Abstract
Test–retest reliability of resting regional cerebral metabolic rate of glucose (rCMR) was examined in selected subcortical structures: the amygdala, hippocampus, thalamus, and anterior caudate nucleus. Findings from previous studies examining reliability of rCMR suggest that rCMR in small subcortical structures may be more variable than in larger cortical regions. We chose to study these subcortical regions because of their particular interest to our laboratory in its investigations of the neurocircuitry of emotion and depression. Twelve normal subjects (seven female, mean age = 32.42 years, range 21–48 years) underwent two FDG‐PET scans separated by approximately 6 months (mean = 25 weeks, range 17–35 weeks). A region‐of‐interest approach with PET‐MRI coregistration was used for analysis of rCMR reliability. Good test–retest reliability was found in the left amygdala, right and left hippocampus, right and left thalamus, and right and left anterior caudate nucleus. However, rCMR in the right amygdala did not show good test–retest reliability. The implications of these data and their import for studies that include a repeat‐test design are considered. Hum. Brain Mapping 10:1–9, 2000. © 2000 Wiley‐Liss, Inc.
Keywords: PET, MRI, amygdala, hippocampus, thalamus, caudate nucleus, reliability
INTRODUCTION
Functional neuroimaging studies often require a repeat test design. For activation studies using functional magnetic resonance imaging (fMRI) or O15 positron emission tomography (PET), the repeated assessments are performed within the same session. Sometimes, such activation measures are repeated at another point in time to examine longitudinal change. For example, clinical studies of regional glucose metabolism often involve examining changes over time as a function of treatment. All studies that involve repeated assessments are predicated on the assumption of there being adequate test–retest reliability of measures of regional brain function, at least in normal subjects where an individual's baseline state is also assumed to remain relatively stable.
[18F]fluorodeoxyglucose (FDG) PET has been an important tool for researchers investigating regional brain glucose metabolism. The intrasubject and intersubject variability of rCMR as measured with FDG‐PET has been previously examined with test–retest studies of healthy normals [Bartlett et al., 1988; Brooks et al., 1987; Camargo et al., 1992; Duara et al., 1987; Gur et al., 1987b; Maquet et al., 1990; Schmidt et al., 1996; Tyler et al., 1988]. These studies have found good overall test–retest stability and low intrasubject variability in metabolism. The variance observed has been shown to be largely because of diffuse changes in metabolism resulting from changes in psychological state such as arousal, habituation, or anxiety [Gur et al., 1987a,b; Mazziota et al., 1982; Risberg et al., 1977; Warach et al., 1992], repositioning problems [Chang et al., 1987], or partial volume effects [Mazziota et al., 1981]. Differences in global cerebral metabolic rate (gCMR) have been found to account for most of the intrasubject and intersubject variance in FDG‐PET studies. Thus, the common practice of normalizing regional activity to gCMR has been shown to substantially reduce interscan variance.
Unfortunately, most of the previous reliability studies were conducted before the recent advances in scanner technology so they utilized PET cameras with relatively poor resolution (most had an in‐plane resolution over 11 mm full width half maximum (FWHM)). This limited resolution may provide inflated estimates of reliability given the lack of discrimination between individual structures and problematic partial voluming in smaller structures. Also, the regions‐of‐interest (ROIs) used in analyses were program‐generated or based on a PET template rather than the individual subject's anatomy; thus, the ROIs had limited anatomical specificity. In addition, the time interval between the test and retest scans was often very short, usually less than 24 h. In fact, only one early study [Gur et al., 1987a,b] had a test–retest range of 7–23 weeks, which just approaches the length of time involved in most treatment studies.
Most previous reliability studies focused on reliability of cortical rCMR. The relatively more recent studies suggest that subcortical rCMR may be more variable than rCMR in most cortical regions [Camargo et al., 1992; Holcomb et al., 1993; Schmidt et al., 1996]. The results of Camargo et al. [1992] suggest that several subcortical regions may be the most variable regions in the brain. They found that rCMR in the caudate nucleus, putamen, and thalamus, along with frontal cortex, temporal cortex, and cingulate gyrus accounted for 70% of the variance in cerebral metabolic rate with a factor analysis of their data. They hypothesized that the variance in rCMR in these structures resulted from the role of these structures in complex functions such as autonomic motor control, visceral information, perception of stimuli, attention, behavior, emotions, and motivation.
Our laboratory has been involved in a program of research on the functional neuroanatomy of emotion and disorders of emotion [see Davidson et al., 1999 for review]. As part of this effort, we are interested in studying changes in the neurocircuitry of emotion in patients with mood disorders as their clinical symptoms remit with treatment. This question requires a test–retest design where both patients and controls are tested on multiple occasions over time. We therefore wished to examine in this study resting rCMR in selected subcortical structures that have been implicated in the neurocircuitry of emotion and depression [Davidson and Irwin, 1999; Davidson et al., 1999; Lindgren et al., 1999]: the amygdala, hippocampus, thalamus, and anterior caudate nucleus. We used an ROI approach that involved coregistration of each individual subject's magnetic resonance imaging (MRI) scan to his/her PET scans. ROIs were drawn on each subject's MRI with criteria for ROI drawing chosen to include exclusively gray matter and to minimize artifact in ROI estimates because of partial volume averaging in the PET scans. The metabolic rate from the exact ROI region was then extracted from the coregistered PET scans. Subjects were tested at time intervals matched to a depressed group receiving antidepressant treatment (between 17–35 weeks). We believe this to be the longest test–retest interval in a study examining rCMR reliability in normals to date. In addition, this study represents the first test of reliability of amygdalar metabolic rate.
MATERIALS AND METHODS
Subjects
Subjects were recruited via advertisements in local media. After the nature of the experimental procedures was explained, informed consent was obtained. Subjects were screened for psychopathology using the Structured Clinical Interview for DSM‐IV Non‐Patient Edition [SCID; First et al., 1995] and had no history of any Axis I disorders or substance abuse in themselves or their first‐degree relatives. All participants were right‐handed as assessed by the Chapman Handedness Inventory [Chapman and Chapman, 1987] and had no history of thyroid problems, diabetes, or brain injury. Twelve subjects (seven female, mean age = 32.42 years, range 21–48 years) underwent two FDG‐PET scans separated by approximately 6 months (mean = 25 weeks, range 17–35 weeks). These subjects were time‐matched to a group of depressed individuals who were similarly tested prior to treatment with an antidepressant and retested after their depression remitted. Data from the depressed subjects will not be presented here.
Procedures
Subjects completed a full day of testing including measurement of resting regional cerebral glucose metabolic rate (CMR) using [18F]‐2‐fluoro‐2‐deoxy‐D‐glucose positron emission tomography (FDG‐PET). Injection of FDG was scheduled between 11:00 and 13:30 hr. Subjects fasted for at least 5 hr prior to injection of the FDG. Two 22‐gauge intravenous catheters were placed, one in the antecubital fossa of the right arm, and the other in a vein in the posterior aspect of the left hand. The left hand was placed into a heated handwarmer to maintain a skin surface temperature of 42°C, which allowed for rapid sequential sampling of “arterialized” venous blood. The right hand was also heated to equate thermal stimulation on both sides of the body.
Subjects were informed that the uptake period would last 30 min and were instructed to remain still (allowing for periodic shifts in position if needed), sit quietly, and relax but stay awake during uptake. Eyes and ears were unoccluded. A 1‐ to 2‐ml blood sample was drawn to obtain an initial plasma glucose level. Approximately 5 millicuries (range of 3.8–5.7 millicuries) of FDG was then administered by bolus injection into the intravenous line in the right arm. Sequential 1‐ to 2‐ml blood samples were collected throughout the 30 min following injection as follows: as quickly as possible for 2 min; every 30 sec for the next 2 min; every minute for the next 4 min; every 2 min for the next 10 min; and every 3 min for the next 12 min. Two additional samples were obtained to measure plasma glucose levels at approximately 15 and 30 min postinjection. A technologist seated to the left of the subject drew blood samples from the left hand, and one to two laboratory assistants recorded the exact time of each draw. Following the uptake period, the subjects were directed to the restroom and encouraged to void their bladders. The subjects were then placed in the PET scanner bed to acquire a 30‐min emission scan.
Each subject repeated this procedure approximately 6 months (mean = 25 weeks, range 17–35 weeks) following his/her initial scan.
PET and MRI Data Acquisition
[18F] fluoride was produced by a CTI RDS Cyclotron (Knoxville, TN, USA). FDG was synthesized using the modified method of Hamacher [Hamacher and Coenen, 1986].
Scans were performed with a GE Advance PET camera (Milwaukee, WI, USA). The head‐holder was the same for each pair of test–retest scans and was both symmetric and uniform along the axial dimension. A laser‐positioning device was used to position the participant's head in the scanner based on external landmarks of the outer compass of the eye and the center of the external meatus. The image planes were positioned parallel to the orbitomeatal line. A 30‐min emission scan with a field of view of 15.2 cm acquired 35 transaxial planes covering the entire brain. In‐plane and axial resolutions were approximately 5 mm FWHM. All images were reconstructed to 256 × 256 × 35 pixels. Pixel dimensions were 1.17 mm × 1.17 mm in plane with a plane thickness of 4.25 mm. Calculated attenuation correction was applied to the data using an unique outline placed by the same experienced PET technician for each individual PET scan.
Structural MRI scans were performed on a 1.5 Tesla GE Signa scanner (Milwaukee, WI, USA) scheduled between the two PET sessions. The MRI protocol consisted of an axial 3D SPGR, with 24 cm FOV, TE = 14, TR = 30, 256 × 192 matrix, NEX = 1, flip angle = 35°, and a 1.2 mm slice thickness, for a total of 124 slices.
Quantification of Emission Data
Pixel absolute radioactivity concentration values and blood radioactivity values were corrected for physical decay back to the time of injection. Previous measurements in our laboratory demonstrated the striking similarity among different individuals, up to a multiplicative scale, of plasma FDG concentration time courses following the initial distribution of tracer into the plasma and extracellular spaces. Therefore, the individual plasma time courses measured over the first 30 min were combined with previously measured normative data to provide tracer concentration time courses over the duration of the PET image data acquisition. Plasma time courses were combined with plasma glucose levels and image pixel values to estimate the rate of glucose utilization in each image pixel according to the Sokoloff method [Sokoloff et al., 1977].
Determination of Metabolic Rate Values
Estimates of global cerebral metabolic rate (gCMR) were obtained using Statistical Parametric Mapping software (SPM 96; Wellcome Department of Cognitive Neurology). Each participant's gCMR value equals the average of all pixels in the image that fell above a threshold of 12.5% of the whole‐volume mean.
Regional cerebral metabolic rate values for each participant for the left and right amygdala, hippocampus, thalamus, and anterior caudate nucleus were obtained using PET‐MRI coregistration. MRI image sets were transformed into the coronal orientation to facilitate identification of the regions‐of‐interest (ROIs). DIP Station, version 1.0.6, (Hayden Image Processing Group) was used to manually draw separate left and right ROIs on coronal MRI planes for each participant. Specific criteria were developed to minimize artifactual ROI metabolic rate estimates because of partial voluming in the PET scans. Therefore, the ROIs were drawn specifically for the purpose of extraction of PET‐MRI coregistration‐derived glucose metabolic rate and were not drawn for the purpose of morphometric analysis (see Table I for ROI volumes). Tracing guidelines were developed through the use of neuroanatomical atlases [DeArmond et al., 1989; Matsui and Hirano, 1978], in consultation with colleagues experienced in neuroanatomy, and based on previous researchers' criteria for ROIs [Potts et al., 1994]. See Figure 1 for illustrations of representative planes from ROIs.
Table I.
Subcortical ROI volumes [mm3; mean (SD); n = 12]
| Brain region | Left hemisphere | Right hemisphere |
|---|---|---|
| Amygdala | 507.67 (37.81) | 478.08 (33.48) |
| Hippocampus | 829.33 (52.17) | 903.25 (59.17) |
| Thalamus | 5436.79 (191.96) | 5353.59 (226.28) |
| Anterior caudate nucleus | 767.92 (43.09) | 758.58 (36.63) |
Figure 1.
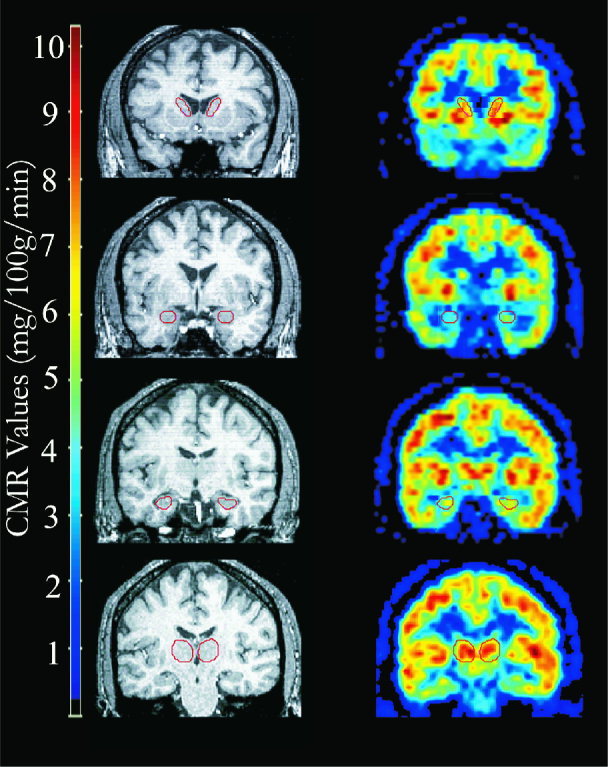
PET‐MRI coregistration and ROI delineation. This figure presents representative image planes, four coronal sections, for one participant. From top to bottom, example right and left ROI of the anterior caudate nucleus, the amygdala, the hippocampus, and the thalamus. The top images correspond to an anterior plane and each successive image corresponds to an increasingly posterior plane. PET image planes are presented to the right of their corresponding co‐registered [using AIR; Woods et al., 1993] MRI plane.
ROIs for the amygdala were drawn on five to eight coronal MRI planes. An almond‐shaped outline was manually traced just inside the gray/white matter interface on every coronal slice on which the amygdala could be visualized. Care was taken not to include the hippocampus in the ROI for the amygdala. Therefore, the posterior boundary of the ROI was always anterior to the basilar artery and the ROI was never drawn lateral to or inferior to the temporal horn of the lateral ventricle.
ROIs for the hippocampus always included 14 coronal MRI planes. The temporal horn of the lateral ventricle was used as a landmark for determination of the anterior boundary of the hippocampus. Once the lateral ventricle formed an arch just caudal to the amygdala, the tissue inferior to the lateral ventricle was outlined. After the anterior boundary was determined, ROIs were then drawn on 13 successive planes. Care was taken to include only tissue inferior to the lateral ventricle and to avoid medial tissue suspected to be the uncus.
ROIs for the anterior caudate nuclei always included the ten most anterior coronal MRI planes on which the head of the caudate was visible. ROIs were drawn within the grey/white matter interface and the lateral ventricle and internal capsule were used as borders. Care was taken to avoid inferior tissue suspected to be the nucleus accumbens.
ROIs for the thalamus were drawn on 25–41 coronal MRI planes. An outline was drawn just inside the gray/white matter interface of the thalamus. Because the thalamus is medially and superiorly bordered by the lateral and third ventricles and includes both white and gray matter, care was taken not to include any cerebral spinal fluid (CSF) or excess white matter. The internal capsule was used as a guide for the lateral and inferior boundaries. The shape of the lateral ventricles typically differentiated the anterior aspect of the thalamus. In other words, the most anterior slice of the thalamus was drawn when the inferior most section of the ventricles became flattened by the appearance of the anterior nucleus of the thalamus. The most posterior slice of thalamus included was typically the slice just anterior to the appearance of the crus of fornix looping down to connect with the hippocampus.
After ROIs were delineated on each MRI scan, the ROIs were copied to a blank image set. The Automated Image Registration package [AIR; Woods et al., 1993] was used to coregister the PET and MRI data. MRI scans were fit to PET scans, and then resliced to match the PET. Each PET‐MRI coregistration was visually inspected to confirm that a good fit was obtained. The same manipulation was then applied to the blank image set containing the ROIs. The transformed ROIs were copied to the PET image and mean rCMR was extracted for the corresponding regions of the ROIs on the coregistered PET scan.
Statistical Analysis
ROI Delineation: Interrater Reliability of rCMR
Intraclass correlation coefficients were computed on absolute rCMR extracted from ROIs drawn by two independent raters. Image sets used in reliability calculations were chosen randomly. Different datasets were used to test interrater reliability of ROI delineation for the amygdala, hippocampus, thalamus, and anterior caudate nucleus, however all were tested on four subjects' data.
Adjustment of Regional Metabolic Rate for Variance in Global Metabolic Rate
Absolute rCMR values were divided by gCMR values to scale rCMR values by whole brain metabolic values. Henceforth, rCMR/gCMR will be referred to as normalized rCMR.
Test–Retest Differences in gCMR and rCMR
Two‐tailed paired t‐tests were performed on gCMR and normalized rCMR values to test for differences in metabolic rate between the two PET scans. Average percent change between each individual's scans was calculated by region with the following formula:
Intraclass correlations were run between the first and second scan's gCMR and normalized rCMR for each of the ROIs.
RESULTS
Interrater Reliability of rCMR
Intraclass correlations indicated reliable extraction of rCMR (n = 4; for the amygdala: right, IC = 0.96, left, IC = 0.95; for the hippocampus: right, IC = 0.97, left, IC = 0.99; for the thalamus: right, IC = 0.95, left IC = 0.97; for the anterior caudate: right, IC = 0.97, left, IC = 0.92).
Test–Retest Differences in gCMR and rCMR
Comparison of test–retest mean gCMR and normalized rCMR revealed no significant differences; however, there was a marginally significant decrease in gCMR at the second scan (t = 1.89, p = 0.09). Mean absolute metabolic rate in each of the ROIs of the test (scan 1) and retest (scan 2) scans is presented in Table II. Mean normalized metabolic rate (rCMR/gCMR) is presented in Table III. The percent change between the two test occasions in normalized rCMR for each of the ROIs is presented in Table IV.
Table II.
Regional metabolic rate in test and retest scans: absolute data [mg glucose/100 g tissue/min; mean (SD); n = 12]
| Brain region | Left hemisphere | Right hemisphere | ||
|---|---|---|---|---|
| Scan 1 | Scan 2 | Scan 1 | Scan 2 | |
| Amygdala | 5.45 (0.22) | 4.93 (0.23) | 5.25 (0.21) | 4.91 (0.19) |
| Hippocampus | 5.88 (0.19) | 5.44 (0.17) | 5.99 (0.19) | 5.45 (0.21) |
| Thalamus | 8.50 (0.32) | 7.71 (0.25) | 8.44 (0.32) | 7.69 (0.25) |
| Anterior caudate nucleus | 10.44 (0.51) | 9.39 (0.36) | 9.95 (0.51) | 9.11 (0.39) |
Table III.
Regional metabolic rate in test and retest scans: normalized data [rCMR/gCMR; mean (SD); n = 12]
| Brain region | Left hemisphere | Right hemisphere | ||
|---|---|---|---|---|
| Scan 1 | Scan 2 | Scan 1 | Scan 2 | |
| Amygdala | 1.02 (0.03) | 1.00 (0.03) | 0.98 (0.02) | 1.00 (0.03) |
| Hippocampus | 1.10 (0.03) | 1.11 (0.02) | 1.12 (0.02) | 1.11 (0.03) |
| Thalamus | 1.58 (0.04) | 1.57 (0.04) | 1.57 (0.04) | 1.57 (0.04) |
| Anterior caudate nucleus | 1.93 (0.03) | 1.91 (0.03) | 1.85 (0.06) | 1.85 (0.05) |
| Global metabolic rate | Scan 1 = 5.40 (0.24) | Scan 2 = 4.93 (0.17) | ||
Table IV.
Percent change in test and retest regional metabolic rate: normalized data [mean (SD); n = 12]
| Brain region | Left hemisphere | Right hemisphere |
|---|---|---|
| Percent (%) change | Percent (%) change | |
| Amygdala | 6.03 (1.80) | 9.85 (1.49) |
| Hippocampus | 4.89 (0.93) | 6.15 (0.95) |
| Thalamus | 2.47 (0.53) | 2.86 (0.49) |
| Anterior caudate nucleus | 3.75 (0.76) | 3.33 (0.53) |
Intraclass correlations revealed no relation between test and retest gCMR (r = 0.16, p = 0.46). Significant intraclass correlations between test and retest normalized rCMR were found for the left amygdala, left and right hippocampus, left and right thalamus, and left and right caudate nucleus (see Table V, Fig. 2).1 There was no relation between right amygdalar normalized rCMR at the two sessions.
Table V.
Intraclass correlations between test and retest scans: normalized metabolic values (scan 1 vs. scan 2; n = 12; *p < 0.01)
| Brain region | Left hemisphere | Right hemisphere |
|---|---|---|
| Amygdala | 0.53* | 0.17 |
| Hippocampus | 0.64* | 0.54* |
| Thalamus | 0.93* | 0.92* |
| Anterior caudate nucleus | 0.65* | 0.93* |
Figure 2.
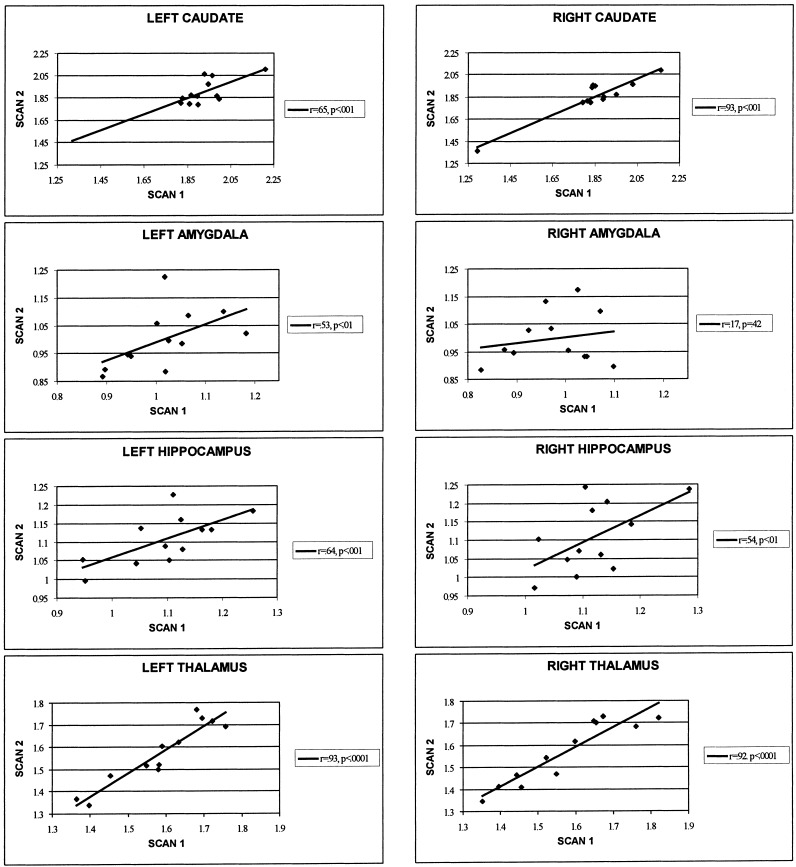
Test–retest correlations. This figure presents scatterplots between the normalized rCMR from scans 1 (test) and 2 (retest) in the left and right anterior caudate nucleus, amygdala, hippocampus, and thalamus.
DISCUSSION
We found good test–retest reliability of resting regional metabolic rate in the left amygdala, left and right hippocampus, left and right thalamus, and left and right anterior caudate nucleus as assessed with intraclass correlations between the test (scan 1) and retest (scan 2) scans. The percent differences observed between the two scans are consistent with reports from previous studies examining test–retest reliability of thalamic, hippocampal, and caudate metabolism in normals, even though the studies were performed under differing conditions, image resolution, and data analysis pathways [Camargo et al., 1992; Maquet et al., 1990; Schmidt et al., 1996]. In fact, our observed differences closely match those reported by Schmidt et al. [1996] in a study of ten healthy normals undergoing sequential FDG‐PET scans while performing an auditory continuous performance task. Schmidt et al. examined reliability of serial FDG‐PET scans for psychopharmacologic studies by testing subjects prior to and following a 30‐min placebo infusion. They found a percent difference of left: 4.8 ± 13.2 and right: 0.1 ± 7.1 in the hippocampus, left: 9 ± 12.6 and right: 6.3 ± 10.2 in the thalamus, and left: 3.5 ± 2.2 and right: 2.1 ± 9.3 in the caudate. We found similar differences even though the Schmidt et al. reliability study measured brain activity prior to and following a placebo infusion, their subjects were scanned while performing a task, and the test–retest scans were sequential, whereas in the present study brain activity was measured at rest approximately 6 months apart. This suggests that measurement of regional brain activity in these subcortical structures is reliable with FDG‐PET over both short and long testing intervals and at both rest and the active state. It also suggests that the variance observed in our two studies might represent variance attributable to causes beyond experimenter control, thus representing the lower achievable limits in terms of variance in FDG‐PET studies.
This study was the first to examine reliability of regional metabolism in the amygdala and, interestingly, we found that metabolism in the right amygdala is not reliable yet metabolism in the left amygdala is reliable. The amygdala is the smallest structure that we examined. The accuracy of measurement of rCMR has been shown to be a function of the spatial resolution of the PET camera, the size and shape of the structure of interest, and method used for extraction of regional metabolic rate [Kuwert et al., 1992]. Using a phantom study, Hoffman et al. [1979] showed that for accurate quantification of isotope concentration in tissue, the size of the structure should be approximately double to the resolution of the scanner. The human amygdala, resembling the shape of an almond, has a volume of approximately 2 cc. Given the GE Advance scanner's in‐plane and axial resolution of approximately 5 mm FWHM, it is theoretically possible to accurately quantify amygdala metabolism. Our strong interrater reliability for delineation of all of the ROIs including the right and left amygdala combined with our ability to reliably measure rCMR in the left amygdala suggests that we are able to accurately measure amygdala rCMR. In addition, there was no significant difference between the volume of our left and right amygdala ROIs (t = −1.01, p = 0.31); therefore, the decreased reliability observed for the right amygdala should not be the result of its decreased volume. Finally, the across subject variability for the right amygdala was virtually identical to that found for the left amygdala, suggesting that our failure to find significant test–retest stability for the right amygdala is not a function of truncation of range.
The amygdala has been consistently identified as playing a crucial role in both the perception of emotional cues and the production of emotional responses, although findings regarding laterality have been confusing at best [see Davidson and Irwin, 1999 for review]. It is quite possible that session effects related to anxiety produced by subjects' initial exposure to the FDG‐PET environment, including the FDG injection, needlesticks, continual blood draws, etc. may be responsible for the lack of reliability seen in right amygdalar metabolism. None of our subjects had been previously exposed to the PET environment prior to his/her first test scan. Normalizing regional activity to global metabolic rate repeatedly has been shown to account for most of the variance in test–retest studies. However, the right amygdala may be differentially activated to the point where normalization does not account for all of the variance. Unfortunately, we do not have adequate self‐report indices of anxiety at the time of the scans to examine this further. Additional studies investigating this lack of reliability in right amygdalar metabolism and its possible relation to changes in anxiety in response to the FDG‐PET environment are needed. Studies that involve assessing possible changes over time in amygdala activation or reactivity should be sensitive to the lack of reliability found here for the right amygdala.
ACKNOWLEDGMENTS
The authors wish to thank Andrea Straus, Joni Hanson, Robert Pyzalski, Dan McGary, Heidi Neudeck, and Corrina Mueller for their sizable contributions in collecting these data. These data were presented at the 5th International Conference on Functional Mapping of the Human Brain in Congress Center Düsseldorf, Germany, June 23–26, 1999. Support for this research was provided by an NIMH center grant to the Wisconsin Center for Affective Science and an NIMH Research Scientist Award to RJD.
Footnotes
The scatterplot of rCMR in the right caudate reveals one outlying data point. When the outlier is left out of the caudate test–retest intraclass correlations, the reliability correlations remain high (n = 11; IC = 0.73 for the right caudate and IC = 0.69 for the left caudate both p < 0. 0005).
REFERENCES
- American Psychiatric Association. (1994): Diagnostic and statistical manual of mental disorders (4th Ed.). Washington, D.C.: American Psychiatric Press. [Google Scholar]
- Bartlett EJ, Brodie JD, Wolf AP, Christman DR, Laska E, Meissner M. (1988): Reproducibility of cerebral glucose metabolic measurements in resting human subjects. J Cereb Blood Flow Metab 8: 502–512. [DOI] [PubMed] [Google Scholar]
- Bartlett EJ, Barouche F, Brodie JD, Wolkin A, Angrist B, Rotrosen J, Wolf AP. (1991): Stability of resting deoxyglucose metabolic values in PET studies of schizophrenia. Psychiat Res: Neuroimaging 40: 11–20. [DOI] [PubMed] [Google Scholar]
- Brooks RA, Di Chiro G, Zukerberg BW, Bairamian D, Larson SM. (1987): Test–retest studies of cerebral glucose metabolism using fluorine‐18‐deoxyglucose: Validation of method. J Nucl Med 28: 53–59. [PubMed] [Google Scholar]
- Camargo EE, Szabo Z, Links JM, Sostre S, Dannals RF, Wagner HN. (1992): The influence of biological and technical factors on the variability of global and regional brain metabolism of 2‐[18F]Fluoro‐2‐deoxy‐D‐glucose. J Cereb Blood Flow Metab 12: 281–290. [DOI] [PubMed] [Google Scholar]
- Chang JY, Barker WW, Apicella A, Emran A, Duara R. (1987): Test–retest paradigms in PET: Errors arising from subject repositioning. J Nucl Med 28: 681–682. [Google Scholar]
- Chapman L, Chapman J. (1987): The measurement of handedness. Brain Cognit 6: 175–183. [DOI] [PubMed] [Google Scholar]
- Davidson RJ, Irwin W. (1999): The functional neuroanatomy of emotion and affective style. Trends Cognit Sci 3: 11–21. [DOI] [PubMed] [Google Scholar]
- Davidson RJ, Abercrombie H, Nitschke JB, Putnam K. (1999): Regional brain function, emotion, and disorders of emotion. Curr Opin Neurobiol 9: 228–234. [DOI] [PubMed] [Google Scholar]
- DeArmond SJ, Fusco MM, Dewey MM. (1989): Structure of the human brain. A photographic atlas (3rd Ed.). Oxford: Oxford University Press. [Google Scholar]
- Duara R, Gross‐Glenn K, Barker WW, Chang JY, Apicella A, Loewenstein D, Boothe T. (1987): Behavioral activation and the variability of cerebral glucose metabolic measurements. J Cereb Blood Flow Metab 7: 266–271. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RI, Gibbon M, et al. (1995): Structured clinical interview for DSM‐IV Axis I disorders. Biometrics Research Department, New York State Psychiatric Institute. [Google Scholar]
- Gur RE, Resnick SM, Gur RC, Alavi A, Carof S, Kushner M, Reivich M. (1987a): Regional brain function in schizophrenia. Arch Gen Psychiat 44: 126–129. [DOI] [PubMed] [Google Scholar]
- Gur RC, Gur RE, Resnick SM, Skolnick BE, Alavi A, Reivich M. (1987b): The effect of anxiety on cortical cerebral blood flow and metabolism. J Cereb Blood Flow Metab 7: 173–177. [DOI] [PubMed] [Google Scholar]
- Hamacher K, Coenen HH. (1986): Efficient stereospecific synthesis of no‐carrier‐added 2‐[18F]‐fluoro‐2‐deoxy‐glucose using aminopolyether supported nucleophilic substitution. J Nucl Med 27: 235–238. [PubMed] [Google Scholar]
- Hoffman EJ, Huang S‐C, Phelps ME. (1979): Quantitation in positron emission computed tomography. 1. Effect of object size. J Comput Assist Tomogr 3: 299–308. [DOI] [PubMed] [Google Scholar]
- Holcomb HH, Cascella NG, Medoff DR, Gastineau EA, Loats H, Thaker GK, Conley RR, Dannals RF, Wagner HN, Tamminga CA. (1993): PET‐FDG Test–retest reliability during a visual discrimination task in schizophrenia. J Comput Assist Tomogr 17: 704–709. [DOI] [PubMed] [Google Scholar]
- Kuwert T, Sures T, Herzog H, Loken M, Hennerici M, Langen K‐J, Feinendegen LE. (1992): On the influence of spatial resolution and of the size and form of regions of interest on the measurement of regional cerebral metabolic rates by positron emission tomography. J Neural Trans S37: 53–66. [DOI] [PubMed] [Google Scholar]
- Lindgren KA, Larson CL, Schaefer SM, Abercrombie HC, Ward RT, Oakes TR, Holden JE, Perlman SB, Benca RM, Davidson RJ. (1999): Thalamic metabolic rate predicts EEG alpha power in healthy control subjects but not in depressed patients. Biol Psychiat 45: 943–952. [DOI] [PubMed] [Google Scholar]
- Maquet P, Dive D, Salmon E, von Frenckel R, Franck G. (1990): Reproducibility of cerebral glucose utilization measured by PET and the [18F]‐2‐fluoro‐2‐deoxy‐d‐glucose method in resting, healthy human subjects. Eur J Nucl Med 16: 267–273. [DOI] [PubMed] [Google Scholar]
- Matsui T, Hirano A. (1978): An atlas of the human brain for computerized tomography. Tokyo, New York: Igaki‐Shoin. [Google Scholar]
- Mazziota JC, Phelps ME, Plummer D, Kuhl DE. (1981): Quantitation in positron emission computer tomography: 5. Physical‐anatomical effects. J Comput Assist Tomogr 5: 734–743. [DOI] [PubMed] [Google Scholar]
- Mazziota JC, Phelps ME, Carson RE, Kuhl DE. (1982): Tomographic mapping of human cerebral metabolism: Sensory deprivation. Ann Neurol 12: 435–444. [DOI] [PubMed] [Google Scholar]
- Potts NLS, Davidson JRT, Krishnan KRR, Doraiswamy PM. (1994): Magnetic resonance imaging in social phobia. Psychiat Res 52: 35–42. [DOI] [PubMed] [Google Scholar]
- Reivich M, Alavi A, Wolf A, Greenberg JH, Fowler J, Christman D, MacGregor R, Jones SC, London J, Shiue C, Yonekura Y. (1982): Use of the 2‐deoxy‐d[1‐11C]glucose for the determination of local cerebral glucose metabolism in humans: Variation within and between subjects. J Cereb Blood Flow Metab 2: 307–319. [DOI] [PubMed] [Google Scholar]
- Risberg J, Maximilian AV, Prohovnik I. (1977): Changes of cortical activity patterns during habituation to a reasoning test. Neuropsychologica 15: 793–798. [DOI] [PubMed] [Google Scholar]
- Schmidt ME, Ernst M, Matochik JA, Maisog JM, Pan B‐S, Zametkin AJ, Potter WZ. (1996): Cerebral glucose metabolism during pharmacologic studies: Test–retest under placebo conditions. J Nucl Med 37: 1142–1149. [PubMed] [Google Scholar]
- Sokoloff L, Reivich M, Kennedy C, et al. (1977): The [14C]deoxyglucose method for the measurement of local cerebral glucose utilization: Theory, procedure, and normal values in the conscious and anesthetized rat. J Neurochem 28: 897–916. [DOI] [PubMed] [Google Scholar]
- Tyler JL, Strother SC, Zatorre RJ, Alivisatos B, Worsley KJ, Diksic M, Yamamoto YL. (1988): Stability of regional cerebral glucose metabolism in the normal brain measured by positron emission tomography. J Nucl Med 29: 631–642. [PubMed] [Google Scholar]
- Warach S, Gur RC, Gur RE, Skolnick BE, Obrist WD, Reivich M. (1992): Decreases in frontal and parietal lobe regional cerebral blood flow related to habituation. J Cereb Blood Flow Metab 12: 546–553. [DOI] [PubMed] [Google Scholar]
- Woods RP, Mazziotta JC, Cherry SR. (1993): MRI‐PET registration with an automated algorithm. J Comput Assist Tomogr 17: 536–546. [DOI] [PubMed] [Google Scholar]


