messenger RNA (mRNA) lifecycle. Here we describe the growing number of studies demonstrating interplay between these spatially disparate processes in eukaryotes. Specifically, cells can maintain mRNA levels by buffering against changes in mRNA stability or transcription, and can also respond to virally induced accelerated decay by reducing RNA polymerase II gene expression. In addition to these global responses, there is also evidence that mRNAs containing a premature stop codon can cause transcriptional upregulation of homologous genes in a targeted fashion. In each of these systems, RNA binding proteins (RBPs), particularly those involved in mRNA degradation, are critical for cytoplasmic to nuclear communication. Although their specific mechanistic contributions to have yet to be fully elucidated, differential trafficking of RBPs between subcellular compartments is likely to a play central role in regulating this gene expression feedback pathway.
Keywords: mRNA decay, transcription, Xrn1, host shutoff, genetic buffering, NITC
Introduction
Gene expression is often described as a linear pathway, beginning with transcription and processing of messenger RNA (mRNA), followed by export to the cytoplasm for translation and ultimately decay. Included in this pathway are many regulatory steps that alter transcript fate, including quality control-based surveillance, splicing, RNA modification and translational control (Glisovic et al. 2008; Popp & Maquat 2013; Herzel et al. 2017). It is intuitive that changing an upstream step of the pathway will influence the cascade of subsequent downstream events. However, evidence emerging over the past decade indicates that the reverse is also true: alterations to cytoplasmic mRNA degradation lead to alterations to mRNA transcription, and vice versa (Sun et al. 2013; Haimovich, Medina, et al. 2013; Abernathy et al. 2015). Thus, these seemingly distal events in the gene expression cascade appear to be coupled, and may coordinate to regulate the overall pool of mRNA in response to perturbations. This RNA abundance-transcription connection has been observed in diverse systems, including yeast, zebrafish and mammalian cells.
Coordination between mRNA degradation and transcription occurs both at a global scale and in a more targeted fashion. Broad changes in mRNA abundance or transcription rates are sensed (Sun et al. 2013; Haimovich, Medina, et al. 2013; Abernathy et al. 2015; Dumdie et al. 2018; Helenius et al. 2011) and degradation of specific transcripts results in selective upregulation of genes with high sequence similarity (Rossi et al. 2015; El-Brolosy et al. 2019; Ma et al. 2019). Given the generally spatial separation of basal mRNA decay and synthesis in the cell, RNA binding proteins (RPBs) that shuttle between the nucleus and the cytoplasm are prime candidates for conveying RNA abundance information between these compartments. That said, many questions remain about the specific proteins involved and their functional roles, and insight into the potentially common nature of pathways between organisms remains elusive.
We begin with a brief overview of the mRNA lifecycle and then summarize the evidence supporting mRNA abundance feedback signaling, focusing on several outstanding questions in the field. These include whether the pathway that links mRNA degradation and transcription is conserved and similarly regulated in yeast and mammals, what factors are involved, and the bi-directional nature of the signaling. We also explore what the functional roles of feedback may be in a cell or organism and address if accelerated decay during viral infection and homeostatic buffering of mRNA levels are converging on the same signaling pathway.
The mRNA lifecycle from synthesis to decay
RNA Polymerase II (Pol II) transcribes mRNA in the nucleus of cells upon its assembly on a gene promoter with a diversity of essential general transcription factors (GTFs) and gene specific transcription factors (Sainsbury et al. 2015). GTF recruitment and regulation determine Pol II escape from the promoter and initiation of productive elongation (Saldi et al. 2016; Chen et al. 2018). What DNA is transcribed is determined by binding of the GTFs to promoter regions, beginning with the TFIID subunit TATA Binding Protein (TBP) on which the subsequent GTFs assemble (Sainsbury et al. 2015). GTF binding and Pol II transcription are dependent upon chromatin accessibility, which can be modified by the cell to alter the transcriptional program. Generally, chromatin remodeling occurs by strengthening or weakening histone-DNA interactions with ATP-dependent changes, with post translational modifications (PTMs) to the histones themselves, or when these PTMs recruit additional remodelers (Venkatesh & Workman 2015). Histone eviction from the DNA or loosening of existing chromatin results in DNA that can be transcribed by Pol II while histone recruitment or chromatin compaction occludes Pol II.
The cycle of initiation and elongation is largely dictated by a series of phosphorylation events on the Pol II C terminal domain (CTD), which contains 52 repeats of the heptad YSPTSTS in mammals and 26 in yeast (Hsin & Manley 2012). The CTD further coordinates processing of the nascent mRNA through interactions with factors that orchestrate co-transcriptional splicing, capping, termination and poly(A) tail addition (Herzel et al. 2017; Hsin & Manley 2012; Proudfoot et al. 2002). As it is transcribed, each mRNA is bound by RBPs that coordinate its export through nuclear pore complexes (NPCs), mark exon-exon junctions, and protect the 5’ cap and poly(A) tail. Nuclear surveillance of proper loading of these RBPs results in nuclear degradation of aberrantly loaded mRNAs, along with unadenylated and 3’ unprocessed mRNAs by the Rrp6p-containing nuclear exosome (Stutz 2003; Burkard & Butler 2000; Libri et al. 2002; Houseley et al. 2006). mRNAs can also undergo a variety of additional modifications; the most prevalent of these is N6-methyladenosine (m6A), which is recognized and bound by m6A “reader” proteins that impact the fate and function of the mRNA in diverse ways (Hailing et al. 2019).
Basal mRNA decay begins with deadenylation in a process that is quite conserved between yeast and mammals. Deadenylation by the major deadenylases Pan2-Pan3 and Ccr4-Not initiate poly(A) tail shortening and removal of the stabilizing Poly(A) Binding Protein (Pabpc) from the 3’ end of transcripts (Mugridge et al. 2018; Parker 2012; Yi et al. 2018; Webster et al. 2018). Protein-protein interactions link deadenylation to the core decapping complex, which is comprised of the Dcp2 decapping enzyme, the decapping activator Dcp1 and the scaffold protein Edc4 (Mugridge et al. 2018, Braun et al. 2012). Interactions of the core decapping complex with the deadenylation machinery stimulate Dcp2 to remove the protective 7mG cap from transcripts. Decapping concludes in rapid degradation of the transcript, primarily by the 5’−3’ exonuclease Xrn1, although 3’−5’ exonucleases such as the exosome and Dis3L2 also participate (Garneau et al. 2007; Schoenberg & Maquat 2012; Lubas et al. 2013; Łabno et al. 2016). 3’−5’ decay can be further stimulated by the nontemplated addition of uridines to the 3’ end of the message by terminal uridylyl transferases (TUTases) (M. Lee et al. 2014; Lim et al. 2014).
In addition to basal mRNA decay, various cytoplasmic pathways exist to degrade aberrant mRNAs or to modulate transcript fate. During translation, mRNAs are surveyed by various quality control factors that detect faulty transcripts, including those with premature termination codons, those lacking termination codons or those with stalled ribosomes (Radhakrishnan & Green 2016; Shoemaker & Green 2012). Aberrant transcripts are generally triggered for rapid clearance through recruitment of specialized endonucleases followed by degradation by exonucleases involved in basal mRNA degradation (Popp & Maquat 2013; Shoemaker & Green 2012; D’Orazio et al. 2019). One of the best-characterized quality control mechanisms is nonsense mediated decay (NMD), in which the presence of a premature termination codon (PTC) directs the mRNA to rapid decay. Central to this pathway is the Upf1 protein, which binds in complex with the SMG1 kinase and the eukaryotic release factors eRF1 and eRF3 to the terminating ribosomes of NMD substrates, delaying termination. Subsequent Upf1 phosphorylation and further remodeling of the RNP complex leads to recruitment of mRNA decay factors to clear the aberrant transcript (Kim & Maquat 2019). The half-life of an individual mRNA can also be influenced by specific sequence elements often found in the 3’ untranslated region (UTR), such as destabilizing AU-rich elements (AREs), which recruit RBPs that increase deadenylation (Barreau 2005), and binding sites for microRNAs, which lead to mRNA cleavage and/or translational repression (Friedman et al. 2008).
Finally, cellular perturbations such as apoptosis and viral infection can result in large-scale alterations to mRNA decay. During the early stages of apoptosis, after mitochondrial outer membrane permeabilization but before DNA fragmentation, caspase 8 activation leads to release of the exoribonuclease Pnpt1 from the mitochondria. Pnpt1 coordinates with Dis3L2 and TUTases 4 and 7 to cause widespread degradation of cytoplasmic mRNA (Thomas et al. 2015; X. Liu et al. 2018).
Multiple diverse viruses, including alpha and gammaherpesviruses, influenza A virus, poxviruses, and coronaviruses, also accelerate basal mRNA decay. These viruses express proteins that promote widespread decapping or endonucleolytic cleavage of mRNA, producing fragments that are degraded by the endogenous RNA decay machinery (Karr & Read 1999; Glaunsinger et al. 2005; Huang et al. 2011; Kamitani et al. 2009; Jagger et al. 2012). This can benefit the virus by decreasing the abundance of immunostimulatory nucleic acids, reducing expression of host immune responses genes, and liberating host translation machinery for viral use (Richner et al. 2011; S. W. Liu et al. 2013; Parrish & Moss 2007; Read 2013; Abernathy & Glaunsinger 2015). For DNA viruses such as alphaherpesviruses, accelerating turnover of virally derived mRNAs also sharpens the kinetic boundaries between stages of viral gene expression (Oroskar & Read 1987; Oroskar & Read 1989).
Given that an array of stimuli can alter the abundance of individual mRNAs or of the overall mRNA pool, what mechanisms exist that enable cells to sense and respond to such perturbations? Over the past decade, numerous studies have established that cells have compensatory mechanisms to deal with these gene expression changes, and research into these pathways has revealed surprising connections between cytoplasmic mRNA turnover and Pol II transcription (Sun et al. 2013; Haimovich, Medina, et al. 2013, Abernathy et al. 2015).
Maintaining consistency: connections between mRNA degradation and synthesis buffer against change
Global mRNA stabilization leads to a compensatory reduction in Pol II transcription
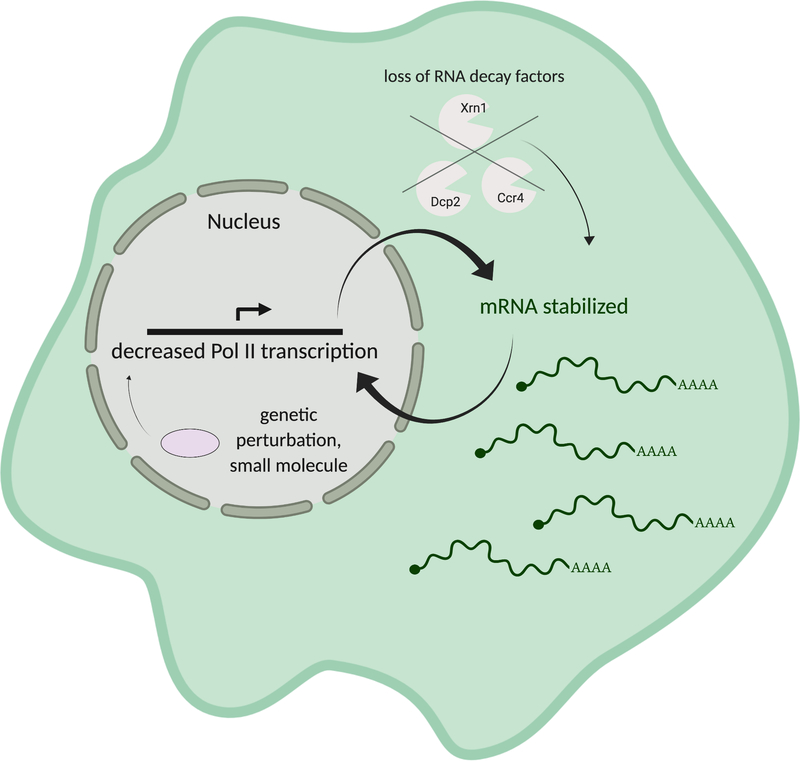
While it might be assumed that slowing mRNA degradation would cause a general increase in transcript abundance, early studies reported that mRNA levels remain relatively constant in S. cerevisiae strains lacking decay factors including the decapping enzyme Dcp1 and the 5’ – 3’ exonuclease Xrn1 (Muhlrad & Parker 1999; He et al. 2003). Indeed, several lines of evidence now indicate that cells respond in a compensatory manner to “buffer” against generalized reductions in either cytoplasmic mRNA degradation or transcription, thereby maintaining constant mRNA levels in the cell (Figure 1).
Figure 1.
The homeostatic model of mRNA decay-transcription feedback proposes that appropriate mRNA levels are maintained by either altering mRNA decay or Pol II transcription. Removal of basal RNA decay factors results in decreased mRNA decay and stabilization of mRNAs; transcription then decreases to buffer against resulting increases in mRNA levels. Similarly, perturbations that cause reductions in transcription and thus less mRNA production are buffered through stabilization of mRNA.
Studies by the Cramer lab comparing transcription rates by 4-thiouridine (4SU) incorporation with computationally inferred RNA decay rates in S. cerevisiae strains deleted for 46 different RNA decay factors including deadenylases, decappers, exonucleases and RNA processing enzymes revealed that cells with reduced mRNA decay generally compensate by decreasing transcriptional output (Sun et al. 2012; Sun et al. 2013). One notable exception was the strain lacking Xrn1, which did not cause a corresponding change in transcription, suggesting a direct role for Xrn1 in this buffering pathway (Sun et al. 2013). The Choder lab also measured RNA stability (by northern blot) and RNA synthesis (using genomic run-on (GRO) analysis) in a panel of decay factor null S. cerevisiae strains, including those lacking Xrn1, Ccr4, Dcp1 and Dcp2. In each of these mutant strains, RNA half-lives were increased while transcription rates were decreased, supporting a buffering model (Haimovich et al. 2013). A recent report further demonstrates reduced elongating Pol II and reduced Pol II speed at GAL genes in an Δxrn1 strain (Begley et al, 2019). Each of these studies implicate Xrn1 in the transcriptional response to alterations in global mRNA stability, although the basis for differing transcriptional effects in the absence of Xrn1 is unclear. They may be attributed to different experimental approaches to measure transcription rates or potential differences in the Δxrn1 strains used. However, the convergence of these reports on Xrn1 highlights its central role in communicating RNA abundance information to the transcriptional machinery.
Experiments in mammalian cells that measure mRNA synthesis upon decay factor knockdown also suggest a buffering phenotype, although the connection is not as marked as it is in S. cerevisiae. Depleting the deadenylase PARN in mouse myoblasts showed a trend of decreased steady state levels and increased half-lives using microarray analysis, but the differences are modest. For the small subset with 2–3 fold increased half-lives, 4SU-based RT-qPCR measurements showed reduced transcription (J. E. Lee et al. 2012). Similarly, depleting Xrn1 from HepG2 cells resulted in reduced transcription for 8 out of 10 transcripts measured by RT-qPCR (Singh et al. 2019). Knockdown of the deadenylase CNOT6 impacted a smaller subset of the examined transcripts, caused less prominent buffering and in some cases increased transcription (Singh et al. 2019). Computational analysis comparing mRNA decay rates upon ActinomycinD treatment to total mRNA levels in induced pluripotent stem cells and human foreskin fibroblasts also provides evidence of a relationship between decay and transcription. Here, genes with faster decay rates were associated with increased steady state mRNA levels, as would be expected in a buffering model, although the genome-wide correlation is modest (Dori-Bachash et al. 2012). Extending these results in mammalian cells to additional global measurements of transcription under conditions of altered mRNA stability will be valuable, as will comparisons of the impact of an expanded set of mammalian mRNA decay pathway components.
Reduced Pol II transcription leads to compensatory increases in mRNA stability
As described above, eukaryotic cells can respond to global increases in mRNA half-life by decreasing transcriptional output. Notably, changes in transcription can be compensated in an analogous manner by altered mRNA stability, supporting the existence of a true feedback loop. In S. cerevisiae, a variety of strategies have been used to reduce transcription. For example, treatment with the magnesium chelator 1,10-phenanthroline stalls all RNA polymerases (Johnston 1994) and phenocopies a genetic knockout of Rpb1–1 (Ray et al. 2013). Microarray-based time course measurements of mRNA levels during 1, 10-phenanthroline treatment were used to calculate decay rates, revealing a negative correlation between RNA stability and mRNA abundance (Shalem et al. 2011). This suggests that mRNAs are stabilized to buffer against the decreasing transcript levels resulting from transcriptional shutdown.
Similar trends were observed upon depletion of the SAGA subunits Sus1 and Spt20, which are key Pol II cofactors (Dori-Bachash et al. 2012; Goler-Baron et al. 2008; Baptista et al. 2017). Measurements of mRNA synthesis by GRO-seq as well as mRNA steady state levels during Sus1 depletion revealed that the stability of nearly all transcripts increased (García-Molinero et al. 2018). Sus1 depletion causes a gradient of reduced transcription rates where highly transcribed genes showed the most pronounced stabilization. Similarly, measurement of 4SU-labeled nascent RNA and total mRNA during loss of Spt20, which causes reduced transcription in close to 90% of genes, also showed that loss of mRNA synthesis was compensated by a decrease in decay rates for the vast majority of transcripts (Baptista et al. 2017). Thus, results from each of these genetic and chemical perturbations in yeast support a buffering model in which increasing mRNA stability is a mechanism used to compensate for reduced transcription, suggesting bidirectional decay/transcription communication.
While there are fewer relevant studies in mammalian cells, existing data largely support the buffering model. In particular, microarray analysis of cells depleted of the TFIIH kinase Mat1, which is involved in serine 5 phosphorylation of the Pol II CTD, showed that despite decreased elongating Pol II in the gene body, most mRNA steady state levels remained unchanged (Helenius et al. 2011). Additional experiments to directly evaluate how altering transcription impacts mRNA half-life will be important to confirm if reduced Pol II output results in reduced cytoplasmic mRNA decay in mammalian cells. One way to do this would be to test mRNA half lives while using a Pol II mutant with reduced elongation speed and correspondingly reduced transcription (Fong et al. 2014), or to globally test mRNA half lives during treatment with a drug that reduces transcription, like α-amanitin.
Gene specific integration of mRNA decay and transcriptional responses
The above examples in yeast and mammals show buffering in situations in which genetic or chemical perturbations globally impact transcription or RNA decay, thus creating an environment where compensation might be particularly pronounced. However, it is also relevant to ask if compensatory mechanisms are at play under steady state conditions — though this might be difficult to detect if the alterations are subtle. Interestingly, in human cell lines from the HapMap Project, RNA decay rates of a small subset of genes (~5%) were indeed anticorrelated with their transcription; higher transcription was associated with faster degradation in approximately half of this subset (Pai et al. 2012). These cell lines were assayed under steady state conditions without perturbation to mRNA decay rates or transcription. While it is difficult to ascertain the directionality of the association, it is intriguing to consider that even in the absence of direct perturbation, mRNA decay and transcription may be coordinated, at least for a subset of transcripts. This raises the possibility that a cell is constantly buffering mRNA levels, which becomes more apparent upon large-scale alternations to rates of decay or transcription.
More compelling evidence has recently emerged related to transcript-specific compensatory responses in mammalian cells and in zebrafish. These targeted changes are known as “genetic compensation” or “nonsense-induced transcriptional compensation” (NITC) (Wilkinson 2019), and have recently been proposed as a way for the cell to respond to the loss of a specific transcript by upregulating sequence-similar transcripts (El-Brolosy et al. 2019; Ma et al. 2019). NITC explains the previously observed discrepancies between gene knockouts and knockdowns and genetic mutants or chemical inhibition in mouse and Arabidopsis, where certain gene knockouts displayed less dramatic phenotypes due to induction of sequence-similar genes that could partially compensate (Rossi et al. 2015; Zhu et al. 2017).
NITC was recently shown to occur for specific premature termination codon bearing transcripts subject to NMD (Figure 2). (Genetic knockouts are most often generated by dsDNA cleavage followed by the error non-homologous end joining repairing the lesion, frequently introducing a PTC (Carroll 2014; Lieber 2010)). Sequence-similar transcripts, often evolutionarily derived from the same precursor gene, become upregulated, as measured by global RNA sequencing of various knockouts (El-Brolosy et al. 2019; Ma et al. 2019). However, mutant genes defective in producing any mRNA are either not upregulated or only modestly so (Rossi et al. 2015; El-Brolosy et al. 2019), suggesting that the mRNA itself is involved in signaling-- consistent with the targeted nature of this transcriptional response.
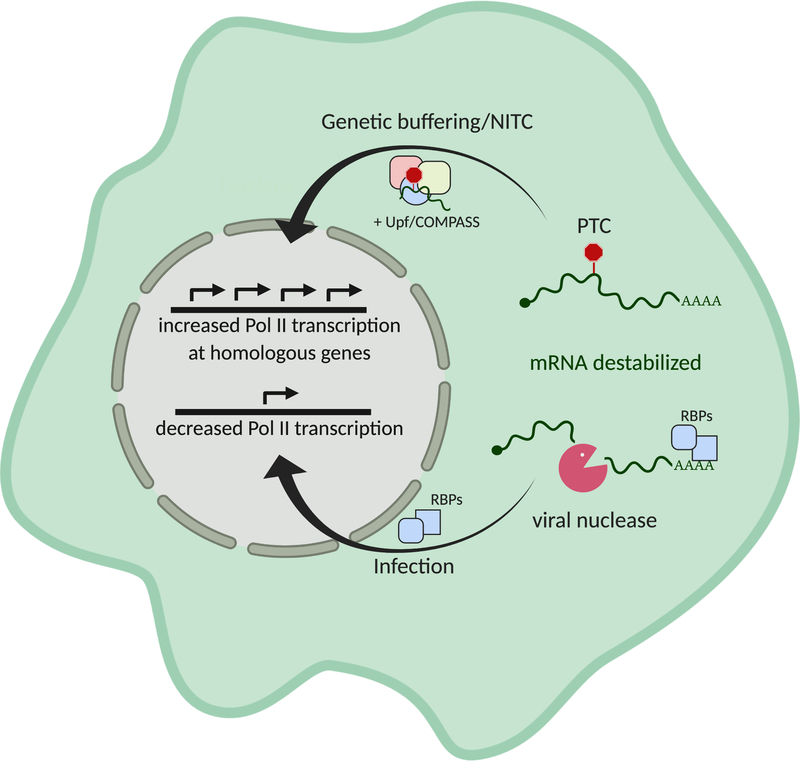
Figure 2.
Cellular responses to mRNA destabilization include nonsense-induced transcriptional compensation (NITC) and decreased overall Pol II occupancy. mRNA with premature termination codons (PTCs) that are destabilized through the nonsense mediated decay pathway can recruit Upf proteins and members of the COMPASS complex. Upon translocation to the nucleus, they promote increased transcription of homologous genes by altering chromatin accessibility. Viral nucleases induce widespread mRNA decay by decapping or endonucleolytically cleaving mRNAs. Degradation of the resulting fragments by host exonucleases liberates previously bound RBPs, which are hypothesized to traffic into the nucleus in an mRNA concentration-dependent manner and decrease Pol II promoter occupancy.
Given the importance of PTCs for eliciting a transcriptional response, it is logical that NMD factors and basal RNA decay enzymes were implicated in the signaling. Indeed, removal of NMD components was found to diminish the transcriptional response, although the studies implicate different proteins in this pathway. El-Brolosy et al. showed that knockdown of Upf1, a central component of NMD, abrogated the transcriptional response, while Ma et al. observed that only knockout of Upf3a, a poorly understood NMD component, was sufficient to block the response (Ma et al. 2019; El-Brolosy et al. 2019). Both studies converged on a role for the COMPASS component Wdr5, which is responsible for deposition of of H3K4me3 at the transcription start sites (TSS) of genes. Wdr5 depletion abolished transcriptional activation at compensating genes (El-Brolosy et al, 2019), and Upf3 was shown to interact with Wdr5 in an RNA-independent manner, providing a potential link between the NMD machinery and gene regulatory epigenetic modification (Ma et al. 2019). The basal RNA decay enzyme Xrn1 was also necessary for transcriptional compensation (El-Brolosy et al. 2019). While the mechanistic link between RNA decay and transcriptional compensation remains unclear, RNA decay proteins could trim and modify RNA fragments, which might then be used to direct RBPs to complementary chromosomal locations.
It remains to be elucidated how fragments of PTC-containing mRNAs are directing chromatin remodeling in the nucleus. For example, how are these small RNAs generated and protected from complete degradation by basal RNA decay enzymes in order to be part of the signaling complex? Secondly, how are these small RNA fragments brought into the nucleus? Chaperoning RNAs could be a potentially novel role for Upf3a, or alternatively may be carried out by another protein, for example a component of the microRNA processing pathway.
Responding to a threat: Transcriptional repression upon accelerated mRNA decay
While it is clear that cells can sense changes to mRNA levels and alter transcription or mRNA stability to buffer against these perturbations, there are circumstances in which a buffering response may not be beneficial. One prominent example is during intracellular pathogen infection, which can cause large-scale alterations to the cellular mRNA pool. Indeed, many diverse viruses accelerate basal mRNA decay as an integral part of their lifecycle. These include gammaherpesviruses such as Epstein-Barr virus (EBV) (Rowe et al. 2007), Kaposi’s sarcoma-associated herpesvirus (KSHV) (Glaunsinger & Ganem 2004; Glaunsinger et al. 2005) and murine gammaherpesvirus 68 (MHV68) (Covarrubias et al. 2011; Richner et al. 2011), alphaherpesvirus such as herpes simplex virus (HSV) (Everly & Read 1997; Smibert et al. 1992; Kwong et al. 1987), influenza A (Jagger et al. 2012; Gaucherand et al. 2019; Desmet et al. 2013), vaccinia virus (Parrish et al. 2007; S. W. Liu et al. 2013), and SARS and MERS coronavirus (Lokugamage et al. 2015; Kamitani et al. 2009; Kamitani et al. 2006). Most of these viruses encode broad acting, mRNA specific endonucleases (EBV BGLF5, KSHV SOX, MHV68 muSOX, HSV-1 vhs, influenza A PA-X) or decapping factors (vaccinia D9, D10), which create mRNA cleavage products that are directly accessed and cleared by the basal mRNA decay machinery such as Xrn1 (Abernathy & Glaunsinger 2015; Gaglia et al. 2012). In addition to the infection context, expression of these viral nucleases alone in mammalian cells is sufficient to drive widespread mRNA decay, reducing cytoplasmic mRNA populations by 50–70% (Glaunsinger & Ganem 2004; Gaglia et al. 2012; Rodriguez et al. 2019; Gaucherand et al. 2019).
A connection between accelerated mRNA decay and transcription was first shown during infection with the gammaherpesviruses MHV68 (Figure 2). Wild type MHV68 causes reduced Pol II recruitment to mammalian promoters. However, infection with a decay-deficient mutant virus does not, as measured by Pol II ChIP qPCR or 4SU pulse labeling at the Gapdh and Rplp0 promoters. Similarly decreased Pol II promoter occupancy at representative loci was observed upon transfection of the gammaherpesvirus endonucleases muSOX and SOX or the alphaherpesvirus endonuclease vhs, indicating that widespread mRNA degradation in the absence of other infection signatures is sufficient to reduce Pol II promoter occupancy. Notably, depletion of Xrn1 or Dis3L2 from these cells restored Pol II occupancy, and the reduction could be reinstated by complementation with wild type but not catalytically dead Xrn1 (Abernathy et al. 2015). This suggests that degradation of the cleaved mRNA fragments (rather than the initial endonucleolytic cleavage) is critical for reducing Pol II occupancy. These data have recently been extended through Pol II ChIP-seq experiments, revealing widespread decreases in Pol II at nearly all host promoters in murine fibroblasts infected with wild type but not RNA decay-deficient MHV68 (Hartenian and Glaunsinger, 2019 bioRxiv). It is notable that infection with the alphaherpesvirus HSV-1 also dramatically reduces Pol II occupancy and nascent mRNA production (Abrisch et al. 2016; Dremel & DeLuca 2019; Rutkowski et al. 2015), although as yet there is no evidence linking this to mRNA degradation.
Although gammaherpesviruses are double-stranded DNA viruses that are transcribed by mammalian Pol II, in the environment of widespread mRNA decay, viral transcription remains robust and the Pol II ChIP signal on the viral genome is not significantly reduced compared to that of an RNA decay-deficient virus (Hartenian and Glaunsinger, 2019 bioRxiv, Abernathy et al. 2015). However, viral promoters become susceptible to decay-induced transcriptional repression when integrated into the host genome, suggesting that either the structure of the replicating viral DNA and/or its location within viral replication compartments protects them from this cellular pathway (Hartenian and Glaunsinger, 2019 bioRxiv).
The viral data suggest that mammalian cells do not attempt to buffer against widespread mRNA decay (Figure 3). Instead, the loss of Pol II from promoters may represent an antiviral response to a pathogen-associated signature, akin to other “patterns of pathogenesis” (Vance et al. 2009) like disruption of the actin cytoskeleton and damage to the plasma membrane, which trigger innate immune responses (Legrand-Poels et al. 2007; Lamkanfi et al. 2009). An observation from apoptotic cells may be in line with this hypothesis: overexpression of the apoptotic RNA decay factor Pnpt1 increases apoptosis. While the link between RNA decay and induction of apoptosis is unclear, cell death is a conserved pathway manipulated by pathogen infection (Jorgensen et al. 2017). Future work exploring the downstream consequences of dramatically accelerated mRNA decay in other contexts, including RNase L activation or non-infectious stresses will help define the generalizable nature of this transcriptional response to globally accelerated RNA decay.
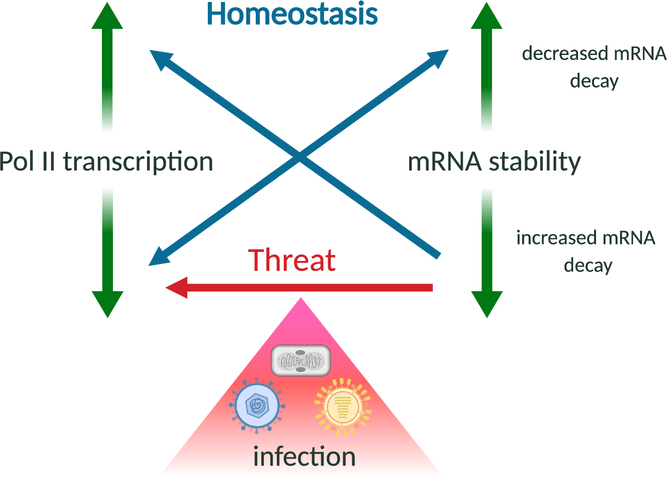
Figure 3.
mRNA stability and Pol II transcription are linked processes, but may be differentially controlled during homeostasis or upon cellular threats. To maintain homeostatic mRNA levels and buffer against global mRNA abundance changes, eukaryotic cells modulate Pol II transcription or mRNA stability in a compensatory manner. However, during infection with mRNA decay-inducing viruses, the loss of mRNA is exacerbated by reduced Pol II transcription, perhaps as a component of a stress or antiviral response.
mRNA decay-dependent transcription silencing during oocyte-to-embryo transition
The maternal to zygotic transition (MZT) is a well-recognized time of maternal mRNA decay followed by zygotic transcriptional activation, when ~35% of maternal mRNAs are destabilized by a series of direct targeting mechanisms (Tadros & Lipshitz 2009). Although there is no evidence that decay elicits transcription during the MZT, there is a connection between mRNA decay and transcription during the oocyte to embryo transition, a stage of development prior to the MZT (Dumdie et al. 2018). The mechanism involves degradation of mRNAs encoding transcriptional regulators by the essential protein Zfp36L2, which is part of a protein family associated with ARE-dependent mRNA decay. Single cell RNA sequencing of a conditional Zfp36L2 knockout revealed that its targets are enriched for chromatin regulation and transcription-associated processes, including histone demethylases. In the absence of Zfp36L2, the oocyte does not undergo transcriptional repression, highlighting the potential role of RNA decay of specific transcripts in regulating transcription (Dumdie et al. 2018).
Role of RNA binding proteins in connecting mRNA decay to transcription
A central question is which factors are involved in sensing mRNA levels and effecting the corresponding changes in transcription and mRNA stability? This remains unclear in both the global and the specific instances of transcriptional or mRNA decay modulation. Multiple reports linking mRNA abundance and transcription implicate RNA binding proteins (RBPs) – particularly RNA decay enzymes like Xrn1, Dis3L2 and Ccr4-Not – as key messengers for conveying information on mRNA abundance between the nucleus and the cytoplasm. RBPs are natural candidates for this role given their ability to shuttle in an RNA-concentration dependent manner (Krecic & Swanson 1999; Piñol-Roma & Dreyfuss 1992; Gilbertson et al. 2018; Timmers & Tora 2018). Furthermore, there is precedent for RBPs having distinct functions in the cytoplasm versus the nucleus. Examples include hnRNPs, whose nuclear binding influences transcriptional activation and splicing while their binding to mRNAs and RBPs in the cytoplasm influences translation, stability and localization (Shyu et al. 2000). The heterodimeric Pol II subunits Rpb4/7 are another example, as they also play a role in determining cytoplasmic mRNA fate (discussed below) (Haimovich, Choder, et al. 2013; Lotan et al. 2005; Lotan et al. 2007; Harel-Sharvit et al. 2010; Shalem et al. 2011; Duek et al. 2018).
Decay factors play a central role in activating mRNA decay-dependent signaling
The majority of proposed models converge on the importance of Xrn1 for signaling to the nucleus to convey information on mRNA decay or abundance, although hypotheses differ as to the specific role it plays in this feedback (Figure 4). The Choder lab found that wild type but not catalytically dead Xrn1 shuttles into the nucleus in an mRNA decay-dependent manner and directly binds chromatin. ChIP-exo analysis further showed that additional decay factors, including Dcp2 and Lsm1, also bind chromatin and elicit a transcriptional response. In the presence of an Xrn1 catalytic mutant, they observed that Pol II elongation is reduced (Haimovich et al. 2013).
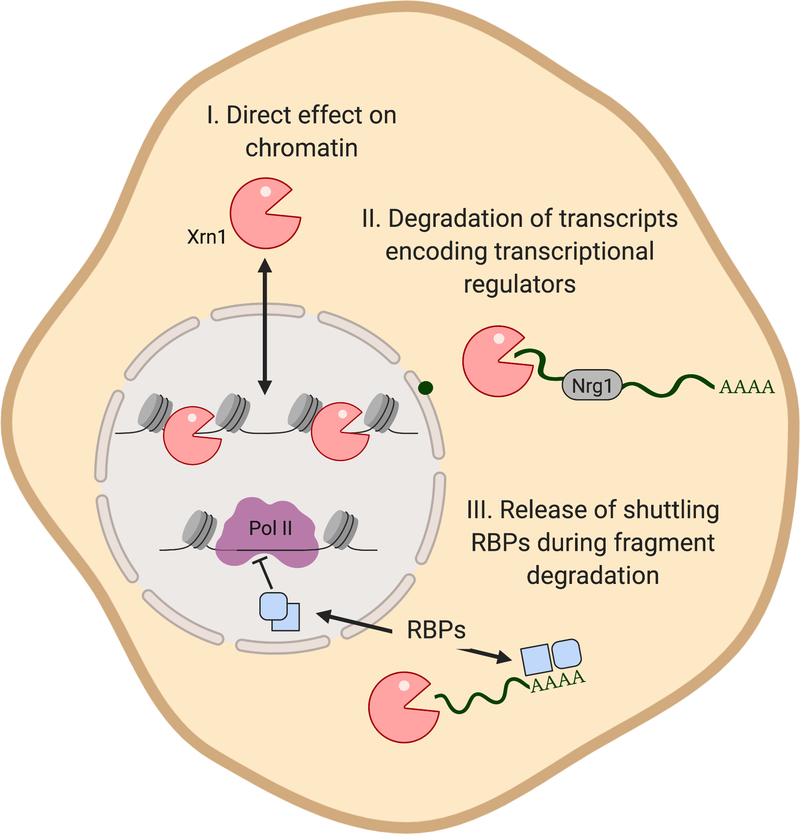
Figure 4.
Multiple models have been proposed to explain how Xrn1 connects mRNA stability with transcription. Xrn1 may act directly by shuttling into the nucleus to bind chromatin and reduce transcription. It may alternatively (or additionally) indirectly impact transcription by degrading transcripts encoding transcriptional regulators such as Nrg1, or by causing release of RNA binding proteins (RBPs) during accelerated mRNA decay, which traffic into the nucleus and result in reduced transcription.
Several recent reports further implicate Xrn1 in transcription elongation at yeast genes. In a Δxrn1 strain less elongating Pol II was found at GAL loci by transcriptional run on (Begley et al, 2019) and genome wide by Native Elongating Transcript (NET) sequencing (Fischer et al. 2019 bioRxiv). Furthermore, the Δxrn1 strain showed increased recruitment of the Pol II release factor TFIIS to the body of the GAL genes (Begley et al, 2019). TFIIS is important for resolving backtracked Pol II, as it stimulates the intrinsic endonucleolytic activity of Pol II on mRNA (Cheung et al, 2011). This raises the possibility that under conditions of reduced mRNA decay, Pol II backtracking and nascent chain release are stimulated.
The Cramer lab did not detect a ChIP signal for Xrn1 on transcriptionally active loci, and proposed an alternate model in which the activity of Xrn1 on specific transcripts, such as the transcriptional repressor Nrg1, leads to broad changes in mRNA synthesis (Sun et al. 2013). There is an additional kinetic component to this model, in which a delay in the mRNA decay-induced transcriptional signaling reflects the time it takes for altered Nrg1 mRNA levels to influence Nrg1 protein abundance. Xrn1 auto-regulation is also a factor, as high Xrn1 protein levels lead to increased decay of its own transcript (presumably dependent on decapping), ultimately lowering Xrn1 protein levels and thus influencing mRNA decay (Sun et al. 2012; Sun et al. 2013). Presumably, Xrn1 decay of its own transcript would be dependent on decapping. How this connection is coordinated is not yet clear, although it has been established that interactions occur between Xrn1 and Pat1 and Xrn1 and Dcp1 (Nissan et al. 2010; Braun et al. 2012). Measurements of the specific transcription, translation and decay rates of Xrn1 and Nrg1 would further bolster these arguments.
Xrn1 is also required for cytoplasmic to nuclear communication in mammalian cells undergoing viral nuclease-driven transcriptional repression (Abernathy et al. 2015). This is notable because in cells expressing a viral endonuclease, mRNA will be cleaved and translationally inactivated prior to exonuclease-mediated degradation of the cleaved fragments. In addition to Xrn1, analysis of a subset of individual mammalian loci suggested that the 3’−5’ decay factor Dis3L2 and the Ccr4/Pan2 deadenylases were also involved in the cytoplasmic decay to transcriptional repression signaling (Abernathy et al. 2015). These data suggest that degradation of the endonuclease-cleaved fragments by multiple mRNA decay factors contributes to decay-transcription signaling.
‘mRNA coordinators’ help direct mRNA fate
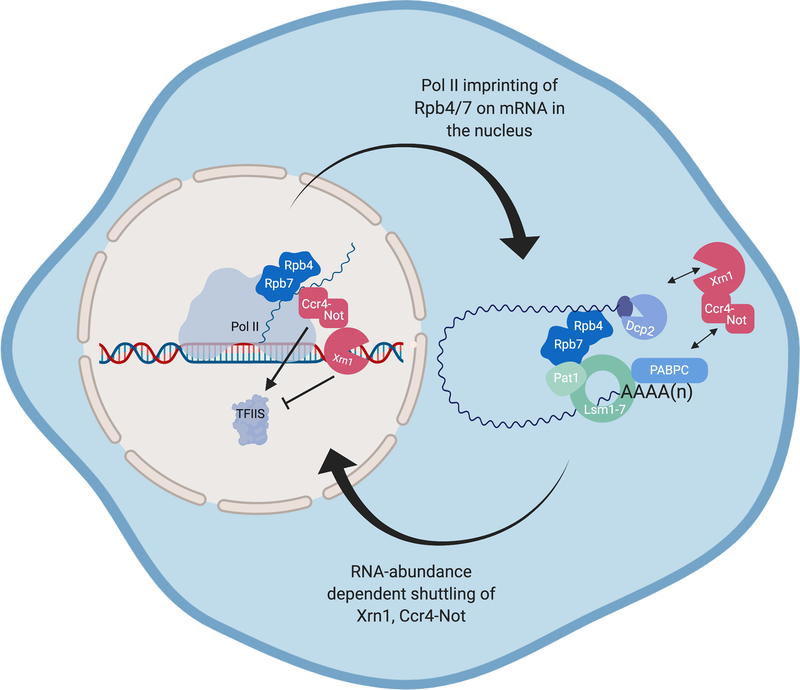
Connections between transcription rates and cytoplasmic mRNA decay have also been linked to the heterodimeric Pol II subunits Rpb4 and Rpb7, which traffic from the nucleus to the cytoplasm and have been termed ‘mRNA coordinators’ (Haimovich, Choder, et al. 2013; Lotan et al. 2005; Lotan et al. 2007; Harel-Sharvit et al. 2010; Shalem et al. 2011; Duek et al. 2018) (Figure 5). Rpb7 interacts with mRNAs as they leave the exit tunnel of the polymerase during in vitro transcription (Újvári & Luse 2005), and Rpb4 binds the primarily cytoplasmic RNA decay proteins Pat1 and Lsm2 (Lotan et al. 2005). Both Rpb4 and Rpb7 can localize to P-bodies (Lotan et al. 2007; Lotan et al. 2005) and loss of Rpb4 is associated with aberrant decay of a subset of mRNAs involved in protein synthesis (Lotan et al. 2005). Given the association of Rpb4 and Rpb7 with multiple stages of the mRNA lifecycle, they have been proposed to act as a platform for regulation, imprinting mRNAs with transcriptional information that can be read out during decay. Although experiments in which covalently linking Rpb4 to a core subunit of Pol II (and thus preventing its trafficking) did not change basal levels of RNA decay (Schulz et al. 2014), it was subsequently shown that a portion of the covalently tethered Rpb4 could still be found in the cytoplasm, potentially retaining the capability to imprint mRNAs (Villanyi & Collart 2015; Duek et al. 2018; Villanyi et al. 2014). Furthermore, a Pol II mutant unable to recruit the Rpb4/7 subunits (rpb6Q100R) does not stabilize mRNA to buffer against reduced transcription (Shalem et al. 2011), consistent with the hypothesis that Rpb4/7 is required for nuclear-cytoplasmic communication during buffering.
Figure 5.
mRNA coordinators Rpb4/7 are implicated in imprinting mRNAs in the nucleus to convey transcription rate to the cytoplasm. Rpb4/7 have been shown to interact with the nascent mRNA chain in the nucleus and Pat1 and members of the Lsm1–7 complex in the cytoplasm. RNA decay factors Ccr4-Not and Xrn1 degrade mRNA in the cytoplasm and may affect Pol II elongation by either coordinating with (Ccr4-Not) or antagonizing (Xrn1) the Pol II co-factor TFIIS, which is responsible for disengaging stalled Pol II.
The Ccr4-Not complex may also be involved in ‘mRNA coordination’, as these factors play multiple roles in transcription and RNA decay (Villanyi & Collart 2015). The Ccr4-Not complex contains 9 subunits in yeast comprising the proteins with deadenylase activity (Ccr4 and Caf1), the Not1 scaffold and additional regulatory subunits. It is these additional subunits that are proposed to coordinate RNA decay and transcription (Villanyi et al. 2015). The Ccr4-Not complex interacts with TBP, TFIIS, SAGA, Pol II and the nascent transcript (Collart et al. 2003; Kruk et al. 2011). Its interaction with TFIIS is proposed to stimulate nascent chain cleavage, causing Pol II backtracking and reengagement of Pol II on stalled transcripts in vitro and in vivo (Kruk et al. 2011; Gupta et al. 2016; Begley et al. 2019). Furthermore, a Δccr4 strain showed a global reduction in TFIIS/Pol II DNA occupancy, which is consistent with Ccr4-Not participation in TFIIS recruitment (Babbarwal et al. 2014). It is worth noting that this same study saw a global increase in TFIIS/Pol ll occupancy in a Δxrn1 strain (Begley et al. 2019), implying a different role for Xrn1 and Ccr4-Not in influencing Pol II occupancy. Future mechanistic studies will be key to understand how these decay factors interface with the transcriptional machinery to integrate decay rates with mRNA synthesis rates.
Additional data provide intriguing links between the Ccr4-Not complex, Xrn1 and the putative mRNA coordinators Rpb4/7. Rescue of arrested Pol II by Ccr4-Not from a yeast reconstitution system requires Rpb4/7 (Babbarwal et al. 2014). It is therefore possible that Rpb4/7 participates in co-transcriptionally recruiting mRNA decay factors such as Ccr4-Not to the nascent transcript. Supporting this model is the observation that Ccr4-Not binding to Pol II is significantly weakened in the absence of Rpb4/7 (Babbarwal et al. 2014). Notably, Ccr4-Not was also shown to bind the C-terminal unstructured domain of Xrn1 in human and drosophila cells (Chang et al. 2019). Thus, one possibility is that through this interaction, both Ccr4-Not and Xrn1 may be recruited to elongating Pol II, thereby connecting transcription and decay. Yet, how the localization of these proteins and their roles in transcription versus mRNA decay are coordinated remains largely enigmatic, particularly in response to stimuli that lead to buffering.
Cytoplasmic poly(A) binding protein accumulates in the nucleus during accelerated mRNA decay
A recent report identified proteins that accumulate in the nucleus during accelerated mRNA decay using a tandem-mass tag (TMT) mass spectrometry approach. mRNA decay was induced with the gammaherpesviral ribonuclease muSOX in the presence or absence of Xrn1 to identify proteins that may shuttle in an mRNA-concentration dependent manner (Gilbertson et al. 2018). The primary class of proteins that accumulated in the nucleus in an Xrn1-dependent manner are associated with RNA 3’ UTRs and poly(A) tails, including Pabpc1, Pabpc4 and Larp4.
Pabpc1 and 4 had previously been identified as indicators of widespread RNA decay and shown to translocate to the nucleus upon viral nuclease-induced mRNA degradation (Kumar & Glaunsinger 2010; Kumar et al. 2011; Glaunsinger & Ganem 2004; Harb et al. 2008; Salaun et al. 2010; Piron et al. 1998; Park et al. 2014). Cytoplasmic to nuclear trafficking of Pabpc is dependent on its RNA-recognition motifs (RRMs). These RRMs contain noncanonical nuclear localization signals (NLS) (Kumar et al. 2011) which are exposed upon release of Pabpc from mRNA. As a result, Pabpc shuttles into the nucleus in an mRNA-concentration dependent manner (Kumar et al. 2011). In cells depleted of Pabpc1 and Pabpc4, Pol II occupancy was no longer reduced in muSOX expressing cells relative to control cells, and over-expression of Pabpc1 in the nucleus was sufficient to decrease Pol II occupancy in the absence of viral nuclease expression (Gilbertson et al. 2018). These data support an RNA-concentration dependent shuttling model and suggest that nuclear accumulation of Pabpc can influence transcription, though the mechanism(s) underlying this phenotype remain unclear.
One can imagine various possible scenarios by which RBPs such as Pabpc might “switch” functions from directing cytoplasmic mRNA fate to altering nuclear processes. As mRNA concentrations change, the proportion of mRNA-bound versus unbound RBPs may change commensurately. The unbound RBP population would then be available to participate in regulating another cellular process, like transcription, by altering the chromatin state either through directly binding to promoters or complexing with other proteins that would perform remodeling. A recent report identified many RBPs with DNA-associated regulatory roles, providing precedence for such a hypothesis (Xiao et al. 2019). Alternatively, RBPs could bind another factor(s) involved in transcription, titrating that factor away from the DNA and thereby regulating transcription in a more indirect fashion or, like Ccr4-Not, RBPs could coordinate directly with the transcription complex. Finally, they might compete with resident nuclear RBPs for binding nascent mRNA, thereby disrupting the mRNA processing environment.
Conclusions and Future Perspectives
In summary, there is consensus across various systems that mRNA decay and transcription are linked processes, such that disruption of one can result in compensatory or antagonistic responses by the other. This is manifest in cells in several ways, including a feedback loop that buffers against changes to overall mRNA levels, NITC that compensates for the loss of specific transcripts, and decreased Pol II promoter occupancy in response to virally-accelerated mRNA decay. However, many questions remain in this relatively young field. It is unclear if global increases and decreases in mRNA half-lives are communicated to the nucleus via the same pathway, either during buffering or potential pathogenic threat responses. Similarly, by which mechanism(s) are increases and decreases in transcription sensed and does the cell respond to both by buffering its mRNA populations? Additional perturbations to each system, namely destabilizing mRNA in yeast and altering transcriptional output in mammalian cells, will be important to determine the extent of convergence of the mammalian and yeast models. Further identification and mechanistic understanding of the factors involved in nuclear to cytoplasmic communication will be key to unraveling the connection between these spatially disparate processes and will provide additional insights into system-to-system commonalities.
Coordination of mRNA abundance appears to be conserved across eukaryotes, highlighting the importance of such communication and suggesting an evolutionary advantage to coupling transcription and decay. It also raises the question of whether mRNA buffering is a continuously active process or if it is specifically initiated upon perturbation. The buffering model has been extrapolated to have benefits in maintaining cellular homeostasis (Haimovich, Medina, et al. 2013; Sun et al. 2013; Timmers & Tora 2018), although there is no evidence indicating the cost to a cell unable to buffer mRNA levels. In the viral context, responding to infection by restricting Pol II recruitment to promoters may be a cellular defense strategy to spare the organism at the cost of an individual cell, akin to antiviral translational shutdown pathways activated by protein kinase R. This hypothesis could be explored by examining whether innate immune or cell-death signaling is initiated downstream of accelerated mRNA decay. While we are still at the early stages of understanding signaling between mRNA decay and transcription, the conservation of this connection across species and contexts indicates that it is an important aspect of gene regulation.
Acknowledgments
All figures were created with Biorender.com
Declaration of Interest
B.A.G. is supported by NIH R01 CA136367 and is an investigator of the Howard Hughes Medical Institute. E.H. is supported by an NSF GFRP predoctoral fellowship.
References
- Abernathy E & Glaunsinger B, 2015. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology, 479–480(C), pp.600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy E et al. , 2015. Viral Nucleases Induce an mRNA Degradation- Transcription Feedback Loop in Mammalian Cells. Cell host & microbe, 18(2), pp.1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrisch RG et al. , 2016. Infection by Herpes Simplex Virus 1 Causes Near-Complete Loss of RNA Polymerase II Occupancy on the Host Cell Genome. Sandri-Goldin RM, ed. Journal of Virology, 90(5), pp.2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbarwal et al. , 2014. The Rpb4/7 module of RNA polymerase II is required for carbon catabolite repressor protein 4-negative on TATA (Ccr4-not) complex to promote elongation. Journal of Biological Chemistry, 289 (48), pp.33125–33130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T et al. , 2017. SAGA Is a General Cofactor for RNA Polymerase II Transcription. Molecular Cell, 68(1), pp.130–143.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Research, 33(22), pp.7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE et al. , 2012. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nature Structural & Molecular Biology, 19(12), pp.1324–1331. [DOI] [PubMed] [Google Scholar]
- Burkard K & Butler JS, 2000. A nuclear 3’−5’ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Molecular and Cellular Biology, 20(2), pp.604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D, 2014. Genome Engineering with Targetable Nucleases. Annual Review of Biochemistry, 83(1), pp.409–439. [DOI] [PubMed] [Google Scholar]
- Chang C et al. , 2019. A low-complexity region in human XRN1 directly recruits deadenylation and decapping factors in 5’−3’ messenger RNA decay. Nucleic Acids Research, 508, pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER & Shilatifard A, 2018. Born to run: control of transcription elongation by RNA polymerase II. Nature Reviews Molecular Cell Biology, pp.1–15. [DOI] [PubMed] [Google Scholar]
- Cheung ACM et al. , 2011. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature, 471 (7337), pp. 249–253. [DOI] [PubMed] [Google Scholar]
- Covarrubias S et al. , 2011. Coordinated Destruction of Cellular Messages in Translation Complexes by the Gammaherpesvirus Host Shutoff Factor and the Mammalian Exonuclease Xrn1 Renne R, ed. PLoS Pathogens, 7(10), pp.e1002339–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene, 313(14), pp1–16. [DOI] [PubMed] [Google Scholar]
- D’Orazio KN et al. , 2019. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. eLife, pp.1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet EA et al. , 2013. Identification of the N-Terminal Domain of the Influenza Virus PA Responsible for the Suppression of Host Protein Synthesis. Journal of Virology, 87(6), pp.3108–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori-Bachash M et al. , 2012. Widespread promoter-mediated coordination of transcription and mRNA degradation. Genome biology, 13(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dremel SE & DeLuca NA, 2019. Genome replication affects transcription factor binding mediating the cascade of herpes simplex virus transcription. Proceedings of the National Academy of Sciences, 116(9), pp.3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek L et al. , 2018. Dissociation of Rpb4 from RNA polymerase II is important for yeast functionality Stoecklin G, ed. PloS one, 13(10), pp.e0206161–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumdie JN et al. , 2018. Chromatin Modification and Global Transcriptional Silencing in the Oocyte Mediated by the mRNA Decay Activator ZFP36L2. Developmental Cell, 44(3), pp.392–402.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA et al. , 2019. Genetic compensation triggered by mutant mRNA degradation. Nature, pp.1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everly DN & Read GS, 1997. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. Journal of Virology, 71(10), pp.7157–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J et al. , 2019. Cytoplasmic mRNA decay factors modulate RNA polymerase II processivity in 5’ and 3’ gene regions in yeast. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N et al. , 2014. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes & Development, 28(23), pp.2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC et al. , 2008. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research, 19(1), pp.92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia MM et al. , 2012. A Common Strategy for Host RNA Degradation by Divergent Viruses. Journal of Virology, 86(17), pp.9527–9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Molinero M et al. , 2018. The SAGA/TREX-2 subunit Sus1 binds widly to transcribed genesa nda ffects mRNA turnover globally. Epigenetics & Chromatin, 11(1) pp.1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J & Wilusz CJ, 2007. The highways and byways of mRNA decay. Nature Reviews Molecular Cell Biology, 8(2), pp.113–126. [DOI] [PubMed] [Google Scholar]
- Gaucherand L et al. , 2019. The Influenza A Virus Endoribonuclease PA-X Usurps Host mRNA Processing Machinery to Limit Host Gene Expression. Cell reports, 27(3), pp.776–791.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson S et al. , Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. eLife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger B & Ganem D, 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Molecular Cell, 13(5), pp.713–723. [DOI] [PubMed] [Google Scholar]
- Glaunsinger B, Chavez L & Ganem D, 2005. The Exonuclease and Host Shutoff Functions of the SOX Protein of Kaposi’s Sarcoma-Associated Herpesvirus Are Genetically Separable. Journal of Virology, 79(12), pp.7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T et al. , 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Letters, 582(14), pp.1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goler-Baron V et al. , 2008. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes & Development, 22(15), pp.2022–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta I et al. , 2016. Translational Capacity of a Cell Is Determined during Transcription Elongation via the Ccr4-Not Complex. Cell reports, 15(8), pp.1782–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailing S et al. , 2019. Where, When and How: Context-Dependent Functions of RNA Methylation Writers, Readers and Erasers. Molecular Cell, 74(4), pp.640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Choder M, et al. , 2013. The fate of the messenger is pre-determined: A new model for regulation of gene expression. BBA - Gene Regulatory Mechanisms, 1829(6–7), pp.643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, et al. , 2013. Gene Expression Is Circular: Factors for mRNA Degradation Also Foster mRNA Synthesis. Cell, 153(5), pp.1000–1011. [DOI] [PubMed] [Google Scholar]
- Harb M et al. , 2008. Nuclear Localization of Cytoplasmic Poly(A)-Binding Protein upon Rotavirus Infection Involves the Interaction of NSP3 with eIF4G and RoXaN. Journal of Virology, 82(22), pp.11283–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel-Sharvit L et al. , 2010. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell, 143(4), pp.552–563. [DOI] [PubMed] [Google Scholar]
- Hartenian E and Glaunsinger BA, 2019. Xrn1 activity broadly repressed RNA polymerase II occypancy at mammalian but not viral promoters during herpesvirus infection. bioRxiv. [Google Scholar]
- He F et al. , 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5’ to 3’ mRNA decay pathways in yeast. Molecular Cell, 12(6), pp.1439–1452. [DOI] [PubMed] [Google Scholar]
- Helenius K et al. , 2011. Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover. Nucleic Acids Research, 39(12), pp.5025–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L et al. , 2017. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nature Reviews Molecular Cell Biology, 18(10), pp.637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley et al. , 2006. RNA-quality control by the exosome. Nature Reviews Molecular Cell Biology, 7(7), pp.529–539. [DOI] [PubMed] [Google Scholar]
- Hsin JP & Manley JL, 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & Development, 26(19), pp.2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C et al. , 2011. SARS Coronavirus nsp1 Protein Induces Template-Dependent Endonucleolytic Cleavage of mRNAs: Viral mRNAs Are Resistant to nsp1-Induced RNA Cleavage Baric RS, ed. PLoS Pathogens, 7(12), pp.e1002433–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger BW et al. , 2012. An Overlapping Protein-Coding Region in Influenza A Virus Segment 3 Modulates the Host Response. Science, 337(6091), pp.199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JR 1994. Molecular genetics of yeast: a practical approach. Oxford University Press, New York, N.Y. [Google Scholar]
- Jorgensen I 2017. Programmed cell death as a defense against infection. Nature Reviews Immunology, 17(30, pp.151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W et al. , 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nature Structural & Molecular Biology, 16(11), pp.1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W et al. , 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proceedings of the National Academy of Sciences, 103(34), pp.12885–12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr BM & Read GS, 1999. The virion host shutoff function of herpes simplex virus degrades the 5’ end of a target mRNA before the 3’ end. Virology, 264(1), pp.195–204. [DOI] [PubMed] [Google Scholar]
- Kim YK & Maquat LE, 2019. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA, 25, pp.407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic AM & Swanson MS, 1999. hnRNP complexes: composition, structure, and function. Current Opinion in Cell Biology, 11(3), pp.363–371. [DOI] [PubMed] [Google Scholar]
- Kruk JA et al. , 2011. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes & Development, 25(6), pp.581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR & Glaunsinger BA, 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Molecular and Cellular Biology, 30(21), pp.4996–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Shum L & Glaunsinger BA, 2011. Importin alpha-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Molecular and Cellular Biology, 31(15), pp.3113–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong AD, Frenkel N, 1987. Herpes simplex virus-infected cells contain a function (s) that destabilizes both host and viral mRNAs. PNAS, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łabno A, Tomecki R & Dziembowski A, 2016. Cytoplasmic RNA decay pathways - Enzymes and mechanisms. BBA - Molecular Cell Research, 1863(12), pp.3125–3147. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M & Dixit VM, 2009. Inflammasomes: guardians of cytosolic sanctity. Immunological reviews, 227(1), pp.95–105 [DOI] [PubMed] [Google Scholar]
- Lee JE et al. , 2012. The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts Lykke-Andersen J, ed. PLOS Genetics, 8(8), pp.e1002901–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Kim B & Kim VN, 2014. Emerging Roles of RNA Modification: m6A and U-Tail. Cell, 158(5), pp.980–987. [DOI] [PubMed] [Google Scholar]
- Legrand-Poels S et al. , 2007. Modulation of Nod2-dependent NF- B signaling by the actin cytoskeleton. Journal of Cell Science, 120(7), pp.1299–1310. [DOI] [PubMed] [Google Scholar]
- Libri D et al. , 2002. Interactions between mRNA export commitment, 3’-end quality control, and nuclear degradation. Molecular and Cellular Biology, 22(23), pp.8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual Review of Biochemistry, 79, pp.181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J et al. , 2014. Uridylation by TUT4 and TUT7 Marks mRNA for Degradation. Cell, 159(6), pp.1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW et al. , 2013. The D10 Decapping Enzyme of Vaccinia Virus Contributes to Decay of Cellular and Viral mRNAs and to Virulence in Mice. Journal of Virology, 88(1), pp.202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X et al. , 2018. PNPT1 Release from Mitochondria during Apoptosis Triggers Decay of Poly(A) RNAs. Cell, pp.1–28. [DOI] [PubMed] [Google Scholar]
- Lokugamage KG et al. , 2015. Middle East Respiratory Syndrome Coronavirus nsp1 Inhibits Host Gene Expression by Selectively Targeting mRNAs Transcribed in the Nucleus while Sparing mRNAs of Cytoplasmic Origin Perlman S, ed. Journal of Virology, 89(21), pp.10970–10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R et al. , 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes & Development, 19(24), pp.3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R et al. , 2007. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. The Journal of Cell Biology, 178(7), pp.1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M et al. , 2013. Exonuclease hDIS3L2 specifies an exosome-independent 3“−5” degradation pathway of human cytoplasmic mRNA. The EMBO journal, 32(13), pp.1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z et al. , 2019. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature, pp.1–25. [DOI] [PubMed] [Google Scholar]
- Mugridge JS, Coller J & Gross JD, 2018. Structural and molecular mechanisms for the control of eukaryotic 5′–3′ mRNA decay. Nature Structural & Molecular Biology, pp.1–9. [DOI] [PubMed] [Google Scholar]
- Muhlrad D & Parker R, 1999. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Fox TD, ed. Molecular Biology of the Cell, 10(11), pp.3971–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T et al. , 2010. Decapping Activators in Saccharomyces cerevisiae Act by Multiple Mechanisms. Molecular Cell, 39(5), pp.773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroskar AA & Read GS, 1987. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. Journal of Virology, 61(2), pp.604–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroskar AA & Read GS, 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. Journal of Virology, 63(5), pp.1897–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA et al. , 2012. The Contribution of RNA Decay Quantitative Trait Loci to Inter-Individual Variation in Steady-State Gene Expression Levels Gibson G, ed. PLOS Genetics, 8(10), pp.e1003000–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park R et al. , 2014. Nuclear Translocation and Regulation of Intranuclear Distribution of Cytoplasmic Poly(A)-Binding Protein Are Distinct Processes Mediated by Two Epstein Barr Virus Proteins Sinclair AJ, ed. PloS one, 9(4), pp.e92593–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, 2012. RNA degradation in Saccharomyces cerevisae. Genetics, 191(3), pp.671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S & Moss B, 2007. Characterization of a Second Vaccinia Virus mRNA-Decapping Enzyme Conserved in Poxviruses. Journal of Virology, 81(23), pp.12973–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Resch W & Moss B, 2007. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proceedings of the National Academy of Sciences, 104(7), pp.2139–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S & Dreyfuss G, 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355(6362), pp.730–732. [DOI] [PubMed] [Google Scholar]
- Piron M et al. , 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. The EMBO journal, 17(19), pp.5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW-L & Maquat LE, 2013. Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay. Annual Review of Genetics, 47(1), pp.139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A & Dye MJ, 2002. Integrating mRNA Processing with Transcription. Cell, 108(4), pp.501–512. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A & Green R, 2016. Connections Underlying Translation and mRNA Stability. Journal of Molecular Biology, pp.1–7. [DOI] [PubMed] [Google Scholar]
- Ray D et al. , 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature, 499(7457), pp.172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read GS, 2013. Virus-encoded endonucleases: expected and novel functions. Wiley Interdisciplinary Reviews: RNA, 4(6), pp.693–708. [DOI] [PubMed] [Google Scholar]
- Richner JM et al. , 2011. Global mRNA Degradation during Lytic Gammaherpesvirus Infection Contributes to Establishment of Viral Latency Speck SH, ed. PLoS Pathogens, 7(7), pp.e1002150–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez W, Srivastav K & Muller M, 2019. C19ORF66 Broadly Escapes Virus-Induced Endonuclease Cleavage and Restricts Kaposi’s Sarcoma-Associated Herpesvirus. Jung JU, ed. Journal of Virology, 93(12), pp.9527–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A et al. , 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature, 524(7564), pp.230–233. [DOI] [PubMed] [Google Scholar]
- Rowe M et al. , 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proceedings of the National Academy of Sciences, 104(9), pp.3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski AJ et al. , 2015. Widespread disruption of host transcription termination in HSV-1 infection. Nature Communications, 6, p.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Bernecky C & Cramer P, 2015. Structural basis of transcription initiation by RNA polymerase II. Nature Reviews Molecular Cell Biology, 16(3), pp.129–143. [DOI] [PubMed] [Google Scholar]
- Salaun C et al. , 2010. Poly(A)-Binding Protein 1 Partially Relocalizes to the Nucleus during Herpes Simplex Virus Type 1 Infection in an ICP27-Independent Manner and Does Not Inhibit Virus Replication. Journal of Virology, 84(17), pp.8539–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T et al. , 2016. Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing. Journal of Molecular Biology, 428(12), pp.2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR & Maquat LE, 2012. Regulation of cytoplasmic mRNA decay. Nature Publishing Group, 13(4), pp.246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D et al. , 2014. Rpb4 subunit functions mainly in mRNA synthesis by RNA polymerase II. The Journal of biological chemistry, 289(25), pp.17446–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O et al. , 2011. Transcriptome Kinetics Is Governed by a Genome-Wide Coupling of mRNA Production and Degradation: A Role for RNA Pol II Barsh GS, ed. PLOS Genetics, 7(9), pp.e1002273–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ & Green R, 2012. Translation drives mRNA quality control. Nature Structural & Molecular Biology, 19(6), pp.594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P et al. , 2019. mRNA levels are buffered upon knockdown of RNA decay and translation factors via adjustment of transcription rates in human HepG2 cells. RNA Biology, 0(0), p.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Johnson DC & Smiley JR, 1992. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. Journal of General Virology, 73 ( Pt 2)(2), pp.467–470. [DOI] [PubMed] [Google Scholar]
- Stutz F, 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends in Cell Biology, 13(6), pp.319–327. [DOI] [PubMed] [Google Scholar]
- Sun M et al. , 2013. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Molecular Cell, 52(1), pp.52–62. [DOI] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Lariviere L, Maier KC, Seizl M, Tresch A & Cramer P, 2012. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Research, 22(7), pp.1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W & Lipshitz HD, 2009. The maternal-to-zygotic transition: a play in two acts. Development, 136(18), pp.3033–3042. [DOI] [PubMed] [Google Scholar]
- Thomas MP et al. , 2015. Apoptosis Triggers Specific, Rapid, and Global mRNA Decay with 3’ Uridylated Intermediates Degraded by DIS3L2. Cell reports, 11(7), pp.1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers HTM & Tora L, 2018. Transcript Buffering: A Balancing Act between mRNA Synthesis and mRNA Degradation. Molecular Cell, 72(1), pp.10–17. [DOI] [PubMed] [Google Scholar]
- Újvári A & Luse DS, 2005. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nature Structural & Molecular Biology, 13(1), pp.49–54. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR & Portnoy DA, 2009. Patterns of Pathogenesis: Discrimination of Pathogenic and Nonpathogenic Microbes by the Innate Immune System. Cell host & microbe, 6(1), pp.10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S & Workman JL, 2015. Histone exchange, chromatin structure and the regulation of transcription. Nature Reviews Molecular Cell Biology, 16(3), pp.178–189. [DOI] [PubMed] [Google Scholar]
- Villanyi Z & Collart MA, 2015. Ccr4-Not is at the core of the eukaryotic gene expression circuitry. Biochemical Society Transactions, 43(6), pp.1253–1258. [DOI] [PubMed] [Google Scholar]
- Villanyi Z et al. , 2014. The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation Choder M, ed. PLOS Genetics, 10(10), pp.e1004569–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW et al. , 2018. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Molecular Cell, 70(6), pp.1089–1100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MF, 2019. Genetic paradox explained by nonsense. Nature, 568(7751), pp.179–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R et al. , 2019. Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription. Cell, 178(1), pp.107–121.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H et al. , 2018. PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Molecular Cell, 70(6), pp.1081–1088.e5. [DOI] [PubMed] [Google Scholar]
- Zhu P et al. , 2017. Short body length phenotype is compensated by the upregulation of nidogen family members in a deleterious nid1a mutation of zebrafish. Journal of Genetics and Genomics, 44(11), pp.553–556. [DOI] [PubMed] [Google Scholar]