Abstract
Angiostrongylus cantonensis (rat lungworm) is the etiological agent of angiostrongyliasis, mainly causing eosinophilic meningitis or meningoencephalitis in human. Although the biology of A. cantonensis is relatively well known, little is understood about the mechanisms of the parasite’s development and survival in definitive hosts, or its adaptation to a broad range of snail intermediate hosts. Here, we generate a high-quality assembly of a well-defined laboratory strain of A. cantonensis from Guangzhou, China, by using Illumina and PacBio sequencing technologies. We undertake comparative analyses with representative helminth genomes and explore transcriptomic data throughout key developmental life-cycles of the parasite. We find that part of retrotransposons and gene families undergo multiple waves of expansions. These include extracellular superoxide dismutase (EC-SOD) and astacin-like proteases which are considered to be associated with invasion and survival of the parasite. Furthermore, these paralogs from different sub-clades based on phylogeny, have different expression patterns in the molluscan and rodent stages, suggesting divergent functions under the different parasitic environment. We also find five candidate convergent signatures in the EC-SOD proteins from flukes and one sub-clade of A. cantonensis. Additionally, genes encoding proteolytic enzymes, involved in host hemoglobin digestion, exhibit expansion in A. cantonensis as well as two other blood-feeding nematodes. Overall, we find several potential adaptive evolutionary signatures in A. cantonensis, and also in some other helminths with similar traits. The genome and transcriptomes provide a useful resource for detailed studies of A. cantonensis-host adaptation and an in-depth understanding of the global-spread of angiostrongyliasis.
Author summary
Angiostrongylus cantonensis, rat lungworm, is a common pathogen that causes human eosinophilic meningitis via eating contaminated food. Human angiostrongyliasis has been reported globally. This worm has a complex life-cycle, which includes an especially wide range of snails as intermediate hosts, making it more difficult to eradicate. In this study, we sequenced the genome and transcriptome, and performed comparative analyses to study the potential genetics of its biology using short-read and long-read sequencing technologies. We revealed some potential adaptive evolution in the genome, such as the expansion of retrotransposons and gene families encoding antioxidant enzymes, invasion, migration and digestion related proteases. Specifically, we found a potential clue suggesting convergent evolution of EC-SODs in Angiostrongylus and flukes, all of which require snails as intermediate hosts. These results provide an abundant data resource to study the biology and evolution of A. cantonensis and showed some potential targets against A. cantonensis and helminths with similar traits.
Introduction
Angiostrongylus cantonensis (rat lungworm) is a parasitic roundworm (nematode) of the superfamily Metastrongyloidea, with a complicated life cycle via a gastropod intermediate host [1]. More than twenty species of Angiostrongylus have been discovered in rodents, carnivores and insectivores, and two of them A. cantonensis and A. costaricensis are human parasites [1]. A. cantonensis is the most common infectious cause of eosinophilic meningitis in humans, causing central nervous system (CNS) angiostrongyliasis [2]. Since the first human CNS angiostrongyliasis case reported in 1945 [3], other clinical symptoms including ocular disease, encephalitis and fever of unknown origin have been reported for this disease [4–6]. While most cases were reported in Asia, the Pacific Basin and Australia, human angiostrongyliasis has been found emerging worldwide in the past decades, including USA, France and the UK [7–10] (Figure S1 in S1 Supporting Information).
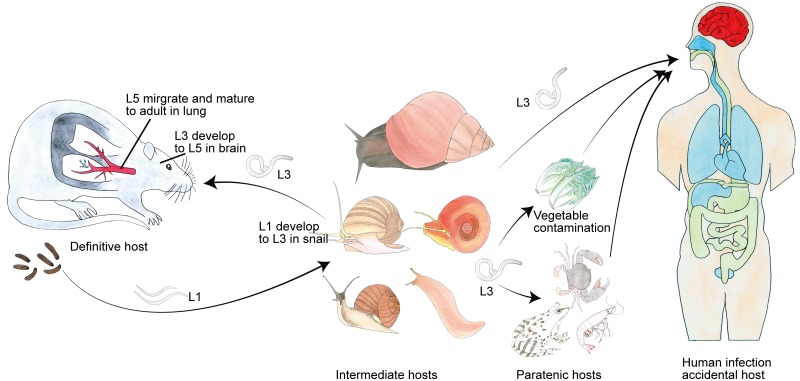
The life cycle of A. cantonensis involves a molluscan intermediate host (various species) and a definitive rodent host (cf. review [11], Fig 1). Briefly, the first-stage larvae (L1) are swallowed by an intermediate host, they molt twice into third-stage larvae (L3). The infective L3 are ingested by a definitive host, then they migrate to the brain and molt twice into young adults (L5). Eventually, the L5 migrate to the lungs where develop to sexual maturity and lay eggs. The eggs embryonate, develop and hatch to L1 and they are excreted in host feces, restarting the life cycle. This worm can infect a very wide range of intermediate hosts, comprising at least 160 species belonging to 44 families of freshwater and land gastropods [12]. The two available assemblies are highly fragmented in nature which has posed as an obstacle to detailed biological and evolutionary investigations [13, 14].
Fig 1. The complex life cycle of A. cantonensis.
The complete life cycle of A. cantonensis requires two different hosts (snail and rat): L1 larvae are excreted in the feces of a definitive host (rat). When ingested by an intermediate host, they develop into infective L3 after molting twice and are maintained at that stage until they are eaten by a definitive host. The L3 are ingested by a rat and invade intestinal tissue and then migrate to the central nervous system (CNS), where they molt twice and develop into L5. Finally, these worms leave the brain and then reach the pulmonary arteries, where they become fully mature adults. Human infections are acquired by eating undercooked snails, paratenic hosts such as frogs, or contaminated vegetables containing L3 of A. cantonensis. Since humans are non-permissive hosts of A. cantonensis, the larvae reach to the brain and cause eosinophilic meningitis.
In the present study, we sequenced and assembled a high-quality reference genome of a well-defined laboratory strain of A. cantonenis from Guangzhou, China. Through analyses of comparative genomics and transcriptome, we explored potential molecules regarding the nematode survival in intermediate host and/or definitive host.
Methods
Ethics statement
Procedures involving animals and their care described here were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (Permit No: 2016–055) and followed the National Guidelines for Experimental Animal Welfare (MOST, China, 2006).
Sample preparation and sequencing
The life-cycle of A. cantonensis was established and maintained in the Department of Parasitology, Zhongshan School of Medicine. L1 larvae were separated from feces of rats. L3 were isolated from experimentally infected snails using the method previously described by Zeng [15]. Sprague Dawley rats were challenged with L3 (200 per animal) via intragastric administration. Procedures for animal care described herein were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. Other developmental worms (L4, L5 and mature adults, including female and male) were harvested from rats at 21, 28, and 48 days post-infection (dpi) respectively. Genomic DNA was extracted from ten adult worms. Seven paired-end and mate-pair whole-genome shotgun libraries (250bp, 500bp, 800bp, 2kb, 5kb, 10kb, and 20kb, Table S1 in S2 Supporting Information) were constructed and then sequenced using the Illumina HiSeq 2000 platform. Another 20 kb library for PacBio sequencing was constructed and sequenced using RSII. RNA was extracted from different developmental stages of A. cantonensis (L1, L3, L4, L5 and mature adults, the Pomacea canaliculata used as an intermediate host), respectively. Seven cDNA libraries were sequenced using the Illumina Hiseq 2000 platform. Another four cDNA libraries (L3 and L4, the Biomphalaria glabrata used as an intermediate host) were constructed and sequenced using the Illumina HiSeq 4000 platform.
Genome assembly and annotation
We employed a hybrid assembly strategy by combining Illumina and PacBio data (Figure S3 in S1 Supporting Information). Illumina reads were first assembled into contigs using the Platanus [16] (v1.2.4) with default parameters. The resulting Illumina contigs and PacBio subreads were further used to assemble with DBG2OLC [17] pipeline (release Jun 2015). Then, correction of the assembly was performed twice with Pilon [18] (v1.22) by using Illumina reads. To further link the corrected contigs, the corrected PacBio reads and Illumina mate-pair reads were employed to extend and link into scaffolds using SSPACE-LongRead [19] (v1–1) and SSPACE [20] (v2.0). Remaining gaps within these scaffolds were filled with GapCloser available in SOAPdenovo2 [21]. CEGMA (Core Eukaryotic Genes Mapping Approach) [22] (v2.4), BUSCO (benchmarking universal single-copy orthologues) [23] (v3.0.1), and de novo assembled transcripts with Trinity [24] (v2.0.6) were used to assess the completeness of the assembly.
Protein-coding gene models were predicted using a strategy to combine the homology-based prediction and RNA-seq data as previously described [25] (cf. S1 Supporting Information, Supplementary Methods and Results sections). The functional annotation of protein-coding genes was performed using BLASTP alignment to databases: Swiss-Prot (release Jun 2019), TrEMBL (release Jun 2019), NCBI NR (release Sep 2017) and KEGG (release 89). InterPro domains and GO terms were assigned with InterProScan [26] (release 5.3).
Repetitive elements (REs) in the assembly were identified using a combination of homology- and ab initio-based approaches. RepeatMasker and RepeatProteinMask (http://www.repeatmasker.org/, version open-4-0-5) were applied to detect homologous REs in the RepBase database (v20.04). PILER [27] (v1.0), RepeatScout [28] (v1.0.5), and LTR-Finder [29] (v1.0.6) were used to build a de novo repeat library. RepeatMasker was run against the de novo library. The same pipeline was employed to predict REs in seven other nematode genomes (Ascaris suum [30], Brugia malayi [31]; Caenorhabditis elegans [32], Haemonchus contortus [33, 34], Necator americanus [35], Meloidogyne hapla [36]). For RTE-RTE transposable elements, proteins of RTE-RTEs deposited in RepeatPep (RepeatMasker-open-4.0.6) were collected and used to search in eight nematode genomes using homology-based prediction pipeline as delineated in gene prediction, except with an alignment rate of more than 50%. Amino acid sequences encoding a reverse transcriptase (RT, PF00078) domain were aligned using MUSCLE [37] (v3.8.31) and then were constructed a phylogenetic tree using FastTree [38] (v2.1).
Genome evolution
The OrthoMCL [39] pipeline was used to determine orthologous groups in A. cantonensis and seven other represented nematode genomes (A. suum, B. malayi; C. elegans, H. contortus, N. americanus, M. hapla and T. spiralis, related data was downloaded from the WormBase [40] (version 246). 788 single-copy orthologous genes were extracted to build a phylogenetic tree. Sequences from each single copy orthologs were aligned using MUSCLE and then filtered with trimAl [41] (v1.2) with default parameters except “-gt 0.5”. RAxML [42] (v8.2) was used to construct gene tree with “GTRGAMMA” model. Finally, ASTRAL [43] (v5.6.1) was used to construct species tree based on 788 gene trees. Phylogenetic relationship among eight nematodes and six flatworms (Schistosoma japonicum [44], S. mansoni [45], S. haematobium [46], Opisthorchis viverrini [47], Clonorchis sinensis [48] and Schmidtea mediterranea [49], related data was downloaded from WormBase Parasite [50], WBPS5) was resolved using the method described above based on 173 single-copy orthologous genes. Species divergence time was estimated using MCMCTREE, which is part of the PAML package [51] (v4.5). Published times for T. spiralis and C. elegans (~428 million years ago, mya), and B. malayi and C. elegans (~241 mya) divergence were used to calibrate divergence time [52]. We investigated the expansion and contraction of gene families using the CAFÉ [53] (Computational Analysis of gene Family Evolution, v2.1), which infers the dynamics of the gene family under a stochastic birth and death model.
Identification, evolution and expression of specific gene or gene families
To avoid systematic biases, for example, different methods in the annotation of the previously published helminth draft genomes, we adopted a uniform strategy to re-annotate and check candidate genes screened from the above comparative analysis. Generally, protein sequences of superoxide dismutase (SOD) genes, astacin-like genes, and several hemoglobin digestion proteases of nematodes deposited in Swiss-Prot or MEROPS [54] (download in Nov 2016) databases were downloaded and mapped to the genomes using the homology-based gene prediction. We also manually checked these putative genes and compared with the original gene annotation (The associations of the gene IDs used in this study and the gene IDs in Wormbase are listed in Table S9 in S2 Supporting Information). Phylogenomic analyses of the gene families studied herein were based on protein sequences. The best model of amino acid replacement was estimated using ProtTest [55] (v3.4.2) software. The phylogenetic trees of these genes were constructed using PhyML [56] (v3.0) software, respectively. For EC-SODs, we also reconstructed the phylogenetic trees using RAxML and IQ-TREE [57] (v1.6.5), and conducted a hypergeometric test site by site at amino acid level to detect the potential convergent evolution [25] between the genus Angiostrongylus and flukes in a broad range of 141 EC-SODs from 62 species (43 nematodes and 19 platyhelminths, Table S10 in S2 Supporting Information).
For RNA-seq analysis, we mapped RNA-seq reads to the genome with Tophat2 [58] (v2.0.8). We quantitated the gene expression level using uniquely mapped reads and measure in reads per kilobase per million reads (RPKM). The expression of EC-SOD and MTP-1 subclade I and II genes were validated using real-time PCR (qPCR, primers are shown in Table S8 in S2 Supporting Information), with pooled larvae/adults isolated from multiple hosts for each developmental stage. β-actin was used as an internal control. The relative changes in gene expression were calculated by equation 2−ΔΔCT, where ΔΔCT = (CT,target—CT,Actin)Time x—(CT,Target—CT, Actin)Time 0. Time x is any time point and Time 0 represents the 1 × expression of the target gene normalized to β-actin [59].
Results
Genome assembly and annotation
The genome of A. cantonensis was sequenced using the Illumina HiSeq 2000 and PacBio RSII platforms, yielding a total of ~267-fold and ~41-fold coverage, respectively (estimated genome size: 290 Mb; Figure S2 in S1 Supporting Information and Table S1 in S2 Supporting Information). The final genome assembly included 282 Mb in 816 contigs and 425 scaffolds, with a contig N50 of 993 kb and a scaffold N50 of 1.8Mb (Table 1). The assembly covered more than 95% (coverage ≥ 70%) of the assembled RNA-seq transcripts, indicating that the gene region was well represented (Table S2 in S2 Supporting Information). In addition, both the CEGMA and BUSCO methods were used, and the results showed that the assembly in this study was more complete than the published fragment assemblies of A. cantonensis in the protein-coding region (Table 1). Taken together, the results showed that the present genome assembly of A. cantonensis represented a substantial part of the whole genome. Combined homology-based and RNA-seq methods, we predicted 13,473 protein-coding gene models (5.16% of the assembly, spanning ~15 Mb) in A. cantonensis genome. Of these genes, 13,114 (97%) were supported by RNA-seq data (RPKM≥1 at least one sample), and 12,407 (92.1%) genes either had homologues in public databases (Swiss-Prot, KEGG, and NCBI NR).
Table 1. Statistics of the A. cantonensis assemblies.
| This study | Yong et al [14]. | Avril et al [13]. | |
|---|---|---|---|
| Assembly size (Mb) | 283 | 260 | 253 |
| Scaffold numbera | 425 | 16,326 | 18,635 |
| Gaps (bp) | 1,480,142 | 25,114,564 | 4,505,497 |
| Contig N50 (kb); Scaffold N50 (kb) | 993; 1,815 | 1.7;42.2 | 27.1;43.9 |
| GC content % | 41.7 | 41.2 | 41.5 |
| Complete BUSCOs[Duplicated]b | 84.3%[1.2%] | 62.5%[0.8%] | 70.0%[1.2%] |
| Fragmented BUSCOs | 7.80% | 12.30% | 10.20% |
| Missing BUSCOs | 7.90% | 25.20% | 19.80% |
| CEGMA completeness | 97.98% | 80.24% | 82.66% |
a, length cut-off: 500bp;
b, Nematoda_odb9 dataset was used
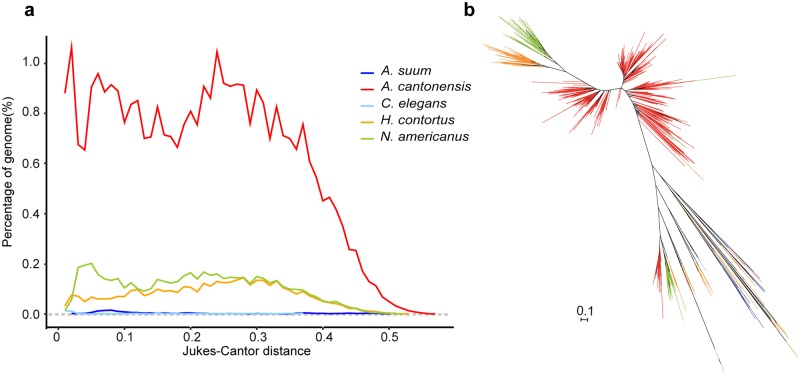
Transposable elements (TEs) represented 54.61% of the assembled genome, representing a greater percentage than most parasitic nematode genomes characterized to date (3.4–32.9% using the same bioinformatics pipeline; Table S3 in S2 Supporting Information). RTE-RTE retrotransposons, belonging to the long interspersed elements (LINEs), were the most abundant group in A. cantonensis genome, representing 39.2% of the genome and 72% of repeats respectively (Table S4 in S2 Supporting Information), a markedly higher percentage than the genomes of other studied nematodes (0~5.1%; Table S3 in S2 Supporting Information). Sequence divergence of extant RTE-RTE copies compared to the repeat consensus showed at least two periods of element expansion in A. cantonensis (Fig 2a). The phylogeny showed that the RTE-RTEs expanded independently in A. cantonensis, N. americanus, and H. contortus, but the A. cantonensis displayed substantially higher divergence and abundance than the other two strongyloids (Fig 2).
Fig 2. RTE-RTE retrotransposons expansion in A. cantonensis.
(a), The genomic portion of the RTE-RTEs in eight nematodes (three of which lack RTE-RTEs using the same criterion) at different given divergence generated by the de novo prediction. The divergence is adjusted for multiple substitutions using the Jukes-Cantor distance. (b), The phylogenetic tree depicting the relationship of RTE-RTEs among five nematodes. The branches of five species of nematodes are colored in blue for A.suum, red for A. cantonensis, light blue for C. elegans, orange for H.contorus and green for N. americanus.
Genome evolution
To better understand the evolution of A. cantonensis genome and to infer genes or gene families associated with parasitism, we performed a comparative analysis with seven other nematode genomes representing clades I, III, IV and V (A. suum, B. malayi; C. elegans, H. contortus, N. americanus, M. hapla and T. spiralis). We identified 788 one-to-one orthologous genes in all eight nematodes and assessed the phylogenetic relationships using a coalescent-based method [43]. The phylogenetic analysis showed that A. cantonensis was genetically closer to H. contortus (Figure S5 in S1 Supporting Information), which was the same as the study of the 50 Helminths Genome Project [13]. We inferred that 26 and 119 gene families respectively underwent significant expansion and contraction in the A. cantonensis lineage (Viterbi P≤0.05; Tables S6 and S7 in S2 Supporting Information, Figure S6 in S1 Supporting Information). Expanded genes included protease (neprilysin-1 and legumain), transporter (sodium-dependent high-affinity dicarboxylate transporter 3), receptor (acetylcholine receptor) and ancylostoma secreted protein. Based on the OrthoMCL cluster result, we also identified 454 genes (159 groups; Table S5 in S2 Supporting Information) that appeared to be unique in A. cantonensis, which were significantly enriched in GO terms of ʺSuperoxide dismutase activityʺ (GO:0004784) and metallopeptidase activityʺ (GO:0008237) (adjusted P-value < 0.05, Figure S4 in S1 Supporting Information). These ʺuniqueʺ genes included astacin-like metalloproteinase (M12A), Aspartic protease (A01) and Extracellular superoxide dismutase (EC-SOD) which are likely related to the invasion, migratory and digestive processes, innate immune of A. cantonensis. Those expanded or specific genes may provide novel clues to A. cantonensis adaptation to hosts.
Expansion of EC-SOD genes related to host adaptation in A. cantonensis
Reactive oxygen species (ROS), such as oxygen radicals and superoxide, are generated by phagocytes (vertebrates) or haemocytes (invertebrates), and represent an innate defense system against pathogens [60–62]. Helminth parasites secrete antioxidant enzymes for defense against host-generated ROS for survival in the host [60]. Extracellular superoxide dismutase (EC-SOD, also known as SOD3) belongs to the SOD gene family, which converts superoxide radical into hydrogen peroxide and represents the first step in the antioxidant enzyme system to reduce ROS [60].
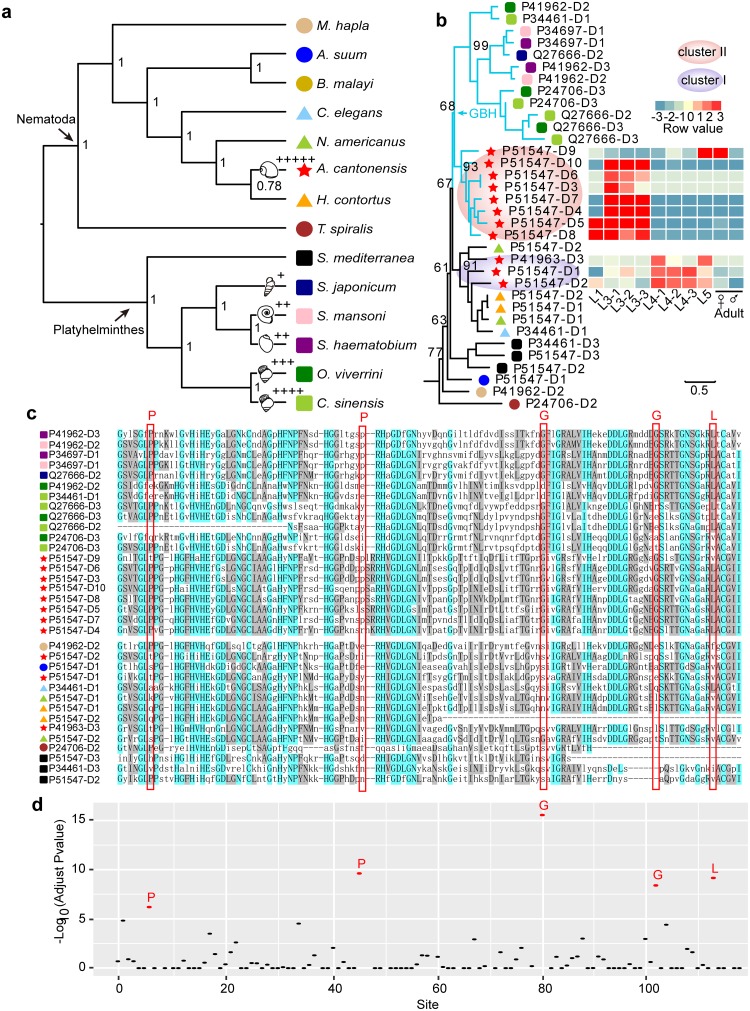
Eleven tandem EC-SOD genes were identified in the A. cantonensis genome, which were also confirmed by PCR using genomic DNA from A. cantonensis (Figure S9 in S1 Supporting Information), which was four times more than the number in the other seven nematodes studied herein (1–2 copies, Fig 3b and Figure S7 in S1 Supporting Information). Phylogenetic analysis revealed that paralogous EC-SOD genes in A. cantonensis might likely arise via two evolutionary events (Fig 3b), reflected by three paralogs (cluster I) in one clade with other nematodes and eight paralogs in another clade (cluster II, Fig 3b and Figure S7 in S1 Supporting Information). In addition, transcription analysis revealed two expression patterns occurred in these two clusters (Fig 3b). The genes in cluster I were relatively highly upregulated in the mammalian stage of A. cantonensis (L4 and L5 in Fig 3b), indicating these genes may be related to defend against definitive host-derived ROS. We examined the gene expression level of EC-SOD in A. cantonensis recovered from its nonpermissive host (mice) and permissive host (rat) using qPCR. We observed significantly higher transcription of the gene in cluster I (termed P51547-D2 herein) in A. cantonensis collected from rats compared with the one harvested from mice using qPCR (Figure S11 in S1 Supporting Information).
Fig 3. Phylogenomic analysis of EC-SOD in different species of nematodes and flatworms.
(a), Phylogenic tree depicts a cladogram of eight nematodes and six flatworms profiled in this study. Number at the node indicates ASTRAL supporting value while the branches with the sketch of snails represents intermediate hosts. Specifically, the number of ʺ+ʺ shows the increasing spectrum of suitable intermediate hosts. (b), Maximum likelihood tree of EC-SODs in 14 species and the mRNA expression patterns of A. cantonensis’s EC-SODs. GBH: the EC-SOD clustered in gastropod-borne helminths. (c), The multiple sequence alignment of EC-SODs from sequences in Fig 3b. (d), Convergent study at amino acids levels in the EC-SODs from gastropod-borne helminths at an extended background of 62 species. The x-axis shows multiple sequence alignment position in Fig 3c.
In contrast, the EC-SODs in cluster II (six out of eight) were transcribed at significantly higher levels in L3, recovered from infected intermediate host compared with the other developmental stages (Fig 3b and Figure S10 in S1 Supporting Information), suggesting that these genes might be related specifically to parasite survival in the intermediate gastropod hosts. Excluding the genus Angiostrongylus from the phylum Nematoda, five well-studied and sequenced digenean trematodes (S. japonicum, S. haematobium, S. mansoni, O. viverrini, and C. sinensis), are gastropod-borne helminths that require snails as the intermediate host or first intermediate host [63]. We then analyzed and compared SODs of these five digenean trematodes and S. mediterranea (a free-living planarian) to determine whether some similarities existed among EC-SODs from the gastropod-borne helminths. Interestingly, the phylogenetic analyses showed that while EC-SOD in cluster II and the EC-SODs from parasitic flukes clustered into one clade, in which only contained the gastropod-borne flatworms and nematode (A. cantonensis) (GBH, Fig 3b and Figure S7 in S1 Supporting Information), which conflicts with the species topology (Fig 3a). Further, both the maximum-likelihood and Bayesian analyses recovered trees in which EC-SOD in cluster II and EC-SODs from digenean trematodes grouped together in one clade based on the conserved domain (“Sod_Cu”, Figure S8 in S1 Supporting Information). The multiple sequence alignment of the EC-SODs from 14 species showed some over-represented amino acid sites existed in the most members in the GBH clade (Fig 3c). Further, we extended the examination in 62 species (43 nematodes including 2 gastropod-borne nematodes, and 19 flatworms including 12 gastropod-borne flukes) and detected five amino acid sites that were significantly enriched in EC-SODs from the gastropod-borne helminths (Fig 3d, adjust P-value < 1e-5). This finding suggests that EC-SOD from cluster II may experience a convergent evolution at some sites with the EC-SODs of flukes.
Proteases related to hemoglobin and tissue digestion in A. cantonensis
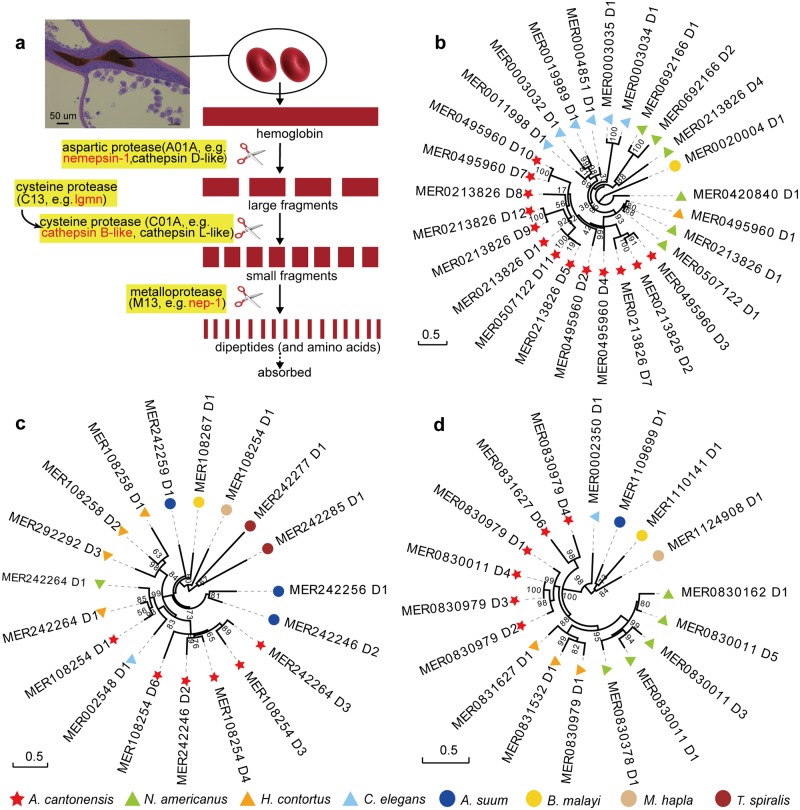
Adults of A. cantonensis dwell in the pulmonary arteries of the definitive host, where worms mature and lay eggs. The parasite digests blood and other tissue components of the host for major protein synthesis [64] (Fig 4a). Hematophagous nematodes employ an ordered pathway with distinct proteases to degrade host hemoglobin or other serum proteins [65] (Fig 4a). Through annotation and comparative analysis of eight nematode genomes, we found that genes encoding proteases such as nematode-specific aspartic proteases (e.g., necepsin-1), cathepsin B-like, legumain (Lgmn) and neprilysin (e.g., NEP-1), inferred to be involved in hemoglobin digestion, are expanded in A. cantonensis (Fig 4 and Figures S13–16 in S1 Supporting Information). For instance, necepsin-1 (known as APR-2 in N. americanus) belongs to the aspartic proteases (MEROPS: A01A), and it is likely involved in the initial cleavage of hemoglobin [66]. Cysteine peptidases (including cathepsin B-like proteases) are likely involved in the second step of digestion. In addition, A. cantonensis has at least six genes encoding legumain, which likely activate cysteine proteases by specific hydrolysis of peptide bonds following asparagine residues [67]. In the final digestion step, metalloproteases, such as NEP-1, likely degrade small peptide fragments to dipeptides. Interestingly, the proteases necepsin-1, cathepsin B-like, legumain and NEP-1 are also expanded in N. americanus and/or H. contortus with respect to the five other nematodes studied herein (Fig 4, Figures S13–16 in S1 Supporting Information).
Fig 4. Phylogenomic analysis of three proteases related to hemoglobin digestion in eight nematodes.
(a), H&E stained longitudinal section of the digestive tract from a female adult (on the upper left) shows the presence of red blood cells. The possible hemoglobin digestion pathway in the nematodes is illustrated on the right panel [65] with subfamily of enzymes (aspartic protease, cysteine protease and metalloprotease) related to hemoglobin and/or tissue digestion highlighted in yellow background. Specifically, the expanded subfamilies of enzymes from A. cantonensis are highlighted in red. Maximum likelihood phylogenies of necepsin-1 (b), Lgmn (c) and Nep-1(d) show expansion of these proteases in A. cantonensis, N. americanus and/or H. contortus, all of which are blood-feeding nematodes.
Expansion of astacin-like genes in A. cantonensis
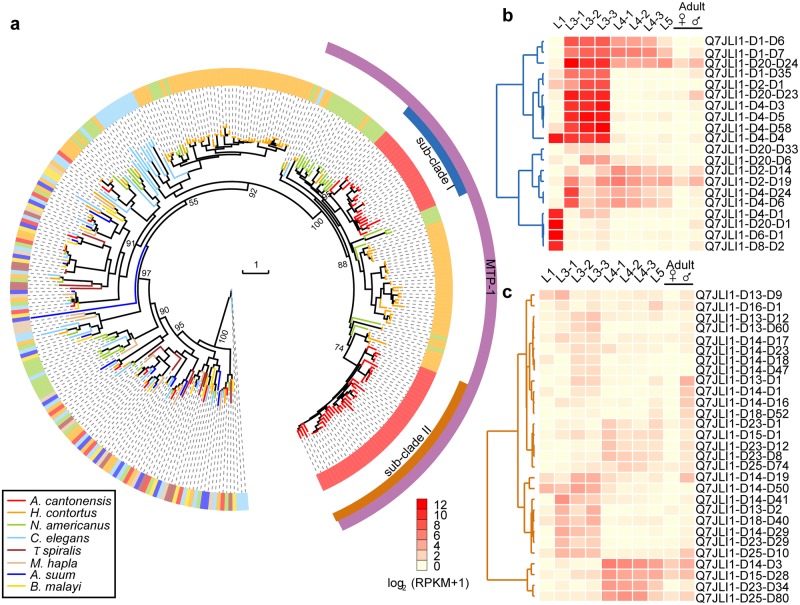
Astacin-like metalloproteases are involved in molting, feeding and/or host tissue penetration in nematodes [68–70]. The number of genes encoding astacin-like protease (M12A, metalloproteases) in A. cantonensis (n = 75) was greater than that in C. elegans (n = 40). One subfamily, with the highest sequence similarity to the MTP-1 of Ancylostoma caninum [71], had only one gene in C. elegans, but had 63 genes in A. cantonensis. The expanded MTP-1 genes of A. cantonensis separated into two large subclades (subclade I: n = 22; subclade II: n = 41; Fig 5a) and ten amino acid sites that were relatively specifically between the two subclades (Figure S19 in S1 Supporting Information). Additionally, genes in subclade I were mainly expressed in the L1 or L3, which was also supported by qPCR (Fig 5b and Figure S18 in S1 Supporting Information). The genes in subclade II had relatively low transcription compared with subclade I (Fig 5 and Figure S18 in S1 Supporting Information). MTP-1 has been reported to be associated with tissue migration in A. caninum [71], suggesting that the expanded MTP-1s may be related to the survival and/or infectivity of A. cantonensis. Additionally, the sequence divergence and distinct RNA expression of these two subclades suggest that A. cantonensis may have acquired multiple related abilities. A study has shown that an expansion of astacin-like genes was also discovered in Strongyloides and Parastrongyloides species [72] (Clade IV). But our phylogenetic analysis showed that the expanded astacin-like genes in S. ratti formed a single and distinct clade that diverged from the above mentioned MTP-1 clade (Figure S20 in S1 Supporting Information). Recently, another study of comparative analysis of the major parasitic worms also identified the expansion of astacin-like genes in the clades IVa, Vc and Vb [13] (including A. cantonensis), which is consistent with our findings.
Fig 5. Evolution of astacin-like genes and expression pattern across in the life-cycles of A. cantonensis.
(a), Phylogenetic analysis of the astacin-like genes containing the astacin domain (PF01400). We named the purple cluster MTP-1 because it shows the best hit with MTP-1 in the MEROPS database. (b), Expression patterns of expanded sub-clade I astacin-like genes in A. cantonensis. The genes in sub-clade I are upregulated in L1 or L3, which are two stages of invasion into intermediate host or mammalian hosts. c, mRNA expression pattern of expanded sub-clade II astacin-like genes in A. cantonensis.
Discussion
Parasite adaptation to the host is a key factor for its success [73]. The increasing genomic data for parasitic worms provides resources to explore biological and genetic differences between free-living and parasitic nematodes of plants and animals, shedding light on genomic adaptions [74]. These resources also offer unique opportunities to explore the fundamental biology of parasitic helminths and to identify potential interventions for diseases caused by worms.
Here we sequenced and assembled a high-quality reference genome of the Guangzhou strain of A. cantonensis, which were superior in quality to previous drafts for this species [13, 14] and published draft genomes for other strongylid nematodes [33–35]. We also employed transcriptomic data from multiple different developmental stages to reliably predict protein-coding genes and to underpin the subsequent analyses. We found that some of the genomic elements experienced multiple waves of expansion in A. cantonensis, including non-coding regions (e.g., RTE-RTE retrotransposons) and protein coding genes (e.g. EC-SOD and astacin-like genes). The paralogs of EC-SODs and astacin-like genes from different sub-clades, have different expression patterns in the molluscan stage and mammalian stage. Thus, the results may partly explain the adaptive evolution of the complex life cycle of A. cantonensis, such as the two different parasitic environments (mollusc and definitive rodent host).
Extracellular/secreted SOD of helminth parasite is one of the main components in excretory-secretory (ES) and plays a key role in fighting against host-produced ROS [60]. Our study showed that the cluster I EC-SODs of A. cantonensis mainly expressed in the mammalian stage, and expressed higher in the permissive host (rat) than in non-permissive host (mice). A previous investigation showed the high activity of EC-SOD in ES from rat-originated A. cantonensis [75]. And another study showed the higher SOD activity in Heligmosomoides polygyrus (mice is non-permissive hosts) than in Nippostrongylus brasiliensis (mice is non-permissive hosts) when they infected the mice [76]. These results suggested that some of parasitic nematode EC-SOD may be important for its survival in permissive mammalian hosts.
In contrast, the cluster II EC-SODs of A. cantonensis showed significant higher expression in the L3 (mollusc-dwelling). Moreover, the cluster II EC-SODs may experience convergent evolution at several amino acids with the EC-SODs of flukes. In Fasciola hepatica, EC-SOD was also identified in ES products from intra-molluscan larval stages [77]. The SOD showed the most significant differential expression patterns of three antioxidant enzymes (SOD, glutathione peroxidase, glutathione-S-transferase) in S. mansoni recovered from the susceptible snail than that from resistant snail [78]. Taken together, sequence convergence and expression similarity suggested the EC-SODs from gastropod-borne helminths might be related to their survival in gastropod species. Further, we observed that the liver flukes (O. viverrini: the family Bithyniidae and C. sinensis: the family Thiaridae and Bithyniidae) [79] with 3–4 copies of EC-SODs have a relatively broad spectrum of intermediate snail hosts than blood flukes (S. japonicum: Oncomelania hupensis, S. haematobium: the genus Bulinus and S. mansoni: the genus Biomphalaria) [63] with 1–2 copies of EC-SODs. While A. cantonensis has 7 paralogs in the GBH clade and has the broadest spectrum of snail hosts (the order Gastropod) [12] among the six species in this study. The largest copy number and diverged EC-SODs in A. cantonensis may provide more resources as well as possibilities for it to escape from host immune attack, which may therefore be a potential explanation for its survival in a variety of intermediate hosts. We also discovered some proteases involved in Hb digestion that were expanded in A. cantonensis and the other two blood-feeding nematodes. This result reveals a comparable spectrum of essential proteases involved in hemoglobin and possible tissue digestion among three haematophagous nematodes (i.e. A. cantonensis, N. americanus and H. contortus). These two instances merit further investigations as they may provide clues to broad-spectrum intervention to not only A. cantonensis control but also other parasites control. The high-quality genome and abundant transcriptomes of A. cantonensis should provide a deeper exploration of the co-evolution in the complex life cycles and host adaptability for helminths, which can be used as a resource to identify regions of genetic diversity in this species and help to deeply understand the global-spread of angiostrongyliasis in order to explore novel anthelmintic agents and/or vaccines.
Supporting information
(DOC)
(XLSX)
Acknowledgments
The authors acknowledge Prof. Robin B. Gasser (the University of Melbourne) for valuable suggestions and revision of the manuscript; Dr Bill Chobotar from Andrews University, Prof. Barry Koehler (University of British Columbia), Prof. Ming-Chiu Fung (The Chinese University of Hong Kong) and Prof. Jerome H.L. Hui (The Chinese University of Hong Kong) for suggestions regarding the manuscript.
Data Availability
The A. cantonensis genome assembly and annotation is accessible at China National GeneBank (CNSA) under accession CNP0000467. Raw sequences (Illumina and PacBio) of the A. cantonensis reference genome have been deposited at NCBI in the SRA under accession number SRP092294 (BioProject number: PRJNA350391). The multiple developmental stages of the A. cantonensis RNA-seq reads have been deposited at NCBI in the SRA under accession number SRP127889 (BioProject number: PRJNA350405).
Funding Statement
ZD-W received grants from National Research and Development Plan of China (2016YFC1200500), the Major Basic Research of Ministry of Science and Technology of China (973 Project) (No.2010CB530000), National Natural Science Foundation of China (No.81261160324, No.81271855, and No.81371836), Science and Technology Planning Project of Guangdong Province (No.2016A050502008), Natural Science Foundation of Guangdong Province, China (No. 2015A030310058) and the 111 Project (Grant No. B12003). ZY-L received a grant from National Natural Science Foundation of China (No. 81572023) and Science and Technology Planning Project of Guangdong Province (2019B030316025). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Spratt DM. Species of Angiostrongylus (Nematoda: Metastrongyloidea) in wildlife: A review. Int J Parasitol Parasites Wildl. 2015;4(2):178–89. 10.1016/j.ijppaw.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flerlage T, Qvarnstrom Y, Noh J, Devincenzo JP, Madni A, Bagga B, et al. Angiostrongylus cantonensis Eosinophilic Meningitis in an Infant, Tennessee, USA. Emerg Infect Dis. 2017;23(10):1756–8. Epub 2017/09/21. 10.3201/eid2310.170978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaver PC, Rosen L. Memorandum on the First Report of Angiostrongylus in Man, by Nomura and Lin, 1945. Am J Trop Med Hyg. 1964;13:589–90. Epub 1964/07/01. 10.4269/ajtmh.1964.13.589 . [DOI] [PubMed] [Google Scholar]
- 4.Foster CE, Nicholson EG, Chun AC, Gharfeh M, Anvari S, Seeborg FO, et al. Angiostrongylus cantonensis Infection: A Cause of Fever of Unknown Origin in Pediatric Patients. Clin Infect Dis. 2016;63(11):1475–8. Epub 2016/09/01. 10.1093/cid/ciw606 . [DOI] [PubMed] [Google Scholar]
- 5.Peng Y, Liu X, Pan S, Xie Z, Wang H. Anti-N-methyl-D-aspartate receptor encephalitis associated with intracranial Angiostrongylus cantonensis infection: a case report. Neurol Sci. 2017;38(4):703–6. Epub 2016/10/26. 10.1007/s10072-016-2718-3 . [DOI] [PubMed] [Google Scholar]
- 6.Sinawat S, Sanguansak T, Angkawinijwong T, Ratanapakorn T, Intapan PM, Sinawat S, et al. Ocular angiostrongyliasis: clinical study of three cases. Eye (Lond). 2008;22(11):1446–8. Epub 2008/06/07. 10.1038/eye.2008.135 . [DOI] [PubMed] [Google Scholar]
- 7.Al Hammoud R, Nayes SL, Murphy JR, Heresi GP, Butler IJ, Perez N. Angiostrongylus cantonensis Meningitis and Myelitis, Texas, USA. Emerg Infect Dis. 2017;23(6):1037–8. Epub 2017/05/19. 10.3201/eid2306.161683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morassutti AL, Thiengo SC, Fernandez M, Sawanyawisuth K, Graeff-Teixeira C. Eosinophilic meningitis caused by Angiostrongylus cantonensis: an emergent disease in Brazil. Mem Inst Oswaldo Cruz. 2014;109(4):399–407. Epub 2014/07/31. 10.1590/0074-0276140023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen Y, Rossi B, Argy N, Baker C, Nickel B, Marti H, et al. Autochthonous Case of Eosinophilic Meningitis Caused by Angiostrongylus cantonensis, France, 2016. Emerg Infect Dis. 2017;23(6):1045–6. Epub 2017/05/19. 10.3201/eid2306.161999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang QP, Lai DH, Zhu XQ, Chen XG, Lun ZR. Human angiostrongyliasis. Lancet Infect Dis. 2008;8(10):621–30. Epub 2008/10/17. 10.1016/S1473-3099(08)70229-9 . [DOI] [PubMed] [Google Scholar]
- 11.Barratt J, Chan D, Sandaradura I, Malik R, Spielman D, Lee R, et al. Angiostrongylus cantonensis: a review of its distribution, molecular biology and clinical significance as a human pathogen. Parasitology. 2016;143(9):1087–118. Epub 2016/05/27. 10.1017/S0031182016000652 . [DOI] [PubMed] [Google Scholar]
- 12.Kim JR, Hayes KA, Yeung NW, Cowie RH. Diverse gastropod hosts of Angiostrongylus cantonensis, the rat lungworm, globally and with a focus on the Hawaiian Islands. PLoS One. 2014;9(5):e94969 Epub 2014/05/03. 10.1371/journal.pone.0094969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Helminth Genomes C. Comparative genomics of the major parasitic worms. Nat Genet. 2019;51(1):163–74. Epub 2018/11/07. 10.1038/s41588-018-0262-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong HS, Eamsobhana P, Lim PE, Razali R, Aziz FA, Rosli NS, et al. Draft genome of neurotropic nematode parasite Angiostrongylus cantonensis, causative agent of human eosinophilic meningitis. Acta Trop. 2015;148:51–7. Epub 2015/04/26. 10.1016/j.actatropica.2015.04.012 . [DOI] [PubMed] [Google Scholar]
- 15.Zeng X, Wei J, Wang J, Wu F, Fung F, Wu X, et al. Angiostrongylus cantonensis: scanning electron microscopic observations on the cuticle of moulting larvae. Korean J Parasitol. 2013;51(6):633–6. Epub 2014/02/12. 10.3347/kjp.2013.51.6.633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24(8):1384–95. Epub 2014/04/24. 10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye C, Hill CM, Wu S, Ruan J, Ma ZS. DBG2OLC: Efficient Assembly of Large Genomes Using Long Erroneous Reads of the Third Generation Sequencing Technologies. Sci Rep. 2016;6:31900 Epub 2016/08/31. 10.1038/srep31900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963 Epub 2014/11/20. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211 Epub 2014/06/22. 10.1186/1471-2105-15-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–9. Epub 2010/12/15. 10.1093/bioinformatics/btq683 . [DOI] [PubMed] [Google Scholar]
- 21.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18 Epub 2012/01/01. 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–7. Epub 2007/03/03. 10.1093/bioinformatics/btm071 . [DOI] [PubMed] [Google Scholar]
- 23.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. Epub 2015/06/11. 10.1093/bioinformatics/btv351 . [DOI] [PubMed] [Google Scholar]
- 24.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. Epub 2011/05/17. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao G, Xu M, Bai C, Yang Y, Li G, Xu J, et al. Comparative genomics and transcriptomics of Chrysolophus provide insights into the evolution of complex plumage coloration. Gigascience. 2018;7(10). Epub 2018/09/08. 10.1093/gigascience/giy113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40. Epub 2014/01/24. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21 Suppl 1:i152–8. Epub 2005/06/18. 10.1093/bioinformatics/bti1003 . [DOI] [PubMed] [Google Scholar]
- 28.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21 Suppl 1:i351–8. 10.1093/bioinformatics/bti1018 . [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(Web Server issue):W265–8. 10.1093/nar/gkm286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, et al. Ascaris suum draft genome. Nature. 2011;479(7374):529–33. Epub 2011/10/28. 10.1038/nature10553 . [DOI] [PubMed] [Google Scholar]
- 31.Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317(5845):1756–60. Epub 2007/09/22. 10.1126/science.1145406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–8. Epub 1998/12/16. [DOI] [PubMed] [Google Scholar]
- 33.Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, et al. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14(8):R88 Epub 2013/08/30. 10.1186/gb-2013-14-8-r88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz EM, Korhonen PK, Campbell BE, Young ND, Jex AR, Jabbar A, et al. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14(8):R89 Epub 2013/08/30. 10.1186/gb-2013-14-8-r89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the human hookworm Necator americanus. Nat Genet. 2014;46(3):261–9. Epub 2014/01/21. 10.1038/ng.2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, et al. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci U S A. 2008;105(39):14802–7. Epub 2008/09/24. 10.1073/pnas.0805946105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. Epub 2004/03/23. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–50. Epub 2009/04/21. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Stoeckert CJ Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44(D1):D774–80. Epub 2015/11/19. 10.1093/nar/gkv1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. Epub 2009/06/10. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. Epub 2014/01/24. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics. 2014;30(17):i541–8. Epub 2014/08/28. 10.1093/bioinformatics/btu462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Zheng H, Chen X, Zhang L, Wang K, Guo J et al. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460(7253):345–51. Epub 2009/07/17. 10.1038/nature08140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–8. Epub 2009/07/17. 10.1038/nature08160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young ND, Jex AR, Li B, Liu S, Yang L, Xiong Z, et al. Whole-genome sequence of Schistosoma haematobium. Nat Genet. 2012;44(2):221–5. Epub 2012/01/17. 10.1038/ng.1065 . [DOI] [PubMed] [Google Scholar]
- 47.Young ND, Nagarajan N, Lin SJ, Korhonen PK, Jex AR, Hall RS, et al. The Opisthorchis viverrini genome provides insights into life in the bile duct. Nat Commun. 2014;5:4378 Epub 2014/07/10. 10.1038/ncomms5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Chen W, Huang Y, Sun J, Men J, Liu H, et al. The draft genome of the carcinogenic human liver fluke Clonorchis sinensis. Genome Biol. 2011;12(10):R107 Epub 2011/10/26. 10.1186/gb-2011-12-10-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robb SM, Gotting K, Ross E, Sanchez Alvarado A. SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis. 2015;53(8):535–46. Epub 2015/07/04. 10.1002/dvg.22872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. WormBase ParaSite—a comprehensive resource for helminth genomics. Mol Biochem Parasitol. 2017;215:2–10. Epub 2016/12/03. 10.1016/j.molbiopara.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. Epub 2007/05/08. 10.1093/molbev/msm088 . [DOI] [PubMed] [Google Scholar]
- 52.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2. Epub 2006/10/06. 10.1093/bioinformatics/btl505 . [DOI] [PubMed] [Google Scholar]
- 53.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–71. Epub 2006/03/18. 10.1093/bioinformatics/btl097 . [DOI] [PubMed] [Google Scholar]
- 54.Rawlings ND, Barrett AJ, Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016;44(D1):D343–50. Epub 2015/11/04. 10.1093/nar/gkv1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–5. Epub 2005/01/14. 10.1093/bioinformatics/bti263 . [DOI] [PubMed] [Google Scholar]
- 56.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–37. Epub 2009/04/21. 10.1007/978-1-59745-251-9_6 . [DOI] [PubMed] [Google Scholar]
- 57.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74. Epub 2014/11/06. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36 Epub 2013/04/27. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 60.Chiumiento L, Bruschi F. Enzymatic antioxidant systems in helminth parasites. Parasitol Res. 2009;105(3):593–603. Epub 2009/05/23. 10.1007/s00436-009-1483-0 . [DOI] [PubMed] [Google Scholar]
- 61.Hahn UK, Bender RC, Bayne CJ. Involvement of nitric oxide in killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata. J Parasitol. 2001;87(4):778–85. Epub 2001/09/06. 10.1645/0022-3395(2001)087[0778:IONOIK]2.0.CO;2 . [DOI] [PubMed] [Google Scholar]
- 62.Hahn UK, Bender RC, Bayne CJ. Killing of Schistosoma mansoni sporocysts by hemocytes from resistant Biomphalaria glabrata: role of reactive oxygen species. J Parasitol. 2001;87(2):292–9. Epub 2001/04/25. 10.1645/0022-3395(2001)087[0292:KOSMSB]2.0.CO;2 . [DOI] [PubMed] [Google Scholar]
- 63.Giannelli A, Cantacessi C, Colella V, Dantas-Torres F, Otranto D. Gastropod-Borne Helminths: A Look at the Snail-Parasite Interplay. Trends Parasitol. 2016;32(3):255–64. Epub 2016/01/08. 10.1016/j.pt.2015.12.002 . [DOI] [PubMed] [Google Scholar]
- 64.Huttemann M, Schmahl G, Mehlhorn H. Light and electron microscopic studies on two nematodes, Angiostrongylus cantonensis and Trichuris muris, differing in their mode of nutrition. Parasitol Res. 2007;101 Suppl 2:S225–32. Epub 2008/01/19. 10.1007/s00436-007-0698-1 . [DOI] [PubMed] [Google Scholar]
- 65.Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 2003;19(9):417–23. Epub 2003/09/06. . [DOI] [PubMed] [Google Scholar]
- 66.Williamson AL, Brindley PJ, Abbenante G, Datu BJ, Prociv P, Berry C, et al. Hookworm aspartic protease, Na-APR-2, cleaves human hemoglobin and serum proteins in a host-specific fashion. J Infect Dis. 2003;187(3):484–94. Epub 2003/01/29. 10.1086/367708 . [DOI] [PubMed] [Google Scholar]
- 67.Oliver EM, Skuce PJ, McNair CM, Knox DP. Identification and characterization of an asparaginyl proteinase (legumain) from the parasitic nematode, Haemonchus contortus. Parasitology. 2006;133(Pt 2):237–44. Epub 2006/05/03. 10.1017/S0031182006000229 . [DOI] [PubMed] [Google Scholar]
- 68.Jing Y, Toubarro D, Hao Y, Simoes N. Cloning, characterisation and heterologous expression of an astacin metalloprotease, Sc-AST, from the entomoparasitic nematode Steinernema carpocapsae. Mol Biochem Parasitol. 2010;174(2):101–8. Epub 2010/07/31. 10.1016/j.molbiopara.2010.07.004 . [DOI] [PubMed] [Google Scholar]
- 69.Park JO, Pan J, Mohrlen F, Schupp MO, Johnsen R, Baillie DL, et al. Characterization of the astacin family of metalloproteases in C. elegans. BMC Dev Biol. 2010;10:14 Epub 2010/01/30. 10.1186/1471-213X-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stepek G, McCormack G, Page AP. Collagen processing and cuticle formation is catalysed by the astacin metalloprotease DPY-31 in free-living and parasitic nematodes. Int J Parasitol. 2010;40(5):533–42. Epub 2009/11/04. 10.1016/j.ijpara.2009.10.007 . [DOI] [PubMed] [Google Scholar]
- 71.Williamson AL, Lustigman S, Oksov Y, Deumic V, Plieskatt J, Mendez S, et al. Ancylostoma caninum MTP-1, an astacin-like metalloprotease secreted by infective hookworm larvae, is involved in tissue migration. Infect Immun. 2006;74(2):961–7. Epub 2006/01/24. 10.1128/IAI.74.2.961-967.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hunt VL, Tsai IJ, Coghlan A, Reid AJ, Holroyd N, Foth BJ, et al. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet. 2016;48(3):299–307. Epub 2016/02/02. 10.1038/ng.3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lively CM, Dybdahl MF. Parasite adaptation to locally common host genotypes. Nature. 2000;405(6787):679–81. Epub 2000/06/23. 10.1038/35015069 . [DOI] [PubMed] [Google Scholar]
- 74.Zarowiecki M, Berriman M. What helminth genomes have taught us about parasite evolution. Parasitology. 2015;142 Suppl 1:S85–97. Epub 2014/12/09. 10.1017/S0031182014001449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morassutti AL, Pinto PM, Dutra BK, Oliveira GT, Ferreira HB, Graeff-Teixeira C. Detection of anti-oxidant enzymatic activities and purification of glutathione transferases from Angiostrongylus cantonensis. Exp Parasitol. 2011;127(2):365–9. Epub 2010/09/03. 10.1016/j.exppara.2010.08.018 . [DOI] [PubMed] [Google Scholar]
- 76.Smith NC, Bryant C. The role of host generated free radicals in helminth infections: Nippostrongylus brasiliensis and Nematospiroides dubius compared. Int J Parasitol. 1986;16(6):617–22. Epub 1986/12/01. 10.1016/0020-7519(86)90029-9 . [DOI] [PubMed] [Google Scholar]
- 77.Gourbal BE, Guillou F, Mitta G, Sibille P, Theron A, Pointier JP, et al. Excretory-secretory products of larval Fasciola hepatica investigated using a two-dimensional proteomic approach. Mol Biochem Parasitol. 2008;161(1):63–6. Epub 2008/06/17. 10.1016/j.molbiopara.2008.05.002 . [DOI] [PubMed] [Google Scholar]
- 78.Zelck UE, Von Janowsky B. Antioxidant enzymes in intramolluscan Schistosoma mansoni and ROS-induced changes in expression. Parasitology. 2004;128(Pt 5):493–501. Epub 2004/06/08. 10.1017/s0031182004004895 . [DOI] [PubMed] [Google Scholar]
- 79.Murell K, Pozio EJGWPP, Part. The liver flukes: Clonorchis sinensis, Opisthorchis spp, and Metorchis spp. 2017;3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLSX)
Data Availability Statement
The A. cantonensis genome assembly and annotation is accessible at China National GeneBank (CNSA) under accession CNP0000467. Raw sequences (Illumina and PacBio) of the A. cantonensis reference genome have been deposited at NCBI in the SRA under accession number SRP092294 (BioProject number: PRJNA350391). The multiple developmental stages of the A. cantonensis RNA-seq reads have been deposited at NCBI in the SRA under accession number SRP127889 (BioProject number: PRJNA350405).