Abstract
Products derived from bacterial members of the gut microbiota evoke immune signaling pathways from the host that promote immunity and barrier function in the intestine. How immune reactions to enteric viruses support intestinal homeostasis is unknown. Recently, we demonstrated that infection by murine norovirus (MNV) reverses intestinal abnormalities upon depletion of bacteria, indicating that an intestinal animal virus can provide cues to the host that are typically attributed to the microbiota. Here, we elucidate mechanisms by which MNV evokes protective responses from the host. We identify an important role for the viral protein NS1/2 in establishing local replication and a type I interferon (IFN-I) response in the colon. We further show that IFN-I acts on intestinal epithelial cells to increase the proportion of CCR2-dependent macrophages and IL-22 producing innate lymphoid cells (ILCs), which in turn promote pSTAT3 signaling in intestinal epithelial cells and protection from intestinal injury. Additionally, we demonstrate that MNV provides a striking IL-22 dependent protection against early life lethal infection by Citrobacter rodentium. These findings demonstrate novel ways in which a viral member of the microbiota fortifies the intestinal barrier during chemical injury and infectious challenges.
INTRODUCTION
The microbiome includes intestinal bacteria that contribute to the development of the immune system, defend against opportunistic infections, and impact the development of autoimmune and inflammatory disease. Bacteriophages, animal viruses, and endogenous viral elements that represent the virome are also numerous, diverse, and prevalent in humans and model organisms1. Alterations in the composition of the enteric virome have been observed in various pathological conditions, suggesting that the presence or absence of certain viruses in the gastrointestinal tract contribute to disease development2. Although many animal viruses have been detected in intestinal specimens, insight into mechanisms by which enteric viruses influence host physiology beyond their roles as pathogens is lacking. An understanding of viral and host factors involved in establishing mutually beneficial relationships could influence the design and implementation of microbiome-based therapies, which currently focus on exploiting bacterial communities.
Studies with murine norovirus (MNV) provide direct evidence that viruses can function similarly to the bacterial members of the microbiome. MNV is a single-stranded RNA virus that frequently establishes a persistent asymptomatic gastrointestinal infection in mice. Like commensal bacteria, MNV induces or exacerbates intestinal inflammation in genetically susceptible backgrounds3–5. In mice with mutation in the autophagy gene Atg16L1, a model of the inflammatory bowel disease (IBD) Crohn’s disease, we showed that MNV infection triggers structural defects in small intestinal Paneth cells similar to patients harboring the disease-associated variant of ATG16L13,6. MNV-infected Atg16L1 mutant mice also display increased mortality following chemical injury to the intestine by dextran sodium sulfate (DSS)3,7. Conversely, we demonstrated in wild-type (WT) antibiotics (ABX)-treated and germ-free (GF) mice that the same MNV strain restores intestinal morphology and lymphocyte differentiation that are defective due to the absence of bacteria8. In addition, and in stark contrast to Atg16L1 mutant mice, MNV protects ABX-treated mice from DSS-induced mortality8. Subsequently, other groups showed that MNV enhances colonization resistance against vancomycin-resistant enterococcus (VRE) after ABX treatment and reduces lung injury following infection with Pseudomonas aeruginosa9,10. This property of MNV, where infection can be beneficial in certain situations yet induces disease in a susceptible background, is reminiscent of the complex symbiotic relationship between host and the bacterial microbiota.
Endogenous viruses and synthetic viral RNA were shown to protect against DSS through production of type I interferon (IFN-I) by dendritic cells (DCs)11,12. Similarly, we demonstrated that MNV-mediated protection against DSS in ABX-treated mice is dependent on the IFN-I receptor (IFNAR1), implicating this conserved antiviral pathway in the protective effects of viral infection8. Ifnar1−/− mice that were not treated with ABX, and thus harbor an intact bacterial microbiota, were similar to WT mice and resistant to DSS under the same conditions. These findings raise the possibility that virus-mediated protection occurs through a mechanism that is distinct from the way an intact bacterial microbiota fortifies the barrier, which we examine here.
RESULTS
Protection from chemical injury is dependent on sequence variation in NS1/2.
We previously showed that the CR6 strain of MNV strain confers greater protection of ABX-treated mice from DSS compared to the CW3 strain8. Although MNV CR6 and CW3 share ~95% amino acid sequence identity, they display significant differences in their virulence. The VP1 major capsid protein of MNV CW3 accounts for the ability of this strain to cause lethality when introduced into immunocompromised Stat1−/− mice13–15. The less virulent CR6 strain evades recognition by lymphocytes to establish persistent infection16, which is attributed to sequence variation in NS1/217. We used recombinant cDNA clones of MNV in which the VP1 and NS1/2 regions were swapped between CR6 and CW3 (Fig. 1a) to determine which of these factors contribute to protection from DSS.
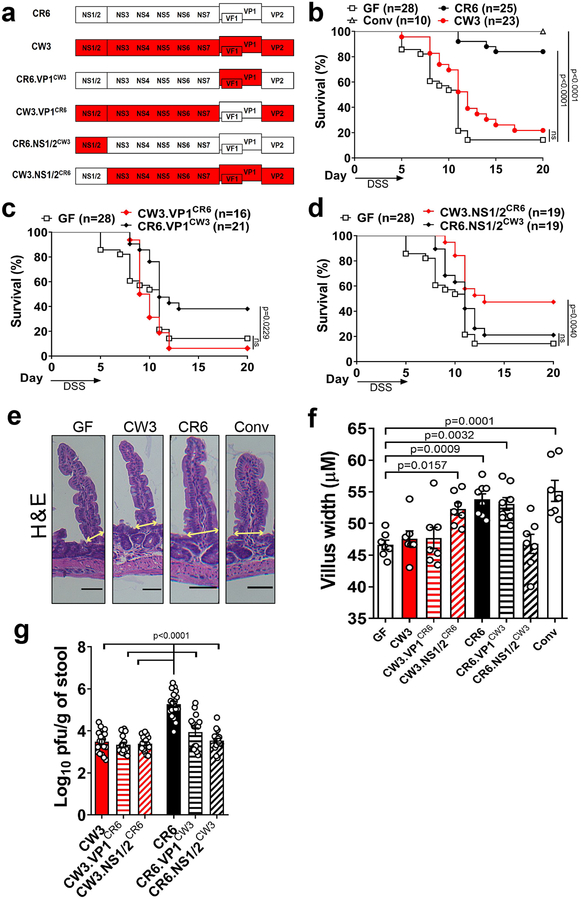
Figure 1: Sequence variation in NS1/2 contributes to protection from intestinal injury.
(a) Illustration of parental and chimeric MNV strains. (b) Survival of uninfected GF, MNV-infected GF mice (CR6 and CW3) and conventional (Conv) mice following administration of 3% DSS in the drinking water for 5 days. Survival of GF mice infected with VP1 (c) or NS1/2 (d) chimeric viruses following DSS. Mice were infected for 10 days prior to DSS administration. Survival experiments with the parental strains and chimeric viruses were conducted concurrently and therefore the uninfected GF survival curve shown in (b), (c) and (d) are identical. Representative H&E images of small intestinal villi (e) and quantification of villus width (f) at 10 dpi. Scale bars represent 50μM. All groups have7 mice except for CR6 and CR6.VP1CW3 that have 8 mice. (g) Infectious MNV in the stool as determined by plaque assay at 10 dpi. The following mice were analyzed per group: CW3, n=18; CW3.VP1CR6, n=15; CW3.NS1/2CR6, n=17; CR6, n=18; CR6.VP1CW3, n=17 and CR6.NS1/2CW3, n=18. Survival curves were analyzed using the log-rank Mantel–Cox test. Villus width and log10 transformed stool titers were analyzed using ANOVA with Dunnett’s multiple comparisons test compared to GF mice or CR6-infected GF mice, respectively. All bars represent mean and error bars represent standard error of the mean. All p-values are shown in the figure.
First, we used GF mice to verify our previous observations made in ABX-treated mice. GF mice infected with MNV CR6, but not CW3, displayed survival following administration of DSS to levels similar as conventional mice, indicating that the CR6 strain has the ability to compensate for the complete absence of bacteria (Fig. 1b and Sup. Fig. 1a). Although no chimeric virus was able to induce the same level of protection as CR6, we observed significant differences. Sequence variation in VP1 did not contribute to protection as the CR6 mutant harboring VP1 from CW3 (CR6.VP1CW3), but not the converse (CW3.VP1CR6), improved the survival of GF mice (Fig. 1c). In contrast, the CW3 mutant harboring NS1/2 from CR6 (CW3.NS1/2CR6) gained the ability to promote survival, while the CR6 mutant harboring NS1/2 from CW3 (CR6.NS1/2CW3) lost this ability (Fig. 1d). We previously showed that protection against DSS correlates with the ability of MNV to restore the width of small intestinal villi in bacterially-depleted mice8. We found that the strains that enhanced survival during DSS treatment (MNV CR6, CR6.VP1CW3, and CW3.NS1/2CR6) increased the villus width of GF mice (Fig. 1e and 1f). Therefore, NS1/2 contributes to the differential response to MNV CR6 and CW3 infection.
We detected all virus strains in the stool at day 10 post-infection indicative of productive infection, albeit at lower levels than conventional mice consistent with the role of bacteria in promoting MNV infection8,18,19. However, the parental CR6 strain replicated to higher titers (Fig. 1g). We did not detect any correlation between extra-intestinal spread and survival following DSS treatment (Sup. Fig. 1b). A more efficient intestinal infection could explain why GF mice infected with CR6 display the highest degree of survival, even when compared with CW3.NS1/2CR6 infection. Importantly, CW3.NS1/2CR6 (which protects against DSS) and the original CW3 strain (which does not protect) display similar degree of shedding indicating that additional factors are important for determining the effect of MNV infection on intestinal injury.
NS1/2 contributes to IFN-I signaling and proliferation of colonic epithelial cells.
NS1/2 sequence variation is associated with persistence in the colon of conventional mice17,20. We hypothesized that the chimeric viruses have differences in the ability to replicate in the colon during injury and drive local viral RNA sensing that is important for the protective IFN-I response. Consistent with this possibility, we observed a correlation between the amount of MNV RNA from the chimeric viruses and expression of the representative IFN-stimulated genes (ISGs) OasL2, Mx2 and Isg15 in the colonic tissue of virally-infected GF mice treated with DSS (Fig. 2a). Although previously shown to primarily infect leukocytes, recent findings indicate that MNV also infects intestinal epithelial cells (IECs), especially tuft cells20,21. Only CR6 and CW3.NS1/2CR6 genomes were detectable in sorted IECs and lamina propria leukocytes (LPLs) indicating that CW3 and CR6.NS1/2CW3 are unable to persist in the colon following DSS treatment (Fig. 2b). The presence of viral RNA in IECs rather than LPLs was a better indicator of ISG expression, and IFN-I expression (Ifnb1) was detectable in LPLs but not IECs (Fig. 2c and Sup. Fig. 1c). These results suggest that, even though the epithelial tropism of MNV is important for persistence in the local niche, immune cells are likely the main producers of IFNβ and IECs are the responding cells under these conditions.
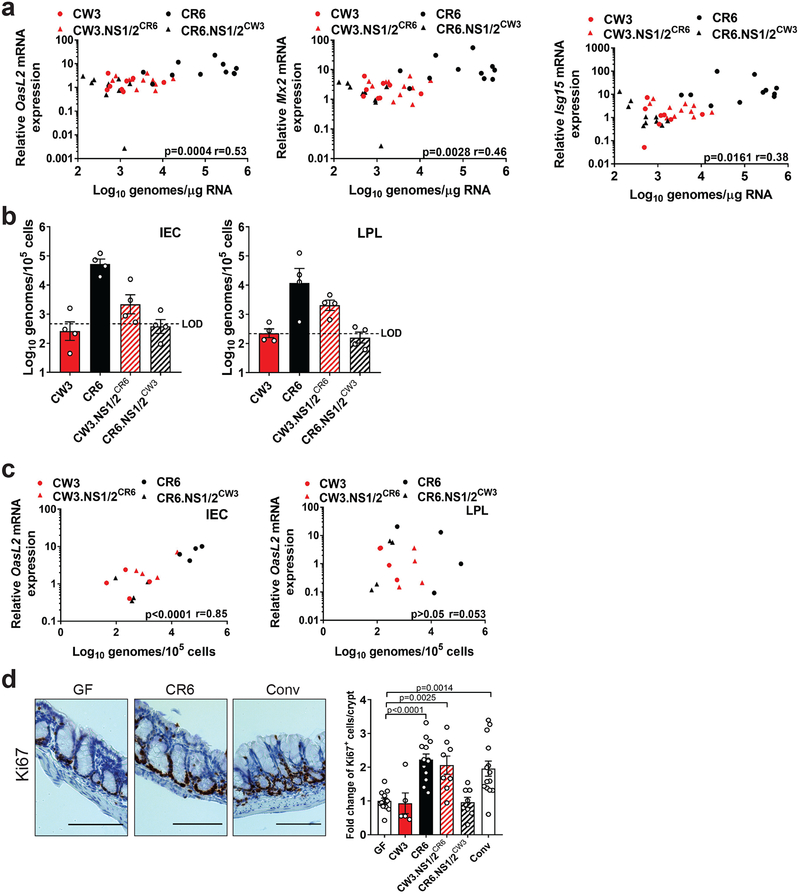
Figure 2: NS1/2 sequence variation contributes to intestinal persistence, IFN-I signaling and epithelial cell proliferation following intestinal injury.
(a) OasL2, Mx2 and Isg15 mRNA expression over uninfected GF mice relative to viral log10 genomes in the colon. Data represents 4 independent experiments. The following mice were analyzed per group: CW3, n=8; CR6, n=11; CW3.NS1/2CR6, n=11 and CR6.NS1/2CW3, n=10. (b) Viral genomes in sorted colonic IECs and LPLs. LOD=limit of detection. Data represents 2 independent experiments each with 2 mice per group. (c) OasL2 mRNA expression over uninfected GF mice relative to viral log10 genomes in IECs and LPLs. Data represents 2 independent experiments each with 2 mice per group. (d) Representative images of the colon stained for Ki67 and corresponding fold change of Ki67+ IECs relative to uninfected GF mice. Scale bars represent 100μM. The following mice were analyzed per group: GF, n=12; CW3, n=5; CR6, n=13; CW3.NS1/2CR6, n=8; CR6.NS1/2CW3, n=8 and Conv, n=14. All samples were analyzed at day 5 of DSS administration. Correlation was analyzed using Pearson r with p-values and r-values as shown. Fold change of Ki67+ IEC/crypt was analyzed using ANOVA with Dunnett’s multiple comparisons test with p-values shown. All bars represent mean and error bars represent standard error of the mean.
Mice deficient in Irgm1, a negative regulator of IFNAR1 signaling, display enhanced wound healing due to increased IEC proliferation22. Similarly, we found that GF mice infected with MNV CR6 and CW3.NS1/2CR6, but not those infected by MNV CW3 or CR6.NS1/2CW3, showed increased staining of the proliferation marker Ki67 in the colon (Fig. 2d). The IFN-I response to MNV is dependent on recognition of viral RNA by cytosolic sensors that signal through the adaptor molecule MAVS23–25. Unlike Mavs+/+ controls, MNV CR6 infection of ABX-treated Mavs−/− mice did not induce ISGs, protect against DSS, or restore the width of small intestinal villi despite productive viral replication (Sup. Fig. 2a–d). MNV CR6 infection of bone marrow-derived DCs induced MAVS-dependent expression of Ifnb1 and Mx2 (Sup. Fig. 2e). Conversely, MNV CR6 induced Mx2 expression without detectable Ifnb1 in both Mavs+/+ and Mavs−/− intestinal organoids, a 3D cell culture in which epithelial lineages are differentiated from primary progenitor cells (Sup. Fig. 2f). These data support a role for MAVS-induced IFN-I production by leukocytes in response to MNV persistence in the colon, which is associated with protective epithelial proliferation.
Protection from DSS by MNV is dependent on IL-22.
We and others have shown that MNV infection increases IL-22-producing group 3 innate lymphoid cells (ILC3s) and IFNγ+ T cells in GF or ABX-treated mice8,9. MNV CR6 retained the ability to improve the survival of ABX+DSS-treated IFN-γ receptor (Ifngr−/−) mice, but was unable to protect similarly-treated Il22−/− mice (Fig. 3a–c). Il22−/− mice that received DSS without ABX displayed 100% survival, indicating that IL-22 is dispensable at this particular dose and duration of DSS treatment when intestinal bacteria are intact, and that this cytokine is especially necessary to overcome the heightened sensitivity to injury when bacteria are depleted. The loss of protection in Il22−/− mice could not be explained by lower intestinal virus levels (Fig. 3e). Increased villus width was observed following CR6 infection in ABX-treated WT and to a lesser extent Ifngr−/− mice, but not IL-22−/− mice (Fig. 3d). These observations support the idea that widening of villi reflects an increase in leukocytes and IEC proliferation downstream of an immune response to local viral replication.
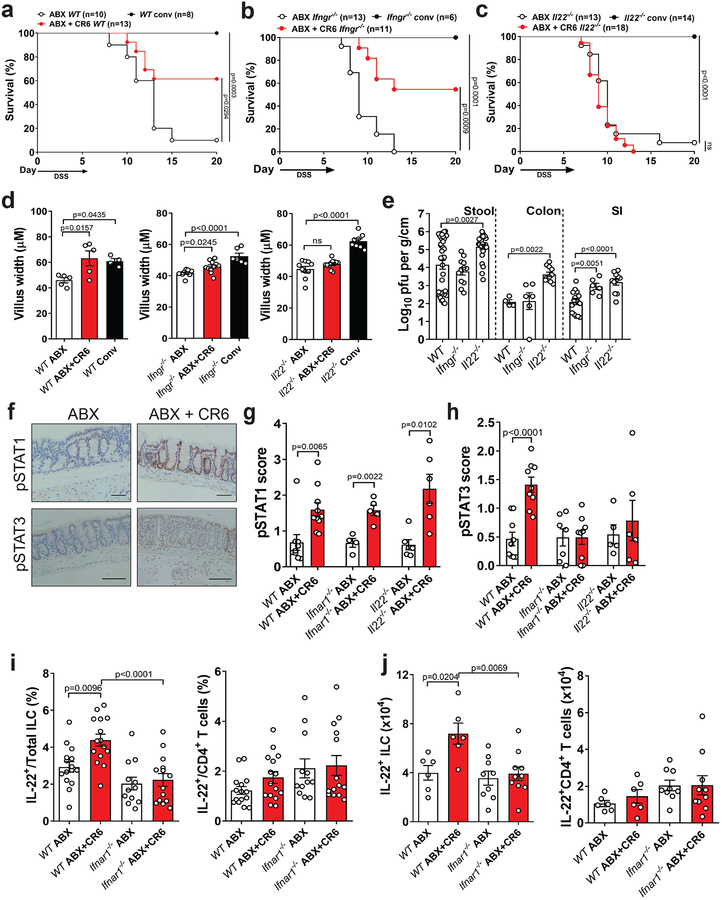
Figure 3: Protection of ABX-treated mice from intestinal injury is dependent on IL-22.
Survival of WT (a), Ifngr−/− (b) and Il22−/− (c) mice following DSS. Mice were treated with ABX for at least 10 days prior to CR6 infection and given DSS at 10 dpi for 6 days. Small intestinal villus width (d) and virus levels in the stool, colon and SI (e) at 10 dpi with CR6. The following ABX, ABX+CR6 and Conv mice were analyzed for villus width: WT n=5,5,4; Ifngr−/−, n=9,11,6 and Il22−/−, n=10,11,8. The following mice were analyzed for stool, colon and SI titer: WT, n=36,4,17; Ifngr−/−, n=12,6,6 and Il22−/−, n=25,11,11. (f) Representative images of pSTAT1 and pSTAT3 expression in the colon of ABX-treated WT mice. Scale bars represent 100μM. pSTAT1 (g) and pSTAT3 (h) expression score in the colon at day 6 post DSS. The following mice were analyzed for pSTAT1 and pSTAT3: WT ABX, n=9,9; WT ABX+CR6, n=10,9; Ifnar1−/− ABX, n=4,8; Ifnar1−/− ABX+CR6, n=5,9; Il22−/− ABX, n=6,5 and Il22−/− ABX+CR6, n=6,6. Proportion (i) and absolute numbers (j) of IL-22 expressing ILC (CD19−CD11b−CD90.2+CD3−TCRβ−) and CD4+ T cells (CD19−CD11b−CD90.2+CD3+TCRβ+CD4+) in the colon of ABX-treated WT and Ifnar1−/− mice at day 6 post DSS. The following mice were analyzed for cell proportions and numbers: WT ABX, n=15,6; WT ABX+CR6, n=15,6; Ifnar1−/− ABX, n=12,9 and Ifnar1−/− ABX+CR6, n=14,10. Survival curves were analyzed using the log-rank Mantel–Cox test. ANOVA with Dunnett’s multiple comparisons test was used to analyze villus width and Log10 transformed MNV titer compared to ABX and WT mice, respectively. A two-tailed t-test was used to analyze STAT expression. ANOVA with Tukey’s multiple comparisons test was used to analyze flow cytometry data. All bars represent mean and error bars represent standard error of the mean. All p-values are shown in the figure.
Next, we examined whether MNV induces phosphorylation of STAT3 (pSTAT3) in the colon because activation of this transcription factor downstream of IL-22 signaling mediates wound repair following DSS treatment26. We detected an IL-22 dependent increase in pSTAT3+ cells in the colon upon infection with MNV CR6, whereas the positive control pSTAT1 (induced by IFN-I, IFN-λ, and IFN-γ) was detectable in all conditions (Fig. 3f–h). Interestingly, we found that MNV was also unable to induce pSTAT3 staining in the colon of Ifnar1−/− mice (Fig. 3h). MNV CR6 infection still induced significant ISG expression in Il22−/− mice (Sup. Fig. 3a), indicating that IFN signaling is not dependent on IL-22. MNV CR6 infection increased the proportion and absolute numbers specifically of IL-22-producing ILCs in the colonic lamina propria in ABX+DSS-treated WT but not Ifnar1−/− mice (Fig. 3i–j, Sup. Fig. 3b–c and 3e). Viral infection increased IFNγ+ CD4+ T cell populations (but not IL-22+ or IL-13+ CD4+ T cells) in an IFNAR1-dependent manner (Fig. 3i and 3j and Sup. Fig. 3d and 3f), but our earlier results (Fig. 3b) show that IFNγ was not involved in protection from injury. MNV CR6 evoked similar responses in GF mice following DSS (Sup. Fig 3g and 3h). Also, IL-22 secretion in colonic explants and pSTAT3 staining in colonic tissue sections were increased in GF mice infected by MNV CR6 (Sup. Fig. 3i and 3j). Together, these results indicate that MNV CR6 increases colonic IL-22 following intestinal damage.
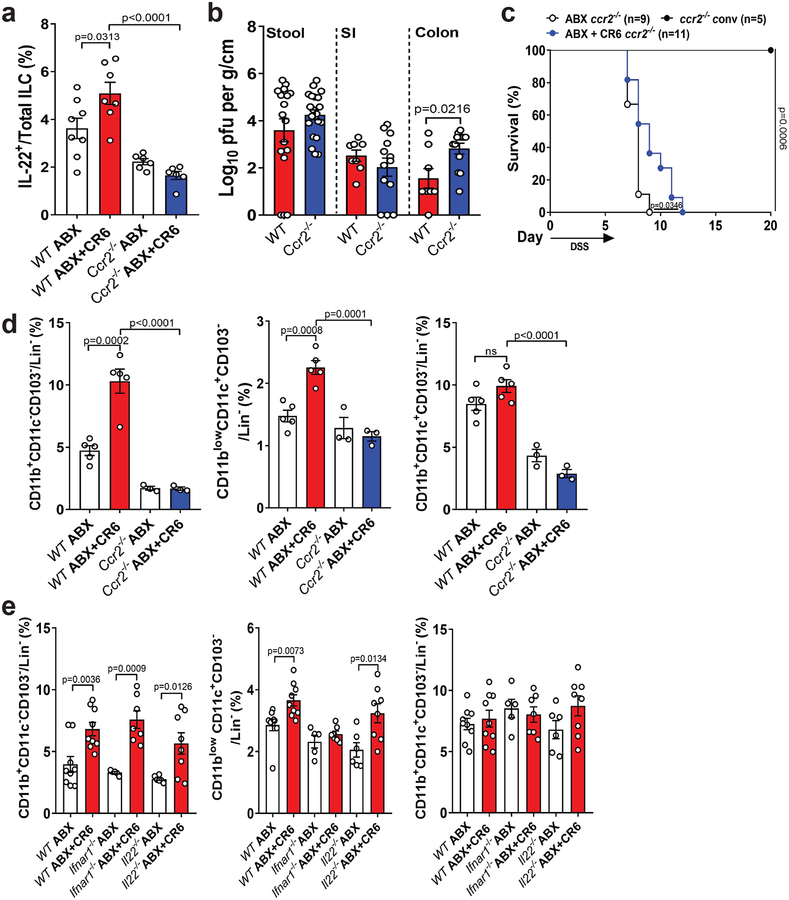
Activation of ILC3s following bacterial infection of the colon is dependent on recruitment of CCR2+ monocytes27. We found that MNV CR6 infection did not increase the proportion of IL-22+ ILCs in the colon of ABX-treated Ccr2−/− mice, indicating that similar mechanisms are necessary for virus-mediated activation of ILC3s (Fig. 4a). Despite equivalent or higher viral burden compared with WT mice, MNV CR6 induced minimal protection from DSS injury in ABX-treated Ccr2−/− mice (Fig. 4b and 4c). We observed an increase in several CCR2-dependent MHCII+ myeloid populations in the colon following infection with MNV CR6 (Sup. Fig. 4a and Fig. 4d). Among these infiltrating cells, MNV-induced increases in CD11blowCD11c+CD103− cells was observed in GF and ABX-treated WT mice but not ABX-treated Ifnar1−/− mice (Fig. 4e and Sup. Fig. 4b). Other MHCII+ populations were not dependent on IFN-I signaling, and recruitment of all subsets following infection remained intact in Il22−/− mice (Fig. 4e). Therefore, MNV-mediated protection from injury is associated with IFN-dependent alterations to myeloid populations upstream of IL-22 expression by ILC3s.
Figure 4: Protection from intestinal injury and IL-22 expression is associated with CCR2-dependent cells.
(a) Proportion of IL-22 expressing ILC in ABX-treated WT and ccr2−/− mice at day 6 post DSS. The following number of mice were analyzed: WT ABX, n=8; WT ABX+CR6, n=7; ccr2−/− ABX, n=6 and ccr2−/− ABX+CR6, n=6. (b) Infectious virus levels in the SI, colon and stool of ABX-treated ccr2−/− mice compared to WT mice. The following number of mice were analyzed for stool titer: WT, n=17 and ccr2−/−, n=22; SI and colon titer: WT, n=8 and ccr2−/−, n=13. (c) Survival of ccr2−/− mice following DSS administration. Mice were treated with ABX for at least 10 days prior to CR6 infection and given DSS at 10 dpi for 6 days. Survival curves were analyzed using the log-rank Mantel–Cox test. Proportion of MHCII+ populations in ABX-treated WT and ccr2−/− (d) and ABX-treated WT, Ifnar1−/− and Il22−/− (e) mice at day 6 post DSS. The following number of mice were analyzed in (d): WT ABX, n=5; WT ABX+CR6, n=5; ccr2−/− ABX, n=3 and ccr2−/− ABX+CR6, n=3. The following number of mice were analyzed in (e): WT ABX, n=9; WT ABX+CR6, n=9; Ifnar1−/− ABX, n=5; Ifnar1−/− ABX+CR6, n=7; Il22−/− ABX, n=6 and Il22−/− ABX+CR6, n=8. Lin− =CD45+CD19−TCRβ−Gr-1low. Figures (a) and (d) were analyzed using ANOVA with Tukey’s multiple comparisons test and figure (b) and (e) were analyzed using two-tailed t-test. All bars represent mean and error bars represent standard error of the mean. All p-values are shown in the figure.
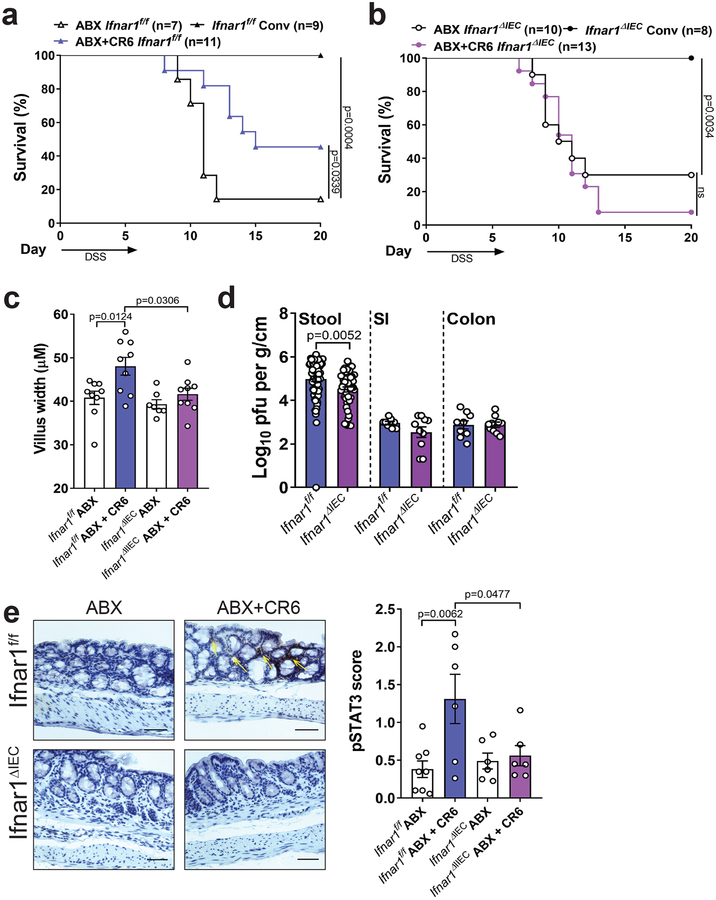
IFN-I signaling in IECs is required for protection.
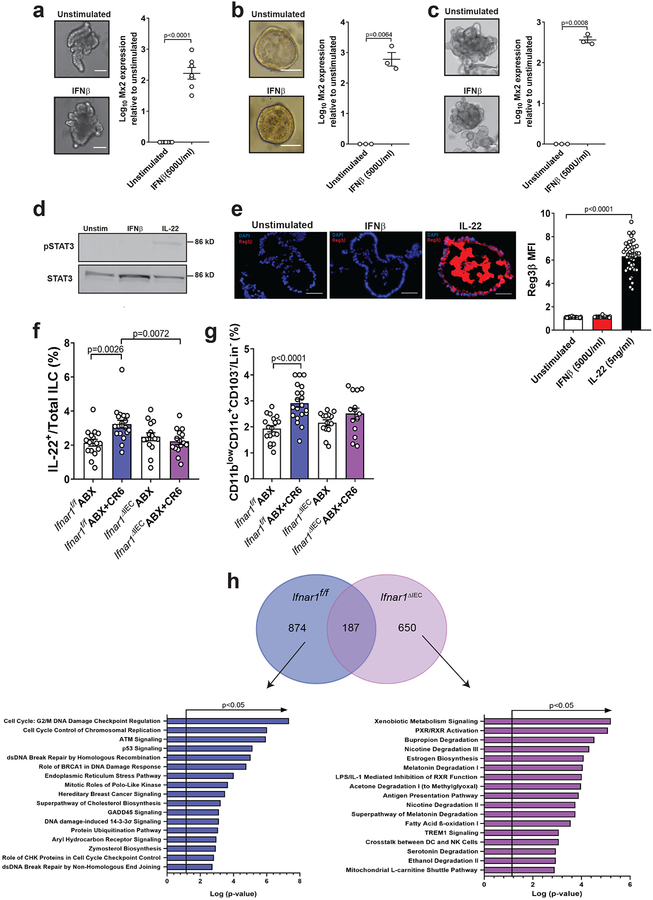
Although Ifnar1 deletion in IECs alters the composition of the bacterial microbiota28, other studies have shown that IECs are unresponsive to IFN-I and implicate a dominant role for IFN-λ signaling in inhibiting enteric RNA viruses19,29–31. In contrast to Ifnar1f/f control mice, MNV infection of ABX-treated Ifnar1f/f;villin-Cre mice in which Ifnar1 is deleted in IECs (Ifnar1ΔIEC) did not enhance survival following DSS injury or restore villi width (Fig. 5a–c). We did not observe an IEC-specific role for IFNAR1 in restricting MNV burden (Fig. 5d). The ability of MNV CR6 to induce pSTAT3 staining was eliminated in Ifnar1ΔIEC but not Ifnar1f/f mice (Fig. 5e). Recombinant IFNβ induced Mx2 expression in human and mouse intestinal organoids (Fig. 6a–c), confirming that IECs can directly respond to IFN-I. According to our model, STAT3 is activated by IL-22 directly and IFN-I indirectly during MNV infection. We found that IL-22, but not IFNβ, induced pSTAT3 and REG3β in organoids (Fig. 6d, 6e and Sup. Fig. 6). These findings show that IECs respond to IFN-I, and that STAT3 phosphorylation is an independent event. MNV-induced increases in IL-22+ and IFNγ+ ILCs and MHCII+CD11blowCD11c+CD103− cells were absent in Ifnar1ΔIEC mice (Fig. 6f–g and Sup. Fig. 4c–d). We then performed RNA-Seq analysis on sorted IECs from Ifnar1f/f and Ifnar1ΔIEC mice following DSS, with or without viral infection. MNV CR6 infection led to increased expression of genes representing cell cycle regulation and DNA damage response pathways in a manner dependent on epithelial IFNAR1 (Fig. 6h). This IFNAR-dependent gene expression pattern is consistent with MNV-induced IEC proliferation (Fig. 2d) and is remarkably similar to a recently described IL-22 induced transcriptional response that prevents genotoxic damage to epithelial stem cells in the colon32. Together, these data indicate that IFNAR signaling in the epithelium mediates the beneficial effect of MNV infection, and decouples the role of IFN-I in antiviral versus injury responses.
Figure 5: IFN-I signaling in IEC is required for protection from intestinal injury.
Survival of Ifnar1f/f (a) and Ifnar1ΔIEC (b) mice following DSS. Mice were treated with ABX for at least 10 days prior to CR6 infection and given DSS at 10 dpi for 6 days. Survival curves were analyzed using the log-rank Mantel–Cox test. Small intestinal villus width (c) and virus levels in the SI, colon and stool (d) at 10 dpi with CR6. The following number of mice were analyzed for villus width: Ifnar1f/f ABX, n=9; Ifnar1f/f ABX+CR6, n=9; Ifnar1ΔIEC ABX, n=7 and Ifnar1ΔIEC ABX+CR6, n=9. The following number of mice were analyzed for stool titer: Ifnar1f/f, n=71 and Ifnar1ΔIEC, n=49; SI and colon titer: Ifnar1f/f, n=9 and Ifnar1ΔIEC, n=10. (e) Representative images of pSTAT3 staining in the colon and pSTAT3 expression score at day 6 post DSS. Scale bars represent 100μM. The following number of mice were analyzed for pSTAT3: Ifnar1f/f ABX, n=8; Ifnar1f/f ABX+CR6, n=6; Ifnar1ΔIEC ABX, n=6 and Ifnar1ΔIEC ABX+CR6, n=6. Figures (c) and (e) were analyzed using ANOVA with Tukey’s multiple comparisons test and figure (d) was analyzed using two-tailed t-test. All bars represent mean and error bars represent standard error of the mean. All p-values are shown in the figure.
Figure 6: IECs react to IFN-I stimulation and promote an IL-22 response.
Representative images of murine SI (a), murine colon (b) or human colon (c) organoids either unstimulated or stimulated with 500U/ml of IFNβ for 48 h and expression of Mx2 mRNA relative to unstimulated organoids. Scale bars represent 50μM. The following number of independent experiments were performed: (a), n=7; (b), n=3 and (c), n=3. Log10 transformed Mx2 expression was analyzed by two-tailed paired t-test. (d) Phosphorylation of STAT3 at 2 h in SI organoids. Representative of 4 independent experiments. (e) Representative images of Reg3β expression and representative mean fluorescence intensity (MFI) of individual organoids of 2 independent experiments at 24 h. Scale bars represent 50μM. The following number of organoids were analyzed: unstimulated, n=34; IFNβ, n=32 and IL-22, n=40. ANOVA with Dunnett’s multiple comparisons test was used to analyze Reg3β expression. Proportion of IL-22 expressing ILC (f) and CD11blowCD11c+CD103− cells (g) in ABX-treated Ifnar1f/f and Ifnar1ΔIEC mice at day 6 post DSS. The following number of mice were analyzed for IL-22: Ifnar1f/f ABX, n=16; Ifnar1f/f ABX+CR6, n=19; Ifnar1ΔIEC ABX, n=15 and Ifnar1ΔIEC ABX+CR6, n=15. The following number of mice were analyzed for CD11blowCD11c+CD103− cells: Ifnar1f/f ABX, n=18; Ifnar1f/f ABX+CR6, n=20; Ifnar1ΔIEC ABX, n=14 and Ifnar1ΔIEC ABX+CR6, n=14. ANOVA with Tukey’s multiple comparisons test was used to analyze cell populations. (h) Gene expression analysis of IECs from MNV CR6-infected Ifnar1f/f (n=3) and Ifnar1ΔIEC (n=3) mice compared to uninfected mice (Ifnar1f/f, n=3 and Ifnar1ΔIEC, n=3) at day 6 post DSS. Circles in Venn diagram represent number of transcripts that are enriched following MNV-infection in the mice with the indicated genotypes. Pathway analysis was performed on the non-overlapping gene set and p-values determined by Ingenuity Pathway Analysis. All bars represent mean and error bars represent standard error of the mean. All p-values are shown in the figure.
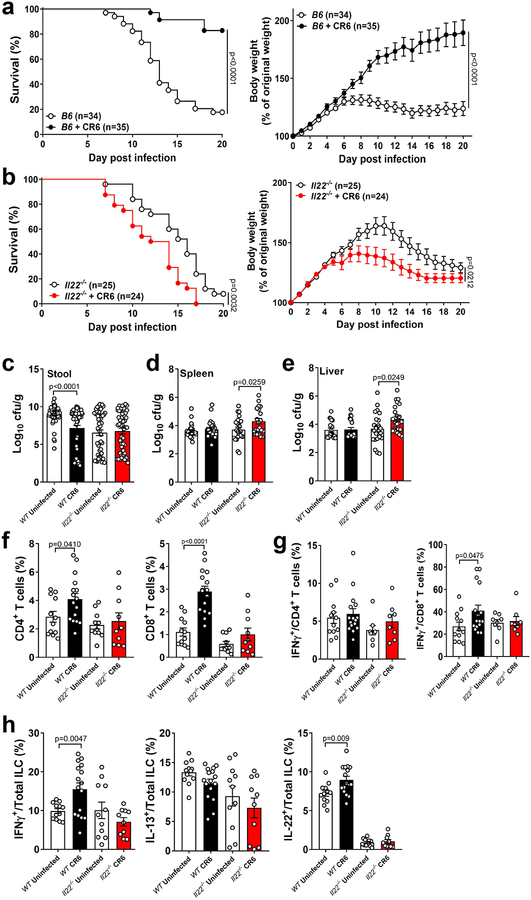
MNV protects against enteric bacterial infection during development.
The gastrointestinal tract becomes exposed to diverse animal viruses over time following birth33. It is unclear how the presence of these viruses affect disease caused by enteric bacterial pathogens, which are also a common occurrence during childhood. To examine the consequence of coinfection, 3-week old WT mice were challenged with the Gram-negative bacterial pathogen Citrobacter rodentium, with or without concurrent MNV CR6 infection. In the absence of the virus, WT mice failed to grow and succumbed to C. rodentium infection, whereas almost all the MNV CR6-infected mice survived and gained weight (Fig. 7a). MNV CR6 did not provide any benefit to 3-week old Il22−/− mice (Fig. 7b). Next, we took advantage of the observation that lymphocyte-deficient Rag1−/− mice have a compensatory increase in IL-22 producing ILC3s during a similar developmental period as the above coinfection model34. We hypothesized that this increase in ILC3s would serve a similar purpose as MNV infection, and thus, Rag1−/− mice will no longer be dependent on MNV for survival during C. rodentium infection. Indeed, Rag1−/− mice were protected from bacterial infection irrespective of MNV infection, which was associated with an overall increase in ILCs (Sup. Fig. 5a–b). Therefore, enhancing IL-22+ ILC3s through viral infection or genetically is associated with protection against secondary bacterial infection in juvenile animals.
Figure 7: MNV protects young mice from enteric bacterial infection.
Survival and weights of B6 (a) and Il22−/− (b) mice infected with C. rodentium at 21 days old. Survival curves were analyzed using the log-rank Mantel–Cox test. For weight curves, AUC was determined for each mouse and difference between uninfected and CR6-infected groups determined by two-tailed t-test. Mean and standard error of the mean is shown for weight analysis. Bacterial titers in the stool (c), spleen (d) and liver (e) at 6 dpi with C. rodentium. The following number of mice were analyzed in (c): WT Uninfected, n=56; WT CR6, n=59; Il22−/− Uninfected, n=48 and Il22−/− CR6, n=47. The following number of mice were analyzed in (d): WT Uninfected, n=25; WT CR6, n=29; Il22−/− Uninfected, n=24 and Il22−/− CR6, n=22. The following number of mice were analyzed in (e): WT Uninfected, n=19; WT CR6, n=21; Il22−/− Uninfected, n=23 and Il22−/− CR6, n=23. (f) Proportion of colonic CD4+ and CD8+ T cells. (g) Proportion of colonic CD4+ and CD8+ T cells expressing IFNγ. (h) Proportion of IFNγ+, IL-13+ and IL-22+ cells in the colonic ILC population. The following number of mice were analyzed for cell proportions: WT Uninfected, n=12; WT CR6, n=16; Il22−/− Uninfected, n=11 and Il22−/− CR6, n=10. Log10 transformed bacterial titers and cell proportions were analyzed using two-tailed t-tests. All bars represent mean and error bars represent standard error of the mean. All p-values are shown in the figure.
IL-22 is more important for reducing intestinal inflammation rather than intestinal C. rodentium burden in adult mice35. Consistent with this observation, MNV CR6 infection of WT mice led to modest or no reduction in bacterial burden (Fig. 7c–e). Furthermore, similar numbers of C. rodentium were recovered from the stool of MNV-infected WT and Il22−/− mice. Instead, we observed that MNV CR6 infection reduced intestinal pathology in an IL-22 dependent manner (Sup. Fig. 5c). Protection from C. rodentium infection was independent of MNV strain, as CW3-infected WT mice showed increased survival and weight gain (Sup. Fig. 5d). Protection from C. rodentium infection was also maintained in Ifnar1−/− mice (Sup. Fig. 5e). Thus, sequence variation in NS1/2 and IFNAR1 signaling do not contribute to protection from bacterially-induced injury.
It is possible that MNV protects young mice by accelerating the development of the microbiota because the compositional shift in intestinal bacterial communities that occurs during neonatal development is key to C. rodentium resistance36. However, we did not detect significant changes in the gut microbiota due to MNV infection or absence of IL-22 (Sup. Fig. 5f). Instead, MNV-mediated protection from C. rodentium was associated with a general increase in the proportion of CD4+ and CD8+ T cells in the colon, which was not observed in MNV-infected Il22−/− mice (Fig. 7f). In addition, MNV CR6 increased the proportion of IFNγ-expressing CD8+ T cells and ILCs in an IL-22 dependent manner, and similar to the DSS model, the presence of the virus was associated with IL-22+ ILCs (Fig. 7g and 7h).
DISCUSSION
The bacterial members of the gut microbiota acquired during development promote the maturation of the immune system and intestinal barrier. This mutualistic host-microbiota relationship can be disrupted in adulthood by ABX, diet, or other factors that alter bacterial community structures. Thus, tremendous effort has been placed in understanding the mechanistic basis of this relationship, and clinical trials are attempting to remodel the microbiota in diseased individuals by transplanting isolated microbes or feces from healthy donors. Despite accumulating evidence of the presence of a dynamic enteric virome that begins to take shape after birth33, it is unknown how intestinal viruses impact the resilience of the intestinal barrier to damage caused by environmental agents. Using two distinct models, we show that MNV infection improves the outcome of intestinal injury through IL-22.
In the chemical injury model, we demonstrate that compensation for the absence of beneficial bacteria by MNV is associated with the NS1/2 region of the viral genome. NS1/2 from mouse and human noroviruses binds the host molecule VAMP-associated protein A (VAPA), which potentially mediates membrane remodeling during viral replication37–39. NS1/2 is structurally disordered and subject to cleavage, suggesting that it can adopt additional conformations with alternate functions that are currently unknown40. NS1/2 from the protective MNV CR6 strain confers the ability to replicate and persist in colonic IECs, leading to a local IFN-I response that is likely downstream of MAVS-dependent IFNβ production in neighboring immune cells, although we cannot rule out an epithelial contribution of IFNβ. Our extensive analysis of Ifnar1ΔIEC mice indicate that IECs are the IFN-responding cell type.
We further found that MNV induces IL-22 producing ILC3s in an IFNAR1 and CCR2-dependent manner. Unlike IFN-λ which acts on neutrophils to protect against DSS41, we identified a role for IFN-I acting on IECs to mediate recruitment or activation of CCR2-dependent cells during MNV infection. Recently, we showed that enhanced IFN-I signaling due to autophagy inhibition in IECs alters the function of CCR2-dependent monocytes42. Thus, it is also plausible that augmenting IFN-I signaling through MNV CR6 infection also induces functional changes to these populations. A role for macrophages in the activation of ILC3 has been previously demonstrated following C. rodentium infection27. Therefore, we hypothesize that alterations in proportions and potentially function of macrophage populations, through IFNAR1-mediated responses in IECs, affects the ability of these cells to promote IL-22 production by ILC3s.
MNV-mediated protection of young mice from C. rodentium is IL-22 dependent as well. The mechanism is distinct from another study which reported that MNV confers resistance to VRE in ABX-treated adult mice involving a virus-dependent reduction in bacterial colonization9. VRE is a Gram-positive bacterium that is sensitive to antimicrobial REG molecules produced in response to IL-22, whereas other functions such as epithelial repair may be more important for protecting against Gram-negative bacteria such as C. rodentium, especially in young mice. Recently it was shown that MNV recruits CCR2-dependent monocytes to the MLN in an IFNAR1-independent manner to support systemic persistence43. Although we identified a subset of myeloid cells that were IFNAR1 dependent, this was not universal across all subsets. Therefore, unlike the DSS model, accumulation of IFNAR1-independent macrophages may be sufficient to provide protection following C. rodentium infection, which would explain why IFNAR1 is dispensable and MNV CW3 is able to provide protection. Due to its increased virulence compared with other strains, infection of mice with MNV CW3 has been used as a surrogate for pandemic human norovirus GII.4 strains. Given that up to 30% of asymptomatic infants and children are positive for norovirus RNA in the stool44, concurrent infection with bacterial pathogens is likely commonplace. Our results raise the possibility that underlying norovirus infections may have a protective role when the virus is not evoking diarrheal disease.
The inter-strain diversity between human noroviruses is far greater than the sequence differences between MNV strains, yet little is known about how different noroviruses impact the mucosal immune system of the human gut. This lack of knowledge applies to other enteric viruses as well. Elegant work combining observations in mice and humans implicate reoviruses and noroviruses in celiac disease onset in a viral strain-dependent manner45,46. CMV can reactivate in the gastrointestinal tract and contribute to IBD flares, but was shown to enhance epithelial turnover in mice, which presumably improves resistance to injury22. These observations resemble some aspects of our findings with MNV showing that an intestinal virus can be beneficial or adverse depending on the strain and properties of the host. Finally, we wish to note that few studies investigating the effect of the bacterial microbiota exclude the possibility that results are driven by the presence of MNV or other enteric viruses such as astroviruses common in mouse facilities. We suggest that the host immune response to enteric viral infections requires attention beyond its antiviral function and needs to be considered in microbiota studies.
MATERIALS AND METHODS
Mice
GF mice on a C57BL/6J background were bred and maintained in flexible-film isolators in the New York University (NYU) School of Medicine Gnotobiotics Animal Facility. Absence of fecal bacteria was confirmed monthly as before8. For experiments, GF mice were housed in Techniplast Bioexclusion cages with access to sterile food and water. Conventional C57BL/6J WT, Ifnar1−/−, Ifngr−/− Ccr2−/−, Mavs−/−, Rag1−/−, Ifnar1fl and Villin-cre mice were purchased from Jackson Laboratories. IL22−/− mice were provided by Sergei Koralov (NYUMC, USA). Ifnar1fl mice were bred to Villin-cre mice to produce cre+/− and cre−/− littermates. Mavs−/− mice on a B6129SF2/J background were bred to C57BL/6J mice to produce Mavs+/− mice that were crossed to each other to produce Mavs−/− and Mavs+/+ littermates. All other knockout strains were on the C57BL/6J background, and C57BL/6J mice were used as WT controls. All mice were bred onsite in an MNV and Helicobacter-negative specific-pathogen free (SPF) animal facility. For DSS experiments, randomly assigned age (8–10 weeks) and gender-matched mice were used in which MNV-infected mice were compared with uninfected littermates to control for potential microbiota differences between mice originating from different breeding pairs. For Citrobacter rodentium experiments, age-matched 12–14-day-old litters were used that were generated from 2 pregnant dams that were co-housed and the subsequent litters separated approximately 2–4 days prior to infection with MNV. Dams were littermates from the previous generation. All young mice were weighed daily throughout the experiment. Sample size for animal experiments was chosen based on prior data generated in the laboratory. All animal studies were performed according to protocols approved by the NYU School of Medicine Institutional Animal Care and Use Committee (IACUC).
Antibiotics and DSS treatment
For ABX treatment, mice were given filter-sterilized water containing ampicillin (1 g l−1; American bioanalytical), vancomycin (0.5 g l−1; Sigma), neomycin (1 g l−1; Sigma), metronidazole (1 g l−1; Sigma) and 1% sucrose (Sigma). ABX-containing water was replaced at least once a week. Mice were treated with this ABX cocktail for at least 10 days prior to infection. Mice were given 3% DSS (TdB consultancy) with or without the ABX cocktail in their drinking water for 6 days and then received regular or ABX-containing drinking water for the remainder of the experiment. GF mice were given 3% DSS in filter-sterilized water for 5 days. Mice were monitored daily for survival.
MNV and bacterial inoculation
Plasmids for chimeric MNV strains were previously described17. Two viruses were on a CR6 backbone containing the indicated gene from CW3: CR6.VP1CW3 and CR6.NS1/2CW3. Two viruses were on a CW3 backbone containing the indicated gene from CR6: CW3.VP1CR6 and CW3.NS1/2CR6. Chimeric viruses, CW3 and CR6 were prepared and titrated as described previously8. Mice were infected orally by pipette with 3 × 106 pfu resuspended in PBS (Corning). C. rodentium DBS100 was grown overnight in Luria-Bertani broth with shaking at 37°C and diluted 1:100 followed by an additional 4 hours of growth until bacteria were at an optical density of 2. Mice were gavaged with 2 × 107 cfu. Bacterial inocula were determined by serial dilution plating on MacConkey plates.
Detection of infectious MNV and Citrobacter rodentium in organs
Stool, 2cm ileum, 2cm of ascending colon, liver, spleen and MLN were harvested from MNV-infected mice at 10 dpi. Organs were weighed and homogenized in serum-free DMEM (Corning). Titers of MNV were determined by plaque assay as previously described8. Titers are shown as pfu/g or pfu/cm. Stools, liver and spleen were harvested from C. rodentium-infected mice at 6 dpi. Organs were weighed and homogenized in PBS and serial dilutions plated onto MacConkey plates. Titers are shown as cfu/g.
Organoid culture
Murine small intestinal and colonic organoids were cultured as described previously7. At day 5 (SI) or day 3 (colon) organoids were stimulated with either 500 U/ml of IFNβ (PBL Assay Science), 5 ng/ml of IL-22 (Biolegend) or left unstimulated. For RNA extraction and immunofluorescence, the organoids were stimulated for 24 or 48 h. For detection of STAT3 phosphorylation, organoids were stimulated for 2h. For human organoids, pinch biopsies were obtained from the colon of adult IBD patients undergoing surveillance colonoscopy using 2.8 mm standard biopsy forceps, after approval by the New York University School of Medicine Institutional Review Board. All tissue was isolated from non-inflamed regions of the colon. Intestinal fragments were incubated in Gentle Cell Dissociation Reagent (Stemcell Technologies) on ice for 30 minutes followed by vigorous pipetting to isolate crypts. Crypt were embedded in Corning Matrigel Growth Factor Reduced Basement Membrane Matrix (Corning), and cultured with human IntestiCult Organoid Growth Medium (Stemcell Technologies). The culture medium was changed every 2–3 days. Human organoids were stimulated with 500 U/ml IFNβ (R&D systems) or left unstimulated for 48 h.
Culture of BMDCs and intestinal organoids in the presence of MNV
Murine BMDCs were derived from bone marrow cells stimulated with 20ng/ml GM-CSF (R&D systems) for 6 days. 5×105 cells were seeded into a 24 well plate and infected with MNV CR6 (MOI 1) for 1h at 37°C. Virus was removed and BMDCs cultured for a further 16h before harvesting RNA. Murine SI organoids were grown for 3–5 days before gentle disruption and re-culturing of approximately 200 organoids/well in the presence of 3×106 pfu of MNV CR6 for 16 h.
Microscopy on intestinal tissue
Small intestinal and colonic tissue was prepared for staining as previously described3. Hematoxylin and eosin (H&E) staining was performed by the NYU Histopathology core. Ki67, phospho-Stat1 and phospho-Stat3 staining were performed by NYU Immunohistochemistry (IHC) core. Formalin fixed paraffin-embedded sections were deparaffinized and antigen retrieved in Ventana Cell Conditioner 2 (Citrate). Endogenous peroxidase activity was blocked with hydrogen peroxide and tissue was incubated with unconjugated rabbit anti-mouse phospho-Stat1 (Tyr701, Cell Signaling Technology), rabbit anti-mouse phospho-Stat3 (Tyr705, Cell Signaling Technology) or rabbit anti-mouse Ki67 (M3062) (Spring Biosciences). Signal was detected with anti-rabbit horseradish peroxidase conjugated multimer and visualized with 3,3 diaminobenzidene. At least 50 small intestinal villi per mouse were measured for villi width at the base of the villi. At least 50 colon crypts were analyzed for the number of Ki67+ cells. At least 20 sections along the length of the colon were scored for pSTAT expression. The scoring criteria for pSTAT staining was as follows: 0 = no staining; 1 = 1 or 2 individual positive cells; 2 = ≤10% positive cells; 3 = 10–30% positive cells; 4= 30–75% positive cells and 5= >75% positive cells. Severity of colon pathology was blindly quantified by a pathologist (A.G.N). Mice received a pathology score between 0 and 4 for inflammatory cell infiltration, inflammatory cell location and depth, edema and necrosis. A score of 0 = negative pathology, 1 = minimal, 2 = mild, 3=moderate and 4= marked pathology for the indicated fields. Crypt hyperplasia was scored by measuring crypt length and given an average score as follows: 0 = <100μM, 1 = 100–150μM, 2 = 150–200μM, 3=200–250μM and 4= >250μM. Mean values were calculated for each mouse and used as individual data points. Sections were imaged either on an Axioplan microscope (Ziess) or Evos FL Auto (Thermo Fisher Scientific). All analyses were performed using ImageJ software.
Microscopy on intestinal organoids
Organoids were frozen in Neg 50 frozen section medium (Thermo Fisher Scientific) and sectioned at 5μm. Tissue was permeabilized with 0.1% (v/v) Tween 20 (Sigma Aldrich) and 0.01% (v/v) TritonX (Sigma Aldrich) followed by blocking with 1% (w/v) BSA (Fisher Scientific). Staining was performed using sheep anti-mouse Reg3β antibody (R&D systems) followed by detection with anti-sheep Ig NL557 (R&D systems) and mounting with Vectashield containing Dapi (Vector Laboratories). Sections were imaged on the Evos FL Auto (Thermo Fisher Scientific). All analyses were performed using ImageJ software. Mean fluorescence intensity values were calculated for each organoid and used as individual data points.
Flow cytometry
Lamina propria cells from colonic tissue were harvested as before8. For intracellular cytokine staining, cells were stimulated using the eBioscience cell stimulation cocktail for 4 h at 37 °C. Cells were fixed and permeabilized using the Biolegend fixation and permeabilization buffer. The following antibodies (clones) were used for staining: CD45 (30-F11), CD19 (6D5), CD11b (M1/70), CD90.2 (53–2.1), CD3 (145–2C11), TCR-β (H57–597), CD4 (RM4–5), CD8 (53–6.7), Gr-1 (RB6–8C5), MHCII (M5/114.15.2), CD11c (N418), CD103 (2E7), IFN-γ (XMG1.2) from Biolegend and IL-22 (1H8PWSR), and IL-13 (eBio13A) from eBioscience. All samples were blocked with Fc block (TruStainfcx) and stained with a fixable live dead stain (Invitrogen). FlowJo v.10 was used to analyze flow cytometry data. Intestinal epithelial cells (IECs) were harvested by incubation at 37°C with 2mM DTT (Sigma) followed by two incubations with 5mM EDTA and then digested with Dispase (Sigma) and DNase (Sigma). IECs were sorted as PI−CD45−EpCam+ using the following antibody clones: CD45 (30-F11) and EpCam (G8.8) from Biolegend on an FACSAria II (BD Biosciences). Purity of IECs was ≥ 95%.
Colon explant culture and IL-22 detection
1cm of colon tissue was opened longitudinally, washed with PBS and cultured for 48 h in 500 μl of complete RPMI containing 2 mM L-glutamine (Corning), penicillin/streptomycin solution (Corning), 20 mM HEPES (Corning), MEM non-essential amino acids (Corning), 1 mM Sodium pyruvate (Corning) and β-mercaptoethanol (Gibco). IL-22 in supernatants was measured using the mouse IL-22 ELISA MAX Deluxe kit (Biolegend) according to the manufacturer’s instructions.
RNA isolation and qPCR
RNA from BMDCs and colonic tissue (1 cm) homogenized using a TissueRuptor (Qiagen) was isolated using the Qiagen RNeasy mini kit with DNase treatment (Qiagen). RNA from sorted IECs and LPLs was isolated using the RNeasy Micro Kit with DNase treatment (Qiagen). RNA from intestinal organoids was isolated with TRIzol reagent (Fisher Scientific) using the manufacturer’s protocol and DNA digested using RNase-free DNase (Thermo Fisher Scientific). cDNA synthesis was conducted with the ProtoScript M-MuLV First Strand cDNA synthesis kit (New England Biolabs). qPCR was performed using SybrGreen (Roche) on a Roche480II Lightcycler using the following primers: mHprt (Accession no. J00423), Fwd 5’-GTCATGCCGACCCGCAGTC-3’, Rev 5’-GTCCTGTCCATAATCAGTCCATGAGGAATAAAC-3’; m18S (Accession no. NR_003278), Fwd 5’-TAGAGGGACAAGTGGCGTTC-3’, Rev 5’-CGCTGAGCCAGTCAGTGT-3’; mIfnb1 (Accession no. X14455), Fwd 5’- TCAGAATGAGTGGTGGTTGC-3’, Rev 5’-GACCTTTCAAATCGAGTAGATTCA-3’; mMx2 (Accession no. KX774217), Fwd 5’- CCAGTTCCTCTCAGTCCCAAGATT-3’, Rev 5’-TACTGGATGATCAAGGGAACGTGG-3’; mOasL2 (Accession no. NM_011854), Fwd 5’-GGATGCCTGGGAGAGAATCG-3’, Rev 5’-TCGCCTGCTCTTCGAAACTG-3’; mIsg15 (Accession no. NM_015783), Fwd 5’-GGTGTCCGTGACTAACTCCAT-3’, Rev 5’-TGGAAAGGGTAAGACCGTCCT-3’; hβ-actin (Accession no. NM_001101), Fwd 5’-TCACCGGAGTCCATCACGAT-3’, Rev 5’-CCCAGCCATGTACGTTGCTA-3’ and hMx2 (Accession no. NM_002463), Fwd 5’- AAACTGTTCAGAGCACGATTGAAG-3’, Rev 5’- ACCATCTGCTCCATTCTGAACTG-3’. Relative expression of the respective genes to either Hprt (murine organoids),18S (murine colon or sorted cells) or β-actin (human organoids) expression was calculated using the ΔΔCT method and values were expressed as fold change from uninfected mice or untreated organoids. Absolute MNV genome copies were determined using a standard curve and the following primers: Fwd 5’-CAGATCACATGCTTCCCAC-3’ and Rev 5’-AGACCACAAAAGACTCATCAC-3’.
Immunoblotting
Organoids were isolated and processed for immunoblotting as previously described7. Briefly, protein was run through a 4–12% gradient protein gel (BioRad), transferred onto a PVDF membrane and blocked using Odyssey Blocking buffer (LI-COR). Membranes were stained with mouse anti–STAT3 and rabbit anti–pSTAT3 (Tyr705) and detected using IRDye 680RD goat anti–rabbit IgG and IRDye 800CW goat anti–mouse IgG (LI-COR) on an Odyssey CLx near-infrared fluorescence imaging system.
RNA deep sequencing
cDNA libraries were prepared with the Ovation Trio Low Input RNA–seq System V2 (NuGEN) and sequenced on Illumina’s HIseq platform using a paired-end protocol at the NYU Genomics Core. On average, each sample yielded 25 million paired reads, which passed FASTQC quality control. Sequencing reads were aligned with STAR v2.6.1d against the GRCm38.p6 genome assembly, resulting in an average of 20 million aligned pairs per sample, and normalized read counts were calculated using Cufflinks v2.2.1 against the same reference genome. Differential gene expression was determined with Cuffdiff and gene ontology analysis performed using Qiagen’s Ingenuity Pathway Analysis (IPA, Qiagen).
16S library preparation and sequencing analysis
DNA was isolated from stool samples using the NucleospinSoil Kit (Macherey-Nagel). Bacterial 16S rRNA gene was amplified at the V4 region using primer pairs and paired-end amplicon sequencing was performed on the Illumina MiSeq system. Sequencing reads were processed using the DADA2 pipeline in the QIIME2 software package47. Beta diversity was calculated using unweighted UniFrac distance48,49. Principle Coordinate Analysis (PCoA) was performed on the UniFrac distance matrix and visualized with EMPeror50.
Statistical Analysis
All data was analyzed using Graphpad Prism v.7. All graphs show the mean and standard error of the mean (SEM). The number of samples per group is indicated in the figure legend. The log-rank Mantel–Cox test was used for comparison of survival curves. An unpaired two-tailed t-test was used to evaluate differences between two groups. Welch’s correction was used when variances were significantly different between groups. An analysis of variance (ANOVA) with Dunnett’s or Tukey’s multiple comparisons test was used to evaluate experiments involving multiple groups. For bacterial and viral titers, log10 transformed data was analyzed. Correlation was analyzed using Pearson r. Weight curves were analyzed using area under the curve (AUC) followed by two-tailed t-test. All p-values are shown in figures.
Supplementary Material
Acknowledgements
We wish to thank the following NYU facilities for use of their instruments and technical assistance: Microcopy Core (RR023704), Histopathology and Immunohistochemistry Core (P30CA016087, NIH S10 OD010584–01A1, and S10 OD018338–01), the Cytometry and Cell Sorting Laboratory (P30CA016087), the Genome Technology Center (P30CA016087), and the Gnotobitoic Facility (Colton Center for Autoimmunity). We also wish to thank Satoru Fujii (Washington University School of Medicine in St. Louis) for his technical support in human organoid culture. This research was supported by US National Institute of Health (NIH) grants R01 HL123340 (K.C.), R01 DK093668 (K.C.), R01 DK103788 (K.C. and P.L.), R01 AI121244 (K.C.), and R01 AI130945 (P.L.). This work was also supported by Vilcek Fellowship (J.A.N.), Sir Keith Murdoch Fellowship (J.A.N.), Crohn’s & Colitis Foundation Research Fellowship Award (Y.M-I); Faculty Scholar grant from the Howard Hughes Medical Institute (K.C.), Advanced Research Grant from the Merieux Institute (K.C.), Rainin Foundation Innovator Award (K.C.), Stony Wold-Herbert Fund (K.C.), PureTech Health (K.C.), Pfizer (K.C. and P.L.), NYU CTSI (NIH/NCATS 1UL TR001445; K.C. and P.L.), and philanthropy from Bernard Levine (K.C. and P.L.); and. K.C. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
Data Availability
The data that support the findings of this study are available from the corresponding author upon request. FASTQ files corresponding to the RNA–seq and 16S rRNA sequencing have been deposited in a public database (RNA–seq accession no. GSE129384, 16S accession no. PRJNA532632).
Competing Financial Interests
K.C. has consulted for PureTech Health and AbbVie Inc. and is an inventor on U.S. patent application 62/608,404.
References
- 1.Cadwell K The virome in host health and disease. Immunity 42, 805–813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neil JA & Cadwell K The Intestinal Virome and Immunity. J Immunol 201, 1615–1624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basic M, et al. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis 20, 431–443 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Seamons A, et al. Obstructive Lymphangitis Precedes Colitis in Murine Norovirus-Infected Stat1-Deficient Mice. Am J Pathol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzawa-Ishimoto Y, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 214, 3687–3705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kernbauer E, Ding Y & Cadwell K An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abt MC, et al. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med 8, 327ra325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thepaut M, et al. Protective role of murine norovirus against Pseudomonas aeruginosa acute pneumonia. Vet Res 46, 91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JY, et al. Enteric Viruses Ameliorate Gut Inflammation via Toll-like Receptor 3 and Toll-like Receptor 7-Mediated Interferon-beta Production. Immunity 44, 889–900 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Vijay-Kumar M, et al. Activation of toll-like receptor 3 protects against DSS-induced acute colitis. Inflamm Bowel Dis 13, 856–864 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Bailey D, Thackray LB & Goodfellow IG A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J Virol 82, 7725–7728 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strong DW, Thackray LB, Smith TJ & Virgin HW Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J Virol 86, 2950–2958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu S, et al. Regulation of Norovirus Virulence by the VP1 Protruding Domain Correlates with B Cell Infection Efficiency. J Virol 90, 2858–2867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomov VT, et al. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol 87, 7015–7031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nice TJ, Strong DW, McCune BT, Pohl CS & Virgin HW A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J Virol 87, 327–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones MK, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346, 755–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldridge MT, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science 347, 266–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, et al. Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine. Cell Host Microbe 22, 449–459 e444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilen CB, et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360, 204–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L, et al. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe 17, 85–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCartney SA, et al. MDA-5 recognition of a murine norovirus. PLoS Pathog 4, e1000108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDuff DA, et al. HOIL1 Is Essential for the Induction of Type I and III Interferons by MDA5 and Regulates Persistent Murine Norovirus Infection. J Virol 92(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickert G, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med 206, 1465–1472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo SU, et al. Intestinal macrophages arising from CCR2(+) monocytes control pathogen infection by activating innate lymphoid cells. Nat Commun 6, 8010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tschurtschenthaler M, et al. Type I interferon signalling in the intestinal epithelium affects Paneth cells, microbial ecology and epithelial regeneration. Gut 63, 1921–1931 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Pott J, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A 108, 7944–7949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldridge MT, et al. Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. J Virol 91, pii: e02079–02016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nice TJ, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gronke K, et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim ES, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21, 1228–1234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao K, et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 14, 282–289 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Kim YG, et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ettayebi K & Hardy ME Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J Virol 77, 11790–11797 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Vega V, et al. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J Virol 78, 4827–4837 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCune BT, et al. Noroviruses Co-opt the Function of Host Proteins VAPA and VAPB for Replication via a Phenylalanine-Phenylalanine-Acidic-Tract-Motif Mimic in Nonstructural Viral Protein NS1/2. MBio 8(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker ES, et al. Inherent structural disorder and dimerisation of murine norovirus NS1–2 protein. PLoS One 7, e30534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broggi A, Tan Y, Granucci F & Zanoni I IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol 18, 1084–1093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin PK, et al. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota. Nat Microbiol 3, 1131–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Winkle JA, et al. Persistence of Systemic Murine Norovirus Is Maintained by Inflammatory Recruitment of Susceptible Myeloid Cells. Cell Host Microbe 24, 665–676 e664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips G, Tam CC, Rodrigues LC & Lopman B Prevalence and characteristics of asymptomatic norovirus infection in the community in England. Epidemiol Infect 138, 1454–1458 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Bouziat R, et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356, 44–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouziat R, et al. Murine Norovirus Infection Induces TH1 Inflammatory Responses to Dietary Antigens. Cell Host Microbe 24, 677–688 e675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28, 2106–2113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozupone C, Lladser ME, Knights D, Stombaugh J & Knight R UniFrac: an effective distance metric for microbial community comparison. ISME J 5, 169–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vazquez-Baeza Y, Pirrung M, Gonzalez A & Knight R EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2, 16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.