Abstract
Requiring only minimal assumptions for validity, nonparametric permutation testing provides a flexible and intuitive methodology for the statistical analysis of data from functional neuroimaging experiments, at some computational expense. Introduced into the functional neuroimaging literature by Holmes et al. ([1996]: J Cereb Blood Flow Metab 16:7–22), the permutation approach readily accounts for the multiple comparisons problem implicit in the standard voxel‐by‐voxel hypothesis testing framework. When the appropriate assumptions hold, the nonparametric permutation approach gives results similar to those obtained from a comparable Statistical Parametric Mapping approach using a general linear model with multiple comparisons corrections derived from random field theory. For analyses with low degrees of freedom, such as single subject PET/SPECT experiments or multi‐subject PET/SPECT or fMRI designs assessed for population effects, the nonparametric approach employing a locally pooled (smoothed) variance estimate can outperform the comparable Statistical Parametric Mapping approach. Thus, these nonparametric techniques can be used to verify the validity of less computationally expensive parametric approaches. Although the theory and relative advantages of permutation approaches have been discussed by various authors, there has been no accessible explication of the method, and no freely distributed software implementing it. Consequently, there have been few practical applications of the technique. This article, and the accompanying MATLAB software, attempts to address these issues. The standard nonparametric randomization and permutation testing ideas are developed at an accessible level, using practical examples from functional neuroimaging, and the extensions for multiple comparisons described. Three worked examples from PET and fMRI are presented, with discussion, and comparisons with standard parametric approaches made where appropriate. Practical considerations are given throughout, and relevant statistical concepts are expounded in appendices. Hum. Brain Mapping 15:1–25, 2001. © 2001 Wiley‐Liss, Inc.
INTRODUCTION
The statistical analyses of functional mapping experiments usually proceeds at the voxel level, involving the formation and assessment of a statistic image: at each voxel a statistic indicating evidence of the experimental effect of interest, at that voxel, is computed, giving an image of statistics, a statistic image or Statistical Parametric Map (SPM). In the absence of a priori anatomical hypotheses, the entire statistic image must be assessed for significant experimental effects, using a method that accounts for the inherent multiplicity involved in testing at all voxels simultaneously.
Traditionally, this has been accomplished in a classical parametric statistical framework. The most commonly used methods are, or are similar to, those originally expounded by Friston et al. (1995b) and Worsley et al. (1992). In this framework, the data are assumed to be normally distributed, with mean parameterized by a general linear model (this flexible framework encompasses t‐tests, F‐tests, paired t‐tests, ANOVA, correlation, linear regression, multiple regression, and ANCOVA, among others). The estimated parameters of this model are contrasted to produce a test statistic at each voxel, which have a Student's t‐distribution under the null hypothesis. The resulting t‐statistic image is then assessed for statistical significance, using distributional results for continuous random fields to identify voxels or regions where there is significant evidence against the null hypothesis (Friston et al., 1994, 1996; Worsley et al., 1995; Worsley, 1996; Poline et al., 1997) [see Appendix B for a glossary of statistical terms].
Holmes et al. (1996) introduced a nonparametric alternative based on permutation test theory. This method is conceptually simple, relies only on minimal assumptions, deals with the multiple comparisons issue, and can be applied when the assumptions of a parametric approach are untenable. Further, in some circumstances, the permutation method outperforms parametric approaches. Arndt (1996), working independently, also discussed the advantages of similar approaches. Subsequently, Grabrowski et al. (1996) demonstrated empirically the potential power of the approach in comparison with other methods. Halber et al. (1997) discussed further by Holmes et al. (1998) also favour the permutation approach. Applications of permutation testing methods to single subject fMRI require modelling the temporal auto‐correlation in the time series. Bullmore et al. (1996) develop permutation based procedures for periodic fMRI activation designs using a simple ARMA model for temporal autocorrelations, though they eschew the problem of multiple comparisons. Locascio et al. (1997) describe an application to fMRI combining the general linear model (Friston et al., 1995b), ARMA modelling (Bullmore et al., 1996), and a multiple comparisons permutation procedure (Holmes et al., 1996). Liu et al. (1998) consider an alternative approach, permuting labels. Bullmore et al. (1999) apply nonparametric methods to compare groups of structural MR images. Applications of these techniques, however, have been relatively scarce (Andreasen et al., 1996; Noll et al., 1996; Locascio et al., 1997).
The aim of this study is to make the multiple comparisons nonparametric permutation approach of Holmes et al. (1996) more accessible, complement the earlier formal exposition with more practical considerations, and illustrate the potential power and flexibility of the approach through worked examples.
We begin with an introduction to nonparametric permutation testing, reviewing experimental design and hypothesis testing issues, and illustrating the theory by considering testing a functional neuroimaging dataset at a single voxel. The problem of searching the brain volume for significant activations is then considered, and the extension of the permutation method to the multiple comparisons problem of simultaneously testing at all voxels is described. With appropriate methodology in place, we conclude with three annotated examples illustrating the approach. Software implementing the approach is available as an extension of the MATLAB based SPM package (see Appendix A for details).
PERMUTATION TESTS
Permutation tests are one type of nonparametric test. They were proposed in the early twentieth century, but have only recently become popular with the availability of inexpensive, powerful computers to perform the computations involved.
The essential concept of a permutation test is relatively intuitive. For example, consider a simple single subject PET activation experiment, where a single subject is scanned repeatedly under “rest” and “activation” conditions. Considering the data at a particular voxel, if there is really no difference between the two conditions, then we would be fairly surprised if most of the “activation” observations were larger than the “rest” observations, and would be inclined to conclude that there was evidence of some activation at that voxel. Permutation tests simply provide a formal mechanism for quantifying this “surprise” in terms of probability, thereby leading to significance tests and p‐values.
If there is no experimental effect, then the labelling of observations by the corresponding experimental condition is arbitrary, because the same data would have arisen whatever the condition. These labels can be any relevant attribute: condition “tags,” such as “rest” or “active”; a covariate, such as task difficulty or response time; or a label, indicating group membership. Given the null hypothesis that the labellings are arbitrary, the significance of a statistic expressing the experimental effect can then be assessed by comparison with the distribution of values obtained when the labels are permuted.
The justification for exchanging the labels comes from either weak distributional assumptions, or by appeal to the randomization scheme used in designing the experiment. Tests justified by the initial randomization of conditions to experimental units (e.g., subjects or scans), are sometimes referred to as randomization tests, or re‐randomization tests. Whatever the theoretical justification, the mechanics of the tests are the same. Many authors refer to both generically as permutation tests, a policy we shall adopt unless a distinction is necessary.
In this section, we describe the theoretical underpinning for randomization and permutation tests. Beginning with simple univariate tests at a single voxel, we first present randomization tests, describing the key concepts at length, before turning to permutation tests. These two approaches lead to exactly the same test, which we illustrate with a simple worked example, before describing how the theory can be applied to assess an entire statistic image. For simplicity of exposition, the methodology is developed using the example of a simple single subject PET activation experiment. The approach, however, is not limited to activation experiments, nor to PET.
Randomization Test
First, we consider randomization tests, using a single subject activation experiment to illustrate the thinking: Suppose we are to conduct a simple single subject PET activation experiment, with the regional cerebral blood flow (rCBF) in “active” (A) condition scans to be compared to that in scans acquired under an appropriate “baseline” (B) condition. The fundamental concepts are of experimental randomization, the null hypothesis, exchangeability, and the randomization distribution.
Randomization
To avoid unexpected confounding effects, suppose we randomize the allocation of conditions to scans before conducting the experiment. Using an appropriate scheme, we label the scans as A or B according to the conditions under which they will be acquired, and hence specify the condition presentation order. This allocation of condition labels to scans is randomly chosen according to the randomization scheme, and any other possible labeling of this scheme was equally likely to have been chosen (see Appendix C for a discussion of the fundamentals of randomization).
Null hypothesis
In the randomization test, the null hypothesis is explicitly about the acquired data. For example, ℋ︁0: “Each scan would have been the same whatever the condition, A or B.” The hypothesis is that the experimental conditions did not affect the data differentially, such that had we run the experiment with a different condition presentation order, we would have observed exactly the same data. In this sense we regard the data as fixed, and the experimental design as random (in contrast to regarding the design as fixed, and the data as a realization of a random process). Under this null hypothesis, the labeling of the scans as A or B is arbitrary; because this labeling arose from the initial random allocation of conditions to scans, and any initial allocation would have given the same data. Thus, we may re‐randomize the labels on the data, effectively permuting the labels, subject to the restriction that each permutation could have arisen from the initial randomization scheme. The observed data is equally likely to have arisen from any of these permuted labelings.
Exchangeability
This leads to the notion of exchangeability. Consider the situation before the data is collected, but after the condition labels have been assigned to scans. Formally, a set of labels on the data (still to be collected) are exchangeable if the distribution of the statistic (still to be evaluated) is the same whatever the labeling (Good, 1994). For our activation example, we would use a statistic expressing the difference between the “active” and “baseline” scans. Thus under the null hypothesis of no difference between the A and B conditions, the labels are exchangeable, provided the permuted labeling could have arisen from the initial randomization scheme. The initial randomization scheme gives us the probabilistic justification for permuting the labels, the null hypothesis asserts that the data would have been the same.
With a randomization test, the randomization scheme prescribes the possible labeling, and the null hypothesis asserts that the labels are exchangeable within the constraints of this scheme. Thus we define an exchangeability block (EB) as a block of scans within which the labels are exchangeable, a definition that mirrors that of randomization blocks (see Appendix C).
Randomization distribution
Consider some statistic expressing the experimental effect of interest at a particular voxel. For the current example of a PET single subject activation, this could be the mean difference between the A and the B condition scans, a two‐sample t‐statistic, a t‐statistic from an ANCOVA, or any appropriate statistic. We are not restricted to the common statistics of classical parametric hypothesis whose null distributions are known under specific assumptions, because the appropriate distribution will be derived from the data.
The computation of the statistic depends on the labeling of the data. For example, with a two‐sample t‐statistic, the labels A and B specify the groupings. Thus, permuting the labels leads to an alternative value of the statistic.
Given exchangeability under the null hypothesis, the observed data is equally likely to have arisen from any possible labeling. Hence, the statistics associated with each of the possible labeling are also equally likely. Thus, we have the permutation (or randomization) distribution of our statistic: the permutation distribution is the sampling distribution of the statistic under the null hypothesis, given the data observed. Under the null hypothesis, the observed statistic is randomly chosen from the set of statistics corresponding to all possible relabelings. This gives us a way to formalize our “surprise” at an outcome: The probability of an outcome as or more extreme than the one observed, the P‐value, is the proportion of statistic values in the permutation distribution greater or equal to that observed. The actual labeling used in the experiment is one of the possible labelings, so if the observed statistic is the largest of the permutation distribution, the P‐value is 1/N, where N is the number of possible labelings of the initial randomization scheme. Because we are considering a test at a single voxel, these would be uncorrected P‐values in the language of multiple comparisons (Appendix E).
Randomization test summary
To summarize, the null hypothesis asserts that the scans would have been the same whatever the experimental condition, A or B. Under this null hypothesis the initial randomization scheme can be regarded as arbitrarily labeling scans as A or B, under which the experiment would have given the same data, and the labels are exchangeable. The statistic corresponding to any labeling from the initial randomization scheme is as likely as any other, because the permuted labeling could equally well have arisen in the initial randomization. The sampling distribution of the statistic (given the data) is the set of statistic values corresponding to all the possible relabeling of the initial randomization scheme, each value being equally likely.
Randomization test mechanics
Let N denote the number of relabel, and let, t i the statistic corresponding to labeling i. The set of t i for all possible relabeling constitutes the permutation distribution. Let T denote the value of the statistic for the actual labeling of the experiment. As usual in statistics, we use a capital letter for a random variable. T is random, because under ℋ︁0 it is chosen from the permutation distribution according to the initial randomization.
Under ℋ︁0, all of the t i are equally likely, so we determine the significance of our observed statistic T by counting the proportion of the permutation distribution as or more extreme than T, giving us our P‐value. We reject the null hypothesis at significance level α if the P‐value is less than α. Equivalently, T must be greater or equal to the 100(1 − α)th percentile of the permutation distribution. Thus, the critical value is the c + 1 largest member of the permutation distribution, where c = ⌊αN⌋, αN rounded down. If T exceeds this critical value then the test is significant at level α.
Permutation Test
In many situations it is impractical to randomly allocate experimental conditions, or perhaps we are presented with data from an experiment that was not randomized. For instance, we can not randomly assign subjects to be patients or normal controls. Or, for example, consider a single subject PET design where a covariate is measured for each scan, and we seek brain regions whose activity appears to be related to the covariate value.
In the absence of an explicit randomization of conditions to scans, we must make weak distributional assumptions to justify permuting the labels on the data. Typically, all that is required is that distributions have the same shape, or are symmetric. The actual permutations that are performed depend on the degree of exchangeability, which in turn depend on the actual assumptions made. With the randomization test, the experimenter designs the initial randomization scheme carefully to avoid confounds. The randomization scheme reflects an implicitly assumed degree of exchangeability (see Appendix C for randomization considerations). With the permutation test, the degree of exchangeability must be assumed post hoc. The reasoning that would have led to a particular randomization scheme can be usually be applied post‐hoc to an experiment, leading to a permutation test with the same degree of exchangeability. Given exchangeability, computation proceeds as for the randomization test.
Permutation test summary
Weak distributional assumptions are made, which embody the degree of exchangeability. The exact form of these assumptions depends on the experiment at hand, as illustrated in the following section and in the examples section.
For a simple single subject activation experiment, we might typically assume the following. For a particular voxel, “active” and “baseline” scans within a given block have a distribution with the same shape, though possibly different means. The null hypothesis asserts that the distributions for the “baseline” and “active” scans have the same mean, and hence are the same. Then the labeling of scans is arbitrary within the chosen blocks, which are thus the exchangeability blocks. Any permutation of the labels within the exchangeability blocks leads to an equally likely statistic.
The mechanics are then the same as with the randomization test. For each of the possible relabeling, compute the statistic of interest; for relabeling i, call this statistic t i. Under the null hypothesis each of the t i are equally likely, so the P‐value is the proportion of the t i greater than or equal to the statistic T corresponding to the correctly labeled data.
Single Voxel Example
To make these concepts concrete, consider assessing the evidence of an activation effect at a single voxel of a single subject PET activation experiment consisting of six scans, three in each of the “active” (A) and “baseline” (B) conditions. Suppose that the conditions were presented alternately, starting with rest, and that the observed data at this voxel are {90.48, 103.00, 87.83, 99.93, 96.06, 99.76} to two decimal places (these data are from a voxel in the primary visual cortex of the second subject in the visual activation experiment presented in the examples section).
As mentioned before, any statistic can be used, so for simplicity of illustration we use the “mean difference,” i.e., T = ⅓∑ (Aj − B j) where Bj and Aj indicate the value of the jth scan at the particular voxel of interest, under the baseline and active conditions respectively. Thus, we observe statistic T = 9.45.
Randomization test
Suppose that the condition presentation order was randomized, the actual ordering of BABABA having being randomly selected from all allocations of three A's and three B's to the six available scans, a simple balanced randomization within a single randomization block of size six. Combinatorial theory, or some counting, tells us that this randomization scheme has twenty (6C3 = 20) possible outcomes (see Appendix D for an introduction to combinatorics).
Then we can justify permuting the labels on the basis of this initial randomization. Under the null hypothesis ℋ︁0: “The scans would have been the same whatever the experimental condition, A or B”, the labels are exchangeable, and the statistics corresponding to the 20 possible labeling are equally likely. The 20 possible labeling are:
| 1. AAABBB | 8. ABBAAB | 15. BABABA |
| 2. AABABB | 9. ABBABA | 15. BABABA |
| 3. AABBAB | 10. ABBBAA | 17. BBAAAB |
| 4. AABBBA | 11. BAAABB | 18. BBAABA |
| 5. ABAABB | 12. BAABAB | 19. BBABAA |
| 6. ABABAB | 13. BAABBA | 20. BBBAAA |
| 7. ABABBA | 14. BABAAB |
Permutation test
Suppose there was no initial randomization of conditions to scans, and that the condition presentation order ABABAB was simply chosen. With no randomization, we must make weak distributional assumptions to justify permuting the labels, effectively prescribing the degree of exchangeability.
For this example, consider permuting the labels freely amongst the six scans. This corresponds to full exchangeability, a single exchangeability block of size six. For this to be tenable, we must either assume the absence of any temporal or similar confounds, or model their effect such that they do not affect the statistic under permutations of the labels. Consider the former. This gives 20 possible permutations of the labels, precisely those enumerated for the randomization justification above. Formally, we're assuming that the voxel values for the “baseline” and “active” scans come from distributions that are the same except for a possible difference in location, or mean. Our null hypothesis is that these distributions have the same mean, and therefore are the same.
Clearly the mean difference statistic under consideration in the current example is confounded with time for labeling such as AAABBB (no. 1) and BBBAAA (no. 20), where a time effect will result in a large mean difference between the and the labeled scans. The test remains valid, but possibly conservative. The actual condition presentation order of BABABA is relatively unconfounded with time, but the contribution of confounds to the statistics for alternative labeling such as no. 1 and no. 20 will potentially increase the number of statistics greater than the observed statistic.
Computation
Let t i be the mean difference for labeling i, as enumerated above. Computing for each of the 20 relabeling:
| t 1 = +4.82 | t 8 = +1.38 | t 15 = −9.45 |
| t 2 = −3.25 | t 9 = −1.10 | t 16 = −6.86 |
| t 3 = −0.67 | t 10 = +1.48 | t 17 = +3.15 |
| t 4 = −3.15 | t 11 = −1.48 | t 18 = +0.67 |
| t 5 = +6.86 | t 12 = +1.10 | t 19 = +3.25 |
| t 6 = +9.45 | t 13 = −1.38 | t 20 = −4.82. |
| t 7 = +6.97 | t 14 = −6.97 |
This is our permutation distribution for this analysis, summarized as a histogram in Figure 1. Each possible labeling was equally likely. Under the null hypothesis the statistics corresponding to these labeling are equally likely. The P‐value is the proportion of the permutation distribution greater than or equal to T. Here the actual labeling (no. 6 with t 6 = +9.45) gives the largest mean difference of all the possible labeling, so the P‐value is 1/20 = 0.05. For a test at given α level, we reject the null hypothesis if the P‐value is less than α, so we conclude that there is significant evidence against the null hypothesis of no activation at this voxel at level α = 0.05.
Figure 1.
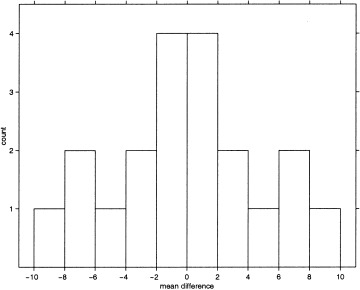
Histogram of permutation distribution for single voxel using a mean difference statistic. Note the symmetry of the histogram about the y‐axis. This occurs because for each possible labeling, the opposite labeling is also possible, and yields the same mean difference but in the opposite direction. This trick can be used in many cases to halve the computational burden.
Multiple Comparisons Permutation Tests
Thus far we have considered using a permutation test at a single voxel. For each voxel we can produce a P‐value, p k, for the null hypothesis ℋ︁, where the superscript k indexes the voxels. If we have an a priori anatomical hypothesis concerning the experimentally induced effect at a single voxel, then we can simply test at that voxel using an appropriate α level test. If we don't have such precise anatomical hypotheses, evidence for an experimental effect must be assessed at each and every voxel. We must take account of the multiplicity of testing. Clearly 5% of voxels are expected to have P‐values less than α = 0.05. This is the essence of the multiple comparisons problem. In the language of multiple comparisons (Appendix E), these P‐values are uncorrected P‐values. Type I errors must be controlled overall, such that the probability of falsely declaring any region as significant is less than the nominal test level α. Formally, we require a test procedure maintaining strong control over image‐wise Type I error, giving adjusted P‐values, P‐values corrected for multiple comparisons.
The construction of suitable multiple comparisons procedures for the problem of assessing statistic images from functional mapping experiments within parametric frameworks has occupied many authors (Friston et al., 1991; Worsley et al., 1992, 1995; Poline and Mazoyer, 1993; Roland et al., 1993; Forman et al., 1995; Friston et al., 1994, 1996; Worsley, 1994; Poline et al., 1997; Cao, 1999). In contrast to these parametric and simulation based methods, a nonparametric resampling based approach provides an intuitive and easily implemented solution (Westfall and Young, 1993). The key realization is that the reasoning presented above for permutation tests at a single voxel rely on relabeling entire images, so the arguments can be extended to image level inference by considering an appropriate maximal statistic. If, under the omnibus null hypothesis, the labels are exchangeable with respect to the voxel statistic under consideration, then the labels are exchangeable with respect to any statistic summarizing the voxel statistics, such as their maxima.
We consider two popular types of test, single threshold and suprathreshold cluster size tests, but note again the flexibility of these methods to consider any statistic.
Single threshold test
With a single threshold test, the statistic image is thresholded at a given critical threshold, and voxels with statistic values exceeding this threshold have their null hypotheses rejected. Rejection of the omnibus hypothesis (that all the voxel hypotheses are true) occurs if any voxel value exceeds the threshold, a situation clearly determined by the value of the maximum value of the statistic image over the volume of interest. Thus, consideration of the maximum voxel statistic deals with the multiple comparisons problem. For a valid omnibus test, the critical threshold is such that the probability that it is exceeded by the maximal statistic is less than α. Thus, we require the distribution of the maxima of the null statistic image. Approximate parametric derivations based on the theory of strictly stationary continuous random fields are given by Friston et al. (1991), Worsley (1994), and Worsley et al. (1992,1995).
The permutation approach can yield the distribution of the maximal statistic in a straightforward manner: Rather than compute the permutation distribution of the statistic at a particular voxel, we compute the permutation distribution of the maximal voxel statistic over the volume of interest. We reject the omnibus hypothesis at level α if the maximal statistic for the actual labeling of the experiment is in the top 100α% of the permutation distribution for the maximal statistic. The critical value is c + 1 largest member of the permutation distribution, where c = ⌊αN⌋, αN rounded down. Furthermore, we can reject the null hypothesis at any voxel with a statistic value exceeding this threshold. The critical value for the maximal statistic is the critical threshold for a single threshold test over the same volume of interest. This test can be shown to have strong control over experiment‐wise Type I error. A formal proof is given by Holmes et al. (1996).
The mechanics of the test are as follows. For each possible relabeling i = 1,…,N, note the maximal statistic t , the maximum of the voxel statistics for labeling i. This gives the permutation distribution for T max, the maximal statistic. The critical threshold is the c + 1 largest member of the permutation distribution for T max, where c = ⌊αN⌋, αN rounded down. Voxels with statistics exceeding this threshold exhibit evidence against the corresponding voxel hypotheses at level α. The corresponding corrected P‐value for each voxel is the proportion of the permutation distribution for the maximal statistic that is greater than or equal to voxel statistic.
Suprathreshold cluster tests
Suprathreshold cluster tests threshold the statistic image at a predetermined primary threshold, and assess the resulting pattern of suprathreshold activity. Suprathreshold cluster size tests assess the size of connected suprathreshold regions for significance, declaring regions greater than a critical size as activated. Thus, the distribution of the maximal suprathreshold cluster size (for the given primary threshold) is required. Simulation approaches have been presented by Poline and Mazoyer (1993) and Roland et al. (1993) for PET, and Forman et al. (1995) for fMRI. Friston et al. (1994) give a theoretical parametric derivation for Gaussian statistic images based on the theory of continuous Gaussian random fields, Cao (1999) gives results for χ2, t, and F fields.
Again, as noted by Holmes et al. (1996), a nonparametric permutation approach is simple to derive. Simply construct the permutation distribution of the maximal suprathreshold cluster size. For the statistic image corresponding to each possible relabeling, note the size of the largest suprathreshold cluster above the primary threshold. The critical suprathreshold cluster size for this primary threshold is the ⌊αN⌋ + 1 largest member of this permutation distribution. Corrected P‐values for each suprathreshold cluster in the observed statistic image are obtained by comparing their size to the permutation distribution.
In general, such suprathreshold cluster tests are more powerful for functional neuroimaging data then the single threshold approach (see Friston et al., 1995b for a fuller discussion). It must be remembered, however, that this additional power comes at the price of reduced localizing power. The null hypotheses for voxels within a significant cluster are not tested, so individual voxels cannot be declared significant. Only the omnibus null hypothesis for the cluster can be rejected. Further, the choice of primary threshold dictates the power of the test in detecting different types of deviation from the omnibus null hypothesis. With a low threshold, large suprathreshold clusters are to be expected, so intense focal “signals” will be missed. At higher thresholds these focal activations will be detected, but lower intensity diffuse “signals” may go undetected below the primary threshold.
Poline et al. (1997) addressed these issues within a parametric framework by considering the suprathreshold cluster size and height jointly. A nonparametric variation could be to consider the exceedance mass, the excess mass of the suprathreshold cluster, defined as the integral of the statistic image above the primary threshold within the suprathreshold cluster (Holmes, 1994; Bullmore et al., 1999). Calculation of the permutation distribution and P‐values proceeds exactly as before.
Considerations
Before turning to example applications of the nonparametric permutation tests described above, we note some relevant theoretical issues. The statistical literature (referenced below) should be consulted for additional theoretical discussion. For issues related to the current application to functional neuroimaging, see also Holmes (1994), Holmes et al. (1996), and Arndt et al. (1996).
Nonparametric statistics
First, it should be noted that these methods are neither new nor contentious. Originally expounded by Fisher (1935), Pitman (1937a–c), and later Edgington (1964, 1969a,b), these approaches are enjoying a renaissance as computing technology makes the requisite computations feasible for practical applications. Had R.A. Fisher and his peers had access to similar resources, it is possible that large areas of parametric statistics would have gone undeveloped! Modern texts on the subject include Good's Permutation Tests (Good, 1994), Edgington's Randomization Tests (Edgington, 1995), and Manly's Randomization, Bootstrap and Monte‐Carlo Methods in Biology (Manly, 1997). Recent interest in more general resampling methods, such as the bootstrap, has further contributed to the field. For a treatise on resampling based multiple comparisons procedures, see Westfall and Young (1993).
Many standard statistical tests are essentially permutation tests. The “classic” nonparametric tests, such as the Wilcoxon and Mann‐Whitney tests, are permutation tests with the data replaced by appropriate ranks, such that the critical values are only a function of sample size and can therefore be tabulated. Fisher's exact test (Fisher and Bennett, 1990), and tests of Spearman and Kendall correlations (Kendall and Gibbons, 1990), are all permutation/randomization based.
Assumptions
For a valid permutation test the only assumptions required are those to justify permuting the labels. Clearly the experimental design, model, statistic and permutations must also be appropriate for the question of interest. For a randomization test the probabilistic justification follows directly from the initial randomization of condition labels to scans. In the absence of an initial randomization, permutation of the labels can be justified via weak distributional assumptions. Thus, only minimal assumptions are required for a valid test.
In contrast to parametric approaches where the statistic must have a known null distributional form, the permutation approach is free to consider any statistic summarizing evidence for the effect of interest at each voxel. The consideration of the maximal statistic over the volume of interest then deals with the multiple comparisons problem.
There are, however, additional considerations when using the non‐parametric approach with a maximal statistic to account for multiple comparisons. For the single threshold test to be equally sensitive at all voxels, the (null) sampling distribution of the chosen statistic should be similar across voxels. For instance, the simple mean difference statistic used in the single voxel example could be considered as a voxel statistic, but areas where the mean difference is highly variable will dominate the permutation distribution for the maximal statistic. The test will still be valid, but will be less sensitive at those voxels with lower variability. So, although for an individual voxel a permutation test on group mean differences is equivalent to one using a two‐sample t‐statistic (Edgington, 1995), this not true in the multiple comparisons setting using a maximal statistic.
One approach to this problem is to consider multi‐step tests, which iteratively identify activated areas, cut them out, and continue assessing the remaining volume. These are described below, but are additionally computationally intensive. Preferable is to use a voxel statistic with approximately homogeneous null permutation distribution across the volume of interest, such as an appropriate t‐statistic. A t‐statistic is essentially a mean difference normalized by a variance estimate, effectively measuring the reliability of an effect. Thus, we consider the same voxel statistics for a non‐parametric approach as we would for a comparable parametric approach.
Pseudo t‐statistics
Nonetheless, we can still do a little better than a straight t‐statistic, particularly at low degrees of freedom. In essence, a t‐statistic is a change divided by the square root of the estimated variance of that change. When there are few degrees of freedom available for variance estimation, this variance is estimated poorly. Errors in estimation of the variance from voxel to voxel appear as high (spatial) frequency noise in images of the estimated variance or near‐zero variance estimates, which in either case cause noisy t‐statistic images. Given that PET and fMRI measure (indicators of) blood flow, physiological considerations would suggest that the variance be roughly constant over small localities. This suggests pooling the variance estimate at a voxel with those of its neighbors to give a locally pooled variance estimate as a better estimate of the actual variance. Because the model is of the same form at all voxels, the voxel variance estimates have the same degrees of freedom, and the locally pooled variance estimate is simply the average of the variance estimates in the neighborhood of the voxel in question. More generally, weighted locally pooled voxel variance estimates can be obtained by smoothing the raw variance image. The filter kernel then specifies the weights and neighborhood for the local pooling. The Pseudo t‐statistic images formed with smoothed variance estimators are smooth. In essence the noise (from the variance image) has been smoothed, but not the signal. A derivation of the parametric distribution of the pseudo t requires knowledge of the variance‐covariance of the voxel‐level variances, and has so far proved elusive. This precludes parametric analyses using a pseudo t‐statistic, but poses no problems for a nonparametric approach.
Number of relabelings and test size
A constraint on the permutation test is the number of possible relabelings. Because the observed labeling is always one of the N possible relabelings, the smallest P‐value attainable is 1/N. Thus, for a level α = 0.05 test to potentially reject the null hypothesis, there must be at least 20 possible labeling.
More generally, the permutation distribution is discrete, consisting of a finite set of possibilities corresponding to the N possible relabelings. Hence, any P‐values produced will be multiples of 1/N. Further, the 100(1 − α)th percentile of the permutation distribution, the critical threshold for a level α test, may lie between two values. Equivalently, α may not be a multiple of 1/N, such that a P‐value of exactly α cannot be attained. In these cases, an exact test with size exactly α is not possible. It is for this reason that the critical threshold is computed as the c + 1 largest member of the permutation distribution, where c = ⌊αN⌋, αN rounded down. The test can be described as almost exact, because the size is at most 1/N less than α.
Approximate tests
A large number of possible labelings is also problematic, due to the computations involved. In situations where it is not feasible to compute the statistic images for all the labelings, a subsample of labelings can be used (Dwass, 1957; Edgington, 1969a). The set of N possible relabelings is reduced to a more manageable N′ consisting of the true labeling and N′ − 1 randomly chosen from the set of N − 1 possible relabelings. The test then proceeds as before.
Such a test is sometimes known as an approximate permutation test, because the permutation distribution is approximated by a subsample, leading to approximate P‐values and critical thresholds (these tests are also known as Monte‐Carlo permutation tests or random permutation tests, reflecting the random selection of permutations to consider).
Despite the name, the resulting test remains exact. As might be expected from the previous section, however, using an approximate permutation distribution results in a test that is more conservative and less powerful than one using the full permutation distribution.
Fortunately, as few as 1,000 permutations can yield an effective approximate permutation test (Edgington, 1969a). For an approximate test with minimal loss of power in comparison to the full test (i.e., with high efficiency), however, one should consider rather more permutations (Jöel, 1986).
Power
Frequently, nonparametric approaches are less powerful than equivalent parametric approaches when the assumptions of the latter are true. The assumptions provide the parametric approach with additional information that the nonparametric approach must “discover.” The more labelings, the better the power of the nonparametric approach relative to the parametric approach. In a sense the method has more information from more labelings, and “discovers” the null distribution assumed in the parametric approach. If the assumptions required for a parametric analysis are not credible, however, a nonparametric approach provides the only valid method of analysis.
In the current context of assessing statistic images from functional neuroimaging experiments, the prevalent Statistical Parametric Mapping techniques require a number of assumptions and involve some approximations. Experience suggests that the permutation methods described here do at least as well as the parametric methods on real (PET) data (Arndt et al., 1996). For noisy statistic images, such as t‐statistic images with low degrees of freedom, the ability to consider pseudo t‐statistics constructed with locally pooled (smoothed) variance estimates affords the permutation approach additional power (Holmes, 1994; Holmes et al., 1996).
Multi‐step tests
The potential for confounds to affect the permutation distribution via the consideration of unsuitable relabelings has already been considered. Recall the above comments regarding the potential for the multiple comparison permutation tests to be differentially sensitive across the volume of interest if the null permutation distribution varies dramatically from voxel to voxel. In addition, there is also the prospect that departures from the null hypothesis influence the permutation distribution. Thus far, our nonparametric multiple comparisons permutation testing technique has consisted of a single‐step. The null sampling distribution (given the data), is the permutation distribution of the maximal statistic computed over all voxels in the volume of interest, potentially including voxels where the null hypothesis is not true. A large departure from the null hypothesis will give a large statistic, not only in the actual labeling of the experiment, but also in other labelings, particularly those close to the true labeling. This does not affect the overall validity of the test, but may make it more conservative for voxels other than that with the maximum observed statistic.
One possibility is to consider step‐down tests, where significant regions are iteratively identified, cut out, and the remaining volume reassessed. The resulting procedure still maintains strong control over family‐wise Type I error, our criteria for a test with localizing power, but will be more powerful (at voxels other that with maximal statistic). The iterative nature of the procedure, however, multiplies the computational burden of an already intensive procedure. Holmes et al. (1996) give a discussion and efficient algorithms, developed further in Holmes (1994), but find that the additional power gained was negligible for the cases studied.
Recall also the motivations for using a normalized voxel statistic, such as the t‐statistic. An inappropriately normalized voxel statistic will yield a test differentially sensitive across the image. In these situations the step‐down procedures may be more beneficial.
Further investigation of step‐down methods and sequential tests more generally are certainly warranted, but are unfortunately beyond the scope of this primer.
WORKED EXAMPLES
The following sections illustrate the application of the techniques described above to three common experimental designs: single subject PET “parametric,” multi‐subject PET activation, and multi‐subject fMRI activation. In each example we will illustrate the key steps in performing a permutation analysis:
-
1
Null Hypothesis Specify the null hypothesis.
-
2
Exchangeability Specify exchangeability of observations under the null hypothesis.
-
3
Statistic Specify the statistic of interest, usually broken down into specifying a voxel‐level statistic and a summary statistic.
-
4
Relabeling Determine all possible relabeling given the exchangeability scheme under the null hypothesis.
-
5
Permutation Distribution Calculate the value of the statistic for each relabeling, building the permutation distribution.
-
6
Significance Use the permutation distribution to determine significance of correct labeling and threshold for statistic image.
The first three items follow from the experimental design and must be specified by the user; the last three are computed by the software, though we will still address them here. When comparable parametric analyses are available (within SPM) we will compare the permutation results to the parametric results.
Single Subject PET: Parametric Design
The first study will illustrate how covariate analyses are implemented and how the suprathreshold cluster size statistic is used. This example also shows how randomization in the experimental design dictates the exchangeability of the observations.
Study description
The data come from a study of Silbersweig et al. (1994). The aim of the study was to validate a novel PET methodology for imaging transient, randomly occurring events, specifically events that were shorter than the duration of a scan. This work was the foundation for later work imaging hallucinations in schizophrenics (Silbersweig et al., 1995). We consider one subject from the study, who was scanned 12 times. During each scan the subject was presented with brief auditory stimuli. The proportion of each scan over which stimuli were delivered was chosen randomly, within three randomization blocks of size four. A score was computed for each scan, indicating the proportion of activity infused into the brain during stimulation. This scan activity score is our covariate of interest, which we shall refer to as DURATION. This is a type of parametric design, though in this context parametric refers not to a set of distributional assumptions, but rather an experimental design where an experimental parameter is varied continuously. This is in contradistinction to a factorial design where the experimental probe is varied over a small number of discrete levels.
We also have to consider the global cerebral blood flow (gCBF), which we account for here by including it as a nuisance covariate in our model. This gives a multiple regression, with the slope of the DURATION effect being of interest. Note that regressing out gCBF like this requires an assumption that there is no interaction between the score and global activity; examination of a scatter plot and a correlation coefficient of 0.09 confirmed this as a tenable assumption.
Null hypothesis
Because this is a randomized experiment, the test will be a randomization test, and the null hypothesis pertains directly to the data, and no weak distributional assumptions are required:
ℋ︁0: “The data would be the same whatever the DURATION.”
Exchangeability
Because this experiment was randomized, our choice of EB matches the randomization blocks of the experimental design, which was chosen with temporal effects in mind. The values of DURATION were grouped into 3 blocks of four, such that each block had the same mean and similar variability, and then randomized within block. Thus we have three EBs of size four.
Statistic
We decompose our statistic of interest into two statistics: one voxel‐level statistic that generates a statistic image, and a maximal statistic that summarizes that statistic image in a single number. An important consideration will be the degrees of freedom. The degrees of freedom is the number of observations minus the number of parameters estimated. We have one parameter for the grand mean, one parameter for the slope with DURATION, and one parameter for confounding covariate gCBF. Hence 12 observations less three parameters leaves just 9 degrees of freedom to estimate the error variance at each voxel.
Voxel‐level statistic
For a voxel‐level statistic we always use some type of t‐statistic. Although the nonparametric nature of the permutation tests allows the use of any statistic at a single voxel (e.g., the slope of rCBF with DURATION) we use the t because it is a standardized measure. It reflects the reliability of a change.
Analyses with fewer than about 20 degrees of freedom tend to have poor variance estimates, variance estimates that are themselves highly variable. In images of variances estimates this variability shows up as “sharpness,” or high frequency noise. This study has just 9 degrees of freedom and shows has the characteristic noisy variance image (Fig. 2). The problem is that this high frequency noise propagates into the t‐statistic image, when one would expect an image of evidence against ℋ︁0 to be smooth (as is the case for studies with greater degrees of freedom) because the raw images are smooth.
Figure 2.
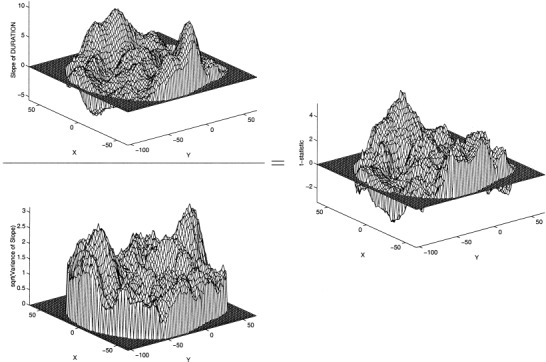
Mesh plots of parametric analysis, z = 0 mm. Upper left: slope estimate. Lower left: standard deviation of slope estimate. Right: t image for DURATION. Note how the standard deviation image is much less smooth than slope image, and how t image is correspondingly less smooth than slope image.
We can address this situation by smoothing the variance images (see section on Pseudo t‐statistics, above), replacing the variance estimate at each voxel with a weighted average of its neighbors. Here we use weights from an 8 mm FWHM spherical Gaussian smoothing kernel. The statistic image consisting of the ratio of the slope and the square root of the smoothed variance estimate is smoother than that computed with the raw variance. At the voxel level the resulting statistic does not have a Student's t‐distribution under the null hypothesis, so we refer to it as a pseudo t‐statistic.
Figure 3 shows the effect of variance smoothing. The smoothed variance image creates a smoother statistic image, the pseudo t‐statistic image. The key here is that the parametric t‐statistic introduces high spatial frequency noise via the poorly estimated standard deviation. By smoothing the variance image we are making the statistic image more like the “signal.”
Figure 3.
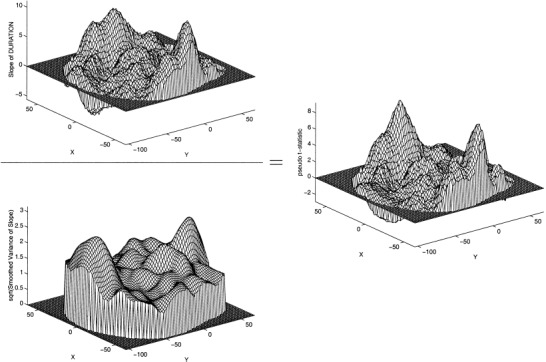
Mesh plots of permutation analysis, z= 0 mm. Upper left: Slope estimate. Lower left: square root of smoothed variance of slope estimate. Right: pseudo t image fot=r DURATION. Note that smoothness of pseudo t image is similar to that of the slope image (c.f. figure 2).
Summary statistic
We have a statistic image, but we need a single value that can summarize evidence against ℋ︁0 for each labeling. For the reasons given in the methods section, we use a maximum statistic, and in this example consider the maximum suprathreshold cluster size (max STCS).
Clusters are defined by connected suprathreshold voxels. Under the ℋ︁0, the statistic image should be random with no features or structure, hence large clusters are unusual and indicate the presence of an activation. A primary threshold is used to define the clusters. The selection of the primary threshold is crucial. If set too high there will be no clusters of any size; if set to low the clusters will be too large to be useful.
Relabeling enumeration
Each of the three previous sections correspond to a choice that a user of the permutation test has to make. Those choices and the data are sufficient for an algorithm to complete the permutation test. This and the next two sections describe the ensuing computational steps.
To create the labeling used in the experiment, the labels were divided into three blocks of four, and randomly ordered within blocks. Taking the division of the labels into the three blocks as given (it is not random), then we need to count how many ways the labels can be randomly permuted within blocks. There are 4! = 4 × 3 × 2 × 1 = 24 ways to permute four labels, and because each block is independently randomized, there are a total of 4!3 = 13,824 permutations of the labels (see Appendix D formulae).
Computations for 13,824 permutations would take a long time, so we consider an approximate test. The significance is calculated by comparing our observed statistic to the permutation distribution. With enough relabeling, a good approximation to the permutation distribution can be made; Here we use 1,000 relabelings. So, instead of 13,824 relabeling, we randomly select 999 relabeling to compute the statistic, giving 1,000 labeling including the actual labeling used in the experiment. The P‐values will be approximate, but the test remains exact.
Permutation distribution
For each of the 1,000 relabeling, the statistic image is computed and thresholded, and the maximal suprathreshold cluster size is recorded. For each relabeling this involves fitting the model at each voxel, smoothing the variance image, and creating the pseudo t‐statistic image. This is the most computationally intensive part of the analysis, but is not onerous on modern computing hardware. See discussion of examples for run times.
Selection of the primary threshold is not easy. For the results to be valid we need to pick the threshold before the analysis is performed. With a parametric voxel‐level statistic we could use its null distribution to specify a threshold by uncorrected P‐value (e.g., by using t table). Here we cannot take this approach because we are using a nonparametric voxel‐level statistic whose null distribution is not known a priori. Picking several thresholds is not valid, as this introduces a new multiple comparisons problem. We suggest gaining experience with similar datasets from post hoc analyses: apply different thresholds to get a feel for an appropriate range and then apply such a threshold to the data on hand. Using data from other subjects in this study we found 3.0 to be a reasonable primary threshold.
Significance threshold
We use the distribution of max STCS to assess the overall significance of the experiment and the significance of individual clusters: The significance is the proportion of labelings that had max STCS greater than or equal to maximum of the correct labeling. Put another way, if max STCS of the correct labeling is at or above the 95th percentile of the max STCS permutation distribution, the experiment is significant at α = 0.05. Also, any cluster in the observed image with size greater than the 95th percentile is the significant at α = 0.05. Because we have 1,000 labeling, 1,000 × 0.95 = 950, so the 950th largest max STCS will be our significance threshold.
Results
The permutation distribution of max STCS under ℋ︁0 is shown in Figure 4a. Most labelings have max STCS less than 250 voxels. The vertical dotted line indicates the 95th percentile. The top 5% are spread from about 500 to 3,000 voxels.
Figure 4.
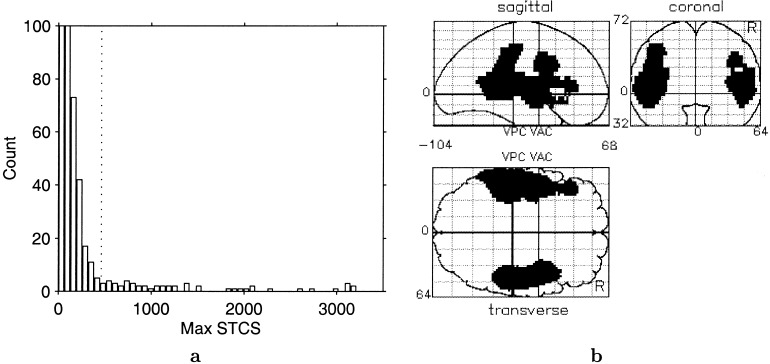
A: Distribution of maximum suprathreshold cluster size, threshold of 3. Dotted line shows 95th percentile. The count axis is truncated at 100 to show low‐count tail; first two bars have counts 579 and 221. B: Maximum intensity projection image of significantly large clusters.
For the correctly labeled data the max STCS is 3,101 voxels. This is unusually large in comparison to the permutation distribution. Only five labelings yield max equal to or larger than 3,101, so the P‐value for the experiment is 5/1,000 = 0.005. The 95th percentile is 462, so any suprathreshold clusters with size greater than 462 voxels can be declared significant at level 0.05, accounting for the multiple comparisons implicit in searching over the brain.
Figure 4b, is a maximum intensity projection (MIP) of the significant suprathreshold clusters. Only these two clusters are significant, that is, there are no other suprathreshold clusters larger than 462 voxels. These two clusters cover the bilateral auditory (primary and associative) and language cortices. They are 3,101 and 1,716 voxels in size, with P‐values of 0.005 and 0.015, respectively. Because the test concerns suprathreshold clusters it has no localizing power: Significantly large suprathreshold clusters contain voxels with a significant experimental effect, but the test does not identify them.
Discussion
The nonparametric analysis presented here uses maximum STCS for a pseudo t‐statistic image. Because the distribution of the pseudo t‐statistic is not known, the corresponding primary threshold for a parametric analysis using a standard t‐statistic cannot be computed. This precludes a straightforward comparison of this nonparametric analysis with a corresponding parametric analysis such as that of Friston et al. (1994).
Although the necessity to choose the primary threshold for suprathreshold cluster identification is a problem, the same is true for parametric approaches. The only additional difficulty occurs with pseudo t‐statistic images, when specification of primary thresholds in terms of upper tail probabilities from a Students' t‐distribution is impossible. Further, parametric suprathreshold cluster size methods (Friston et al., 1994; Poline et al., 1997) utilize asymptotic distributional results, and therefore require high primary thresholds. The nonparametric technique is free of this constraint, giving exact P‐values for any primary threshold (although very low thresholds are undesirable due to the large suprathreshold clusters expected and consequent poor localization of an effect).
Although only suprathreshold cluster size has been considered, any statistic summarizing a suprathreshold cluster could be considered. In particular an exceedance mass statistic could be employed.
Multi‐Subject PET: Activation
For the second example consider a multi‐subject, two condition activation experiment. We will use a standard t‐statistic with a single threshold test, enabling a direct comparison with the standard parametric random field approach.
Study description
Watson et al. (1993) localized the region of visual cortex sensitive to motion, area MT/V5, using high resolution 3D PET imaging of 12 subjects. These the data were analyzed by Holmes et al. (1996), using proportional scaling global flow normalization and a repeated measures pseudo t‐statistic. We consider the same data here, but use a standard repeated measures t‐statistic, allowing direct comparison of parametric and nonparametric approaches.
The visual stimulus consisted of randomly placed rectangles. During the baseline condition the pattern was stationary, whereas during the active condition the rectangles smoothly moved in independent directions. Before the experiment, the 12 subjects were randomly allocated to one of two scan condition presentation orders in a balanced randomization. Thus six subjects had scan conditions ABABABABABAB, the remaining six having ABABABABABAB, which we'll refer to as AB and BA orders, respectively.
Null hypothesis
In this example the labels of the scans as A and B are allocated by the initial randomization, so we have a randomization test, and the null hypothesis concerns the data directly:
ℋ︁0: For each subject, the experiment would have yielded the same data were the conditions reversed.
Note that it is not that the data itself is exchangeable, as the data is fixed. Rather, the labels are the observed random process and, under the null hypothesis, the distribution of any statistic is unaltered by permutations of the labels.
Exchangeability
Given the null hypothesis, exchangeability follows directly from the initial randomization scheme. The experiment was randomized at the subject level, with six AB and six BA labels randomly assigned to the 12 subjects. Correspondingly, the labels are exchangeable subject to the constraint that they could have arisen from the initial randomization scheme. Thus we consider all permutations of the labels that result in six subjects having scans labeled AB, and the remaining six AB. The initial randomization could have resulted in any six subjects having the AB condition presentation order (the remainder being BA), and under the null hypothesis the data would have been the same, hence exchangeability.
Statistic
Note that the permutations arrived at above permute across subjects, such that subject‐to‐subject differences in activation (expressed through the as yet unspecified statistic) will be represented in the permutation distribution. Because subject‐to‐subject differences in activation will be present in the permutation distribution, we must consider a voxel statistic that accounts for such inter‐subject variability, as well as the usual intra‐subject (residual) error variance. Thus we must use a random effects model incorporating a random subject by condition interaction term (many published analyses of multi‐subject and group comparison experiments have not accounted for variability in activation from subject‐to‐subject, and used fixed effects analyses).
Voxel‐level statistic
Fortunately, a random effects analysis can be easily effected by collapsing the data within subject and computing the statistic across subjects (Worsley et al., 1991; Holmes and Friston, 1999). In this case the result is a repeated measures t‐statistic after proportional scaling global flow normalization: Each scan is proportionally scaled to a common global mean of 50; each subjects data is collapsed into two average images, one for each condition; a paired t‐statistic is computed across the subjects' “rest”–“active” pairs of average images. By computing this paired t‐statistic on the collapsed data, both the inter‐subject and intra‐subject (error) components of variance are accounted for appropriately. Because there are 12 subjects there are 12 pairs of average condition images, and the t‐statistic has 11 degrees of freedom. With just 11 degrees of freedom we anticipate the same problems with noisy variance images as in the previous examples, but to make direct comparisons with a parametric approach, we will not consider variance smoothing and pseudo t‐statistics for this example.
Summary statistic
To consider a single threshold test over the entire brain, the appropriate summary statistic is the maximum t‐statistic.
Relabeling enumeration
This example is different from the previous one in that we permute across subjects instead of across replications of conditions. Here our EB is not in units of scans, but subjects. The EB size here is 12 subjects, because the six AB and six BA labels can be permuted freely amongst the 12 subjects. There are
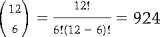 ways of choosing six of the 12 subjects to have the AB labeling. This is a sufficiently small number of permutations to consider a complete enumeration.
ways of choosing six of the 12 subjects to have the AB labeling. This is a sufficiently small number of permutations to consider a complete enumeration.
Note that although it might be tempting to consider permuting labels within subjects, particularly in the permutation setting when there is no initial randomization dictating the exchangeability, the bulk of the permutation distribution is specified by these between‐subject permutations. Any within‐subject permutations just flesh out this framework, yielding little practical improvement in the test at considerable computational cost.
Permutation distribution
For each of the 924 labelings we calculate the maximum repeated measures t‐statistic, resulting in the permutation distribution shown in Figure 5a. Note that for each possible labeling and t‐statistic image, the opposite labeling is also possible, and gives the negative of the t‐statistic image. Thus, it is only necessary to compute t‐statistic images for half of the labelings, and retain their maxima and minima. The permutation distribution is then that of the maxima for half the relabeling concatenated with the negative of the corresponding minima.
Figure 5.
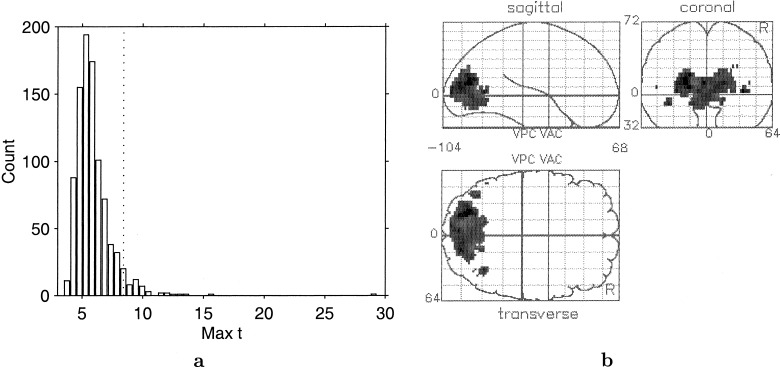
A: Permutation distribution of maximum repeated measures t‐statistic. Dotted line indicates the 5% level corrected threshold. B: Maximum intensity projection of t‐statistic image, thresholded at critical threshold for 5% level permutation test analysis of 8.401.
Significance threshold
As before, the 95th percentile of the maximum t distribution provides both a threshold for omnibus experimental significance and a voxel‐level significance threshold appropriate for the multiple comparisons problem. With 924 permutations, the 95th percentile is at 924 × 0.05 = 46.2, so the critical threshold is the 47th largest member of the permutation distribution. Any voxel with intensity greater than this threshold can be declared significant at the 0.05 level.
Results
Figure 5a shows the permutation distribution of the maximum repeated measures t‐statistic. Most maxima lie between about 4 and 9, though the distribution is skewed in the positive direction.
The outlier at 29.30 corresponds to the observed t‐statistic, computed with correctly labeled data. Because no other labelings are higher, the P‐value is 1/924 = 0. 0011. The 47th largest member of the permutation distribution is 8.40, the critical threshold (marked with a dotted vertical line on the permutation distribution). The t‐statistic image thresholded at this critical value is shown in Figure 5b. There is a primary region of 1,424 significant voxels covering the V1/V2 region, flanked by two secondary regions of 23 and 25 voxels corresponding to area V5, plus six other regions of 1 or 2 voxels.
For a t‐statistic image of 43,724 voxels of size 2 × 2 × 4 mm, with an estimated smoothness of 7.8 × 8.7 × 8.7 mm, the parametric theory gives a 5% level critical threshold of 11.07, substantially higher than the corresponding 4.61 of the nonparametric result. The thresholded image is shown in Figure 6b. The image is very similar to the nonparametric image (Fig. 5b), with the primary region having 617 voxels, with two secondary regions of 7 and 2 voxels. Another parametric result is the well‐known, but conservative Bonferroni correction; here it specifies a α‐0.05 threshold of 8.92 that yields a primary region of 1,212 voxels and 5 secondary regions with a total of 48 voxels. In Figure 6a we compare these three approaches by plotting the significance level vs. the threshold. The critical threshold based on the expected Euler characteristic (Worsley et al., 1995) for a t‐statistic image is shown as a dot‐dash line and the critical values for the permutation test is shown as a solid line. For a given test level (a horizontal line), the test with the smaller threshold has the greater power. At all thresholds in this plot the nonparametric threshold is below the random field threshold, though it closely tracks the Bonferroni threshold below the 0.05 level. Thus the random field theory appears to be quite conservative here.
Figure 6.
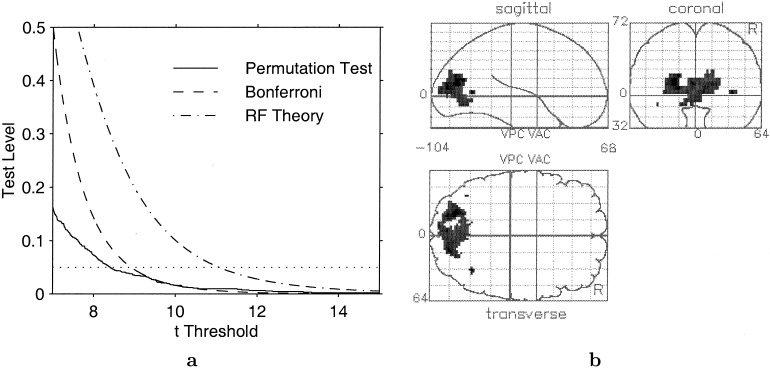
A: Test significance (α) levels plotted against critical thresholds, for nonparametric and parametric analyses. B: Maximum intensity projection of t image, thresholded at parametric 5% level critical threshold of 11.07.
Discussion
This example again demonstrates the role of the permutation test as a reference for evaluating other procedures, here the parametric analysis of Friston et al. (1995b). The t field results are conservative for low degrees of freedom and low smoothness (Holmes, 1994; Stoeckl et al., 2001); the striking difference between the nonparametric and random field thresholds makes this clear.
Figure 6a provides a very informative comparison between the two methods. For all typical test sizes (α ≤ 0.05), the nonparametric method specifies a lower threshold than the parametric method. For these data, this is exposing the conservativeness of the t field results. For lower thresholds the difference between the methods is even greater, though this is anticipated because the parametric results are based high threshold approximations.
Multi‐Subject fMRI: Activation
For this third and final example, consider a multi‐subject fMRI activation experiment. We will perform a permutation test so that we can make inference on a population, in contrast to a randomisation test. We will use a smoothed variance t‐statistic with a single threshold test and will make qualitative and quantitative comparisons with the corresponding parametric results.
Before discussing the details of this example, we note that fMRI data presents a special challenge for nonparametric methods. Because fMRI data exhibits temporal autocorrelation (Smith et al., 1999), an assumption of exchangeability of scans within subject is not tenable. To analyze a group of subjects for population inference, however, we need only assume exchangeability of subjects. Therefore, although intrasubject fMRI analyses are not straightforward with the permutation test, multisubject analyses are.
Study description
Marshuetz et al. (2000) studied order effects in working memory using fMRI. The data were analyzed using a random effects procedure (Holmes and Friston, 1999), as in the last example. For fMRI, this procedure amounts to a generalization of the repeated measures t‐statistic.
There were 12 subjects, each participating in eight fMRI acquisitions. There were two possible presentation orders for each block, and there was randomization across blocks and subjects. The TR was two seconds, with a total of 528 scans collected per condition. Of the study's three conditions we only consider two, item recognition and control. For item recognition, the subject was presented with five letters and, after a two second interval, presented with a probe letter. They were to respond “yes” if the probe letter was among the five letters and “no” if it was not. In the control condition they were presented with five X's and, two seconds later, presented with either a “y” or a “n”; they were to press “yes” for y and “no” for n.
Each subject's data was analyzed, creating a difference image between the item recognition and control effects. These images were analyzed with a one‐sample t‐test, yielding a random effects analysis that accounts for intersubject differences.
Null hypothesis
This study used randomization within and across subject and hence permits the use of a randomization test. Although randomization tests require no distributional assumptions, they only make a statement about the data at hand. To generalize to a population we need to use a permutation test.
The permutation test considers the data to be a random realization from some distribution, which is the same approach used in a parametric test (except that a particular parametric distribution, usually a normal, is specified). This is in distinction to the randomization test used in the last two examples, where the data is fixed and we use the randomness of the experimental design to perform the test. Although the machinery of the permutation and randomization tests are the same, the assumptions and scope of inference differ.
Each subject has an image expressing the item recognition effect, the difference of the item and control condition estimates. We make the weak distributional assumption that the values of the subject difference images at any given voxel (across subjects) are drawn from a symmetric distribution (the distribution may be different at different voxels, provided it is symmetric). The null hypothesis is that these distributions are centered on zero:
ℋ︁0: The symmetric distributions of the (voxel values of the) subjects' difference images have zero mean.
Exchangeability
The conventional assumption of independent subjects implies exchangeability, and hence a single EB consisting of all subjects.
We consider subject labels of “+1” and “−1,” indicating an unflipped or flipped sign of the data. Under the null hypothesis, we have data symmetric about zero, and hence for a particular subject the sign of the observed data can be flipped without altering its distribution. With exchangeable subjects, we can flip the signs of any or all subjects' data and the joint distribution of all of the data will remain unchanged.
Statistic
In this example we use a single threshold test.
Voxel‐level statistic
As noted above, this analysis amounts to a one‐sample t‐test on the first level images, testing for a zero‐mean effect across subjects. Because we will have only 11 degrees of freedom we will use a pseudo t‐test. We used a variance smoothing of 4 mm FWHM, comparable to the original within subject smoothing. In our experience, the use of any variance smoothing is more important than the particular magnitude (FWHM) of the smoothing.
Summary statistic
Again we are interested in searching over the whole brain for significant changes, hence we use the maximum pseudo t.
Relabeling enumeration
Based on our exchangeability under the null hypothesis, we can flip the sign on some or all of our subjects' data. There are 212 = 4,096 possible ways of assigning either “+1” or “−1” to each subject.
Permutation distribution
For each of the 4,096 relabelings, we computed a pseudo t‐statistic image and noted the maximum over the image, yielding the distribution in Figure 7a. As in the last example, we have a symmetry in these labels; we need only compute 2,048 statistic images and save both the maxima and minima.
Figure 7.
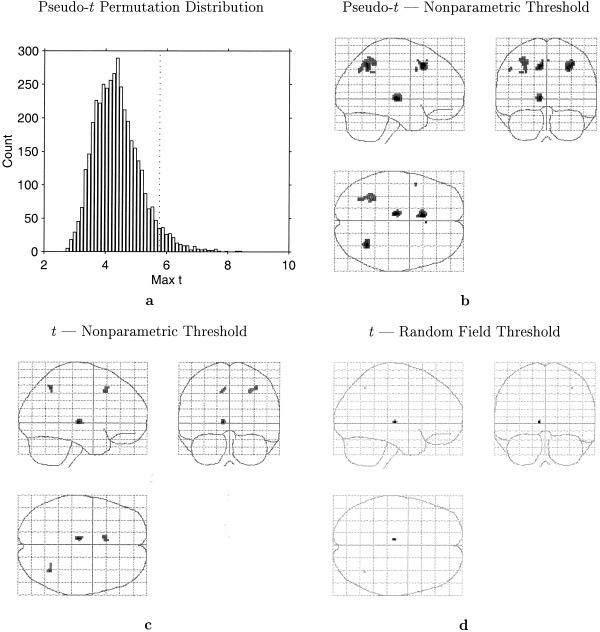
A: Permutation distribution of maximum repeated measures t statistic. Dotted line indicates the 5% level corrected threshold. B: Maximum intensity projection of pseudo t statistic image threshold at 5% level, as determined by permutation distribution. C: Maximum intensity projection of t statistic image threshold at 5% level as determined by permutation distribution. D: Maximum intensity projection of t statistic image threshold at 5% level as determined by random field theory.
Significance threshold
With 4,096 permutations the 95th percentile is 4,096 × 0.05 = 452.3, and hence the 453rd largest maxima defines the 0.05 level corrected significance threshold.
Results
The permutation distribution of the maxim pseudo‐t‐statistics under ℋ︁0 is shown in Figure 7a. It is centered around 4.5 and is slightly skewed positive; all maxima are found between about 3 and 8.
The correctly labeled data yielded the largest maximum, 8.471. Hence the overall significance of the experiment is 1/4,096 = 0. 0002. The dotted line indicates the 0.05 corrected threshold, 5.763. Figure 7b shows the thresholded MIP of significant voxels. There are 312 voxels in 8 distinct regions; in particular there is a pair of bilateral posterior parietal regions, a left thalamus region and an anterior cingulate region; these are typical of working memory studies (Marshuetz et al., 2000).
It is informative to compare this result to the traditional t‐statistic, using both a nonparametric and parametric approach to obtain corrected thresholds. We reran this nonparametric analysis using no variance smoothing. The resulting thresholded data is shown in Figure 7c; there are only 58 voxels in 3 regions that exceeded the corrected threshold of 7.667. Using standard parametric random field methods produced the result in Figure 7d. For 110,776 voxels of size 2 × 2 × 2 mm, with an estimated smoothness of 5.1 × 5.8 × 6.9 mm, the parametric theory finds a threshold of 9.870; there are only 5 voxels in 3 regions above this threshold. Note that only the pseudo‐t‐statistic detects the bilateral parietal regions. Table I summaries the three analyses along with the Bonferroni result.
Table I.
Comparison of four inference methods for the item recognition fMRI data*
| Statistic | Inference method | Minimum corrected P value | Number of significant voxels | Corrected threshold | |
|---|---|---|---|---|---|
| t | Pseudo‐t | ||||
| t | Random field | 0. 0062646 | 5 | 9.870 | |
| t | Bonferroni | 0. 0025082 | 5 | 9.802 | |
| t | Permutation | 0. 0002441 | 58 | 7.667 | |
| Pseudo‐t | Permutation | 0. 0002441 | 312 | 5.763 | |
The minimum corrected P‐value and number of significant voxels give an overall measure of sensitivity; corrected thresholds can only be compared within statistic type. For this data, the Bonferroni and random field results are very similar, and the nonparametric methods are more powerful. The nonparametric t method detects 10 times as many voxels as the parametric method, and the nonparametric pseudo‐t detects 60 times as many.
Discussion
In this example we have demonstrated the utility of the nonparametric method for intersubject fMRI analyses. Based solely on independence of the subjects and symmetric distribution of difference images under the null hypothesis, we can create a permutation test that yields inferences on a population.
Intersubject fMRI studies typically have few subjects, many fewer than 20 subjects. By using the smoothed variance t‐statistic we have gained sensitivity relative to the standard t‐statistic. Even with the standard t‐statistic, the nonparametric test proved more powerful, detecting 5 times as many voxels as active. Although the smoothed variance t can increase sensitivity, it does not overcome any limitations of the face validity of an analysis based on only 12 subjects.
We note that this relative ranking of sensitivity (nonparametric pseudo‐t, nonparametric t, parametric t) is consistent with the other second level datasets we have analyzed. We believe this is due to a conservativeness of the random field method under low degrees of freedom, not just to low smoothness.
Discussion of Examples
These examples have demonstrated the nonparametric permutation test for PET and fMRI with a variety of experimental designs and analyses. We have addressed each of the steps in sufficient detail to follow the algorithmic steps that the software performs. We have shown how that the ability to utilize smoothed variances via a pseudo t‐statistic can offer an approach with increased power over a corresponding standard t‐statistic image. Using standard t‐statistics, we have seen how the permutation test can be used as a reference against which parametric random field results can be validated.
Note, however, that the comparison between parametric and nonparametric results must be made very carefully. Comparable models and statistics must be used, and multiple comparisons procedures with the same degree of control over image‐wise Type I error used. Further, because the permutation distributions are derived from the data, critical thresholds are specific to the data set under consideration. Although the examples presented above are compelling, it should be remembered that these are only a few specific examples and further experience with many data sets is required before generalizations can be made. The points noted for these specific examples, however, are indicative of our experience with these methods thus far.
Finally, although we have noted that the nonparametric method has greater computational demands than parametric methods, they are reasonable on modern hardware. The PET examples took 35 min and 20 min, respectively, on a 176 MHz Sparc Ultra 1. The fMRI example took 2 hr on a 440 MHz Sparc Ultra 10. The fMRI data took longer due to more permutations (2,048 vs. 500) and larger images.
CONCLUSIONS
In this paper, the theory and practicalities of multiple comparisons non‐parametric randomization and permutation tests for functional neuroimaging experiments have been presented, and illustrated with worked examples.
As has been demonstrated, the permutation approach offers various advantages. The methodology is intuitive and accessible. By consideration of suitable maximal summary statistics, the multiple comparisons problem can easily be accounted for; only minimal assumptions are required for valid inference, and the resulting tests are almost exact, with size at most 1/N less than the nominal test level α, where N is the number of relabelings.
The nonparametric permutation approaches described give results similar to those obtained from a comparable Statistical Parametric Mapping approach using a general linear model with multiple comparisons corrections derived from random field theory. In this respect these nonparametric techniques can be used to verify the validity of less computationally expensive parametric approaches (but not prove them invalid). When the assumptions required for a parametric approach are not met, the non‐parametric approach described provides a viable alternative analysis method.
In addition, the approach is flexible. Choice of voxel and summary statistic are not limited to those whose null distributions can be derived from parametric assumptions. This is particularly advantageous at low degrees of freedom, when noisy variance images lead to noisy statistic images and multiple comparisons procedures based on the theory of continuous random fields are conservative. By assuming a smooth variance structure, and using a pseudo t‐statistic computed with smoothed variance image as voxel statistic, the permutation approach gains considerable power.
Therefore we propose that the nonparametric permutation approach is preferable for experimental designs implying low degrees of freedom, including small sample size problems, such as single subject PET/SPECT, but also PET/SPECT and fMRI multi‐subject and between group analyses involving small numbers of subjects, where analysis must be conducted at the subject level to account for inter‐subject variability. It is our hope that this paper, and the accompanying software, will encourage appropriate application of these non‐parametric techniques.
Acknowledgements
We thank the authors of the three example data sets analyzed for permission to use their data; to Karl Friston and Ian Ford for statistical advice; to the two anonymous reviewers for their insightful comments, and to Mark Mintun, David Townsend, and Terry Morris for practical support and encouragement in this collaborative work. Andrew Holmes was funded by the Wellcome Trust for part of this work. Thomas Nichols was supported by the Center for the Neural Basis of Cognition.
APPENDIX A.
STATISTICAL NONPARAMETRIC MAPPING
Statistical Parametric Mapping refers to the conceptual and theoretical framework that combines the general linear model and Gaussian random field (GRF) theory to construct, and make inferences about statistic maps respectively. The approach depends on various image processing techniques for coregistering, smoothing and spatially normalizing neuroimages. As the name suggests, the statistical technology employed is parametric. The data are modeled using the general linear model, and the resulting statistic images are assessed using approximate results from the theory of continuous random fields. The methodologies are presented in the peer reviewed literature (Friston et al., 1995a,b; Worsley and Friston, 1995). A complete and more accessible exposition, together with examples, is presented in Human Brain Function (Frackowiak et al., 1997).
The Statistical Parametric Mapping approach is implemented in a software package known as SPM, which runs within the commercial MATLAB (http://www.mathworks.com/) numerical computing environment. The SPM software is freely available to the functional neuroimaging community, and may be retrieved from the SPM web site at http://www.fil. ion.ucl.ac.uk/spm.
The nonparametric permutation methods described in this study build on the Statistical Parametric Mapping framework and may be referred to as Statistical nonParametric Mapping. The computer programs used in this article are available as a “toolbox” built on top of SPM, hence SnPM. SnPM is available from the SPM website, at http://www.fil.ion.ucl.ac.uk/spm/snpm, where additional resources complementing this article can be found. These include an example data set and a step‐by‐step description of the analysis of these data using the SnPM toolbox.
APPENDIX B.
STATISTICAL HYPOTHESIS TESTING
Statistical hypothesis testing formulates the experimental question in terms of a null hypothesis, written ℋ︁0, hypothesizing that there is no experimental effect. The test rejects this null hypothesis in favor of an alternative hypothesis, written ℋ︁A, if it is unlikely that the observed data could have arisen under the null hypothesis, where the data are summarized by a statistic, and appropriate assumptions are made. Thus, the probability of falsely rejecting a true null hypothesis, a Type I error, is controlled. The test level, usually denoted by α, is the accepted “risk” of the test, the probability of committing a Type I error. Formally, we compute the P‐value as the probability that the statistic would exceed (or equal) that observed under the null hypothesis, given the assumptions. If the P‐value is smaller than the chosen test level α, then the null hypothesis is rejected. Rigorously we say “there is evidence against the null hypothesis at level α,” or “at the 100α% level.” Hence, the P‐value is the smallest test level α at which the null hypothesis would be rejected. The value of the statistic with P‐value equal to α is the critical value because more extreme values lead to rejection of the null hypothesis. Commonly α is chosen to be 0.05, corresponding to an expected false positive rate of one in every 20 applications of the test (5%).
Frequently the computation of the P‐value involves approximations, either direct mathematical approximations, or indirectly via reliance on results or assumptions that are only approximately true. The size of a test is the actual probability of a Type I error. A test is valid if the size is at most the specified test level α, that is the true probability of a Type I error is less than α. If approximate P‐values are under‐estimated (overestimating the significance), the size exceeds α, and the test is invalid. If the approximate P‐values are over‐estimated (underestimating the significance), then the test is said to be conservative, because the size of the test is less than the allowed α. A test with size equal to the specified level of α is said to be exact.
A Type II error is a false‐negative, the error of not rejecting a false null‐hypothesis. The probability of a Type II error, obviously dependent on the degree of departure from the null hypothesis, is often denoted by β. The power of a test, for a given departure from ℋ︁0, is given by (1 − β). Frequently power is discussed generically. A conservative test is usually, but not necessarily, a less powerful test than an exact test. In functional neuroimaging the emphasis has been on avoiding false positives at all costs, concentrating on Type I errors, frequently at the expense of power. This has led to testing procedures with a high probability of Type II error, for even a fairly robust departure from the null hypothesis. In this study, we shall consider the traditional testing framework, focusing on Type I error.
Lastly, hypothesis tests may be two‐sided, in which the alternative hypothesis ℋ︁A specifies any departure from the null; or one‐sided, in which the alternative hypothesis is directional. For instance a two‐sided two sample t‐test would assume normality and equal variance of the two groups, and assess the null hypothesis ℋ︁0: “equal group means” against the alternative ℋ︁A: “group means differ.” A one‐sided test would have alternative ℋ︁A: “Group 1 mean is greater than Group 2 mean,” or vice‐versa.
APPENDIX C.
EXPERIMENTAL DESIGN AND RANDOMIZATION
Randomization is a crucial aspect of experimental design. The basic idea is to randomly allocate subjects to treatments, or in our case conditions to scans, so that any unforeseen confounding factors are randomly distributed across all treatments/conditions, and are thereby accounted for as error. In the absence of random allocation, unforeseen factors may bias the results.
For instance, consider the example of a simple PET activation experiment, where a single subject is to be scanned under two conditions, A and B, with six replications of each condition. We must choose a condition presentation order for the 12 scans. Clearly BBBBBBAAAAAA is unsatisfactory, because comparing the A's with the B's will reveal changes over time as well as those due to condition. The condition effect is confounded with time. Even the relatively innocuous and widely employed ABABABABABAB paradigm, however, may be confounded with time. Indeed, principal component analysis of datasets often indicates that time is a serious confound, whose effect may not be easy to model, and temporal effects are only one example of possible confounds. Thus, some form of randomization is almost always required.
The simplest scheme would be to decide the condition for each scan on the toss of a fair coin. This unrestricted randomization, however, may not result in six scans for each condition, and is therefore unsatisfactory. We need a restricted randomization scheme that allocates equal A's and B's across the 12 scans. A simple balanced randomization would allocate the six A's and six B's freely amongst the 12 scans. This is obviously unsatisfactory, because BBBBBBAAAAAA & ABABABABABAB are possible outcomes, unacceptable due to temporal confounding. A block randomization is required.
In a block randomization scheme, the scans are split up into blocks, usually contiguous in time, and usually of the same size. Conditions are then randomly allocated to scans within these randomization blocks, using a simple restricted randomization scheme. For instance, consider our simple PET activation experiment example. The 12 scans can be split up into equally sized randomization blocks in various ways: two blocks of six scans; three blocks of four scans; or six blocks of two scans. The size of the randomization blocks in each case is a multiple of the number of conditions (two), and a divisor of the number of scans (12). Within randomization blocks, we assign equal numbers of A's and B's at random. So, a randomization block of size 2 could be allocated in two ways as AB or BA; blocks of size four in six ways as AABB, ABAB, ABBA, BAAB, BABA, or BBAA; and for randomization blocks of size six there are 20 possible allocations. The implicit assumption is that the randomization blocks are sufficiently short that confounding effects within blocks can be ignored. That is, the different allocations within each block are all assumed to be free from confound biases, such that the distribution of a statistic comparing the A's and B's will be unaffected by the within‐block allocation. This parallels the properties of the exchangeability blocks.
APPENDIX D.
COMBINATORICS
Combinatorics is the study of permutations and combinations, usually expressed generically in terms of “drawing colored balls from urns.” Fortunately we only need a few results:
There are n! ways of ordering n distinguishable objects. Read “n‐factorial,” n! is the product of the first n natural numbers: n! = 1 × 2 × … × (n − 1( × n Example: In the current context of functional neuroimaging, a parametric design provides an example. Suppose we have 12 scans on a single individual, each with a unique covariate. There are 12! ways of permuting the 12 covariate values amongst the 12 scans.
- There are nCr ways of drawing r objects (without replacement) from a pool of n distinguishable objects, where the order of selection is unimportant. Read “n‐choose‐r,” these are the Binomial coefficients. Also written
), nCr is a fraction of factorials: n C r =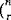
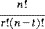 Example: Consider a balanced randomization of conditions A and B to scans within a randomization block of size four. Once we choose two of the four scans to be condition A, the remainder must be B, so there are 4C2 = 6 ways of ordering two A's and two B's.
Example: Consider a balanced randomization of conditions A and B to scans within a randomization block of size four. Once we choose two of the four scans to be condition A, the remainder must be B, so there are 4C2 = 6 ways of ordering two A's and two B's. There are n r ways of drawing r objects from a pool of n distinguishable objects, when the order is important and each drawn object is replaced before the next selection. Example: Suppose we have a simple single subject activation experiment with two conditions, A and B, to be randomly allocated to 12 scans using a balanced randomization within blocks of size four. From above, we have that there are 4C2 = 6 possibilities within each randomization block. Because there are three such blocks, the total number of possible labeling for this randomization scheme is 63 = 216.
APPENDIX E.
MULTIPLE COMPARISONS
For each voxel k in the volume of interest W, k ∈ W, we have a voxel level null hypothesis ℋ︁, and a test at each voxel. In the language of multiple comparisons (Hochberg and Tamhane, 1987), we have a family of tests, one for each voxel, a “collection of tests for which it is meaningful to take into account some combined measure of errors.” The probability of falsely rejecting any voxel hypothesis is formally known as the family‐wise or experiment‐wise Type I error rate. For the current simultaneous testing problem of assessing statistic images, experiment‐wise error is better described as image‐wise error.
If the voxel hypotheses are true for all voxels in the volume of interest W, then we say the omnibus hypothesis ℋ︁ is true. The omnibus hypothesis is the intersection of the voxel hypotheses, a hypothesis of “no experimental effect anywhere” within the volume of interest. Rejecting any voxel hypothesis implies rejecting the omnibus hypothesis. Rejecting the omnibus hypothesis implies rejecting some (possibly unspecified) voxel hypotheses. Image‐wise error is then the error of falsely rejecting the omnibus hypothesis.
Clearly a valid test must control the probability of image‐wise error. Formally, a test procedure has weak control over experiment‐wise Type I error if the probability of falsely rejecting the omnibus hypothesis is less than the nominal level α:
Such a test is known as an omnibus test. A significant test result indicates evidence against the omnibus null hypothesis, but because the Type I error for individual voxels is not controlled the test has no localizing power to identify specific voxels. We can only declare “some experimental effect, somewhere.”
For a test with localizing power we must consider a further possibility for Type I error, namely that of attributing a real departure from the omnibus null hypothesis to the wrong voxels. If we are to reject individual voxel hypotheses, then in addition to controlling for image‐wise Type I error, we must also control the probability of Type I error at the voxel level. This control must be maintained for any given voxel even if the null hypothesis is not true for voxels elsewhere. A test procedure has strong control over experiment‐wise Type I error if the tests are valid for any set of voxels where the null hypothesis is true, regardless of the veracity of the null hypothesis elsewhere. Formally, for any subset U of voxels in the volume of interest, U ⊆ W, where the corresponding omnibus hypothesis ℋ︁ is true, strong control over experiment‐wise Type I error is maintained if and only if
In other words, the validity of a test in one region is unaffected by the veracity of the null hypothesis elsewhere. Such a test has localizing power: A departure from the null hypothesis in one region will not cause the test to pick out voxels in another region where the null hypothesis is true. Clearly strong control implies weak control.
A multiple comparisons procedure with strong control over experiment‐wise Type I error can yield corrected or adjusted P‐values. Considering a test at a single voxel, the P‐value is the smallest test level α at which the null hypothesis is rejected. In the context of the multiple comparisons problem of assessing the statistic image, these are uncorrected P‐values, because they do not take into account the multiplicity of testing. By analogy, a corrected P‐value for the null hypothesis at a voxel is the smallest test level α at which an appropriate multiple comparisons procedure with strong control over experiment‐wise Type I error rejects the null hypothesis at that voxel. Thus, corrected P‐values, denoted ˜, account for the multiplicity of testing.
REFERENCES
- Andreasen NC, O'Leary DS, Cizadlo T, Arndt S, Rezai K, Ponto LL, Watkins GL, Hichwa RD (1996): Schizophrenia and cognitive dysmetria: a positron‐emission tomography study of dysfunctional prefrontal‐thalamic‐cerebellar circuitry. Proc Natl Acad Sci USA 93: 9985–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt S, Cizadlo T, Andreasen NC, Heckel D, Gold S, O'Leary DS (1996): Tests for comparing images based on randomization and permutation methods. J Cereb Blood Flow Metab 16: 1271–1279. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Brammer M, Williams SCR, Rabe‐Hesketh S, Janot N, David A, Mellers J, Howard R, Sham P (1996): Statistical methods of estimation and inference for functional MR image analysis. Magn Reson Med 35: 261–277. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Suckling J, Overmeyer S, Rabe‐Hesketh S, Taylor E, Brammer MJ (1999): Global, voxel, and cluster tests, by theory and permutation, for difference between two groups of structural MR images of the brain. IEEE Trans Med Imaging 18: 32–42. [DOI] [PubMed] [Google Scholar]
- Cao J (1999): The size of the connected components of the excursion sets of χ2, t, and F fields. Adv Appl Probability 51: 579–595. [Google Scholar]
- Dwass M (1957): Modified randomization tests for nonparametric hypotheses. Ann Math Stat 28: 181–187. [Google Scholar]
- Edgington ES (1964): Randomization tests. J Psychol 57: 445–449. [DOI] [PubMed] [Google Scholar]
- Edgington ES (1969a): Approximate randomization tests. J Psychol 72: 143–149. [Google Scholar]
- Edgington ES (1969b): Statistical inference: the distribution free approach. New York: McGraw‐Hill. [Google Scholar]
- Edgington ES (1995): Randomization tests, 3rd ed. New York: Marcel Dekker. [Google Scholar]
- Fisher RA (1990): Statistical methods, experimental design, and scientific inference In: Bennett JH. ??: Oxford University Press. [Google Scholar]
- Fisher RA (1935): The design of experiments. Edinburgh: Oliver Boyd. [Google Scholar]
- Forman SD, Cohen JD, Fitzgerald M, Eddy WF, Mintun MA, Noll DC (1995): Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): use of a cluster‐size threshold. Magn Reson Med 33: 636–647. [DOI] [PubMed] [Google Scholar]
- Frackowiak RSJ, Friston KJ, Frith CD, Dolan RJ, Mazziotta JC (1997): Human brain function. San Diego: Academic Press. [Google Scholar]
- Friston KJ, Frith CD, Liddle PF, Frackowiak RSJ (1991): Comparing functional (PET) images: the assessment of significant change. J Cereb Blood Flow Metab 11: 690–699. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak RSJ, Mazziotta JC, Evans AC (1994): Assessing the significance of focal activations using their spatial extent. Hum Brain Mapp 1: 214–220. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Poline JB, Grasby PJ, Williams SCR, Frackowiak RSJ, Turner R (1995a): Analysis of fMRI time series revisited. Neuroimage 2: 45–53. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline J‐B, Frackowiak RSJ (1995b): Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp 2: 189–210. [Google Scholar]
- Friston KJ, Holmes AP, Poline J‐B, Price CJ, Frith CD (1996): Detecting activations in PET and fMRI: levels of inference and power. Neuroimage 4: 223‐235. [DOI] [PubMed] [Google Scholar]
- Good P (1994): Permutation tests. A practical guide to resampling methods for testing hypotheses. ??: Springer‐Verlag. [Google Scholar]
- Grabowski TJ, Frank RJ, Brown CK, Damasio H, Boles Ponto LL, Watkins GL, Hichwa RD (1996): Reliability of PET activation across statistical methods, subject groups, and sample sizes. Hum Brain Mapp 4: 23–46. [DOI] [PubMed] [Google Scholar]
- Halber M, Herholz K, Wienhard K, Pawlik G, Heiss W‐D (1997): Performance of randomization test for single‐subject 15‐O‐water PET activation studies. J Cereb Blood Flow Metab 17: 1033–1039. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Tamhane AC (1987): Multiple comparison procedures. New York: Wiley. [Google Scholar]
- Holmes AP, Watson JDG, Nichols TE (1998): Holmes and Watson on ‘Sherlock’. J Cereb Blood Flow Metab 18: S697. [DOI] [PubMed] [Google Scholar]
- Holmes AP (1994): Statistical issues in functional brain mapping, PhD thesis. University of Glasgow. http://www.fil.ion.ucl.ac.uk/spm/papers/APH_thesis
- Holmes AP, Friston KJ (1999): Generalizability, random effects, and population inference. Proceedings of the Fourth International Conference on Functional Mapping of the Human Brain, June 7–12, 1998, Montreal, Canada. Neuroimage 7: S754. [Google Scholar]
- Holmes AP, Blair RC, Watson JDG, Ford I (1996): Nonparametric analysis of statistic images from functional mapping experiments. J Cereb Blood Flow Metab 16: 7–22. [DOI] [PubMed] [Google Scholar]
- Jöckel K‐H (1986): Finite sample properties and asymptotic efficiency of Monte‐Carlo tests. Ann Stat 14: 336–347. [Google Scholar]
- Kendal M, Gibbons JD (1990): Rank correlation methods, 5th ed. ??: Edward Arnold. [Google Scholar]
- Liu C, Raz J, Turetsky B (1998): An estimator and permutation test fro single‐trial fMRI data. In: Abstracts of ENAR meeting of the International Biometric Society. International Biometric Society.
- Locascio JJ, Jennings PJ, Moore CI, Corkin S (1997): Time series analysis in the time domain and resampling methods for studies of functional magnetic resonance brain imaging. Hum Brain Mapp 5: 168–193. [DOI] [PubMed] [Google Scholar]
- Manly BFJ (1997): Randomization, bootstrap, and Monte‐Carlo methods in biology. London: Chapman and Hall. [Google Scholar]
- Marshuetz C, Smith EE, Jonides J, DeGutis J, Chenevert TL (2000): Order information in working memory: fMRI evidence for parietal and prefrontal mechanisms. J Cogn Neurosci 12: 130–144. [DOI] [PubMed] [Google Scholar]
- Noll DC, Kinahan PE, Mintun MA, Thulborn KR, Townsend DW (1996): Comparison of activation response using functional PET and MRI. Proceedings of the Second International Conference on Functional Mapping of the Human Brain, June 17–21, 1996, Boston, MA. Neuroimage 3: S34. [Google Scholar]
- Pitman EJG (1937a): Significance tests which may be applied to samples from any population. J R Stat Soc 4(Suppl): 119–130. [Google Scholar]
- Pitman EJG (1937b): Significance tests which may be applied to samples from any population. II. The correlation coefficient test. J R Stat Soc 4(Suppl): 224–232. [Google Scholar]
- Pitman EJG (1937a): Significance tests which may be applied to samples from any population. III. The analysis of variance test. Biometrika 29: 322–335. [Google Scholar]
- Poline JB, Mazoyer BM (1993): Analysis of individual positron emission tomography activation maps by detection of high signal‐to‐noise‐ratio pixel clusters. J Cereb Blood Flow Metab 13: 425–437. [DOI] [PubMed] [Google Scholar]
- Poline JB, Worsley KJ, Evans AC, Friston KJ (1997): Combining spatial extent and peak intensity to test for activations in functional imaging. Neuroimage 5: 83–96. [DOI] [PubMed] [Google Scholar]
- Roland PE, Levin B, Kawashima R, Akerman S (1993): Three‐dimensional analysis of clustered voxels in 15‐O‐butanol brain activation images. Hum Brain Mapp 1: 3–19. [Google Scholar]
- Silbersweig DA, Stern E, Schnorr L, Frith CD, Ashburner J, Cahill C, Frackowiak RSJ, Jones T (1994): Imaging transient, randomly occurring neuropsychological events in single subjects with positron emission tomography: an event‐related count rate correlational analysis. J Cereb Blood Flow Metab 14: 771–782. [DOI] [PubMed] [Google Scholar]
- Silbersweig DA, Stern E, Frith C, Cahill C, Holmes A, Grootoonk S, Seaward J, McKenna P, Chua SE, Schnorr L, Jones T, Frackowiak RSJ (1995): A functional neuroanatomy of hallucinations in schizophrenia. Nature 378: 169–176. [DOI] [PubMed] [Google Scholar]
- Smith AM, Lewis BK, Ruttimann UE, Ye FQ, Sinnwell TM, Yang Y, Duyn JH, Frank JA (1999): Investigation of low frequency drift in fMRI signal. Neuroimage 9: 526–533. [DOI] [PubMed] [Google Scholar]
- Stoeckl J, Poline J‐B, Malandain G, Ayache N, Darcourt J (2001): Smoothness and degrees of freedom restrictions when using SPM99. NeuroImage 13: S259. [Google Scholar]
- Watson JDG, Myers R, Frackowiak RSJ, Hajnal JV, Woods RP, Mazziotta JC, Shipp S, Zeki S (1993): Area V5 of the human brain: evidence from a combined study using positron emission tomography and magnetic resonance imaging. Cereb Cortex 3: 79–94. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young SS (1993): Resampling‐based multiple testing: examples and methods for P‐value adjustment. New York: Wiley. [Google Scholar]
- Worsley KJ (1994): Local maxima and the expected Euler characteristic of excursion sets of χ2, F, and t fields. Adv Appl Prob 26: 13–42. [Google Scholar]
- Worsley KJ, Evans AC, Strother SC, Tyler JL (1991): A linear spatial correlation model, with applications to positron emission tomography. J Am Stat Assoc 86: 55–67. [Google Scholar]
- Worsley KJ (1996): The geometry of random images. Chance 9: 27–40. [Google Scholar]
- Worsley KJ, Friston KJ (1995): Analysis of fMRI time‐series revised–again. Neuroimage 2: 173–181. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Evans AC, Marrett S, Neelin P (1992): A three‐dimensional statistical analysis for CBF activation studies in human brain. J Cereb Blood Flow Metab 12: 1040–1042. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC (1995): A unified statistical approach for determining significant signals in images of cerebral activation. Hum Brain Mapp 4: 58–73. [DOI] [PubMed] [Google Scholar]


