Abstract
S-palmitoylation is a reversible lipid post-translational modification that has been observed on mitochondrial proteins, but both the regulation and functional consequences of mitochondrial S-palmitoylation are poorly understood. Here, we show that perturbing the “erasers” of S-palmitoylation, acyl protein thioesterases (APTs), with either pan-active inhibitors or a new mitochondrial-targeted APT inhibitor, diminishes the antioxidant buffering capacity of mitochondria. Surprisingly, this effect was not mediated by the only known mitochondrial APT, but rather by a resident mitochondrial protein with no known endogenous function, ABHD10. We show that ABHD10 is a new member of the APT family of regulatory proteins and identify peroxiredoxin 5 (PRDX5), a key antioxidant protein, as the first target of ABHD10 S-depalmitoylase activity. We then discover that ABHD10 regulates the S-palmitoylation status of the nucleophilic active site residue of PRDX5, providing a direct mechanistic connection between ABHD10-mediated S-depalmitoylation of PRDX5 and its antioxidant capacity.
Protein cysteine-acylation (S-acylation) is an abundant post-translational modification (PTM) in mammals1–3, with modification with the saturated C16 lipid palmitate (S-palmitoylation) being the most abundant S-acylation modification. The addition of hydrophobic palmitate alters the biochemical and biophysical properties of modified proteins, affecting their stability, trafficking, localization, activity, and signaling4–7, with consequences for the pathogenesis of cancer8, neurodegenerative diseases9, and microbial infection10. Unlike other protein lipid modifications, S-palmitoylation is reversible, due to the lability of the thioester bond linking the palmitate to the sulfur atom of the cysteine residue2. S-palmitoylation of target proteins is regulated by the competitive enzymatic interplay of “writer” (protein acyltransferases, PATs) and “eraser” (acyl protein thioesterases, APTs) proteins. In humans, there are 23 known Asp-His-His-Cys (DHHC) domain-containing PATs11, thought to be associated with the Golgi, endoplasmic reticulum (ER), and plasma membranes, while the seven identified APTs are localized primarily in lysosomes (PPT1 and 2) and the cytosol (APT1 and 2; ABHD17A, B, and C)12–15.
Unbiased mass spectrometry-based protein profiling approaches have revealed that many resident mitochondrial proteins are S-palmitoylated, including those involved in fatty acid transport and metabolism, the electron transport chain, ATP synthesis, and antioxidant defense and redox signaling, such as the peroxiredoxins (PRDXs)1, 16, 17. The presence of S-palmitoylated mitochondrial proteins is particularly interesting because the enzymatic machinery for the installation and removal of protein lipidation in the mitochondria is, to date, mostly undescribed. Aside from reports implicating DHHC8 and 13 in mitochondrial function, data regarding DHHC activity and localization in mitochondria remains limited18, 19. Until very recently, there were no reports of enzyme-mediated S-depalmitoylation in this organelle. To look for potential mitochondrial APTs, we generated mitochondrial-targeted S-depalmitoylation probes (mitoDPPs), a toolkit of small molecule-based fluorescent probes that are selectively targeted to the mitochondria and measure S-deacylase activity within this compartment20, 21. Using mitoDPPs, we uncovered S-depalmitoylation activity in mitochondria and discovered that APT1, which was previously annotated primarily as a cytosolic protein12, is also localized to this organelle – revealing a potential for enzymatic regulation of mitochondrial protein lipidation20. However, although we observed mitochondrial APT (peptide S-deacylation) activity in mitochondria, and despite the multitude of mitochondrial proteins that appear in the mass spectrometry-based S-palmitoylation catalogs, no function of mitochondrial S-depalmitoylation had yet been identified.
Here, we sought to determine if there is a functional consequence of protein S-depalmitoylation in mitochondria. Using a pan-APT inhibitor, we first observed that blocking global S-depalmitoylation diminishes the antioxidant capacity of mitochondria. To confirm that the diminished antioxidant capacity was not mediated by a cytosolic target, we then synthesized and validated a spatially constrained mitochondrial pan-APT inhibitor, mitoFP, which strongly suggested that the phenotype was due to a mitochondrial-localized protein. Surprisingly, we determined that this new regulatory function was not modulated by APT1, the only annotated mitochondrial APT, but by ABHD10, a putative mitochondrial resident protein related to APT1 that belongs to the metabolic serine hydrolase (mSH) superfamily14. We characterized ABHD10 through in vitro biochemical and cell-based assays, as well as high-resolution structural studies, which revealed that ABHD10 has peptide S-depalmitoylase activity. Finally, we discovered that the antioxidant activity of PRDX5 (ref. 22), a key player in the mitochondrial antioxidant machinery, is regulated by ABHD10 via S-depalmitoylation of its active site, providing a connection between lipidation-regulated ABHD10 activity and mitochondrial redox homeostasis. Collectively, these results provide the first example of a cellular function mediated by mitochondrial S-depalmitoylation, expand the family of S-depalmitoylases with the discovery of a new APT family member, and reveal a new mode of redox regulation by PRDX lipidation.
Results
APT inhibition impairs mitochondrial antioxidant capacity
Although we recently discovered that APT1 is enriched in mitochondria and multiple mitochondrial proteins appear in unbiased S-palmitoylation datasets, no functional role for S-depalmitoylation has been described in mitochondria. However, perturbing DHHC writer S-palmitoylases has been shown to influence mitochondrial function18, 19, including redox buffering, oxidative phosphorylation, and apoptosis23, indicating protein lipidation is important for mitochondrial physiology. We reasoned that alterations in mitochondrial function resulting from treatment with APT inhibitors would offer clues as to which S-palmitoylated mitochondrial targets are functionally relevant.
Mitochondria are a major source of reactive oxygen species (ROS)24, and a multitude of mitochondrial ROS regulatory proteins have been identified in S-palmitoylation datasets1, 25. To test whether S-depalmitoylation plays a role in the regulation of mitochondrial redox status, we first used a pan-APT inhibitor, Palmostatin B (PalmB)4, to inhibit all APTs. To measure mitochondrial antioxidant buffering capacity, we used mitoPY1, a turn-on fluorescent probe, whose signal reflects H2O2 levels in mitochondria26. If APTs regulate the redox buffering capacity of mitochondria, then inhibiting APTs with PalmB would alter the ability of cells to combat oxidative stress, resulting in an enhanced mitoPY1 signal. We treated HEK293T cells with either 10 μM PalmB or DMSO vehicle in the presence of 2 μM mitoPY1, followed by induction of oxidative stress (100 μM H2O2 for 10 minutes). We observed enhanced mitoPY1 signal following H2O2 stimulation in cells treated with PalmB as compared to the DMSO-pretreated controls (Fig. 1a,b and Supplementary Fig. 1), suggesting that inhibition of S-depalmitoylation is connected to deactivation of the cellular machinery that regulates mitochondrial H2O2 levels.
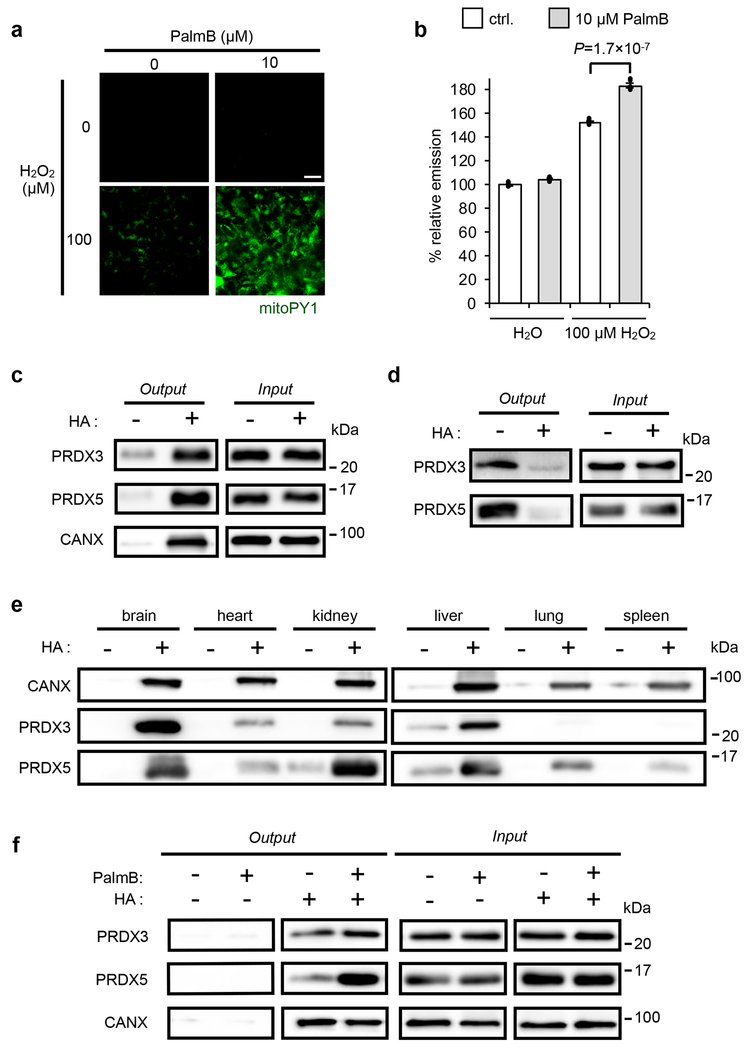
Fig. 1. Connections between S-depalmitoylation and mitochondrial redox buffering capacity.
(a) Representative images showing that PalmB treatment diminishes the mitochondrial redox buffering capacity as measured by mitoPY1 (mitochondrially targeted dye that generates H2O2-dependent fluorescence) in HEK293T cells when exposed to H2O2. 25 μm scale bar shown. (b) Quantification of the relative fluorescence intensity from mitoPY1 in each set of conditions shown in a. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 6 images). Data expressed as mean ± s.e.m. and normalized to control cells treated with DMSO. Dots represent individual data points. (c) ABE assay confirms S-acylation of PRDX3 and PRDX5 in HEK293T cells, with calnexin (CANX) as a known S-palmitoylated protein control. Input: total protein before enrichment; Output: S-acylated protein enriched via biotin labeled thiols upon hydroxyl amine (HA) treatment which selectively cleaves thioester bonds to expose acylated thiols. (d) Metabolic labeling with the palmitic acid analogue, 17-ODYA (50 μM) further validates PRDX3 and PRDX5 are S-palmitoylated in HEK293T cells. 17-ODYA is metabolically incorporated into palmitoylated proteins and is clicked with biotin-azide for enrichment. The signal difference between −HA and +HA lanes indicates the palmitoylation modification occurs on cysteine residues. (e) PRDX3 and PRDX5 are prone to S-acylation in vivo as determined by an ABE assay performed on mouse tissues. (f) Pan-APT inhibitor PalmB treatment increases the S-palmitoylation level of PRDX3 and PRDX5 as determined by an ABE assay in HEK293T cells. Two biological replicates performed for b-d and f.
To further underscore the connection between APT activity and mitochondrial redox regulation, we looked to see if treatment with PalmB influences the lipidation levels of key mitochondrial H2O2 first-responder antioxidant proteins, PRDX3 and PRDX5 (ref. 27). First, we used an acyl-biotin exchange (ABE) assay28 (Supplementary Fig. 2) to confirm cysteine acylation and then 17-octadecynoic acid (17-ODYA) metabolic labeling3, 29 (Supplementary Fig. 3) to confirm that PRDX3 and PRDX5 are susceptible to S-palmitoylation (Fig. 1c,d). Additionally, we performed ABE on various mouse tissues to validate PRDX3 and PRDX5 lipidation in vivo (Fig. 1e and Supplementary Fig. 4). After establishing PRDX3 and PRDX5 S-palmitoylation, we then observed that inhibiting APTs with PalmB resulted in an increase in the lipidation status of PRDX5 (Fig. 1f), providing a potential connection between APT inhibition, mitochondrial PRDX lipidation, and redox buffering capacity in the mitochondria. However, although the PalmB-mediated mitochondrial ROS effect phenotype is intriguing, PalmB inhibits S-depalmitoylation activity in all cellular compartments, including the cytosol30 and mitochondria20. As a result, we cannot attribute the diminished mitochondrial antioxidant capacity specifically to either cytosolic or mitochondrial APTs.
Design and synthesis of mitoFP
To elucidate the direct role of mitochondrial APTs in the observed phenotype, we set out to develop a spatially-constrained pan-APT inhibitor. We based our inhibitor on hexadecyl fluorophosphonate (HDFP)3, which relies on a serine-reactive electrophilic fluorophosphonate moiety coupled with a lipid to target lipid-active mSHs, including all known APTs (Fig. 2a). We reasoned that a derivative of HDFP could be targeted to mitochondria with the addition of a triphenylphosphonium (TPP) group31, which has been successfully used to deliver a range of cargo to the mitochondria based on the electrochemical gradient (Fig. 2a). We also shortened the lipid chain to enhance membrane permeability. With this design in hand, we synthesized mitoFP in three steps (Fig. 2b). Briefly, condensation of (4-iodobutyl)triphenylphosphonium with triethyl phosphite yielded intermediate 1. The Lewis acid TMSBr facilitated hydrolysis of 1, resulting in the dihydroxy intermediate 2. Difluorination by DAST and subsequent nucleophilic monosubstitution in the presence of octanol produces mitoFP (3) with a 36% overall yield.
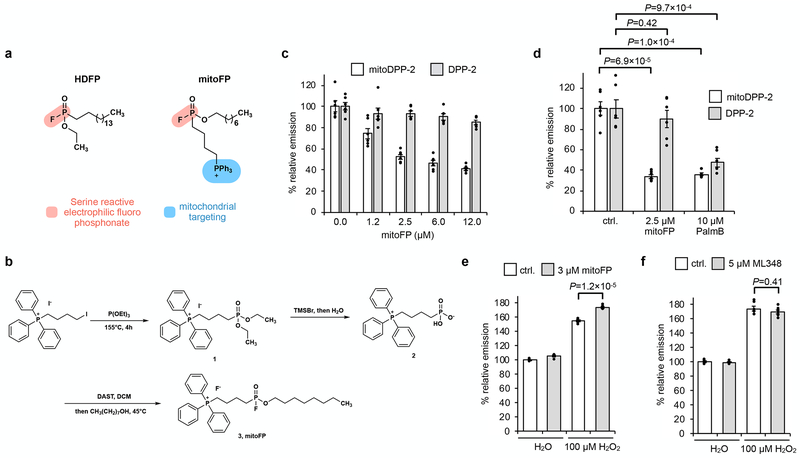
Fig. 2. Selective inhibition of mitochondrial APTs reduces mitochondrial redox buffering capacity.
(a) Design of mitoFP (right), based on the known pan-lipase inhibitor, HDFP (left). The electrophilic fluorophosphonate (red) reacts with active site serine in serine hydrolases, while the triphenylphosphonium group (blue) selectively delivers mitoFP to mitochondria. (b) Schematic for chemical synthesis of mitoFP. (c) MitoFP dose-dependent response in HEK293T cells of mitochondrial and cytosolic APT activity as measured by epifluorescence microscopy using the mitochondrial-localized APT probe, mitoDPP-2 (black), and the cytosolic APT probe, DPP-2 (grey). Data expressed as mean ± s.e.m. (n = 6 images) and normalized to control cells treated with DMSO alone. (d) Quantification of the relative fluorescence intensity from mitoDPP-2 (black) and DPP-2 (grey) in HEK293T cells comparing deactivation of mitochondrial and cytosolic APT activity, respectively, by mitoFP and PalmB. (e) Quantification of the relative fluorescence intensity from mitoPY1 showing that mitoFP diminishes the mitochondrial redox buffering capacity in HEK293T cells exposed to H2O2. (f) Quantification of the relative fluorescence intensity from mitoPY1 showing that ML348 (5 μM) does not alter mitochondrial redox buffering capacity in HEK293T cells exposed to H2O2. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 6 images for d and 5 images for each e and f). Data expressed as mean ± s.e.m. and normalized to control cells treated with DMSO. Dots represent individual data points. Two biological replicates were performed for c-f.
Characterization of mitoFP
Once synthesized, we confirmed in vitro that mitoFP inhibits APT1 using our established fluorescent S-depalmitoylation substrate DPP-5 (peptide S-depalmitoylase probe, 5; Supplementary Fig. 5)21, 32; as expected, we observed a dose-dependent inhibition of APT1 (Supplementary Fig. 6a). Next, we tested if mitoFP was a mitochondrial-specific APT inhibitor in live cells by measuring APT activity using our previously-developed APT probes, DPP-2 (peptide S-deacylase probe, 4)30 and mitoDPP-2 (mitochondrial-targeted peptide S-deacylase probe, 6)20, to assess the effects of mitoFP on cytosolic and mitochondrial APT activity, respectively (Supplementary Fig. 5). We performed a mitoFP dose-response experiment in HEK293T cells and measured DPP-2 and mitoDPP-2 signal via epifluorescence microscopy (Fig. 2c). Even at the highest concentration tested (12 μM), mitoFP had a minimal effect on DPP-2 signal, indicating that the cytosolic APTs are relatively unperturbed by the inhibitor. In contrast, treatment with mitoFP caused a dose-dependent decrease in the mitoDPP-2 signal, validating its efficient, spatially-specific inhibition of mitochondrial APTs. Treatment with 2.5 μM mitoFP blocked the mitoDPP-2 signal as efficiently as 10 μM PalmB, indicating robust inhibition of mitochondrial APTs with minimal effect on the cytosolic DPP-2 signal (Fig. 2d and Supplementary Fig. 7). We obtained similar results using hepatocellular carcinoma HepG2 cells, confirming the general applicability of mitoFP as a mitochondrial-specific APT inhibitor (Supplementary Figs. 8 and 9). Together, these experiments establish mitoFP as a new tool for perturbing mitochondrial APT activities for functional studies.
A mitochondrial APT regulates the antioxidant capacity
We next investigated if pretreating cells with mitoFP results in a reduction in mitochondrial antioxidant capacity, as seen with PalmB treatment. We pretreated HEK293T cells with either 3 μM mitoFP or DMSO vehicle in the presence of 2 μM mitoPY1, followed by H2O2 stimulation. Cells pretreated with mitoFP showed significantly higher fluorescence signal – and thus higher H2O2 levels – than control cells (Fig. 2e and Supplementary Fig. 10). We repeated the experiment in HepG2 cells, which yielded similar results and suggests a role for mitochondrial APTs on mitochondrial redox capacity across cell types (Supplementary Fig. 11). The recapitulation of PalmB-triggered diminishment of mitochondrial H2O2 buffering capacity with mitoFP points to a direct connection between mitochondrial redox regulation and local APTs, which we presumed was via mitochondrial pools of APT1.
APT1 does not regulate mitochondrial redox
We have demonstrated that the APT1-specific inhibitor ML348 (refs. 33, 34), previously shown to inhibit cytosolic APT1, can efficiently inhibit mitochondrial APT1 as well20. Therefore, we tested whether inhibition of APT1 with ML348 decreases mitochondrial antioxidant capacity, which would suggest that mitochondrial pools of APT1 affect the response observed with PalmB and mitoFP. We pretreated HEK293T cells with either 5 μM ML348 or DMSO vehicle in the presence of 2 μM mitoPY1 and stimulated with H2O2 and, intriguingly, found that pharmacological inhibition of APT1 had no effect on the H2O2-induced mitoPY1 signal (Fig. 2f and Supplementary Fig. 12). Genetic perturbation of APT1 via RNAi (Supplementary Fig. 13a) also showed no significant difference in mitochondrial redox buffering capacity during oxidative stress (Supplementary Fig. 14). Taken together, these results suggest that there is a mitochondrial enzyme other than APT1 that is targeted by both PalmB and mitoFP, which regulates mitochondrial redox buffering capacity.
ABHD10 is a novel mitochondrial APT candidate
To identify the potentially new S-depalmitoylase, we used our S-depalmitoylation probe DPP-2 (ref. 30) to perform a fluorescence imaging-based screen in cells overexpressing a library of α/β-hydrolase domain containing protein (ABHD) family members15 related to APT1 and APT2 (Fig. 3a)14. As expected, we observed an increase in the DPP-2 fluorescence signal in cells overexpressing known S-depalmitoylases, such as APT1, APT2, and PPT1 (Fig. 3b). Strikingly, overexpression of ABHD10 resulted in an enhancement of DPP-2 fluorescence similar to that of APT1 – suggesting ABHD10 has peptide S-deacylase activity (Fig. 3b). ABHD10 is a putative mitochondrial protein35 annotated as a lipase for acyl glucuronide (AcMPAG) deglucuronidation36, 37, but as yet has no identified endogenous substrates. Thus, given that ABHD10 possessed peptide S-deacylase activity in live cells and is purportedly mitochondrial, it became the candidate for further investigation.
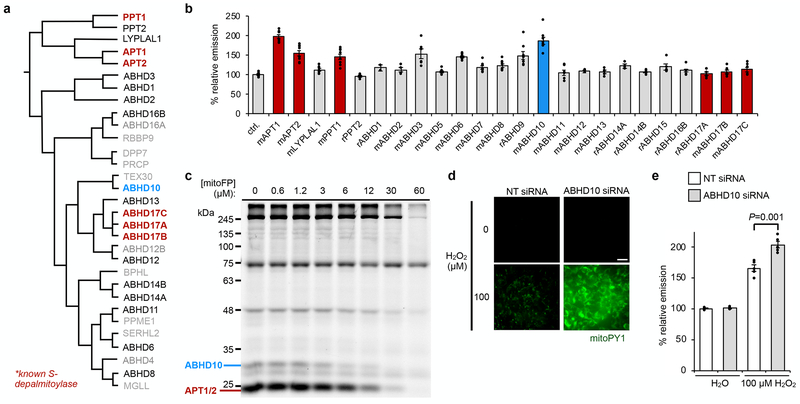
Fig. 3. ABHD10 regulates mitochondrial redox buffering capacity.
(a) Dendrogram adapted from literature14 depicting known depalmitoylases (bold) and members of the mSH family screened for potential peptide S-deacylase activity in HEK293T cells. Additionally, ABHD5, ABHD7, ABHD9 and ABHD15 were included in the screen. (b) Epifluorescence-based screening with S-deacylase probe DPP-2 in cells overexpressing a library of proteins related to the known APTs. ‘m’ and ‘r’ indicate mouse and rat, respectively. Data expressed as mean ± s.e.m. (n ≥ 2 images from two biological replicates) and normalized to control cells treated with empty vector. ABHD10 (blue), a putative mitochondrial protein35, enhances DPP-2 signal to a similar extent as APT1 (red). (c) Competitive activity-based protein profiling (ABPP) of serine hydrolases in HEK293T cells treated with indicated concentrations of mitoFP shows a dose-dependent inhibition of ABHD10 activity. After treatment with the inhibitor, the cell lysates were reacted with FP-TAMRA and the analyzed by in-gel fluorescence. Decreased fluorescence signal indicates proteins inhibited by mitoFP. Full gel images are shown in Supplementary Figures 15 and 46. (d) Representative images showing that ABHD10 knockdown diminishes the mitochondrial redox buffering capacity in HEK293T cells exposed to H2O2 as measured by epifluorescence microscopy using mitoPY1. 25 μm scale bar shown. (e) Quantification of the relative fluorescence intensity from mitoPY1 in each set of conditions shown in d. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 5 images). Data expressed as mean ± s.e.m. and normalized to control cells treated with NT siRNA. Three biological replicates were performed.
We assayed whether ABHD10 is inhibited by mitoFP using competitive activity-based protein profiling (ABPP)33 with the established serine hydrolase ABPP probe, FP-TAMRA, in both HEK293T and HepG2 cells. We observed a dose-dependent decrease of fluorescence signal from a band at ~25 kDa, signifying inhibition of APT1 activity by mitoFP (Fig. 3c). APT1 is not entirely inhibited by mitoFP at low concentrations, as expected, because APT1 is also present in the cytosol. Notably, an additional target at ~28 kDa was inhibited by mitoFP (Fig. 3c), which we identified via RNAi-mediated knockdown as the mitochondrial targeting peptide-cleaved, mature ABHD10 (Supplementary Figs. 15 and 16). We also performed ABPP in both enriched mitochondrial and cytosolic fractions prepared from mitoFP-treated HepG2 cells, which further confirmed that ABHD10 is a target of mitoFP and that ABHD10 is highly enriched in mitochondria (Supplementary Fig. 17).
ABHD10 modulates mitochondrial H2O2 buffering
We investigated whether ABHD10 is the enzyme responsible for the effect on mitochondrial redox homeostasis observed with mitoFP treatment. While the basal mitoPY1 signal was unaffected in cells with RNAi knockdown of ABHD10 (Supplementary Fig. 13b), oxidative stress resulted in a striking increase in mitochondrial H2O2 levels compared to control cells (Fig. 3d,e and Supplementary Fig. 18). In contrast, overexpression of ABHD10 decreases mitoPY1 signal in both basal and H2O2 stimulation conditions (Supplementary Fig. 19). Consistent tetramethylrhodamine methyl ester (TMRM) signal between control and various ABHD10 perturbation conditions confirmed that this effect is not due to the disruption of the mitochondrial membrane potential (Supplementary Fig. 20 and 21). Additionally, no significant changes in signal from PY1, an indicator of cytosolic H2O2 levels38, on H2O2 stimulation confirmed that the diminished mitochondrial antioxidant capacity was not due to alterations in cytoplasmic redox buffering machinery upon ABHD10 perturbation (Supplementary Fig. 22). Given that ABHD10 has a role in regulating mitochondrial redox buffering capacity, we next looked to characterize it biochemically and structurally.
ABHD10 has mitochondrial S-deacylase activity in cellulo
To confirm ABHD10 is a mitochondrial S-deacylase in live cells, we expressed ABHD10 in HEK293T cells and measured mitochondrial APT activity using mitoDPP-2, the mitochondria-targeted APT activity probe (Supplementary Fig. 5). Overexpression of wild-type human ABHD10 results in increased mitoDPP-2 signal, signifying higher S-deacylase activity, as compared with both overexpression of the catalytically inactive ABHD10 (S152A) mutant (Fig. 4a,b and Supplementary Fig. 23) or the empty vector (Supplementary Fig. 24).
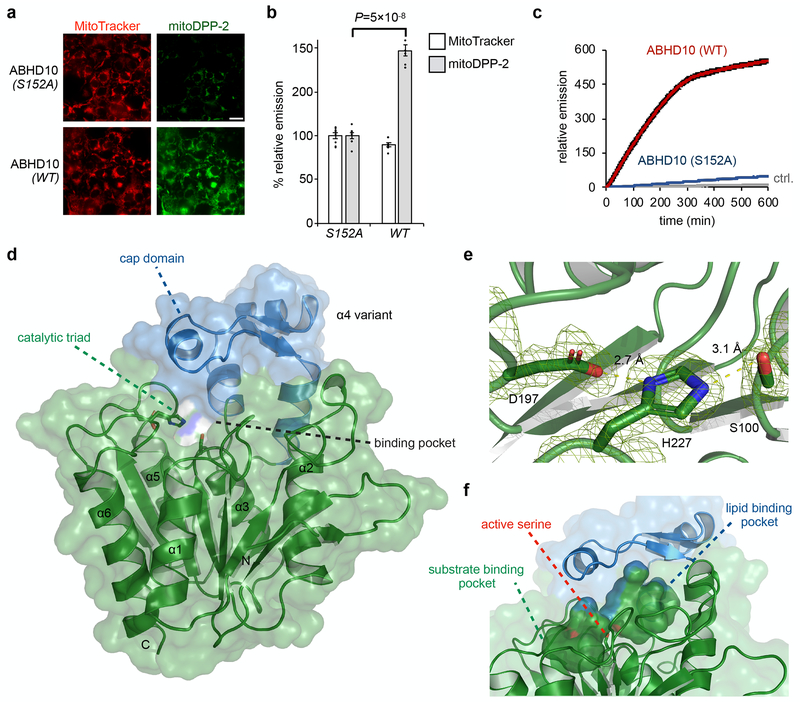
Fig. 4. Biochemical and structural characterization of ABHD10.
(a) Representative images confirming mitochondrial S-deacylation activity of ABHD10 (WT vs S152A) in HEK293T cells as measured using mitoDPP-2, a mitochondrial APT probe. 25 μm scale bar shown. (b) Quantification of relative fluorescence intensities from mitochondrial marker MitoTracker (black) and mitoDPP-2 (grey) in each set of conditions shown in a. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 8 images from two biological replicates), Data expressed as mean ± s.e.m. and normalized to cells expressing ABHD10 (S152A). Dots represent individual data points. (c) In vitro kinetic assay showing S-depalmitoylation activity of recombinant mature ABHD10 (500 nM, red) and S152A variant (500 nM, blue) compared to control without added enzyme (grey) as measured using the peptide S-depalmitoylase probe DPP-5 (5 μM). Data expressed as mean ± s.e.m (n = 4 biological replicates) and normalized to relative emission of control (grey) at t = 0. (d) X-ray diffraction structure of mature ABHD10 from Mus musculus (PDB: 6NY9). The “cap” domain (blue) sits above the catalytic triad (shown as sticks) and forms pockets. (e) Zoomed-in view of the Asp–His–Ser catalytic triad shown with weighted 2Fo–Fc electron density map (carve = 1.5). Length of hydrogen bonds between Asp and His (2.7 Å) and His and Ser (3.1 Å) is shown. (f) The surface of two major cavities within mouse ABHD10 crystal structure is shown, along with the hydroxyl group of the active serine (shown in sticks), which points towards the junction of the two cavities.
ABHD10 has S-depalmitoylase activity in vitro
Given that ABHD10 expression resulted in enhanced APT activity in cellulo, as measured by DPP fluorescence, we tested whether it had S-depalmitoylase activity in vitro. We purified the mature form of human ABHD1037 (Supplementary Fig. 25) and its active site-mutated variant (S152A) for in vitro S-depalmitoylation assays with DPP-5 (ref. 32) (Supplementary Fig. 5). Unlike DPP-2, which has a surrogate octanoyl lipid rather than a natural palmitoyl lipid, DPP-5 uses a physiologically relevant cysteine S-palmitoyl substrate and therefore directly reports on peptide S-depalmitoylase activity (Supplementary Fig. 5). Enzymatic assays with DPP-5 revealed that ABHD10 does indeed possess S-depalmitoylase activity (Fig. 4c). Kinetic analysis showed that ABHD10 has a slower turnover rate compared to APT1, but a lower KM (Supplementary Fig. 26). Consistent with the ABPP results, the S-depalmitoylation activity of ABHD10 was subject to inhibition by both mitoFP and PalmB (Supplementary Fig. 6b). Interestingly, ABHD10 is even more sensitive to mitoFP inhibition in vitro (Supplementary Fig. 6c,d). Taken together, these in vitro and in cellulo assays indicate that ABHD10 possesses mitochondrial S-depalmitoylase activity.
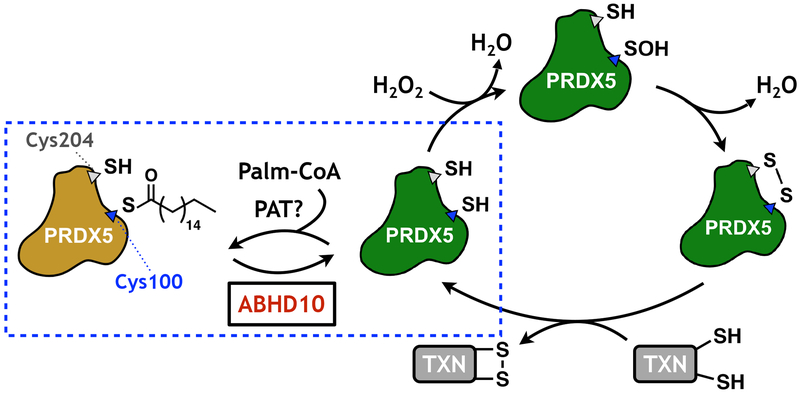
Fig. 6. Schematic of PRDX5 regulation by ABHD10-mediated S-depalmitoylation.
The active site cysteine of PRDX5 is regulated by S-palmitoylation, which then blocks the H2O2-quenching antioxidant activity of the protein. ABHD10-mediated S-depalmitoylation releases the active form of PRDX5, which then reacts with H2O2 in a disulfide-mediated redox cycling mechanism. TXN: thioredoxin.
Structural characterization of ABHD10
In acquiring kinetic parameters for ABHD10, we observed slower kinetic parameters (kcat/KM) for ABHD10 (1.1x103 M−1 s−1), as compared with APT1 (2.8x104 M−1 s−1) in vitro (Fig. 4c and Supplementary Fig. 26). This contrasts with the robust activity observed in live cells (Fig. 4a,b), suggesting additional regulatory controls for ABHD10 activity in cellulo may exist. To gain insights into the molecular basis of ABHD10 S-depalmitoylase activity, we sought to structurally characterize this mitochondrial APT.
We purified the mature form of both human and mouse ABHD10 (Supplementary Fig. 25) to a quality suitable for crystallization. However, only mouse ABHD10, which has 78.6% identity with the human version (Supplementary Fig. 27) and also exhibited S-depalmitoylation activity in vitro (Supplementary Fig. 28), yielded suitable crystals in our screens. These crystals produced X-ray diffraction patterns at a resolution of 1.66 Å, and initial phases were determined by molecular replacement, using a predicted structure as a search model (Online Methods). The structure was refined to Rwork and Rfree of 0.195 and 0.218, respectively (Supplementary Table 1). As expected, mature ABHD10 possesses a canonical α/β hydrolase domain with a catalytic triad formed by Ser100–His227–D197 (Fig. 4d,e). Notably, the catalytic serine points directly toward the junction of two pockets, one of which is covered by a “cap” domain and is presumably for lipid chain insertion based on the hydrophobicity of its interior surface, the presence of an methyl pentane diol molecule from the crystallization liquor and comparison with inhibitor-bound APT1 (Supplementary Fig. 29 and 30)34, while the other pocket is open to the solvent for substrate binding (Fig. 4f). Additionally, alignment of the structures for ABHD10 and APT1 shows that the typical lipase “cap” domain of ABHD10 is replaced by a loop in APT1 (Supplementary Fig. 31)34, 39. The “cap” vs. loop size difference may affect the accessibility of the catalytic serine and explain the slower turnover rate we observed. Overall, these structural elements confirm that ABHD10 is suited for S-depalmitoylase activity.
PRDX5 is a target of ABHD10 S-depalmitoylation activity
Having characterized the S-depalmitoylase activity of ABHD10 and confirmed that key mitochondrial H2O2-reactive antioxidant proteins, such as PRDX3 and PRDX5, are palmitoylated (Fig 1c,d), we tested whether ABHD10 regulates their lipidation levels. In HEK293T cells, we found that ABHD10 knockdown had no significant effect on the lipidation levels of PRDX3 or ALDH6A1, another lipidated mitochondrial protein (Fig. 5a,b). However, for PRDX5 the ABE assay output signal increased when ABHD10 was knocked down (Fig 5a,b) and decreased when ABHD10 is overexpressed, demonstrating that ABHD10 regulates PRDX5 lipidation levels (Supplementary Fig. 32). Similarly, ABHD10 knockdown in HeLa cells also increased the lipidation of PRDX5 (Supplementary Fig. 33). APT1 knockdown did not yield any changes in the S-palmitoylation levels of either PRDX3 or PRDX5 (Supplementary Fig. 34). Immunoprecipitation ABE further confirmed that the lipidation levels of overexpressed PRDX5-FLAG were increased upon ABHD10 knockdown (Supplementary Fig. 35). We therefore hypothesized that the ABHD10-mediated changes in PRDX5 lipidation have functional consequences for the antioxidant activity of PRDX5.
Fig. 5. ABHD10 modulates mitochondrial redox buffering capacity by regulating S-palmitoylation of PRDX5 active site.
(a) ABE assay in HEK293T cells demonstrates that PRDX5 S-palmitoylation level increases upon ABHD10 knockdown, whereas no changes are observed for PRDX3 and ALDH6A1. Calnexin (CANX) is shown as a stable S-palmitoylated protein control. (b) Quantification of relative S-palmitoylation levels of PRDX3 and PRDX5 upon ABHD10 knockdown as shown in a. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 5 biological replicates). Data expressed as mean ± s.e.m. (c) Protein map showing the three cysteine residues in mature PRDX5. (d) ABE assay in HEK293T cells expressing various PRDX5-Flag constructs (WT and three Cys to Ser mutants) shows that S-palmitoylation occurs at the catalytic site Cys100. Two biological replicates performed. (e) HEK293T cells transfected with either NT siRNA or ABHD10 siRNA were challenged with varying concentrations of H2O2 and then analyzed for viability by the MTS assay. (f) HEK293T cells were transfected with either NT siRNA or ABHD10 siRNA were challenged with varying concentrations of paraquat and then analyzed for viability by the MTS assay. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n ≥ 6 biological replicates in e and f). Data expressed as mean ± s.e.m. and cells with various concentrations of H2O2 (e) or paraquat (f) is normalized to respective cells treated with NT or ABHD10 siRNA and H2O. Dots represent individual data points.
ABHD10 modulates PRDX5 antioxidant activity
The mature form of PRDX5 contains three cysteine residues at amino acid positions 100, 125, and 204 (Fig. 5c). Of these, Cys100 and Cys204 participate in the catalytic cycle of PRDX5 in mitochondria22. To ascertain which of the cysteine residue(s) on PRDX5 are S-palmitoylated, we expressed wild-type PRDX5-Flag and all three cysteine to serine single point mutants in HEK293T cells and analyzed their S-acylation levels by ABE (Fig. 5d). While PRDX5WT, PRDX5C125S, and PRDX5C204S all showed comparable S-palmitoylation levels, we observed a complete abrogation of signal for PRDX5C100S, indicating that Cys100 is the primary S-acylation site of PRDX5. We confirmed the S-palmitoylation using metabolic labeling with clickable lipids, again demonstrating complete abrogation of signal enrichment for PRDX5C100S (Supplementary Fig. 36). Intriguingly, Cys100 is the catalytic residue of PRDX5 and hence is required for nucleophilic attack on H2O2, which only occurs when Cys100 is in the free thiol form. S-palmitoylation therefore masks the reactivity of this critical residue, and thus, ABHD10 can directly modulate mitochondrial antioxidant ability through depalmitoylation of PRDX5.
ABHD10 knockdown amplifies ROS-induced cell death
Finally, to test for the functional relevance of ABHD10 in mitochondrial redox homeostasis, we examined whether perturbation of ABHD10-dependent redox regulation affected cell viability under various oxidative stress conditions. ABHD10 knockdown resulted in a reduction of cell viability in both HEK293T or HeLa cells under H2O2-mediated oxidative stress conditions (Fig. 5e and Supplementary Fig. 37). Additionally, treatment of HEK293T cells with paraquat, a potent mitochondrial ROS inducer40, yields diminished cell viability at concentrations as low as 50 μM in ABHD10 knockdown cells, with minimal effect on control cells (Fig. 5f). We also tested whether the influence of ABHD10 knockdown on cell viability upon H2O2 stimulation is affected by PRDX5 perturbation by PRDX5 knockdown, which does not affect cell viability under normoxia41. We found that PRDX5 knockdown (Supplementary Fig. 38) abrogates H2O2-induced cell toxicity upon ABHD10 knockdown in both HEK293T and HeLa cells (Supplementary Fig. 39). Together, these data show that reduced ABHD10 levels with concomitant enhanced PRDX5 lipidation increases cell death under oxidative stress conditions and establishes that ABHD10 regulates mitochondrial redox homeostasis (Fig. 6).
Discussion
ABHD10 is different from the other APTs thus far identified. While PPT1 is located in lysosomes, APT2 in the cytosol and the golgi15, ABHD17A, B, and C in membranes15, and APT1 in the cytosol, membrane, golgi and the mitochondria12, 42, ABHD10 appears to be exclusively located in the mitochondria. Additionally, unlike APT1, our structural and biochemical data indicate that ABHD10 has an extensive “cap” domain and also possesses slower kinetic parameters than APT1 in vitro, suggesting additional regulatory mechanisms. If so, this would not be entirely novel, as we have previously observed growth factor-mediated dynamic regulation of cytosolic APTs30 and helped identify the first phosphorylation-mediated regulatory site on APT143. While we identified one substrate for ABHD10, it potentially has additional mitochondrial substrates. Recently developed libraries of synthetic palmitoylated peptide substrates could provide powerful methods to study the substrate specificity of ABHD1044, but assigning natural substrates to APTs is often a challenge due to compensation between APTs and an incomplete understanding of regulatory elements. However, the addition of ABHD10 to the APT family makes assigning more substrates and functions now possible. Indeed, an ABHD10 inhibitor is already available, which will aid in studying its APT function in live cells with temporal control45.
Mitochondria generate reactive oxygen species (ROS) byproducts, which can be both advantageous and deleterious46. Therefore, maintaining mitochondrial redox homeostasis is crucial for cellular health. Peroxiredoxins (PRDXs) are the major regulators of H2O2 levels, due to their high reactivity towards peroxides. Since H2O2 is both necessary at low levels and toxic at high levels, the PRDXs are tightly regulated by a variety of processes27, such as phosphorylation, oxidation, and nitrosylation. Our discovery that PRDX5 is regulated by S-palmitoylation modification at the active site cysteine not only represents a new mechanism for regulating the activity of a peroxiredoxin, but is also a rare instance of S-palmitoylation directly modifying an enzyme active site47. Future work will involve identifying additional consequences of PRDX5 lipidation, including its impact on PRDX5 stability, trafficking, and membrane localization. Additionally, assessing whether other peroxiredoxins are similarly regulated, both in the mitochondria and in other cellular compartments, will help further elucidate the role of PRDX lipidation. The redox buffering capacity of compartments within the cell are interconnected, so there is potentially feedback with other PRDXs. The data presented here suggests that other targets of PRDX5 also contribute to cell survival in oxidative stress conditions. Moreover, we showed that PRDX3, another mitochondrial peroxiredoxin, is also lipidated, but is not regulated by ABHD10. This suggests that additional regulatory APTs exist along these pathways, warranting extensive further interrogation.
Determining localization-based enzymatic activities via genetic approaches, i.e. knocking down or out a target gene product, is challenging, especially when protein targets reside in several cellular compartments. In this work, we supplemented genetic approaches with the development of a novel spatially-constrained APT inhibitor, mitoFP. While synthesizing TPP-tagged inhibitors is not challenging, confirming the proper localization of a novel inhibitor is often not possible. Here, due to our previous development of spatially-constrained activity probes for APTs20, coupled with organelle-specific ABPP3, 14, 48–50, we were able to validate the targeting and potency of mitoFP in live cells. Using mitoFP, we conclude that ABHD10 activity in the mitochondria mediates the antioxidant stress phenotype observed with pan-active, non-targeted APT inhibitors. Further use of mitoFP will help us and others continue to illuminate mitochondrial S-depalmitoylation function and regulation.
This work focuses entirely on the erasers of S-palmitoylation, the APTs. However, equally important to the regulation of PRDX5 is the lipid installation. Although perturbation of both DHHC8 and 13 disrupts mitochondrial metabolic functions18, 19, no known writers have definitively been localized to the mitochondria. It is possible that PRDX5 is lipidated prior to mitochondrial import and then released in an active form once in the mitochondria. In this scenario, the lipid modification could be regulating PRDX5 trafficking to the mitochondria, as was previously observed for BAX23. However, there also may be DHHCs, or as-yet undiscovered acyltransferases, present in or at the mitochondria, that mediate PRDX5 lipidation. Future work exploring acyltransferase activity on these mitochondrial targets, likely involving the development of better chemical probes and inhibitors, will help complete the regulatory picture of mitochondrial proteome S-palmitoylation.
Online Methods
Reagents.
DMEM GlutaMAX (Gibco), Fetal Bovine Serum (FBS; Gibco/Life Technologies, Qualified US origin or Gemini Benchmark™ 100-106), Live Cell Imaging Solution (Molecular Probes), Opti-MEM (Gibco), Lipofectamine 3000 reagent (Invitrogen), Lipofectamine RNAiMAX transfection reagent (Invitrogen), polyethylenimine (PEI) (Sigma; average Mw ~25,000 by LS, average Mn ~10,000 by GPC, branched), Dynabead Protein G magnetic beads (Invitrogen) MitoTracker Deep Red FM (Invitrogen), Hoechst 33342 (Fisher), 2-BP (Sigma), MitoPY1 (Sigma), PY1 (Sigma), Palmostatin B (PalmB) (EMD Millipore), Charcoal-filter FBS (A3382101, Thermo Fischer), MTS (BioVision), Phenazine methosulfate (Sigma) were purchased as mentioned in parenthesis. siRNAs targeting human ABHD10 (SI04229519), human APT1/LYPLA1 (SI03246586) and human PRDX5 (SI00096971, SI02638888, SI02638902), as well as non-targeting (NT) control siRNA (SI03650325), were purchased from Qiagen. Silica gel P60 (40-63 μm, 230-400 mesh; SiliCycle) was used for column chromatography. Analytical thin layer chromatography was performed using precoated 60 F254 silica gel sheets (0.25 mm thick; SiliCycle). DPP-230, DPP-532 and mitoDPP-220 were synthesized as previously reported. All chemicals for synthesis were purchased from Sigma-Aldrich or Fisher Scientific and used as received. ML348 was purchased from Tocris (Bristol, UK).
Spectrometry.
1H- and 13C-NMR spectra were collected in the NMR solvent CDCl3 (Sigma-Aldrich, St. Louis, MO) at 25 ºC using a 500 MHz Bruker Avance II+ spectrometer with 5 mm QNP probe at the Department of Chemistry NMR Facility at the University of Chicago. 1H-NMR chemical shifts are reported in parts per million (ppm) relative to the peak of residual proton signals (CDCl3: 7.26 ppm). Multiplicities are given as: t (triplet), q (quartet), br (broad), m (multiplet). 13C-NMR chemical shifts are reported in ppm relative to the peak of residual proton signals (CDCl3: 77.16 ppm). NMR analysis was done in TopSpin 3.5pl7. High resolution mass data was obtained from an Agilent 6224 TOF High Resolution Accurate Mass Spectrometer (HRA-MS) using a combination of APCI and ESI at the Department of Chemistry Mass Spectrometry Facility at the University of Chicago. Low resolution mass spectral and liquid chromatography analyses were carried out on an Advion Expression-L mass spectrometer (Ithaca, NY) coupled with an Agilent 1220 Infinity LC System (Santa Clara, CA).
Imaging.
For fluorescence microscopy, an inverted epifluorescence microscope (Lieca DMi8) equipped with a Hamamatsu Orca-Flash 4.0 camera, a 63x oil objective (N/A 1.4), and a 300 W Xenon light source (Sutter Lamda XL) was used. Leica LASX software was used to obtain images for mitoPY1/DPPs (YFP filter cube 1525306), Hoechst 33342 (ET 402/15x, Quad-S, ET 455/50m), MitoTracker (ET 645/30x, Quad-S, ET 705/72m) and brightfield.
Western Blots.
After SDS-PAGE, proteins were transferred onto methanol-preactivated Immobilon-P PVDF membranes (pore size 0.45 μm; Milipore) using a semi-dry transfer cell (Bio-Rad). After transfer, the membranes were blocked for 1 hr with TBST wash buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween-20) containing 3% Bovine Serum Albumin (BSA; Fischer or Thermo Scientific). Considering the long procedure of the assay, we cut each membrane into several small membranes containing the target protein band based on the molecule weight. Each membrane was then incubated with primary antibody in 3% BSA-TBST. The membrane was washed with TBST 5 times for 5 min (5 x 5min) followed by 1 hr incubation with either anti-rabbit IgG HRP or anti-mouse IgGκ BP-HRP in 3% BSA-TBST, washed 5x5min with TBST, and then visualized using SuperSignal™ West Pico PLUS chemiluminescent substrate (Thermo Scientific) and recorded on a chemiluminescent western blot imaging system (Azure Biosystems C300). Additional Western blots in this study were performed identically. For antibody dilutions and vendor information, see Supplementary Table 2. The uncroped membrane figures are shown as Supplementary Figs 42–49.
Cell culture.
Unless otherwise stated, HEK293T (ATCC), HeLa (from Prof. Chuan He, University of Chicago) and HepG2 cells (from Cellular Screening Center, University of Chicago) were plated and maintained in DMEM GlutaMAX (supplemented with 10% Fetal Bovine Serum, and 1% Penicillin/Streptomycin) at 37 °C and 5% CO2. For all experiments, cells had undergone fewer than 25 passages and hence mycoplasma contamination was not tested. HEK293T, HeLa, and HepG2 cells are not listed in the database of commonly misidentified cell lines maintained by ICLAC (http://iclac.org/databases/crosscontaminations/). Cells were plated the day before all experiments. For all treatments, cells were first washed with fresh growth medium, which was then replaced with fresh growth medium containing the drug and/or probe. Where indicated, treatments also included Hoechst 33342 (1 μM) and MitoTracker Deep Red (100 nM) for nuclear and mitochondrial visualization, respectively. Transfections were conducted with either PEI (for plasmids), Lipofectamine 3000 (for plasmids) or Lipofectamine RNAiMAX (for siRNAs) according to the manufacturer’s recommended protocols, which were scaled according to the volume of the culture dish.
Plasmid cloning.
All plasmids were constructed by Gibson Assembly from PCR products generated using Q5 Hot Start DNA Polymerase (New England Biolabs) or Phusion Polymerase (generated in-house). The pcDNA3 vector was a gift from Prof. Chuan He and pET-30a was used as described previously30. Genes for human PRDX5, human ABHD10, mouse ABHD10 and primers were synthesized by Integrated DNA Technologies, Inc (IDT). The cDNAs for PRDX5 and ABHD10 were cloned into the pcDNA3 vector for mammalian expression. In addition, both human and mouse ABHD10 were truncated to remove the mitochondria localization signal peptide (residues 1-52 for human ABHD10 (Q9NUJ1-1) and 1-43 for mouse (Q6PE15-1), optimized using IDT Codon Optimizing Tool, and cloned into the pET-30a vector containing a His-tag for E. coli expression. All newly constructed plasmids were sequence-verified at the University of Chicago Comprehensive Cancer Center DNA Sequencing and Genotyping Facility and are available upon request.
Assessment of mitochondrial H2O2 following depalmitoylation inhibition.
300,000 HEK293T cells/well or 125,000 HepG2 cells/well were plated in four-well chamber slides (D35C4-20-1.5-N, Cellvis) precoated with 5 μg Poly-D-lysine (30-70 KDa, Alfa Aesar). After 20-24 hr, cells were pretreated with 10 μM PalmB, 5 μM ML348, or 2.5 μM mitoFP for 30 min at 37 °C. Control cells were pretreated with vehicle (DMSO). Hoechst 33342 and MitoTracker Deep Red were included for respective nuclear and mitochondrial visualization, as was 2 μM of the mitochondrial-targeted H2O2 probe, mitoPY1 (Sigma). After pretreatment, cells were briefly washed with DPBS, and treated with 100 μM H2O2 in fresh DPBS (400 μL) for 10 min at 37 °C. Control cells were untreated. Cells were then imaged on an inverted epifluorescence microscope. Analyses were performed in ImageJ (Wayne Rasband, NIH). For data analysis, the average fluorescence intensity per image in each experimental condition was obtained by gating cells using the brightfield image and applying that mask in the corresponding mitoPY1 image. All data was normalized to the average fluorescence intensity of the DMSO-pretreated control that was not exposed to H2O2. Each experiment was repeated in at least two biological replicates with identical results.
Acyl Biotin Exchange (ABE) of cell culture samples.
Volumes here are representative of an experiment using cells from a 10 cm plate. HEK293T cells were washed with DPBS, lysed with 1 mL RIPA lysis buffer containing protease inhibitors (1 mM PMSF and 50 mM N-Ethylmaleimide (NEM, Acros)), and subject to end-over-end rotation overnight at 4 °C. The cell debris was pelleted by centrifugation at 13000 xg for 15-30 min at 4 °C and the supernatant collected. Protein concentration was measured using BCA assay and equal amounts of total protein from each sample was subjected to acetone precipitation for 2 hr at −20 °C. The resulting pellet was dissolved by sonication in 100 μL/mg protein of 4% SDS buffer (resuspension buffer; 150 mM NaCl, 50 mM HEPES, 5 mM EDTA, pH 7.4) containing 50 mM NEM. 250 μL/mg protein of 0.2% Triton buffer (150 mM NaCl, 50 mM HEPES, 5 mM EDTA, pH 7.4) containing 50 mM NEM was added and protein solution was rotated end-over-end for 3 hr at 25 °C. Two subsequent acetone precipitations were performed to remove excess NEM. The resulting protein pellet was dissolved in 105 μL of resuspension buffer by sonication. The protein sample was divided into two equal parts for ± HA (hydroxylamine (Acros)) treatment in fresh 50 μL tubes. Each sample was treated with 150 μL of either –HA buffer (0.2% Triton buffer) or +HA buffer (0.2% Triton buffer containing 1.33 mM HA, pH ~ 7.3). Samples were first incubated at room temperature with shaking for 1 hr, and then proteins were precipitated by chloroform-methanol precipitation to remove the excess hydroxylamine. Protein pellets were dried for 20-30 min at room temperature and resuspended by sonication in 60 μL of resuspension buffer containing 10 μM EZ-Link HPDP-Biotin (Themo, #21341). Protein solutions were diluted with 240 μL Biotin buffer (0.2% Triton buffer, 10 μM EZ-Link HPDP-Biotin) and incubated for 2 hr at room temperature with shaking. Excess biotin was removed via acetone precipitation. Protein pellets were dissolved in 30 μL/mg protein of resuspension buffer by sonicating and total volume was brought to 1200 μL with 0.2% Triton buffer. Undissolved residues were removed via centrifugation at 14000 rpm for 5 min at 4 °C. Protein concentrations were measured using BCA assay. 35 μg of protein was transferred to a fresh tube to serve as the loading control for normalizing levels of protein of interest (“input”). 350-400 ug protein was transferred to a new tube and diluted to 0.6-0.8 mg/ml with wash buffer (0.1% SDS, 0.2% Triton X-100, 150 mM NaCl, 50 mM HEPES, 5 mM EDTA, pH 7.4). 80 μL of streptavidin-agarose beads (Thermo, #20361) per mg of protein were added to the protein solution, which was then incubated at 4 °C overnight with end-over-end rotation. Unbound proteins were removed by washing 6x with 1 mL washing buffer and spinning for 1 min at 6000 rpm each time. Bound proteins were eluted by boiling the beads for 10 min at 95 °C with 1X Laemelli sample buffer (containing 20-30 mM DTT). The protein was resolved on 10% or 12% SDS-PAGE gels and subjected to Western Blotting protocol using the protocol described above.
Mouse tissues ABE assay.
The use of vertebrate animals (M. musculus, mice) in the laboratory of Bryan Dickinson has been approved by the Institutional Animal Care and Use Committee (IACUC) under the Animal Care and Use Protocol #72531, “Mouse models of lipid signaling regulation.” For ABE performed on mouse tissue samples, organs were isolated from 7-week-old female C57BL/6J mice, quickly washed with Hanks’ Balanced Salt solution (HBSS), and immediately flash frozen. Following mechanical homogenization, samples were lysed in 2 to 5 mL (depending on organ mass) of HEPES lysis buffer (150 mM NaCl, 40 mM HEPES, 0.5% Triton, 3 mM EDTA, pH = 7.4 with protease inhibitors and 1 mM PMSF) containing 50 mM NEM, vortexed vigorously, and rotated end-over-end for 8 hr. After centrifugation at 15000 xg, the supernatant was removed and subjected to sequential acetone and chloroform/methanol precipitations to yield dry protein. The resulting protein pellet was then processed as described above.
Palmostatin B (PalmB) ABE assay.
A solution of PalmB (10 μM) or DMSO in DMEM GlutaMAX (10% charcoal-filtered FBS and 1% fatty acid-free BSA) was added to HEK293T cells grown to 90% confluency. Cells were incubated for 1.5 hr, washed twice with ice-cold PBS, and processed as described in the ABE protocol.
Metabolic Labeling.
Volumes here are representative of an experiment using cells from a 10cm plate. HEK293T cells were washed with DPBS and then treated with 50 μM 17-ODYA absorbed on 5% BSA in DMEM GlutaMAX supplemented with 10% charcoal filtered FBS. Cells were then incubated for 6 hr unless noted differently at 37 °C with supply of 5% CO2. Cells were washed once in DPBS, lysed with 1 mL HEPES Lysis buffer (150 mM NaCl, 50 mM HEPES, 0.2% SDS, 1% Triton-100, pH 7.4, with EDTA-free Protease Inhibitor and 1mM PMSF) and subject to end-over-end rotation overnight at 4°C. The cell debris was pelleted by centrifugation at 13000 xg for 15-30 min at 4°C and the supernatant collected. Protein concentration was measured using BCA assay and 1 mg of protein was placed into a new tube. First, the same volume of HEPES buffer (150 mM NaCl, 50 mM HEPES, pH 7.4) was added in order to adjust the SDS concentration to 0.1% and then all samples were diluted with the click reaction buffer (150 mM NaCl, 50 mM HEPES, 0.1% SDS, 0.5% Triton-100, pH7.4) for a final protein concentration of 1 mg/ml. 88 μL of master mix made from 22 μL of each 5 mM Azide-PEG3-biotin (Sigma), 5 mM TBTA (Combi-Blocks), 50 mM CuSO4 and 5 mM TCEP was added for a 1 mL “Click” reaction. The resulting solution was incubated for 1 hr at room temperature with shaking. 121 μL of 100 mM EDTA was added to quench the “Click” reaction and proteins were precipitated by chloroform-methanol precipitation. Protein pellet was dissolved in 50 μL of resuspension buffer by sonicating and 150 μL of 0.2% triton buffer added. Protein was further precipitated by chloroform-methanol precipitation. The resulting protein pellet was dissolved in 105 μL of resuspension buffer by sonication. Protein samples were divided into two equal parts for ± HA (hydroxylamine) treatment by placing 50 μL into fresh tubes. Each sample was treated with 150 μl of either –HA buffer (0.2% Triton buffer) or +HA buffer (0.2% Triton buffer containing 1.33 mM HA, pH ~ 7.3). Samples were first incubated at room temperature with shaking for 1 hr, and then proteins were precipitated by Chloroform-Methanol precipitation to remove the excess hydroxylamine. Protein pellets were dried for 20-30 min at room temperature and resuspended in 30 μL of resuspension buffer per mg protein by sonicating and total volume was brought to 1200 μL with 0.2% Triton buffer. Undissolved residues were removed by centrifugation at 14000 rpm for 5 min at 4°C. Protein concentrations were measured using BCA assay. 35 μg of protein was transferred to a fresh tube to serve as the “input” sample, as described above. 450-600 μg protein was transferred to new tube and treated as described for the ABE assay.
In cellulo dose curve of mitoFP.
22,000 HEK293T cells/well or 10,000 HepG2 cells/well were plated in 10 wells of a 96 well plate (P96-1-N, Cellvis) that were either precoated with 0.7 μg Poly-D-lysine (for HEK293T cells) or used as-is (for HepG2 cells). After 20-24 hr, cells were pretreated with 1.2, 2.5, 6, or 12 μM mitoFP for 30 min at 37°C. Control cells were treated with DMSO vehicle. All treatments included Hoechst 33342 and MitoTracker Deep Red. Cells were then washed with imaging buffer, and probed for 10 min at 37 °C with either 1 μM DPP-2 or 500 nM mitoDPP-2 in fresh imaging buffer (100 μL). Cells were imaged on an inverted epifluorescence microscope. Six images from two biological replicates were used for analysis. Data analysis was performed as described above, and the data was normalized to the average fluorescence intensity of the DMSO-treated control.
Comparison of PalmB- and mitoFP inhibition of APTs activity.
30,000 HEK293T cells/well or 10,000 HepG2 cells/well were plated in 10 wells of a 96 well plate in the same manner as for the in cellulo mitoFP dose curve. After 20-24 hr, cells were treated with 2.5 μM mitoFP or 10 μM PalmB for 30 min at 37 °C. Control cells were treated with DMSO vehicle. Cells were then washed with imaging buffer and probed for 13 min at 37 °C with etiher 1 μM DPP-2 or 500 nM mitoDPP-2 in fresh imaging buffer (100 μL). Images were obtained on an inverted epifluorescence microscope. Six images from two biological replicates were used for analysis. Data analysis was performed as mentioned above, and the data was normalized to the average fluorescence intensity of the DMSO-treated control.
Assessment of APT1, ABHD10 and PRDX5 knockdown efficiency.
550,000 HEK293T cells/well were plated in 2 mL DMEM GlutaMAX (10% FBS) into 6-well dish. After 20-22 hr, cells were transfected with 44 pmol of siRNA targeting either APT1 or ABHD10, or a non-targeting (NT) siRNA control. 44 hr post-transfection, cells were washed with 1 mL DPBS, and lysed with 500 μL of RIPA buffer (50 mM Tris, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1.0% TritonX-100, pH 7.4, PMSF and Protease inhibitor cocktail). Lysed cells were collected and vortexed for 5 sec and end-over-end rotated at 4 °C overnight. Protein concentration of each sample was measured by BCA assay (Thermo Scientific). 6X Laemmli SDS sample buffer (Alfa Aesar) containing 180 mM DTT was added, and proteins were denatured by heating at 90 °C for 10 min. An equal amount of protein from each sample was resolved on 12% SDS-PAGE gels and subjected to Western Blotting protocol using the protocol described above. For checking PRDX5 knockdown efficiency, 500,000 Hela cells/well were plated and 25 pmol of siRNAs is used.
Assessment of mitochondrial H2O2 following APT1 or ABHD10 knockdown, and ABHD10 overexpression.
140,000 HEK293T cells/well were plated in four well chamber slides, as detailed above. After 18-20 hr, 150 μL of media was removed from each well and the cells were transfected with 12.5 pmol of siRNA targeting either APT1 or ABHD10. Control cells were transfected with NT siRNA following manufacture’s conditions. 44-48 hr post-transfection, cells were pretreated with 2 μM mitoPY1 for 30 min at 37 °C. Cells were then washed with DPBS and treated with fresh DPBS with or without 100 μM H2O2 for 10 min at 37 °C. Images were obtained on an inverted epifluorescence microscope. Data analysis was performed as described above. Data was normalized to the average fluorescence intensity of the NT siRNA-transfected control that was not treated with H2O2. Each experiment was repeated in at least two biological replicates with identical results. Similar experiments were also done in HEK293T cells transfected with 600 ng of ABHD10 vector or control vector.
Assessment of cytoplasmic H2O2 following knockdown of ABHD10.
150,000 HEK293T cells/well were plated in four well chamber slides, as detailed above. After 18-20 hr, 150 μL of media was removed from each well and the cells were transfected with 12.5 pmol of siRNA targeting either ABHD10 or non-targeting (NT) siRNA control following manufacturer’s conditions. 48 hr post-transfection, cells were pretreated with 5 μM PY1 for 30 min at 37 °C. Cells were then washed with DPBS (400 μL) and treated with fresh DPBS (400 μL) with or without 100 μM H2O2 for 13 min at 37 °C. Images were obtained on an inverted epifluorescence microscope. Data analysis was performed as described above. Data was normalized to the average fluorescence intensity of the NT siRNA-transfected control that was not treated with H2O2.
Epifluorescence-based genetic overexpression screen.
16,000-18,000 HEK293T cells/well were plated in 96 well plates as described above. After 18-22 hr, cells were transfected with 80 ng of either empty vector plasmid or individual plasmids containing the protein targets. 40-44 hr post-transfection, the media was replaced with 100 μL of fresh media containing 1 μM Hoechst 33342 and 100 nM MitoTracker Deep Red. After 30 min, the cells were washed with 100 μL of Imaging Buffer and 70 μL of 1 μM DPP-2 in fresh Imaging Buffer was added to the cells. Cell were incubated for 10 min at 37 °C and then imaged from 25 min onwards on an inverted epifluorescence microscope set on automated focus mode. Multiple images from two biological replicates were used for analysis. Data analysis was performed as described above, and the data was normalized to the average fluorescence intensity of the empty vector plasmid transfected control cells.
Activity-Based Protein Profiling (ABPP).
HEK293T or HepG2 cells were incubated with various concentrations of mitoFP for 2 hr, and lysed in SDS-free RIPA lysis buffer. Knockdown lysates from APT1 or ABHD10 siRNA transfected cells were obtained by treating cells with SDS-free RIPA lysis buffer. After 30 min of lysis on ice, 50 μg of protein from total cell, cytosolic, and mitochondrial fractions were incubated with 2 μM fluorophosphonate (FP)-TAMRA (Thermo) 3 for 1 hr at RT, then quenched with 6X Laemelli sample buffer (containing 60 mM DTT). Proteins were resolved with 12% SDS-PAGE and visualized with Chemidoc XRS (Biorad) using the Cy3 channel. As a loading control, Coomassie blue was used to stain total protein.
Mitochondria isolation.
Mitochondrial and cytosolic fractions from HepG2 cells were performed according to the manufacturer’s recommended protocol with Mitochondria Isolation Kit for Cultured Cells (Pierce, 89874).
Imaging with mitoDPP-2.
140,000 HEK293T cells/well were plated in four-well chamber slides as described above. After 18–20 h, the cells were transfected with 600 ng of either pcDNA3-ABHD10WT or -ABHD10S152A. Control cells were transfected with the empty pcDNA3 vector backbone. 32–35 hr post-transfection, nuclei and mitochondria were stained with Hoechst 33342 and MitoTracker Deep Red, respectively, for 30 min at 37 °C. Cells were then briefly washed with imaging solution, and probed with 500 nM mitoDPP-2 in fresh imaging solution. After 10 min at 37 °C, images were obtained on an inverted epifluorescence microscope, and data analyses were performed as described above. For data analysis, the average fluorescence intensity per image in each experimental condition was obtained by gating cells using the brightfield image, and applying that mask to the corresponding MitoTracker and mitoDPP-2 image. Eight images from two biological replicates were used to quantify the images, and the data was normalized to the average fluorescence intensity of the MitoTracker channel of cells transfected with the empty vector plasmid.
For assessing mitochondrial membrane integrity in HEK293T, cells were plated, treated as described above and replaced with fresh growth media containing 1 μM Hoechst 33342 and 100 nM TMRM. Cells were then incubated at 37 °C for 30 mins, washed and replaced with 400 μL imaging solution. Images were obtained on an inverted epifluorescence microscope using TRITC/RFP filter settings, and data analyses were performed as described above. Eight images from two biological replicates were used to quantify the images, and the data was normalized to the average fluorescence intensity of control cells.
In vitro kinetic assay of APTs following mitoFP-mediated inhibition.
In vitro biochemical assays with purified APT1 or ABHD10 enzymes were performed on a Biotek synergy Neo2 plate reader. 100 nM APT1 or ABHD10 was incubated for 30 min at 37°C with 1.2, 2.4, 6, 12, 24, 60, or 120 μM mitoFP equal in HEPES buffer (20 mM, pH = 7.8, 150 mM NaCl). Controls were incubated with an equal volume of DMSO. At the end of 30 min, the incubated APT1 or ABHD10 solutions (150 μL) were added to a 96-well optical bottom plate (Nunc 265301, Thermo Scientific) at room temperature. Wells containing HEPES buffer alone served as negative controls. Using a multi-channel pipette, 150 μL of a solution of 2 μM DPP-5 in HEPES buffer was added, for a final concentration of: 1 μM DPP-5; 50 nM enzyme; and 0, 0.6, 1.2, 3, 6, 12, 30, or 60 μM mitoFP. Fluorescence intensities (λex 490/20 nm, λem 545/20 nm, Gain 80, read from bottom with height 4.5 mm, and sweep method) were measured at 15-sec time intervals for 30 min at 37 °C. Incubations of 12 μM PalmB with 100 nM APT1, and 6 μM PalmB with 100 nM ABHD10 were run in parallel and served as positive controls. The initial velocity is calculated from linear regression of first 20 data points of APT1 and first 30 data points for ABHD10 (n=4) and the one phase exponential decay analysis (Y=Span*exp(−K*X) + Plateau) was done using GraphPad Prism version 6.0e.
Purification of recombinant ABHD10.
BL21 competent E. coli were transformed with both the pGro7 chaperone plasmid and plasmids encoding transit peptide cleaved human ABHD10 (either WT or S152A) or mouse ABHD10. The cells were cultured in 500 mL 2x YT media with 33 μg/ml chloramphenicol, 40 μg/ml kanamycin and 0.5 mg/ml L-arabinose in a 2 L flask shaking at 37 °C until OD600 ≈ 0.6, at which point 1 mM IPTG was added. Cultures were then grown at 25 °C for an additional 20 hr. Bacteria was harvested by centrifugation and lysed by sonication in 25 mL lysis buffer (50 mM Tris, 1 M NaCl, 20% glycerol, 10 mM TCEP, pH 7.5). Cell debris was discarded and the lysis supernatant was incubated with 1 mL Takara His60 Ni Superflow Resin with gentle rotation at 4 °C for 1 hr. The His-tagged proteins were purified using a standard protocol of washing and eluting the resin with wash buffer-1 (20 mM imidazole, 50 mM Tris, 1 M NaCl, 20% glycerol, 10 mM TCEP, pH 7.5), wash buffer-2 (40 mM imidazole, 50 mM Tris, 1 M NaCl, 20% glycerol, 10 mM TCEP, pH 7.5) and elution buffer (300 mM imidazole, 50 mM Tris, 1 M NaCl, 20% glycerol, 10 mM TCEP, pH 7.5). The purified proteins were then desalted on GE Disposable PD-10 Desalting Columns and stored in the protein storage buffer (50 mM Tris, 100 mM NaCl, 2 mM TCEP, 50% Glycerol, pH 7.5). The purity of the elution fractions was validated by 12% SDS–PAGE gel. Purified proteins were stored in individual aliquots in protein storage buffer at −20 °C (for more immediate use) or −80 °C (for long-term storage).
In vitro kinetics of recombinant human and mouse ABHD10.
150 μL of 10 μM DPP-5 in HEPES (20 mM, pH = 7.8, 150 mM NaCl) were added to the 96-well optical bottom plate. 150 μL of either HEPES buffer alone or HEPES buffer containing 1 μM proteins (human, mouse or S152A variant ABHD10, purified as described above) were added using a multi-channel pipette, resulting in a final concentration of 5 μM DPP-5 and 500 nM proteins. Fluorescence intensities (λex 490/20 nm, λem 545/20 nm, Gain 50, read from bottom with height 4.5 mm, and sweep method) were measured at 1 min time intervals for 12 h at 37 °C. The Michaelis–Menten kinetics regression analysis was done using GraphPad Prism version 6.0e.
Crystallization of truncated mouse ABHD10.
Mouse ABHD10 was purified from Ni-resin as described above. Mouse ABHD10 was solubilized in Buffer A (20 mM Tris, pH 8.0) with PD-10 Desalting Columns and loaded onto a Q Sepharose Ion-Exchange column for gradient-elution with Buffer B (1 M NaCl, 20 mM Tris, pH 8.0). Fractions were collected and verified by 12% SDS-PAGE. Pure fractions were combined and loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences). The fractions eluted by SEC Buffer (10 mM Tris, 50 mM NaCl, pH 8.0) were again verified by SDS-PAGE and concentrated to 35 mg/ml. The pure protein was then aliquoted, flash frozen in liquid nitrogen and stored at −80 °C. The crystallization was performed using the hanging drop vapor diffusion technique. The high-throughput screening of crystallization conditions was set up by a Mosquito Crystallization Robot (TTP Labtech) at 4 °C using commercially available screening kits from Hampton Research Corp. The best crystals were obtained from 25 mg/mL proteins mixed with the precipitant solution from Natrix 2 (0.08 M strontium chloride hexahydrate, 0.04 M sodium cacodylate trihydrate pH 6.0, 35% v/v (+/−)-2-methyl-2,4-pentanediol (MPD), 0.012 M spermine tetrahydrochloride). Following optimization of pH at 6.4 and precipitant concentration at 25%, crystals were grown in the 2 μL of 1:1 protein and buffer solution mixture drops hanging on siliconized glass slides, harvested by soaking the cryo buffer (same as precipitant buffer except that the MPD concentration increased to 50%), and flash-frozen in liquid nitrogen.
X-ray diffraction data collection and refinement.
The X-ray diffraction data were collected at the Advanced Photon Source NE-CAT section beamline 24-ID-C at 100K in Argonne National Laboratory (X-ray wavelength 0.97910 Å). All the datasets were then integrated and scaled using its on-site RAPD automated programs (https://rapd.nec.aps.anl.gov/rapd/). The asymmetric unit contained one molecule and the structure was solved by Molecular Replacement (MR). Initial phases were obtained with Phaser51 using a structure predicted by RaptorX (http://raptorx.uchicago.edu/)52, 53, based on template 3LLC.pdb. The structure building and refinement were completed by AutoBuild and phenix.refine in Phenix54 and by manually modeling in the program COOT55 according to weighted |2Fo|-|Fc| and |Fo|-|Fc| maps. Thermal parameters were handled as individual isotropic B-factors with one overall TLS (translation, libration and screw) group. One well-ordered MPD molecule was found within a pocket in the protein, and 4 strontium ions were found at the surface, 3 of them mediating crystal packing. The identity of the strontium ions was confirmed with an anomalous difference Fourier. The Ramachandran favored residues are 98% with 0% outliers.
Assessment of S-palmitoylation levels of PRDX3 and PRDX5 upon ABHD10 knockdown by ABE assay.
HEK293T cells plated in 10 cm dishes were transfected when 30-40% confluent with control siRNA (NT) or ABHD10 siRNA. After 44-48 hrs, cells were lysed and processed as described in the ABE protocol. For quantification, 5 biological replicates in HEK293T cells were used and S-palmitoylation levels of PRDX3 and PRDX5 were normalized by following equation using output signal for corresponding proteins and calnexin from western blots:
S(PRDX-10) = Chemiluminescence Signal of PRDX upon ABHD10 knockdown
S(PRDX-NT) = Chemiluminescence Signal of PRDX in control cells
S(CANX-10) = Chemiluminescence Signal of calnexin upon ABHD 10 knockdown in control cells
S(CANX-NT) = Chemiluminescence Signal of calnexin in control cells
Similar experiments in HEK293T cells were performed in 3 biological replicates to assess S-palmitoylation levels of PRDX3 and PRDX5 upon APT1 knockdown. For assessing S-palmitoylation levels of PRDX3 and PRDX5 in HeLa cells upon ABHD10 knockdown, we took 24 mg total protein as input, and use 211 mg total protein for biotin enrichment and measured percentages of S-palmitoylation as follows:
S(Output) = Chemiluminescence Signal of protein in output sample
S(Input) = Chemiluminescence Signal of protein in input sample
PRDX5 immunoprecipitation ABE assay.
HEK293T cells plated in 10 cm culture dishes were transfected with 10 μg of pcDNA3-PRDX5-Flag WT plasmid and PEI (PEI:DNA=4:1)56 when 30-40% confluent. After 16 hr, the media was removed and transfected with 150 pmol of either ABHD10 or NT siRNA for another 24 hr. Cells were then lysed in SDS-free RIPA lysis buffer and rotated end-to-end for 2 hr at 4°C before insoluble material was removed by centrifugation at 14,000 rpm for 15 min at 4°C. Immunoprecipitation was performed overnight at 4°C with anti-FLAG antibody-incubated protein G magnetic beads (prepared according to the manufacturer’s recommended protocol). Inputs were taken from lysates (10% of the lysate volume) before immunoprecipitation. Beads were washed once with SDS-free RIPA lysis buffer and then incubated with 500 μL SDS-free RIPA lysis buffer containing 50 mM NEM for 2 hr at 25 °C with end-to-end rotation. Beads were washed three times with SDS-free RIPA lysis buffer then equally separated into two new tubes. One tube was incubated with SDS-free RIPA lysis buffer containing 0.8 M Hydroxylamine (pH 7.4), and another incubated with SDS-free RIPA lysis buffer, and rotated end-to-end for 1 hr at room temperature. Beads were washed two times with SDS-free RIPA lysis buffer before incubating with 10 μM EZ-Link HPDP-Biotin for 2 hr at 25 °C with end-to-end rotation. Beads were washed twice and eluted with 50 mM Glycine solution (pH 2.8). The supernatant was neutralized with 1 M Tris before boiling for 10 min at 95 °C with 6X Laemelli sample buffer. The protein was resolved on 12% SDS-PAGE gels and Western Blotting was conducted using the protocol described above.
PRDX5 palmitoylation site identification by ABE.
HEK293T cells plated in 10 cm culture dishes were transfected when 30-40% confluent with 15 μg of either the pcDNA3-PRDX5-Flag WT plasmid (control) or individual plasmids containing each Cysteine-to-Serine point mutant. After 42-48 hr, cells were washed with DPBS, lysed, and processed as described above for the ABE protocol. Input and output samples for C100S are taken three times more than corresponding samples for WT, C125S and C204S.
PRDX5 palmitoylation site identification by metabolic labeling.
HEK293T cells plated in 10 cm culture dishes were transfected when 30-40% confluent with 15 μg of either the pcDNA3-PRDX5-Flag WT or C100S mutant plasmid. After 44 hr, cells were metabolically labeled with 50 μM 17-ODYA for 3h, and then processed using the metabolic labeling protocol described above.
Cell viability experiment.
For the H2O2 experiment, HEK293T or HeLa cells transfected with NT siRNA or ABHD10 siRNA for 24 hr were replated into a 96-well plate. After 24 hr, the media was replaced with 200 μL pyruvate-free DMEM GlutaMAX (10% FBS) containing either H2O or 25 or 50 μM H2O2. After 36 hr incubation at 37°C, the media was replaced with 200 μL 1x MTS reagent (10x PBS stock solution containing 0.6 mM MTS and 0.033 mM Phenazine methosulfate, pH 5.2-5.4)57 in DMEM GlutaMAX, and incubated at 37 °C for 1-2 hr. The plate was then gently shaken and absorbance was measured at 490 nm using a plate reader. Cells treated with NT or ABHD10 siRNA and no H2O2 was set to 100% viability. Each group of cells with various concentrations of H2O2 is normalized to respective cells treated with H2O as follows:
A(c) = Average absorbance at concentration c
A(0) = Average absorbance at concentration 0
For the H2O2 experiment with PRDX5 knockdown, HEK293T or HeLa cells were transfected with NT siRNA, PRDX5 siRNA, ABHD10 siRNA or co-transfected with ABHD10 siRNA and PRDX5 siRNA (For the non-co-transfected group, NT siRNA was added to make the total amount of siRNA consistent between groups) for 24 h (for the HeLa cells, we replaced the media without siRNA after 12 h in order to decrease toxicity stemming from the transfection reagent), and then were replated into a 96-well plate. The rest of protocol is the same as described above.
For the paraquat experiment, HEK293T cells transfected with NT siRNA or ABHD10 siRNA for 24 h were replated into a 96-well plate. After 24 hr, the media was replaced with 200 μL pyruvate-free DMEM GlutaMAX (10% FBS) containing either H2O or 50, 100, or 200 μM aqueous paraquat. After 12 h at 37 °C, the media was replaced with 200 μL of pyruvate-free DMEM GlutaMAX (10% FBS). 24 h later, the media was replaced with 200 μL 1x MTS reagent (10x PBS stock solution containing 0.6 mM MTS and 0.033 mM Phenazine methosulfate, pH 5.2-5.4) in DMEM GlutaMAX, and incubated at 37 °C for 1 h. The plate was then gently shaken, and absorbance was measured at 490 nm using a plate reader. Quantification was performed as described above.
Synthetic Methods
Synthesis of 1.
(4-Iodobutyl)triphenylphosphonium58 (1.2 g, 1.0 eq, 2.10 mmol) was dissolved in excess triethyl phosphite (1.80 mL, 5.0 eq, 10.5 mmol) and heated at 155 °C for 4 hr. Excess triethyl phosphite was removed at 80 °C under vacuum to yield 1 as a colorless oil (0.79 g), which was used without further purification.
Synthesis of mitoFP (3).
Compound 1 (0.79 g, 1.0 eq, 1.36 mmol) was dissolved in DCM (30 mL) and cooled to 0 °C. TMSBr (1.10 mL, 6.0 eq, 8.33 mmol) was added and the reaction was stirred first for 30 min at 0 °C and then for 2 hr at room temperature. Then 15 mL water was added to quench the reaction. The aqueous phase was separated, and an additional 10 mL water was used to extract the product from the organic phase. The combined aqueous phase was washed one time with 15 mL DCM. The aqueous solution was azeotroped with toluene (3x) to yield 2 as a yellow oil, which was used without further purification. Compound 2 was dissolved in 25 mL dry DCM, cooled to 0 °C, followed by dropwise addition of DAST (2.1 mL, 11.7 eq, 15.9 mmol). The reaction was first stirred at 0 °C for 15 min, and then at room temperature for another 15 min to generate the difluoro intermediate. Octanol (5 mL in total) was added dropwise at 45 °C until no difluoro intermediate was detected by LC-MS. The reaction was quenched with water and extracted with DCM. The organic layer was washed with water and then brine. The organic layer was then dried over Na2SO4, and then concentrated and purified by column chromatography (Silica; 0–5% MeOH in 1:1 DCM:EtOAc) to yield mitoFP (3) (0.264 g, 36%). Rf: 0.45 (Silica; 10% MeOH in 1:1 DCM:EtOAc). Purity of mitoFP was ascertained by LCMS (Supplementary Fig. 40) and NMR (Supplementary Fig. 41) 1H-NMR (500 MHz; CDCl3): δ 7.82–7.68 (m, 15H), 4.11 (q, J = 7Hz, 2H), 3.54–3.48 (m, 2H), 2.07–1.99 (m, 4H), 1.88–1.82 (m, 2H), 1.66–1.62 (m, 2H), 1.31–1.25 (br, 10H), 0.86 (t, J = 6.8 Hz, 3H). 13C-NMR (126 MHz; CDCl3): δ 135.4, 135.3, 133.7, 133.6, 130.8, 130.7, 118.3, 117.6, 67.6, 67.5, 31.8, 30.4, 30.3, 29.2, 29.1, 25.3, 22.7, 14.2. HRA-MS(+): Calculated for C30H40FO2P2 [M+] 513.2482; found 513.2587.
Data availability
All structural data has been deposited in the PDB (PDB ID: 6NY9). Additional data supporting the findings of this manuscript are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This work was supported by the University of Chicago, the National Institute of General Medical Sciences of the National Institutes of Health (R35 GM119840) to B.C.D. and a “Catalyst Award” to B.C.D. from the Chicago Biomedical Consortium, with support from the Searle Funds at The Chicago Community Trust. The crystallographic work is based on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamline, 24-ID-C, which is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403) from the National Institutes of Health. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We would like to thank the staff of the Advanced Photon Source at Argonne National Laboratory for providing technical advice during data collection, Dr. Lulu Hu (University of Chicago) for providing advice on crystal growth, Dr. Deepak Koirala (University of Chicago) for assistance with X-ray diffraction data collection, Dr. Yaming Shao (University of Chicago) for advice on structure refinement, and Dr. Somayeh Ahmadiantehrani (University of Chicago) for assistance proofing the manuscript.
Footnotes
Competing interests
B.C.D. and R.S.K. have a patent (US20180147250A1) on the DPPs.
References
- 1.Blanc M et al. SwissPalm: Protein Palmitoylation database. F1000Res 4, 261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanyon-Hogg T, Faronato M, Serwa RA & Tate EW Dynamic Protein Acylation: New Substrates, Mechanisms, and Drug Targets. Trends Biochem Sci 42, 566–581 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Martin BR, Wang C, Adibekian A, Tully SE & Cravatt BF Global profiling of dynamic protein palmitoylation. Nat Methods 9, 84–89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker FJ et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol 6, 449–456 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Brownlee C & Heald R Importin alpha Partitioning to the Plasma Membrane Regulates Intracellular Scaling. Cell 176, 805–815 e808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan P et al. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol 12, 282–289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature 549, 399–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko PJ & Dixon SJ Protein palmitoylation and cancer. EMBO Rep 19, e46666 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zareba-Koziol M, Figiel I, Bartkowiak-Kaczmarek A & Wlodarczyk J Insights Into Protein S-Palmitoylation in Synaptic Plasticity and Neurological Disorders: Potential and Limitations of Methods for Detection and Analysis. Front Mol Neurosci 11, 175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobocinska J, Roszczenko-Jasinska P, Ciesielska A & Kwiatkowska K Protein Palmitoylation and Its Role in Bacterial and Viral Infections. Front Immunol 8, 2003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb CD & Linder ME Structure and function of DHHC protein S-acyltransferases. Biochem Soc Trans 45, 923–928 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Duncan JA & Gilman AG A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J Biol Chem 273, 15830–15837 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Lin DT & Conibear E ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife 4, e11306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long JZ & Cravatt BF The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev 111, 6022–6063 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoi N et al. Identification of PSD-95 Depalmitoylating Enzymes. J Neurosci 36, 6431–6444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostiuk MA et al. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J 22, 721–732 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang M, Lu L, Huang Z & Chen L Palmitoylation signaling: a novel mechanism of mitochondria dynamics and diverse pathologies. Acta Biochim Biophys Sin (Shanghai) 50, 831–833 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Maynard TM et al. Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes. Mol Cell Neurosci 39, 439–451 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen LF et al. Role of S-Palmitoylation by ZDHHC13 in Mitochondrial function and Metabolism in Liver. Sci Rep 7, 2182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathayat RS et al. Active and dynamic mitochondrial S-depalmitoylation revealed by targeted fluorescent probes. Nat Commun 9, 334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathayat RS & Dickinson BC Measuring S-Depalmitoylation Activity In Vitro and In Live Cells with Fluorescent Probes. Methods Mol Biol 2009, 99–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knoops B, Goemaere J, Van der Eecken V & Declercq JP Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid Redox Signal 15, 817–829 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Frohlich M, Dejanovic B, Kashkar H, Schwarz G & Nussberger S S-palmitoylation represents a novel mechanism regulating the mitochondrial targeting of BAX and initiation of apoptosis. Cell Death Dis 5, e1057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zorov DB, Juhaszova M & Sollott SJ Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev 94, 909–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thinon E, Fernandez JP, Molina H & Hang HC Selective Enrichment and Direct Analysis of Protein S-Palmitoylation Sites. J Proteome Res 17, 1907–1922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickinson BC & Chang CJ A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J Am Chem Soc 130, 9638–9639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee SG & Kil IS Multiple Functions and Regulation of Mammalian Peroxiredoxins. Annu Rev Biochem 86, 749–775 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Wan J, Roth AF, Bailey AO & Davis NG Palmitoylated proteins: purification and identification. Nat Protoc 2, 1573–1584 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Martin BR & Cravatt BF Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods 6, 135–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kathayat RS, Elvira PD & Dickinson BC A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling. Nat Chem Biol 13, 150–152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zielonka J et al. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem Rev 117, 10043–10120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu T, Kathayat RS, Cao Y, Beck MW & Dickinson BC A Fluorescent Probe with Improved Water Solubility Permits the Analysis of Protein S-Depalmitoylation Activity in Live Cells. Biochemistry 57, 221–225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adibekian A et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc 134, 10345–10348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won SJ et al. Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem Biol 11, 3374–3382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee HW et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito Y, Fukami T, Yokoi T & Nakajima M An orphan esterase ABHD10 modulates probenecid acyl glucuronidation in human liver. Drug Metab Dispos 42, 2109–2116 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Iwamura A, Fukami T, Higuchi R, Nakajima M & Yokoi T Human alpha/beta hydrolase domain containing 10 (ABHD10) is responsible enzyme for deglucuronidation of mycophenolic acid acyl-glucuronide in liver. J Biol Chem 287, 9240–9249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickinson BC, Huynh C & Chang CJ A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J Am Chem Soc 132, 5906–5915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devedjiev Y, Dauter Z, Kuznetsov SR, Jones TL & Derewenda ZS Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure 8, 1137–1146 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Castello PR, Drechsel DA & Patel M Mitochondria are a major source of paraquat-induced reactive oxygen species production in the brain. J Biol Chem 282, 14186–14193 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald C, Muhlbauer J, Perlmutter G, Taparra K & Phelan SA Peroxiredoxin proteins protect MCF-7 breast cancer cells from doxorubicin-induced toxicity. Int J Oncol 45, 219–226 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto H, Hayashi H & Yamashita S Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J Biol Chem 271, 7705–7711 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Sadeghi RS et al. Wnt5a signaling induced phosphorylation increases APT1 activity and promotes melanoma metastatic behavior. Elife 7, e34362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amara N, Foe IT, Onguka O, Garland M & Bogyo M Synthetic Fluorogenic Peptides Reveal Dynamic Substrate Specificity of Depalmitoylases. Cell Chem Biol 26, 35–47 e37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuhl AM et al. Competitive activity-based protein profiling identifies aza-beta-lactams as a versatile chemotype for serine hydrolase inhibition. J Am Chem Soc 134, 5068–5071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parvez S, Long MJC, Poganik JR & Aye Y Redox Signaling by Reactive Electrophiles and Oxidants. Chem Rev 118, 8798–8888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corvi MM, Soltys CL & Berthiaume LG Regulation of mitochondrial carbamoyl-phosphate synthetase 1 activity by active site fatty acylation. J Biol Chem 276, 45704–45712 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Garland M et al. Development of an activity-based probe for acyl-protein thioesterases. PLoS One 13, e0190255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Patricelli MP & Cravatt BF Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A 96, 14694–14699 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogasawara D et al. Selective blockade of the lyso-PS lipase ABHD12 stimulates immune responses in vivo. Nat Chem Biol 14, 1099–1108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng J & Xu J RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins 79 Suppl 10, 161–171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kallberg M et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu CY & Uludag H A simple and rapid nonviral approach to efficiently transfect primary tissue-derived cells using polyethylenimine. Nat Protoc 7, 935–945 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Buttke TM, McCubrey JA & Owen TC Use of an aqueous soluble tetrazolium/formazan assay to measure viability and proliferation of lymphokine-dependent cell lines. J Immunol Methods 157, 233–240 (1993). [DOI] [PubMed] [Google Scholar]
- 58.Lin TK et al. Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. J Biol Chem 277, 17048–17056 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All structural data has been deposited in the PDB (PDB ID: 6NY9). Additional data supporting the findings of this manuscript are available from the corresponding author upon reasonable request.