Abstract
Individuals with a constitutional chromosome abnormality consisting of a deletion of a portion of the long arm of chromosome 18 (18q−) have a high incidence (∼95%) of dysmyelination. Neuroradiologic findings in affected children report a smaller corpus callosum, but this finding has not been quantified. This is in part due to the large intersubject variability of the corpus callosum size and shape and the small number of subjects with 18q−, which leads to low statistical power for comparison with typically developing children. An analysis method called targetless spatial normalization (TSN) was used to improve the sensitivity of statistical testing. TSN converges all images in a group into what is referred as group common space. The group common space conserves common shape, size, and orientation while reducing intragroup variability. TSN in conjunction with a Witelson vertical partitioning scheme was used to assess differences in corpus callosum size between 12 children with 18q− and 12 age‐matched normal controls. Significant global and regional differences in corpus callosum size were seen. The 18q− group showed an overall smaller (25%) corpus callosum (P < 10−7), even after correction for differences in brain size. Regionally, the posterior portions of corpus callosum (posterior midbody, isthmus, and splenium), which contain heavily myelinated fibers, were found to be 25% smaller in the population with 18q−. Hum Brain Mapping 24:325–331, 2005. © 2005 Wiley‐Liss, Inc.
INTRODUCTION
Caused by a partial deletion of the long arm (q arm) of chromosome 18, 18q− is a rare (∼1/40,000 live births) disorder. Affected individuals have a broad spectrum of phenotypic findings [Cody et al.,1999] including dysmyelination of central nervous system (CNS) and mental retardation [Felding et al.,1987; Kline et al.,1993; Miller et al.,1990; Rodichock et al.,1992; Vogel et al.,1990]. Although 95% of all individuals with an 18q deletion have dysmyelination, 100% of the individuals with a deletion of a specific 2‐megabase region of 18q have dysmyelination. This region of the chromosome contains seven known genes and several hypothetical genes. One of the known genes is myelin basic protein (MBP), a logical candidate gene associated with dysmyelination, and it is hypothesized that the abnormal CNS myelination is the result of its reduced gene‐copy number (one copy instead of two) [Brkanac et al.,1998; Gay et al.,1997; Ono et al.,1994; Strathdee et al.,1995]. MBPs are required for normal myelination because they play an essential role in stabilization of the fusion of cytoplasmic membrane leaflets [Kamholz et al.,1987]. The primary goal of this study was to investigate if CNS dysmyelination due to the absence of one copy of the region containing the MBP gene affects the normally heavily myelinated regions of corpus callosum.
The corpus callosum (CC) is the largest of cortical commissure systems. Diffuse or focal abnormalities of bilaterally connected cerebral structures may produce secondary effects in corpus callosum, observed as global size changes, regional size changes, or both. For years, analysis of regional CC neuroanatomy has been a key factor in radiologic assessment of a wide variety of degenerative, neurologic, psychiatric, and developmental disorders [reviewed in Thompson et al.,2002]. Most CC studies were carried out using regions of interest defined by vertical partitioning of a midsagittal section of CC [Thompson et al.,2002], such as the Witelson partitioning (WP) scheme [Witelson,1989]. This approach was originated in cadaver studies and is based on topographic distribution of CC fibers [Aboitiz et al.,1992,1996].
The WP scheme defines five callosal regions based on dividing the CC along its anterior–posterior dimension. The WP scheme is based on a general rule: that the anterior third of the CC contains fibers connecting the bilateral prefrontal cortices; the midbody (middle third) primarily contains crossing fibers for motor, somatosensory, and auditory cortices; the splenium (posterior fifth) carries fibers mainly to temporal, parietal, and occipital lobes; and the isthmus (in between the midbody and splenium) is thought to carry fibers connecting perisylvian regions [Witelson,1989].
Manual WP measurements are easy to carry out, but suffer from the intrinsically large variability in CC shape and size. Many subjects (∼100) are therefore often needed to detect small (<10%) statistically significant (P < 0.05) changes [Thompson et al.,2002]. Due to the limited number of subjects in our study (<20 per group), we opted to use targetless spatial normalization (TSN), which can sometimes detect small anatomic changes in much smaller groups with statistical precision similar to the large group studies [Lancaster et al.,2003]. TSN was developed recently [Kochunov et al.,2001; Woods et al.,1998; Woods,2003] to build population‐optimized target templates by incorporating global and regional features that are common to all the images. Woods et al. [1998] implemented an algorithm for global TSN (gTSN), which is distributed as part of the automated image registration (AIR) package [Woods et al.,1998; Woods,2003]. Regional TSN (rTSN) was introduced in Kochunov et al. [2001]. Similar to gTSN, rTSN transforms images into group space, preserving common features for each group while further reducing within‐group variability [Kochunov et al.,2001] by the use of a high degree‐of‐freedom warping algorithm.
SUBJECTS AND METHODS
Subjects
Anatomic magnetic resonance imaging (MRI) studies for 12 children with 18q− (8 females, 4 males) and 12 normally developing children (8 females and 4 males) were acquired on the 1.9‐T Elscient/GE scanner. A T1‐weighted, 3‐D, gradient recalled echo (repetition time [TR] = 35 ms; echo time [TE] = 7 s; flip angle = 60 degrees) MRI sequence was used. Three patients and four controls were imaged in axial plane with 1 × 1 mm in‐plane resolution and slice thickness of 1.2 mm, four patients and three controls were imaged in sagittal plane with an 1 × 1 mm in‐plane resolution and slice thickness of 1.2 mm, and the rest of the images were acquired in sagittal plane with an isotropic 1 mm3 resolution.
The average age for children in the 18q− group was 8.3 ± 2.3 years (age range, 4.1–12.3 years). The average age for children in the normal control (NC) group was 9.9 ± 2.6 years (age range, 4.5–12.7 years). Three NC subjects were unaffected siblings of two 18q− patients. All children with 18q− had a confirmed deletion of the chromosomal region containing the MBP gene. All 18q− anatomic MR studies were examined by two pediatric neuroradiologists who reported finding of CNS dysmyelination in every subject [Gay et al.,1997].
Preprocessing
Preprocessing (Fig. 1A–C) prepared images for TSN and included the following steps: brain segmentation; registration to the Talairach frame [Talairach and Tournoux,1988]; manual segmentation of the corpus callosum; and collective alignment of CC images.
Figure 1.
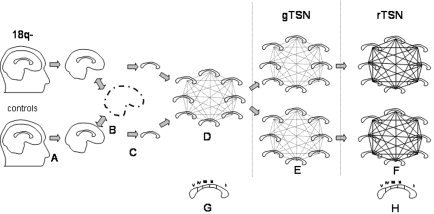
TSN processing stream. Preprocessing: A: Brain segmentation. B: Brain normalization to Talairach space. C: Segmentation of CC. D: Collective alignment of images. TSN processing consisted of gTSN (E) and rTSN (F) processing. G: WP measurements before TSN processing. H: WP measurements after TSN processing.
Brain segmentation
The non‐brain brain tissues such as skin, muscle, and fat were removed using an automated skull stripping procedure called BET [Smith,2002], which is provided as an add‐on for MEDx (Sensor Systems; Fig. 1A). Segmented brain images were cropped at the level of the brain stem.
Registration to Talairach Frame
All brain images were globally spatially normalized to the Talairach coordinate system to remove gross differences in brain size, position, and orientation (Fig. 1B). Automated, nine‐parameter (three rotations, three scales, and three translations) Convex Hull global spatial normalization software [Lancaster et al.,1999] was used to register all images into the Talairach reference frame. The midline and anterior commissure–posterior commissure (AC–PC) alignments were verified manually by two experienced neuroanatomists. All images were resliced at isotropic 0.5‐mm3 resolution using a sinc interpolation kernel.
Segmentation of the CC
An 8 mm‐thick median band of CC was segmented manually from the 0.5‐mm3 images (Fig. 1C). Two skilled neuroanatomists carried out CC segmentation using an interactive 3D painting/segmentation tool called MNI‐Display [MacDonald,1996]. The 16 medial sagittal slices of median CC were segmented. Axial and coronal views of segmented CC slices were used to delineate better the medial CC/lateral ventricle borders and the inferior CC/fornix division.
Collective alignment of CC images
During collective alignment (Fig. 1D), CC images from both groups were transformed to consistent orientation and position. A six‐parameter (three translations and three rotations) linear affine transformation version of the AIR software [Woods et al.,1998] was used to compute all pair‐wise transformation matrices between all the images in this study. The software application reconcile_air (part of the AIR distribution) was used to transform each image into a collectively consistent position and orientation.
TSN Processing Stream
The TSN processing stream transformed NC and 18q− subject groups into their common space (illustrated in Fig. 1E,F). It consisted of a gTSN step followed by an rTSN step.
Global TSN (gTSN)
This process reduces the variability in orientation, position, and size of individual CC structures by transforming them into a common global reference frame that conserves the average size and shape of the group [Woods et al.,1998; Woods,2003]. During this step, the 18q− and control groups were processed separately (Fig. 1E). The goal was to separately reduce variability in size, orientation, and position of CC images within each of the two groups. A nine‐parameter (three translations, three rotations, and three scales) model of AIR and reconcile_air were used to transform each group CC images.
The transform matrices obtained from the steps D and E in Figure 1 were concatenated to a single transform for collective alignment for gTSN to interpolate only once. This transform was applied to the original CC images using a nearest neighbor interpolation followed by 33 median filtering to remove edge artifacts.
Regional TSN (rTSN)
The rTSN processing transforms all CC images in a group toward a common shape by iteratively warping each image to the middle of the group's space (Fig. 1F). Similar to the gTSN step, the rTSN processing carries out all pair‐wise transformations and calculates all pair‐wise transformations for each image that are stored as a high degree‐of‐freedom deformation field (DF). Next, for each image, a DF that transforms each voxel to the middle of the group's space was calculated by averaging the DFs. The rTSN is an iterative process where processing is repeated until the average deformation per voxel (DPV) is reduced to 0.5 mm/voxel.
Standard Error of the Mean Estimation
Because the relationship between group sizes and standard deviation (SD) for a group is not well characterized for TSN, the standard error of the mean (SEM) had to be estimated. To estimate SEM for WP volumes after TSN processing, subjects in both the 18q− and control groups were subdivided into several smaller groups where the male‐to‐female ratio and mean age of each group was maintained as close to the original as possible. The control group of 12 subjects was subdivided into four groups of 3, three groups of 4, and two groups of 6. No other reasonable subgroupings were possible with reasonable matching of gender and age. The SEM was determined for each of these group sizes for each of WP regions and for the total CC. A power curve was fitted to SEM versus group size for each WP region and used to estimate the SEM for the full group size (n = 12). The 18q− group was processed in a similar manner. To remove global effects in the regional WP analysis of CC, the total volume in the 18q− group was adjusted be equal to that in the control group.
Computation
TSN processing was executed on a cluster of Linux computers, consisting of 20 nodes managed by the Sun ONE cluster grid engine. Each cluster node is an Intel Pentium 4, 2.4‐GHz workstation with 1 GB of RAM, running the RedHat 8.0 Linux operation system. The Sun ONE Grid engine (online at http://www.sun.com) is a scheduling and load‐balancing system that spreads the processing among nodes in the cluster. Distributing TSN processing over multiple workstations reduced computation time by roughly a factor equal to the number of nodes in the cluster. The three iterations of rTSN processing for both groups took less than 4 hr.
RESULTS
TSN Processing
After collective alignment and the gTSN steps, the CC images were in a consistent location and orientation, and after the rTSN step, intrinsic individual regional variability in CC shapes was reduced further. This reduction r is illustrated in Figure 2, where the midsagittal slices of the intragroup mean and SD images are shown after gTSN and rTSN steps for both groups. Figure 2 also shows that the principal outline for the average image was not changed by rTSN processing for either group, whereas the SD images show reduction in variability, especially in the isthmus area of the 18q− group.
Figure 2.

Mean and standard deviation images for 18q− and control groups after gTSN and rTSN showed marked reduction of within‐group regional variability after the rTSN step.
Volumetric Analysis of CC
SEM extrapolation
All power curves fit these data well with R2 values more than 0.85. Although there was appreciable variation in SEM values for different regions, all had decreasing SEMs as a function of group size, as was anticipated. Several other model curves (polynomial, exponential, and log) were tested but did not fit as well as the power curve model did. The power for the total CC volume was −1.73, nearing the theoretical value (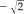 ) often used to account for group size effect.
) often used to account for group size effect.
Total CC volume difference
The comparison of CC volumes before TSN processing (Fig. 1G) showed that the average CC volumes in the 18q− group were about 25% smaller than that in the control group and this difference was statistically significant (P < 10−3). This is an important finding because the difference in brain size was accounted for already by Talairach spatial normalization (Fig. 1C). After TSN processing, the overall intergroup global difference in CC volumes remained at 25%, but became even more statistically significant (P < 10−7) due to lower within‐group variability.
Regional CC volume difference
To estimate regional intergroup differences, the average total CC volumes were adjusted to be the same for both groups. Measurements of WP regional volumes carried out before TSN processing, which corresponds to the classic WP analysis (Fig. 1G) are shown in Figure 3A. No significant intergroup difference for any of the WP regions was discovered (Fig. 3A), although volume differences for isthmus and splenium came close to reaching statistical significance (two‐tailed t test, P ∼ 0.07).
Figure 3.
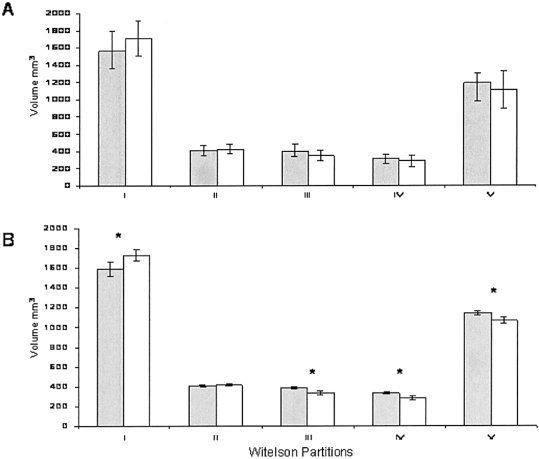
Average volume for WP regions after rTSN corrected for global volume: A: Classic WP measurements. B: WP measurements after TSN processing. *Statistically significant difference between groups (P < 0.05). Gray bars, controls; white bars, 18q.
After TSN (Fig. 1H) processing, significant intergroup differences in the volumes of WP regions were detected. WP region I (genu) was significantly larger for the 18q− group, whereas regions III, IV, and V were significantly smaller than corresponding regions in the control group (P < 0.05) (Fig. 3B). This finding indicates that there are regional CC differences associated with 18q−, with posterior midbody, isthmus and splenium being smaller in children with 18q−.
DISCUSSION
Significant global and regional differences in CC volume were observed between 18q− and NC groups. Global CC volumes were ∼25% smaller in the 18q− group, and this difference was highly significant (P < 10−7). One possible explanation for the smaller global CC volumes in 18q− children is reduced myelin levels resulting from haploinsufficiency of the MBP gene. All 18q− children in this study were haploinsufficient for the MBP gene. MBP comprises about 30% of the protein in the cerebral white matter and is required for normal myelination. Postmortem autopsies and quantitative MRI measurements indicate that myelin production in participants with 18q− does not fully support the myelination demands, resulting in reduced myelin levels [Felding et al.,1987; Lancaster et al.,2004; Vogel at al.,1990]. Other factors, such as fewer axonal fibers might lead to a smaller CC, but autopsies in18q− patients presented no evidence to support this.
Volumetric intergroup regional differences are also consistent with the hypothesis that reduced levels of myelin in 18q− led to smaller CC volumes. Although the CC is composed of many myelinated tracks, its regional composition with respect to fiber myelination is not uniform.
The largest intergroup regional differences were observed in areas III and IV (posterior midbody and isthmus). Here, the regional CC volumes for 18q− group were about 16% smaller than that for the NC group (P < 0.05), even after normalization for global CC size. Regions III and IV have the highest regional density of heavily myelinated fibers (diameter >3 micron), and thus would be most affected by reduced myelination capacity [Aboitiz et al.,1992,1996; Rakic and Yakovlev,1968]. Region V (splenium) is composed of a mixture of fiber types and it was intermediately smaller in the 18q− group. Region I (genu) was relatively larger in the 18q− group after normalization for global CC size. This region is composed mostly of thinly myelinated fibers (diameter <1 micron) and thus seemed less affected by lower myelin levels. Normalization for global CC size scaled and made genu volumes relatively bigger for children with 18q−, whereas regions III and IV remained smaller, thus emphasizing the difference between thinly and heavily myelinated regions of CC. The interhemispheric commissure tracks for motor and sensory information decussate in the regions III and IV, and reduced CC volumes in these regions might have a physiologic manifestation. Lower myelin content of such commissure tracks in children with 18q− is believed to be cause of longer interhemispheric conduction times for motor and sensory tracks, as reported in Lancaster et al. [2004].
Methods and Limitations
To estimate variance for TSN processing, SEMs were extrapolated through power curve analysis. This processing can be thought of as a permutation or a random analysis. A full‐scale permutation analysis was not practical due to restrictions on group subdivisions imposed by the need to control for external variables such as age and gender. These factors influence the regional morphology of CC [Allen et al.,1991; Bookstein,1997; Davatzikos et al.,1996; Witelson,1989,1991] and do not allow for completely unconstrained (i.e., random) subdivision of subjects into groups as required by permutation analysis.
We consider the difference in the average age between 18q− and NC groups as a potential problem for this analysis. The average age for the NC group was older than was that for the 18q− group (ageNC = 9.9 ± 2.6 years vs. age18q− = 8.3 ± 2.3 years). Although the age difference was not highly statistically significant (two‐tailed t test, P < 0.07), a question arises as to whether the regional finding between groups was influenced partially by difference in age‐related maturation of the CC in the two groups. To investigate this, linear regression analysis was carried out to study age‐related trends in CC volume. A positive but not statistically significant correlation of global CC size in Talairach frame with subject age was observed. The correlation coefficients were similar for both NC and 18q− groups (r NC = 0.25 and r18q− = 0.23), and neither reached statistical significance (P ∼ 0.45). The groups also had similar slopes (S) for the global CC volume versus age trends (SNC = 18.1 ± 20.0 mm3/year vs. S18q− = 15.3 ± 22.1 mm3/year) but there was a significant difference (P < 0.05) in the intercept (I) values (INC = 17,120 ± 1,630 mm3 vs. I18q− = 13,643 ± 1,937mm3). It seems that the age difference between NC and 18q− group should have a minimal effect on the outcome of the regional comparison. The CC maturation trends of 15.3 and 18.1 mm3/year would predict that the global CC size effect from the intergroup age difference of 1.6 years to be about 30 mm3. The regional effects due to age differences are estimated to range from 5 to 15 mm3 for the genu, posterior midbody, isthmus, and splenium volumes. This predicted change in volumes is much smaller than are the regional differences observed between two groups (∼100 mm3).
REFERENCES
- Aboitiz F, Scheibel AB, Fisher RS, Zaidel E (1992): Fiber composition of the human corpus callosum. Brain Res 598: 143–153. [DOI] [PubMed] [Google Scholar]
- Aboitiz F, Rodriguez E, Olivares R, Zaidel E (1996): Age‐related changes in fibre composition of the human corpus callosum: sex differences. Neuroreport 7: 1761–1764. [DOI] [PubMed] [Google Scholar]
- Allen LS, Richey MF, Chai YM, Gorski RA (1991): Sex differences in the corpus callosum of the living human being. J Neurosci 11: 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein FL (1997): Landmark methods for forms without landmarks: morphometrics of group differences in outline shape. Med Image Anal 1: 225–243. [DOI] [PubMed] [Google Scholar]
- Brkanac Z, Cody JD, Leach RJ, DuPont BR (1998): Identification of cryptic rearrangements in patients with 18q− deletion syndrome. Am J Hum Genet 62: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody JD, Ghidoni PD, DuPont B, Hale DE, Hilsenbeck SG, Stratton RF, Hoffman DS, Muller S, Schaub RL, Leach RJ, Kaye CI (1999): Congenital anomalies and anthropometry of 42 individuals with deletions of 18q. Am J Med Genet 85: 455–462. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Vaillant M, Resnick SM, Prince JL, Letovsky S, Bryan RN (1996): A computerized approach for morphological analysis of the corpus callosum. J Comput Assist Tomogr 20: 88–97. [DOI] [PubMed] [Google Scholar]
- Evans A, Collins L, Mills S, Brown E, Kelly L, Peters T (1993): 3D statistical neuroanatomical models from 305 MRI volumes. IEEE Nuclear Science Symposium and Medical Imaging Conference, San Francisco. p 1813–1817.
- Felding I, Kristoffersson U, Sjostrom H, Noren O (1987): Contribution to the 18q− syndrome. A patient with del(18)(q22.3qter). Clin Genet 31: 206–210. [DOI] [PubMed] [Google Scholar]
- Gay CT, Hardies LJ, Rauch RA, Lancaster JL, Plaetke R, DuPont BR, Cody JD, Cornell JE, Herndon RC, Ghidoni PD, Schiff JM, Kaye CI, Leach RJ, Fox PT (1997): Magnetic resonance imaging demonstrates incomplete myelination in the 18q− syndrome: evidence for myelin basic protein haploinsufficiency. Am J Med Genet 74: 422–431. [DOI] [PubMed] [Google Scholar]
- Kamholz J, Spielman R, Gogolin K, Modi W, O'Brien S, Lazzarini R (1987): The human myelin‐basic‐protein gene: chromosomal localization and RFLP analysis. Am J Hum Genet 40: 365–373. [PMC free article] [PubMed] [Google Scholar]
- Kline AD, White ME, Wapner R, Rojas K, Biesecker LG, Kamholz J, Zackai EH, Muenke M, Scott CI Jr, Overhauser J (1993): Molecular analysis of the 18q− syndrome and correlation with phenotype. Am J Hum Genet 52: 895–906. [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson P, Woods R, Mazziotta J, Hardies J, Fox P (2001): Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr 25: 805–816. [DOI] [PubMed] [Google Scholar]
- Lancaster J, Cody J, Andrews T, Hardies J, Hale D, Fox P (in press): Myelination in children with partial deletions of chromosome 18q. Am J Neuroradiol. [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Fox PT, Downs H, Nickerson DS, Hander TA, El Mallah M, Kochunov PV, Zamarripa F (1999): Global spatial normalization of human brain using convex hulls. J Nucl Med 40: 942–955. [PubMed] [Google Scholar]
- Lancaster J, Kochunov P, Thompson P, Fox P (2003): Asymmetry of the brain surface from deformation field analysis. Hum Brain Mapp 19: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mowrey PN, Hopper KD, Frankel CA, Ladda RL (1990): Neurologic manifestations in 18q− syndrome. Am J Hum Genet 37: 128–132. [DOI] [PubMed] [Google Scholar]
- MacDonald D (1996): MNI‐Display: program for display and segmentation of surfaces and volumes (http://www.bic.mni.mcgill.ca/software/Display/Display.html). Technical report. Montreal: McConnell Brain Imaging Center, Montréal Neurological Institute, McGill University. [Google Scholar]
- Ono J, Harada K, Yamamoto T, Onoe S, Okada S (1994): Delayed myelination in a patient with 18q− syndrome. Pediatr Neurol 11: 64–67. [DOI] [PubMed] [Google Scholar]
- Rakic P, Yakovlev PI (1968): Development of the corpus callosum and cavum septi in man. J Comp Neurol 26: 100–104. [DOI] [PubMed] [Google Scholar]
- Rodichok L, Miller G (1992): A study of evoked potentials in the 18q− syndrome which includes the absence of the gene locus for myelin basic protein. Neuropediatrics 23: 218–220. [DOI] [PubMed] [Google Scholar]
- Smith SM (2002): Fast robust automated brain extraction. Hum Brain Mapp 17: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee G, Zachai EH, Shapiro R, Kamholz J, Overhauser J (1995): Analysis of clinical variation seen in patients with 18q terminal deletions. Am J Med Genet 59: 476–483. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P (1988): Co‐planar stereotaxic atlas of the human brain. New York: Thieme. [Google Scholar]
- Thompson PM, Narr KL, Blanton RE, Toga AW (2002): Mapping structural alterations of the corpus callosum during brain development and degeneration In: Zaidel E, Iacoboni M, editors. The parallel brain: the cognitive neuroscience of the corpus callosum. Cambridge, MA: MIT Press; p 93–130. [Google Scholar]
- Vogel H, Urich H, Horoupian DS, Wertelecki W (1990): The brain in the 18q− syndrome. Dev Med Child Neurol 32: 732–737. [DOI] [PubMed] [Google Scholar]
- Witelson SF (1989): Hand and sex differences in the isthmus and genu of the human corpus callosum. A postmortem morphological study. Brain 112: 799–835. [DOI] [PubMed] [Google Scholar]
- Witelson SF (1991): Sex differences in neuroanatomical changes with aging. N Engl J Med 325: 211–212. [DOI] [PubMed] [Google Scholar]
- Woods RP (2003): Characterizing volume and surface deformations in an atlas framework: theory, applications, and implementation. Neuroimage 18: 769–788. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC (1998): Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Assist Tomogr 22: 153–165. [DOI] [PubMed] [Google Scholar]


