SUMMARY
The size of dendrite arbors shapes their function and differs vastly between neuron types. The signals that control dendritic arbor size remain obscure. Here, we find that in the retina, starburst amacrine cells (SACs) and rod bipolar cells (RBCs) express the homophilic cell-surface protein AMIGO2. In Amigo2 knockout (KO) mice, SAC and RBC dendrites expand while arbors of other retinal neurons remain stable. SAC dendrites are divided into a central input region and a peripheral output region that provides asymmetric inhibition to direction-selective ganglion cells (DSGCs). Input and output compartments scale precisely with increased arbor size in Amigo2 KO mice, and SAC dendrites maintain asymmetric connectivity with DSGCs. Increased coverage of SAC dendrites is accompanied by increased direction selectivity of DSGCs without changes to other ganglion cells. Our results identify AMIGO2 as a cell-type-specific dendritic scaling factor and link dendrite size and coverage to visual feature detection.
Graphical Abstract
In Brief
Soto et al. find that two retinal interneurons express the cell-surface protein AMIGO2. Deletion of Amigo2 causes dendrites of these neurons, but not others, to expand, preserving branching patterns and connectivity. Increased interneuron dendrite coverage is accompanied by enhanced response selectivity of retinal output neurons.
INTRODUCTION
Throughout the nervous system, dendritic arbor size is critical for neuronal function (Lefebvre et al., 2015; Wong and Ghosh, 2002). In the retina, dendrites prescribe the region from which photoreceptor signals are collected and shape receptive fields (Brown et al., 2000; Field et al., 2010; Schwartz et al., 2012). The dendrites of each neuron type cover the retina evenly to represent visual space homogeneously. Dendrite size and overlap (i.e., coverage) vary widely between retinal neurons, and differences in coverage determine the ratios in which circuit components are combined (Keeley et al., 2014; Lefebvre et al., 2015). Whether dendrite size is controlled independent of other morphological features, what molecular mechanisms determine the overlap of low- and high-coverage neurons, and how dendrite coverage shapes specific retinal computations is unknown.
Rod bipolar cells (RBCs) are conserved from rodents to primates (Grünert and Martin, 1991; Peng et al., 2019), receive input from rod photoreceptors, and mediate vision near the threshold (Field et al., 2005). The dendrites of RBCs have low coverage (~2) (Tsukamoto and Omi, 2013). Their arbor size varies inversely with RBC density across mouse strains (Keeley et al., 2014) and increases when neighbors are removed during development (Johnson et al., 2017), indicating that homotypic signals constrain RBC dendrite growth. The cell-adhesion molecule DSCAML1 mediates repulsive interactions between RBCs, and their dendrites fasciculate in Dscaml1-null mutants (Fuerst et al., 2009). However, RBC arbor size is reduced, rather than increased, in these mutants (Fuerst et al., 2009). The homotypic signals that limit RBC dendrite growth, therefore, remain to be identified.
Starburst amacrine cells (SACs) are conserved from rodents to primates (Peng et al., 2019; Yonehara et al., 2016) and have the highest coverage (>40) of all cells in the retina (Keeley et al., 2007; MacNeil and Masland, 1998). The cell bodies of ON and OFF SACs are distributed regularly (i.e., mosaics) in the ganglion cell and inner nuclear layer, respectively (Keeley et al., 2007; Rockhill et al., 2000). Soma mosaics facilitate even dendrite coverage of the retina. Cell death initiated by purinergic signaling (Resta et al., 2005) and repulsive signals from two cell-surface proteins (MEGF10 and MEGF11) (Kay et al., 2012) organize SAC mosaics. However, SAC arbor size is reduced, rather than increased, by deletion of Megf10, and dendrite territories do not vary with SAC density from the center to the periphery of the retina or across different mouse strains (Keeley et al., 2007; Ray et al., 2018). Thus, unknown signals control SAC dendrite size independent of cell body mosaics.
The dendrites of ON and OFF SACs stratify in two narrow bands in the inner plexiform layer (IPL). Within each band, SACs elaborate radially symmetric arbors with central input and peripheral output regions (Briggman et al., 2011; Ding et al., 2016; Famiglietti, 1991; Greene et al., 2016; Kim et al., 2014). The four to six primary dendrites of SAC arbors with their daughter branches function as independent motion sensors, each preferring motion away from the soma (Euler et al., 2002; Morrie and Feller, 2018; Poleg-Polsky et al., 2018). Centrifugal motion preference is shaped by the distribution of input and output regions in the SAC arbor (Ding et al., 2016; Greene et al., 2016; Kim et al., 2014; Vlasits et al., 2016) and translated into direction-selective inhibition of direction-selective ganglion cells (DSGCs) through asymmetric connectivity (Briggman et al., 2011; Fried et al., 2002; Wei et al., 2011; Yonehara et al., 2011). Thus, as for many neurons, the circuit function of SACs relies on dendrite stratification, branching, subcellular compartmentalization, and synaptic specificity in addition to arbor size. Signals that control arbor size independent of other features of dendritic morphology and connectivity have not yet been identified. In addition, how dendrite arbor size and coverage shape the detection of specific visual features is unknown.
Here, we discover that RBCs and SACs express the homophilic cell-surface protein AMIGO2. We show that AMIGO2 selectively controls RBC and SAC dendrite size and coverage and shapes the encoding of motion direction in the retina.
RESULTS
Expression of Amigo2 in the Retina
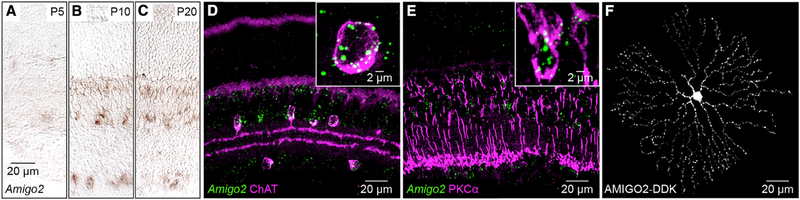
Cell-surface proteins with extracellular leucine-rich repeat (LRR) domains guide many processes in neural development (de Wit and Ghosh, 2014). In an in situ hybridization screen, we found that the LRR-containing cell-surface protein AMIGO2 is expressed by cells on either side of the IPL and in a band of cells near the outer margin of the inner nuclear layer (Figures 1A–1C). Transcripts were abundant by postnatal day 10 (P10), when retinal circuits are forming, and persisted in mature neurons (P20) (Hoon et al., 2014). In combined in situ hybridization and immunohistochemistry experiments, we found that Amigo2-positive cells on either side of the IPL stained for choline acetyltransferase (ChAT), identifying these neurons as SACs (Figure 1D). Similarly, combined Amigo2 in situ hybridization and protein kinase Cɑ (PKCɑ) immunohistochemistry identified the Amigo2-positive band of cells at the outer margin of the inner nuclear layer as RBCs (Figure 1E). Analysis of single-cell RNA-sequencing (scRNA-seq) data profiling amacrine cells (Macosko et al., 2015) confirmed strong expression of Amigo2 in SACs as well as a small population of Slc35d3-positive cells (Figure S1). A separate analysis of scRNA-seq data profiling bipolar cells (Shekhar et al., 2016) confirmed strong expression in Prkca-positive RBCs (Figure S1).
Figure 1. Amigo2 Expression in the Retina.
(A–C) In situ hybridization for Amigo2 in postnatal day 5 (P5; A), P10 (B), and P20 (C) retinas.
(D and E) Combined in situ hybridization for Amigo2 (green) with immunohistochemistry for ChAT (D; magenta) and PKCɑ (E; magenta) in sections of P20 retinas.
(F) Representative SAC biolistically labeled with AMIGO2-DDK in a flat-mounted P20 retina. The cell was digitally isolated in Amira for visual clarity
See also Figure S1.
Our efforts to raise specific antibodies against AMIGO2 failed and commercially available antibodies indistinguishably labeled wild-type and Amigo2 knockout (KO) retinas (data not shown). To evaluate the subcellular distribution of AMIGO2, we used a gene gun (i.e., biolistics) to deliver a DDK-tagged construct to SACs (STAR Methods). This technique cannot label RBCs (Morgan and Kerschensteiner, 2011). AMIGO2-DDK was distributed in puncta across SAC arbors (Figure 1F). Thus, Amigo2 is expressed in SACs and RBCs in the developing and mature retina, with the protein covering dendrite arbors of the former.
Cell Density and Neurite Stratification of SACs and RBCs in Amigo2 KO Mice
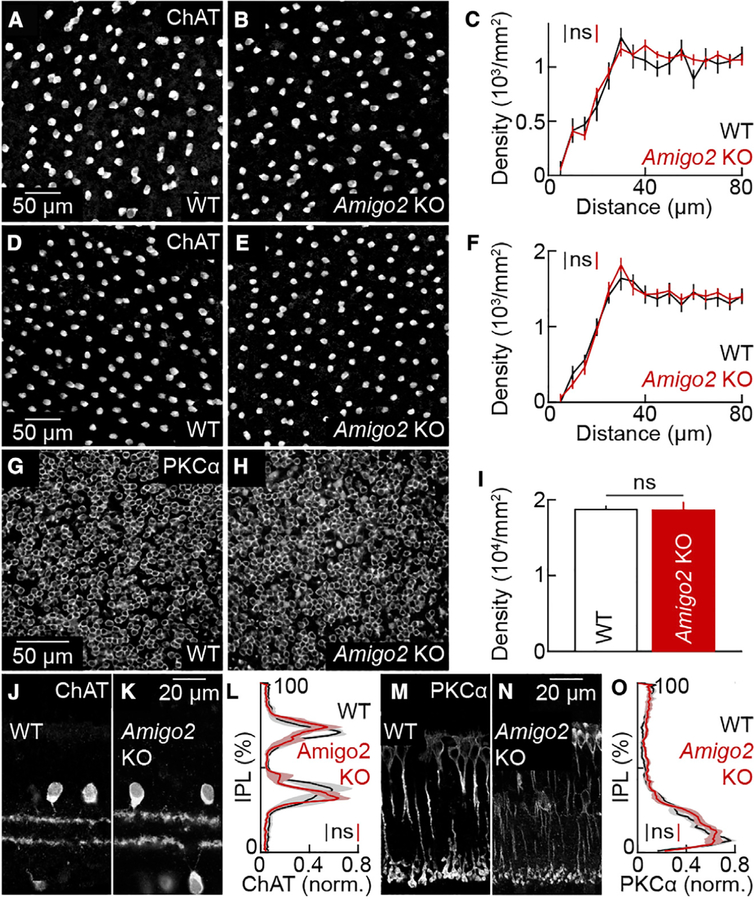
To study the function of AMIGO2 in development, we generated Amigo2 KO mice with transcription activator-like effector nucleases (TALENs; STAR Methods). ON and OFF SACs form independent mosaics in the ganglion cell and inner nuclear layer, respectively (Keeley et al., 2007; Rockhill et al., 2000). The density of ON SACs and their distribution in the ganglion cell layer measured by density recovery profiles (Rodieck, 1991) were unchanged in Amigo2 KO compared to wild-type mice (Figures 2A–2C). OFF SACs were more abundant than ON SACs, but their density and distributions in the inner nuclear layer were indistinguishable between wild-type and Amigo2 KO littermates (Figures 2D–2F). RBCs are the most numerous bipolar cell type and are packed near the outer margin of the inner nuclear layer (Keeley et al., 2014; Wässle et al., 2009). The density of RBCs was not significantly different between wild-type and Amigo2 KO mice (Figure 2G–2I). In addition, the overall area of the retina was the same in Amigo2 KO and wild-type mice (Figure S2). Matching cell densities, therefore, reflect preservation of total SAC and RBC numbers.
Figure 2. Soma and Neurite Distributions of SACs and RBCs in Wild-Type and Amigo2 KO Mice.
(A and B) Images of the ganglion cell layer in retinal flat mounts from wild-type (A) and Amigo2 KO (B) retinas stained for ChAT.
(C) Density recovery profiles (mean ± SEM) of SACs in the ganglion cell layer of wild-type (n = 7 retinas) and Amigo2 KO (n = 12 retinas) mice; p = 0.74 by bootstrapping. The overall density of SACs in the ganglion cell layer was not significantly different between wild-type (1,143 ± 70 cells mm−2) and Amigo2 KO retinas (1,108 ± 38 cells mm−2; p = 0.89 by Mann-Whitney U test.
(D and E) Images of the inner nuclear layer in retinal flat mounts from wild-type (D) and Amigo2 KO (E) retinas stained for ChAT.
(F) Density recovery profiles (mean ± SEM) of SAC cell bodies in the inner nuclear layer of wild-type (n = 8 retinas) and Amigo2 KO (n = 11 retinas) mice; p = 0.98 by bootstrapping. The overall density of SACs in the nuclear layer was not significantly different between wild-type (1,472 ± 108 cells mm−2) and Amigo2 KO retinas (1,453 ± 79 cells mm−2; p = 0.97 by Mann-Whitney U test.
(G and H) Images of the inner nuclear layer in retinal flat mounts from wild-type (G) and Amigo2 KO (H) retinas stained for PKCɑ.
(I) Density (mean ± SEM) of RBCs in wild-type (18,738 ± 435 cells mm−2, n = 4 retinas) and Amigo2 KO (18,629 ± 1,036 cells mm−2, n = 5 retinas) mice; p = 0.91 by Mann-Whitney U test.
(J and K) Sections of P20 wild-type (J) and Amigo2 KO (K) retinas stained for ChAT.
(L) Lines (shaded areas) indicate the mean (± SEM) ChAT lamination patterns in the inner plexiform layer (IPL) in wild-type (n = 4 retinas) and Amigo2 KO (n = 8 retinas) mice; p = 0.43 by bootstrapping.
(M and N) Sections of P20 wild-type (M) and Amigo2 KO (N) retinas stained for PKCɑ.
(O) Lines (shaded areas) indicate the mean (± SEM) PKCɑ lamination patterns in the IPL in wild-type (n = 8 retinas) and Amigo2 KO (n = 9 retinas) mice; p = 0.40 by bootstrapping.
Throughout the figure, ns indicates no significant differences for statistical comparisons.
See also Figure S2.
Bipolar cell axons and amacrine cell dendrites target particular depths of the retina’s IPL to form specific circuits (Masland, 2001). Neurite stratification is regulated by cell-adhesion molecules (Duan et al., 2018; Peng et al., 2017; Yamagata and Sanes, 2008). However, vibratome sections stained for ChAT and PKCɑ revealed that stratification patterns of SACs and RBCs in the IPL were unaffected by Amigo2 deletion (Figures 2J–2O). Thus, SACs and RBCs are generated and survive in appropriate numbers, are evenly distributed in the right layers, and target their neurites correctly independent of AMIGO2.
Precisely Scaled Expansion of ON SAC Arbors in Amigo2 KO Mice
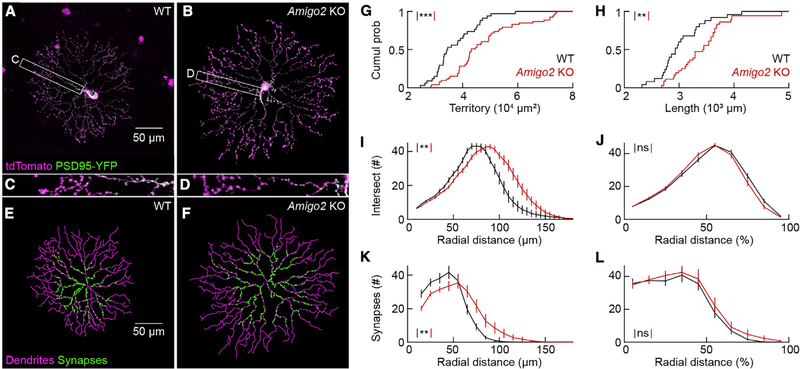
To analyze the effects of AMIGO2 on individual neurons, we biolistically labeled ON SACs with a cytosolic fluorophore (tdTomato) and PSD95-YFP, a marker of excitatory input synapses (Kerschensteiner et al., 2009; Morgan et al., 2008; Figures 3A–3D). We manually traced neurites and automatically identified synapses (Kerschensteiner et al., 2009; Morgan et al., 2008; Figures 3E and 3F). The dendrite size of many neurons, although not ON SACs (Keeley et al., 2007), varies with retinal eccentricity (Wässle and Boycott, 1991). We therefore restricted our analysis throughout this study to neurons at mid-eccentricity (i.e., between one-third and two-thirds of the distance from the optic nerve head to the edge of the retina) in P20–P35 mice. In Amigo2 KO mice, ON SACs had longer dendrites that occupied larger territories than in wild-type mice (Figures 3G and 3H). SAC branching is sparse in the central input region of the arbor and increases toward the arbor periphery where neurotransmitters are released (Briggman et al., 2011; Ding et al., 2016; Famiglietti, 1991; Greene et al., 2016; Kim et al., 2014). Branches of individual SACs avoid each other to establish space-filling arbors (i.e., dendritic self-avoidance). The numbers of ON SAC self-crossings were not significantly different between Amigo2 KO and wild-type mice (Amigo2 KO: 6.8 ± 0.4 cell−1, n = 29; wild-type: 7 ± 0.6 cell−1, n = 22; p = 0.85 by Mann-Whitney U test). Branch distributions were shifted away from the soma in Amigo2 KO compared to wild-type ON SACs (Figure 3I), but, when arbor expansion was taken into account, branch distributions of Amigo2 KO and wild-type ON SACs were indistinguishable (Figure 3J). Similarly, the distribution of input synapses was right-shifted in Amigo2 KO ON SACs for absolute radial distances (Figure 3K) but matched wild-type ON SAC distributions exactly when this distance was normalized to the maximal arbor extent (Figure 3L). Thus, the dendrites of Amigo2 KO ON SACs are precisely scaled versions of their wild-type counterparts with intact self-avoidance and accurately preserved input and output divisions.
Figure 3. ON SAC Arbors Expand but Maintain Branching Patterns and Subcellular Compartmentalization in Amigo2 KO Mice.
(A and B) ON SACs biolistically labeled with cytosolic tdTomato (magenta) and PSD95-YFP (green), a marker of excitatory synapses, in flat-mounted P20 wild-type (A) and Amigo2 KO (B) retinas. Cells were digitally isolated in Amira for visual clarity.
(C and D) Higher magnification view of the insets in (A) (shown in C) and (B) (shown in D).
(E and F) Dendrite tracings (magenta) and output of synapse identification (green) for the ON SACs in (A) (shown in E) and in (B) (shown in F).
(G) Cumulative distributions of ON SAC dendrite territories in wild-type (37,259 ± 1,361 μm2, n = 34 cells, n = 12 retinas) and Amigo2 KO (47,584 ± 1,712 μm2, n = 53 cells, n = 16 retinas) mice; p = 5.5 3 10−5 by Mann-Whitney U test.
(H) Cumulative distributions of ON SAC dendrite lengths in wild-type (3,028 ± 86 μm, n = 25 cells, n = 12 retinas) and Amigo2 KO (3,422 ± 85 μm, n = 36 cells, n = 16 retinas) retinas; p = 0.0021 by Mann-Whitney U test.
(I and J) Summary data of Sholl analyses for ON SAC branching patterns in wild-type (n = 16 cells, n = 10 retinas) and Amigo2 KO (n = 18 cells, n = 10 retinas) retinas. Distributions of branches as a function of absolute distance from the soma; p = 0.0056 by bootstrapping (I). (J) Distributions of branches as a function of normalized radial distance; p = 0.21 by bootstrapping (J).
(K and L) Summary data of the radial distribution of excitatory synapses in ON SAC dendrites in wild-type (n = 16 cells, n = 10 retinas) and Amigo2 KO (n = 16 cells, n = 9 retinas) retinas. Distributions of synapses as a function of absolute distance from the soma; p = 0.012 by bootstrapping (K). Distributions of synapses as a function of normalized radial distance; p = 0.79 by bootstrapping (L).
Throughout the figure, ***p < 0.001, **p < 0.01, and ns indicates no significant differences for statistical comparisons.
Precisely Scaled Expansion of OFF SAC Arbors in Amigo2 KO Mice
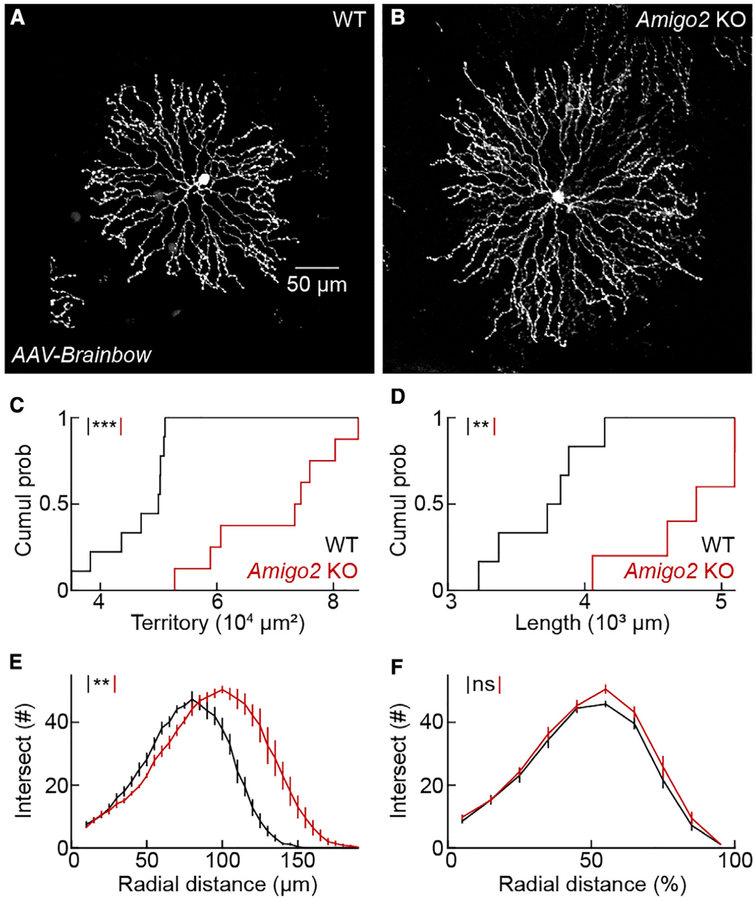
OFF SACs in the inner nuclear layer are inaccessible to biolistic labeling (Morgan and Kerschensteiner, 2011). To test the influence of AMIGO2 on OFF SAC dendrites, we labeled these cells by injecting adeno-associated viruses expressing varying ratios of fluorescent proteins (i.e., AAV-Brainbow; Cai et al., 2013) into the vitreous of ChAT-Cre mice on an Amigo2 KO or wild-type background (Figures 4A and 4B). The dendrites of OFF SACs in Amigo2 KO retinas were longer and covered larger territories than in wild-type retinas (Figures 4C and 4D). As for ON SACs, branch distributions were right-shifted for absolute radial distances in Amigo2 KO compared to wild-type OFF SACs (Figure 4E) but overlapped precisely when branching was analyzed relative to the maximal arbor extent (Figure 4F). Thus, AMIGO2 controls ON and OFF SAC arbor size without affecting the branching patterns or compartmentalization of dendrites.
Figure 4. OFF SAC Arbors Expand but Maintain Branching Patterns in Amigo2 KO Mice.
(A and B) OFF SACs labeled by AAV-Brainbow injections into the vitreous of ChAT-Cre mice on a wild-type (A) or Amigo2 KO (B) background.
(C) Cumulative distributions of OFF SAC dendrite territories in wild-type (46,283 ± 2,003 μm2,n =9 cells, n = 3 retinas) and Amigo2 KO (70,070 ± 3,972 μm2, n = 8 cells, n = 3 retinas) mice; p = 8.2 3 10−5 by Mann-Whitney U test.
(D) Cumulative distributions of OFF SAC dendrite lengths in wild-type (3,696 ± 139 μm, n = 6 cells, n = 3 retinas) and Amigo2 KO (4,733 ± 193 μm, n = 5 cells, n = 3 retinas) mice; p = 0.0087 by Mann-Whitney U test.
(E and F) Summary data of Sholl analyses for OFF SAC branching patterns in wild-type (n = 6 cells, n = 3 retinas) and Amigo2 KO (n = 5 cells, n = 3 retinas) mice. Distributions of branches as a function of absolute distance from the soma; p = 0.0051 by bootstrapping (E). Distributions of branches as a function of normalized radial distance; p = 0.32 by bootstrapping (F).
Throughout the figure, ***p < 0.001, **p < 0.01, and ns indicates no significant differences for statistical comparisons.
Asymmetric Connectivity of SACs with DSGCs in Amigo2 KO Mice
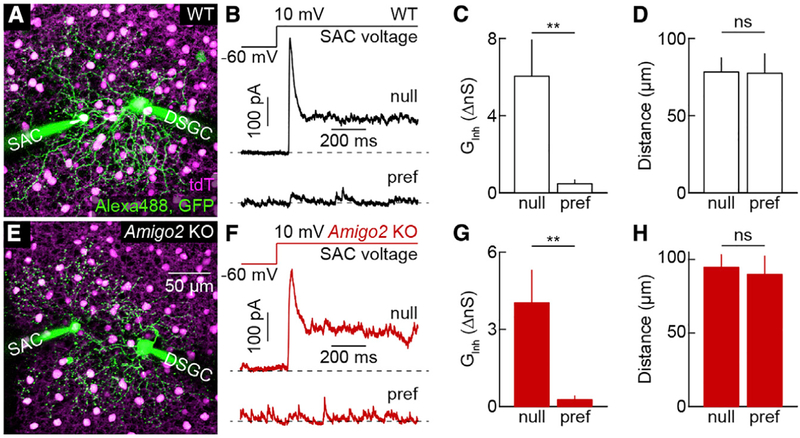
Different dendrites of SACs synapse onto different DSGCs (Briggman et al., 2011; Fried et al., 2002; Wei et al., 2011; Yonehara et al., 2011). SAC dendrites pointing toward the temporal retina provide GABAergic input selectively to DSGCs that prefer motion in the nasal direction (nDSGCs). Combined with the centrifugal motion preference of SAC dendrites, asymmetric connectivity results in direction-selective inhibition of DSGCs (Mauss et al., 2017). To probe whether AMIGO2 regulates the connectivity of SACs with DSGCs, we performed paired patch-clamp recordings in ChAT-Cre Ai9 DRD4-EGFP mice on wild-type or Amigo2 KO backgrounds. In these mice, all SACs express tdTomato and nDSGCs express EGFP (Huberman et al., 2009; Kay et al., 2011; Pei et al., 2015). We targeted nDSGCs and SACs on their nasal (null) or temporal (preferred) side under two-photon guidance (Figures 5A and 5E). We isolated inhibitory signals pharmacologically (30 μM D-AP5, 40 μM NBQX, and 5 μM DHμE) and clamped the voltage of nDSGCs to the reversal potential of excitatory conductances (~0 mV). Consistent with previous observations (Brombas et al., 2017; Fried et al., 2002; Lee et al., 2010; Wei et al., 2011; Yonehara et al., 2011), we found that on a wild-type background, depolarization of null-side SACs elicited robust inhibitory post-synaptic currents in nDSGCs, whereas depolarization of equidistant preferred-side SACs elicited weak or no input (Figures 5B–5D). Asymmetric inhibitory connectivity of null- and preferred-side SACs with nDSGCs was preserved in the Amigo2 KO background (Figures 5F–5H). Thus, AMIGO2 regulates neither the subcellular compartmentalization of SACs’ input synapses (Figure 3) nor the asymmetric target preferences of their output connections.
Figure 5. Asymmetric Connectivity between SACs and DSGCs in Wild-Type and Amigo2 KO Mice.
(A) Maximum intensity projection of a two-photon image stack acquired at the end of a paired recording from a DSGC and a null-side SAC in a ChAT-Cre Ai9 DRD4-EGFP mouse on a wild-type background.
(B) Representative inhibitory postsynaptic currents (IPSCs) elicited in DSGCs by depolarizing SACs on the null (top trace) and preferred side (bottom trace) in a wild-type background.
(C and D) Summary data (mean ± SEM) comparing the inhibitory conductances activated in DSGCs by stimulation of SACs on the null versus preferred side (C; null side: 6.0 ± 1.9 nS, n = 6 pairs, n = 4 retinas; preferred side: 0.47 ± 0.20 nS n = 4 pairs, n = 4 retinas; p = 0.0095 by Mann-Whitney U test) and the soma-soma distance of SAC-DSGC pairs (D, null side: 78 ± 9.2 μm; preferred side: 78 ± 13 μm; p = 0.91 by Mann-Whitney U test) in a wild-type background.
(E) Maximum intensity projection of a two-photon image stack acquired at the end of a paired recording from a DSGC and a null-side SAC in a ChAT-Cre Ai9 DRD4-EGFP mouse on an Amigo2 KO background.
(F) Representative IPSCs elicited in DSGCs by depolarizing SACs on the null (top trace) and preferred side (bottom trace) in an Amigo2 KO background.
(G and H) Summary data (mean ± SEM) comparing the inhibitory conductances activated in DSGCs by stimulation of SACs on the null versus preferred side (G; null side: 4.0 ± 1.3 nS, n = 7 pairs, n = 4 retinas; preferred side: 0.26 ± 0.15 nS, n = 6 pairs, n = 4 retinas; p = 0.0012 by Mann-Whitney U test) and the soma-soma distance of SAC-DSGC pairs (H; null side: 94.7 ± 8.6 μm; preferred side: 90 ± 13 μm; p = 1 by Mann-Whitney U test) in an Amigo2 KO background. Inhibitory conductances activated by stimulation of individual null-side SACs were not significantly different between wild-type and Amigo2 KO backgrounds (p = 0.37).
Throughout the figure, **p < 0.01 and ns indicates no significant differences for statistical comparisons.
Enhanced Selectivity of DSGCs in Amigo2 KO Mice
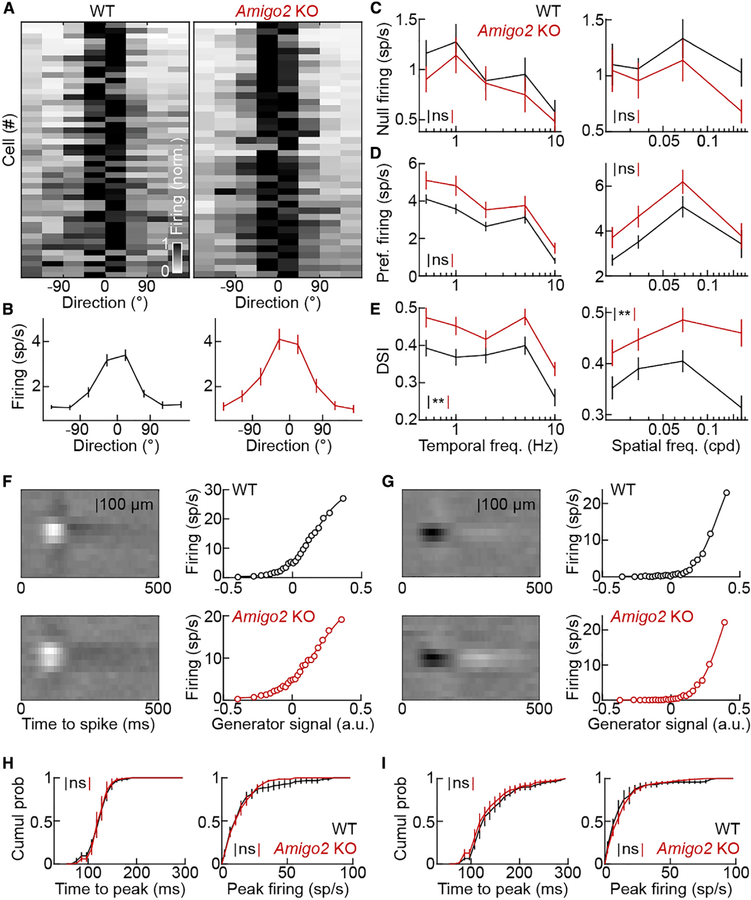
Because SAC density remains constant as arbors expand, dendrite coverage is higher in Amigo2 KO than wild-type mice. SAC arbor size increases in Amigo2 KO retinas without other changes in morphology or connectivity, allowing us to test the influence of dendrite coverage on circuit function. We recorded large ensembles of retinal ganglion cells on multielectrode arrays and identified DSGCs by their responses to square-wave gratings drifting in eight different directions (Figures 6A and 6B; STAR Methods). We presented drifting grating stimuli at a range of temporal and spatial frequencies. SACs suppress null-direction firing of DSGCs by GABAergic inhibition (Fried et al., 2002; Taylor and Vaney, 2002; Vlasits et al., 2014; Yoshida et al., 2001) and enhance preferred-direction firing of DSGCs via cholinergic volume transmission (Brombas et al., 2017; Chen et al., 2016; Lee et al., 2010; Sethuramanujam et al., 2016; Yonehara et al., 2011). In Amigo2 KO mice, null-direction firing rates of DSGCs tended to be lower (Figure 6C) and preferred-direction firing rates higher (Figure 6D) than in their wild-type littermates, but neither trend alone reached statistical significance. However, when responses to all stimulus directions were taken into account, the direction selectivity of DSGCs was enhanced robustly across temporal and spatial stimulus frequencies in Amigo2 KO compared to wild-type retinas (Figure 6E).
Figure 6. Response Selectivity of DSGCs Is Enhanced in Amigo2 KO Retinas.
(A) Grayscale plots of the DSGC responses in wild-type (left panel) and Amigo2 KO (right panel) mice to drifting grating stimuli. Each row depicts average responses of one cell to 0.023–0.034 cycles per degree (cpd) gratings drifting at 1–2 cycles s−1 (wild-type: n = 46 cells, n = 6 retinas; Amigo2 KO: n = 40 cells, n = 8 retinas). Responses of each cell were centered on the direction eliciting the maximal response and its highest-response neighbor.
(B) Summary data (mean ± SEM) of DSGC responses in (A) for wild-type (left panel) and Amigo2 KO (right panel) mice.
(C) Responses of DSGCs (mean ± SEM) to null-direction drifting grating stimuli of varying temporal (left panel) and spatial (right panel) frequencies in wild-type (n = 46 cells, n = 6 retinas) and Amigo2 KO (n = 40 cells, n = 8 retinas) mice. Null-direction firing rates tended to be lower in Amigo2 KO compared to wild-type mice, but this trend did not reach statistical significance; p = 0.39 (left panel) and p = 0.52 (right panel) by bootstrapping.
(D) Responses of DSGCs (mean ± SEM) to preferred-direction drifting grating stimuli of varying temporal (left panel) and spatial (right panel) frequencies in wild-type (n = 46 cells, n = 6 retinas) and Amigo2 KO (n = 40 cells, n = 8 retinas) mice. Preferred-direction firing rates tended to be higher in Amigo2 KO compared to wild-type mice, but this trend did not reach statistical significance; p = 0.15 (left panel) and p = 0.056 (right panel) by bootstrapping.
(E) Direction selectivity indices (DSIs; mean ± SEM) of DSGC responses to drifting grating stimuli of varying temporal (left panel) and spatial (right panel) frequencies in wild-type (n = 46 cells, n = 6 retinas) and Amigo2 KO (n = 40 cells, n = 8 retinas) mice. DSIs were consistently higher in Amigo2 KO compared to wild-type mice; p = 0.0032 (left panel) and p = 0.0085 (right panel) by bootstrapping.
(F and G) Spatiotemporal receptive field maps (left panels) and static nonlinearities (right panels) of representative ON (F) and OFF (G) ganglion cells in wild-type (top panels) and Amigo2 KO (bottom panels) mice.
(H) Cumulative distributions of time to peak sensitivity (left panel) and peak firing rates (right panel) of ON ganglion cells in wild-type (n = 143 cells, n = 5 retinas) and Amigo2 KO (n = 156 cells, n = 6 retinas) mice; p = 0.94 for time to peak sensitivity and p = 0.54 for peak firing rates by bootstrapping.
(I) Cumulative distributions of time to peak sensitivity (left panel) and peak firing rates (right panel) of OFF ganglion cells in wild-type (n = 185 cells, n = 5 retinas) and Amigo2 KO (n = 233 cells, n = 6 retinas) mice; p = 0.67 for time to peak sensitivity and p = 0.59 for peak firing rates by bootstrapping.
Throughout the figure, **p < 0.01 and ns indicates no significant differences for statistical comparisons.
See also Figures S3 and S6.
In the same recordings, we analyzed the responses of non-DSGCs to spatiotemporal white noise stimuli with a linear-nonlinear cascade model (Chichilnisky, 2001; Pearson and Kerschensteiner, 2015; Figures 6F and 6G). Spatiotemporal receptive fields were not significantly different in their time to peak sensitivity (Figures 6F and 6G) or size (Figure S3) for ON and OFF ganglion cells in Amigo2 KO compared to wild-type retinas. Equally, the peak firing rates of ON and OFF ganglion cells in response to white noise stimuli were unchanged in Amigo2 KO compared to wild-type retinas (Figures 6F and 6G). Thus, increased dendrite coverage of SACs in Amigo2 KO retinas enhances the feature selectivity of DSGCs across a wide range of spatial and temporal stimulus frequencies without affecting the responses of other ganglion cell types.
Selective Expansion of RBC Dendrites in Amigo2 KO Mice
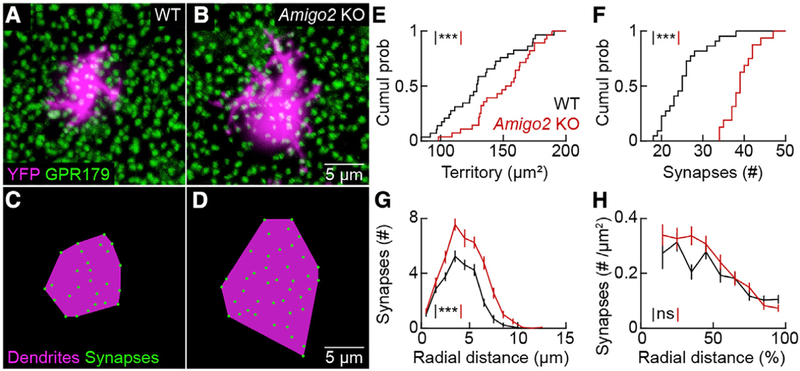
To probe whether the function of AMIGO2 is conserved between SACs and RBCs, we sparsely labeled the latter by intravitreal injection of AAV-Grm6-YFP (Johnson et al., 2017). In retinal flat mounts stained for GPR179, a component of the postsynaptic receptor complex (Orlandi et al., 2013; Ray et al., 2014; Sarria et al., 2016), we found that RBC dendrites expanded and formed more synapses in Amigo2 KO than wild-type mice (Figures 7A–7F). Similar to SACs, the distribution of input synapses on RBC dendrites was shifted to the right when measured as a function of absolute distance from their territory centers (Figure 7G), but when arbor expansion was taken into account, the radial distributions of RBC input synapse densities in Amigo2 KO and wild-type mice were brought into register (Figure 7H).
Figure 7. RBC Dendrites Expand and Form More Synapses in Amigo2 KO Mice.
(A and B) Maximum intensity projections of confocal image stacks of the outer plexiform layer of wild-type (A) and Amigo2 KO (B) retinas. RBC dendrites are labeled by AAV-Grm6-YFP and postsynaptic specializations are marked by GPR179 clusters.
(C and D) Schematic representation of dendrite territories (magenta) and synapses (green) of the RBCs in (A) (shown in C) and (B) (shown in D), respectively.
(E) Cumulative distributions of RBC dendrite territories in wild-type (131.4 ± 5.2 μm2,n= 29 cells, n =5 retinas) and Amigo2 KO (149.9 ± 4.5 μm2, n = 28 cells, n = 4 retinas) mice; p = 0.0075 by Mann-Whitney U test.
(F) Cumulative distributions of RBC dendritic synapses in wild-type (25.2 ± 1.1, n = 22 cells, n = 5 retinas) and Amigo2 KO (39.13 ± 0.89, n = 16 cells, n = 4 retinas) mice; p = 6.1 3 10−7 by Mann-Whitney U test.
(G) Summary data (mean ± SEM) of the distribution of synapses onto RBCs as a function of the absolute distance from the center of their dendritic territory in wild-type (n = 22 cells, n = 5 retinas) and Amigo2 KO (n = 16 cells, n = 4 retinas) mice; p = 1.0 3 10−5 by bootstrapping.
(H) Summary data (mean ± SEM) of the synapse density across RBC dendrite territories as a function of the relative distance from the center of the territory in wild-type (n = 22 cells, n = 5 retinas) and Amigo2 KO (n = 16 cells, n = 4 retinas) mice; p = 0.16 by bootstrapping.
Throughout the figure, ***p < 0.001 and ns indicates no significant differences for statistical comparisons.
See also Figures S4 and S5.
Unlike SACs, RBCs have separate axon arbors. The territories of RBC axons were indistinguishable between Amigo2 KO and wild-type mice (Figure S4), indicating that AMIGO2 controls dendrite size selectively. RBC dendrites contact rod photoreceptors together with horizontal cells (Hoon et al., 2014), which do not express AMIGO2. We sparsely labeled horizontal cells by intravitreal injection of AAV-CAG-YFP (Soto et al., 2018). Neither horizontal cell axons, which contact rods, nor horizontal cell dendrites, which contact cones, differed in size between Amigo2 KO and wild-type mice (Figure S5). Thus, the influence of AMIGO2 on arbor size is conserved between SACs and RBCs, selective for dendrites versus axons, and restricted to neurons expressing AMIGO2.
DISCUSSION
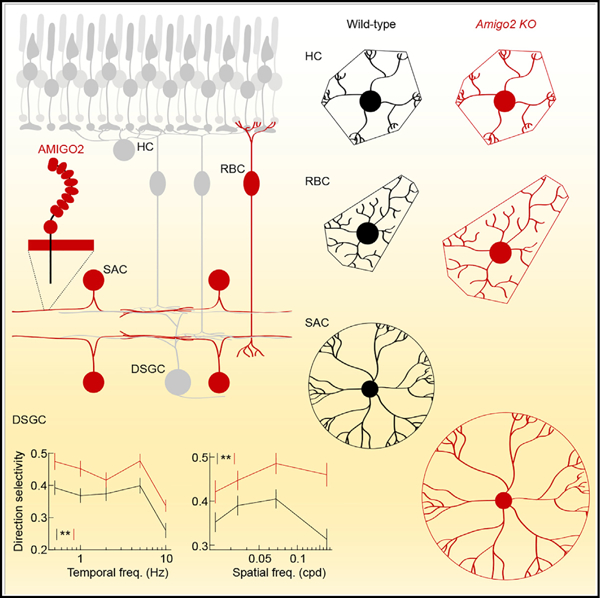
Here, we discover that AMIGO2 controls the size and coverage of SAC and RBC dendrites and shapes direction-selective signals from the retina to the brain. AMIGO2 is one of three related type I transmembrane proteins (AMIGO1–AMIGO3) (Kuja-Panula et al., 2003). The three AMIGOs contain six conventional and two cysteine-rich LRR domains followed by an immunoglobulin domain in their extracellular N terminus and interact homo- and heterophilically (Kuja-Panula et al., 2003). AMIGO1 promotes axon development in cultured neurons and zebrafish (Kuja-Panula et al., 2003; Zhao et al., 2014). We show that AMIGO2 regulates dendrite development in the retina (Figures 3, 4, and 7). The function of AMIGO3 remains unknown.
Because AMIGO2 interacts homophilically (Kuja-Panula et al., 2003), the most parsimonious explanation for the increased dendrite size in KO mice is that AMIGO2 mediates growth-inhibiting signals between neurons of the same type (i.e., homotypic interactions). The effects of AMIGO2 are conserved between RBCs and SACs, indicating that the same cue can control the dendrite size of low-coverage (RBCs) and high-coverage (SACs) neurons. Homotypic repulsion constrains dendrite growth of strictly territorial neurons (i.e., coverage = 1) in the retina and other sensory systems (Grueber and Sagasti, 2010; Grueber et al., 2003; Lefebvre et al., 2015; Millard et al., 2007). We propose that, unlike the absolute stop signals of homotypic repulsion, AMIGO2-mediated interactions limit dendrite growth in a cumulative manner. We speculate that differences in AMIGO2 expression levels and downstream cascades amplify and attenuate signals to establish cell-type-specific coverage of RBC and SAC dendrites.
In many instances, dendrites of a neuron type vary selectively in size across tissue topography (e.g., retinal eccentricity) and species (e.g., mouse versus macaque) (Rodieck, 1989; Wässle and Boycott, 1991). These observations suggest that dendrite size can be controlled independent of other morphological features. However, all previously identified molecular cues co-regulate dendrite size with branching patterns and/or arbor shapes (Fuerst et al., 2009; McAllister et al., 1995; Shen et al., 2009; Soto et al., 2013; Sun et al., 2013). Here, we find that dendrite arbors of ON and OFF SACs and RBCs in Amigo2 KO mice are precisely scaled versions of their wild-type counterparts (Figures 3, 4, and 7). This suggests that AMIGO2 selectively controls dendrite size. We, therefore, propose to categorize AMIGO2 as a dendritic scaling factor, the first of its kind. We hypothesize that signals mediated by AMIGO2 and other scaling factors contribute to cell-type-specific, topographic, and species-dependent differences in dendrite size.
Some effects of dendrite size on neuronal function are easy to predict. In the retina, receptive fields are approximately congruent with dendrites, and receptive field size, therefore, scales with dendritic arbor size (Bleckert et al., 2014; Brown et al., 2000; Crook et al., 2008; Schwartz et al., 2012). By contrast, how dendrite size and coverage shape circuit functions emerging from complex interactions of multiple components is less obvious and remains unknown. SACs serve at least two circuit functions. In developing retina, SACs generate and propagate cholinergic waves of activity that pattern projections from the retina to the brain (Kerschensteiner, 2014; Kirkby et al., 2013). In the mature retina, SACs generate direction-selective responses of DSGCs by cholinergic volume transmission and asymmetric GABAergic inhibition (Diamond, 2017; Lee et al., 2010; Mauss et al., 2017; Sethuramanujam et al., 2016; Wei, 2018). Cholinergic waves were indistinguishable between Amigo2 KO mice and wild-type littermates (Figure S6), likely because the expression of AMIGO2 and its effects on SAC arbors size begin after the respective period of development (Figure 1). In the mature retina, we found that the connectivity of individual SACs with DSGCs was unchanged (Figure 5), but the direction selectivity of DSGC responses was robustly enhanced (Figure 6). We speculate that this is because increased SAC coverage and convergence onto DSGCs increase cholinergic and GABAergic input from the SAC population. A recent study found that direction selectivity is decreased in Sema6A KO mice, in which SAC coverage is reduced (Morrie and Feller, 2018). Together, these studies indicate the feature-selective signals from the retina to the brain are controlled bidirectionally by the dendrite size and coverage of an interneuron.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. Information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Kerschensteiner (kerschensteinerd@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
We used Transcription Activator-like Effector Nucleases (TALENs) to generate Amigo2 knockout (Amigo2 KO) mice. Genomic DNA sequences of Amigo2 were obtained from the GenBank Database (NM_178114). TALEN target sequences were: left 5′-TCAGG AATGTGCCCCACTGC-3′ and right 50-TTGGTGCAGCTGACAATGTC-3′ separated by a 16-bp spacer containing a SfaNI restriction site. Target specificity of TALENs was validated in N2A cells with Xtremegene HP (Roche), followed by a T7E1 assay (NEB). T7 TALEN templates for in vitro transcription were EcoRI digested, purified, and in-vitro transcribed with the T7 mMessage mMachine Ultra kit (Life Technologies). After transcription, both RNAs were purified with the Megaclear kit (Life Technologies). We obtained mouse zygotes by mating C57Bl6/J DBA2 stud males (Jackson Labs) to super-ovulated C57Bl6/J DBA2 females at a 1:1 ratio. Fertilized one-cell embryos were injected with 50 ng nL−1 (25 ng nL−1 of each TALEN) into the pronucleus and cytoplasm of each zygote and transferred into pseudo-pregnant females. With this procedure, we obtained 22 live F0 mice. We extracted genomic DNA from the tails of F0 mice and amplified a 299-bp PCR product using Amigo2-F1: 5′-ATT GGT GGG AGA CTG AGC TGA TGA GAA GCG-30 and Amigo2-R1: 5′-GTC CGA TTC TGT TAT AGC TCA GAT CCA GTC-3′ oligonucleotides, Klentaq LA (DNA Polymerase Technology) and a Biometra PCR machine (94°C for 3 min, then 40 cycles of 94○C for 1 min and 68°C for 2.5 min, followed by a final extension step at 68°C for 8 min). PCR products were digested with SfaNI (Biolabs) and run on 2% agarose gels. Restriction yielded 101-bp and 198-bp fragments for wild-type animals, whereas PCR fragments of potential Amigo2 KO animals remained uncut (i.e., 299 bp). Our results revealed that 12 of the 22 F1 mice animals had lost the targeted SfaNI site in one or both Amigo2 alleles. Litters obtained by crossing the 12 F1 founders with C57Bl6/J mice were used to analyze the corresponding mutations by sequencing with the Amigo2-F1 oligo. Five deletions at the TALENs target resulted in frameshifts mutations in the Amigo2 gene. Four lines (2-bp, 8-bp, 22-bp, and 43-bp deletions) were crossed to C57Bl6/J for at least four more generations. After confirming that results from the four lines were identical, we combined them and present them as Amigo2 KO data throughout this paper.
For paired recordings, we first crossed ChAT-Cre mice (Rossi et al., 2011) to the Ai9 reporter strain (Madisen et al., 2010) to label SACs with tdTomato. We then paired ChAT-Cre Ai9 mice with DRD4-EGFP mice line, in which nasal-motion-preferring DSGCs (nDSGCs) express EGFP (Huberman et al., 2009; Rivlin-Etzion et al., 2011). All mouse lines were crossed onto a C57Bl6/J background for more than five generations. Except for developmental in situ hybridization experiments, we used postnatal day 20 to 35 (P20–35) mice of both sexes throughout our study. All procedures were approved by the Animal Studies Committee of Washington University School of Medicine (Protocol # 20170033) and performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
METHOD DETAILS
Adeno-associated viruses
To label OFF SACs, we injected 250 nL of AAV-Brainbow (Cai et al., 2013) into the vitreous of newborn (postnatal day 0, P0) ChAT-Cre and ChAT-Cre Amigo2 KO mice. To label RBCs and horizontal cells, we injected 250 nL of AAV-Grm6-YFP (Johnson et al., 2017) and AAV-CAG-YFP (Soto et al., 2018), respectively, into the vitreous of newborn wild-type and Amigo2 KO mice.
Tissue preparation
Mice were euthanized with CO2 followed by decapitation and enucleation. For in situ hybridization, immunohistochemistry, and biolistic labeling, eyes were transferred into oxygenated mouse artificial cerebrospinal fluid (mACSFHEPES) containing (in mM): 119 NaCl, 2.5 KCl, 1 NaH2PO4, 2.5 CaCl2, 1.3 MgCl2, 20 HEPES, and 11 glucose (pH adjusted to 7.37 using NaOH). Retinas were either isolated and flat-mounted on filter paper (HABG01300, Millipore), or left in the eyecup for 30 min fixation with 4% paraformaldehyde in mACSFHEPES. For patch-clamp and multielectrode array recordings, mice were dark-adapted for at least 2 hr before their retinas were isolated under infrared illumination (> 900 nm) in mACSFNaHCO3 containing (in mM) 125 NaCl, 2.5 KCl, 1 MgCl2, 1.25 NaH2PO4, 2 CaCl2, 20 glucose, 26 NaHCO3 and 0.5 L-glutamine equilibrated with 95% O2 5% CO2. Retinas were then flat mounted on membrane disks (Anodisc13, Whatman).
In situ hybridization
We followed previously described in situ hybridization methods (Soto et al., 2013; Yamagata et al., 2002). We prepared the DNA template for riboprobes by PCR from an MGC clone obtained from Horizon/Dharmacon using the following primers: Amigo2-ST3: 5′-GCAATTAACCCTCACTAAAGCCCCAGCGCCTCAGGAATGTGC-30 and Amigo2-RT7: 5′-TAATACGACTCACTATAGGAGCTGGTG GGAGTGCCTGGAGTC-3′. We synthesized the antisense RNA probes using the DIG RNA labeling kit (Roche) from T7 sites incorporated by PCR in the DNA template. Fixed eyecups (s. Tissue preparation) were cryoprotected and sliced (thickness: 20 μm) with a cryotome (Leica). Retinal sections were pretreated using proteinase K, postfixed, permeabilized using Triton X-100, and prehybridized for 4 hr at 65°C. Hybridization was performed overnight at 65°C using 1–2 μg mL−1 antisense RNA. The hybridized riboprobe was detected using anti-DIG alkaline phosphatase labeled antibodies and BCIP/NBT (Roche) overnight. For combined in situ hybridization and immunohistochemistry, the in situ signal was detected using anti-DIG peroxidase-conjugated antibodies with Cy3-Tyramide as a substrate (PerkinElmer), followed by staining with an antibody against ChAT (goat anti-ChAT, EMD Millipore, 1:1000) or PKCα (mouse anti-PKAα, Sigma, 1:1000) (Kay et al., 2012; Soto et al., 2013).
Immunohistochemistry
Vibratome slices (thickness: 60 μm) were blocked for 2 hr with 5% Normal Donkey Serum in PBS, embedded in 4% agarose (Sigma) and incubated overnight at 4°C with primary antibodies. Slices were then washed in PBS (3 3 20 min) and incubated in secondary antibodies for 2 hr. Flat-mount preparations were frozen and thawed three times after cryoprotection (1 hr 10% sucrose in PBS, 1 hr 20% sucrose in PBS, and overnight 30% sucrose in PBS at 4°C), blocked with 5% Normal Donkey Serum in PBS for 2 hr, and then incubated with primary antibodies for five days at 4○C and washed in PBS (3 × 1 hr). Subsequently, flat mounts were incubated with secondary antibodies for one day at 4○C and washed in PBS (3 × 1 hr). The following primary antibodies were used in this study: goat anti-ChAT (1:1000, EMD Millipore), mouse anti-PKAɑ (1:1000, Sigma),mouse anti-DDK (1:1000, Origene), rabbit anti-GFP (1:500, Invitrogen), and mouse anti-GPR179 (1:1000, EDM Millipore). Secondary antibodies were Alexa 488- and Alexa 568 conjugates (1:1000, Invitrogen).
Biolistic labeling
We coated gold particles (diameter: 1.6 μm, Bio-Rad) with plasmids encoding cytosolic tdTomato and postsynaptic density protein 95 (PSD95) fused at its C terminus to YFP (Kerschensteiner et al., 2009), or AMIGO2-DDK (Origene). We used a helium-pressurized gun (40 psi, Bio-Rad) to deliver particles to a sparse population of cells in the ganglion cell layer and incubated the transfected retinas in mACSFHEPES in a humid oxygenated chamber at 33°C for 16–18 hr (Morgan and Kerschensteiner, 2011). We identified ON SACs by their characteristic arbor morphology.
Confocal imaging
We acquired confocal image stacks on an Fv1000 laser-scanning microscope (Olympus) or an LSM 800 microscope (Zeiss) with an AiryScan detector array. Voxel sizes varied from 0.043–0.1 μm (x/y-z) to 0.309–0.5 μm (x/y-z). We traced neurite arbors of SACs using Simple Neurite Tracer in Fiji (Schindelin et al., 2012) and performed Sholl analysis on the tracings (Ferreira et al., 2014). SAC and RBC arbor territories were measured as the smallest convex polygons to encompass the dendrites of a given cell in a 2D-projection. PSD95-YFP clusters marking excitatory input synapses on SAC dendrites were identified in Fiji. RBC dendrite tips that overlapped with staining for GPR179 were counted as synapses (Johnson et al., 2017). Radial distributions of neurites and synapses were calculated using scripts written in MATLAB. The surface area of RBC axons was measured from iso-intensity surfaces in Amira (FEI) (Johnson et al., 2017). The density of SACs and RBCs changes with retinal eccentricity (Keeley et al., 2007). Therefore, to minimize variation, we restricted our analysis to the middle third of the retina (i.e., >⅓ and <⅔ the distance from the optic nerve head to the margins of the retina).
Patch-clamp recordings
Dual whole-cell voltage-clamp recordings from SACs and DSGCs were performed in flat-mounted retinas superfused (5–7 mL min−1) with warm (30–33°C) mACSFNaHCO3 equilibrated with 95% O2 5% CO2. The somata of EGFP-expressing nDSGCs and tdTomato-expressing SACs were targeted under two-photon guidance in ChAT-Cre Ai9 DRD4-EGFP mice on a wild-type or Amigo2 KO background, and correct targeting was confirmed by morphologies revealed by two-photon imaging of Alexa 488 (0.1 mM) included in the intracellular solution containing (in mM) 120 Cs-gluconate, 1 CaCl2, 1 MgCl2, 10 Na-HEPES, 11 EGTA, 10 TEA-Cl, 2 Qx314, ATP-Na2, and 0.1 GTP-Na (pH adjusted to 7.2 with CsOH). Patch pipettes had resistances of 5–8 MΩ (borosilicate glass). The preferred direction of nDSGCs was inferred by the coordinates of retinas. SACs located on the nasal side of the nDSGCs were defined as null-side SACs, whereas SACs located on the temporal side were defined as preferred-side SACs. Inhibitory postsynaptic currents (IPSCs) of DSGCs were measured at the reversal potential of excitatory conductances (0 mV) in response to depolarization of paired SACs from −60 to 10 mV, in the presence of D-AP5 (30 μM, Tocris), NBQX (40 mM, Tocris) and DHβE (5 μM, Tocris). Liquid junction potentials were corrected offline. Signals were amplified with a Multiclamp 700B amplifier (Molecular Devices), filtered at 3 kHz (8-pole Bessel low-pass), and sampled at 10 kHz (Digidata 1440A, Molecular Devices).
Multielectrode array recordings
We recorded large ensembles of retinal ganglion cells on planar arrays with 252 electrodes arranged in a 16 × 16 grid with the corner positions empty (30 μm electrode size, 100 μm center-center spacing, Multi Channel Systems). During recordings, retinas were perfused with warm (30–33°C) mACSFNaHCO3 equilibrated with 95% O2 5% CO2 at 5–7 mL min-1. Signals of each electrode were filtered (300–3,000 Hz) and digitized at 10 kHz. Signal cut-outs from 1 ms before to 2 ms after crossings of negative thresholds (set manually for each channel) were recorded to hard disk together with the time of threshold crossing (i.e., the spike time). We sorted spikes into trains representing the activity of individual neurons by principal component analysis of spike waveforms (Offline Sorter, Plexon). We used refractory periods to assess the quality of the sorting and retained only spike trains in which < 0.2% of interspike intervals were < 2 ms. When the activity of a single neuron had been recorded on more than one electrode (identified by cross-correlation), we used only the train with the most spikes in our subsequent analysis.
Visual stimuli were presented on an organic light-emitting display (OLED-XL, eMagin) and focused on the retina through a 20X 0.5 NA water immersion objective (Olympus) covering a ~1.7 × 2.3 mm rectangular area. Stimuli were generated in MATLAB using the Cogent Graphics toolbox extensions developed by John Romaya at the LON at the Wellcome Department of Imaging Neuroscience. The display output was linearized using custom-written scripts. All recordings were from the dorsal retina where M-opsin dominates (Wang et al., 2011; Wei et al., 2010), and the average intensity of each stimulus was 1000 M-opsin isomerizations per cone per second (1000 R* cone−1 s−1). To evaluate direction selectivity, we presented four repeats of full-field square-wave gratings of varying spatial and temporal frequencies (spatial frequency: 0.023–0.17 cycles per degree or cpd, temporal frequency: 0.5–10 cycles s−1) drifting in eight directions at 45° intervals. Stimuli were shown in pseudorandom orders. Each stimulus repeat lasted 5 s. Direction selectivity indices (DSIs) were calculated based on the circular variance of the response (Pearson and Kerschensteiner, 2015; Piscopo et al., 2013). Cells with DSI ≥ 0.3 at temporal and spatial stimulus frequencies eliciting preferred-direction average firing rates >4 Hz were considered direction selective. To map spatiotemporal receptive fields, the stimulus display was divided into vertical bars (width: ~50 μm, height: ~1.7 mm). The intensity of each bar was randomly chosen from a Gaussian distribution (RMS contrast: 40%) and updated every 33 ms (refresh rate: 30 Hz) for 30 min. A linear-nonlinear cascade model was used to analyze the responses of ganglion cells to this stimulus (Chichilnisky, 2001; Pearson and Kerschensteiner, 2015). We mapped linear spatiotemporal receptive fields by reverse correlation of the spike response with the stimulus (i.e., the spike-triggered stimulus average or STA). In a separate part of the recording, we then analyzed the dependence of the spike rate on the match between the stimulus and the STA (i.e., the generator signal) to compute the static nonlinearity.
Cholinergic waves were recorded in P7 retinas for >1 hr in darkness. Waves were detected as peaks in the population activity that exceeded a threshold of 1.5 times the Loess-filtered (f = 0.67) running average (Demas et al., 2003). Spike time tiling coefficients of ganglion cell pairs were calculated as defined by Cutts and Eglen (2014).
scRNA-seq Analysis
Count matrices were downloaded from GEO accessions GSE63473 [whole retina/amacrine cell (Macosko et al., 2015)] and GSE81905 [bipolar cell (Shekhar et al., 2016)]. Data were loaded into R and analyzed using Monocle 3 (version 0.1.3) (Cao et al., 2019; Qiu et al., 2017a, 2017b; Trapnell et al., 2014). Preliminary steps included removal of dead cells and doublets (cells with low or high read counts). High variance genes were used as ordering genes for dimension reduction using tSNE (Clark et al., 2019). Normalized expression values were calculated for each cell and all plotting was done using ggplot2 (version 3.2.1).
Whole retina data were first processed and amacrine cells identified by Pax6 expression. Cells within these clusters were reanalyzed separately to optimize visualization of discrete subtypes. Genes identified as cell type specific markers (Macosko et al., 2015) were plotted and co-expression within Amigo2 was determined for both Chat and Slc35d3. Bipolar cells were analyzed in a similar manner; data were first cleaned for contaminating cells (e.g., rods) and putative bipolar cells (clusters marked by Vsx2 and Otx2) were re-processed separately to optimize dimension reduction and separation of cell types. Genes identified as cell type specific markers (Shekhar et al., 2016) were plotted and co-expression with Amigo2 was determined for Prkca.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed using scripts written in MATLAB. Summary data are given as mean ± SEM. Non-parametric tests (Mann-Whitney U, Wilcoxon signed-rank) and bootstrapping were used to compare data from different experimental groups as specified in the figure legends. Statistical significance was considered when p < 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-ChAT | Abcam | RRID:AB_2079595 |
| Mouse anti-PKCɑ | Sigma | RRID:AB_477375 |
| Mouse anti-DDK | Origene Technologies | RRID:AB_2622345 |
| Rabbit anti-GFP | Invitrogen | RRID:AB_221569 |
| Mouse anti-GPR179 | Millipore | RRID:AB_2069582 |
| Donkey anti-rabbit IgG Alexa 488 | ThermoFisher | RRID:AB_2535792 |
| Donkey anti-rabbit IgG Alexa 568 | ThermoFisher | RRID:AB_253401 |
| Donkey anti-goat IgG Alexa 488 | ThermoFisher | RRID:AB_2534104 |
| Donkey anti-goat IgG Alexa 568 | ThermoFisher | RRID:AB_2534104 |
| Bacterial and Virus Strains | ||
| AAV-EF1a-BbTagBY (AAV-Brainbow) | Addgene | Cat# 45185-AAV9 |
| AAV-EF1a-BbChT (AAV-Brainbow) | Addgene | Cat# 45186-AAV9 |
| AAV-Grm6-YFP | Johnson et al., 2017 | N/A |
| AAV-CAG-YFP | Soto et al., 2018 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | ThermoFisher | RRID:AB_2629482 |
| DIG-RNA labeling kit | Roche | Cat# 11277073910 |
| T7 RNA polymerase | Promega | Cat# P2075 |
| RNasin | Promega | Cat# N2111 |
| SfaNI | NEB | Cat# R0172S |
| Xtremegene HP | Roche | Cat# 6366244001 |
| T7E1 | NEB | Cat# E3321 |
| T7 mMessage mMachine Ultra Kit | Life Technologies | Cat# AM1345 |
| Megaclear Kit | Life Technologies | Cat# AM1908 |
| Microcarrier gold 1.6 mm | BioRad | Cat# 1652264 |
| Tissue-Tek OCT compound | Sakura | Cat# M71484 |
| TSA Cyanine 3 (Cy3) | Perkin Elmer | Cat# SAT704A001EA |
| NBT/BCIP | Roche | Cat# 11681451001 |
| Experimental Models: Organisms/Strains | ||
| Wild-type (C57Bl6/J) | Jackson Laboratory | RRID:IMSR_JAX:000664 |
| B6;Cg-Gt(ROSA)26Sortm9(CAG-tdTomato) Hze/J (Ai9) | Jackson Laboratory | RRID:IMSR_JAX:007909 |
| B6;129S6-Chattm2(cre)Lowl/J | Jackson Laboratory | RRID:IMSR_JAX:006410 |
| Tg(Drd4-EGFP)W18Gsat | MMRRC | Cat# MGI:3839370 |
| Amigo2 KO | This study | N/A |
| Recombinant DNA | ||
| CMV-tdTomato | Clontech | 632534 |
| CMV-PSD95-YFP | Ann Marie Craig | |
| Mouse Amigo2 | Horizon Dharmacon | 105827 |
| Software and Algorithms | ||
| MATLAB | The Mathworks | RRID:SCR_001622 |
| Cogent Graphics Toolbox | Laboratory of Neurobiology | www.vislab.ucl.ac.uk |
| Fiji | Schindelin et al.,. 2012 | RRID:SCR_002285 |
| Amira | ThermoFisher | RRID:SCR_014305 |
| Offline Sorter | Plexon Inc | RRID:SCR_000012 |
| Monocle 3 | Trapnell Laboratory | https://cole-trapnell-lab.github.io/monocle3 |
Highlights.
Starburst amacrine cells (SACs) and rod bipolar cells (RBCs) express AMIGO2
Dendrites of SACs and RBCs expand in a precisely scaled manner in Amigo2 KO mice
Dendrites of other retinal neurons and connectivity of SACs and RBCs are unchanged
Enhanced direction selectivity accompanies increased SAC dendrite coverage
ACKNOWLEDGMENTS
We thank Renate Lewis (Hope Center Transgenic Vector Core) and Mia Wallace (Mouse Genetic Core) for help in generating Amigo2 KO mice. We are grateful to Mike Casey (Molecular Genetics Service Vision Core) for help cloning cDNA constructs. We thank the members of the Kerschensteiner lab for helpful discussion throughout the project and for critical reading of the manuscript. This work was supported by the NIH (grant EY027411 to F.S. and D.K., grants EY023341 and EY026978 to D.K., and grant EY002687 to the Department of Ophthalmology and Visual Sciences), the Grace Nelson Lacy Glaucoma Research Fund (to D.K.) and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.09.085.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
The code supporting the current study is available at https://github.com/kerschensteinerd/CellRep_2019. The published article includes all datasets generated or analyzed during this study.
REFERENCES
- Bleckert A, Schwartz GW, Turner MH, Rieke F, and Wong ROL (2014). Visual space is represented by nonmatching topographies of distinct mouse retinal ganglion cell types. Curr. Biol 24, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, and Denk W (2011). Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188. [DOI] [PubMed] [Google Scholar]
- Brombas A, Kalita-de Croft S, Cooper-Williams EJ, and Williams SR (2017). Dendro-dendritic cholinergic excitation controls dendritic spike initiation in retinal ganglion cells. Nat. Commun 8, 15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, He S, and Masland RH (2000). Receptive field microstructure and dendritic geometry of retinal ganglion cells. Neuron 27, 371–383. [DOI] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, and Sanes JR (2013). Improved tools for the Brainbow toolbox. Nat. Methods 10, 540–547. [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Pei Z, Koren D, and Wei W (2016). Stimulus-dependent recruitment of lateral inhibition underlies retinal direction selectivity. eLife 5, e21053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichilnisky EJ (2001). A simple white noise analysis of neuronal light responses. Network 12, 199–213. [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, et al. (2019). Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook JD, Peterson BB, Packer OS, Robinson FR, Gamlin PD, Troy JB, and Dacey DM (2008). The smooth monostratified ganglion cell: evidence for spatial diversity in the Y-cell pathway to the lateral geniculate nucleus and superior colliculus in the macaque monkey. J. Neurosci 28, 12654–12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutts CS, and Eglen SJ (2014). Detecting pairwise correlations in spike trains: an objective comparison of methods and application to the study of retinal waves. J. Neurosci 34, 14288–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A (2014). Control of neural circuit formation by leucinerich repeat proteins. Trends Neurosci 37, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demas J, Eglen SJ, and Wong ROL (2003). Developmental loss of synchronous spontaneous activity in the mouse retina is independent of visual experience. J. Neurosci 23, 2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond JS (2017). Inhibitory interneurons in the retina: types, circuitry, and function. Annu. Rev. Vis. Sci 3, 1–24. [DOI] [PubMed] [Google Scholar]
- Ding H, Smith RG, Poleg-Polsky A, Diamond JS, and Briggman KL (2016). Species-specific wiring for direction selectivity in the mammalian retina. Nature 535, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, Laboulaye MA, Liu J, Peng YR, Yamagata M, Toma K, and Sanes JR (2018). Cadherin combinations recruit dendrites of distinct retinal neurons to a shared interneuronal scaffold. Neuron 99, 1145–1154.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T, Detwiler PB, and Denk W (2002). Directionally selective calcium signals in dendrites of starburst amacrine cells. Nature 418, 845–852. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV (1991). Synaptic organization of starburst amacrine cells in rabbit retina: analysis of serial thin sections by electron microscopy and graphic reconstruction. J. Comp. Neurol 309, 40–70. [DOI] [PubMed] [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjöström PJ, and van Meyel DJ (2014). Neuronal morphometry directly from bitmap images. Nat. Methods 11, 982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field GD, Sampath AP, and Rieke F (2005). Retinal processing near absolute threshold: from behavior to mechanism. Annu. Rev. Physiol 67, 491–514. [DOI] [PubMed] [Google Scholar]
- Field GD, Gauthier JL, Sher A, Greschner M, Machado TA, Jepson LH, Shlens J, Gunning DE, Mathieson K, Dabrowski W, et al. (2010). Functional connectivity in the retina at the resolution of photoreceptors. Nature 467, 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried SI, Münch TA, and Werblin FS (2002). Mechanisms and circuitry underlying directional selectivity in the retina. Nature 420, 411–414. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, and Burgess RW (2009). DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 64, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene MJ, Kim JS, and Seung HS; EyeWirers (2016). Analogous convergence of sustained and transient inputs in parallel on and off pathways for retinal motion computation. Cell Rep 14, 1892–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, and Sagasti A (2010). Self-avoidance and tiling: mechanisms of dendrite and axon spacing. Cold Spring Harb. Perspect. Biol 2, a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, and Jan YN (2003). Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr. Biol 13, 618–626. [DOI] [PubMed] [Google Scholar]
- Grünert U, and Martin PR (1991). Rod bipolar cells in the macaque monkey retina: immunoreactivity and connectivity. J. Neurosci 11, 2742–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon M, Okawa H, Della Santina L, and Wong ROL (2014). Functional architecture of the retina: development and disease. Prog. Retin. Eye Res 42, 44–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, and Barres BA (2009). Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron 62, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Tien NW, Shen N, Pearson JT, Soto F, and Kerschensteiner D (2017). Homeostatic plasticity shapes the visual system’s first synapse. Nat. Commun 8, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, and Sanes JR (2011). Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J. Neurosci 31, 7753–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Chu MW, and Sanes JR (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Whitney IE, Raven MA, and Reese BE (2007). Dendritic spread and functional coverage of starburst amacrine cells. J. Comp. Neurol 505, 539–546. [DOI] [PubMed] [Google Scholar]
- Keeley PW, Whitney IE, Madsen NR, St John AJ, Borhanian S, Leong SA, Williams RW, and Reese BE (2014). Independent genomic control of neuronal number across retinal cell types. Dev. Cell 30, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner D (2014). Spontaneous network activity and synaptic development. Neuroscientist 20, 272–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner D, Morgan JL, Parker ED, Lewis RM, and Wong ROL (2009). Neurotransmission selectively regulates synapse formation in parallel circuits in vivo. Nature 460, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, Turaga SC, Purcaro M, Balkam M, Robinson A, Behabadi BF, et al. ; EyeWirers (2014). Space-time wiring specificity supports direction selectivity in the retina. Nature 509, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkby LA, Sack GS, Firl A, and Feller MB (2013). A role for correlated spontaneous activity in the assembly of neural circuits. Neuron 80, 1129–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuja-Panula J, Kiiltomäki M, Yamashiro T, Rouhiainen A, and Rauvala H (2003). AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J. Cell Biol 160, 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim K, and Zhou ZJ (2010). Role of ACh-GABA cotransmission in detecting image motion and motion direction. Neuron 68, 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Sanes JR, and Kay JN (2015). Development of dendritic form and function. Annu. Rev. Cell Dev. Biol 31, 741–777. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, and Masland RH (1998). Extreme diversity among amacrine cells: implications for function. Neuron 20, 971–982. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH (2001). The fundamental plan of the retina. Nat. Neurosci 4, 877–886. [DOI] [PubMed] [Google Scholar]
- Mauss AS, Vlasits A, Borst A, and Feller M (2017). Visual circuits for direction selectivity. Annu. Rev. Neurosci 40, 211–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, and Katz LC (1995). Neurotrophins regulate dendritic growth in developing visual cortex. Neuron 15, 791–803. [DOI] [PubMed] [Google Scholar]
- Millard SS, Flanagan JJ, Pappu KS, Wu W, and Zipursky SL (2007). Dscam2 mediates axonal tiling in the Drosophila visual system. Nature 447, 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, and Kerschensteiner D (2011). Shooting DNA, dyes, or indicators into tissue slices using the gene gun. Cold Spring Harb. Protoc 2011, 1512–1514. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Schubert T, and Wong ROL (2008). Developmental patterning of glutamatergic synapses onto retinal ganglion cells. Neural Dev 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrie RD, and Feller MB (2018). A dense starburst plexus is critical for generating direction selectivity. Curr. Biol 28, 1204–1212.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi C, Cao Y, and Martemyanov KA (2013). Orphan receptor GPR179 forms macromolecular complexes with components of metabotropic signaling cascade in retina ON-bipolar neurons. Invest. Ophthalmol. Vis. Sci 54, 7153–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JT, and Kerschensteiner D (2015). Ambient illumination switches contrast preference of specific retinal processing streams. J. Neurophysiol 114, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Chen Q, Koren D, Giammarinaro B, Acaron Ledesma H, and Wei W (2015). Conditional knock-out of vesicular GABA transporter gene from starburst amacrine cells reveals the contributions of multiple synaptic mechanisms underlying direction selectivity in the retina. J. Neurosci 35, 13219–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Tran NM, Krishnaswamy A, Kostadinov D, Martersteck EM, and Sanes JR (2017). Satb1 regulates contactin 5 to pattern dendrites of a mammalian retinal ganglion cell. Neuron 95, 869–883.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo DM, El-Danaf RN, Huberman AD, and Niell CM (2013). Diverse visual features encoded in mouse lateral geniculate nucleus. J. Neurosci 33, 4642–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleg-Polsky A, Ding H, and Diamond JS (2018). Functional compartmentalization within starburst amacrine cell dendrites in the retina. Cell Rep 22, 2898–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017a). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C (2017b). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Heath KM, Hasan N, Noel JM, Samuels IS, Martemyanov KA, Peachey NS, McCall MA, and Gregg RG (2014). GPR179 is required for high sensitivity of the mGluR6 signaling cascade in depolarizing bipolar cells. J. Neurosci 34, 6334–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Roy S, Kozlowski C, Wang J, Cafaro J, Hulbert SW, Wright CV, Field GD, and Kay JN (2018). Formation of retinal direction-selective circuitry initiated by starburst amacrine cell homotypic contact. eLife 7, e34241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resta V, Novelli E, Di Virgilio F, and Galli-Resta L (2005). Neuronal death induced by endogenous extracellular ATP in retinal cholinergic neuron density control. Development 132, 2873–2882. [DOI] [PubMed] [Google Scholar]
- Rivlin-Etzion M, Zhou K, Wei W, Elstrott J, Nguyen PL, Barres BA, Huberman AD, and Feller MB (2011). Transgenic mice reveal unexpected diversity of on-off direction-selective retinal ganglion cell subtypes and brain structures involved in motion processing. J. Neurosci 31, 8760–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill RL, Euler T, and Masland RH (2000). Spatial order within but not between types of retinal neurons. Proc. Natl. Acad. Sci. USA 97, 2303–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW (1989). Starburst amacrine cells of the primate retina. J. Comp. Neurol 285, 18–37. [DOI] [PubMed] [Google Scholar]
- Rodieck RW (1991). The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis. Neurosci 6, 95–111. [DOI] [PubMed] [Google Scholar]
- Rossi J, Balthasar N, Olson D, Scott M, Berglund E, Lee CE, Choi MJ, Lauzon D, Lowell BB, and Elmquist JK (2011). Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab 13, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria I, Orlandi C, McCall MA, Gregg RG, and Martemyanov KA (2016). Intermolecular interaction between anchoring subunits specify subcellular targeting and function of RGS proteins in retina ON-bipolar neurons. J. Neurosci 36, 2915–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz GW, Okawa H, Dunn FA, Morgan JL, Kerschensteiner D, Wong RO, and Rieke F (2012). The spatial structure of a nonlinear receptive field. Nat. Neurosci 15, 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuramanujam S, McLaughlin AJ, deRosenroll G, Hoggarth A, Schwab DJ, and Awatramani GB (2016). A central role for mixed acetyl-choline/GABA transmission in direction coding in the retina. Neuron 90, 1243–1256. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Da Silva JS, He H, and Cline HT (2009). Type A GABA-receptor-dependent synaptic transmission sculpts dendritic arbor structure in Xenopus tadpoles in vivo. J. Neurosci 29, 5032–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto F, Watkins KL, Johnson RE, Schottler F, and Kerschensteiner D (2013). NGL-2 regulates pathway-specific neurite growth and lamination, synapse formation, and signal transmission in the retina. J. Neurosci 33, 11949–11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto F, Zhao L, and Kerschensteiner D (2018). Synapse maintenance and restoration in the retina by NGL2. eLife 7, e30388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Jiang Z, Rivlin-Etzion M, Hand R, Brady CM, Matsuoka RL, Yau KW, Feller MB, and Kolodkin AL (2013). On and off retinal circuit assembly by divergent molecular mechanisms. Science 342, 1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, and Vaney DI (2002). Diverse synaptic mechanisms generate direction selectivity in the rabbit retina. J. Neurosci 22, 7712–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, and Omi N (2013). Functional allocation of synaptic contacts in microcircuits from rods via rod bipolar to AII amacrine cells in the mouse retina. J. Comp. Neurol 521, 3541–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasits AL, Bos R, Morrie RD, Fortuny C, Flannery JG, Feller MB, and Rivlin-Etzion M (2014). Visual stimulation switches the polarity of excitatory input to starburst amacrine cells. Neuron 83, 1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasits AL, Morrie RD, Tran-Van-Minh A, Bleckert A, Gainer CF, DiGregorio DA, and Feller MB (2016). A role for synaptic input distribution in a dendritic computation of motion direction in the retina. Neuron 89, 1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YV, Weick M, and Demb JB (2011). Spectral and temporal sensitivity of cone-mediated responses in mouse retinal ganglion cells. J. Neurosci 31, 7670–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wässle H, and Boycott BB (1991). Functional architecture of the mammalian retina. Physiol. Rev 71, 447–480. [DOI] [PubMed] [Google Scholar]
- Wässle H, Puller C, Müller F, and Haverkamp S (2009). Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J. Neurosci 29, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W (2018). Neural mechanisms of motion processing in the mammalian retina. Annu. Rev. Vis. Sci 4, 165–192. [DOI] [PubMed] [Google Scholar]
- Wei W, Elstrott J, and Feller MB (2010). Two-photon targeted recording of GFP-expressing neurons for light responses and live-cell imaging in the mouse retina. Nat. Protoc 5, 1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Hamby AM, Zhou K, and Feller MB (2011). Development of asymmetric inhibition underlying direction selectivity in the retina. Nature 469, 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ROL, and Ghosh A (2002). Activity-dependent regulation of dendritic growth and patterning. Nat. Rev. Neurosci 3, 803–812. [DOI] [PubMed] [Google Scholar]
- Yamagata M, and Sanes JR (2008). Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 451, 465–469. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Weiner JA, and Sanes JR (2002). Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110, 649–660. [DOI] [PubMed] [Google Scholar]
- Yonehara K, Balint K, Noda M, Nagel G, Bamberg E, and Roska B (2011). Spatially asymmetric reorganization of inhibition establishes a motion-sensitive circuit. Nature 469, 407–410. [DOI] [PubMed] [Google Scholar]
- Yonehara K, Fiscella M, Drinnenberg A, Esposti F, Trenholm S, Krol J, Franke F, Scherf BG, Kusnyerik A, Müller J, et al. (2016). Congenital nystagmus gene FRMD7 is necessary for establishing a neuronal circuit asymmetry for direction selectivity. Neuron 89, 177–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Watanabe D, Ishikane H, Tachibana M, Pastan I, and Nakanishi S (2001). A key role of starburst amacrine cells in originating retinal directional selectivity and optokinetic eye movement . Neuron 30, 771–780. [DOI] [PubMed] [Google Scholar]
- Zhao X, Kuja-Panula J, Sundvik M, Chen YC, Aho V, Peltola MA, Porkka-Heiskanen T, Panula P, and Rauvala H (2014). Amigo adhesion protein regulates development of neural circuits in zebrafish brain. J. Biol. Chem 289, 19958–19975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.