Abstract
An automated coordinate‐based system to retrieve brain labels from the 1988 Talairach Atlas, called the Talairach Daemon (TD), was previously introduced [Lancaster et al., 1997]. In the present study, the TD system and its 3‐D database of labels for the 1988 Talairach atlas were tested for labeling of functional activation foci. TD system labels were compared with author‐designated labels of activation coordinates from over 250 published functional brain‐mapping studies and with manual atlas‐derived labels from an expert group using a subset of these activation coordinates. Automated labeling by the TD system compared well with authors' labels, with a 70% or greater label match averaged over all locations. Author‐label matching improved to greater than 90% within a search range of ±5 mm for most sites. An adaptive grey matter (GM) range‐search utility was evaluated using individual activations from the M1 mouth region (30 subjects, 52 sites). It provided an 87% label match to Brodmann area labels (BA 4 & BA 6) within a search range of ±5 mm. Using the adaptive GM range search, the TD system's overall match with authors' labels (90%) was better than that of the expert group (80%). When used in concert with authors' deeper knowledge of an experiment, the TD system provides consistent and comprehensive labels for brain activation foci. Additional suggested applications of the TD system include interactive labeling, anatomical grouping of activation foci, lesion‐deficit analysis, and neuroanatomy education. Hum. Brain Mapping 10:120–131, 2000. © 2000 Wiley‐Liss, Inc.
INTRODUCTION
It is common practice in brain mapping experiments to report locations of functional and anatomical sites using standardized x‐y‐z coordinates [Fox et al., 1985; Friston et al., 1989, 1991; Fox 1995]. Widespread use of Talairach coordinates [Talairach et al., 1988] fostered the development of the BrainMap® database that encodes and queries the locations of functional neuroimaging findings using these coordinates [Fox et al., 1994]. The Talairach Daemon (TD) system [Lancaster et al., 1997] expands this concept by providing easy Internet access to a 3‐dimensional (3‐D) database of brain labels accessed by Talairach coordinates. The Talairach labels database uses a volume‐filling hierarchical naming scheme to organize labels for brain structures ranging from hemispheres to cytoarchitectural regions [Freitas et al., 1996, Lancaster et al., 1997]. This scheme is reflected in the database name, Volume Occupancy Talairach Labels (VOTL). The focus of this report is an evaluation of the accuracy of the TD system for obtaining Talairach labels for functional activation sites.
Brain atlases define and catalog rudimentary spatial features of brain structure using traditional nomenclature. Atlas labels provide a consistent terminology for qualitative description of regional brain structures. This is exemplified by the broad use of the 1988 Talairach atlas by the human brain mapping community [Steinmetz et al., 1989; Fox 1995]. Subcortical structures such as thalamus, caudate, and lentiform are easily identified by visual comparison of atlas sections with high‐resolution 3‐D MR images. Unlike the subcortical region, however, visual labeling in the cortex is highly problematic. Visual identification of gyri in serial MR section images is tedious, subject to reproducibility problems [Sobel et al., 1993], and subject to failure for secondary and tertiary sulci, that are not always present [Ono et al., 1990]. Gyri identification can be improved by use of surface rendering [Watson et al., 1993] and/or surface flattening [Dale and Sereno, 1993; Drury and VanEssen, 1997] to create images with better definition of sulcal and gyral boundaries. These processing methods, however, are neither time‐efficient nor readily available and provide little support for standardized labeling of individual brains. The coordinate‐based labeling scheme of the TD system, with concise gyral definitions, provides a consistent labeling alternative to visual labeling of gyri.
Visual labeling is hampered by vague boundary definitions for many brain structures. For example, complete boundaries of lobes and Brodmann areas are not found in popular atlases due to lack of consensus for their definition, including the 1988 Talairach atlas. Users must therefore select among several possible labels near non‐delineated boundaries, increasing inter‐user variability and making labeling susceptible to observer expectancy‐based bias. The VOTL database provides explicit boundaries for each labeled brain structure to manage this problem (see Methods).
Visual labeling is problematic even when explicit atlas labels are available for lobes, gyri and Brodmann areas. Boundaries for these regions are not easy to identify in high‐resolution MR images, more difficult in group‐average MR images, and practically impossible to identify in low resolution images (PET, SPECT, and fMRI), whether from individual subjects or group averages. Numerous automated methods, based on computerized mapping of atlas region labels onto medical images, have been developed to facilitate labeling [Bohm et al., 1983, 1991; Evans et al., 1991; Roland et al., 1994; Collins et al., 1994, 1995]. However, none of these atlas‐to‐image label‐mapping methods fully label the brain volume, nor are they simple to use or widely distributed. Alternatively, an efficient image‐to‐atlas mapping method is provided in the TD system to retrieve 1988 Talairach atlas labels for activation coordinates [Lancaster et al., 1997].
A coordinate‐based automated labeling system for functional activation sites should provide appropriate labels regardless of methodological differences. To test this capability in the TD system, it was compared with a large set of published functional activations that reported both coordinates and labels.
METHODS
The methods section is divided into development and evaluation subsections. The development subsection describes: (1) creation of a 3‐D Talairach Atlas from the published atlas, (2) formulation of the VOTL labeling scheme for this 3‐D atlas, and (3) features of the TD system software. The evaluation subsection focuses on three important areas: (1) labeling by the TD system vs. Talairach labels from published functional brain studies, (2) labeling by the TD system vs. knowledgeable users of the Talairach atlas, and (3) labeling accuracy of the TD system in a PET activation study.
TD System Development
3‐D Talairach Atlas
The 1988 Talairach Atlas contains a series of high‐detail color tracings (coronal, axial, and sagittal sections) derived from MR images of a 60‐year old right‐handed European female. Axial section images were chosen for developing the 3‐D atlas because this section format is the most common acquired by tomographic medical imagers. There are twenty‐seven axial sections ranging from z = + 65 mm and to z = ‐ 40 mm. The section spacing ranges from 2 mm near the AC to 5 mm for the slices near the top of the brain. Each section image was digitized with x‐y resolution of 0.43 mm, and each major structure segmented using a combination of color discrimination (Adobe PhotoShop™, San Jose, CA) and thresholding (Alice™, PAREXEL, Boston, MA). The digitized section images served as reference planes to interpolate a continuous 3‐D brain atlas using pixel (x & y) and section (z) locations relative to the AC or origin. Section images were assigned the z‐coordinate designated in the atlas. All images were carefully registered during digitization, to guarantee that the x‐y coordinate of the origin was maintained at a consistent location and that the x‐ and y‐axes were properly aligned. A contiguous 1‐mm 3‐D Talairach atlas volume was created from the reference images using resampling in the x and y directions and nearest neighbor interpolation in the z direction. Care was taken to guarantee that the brain dimensions matched that previously published for the atlas brain [Lancaster et al., 1995] (L‐R = 136 mm, A‐P = 172 mm, and S‐I = 118 mm). Small regional differences were seen, but these were usually less than 1 mm. Since most axial sections of the 1988 Talairach atlas were incomplete on the right, right‐side data was made by reflecting left‐side data about the y‐axis for all images.
Volume Occupancy Talairach Labels Database (VOTL)
The anatomical structure‐naming scheme devised for organizing the numerous 3‐D anatomical regions (volumes of interest ‐ VOIs) from the 3‐D Talairach atlas is based on volume occupancy (Fig. 1). More specifically a volume‐filling, hierarchical, anatomical‐labeling scheme is used, wherein each VOI is defined using 3‐D coordinates (for location) and a unique code (for anatomical label). The VOIs in the computerized 3‐D Talairach atlas were organized into five hierarchical levels: Hemisphere, Lobe, Gyrus, Tissue type, and Cell type (Table I). Rules and procedures were adopted for the VOTL labeling scheme to explicitly define boundaries for labeled brain structures [Freitas et al., 1996, Lancaster et al., 1997], and the entire brain volume fully labeled at each hierarchical level (Fig. 1). The rules and procedures for the VOTL labeling scheme are outlined below:
Figure 1.
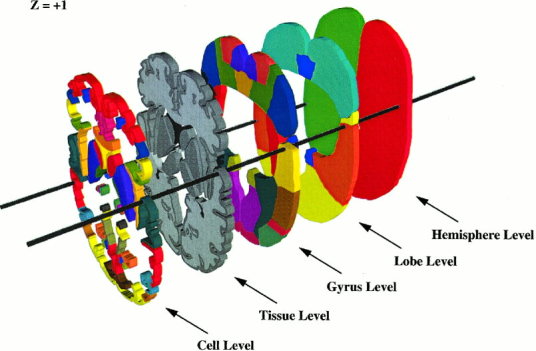
A typical set of 3‐D data used to create the volume occupancy (VOTL) database around the z = +1 level. Openings in Lobe through Cell levels were provided to emphasize the 3‐D nature of data at each level.
Table I.
Volume‐occupancy talairach labels
| Level | Label | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hemisphere (Level 1) | Cerebrum (R/L) | Cerebellum (R/L) | Brainstem (R/L) | |||||||
| Lobe (Level 2) | Lobes | Sub‐Lobar | In Progress | In Progress | ||||||
| Gyrus (Level 3) | Gyri | Sub‐Gyral | Nuclei | Extra‐Nuclear | “ | “ | ||||
| Tissue Type (Level 4) | GM | WM | WM | CSF | GM | WM | WM | CSF | “ | “ |
| Cell Type (Level 5) | BA | — | — | — | Sub‐Nuc. | — | — | Space | “ | “ |
BA ‐ Brodmann Area; WM ‐ White Matter; GM ‐ Gray Matter; CSF ‐ Cerebral Spinal Fluid
Hemisphere Level.
The largest anatomical structures in the brain (cerebrum, cerebellum, and brainstem) are assigned to the hemisphere level. The outer borders of hemisphere structures were extended slightly to include small invaginations (Fig. 2). The TD system returns “*” or “interhemispheric” labels for coordinates falling outside hemisphere level structures. The left‐ and right‐side attribute is assigned at this level.
Figure 2.
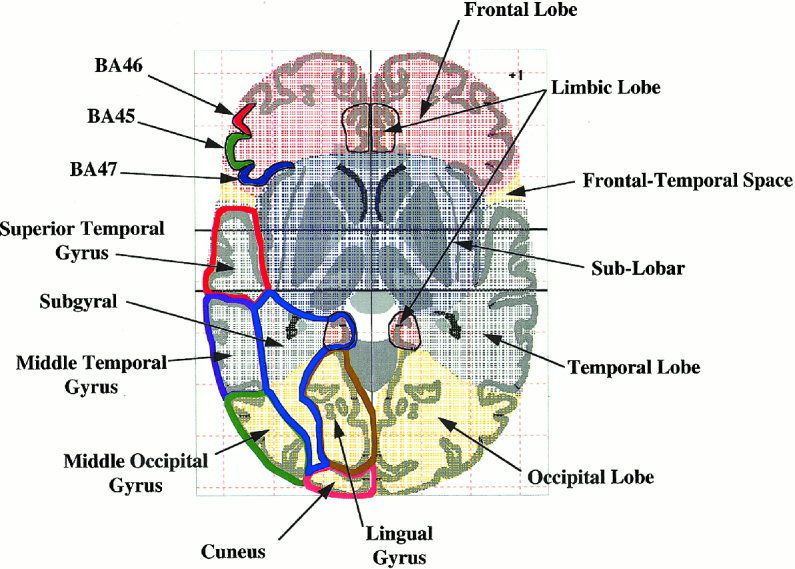
Example of VOTL region labels for a Talairach atlas section image at the z = + 1 level. Lobe labels are illustrated with patterned color fills. Several Brodmann areas (cell level) are illustrated on the top left using solid color fills. Several gyral level structures are illustrated on the bottom left using bold color outlines.
Lobe Level.
For the cerebrum, the lobe level consists of the four main lobes (Frontal, Temporal, Parietal, and Occipital), a single lobular equivalent (Limbic lobe) deeper within the brain, and a sub‐lobar region. Using this labeling scheme, cingulate is within the limbic lobe and insula within the sub‐lobar region. Outer boundaries of lobes were defined following cortical boundaries. Since explicit inner lobe boundaries were not present in the Talairach atlas, inner boundaries were defined using lines connecting the deepest extent of bounding sulci in axial section images (Fig. 2). The sub‐lobar region was assigned to the volume of the cerebrum not specifically designated as a lobe, completing the lobe‐level volume‐occupancy labeling for the cerebrum. The cerebellum and brainstem labeling (in progress) will follow this general scheme.
Gyrus Level.
This level includes gyri for lobes and various deep gray‐matter structures within the sub‐lobar region. Outer gyral boundaries follow lobar boundaries, while inner boundaries were defined using lines drawn between bounding sulcal pits in axial section images. Since inner lobar boundaries often extended deeper than inner gyral boundaries a sub‐gyral label was assigned to the region between the inner boundary of a gyrus and the inner boundary of its lobe (Fig. 2). Boundaries for sub‐lobar gyrus‐level structures (Caudate, Thalamus, Lentiform, etc.) were well delineated in the Talairach Atlas and used without modification. There are forty‐eight different gyrus level labels.
Tissue Level.
This level provides labels for gray matter, white matter, and cerebral spinal fluid (CSF). The lateral, third, and fourth ventricles were the only CSF‐designated regions since non‐ventricular CSF was not labeled in the Talairach atlas and is too variable to label consistently. Brain regions normally associated with gray matter (cortex and nuclei) were labeled as gray matter. Brain regions not labeled as gray matter or CSF were designated as white matter.
Cell Level.
The brain was labeled by cell‐type in the cortex using Brodmann's scheme [Garey, 1994] and tracts, spaces, and sub‐nuclear regions for other portions of the brain. Forty‐seven Brodmann areas (BA) labels were defined. Boundaries between adjacent Brodmann areas are not defined in the Talairach Atlas, so explicit BA boundaries were established that conformed to their original descriptions as closely as possible [Garey, 1994]. The BA boundaries were set at a sulcal pit or gyral crown when possible (Fig. 2). Care was taken to provide continuity of BA boundaries between adjacent sections [Freitas et al., 1996]. Other cell‐level labels (e.g., for sub‐lobar regions) closely follow explicit Talairach Atlas labels. Labels for tracts and spaces have not yet been created. The insular region had not been assigned a Brodmann area, so it was designated as BA 13. There are sixty‐six different cell level labels.
Cerebellum Labels
Several sources [Schmahmann et al., 1999; Courchesne et al., 1989; Press et al., 1989, 1990; Angevine et al., 1961] are being used for development of cerebellar labels. The cerebellum labels will not come from a single brain, but rather from a consensus based on numerous sources. A working version of cerebellar labels is in place. Since numerous refinements are anticipated cerebellar labels were not evaluated in the present study.
Talairach Daemon System Software
The general operating scheme of the TD system is that a user sends Talairach coordinates to the TD system server via a TD client. The server looks up labels in the VOTL database and sends coordinates with labels back to the user. The TD server is Internet accessible and supports multiple concurrent requests. It was implemented as a multi‐threaded application, with network communication using Berkeley Sockets, with a query/response protocol using ASCII strings, and a streamlined query/response processing method. A variety of freely distributed client applications, including Java versions, provide Internet access to TD databases (http://ric.uthscsa.edu/projects/talairachdaemon.html). The client‐server database environment was named the Talairach Daemon for the atlas space it references and the fact that the server is a UNIX daemon process [Freitas et al., 1996; Lancaster et al., 1997]. A disk‐based version of the TD system (client & VOTL database) has also been developed to support high‐speed labeling for large groups of coordinates.
Newer TD clients provide an adaptive range‐search utility to find nearby GM labels. Searching is performed in a cubic region centered on the coordinate of interest. The search is started using a 3x3x3‐mm3 cube, that is, with a search range of +/‐ 1‐mm along major coordinate directions. If no GM label is found, the search range is expanded to 5×5×5 mm3 (+/‐ 2 mm). Expansion of the search range is continued in this manner to a maximum range of 11×11×11 mm3 (+/‐ 5 mm). If a single GM label is found at any search range, its VOTL label and search range (+/‐ mm) are returned, and the search terminated. If multiple GM labels are encountered within a search range, the label with the highest frequency of occurrence is used. In cases of ties the search range is enlarged until the tie is broken. If no GM label is found, “No GM” is returned. This GM search algorithm has been successful for all Talairach coordinates tested to date, and there have been no cases of ties.
TD System Evaluation
TD vs. Author Labels
A data set containing author‐designated labels and Talairach coordinates for activation sites widely distributed throughout the brain was compared with labels obtained from the VOTL database by the TD system. Mostly group‐mean data (>5000 points; published prior to 1996) from numerous laboratories around the world were retrieved from the BrainMap® database [Fox et al., 1994; Fox and Lancaster, 1998]. All data from our research group were removed to eliminate potential bias. Additionally, all points with missing or incompatible label descriptions were removed resulting in a refined data set of approximately 2500 points. The refined data set (113 authors and > 250 studies) provided a wide range of TD system labels for comparison: all lobes, 33 gyri, 42 Brodmann areas, and 17 sublobar labels (nuclei, ventricles, and corpus callosum).
Multiple author‐designated labels were often seen for the same anatomical region, so an author‐to‐VOTL label‐mapping scheme was designated to support like‐label comparisons (Table II). This table indicates that author's labels paired reasonably well with gyrus and cell level labels. All label comparisons were therefore made at the “Gyrus” or “Cell” levels since hemisphere, lobe, and tissue labels can generally be inferred from these. A 670‐coordinate subset of labels for ten commonly referenced brain regions was selected for a detailed comparison with the TD system (Fig. 3, Legend). To characterize distances from coordinates to author‐designated structures, label‐matching scores were calculated as a function of search range from 0 to +/‐ 5mm.
Table II.
Author‐to‐TD label mapping
| Author label | Corresponding TD label | Hierarchical level |
|---|---|---|
| Cuneate | Cuneus | Gyrus |
| Cuneus | ||
| Cuneus‐striate junction | ||
| Precuneate cortex | Precuneus | Gyrus |
| Precuneus | ||
| Calcarine (sulcus) | Brodmann area 17 | Cell |
| Primary visual (area or cortex) | ||
| Striate | ||
| Primary motor | Brodmann area 4 | Cell |
| Motor area | ||
| Motor cortex | ||
| Motor hand area | Brodmann areas 4 or 6 | |
| Insula | Insula (BA 13) | Gyrus (cell) |
| Insular cortex | ||
| Insular gyrus | ||
| Insular region | ||
| Sylvian‐insular | ||
| Globus pallidus | Lentiform | Gyrus |
| Lenticular nucleus | ||
| Lenticulate | ||
| Lentiform nucleus | ||
| Putamen | ||
| Pallidum | ||
| Caudate | Caudate | Gyrus |
| Caudate nucleus | ||
| Caudate, head | ||
| Caudatum | ||
| Caudatus | ||
| Anterior cingulate | Anterior cingulate (BA 24,32) | Gyrus (cell) |
| Anterior cingulate cortex | ||
| Primary auditory | Brodmann area 41, 42 | Cell |
| Heschl's gyrus | ||
| Frontal eye fields | Brodmann area 8 | Cell |
| Wernicke's area | Brodmann area 22 | Cell |
| Broca's area | Brodmann area 44, 45 | Cell |
| Somatosensory | Brodmann area 1, 2, 3 | Cell |
| Thalamus | Thalamus | Gyrus |
| Amygdala | Amygdala | Gyrus |
| SMA | Brodmann area 6 | Cell |
| Parahippocampal gyrus | Parahippocampal gyrus | Gyrus |
| Uncus | Brodmann area 34 | Cell |
Figure 3.
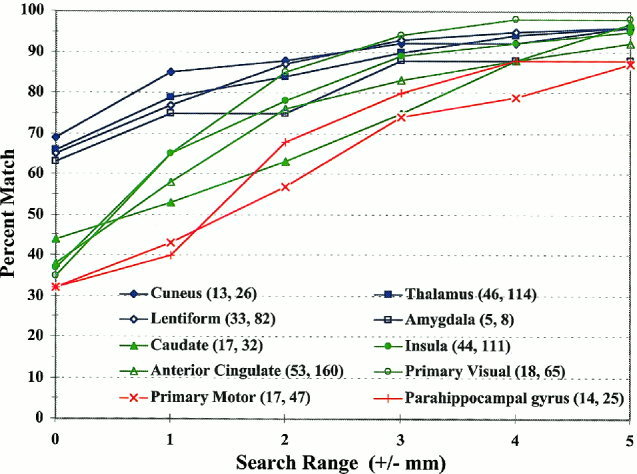
Percent match between TD and ten author‐designated labels as a function of the search range in mm. Data was calculated from 670 points reported by numerous authors. The legend data is ordered by initial percent match (search range = 0 mm). The numbers after each label are organized as follows: (# of different first authors, # of points for the label).
TD vs. Human for Atlas Labels
It was proposed that the TD system would provide a reliable method to obtain Talairach labels for functional activation studies, and that automated labeling could be done as well as or better than a user with atlas in hand. To test these propositions TD‐derived labels were compared with labels looked up in the 1988 Talairach atlas by three knowledgeable users (ML, LP, and MW). A set of approximately 100 labels was targeted for testing by this group. The set of 670 author‐labeled coordinates (See Previous Section) were sorted by author label and like labels grouped using Table II as a guide. A subset of widely distributed structures was selected including motor, sensory, association, and limbic areas, encompassing numerous Brodmann areas, as well as several deep gray structures. Coordinates were equally distributed between left and right hemispheres, and labels distributed across all lobes. Data for each structure were selected to include as many authors as possible to minimize bias. The resulting test set from 51 authors contained 106 labels in 16 different gyrus‐level and 19 different cell‐level structures.
Since the TD system was based on atlas axial section images, human labelers (testers) were instructed to initially use axial sections to determine a label for each point's Talairach coordinate. They then reviewed coronal and sagittal section images to determine if the same label would be obtained. Testers were instructed to indicate the nearest gray matter label if a coordinate fell outside gray matter. The testers were provided with a 1988 Talairach atlas axial section image similar to Figure 2 as an example of the VOTL scheme for lobe and gyrus labeling (included lobe, sub‐lobar, gyral and sub‐gyral region bounds outlined in color). Testers were also briefed on how Brodmann area boundaries were determined for the VOTL scheme. The method for looking up labels using x‐y‐z coordinates was left to the testers. Each tester indicated a level of difficulty (easy, medium, and hard) for each label and the total labeling time required.
Brodmann Area Labels for Cortical Activations
The accuracy of the TD system was tested using Talairach coordinates taken from a previously reported O‐15 PET water study designed to activate M1 mouth motor areas [Fox et al., 1997, 1999]. The change distribution analysis (CDA) method [Fox et al., 1988] was used to determine significant activation foci in each of 30 subjects [Fox et al., 1999]. All subjects were healthy, right‐handed, native English speakers between the ages of 21 and 49 (mean = 32; SD = 7). Statistically significant activations (uncorrected p < 0.001) for BA 4 or 6 were isolated for testing using visual inspection (PTF). Fifty‐two different x‐y‐z coordinates were selected, 24 on the right and 28 on the left side. Talairach coordinates for each activation site were submitted to the TD system for labeling. If no GM label was found, the adaptive range‐search utility was invoked to provide a nearby GM label.
RESULTS
TD vs. Author Labels
Label matching between TD system (VOTL) labels and author‐designated labels for ten regions (670 activation sites) is presented in Figure 3. The graph in this figure indicates the probability of matching the label designated by authors as a function of search range. For the Cuneus and Thalamus (gyrus‐level labels) the initial match (search range = 0 mm) was about 70%. Label matching rose to 80% for a search range of ±2 mm and to over 90% for a ±3‐mm range. Label matching for Lentiform (cell‐level) and Amygdala (gyrus‐level) labels followed a similar trend. The primary motor area (BA 4) and the parahippocampal gyrus had considerably lower initial label matching scores (32%). Label matching showed little improvement for a ± 1‐mm range but approached 80% for a ± 3‐mm range. For the four remaining regions (caudate, insula, anterior cingulate, and primary visual – BA 17), label matching scores fell between the values for the worst (primary motor area) and the best (Thalamus) label matches. Author‐TD label matching was above 90% for half of the regions evaluated within a search range of ± 4 mm. For eight of ten of the regions, label matching was greater than 90% using a search range of ± 5 mm. These results are for group‐mean data, and individual subject results are presented in the Brodmann Area Labels section below.
TD vs. Human for Atlas Labels
Difficulty ratings provided by the human labelers (testers) showed that similar numbers of sites were of low (47%) and medium (40%) difficulty. Tester's impression of difficulty was mirrored by their author label matching scores (Table III, columns 1 & 2). Categorization of difficulty by level was not obviously associated with any specific label, nor did it appear to be consistent among testers. Interestingly, the TD system (Raw – without range search) had its highest matching score for sites in the tester's medium‐difficulty category. Both the testers and the TD system had lowest matching scores for sites in the high‐difficulty category. Testers' author‐matching scores for labels derived from atlas axial sections were similar to TD raw scores, which were also from axial sections (Table III, columns 1 & 3).
Table III.
Manual vs. TD system matching to author labels using talairach coordinates
| Difficulty rating | Percentage match to authors' labels | |||
|---|---|---|---|---|
| Testers | TD system | |||
| Axial section* | All sections** | Raw | Range search | |
| All sites (regardless of rating) | 73% | 80% | 71% | 90% |
| Low (47% of sites) | 83% | 88% | 72% | 92% |
| Medium (40% of sites) | 72% | 76% | 77% | 89% |
| High (13% of sites) | 40% | 64% | 47% | 84% |
Average scores for three experts manually labeling 106 coordinates using axial section images from the 1988 Talairach Atlas.
Average scores of expert's best label using all (axial, coronal, and sagittal) section images.
Testers' scores improved somewhat when all atlas sections (axial, sagittal, and coronal) were used to determine a best label, with the score averaged across all sites increasing from 73% to 80%. Author‐matching scores for the TD system improved dramatically when the range‐search utility was used to find nearest GM labels, with scores rising by as much as 37% for the high difficulty sites. The score averaged across all sites rose from 71% to 90%. This trend is similar to that found for the TD‐to‐author evaluation with 670 points (Fig. 3). Percentage matching to author's labels by the TD system, using the adaptive GM range‐search utility, exceeded that by the tester group in all difficulty rating categories (Table III, columns 4 vs. 2). The time to obtain the axial labels for the 106 sites was over three hours for each tester, while the TD system, with the GM range‐search utility, completed this task in less than one minute.
Median scores for label agreement between testers and the TD system (without range search) dropped level‐by‐level (hemisphere: 98%, lobe: 83%, gyrus: 63%, and cell: 55%). Label consistency among testers (all agreed) dropped similarly (hemisphere: 96%, lobe: 72%, gyrus: 42%, and cell: 53%). Incomplete agreement at the hemisphere level and diminished agreement at the gyrus level were due to inconsistent nomenclature among users. Full label matching disparity (all testers disagreed) was seen only at the gyrus (13%) and cell (10%) levels. These data demonstrate a potentially high level of variability among users for manually obtaining labels for activation sites from the Talairach atlas. There were no reproducibility problems observed with the TD system.
A consensus (at least 2 agreed) among testers' best labels was seen for 94% of the sites. Consensus labels are less sensitive to task‐related user variability and provide a good data set to evaluate the potential performance for the manual‐labeling task. The consensus labels compared well with labels from the TD range search (89% agreement) and provided a better overall match with author designated labels (86%). However, the TD system (with adaptive range‐search utility) had a slightly higher author label‐matching score (90%) than that of testers' consensus labeling.
Brodmann Area Labels for Cortical Activations
The TD system returned matching Brodmann labels (BA 4 or 6) for 40% of the coordinates without using the range search. The label match increased rapidly to 56%, 79%, and 85% with search ranges of ±1, ±2, and ±3 mm. It rose slightly after that with a final label match of 87% at ±5 mm. Seven errant labels were seen, all postcentral gyrus (BA 2 and 3), and five of these occurred at a search range of zero. Similar numbers of errant labels were seen for left (4) and right (3) sides. Label matching as a function of search range was better than that recorded for the 670 group‐mean coordinates taken from the BrainMap database (Fig. 3, Primary Motor).
DISCUSSION
The large set of author labels selected for testing provided reasonable anatomical labeling accuracy, a diversity of sites distributed throughout the brain, and numerous methodological challenges, all important for evaluating automated labeling of functional activation sites. Authors were not assumed to all correctly label sites. However, their labels were considered to be more nearly correct than labels obtained from Talairach coordinates alone [Roland, et al., 1997]. This assumption was based on additional information available to authors (i.e., experimental design, overlays of functional maps on MR images, etc.).
Automated labeling by the TD system performed as well as or better than our experts, or even the consensus of 2 of 3 experts, when compared with author's labels. It should be noted, however, that neither achieved a 100% match with author‐designated labels. The fact that the TD system can achieve an author match of 90% for many labels indicates that it can be a valuable asset to brain mappers seeking to standardize labeling of activation foci. However, it is recommended that the TD system be used with caution, and that users treat labels as candidate labels to be reviewed for appropriateness. Questionable labels should be resolved by inspecting overlays of functional maps on high‐resolution MR images. This is especially important for sites in highly variable regions such as the occipital‐cerebellar boundary. For example, if the TD system reports labels from the cerebellum that are obviously in the occipital lobe (based on overlays on MR images), it is useful to expand the search range to produce an extended list of label options. This approach is similar to how authors use the Talairach atlas when such confusion arises, to review nearby labels and select the most likely candidate.
Brain Labeling Accuracy
The degree of feature correspondence between individual brain images and an atlas varies throughout the brain [Ono et al., 1990; Mazziotta et al., 1995; Zilles et al., 1997; Roland et al., 1997]. For visual labeling, a more informed choice can be made after a survey of nearby features in both brain image and atlas. A label is then selected, along with a qualitative estimate of confidence level. However, there is no guarantee that multiple users will choose the same label given identical information or that the label will be correct. Accordingly, the Talairach atlas labels provided by the TD system should be treated as candidate labels. Intrinsic coordinate‐based TD‐system labeling errors are expected to be smallest for larger structures seen at the hemisphere and lobe level and largest for smaller structures and near boundaries (Fig. 3). However, using the adaptive GM range‐search utility, the TD‐system matching with author's labels exceeded 90% for many labels (Fig. 3 and Table III).
Label errors for coordinate‐based labeling methods can come from incomplete anatomical matching by global spatial normalization as well as methodological differences [Strother et al., 1994]. While several non‐linear, high degree‐of‐freedom 3‐D regional spatial normalization algorithms have been developed [Collins et al., 1994; Friston et al., 1995; Woods et al., 1998, 1998a, Kochunov et al., 1999], their accuracy remains unproved, and they have not been implemented to regionally match the standard 1988 Talairach atlas. A design goal of the adaptive GM range‐search utility was to accommodate residual errors following spatial normalization. The notable improvement in label accuracy (56 to 87%) for M1 hand motor PET activation studies, using linear affine spatial normalization [Lancaster, et al., 1995], shows that this strategy can work reasonably well. While label errors were reduced, more accurate spatial normalization (bringing coordinates closer to Talairach labels), coupled with the range‐search utility, should produce even better results. Another design goal of the GM range‐search utility was to accommodate methodological difference between different laboratories. The improved author matching (71 to 90%) for all labels (51 authors) using the adaptive GM range‐search utility (Table III, column 4 vs. 3) shows that this strategy works reasonably well.
Common Talairach Daemon System Uses
BrainMap®
The BrainMap® database provides access to brain findings from more than 200 research papers, 700 experiments, and 7000 locations in the human brain [Fox et al., 1994; Fox and Lancaster, 1998]. Data for the evaluation of the TD system was taken from this database. Access to TD labels is provided in the BrainMap® Search & View client, where a user can quickly review candidate Talairach labels for coordinates from published brain function studies (Fig. 4). A future enhancement of the BrainMap® database is automatic entry of VOTL labels of each coordinate to support searching using VOTL labels.
Figure 4.
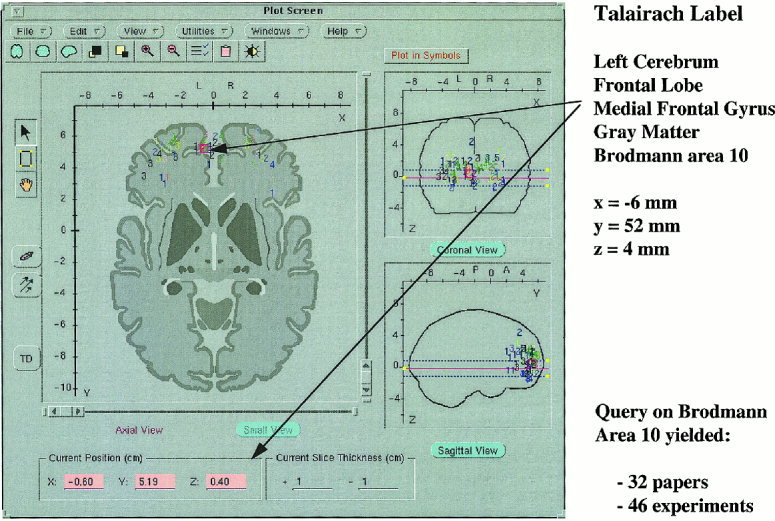
The BrainMap™ Search & View 4.1 application provides direct access to the VOTL (Talairach labels) database to obtain anatomical labels. A coordinate returned following a search on Brodmann area 10 was used to automatically retrieve its hierarchical label.
Anatomical Organization of Coordinate Data
Statistical analyses of brain function studies often result in a long list of coordinates for activation foci. For software that provides Talairach coordinates in an x‐y‐z list format (SPM; MEDx, Sensor Systems, Sterling, VA) it is helpful to retrieve VOTL database labels using the TD system and to reorganize the list anatomically. This sorting feature was used to categorize activation sites for testing in this report.
Lesion‐Deficit Analysis
The TD system provides a means to tabulate and standardize labels for the volume occupied by a brain lesion in Talairach space. This standardization supports correlation of standard anatomical nomenclature with measures of neurological deficit (NIH/NINDS 2 RO1 NS 21889‐16).
Education
A Java applet called the Talairach Daemon was developed as an educational tool (http://ric.uthscsa.edu/projects/talairachdaemon.html). It contains a complete hand‐detailed version of the 1988 Talairach atlas axial sections and overlays for all VOTL structures. The interactive labeling features of the TD applet provide excellent examples for teaching brain anatomy.
CONCLUSIONS
The Talairach Daemon system provides rapid access to Talairach atlas labels for functional activation foci using Talairach coordinates. Many gyri and cell‐level labels in the volume occupancy Talairach labels (VOTL) database directly map to common author labels. VOTL labels matching author's labels were found within ±5 mm of author‐designated coordinates for most activation foci. The GM range‐search utility greatly enhanced the TD system, enabling it to perform better than a group of three knowledgeable users, when attempting to match published Talairach coordinates with labels. Using the GM range‐search utility, Brodmann area labels for the M1 hand motor area activations (BA 4 and 6) were correctly identified in 87% of the subjects. The TD system, with its numerous Internet distributed client applications, provides an extensive informatics resource for the human functional brain mapping community.
Web address for this project: http://nc.uthscsa.edu/projects/tnlairachdaemon.html
REFERENCES
- Angevine JB, Mancall EL, Yakovlev cPI. (1961): The human cerebellum: An atlas of gross topography in serial sections. Little, Brown, and Co., Boston. [Google Scholar]
- Bohm C, Greitz T, Kingsley D, Berggren, Olsson L. (1983): Adjustable computerized stereotaxic brain atlas for transmission and emission tomography. AJNR 4: 731–733. [PMC free article] [PubMed] [Google Scholar]
- Bohm C, Greitz T, Seitz R, Eriksson L. (1991): Specification and selection of regions of interest (TOIs) in a computerized brain atlas. J Cereb Blood Flow and Metab 11: A64–A68. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. (1994): Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 18: 192–205. [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AE. (1995): Automatic 3‐D model‐based neuroanatomical segmentation. Hum Brain Mapp 3: 209–223. [Google Scholar]
- Courchesne E, Press GA, Murakami J, Berthoy D, Grafe M, Wiley CA, Hesselink JR. (1989): The cerebellum in sagittal plane‐Anat‐ omic‐MR correlation: The vermis. Am J of Neurol Res 10: 659–665. [Google Scholar]
- Drury HA and Van Essen DC. (1997): Functional specilizations in human cerebral cortex analyzed using the visible man surface‐based atlas. Hum Brain Mapp 5: 233–237. [PubMed] [Google Scholar]
- Evans AC, Marrett S, Torrescorozo J, Ju S, Collins L. (1991): MRI‐PET correlation in three dimensions using a volume‐of‐interest (VOI) atlas. J Cereb Blood Flow and Metab 11: A69–A78. [DOI] [PubMed] [Google Scholar]
- Fox PT, Perlmutter JS, Raichle ME. (1985): A stereotactic method of anatomical localization for positron emission tomography. J Comput Assist Tomogr 9: 141–153. [DOI] [PubMed] [Google Scholar]
- Fox PT, Mintum MA, Reiman EM, Raichle ME. (1988): Enhanced detection of focal brain responses using intersubject averaging and change distribution analysis of subtracted PET images. J Cereb Blood Flow Metab 8: 642–653. [DOI] [PubMed] [Google Scholar]
- Fox PT. (1995): Spatial Normalization: Origins, objectives, applications and alternatives. Hum Brain Mapp 3: 161–164. [Google Scholar]
- Fox PT, Mikiten S, Davis G, Lancaster JL. (1994): BrainMap: A database of human functional brain mapping In: Thatcher RW, Zeffiro T, Huerta M. (eds): Advances in Functional Neuroimaging: Technical Foundations. Orlando: Academic Press, 95–106. [Google Scholar]
- Fox PT, Ingham TJ, Ingham JC, Hirsch TB, Downs JH, Martin C, Jerabek P, Glass T, Lancaster JL. (1996): A PET study of the neural systes of stuttering. Nature 382: 158–162. [DOI] [PubMed] [Google Scholar]
- Fox PT, Lancaster JL, Parsons LM, Xiong J, Zamarripa E. (1997): Functional volumes modeling: Theory and preliminary assessment. Hum Brain Mapp 5(4): 306–311. [DOI] [PubMed] [Google Scholar]
- Fox PT, Lancaster JL. (1998): BrainMap®: Electronic integration of mind and brain In: Positron Emisssion Tomography: A critical assessment of research trends, Gulyas B. and Muller‐Gartner HW, eds., Kluwer Academic Publishers. [Google Scholar]
- Fox PT, Huang A, Parsons LM, Xiong J, Zamarripa F, Rainey L, Lancaster JL. (2000): Location‐probabiolity profiles for the mouth region of human primary motor cortex: Metanalysis and validation, NeuroImage: in process. [DOI] [PubMed] [Google Scholar]
- Freitas CS, Summerlin JL, Lancaster JL, Fox PT. (1996): Soc Neurosci Ann Mtg V–101. [Google Scholar]
- Friston KJ, Passingham RE, Nutt JG, Heather JD, Sawle GV, Frackowiak RSJ. (1989): Localization of PET images: Direct fitting of the intercommissural (AC‐PC) line. J Cereb Blood Flow Metab 9: 690–695. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Frith CD, Liddle PF, Frakowiak RSJ. (1991): Comparing functional (PET) images: The assessment of significant change. J Cereb Blood Flow Metab 11: 690–699. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Poline JB, Heather JD, Frakowiak RSJ. (1995): Spatial registration and normalization of images. Hum Brain Mapp 3: 165–189. [Google Scholar]
- Garey LJ. (1994): Brodmann's Localisation in the Cerebral Cortex. London: Smith‐Gordon, p 109–129. [Google Scholar]
- Kochunov PV, Lancaster JL, Fox PT. (2000): Accurate high‐speed spatial normalization using an octree method. NeuroImage: in press. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Glass TG, Lankipalli BR, Downs H, Mayberg H, Fox PT. (1995): A modality‐independent approach to spatial normalization. Hum Brain Mapp 3: 209–223. [Google Scholar]
- Lancaster JL, Rainey LH, Summerlin JL, Freitas CS, Fox PT, Evans AE, Toga AW, Mazziotta JC. (1997): Automated labeling of the human brain: A preliminary report on the development and evaluation of a forward‐transform method. Hum Brain Mapp 5: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster JL, Fox PT, Downs H, Nickerson D, Hander T, El Mallah M, Zamarripa F. (1999): Global spatial normalization of the human brain using convex hulls, J of Nucl Med 40: 942–955. [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Lancaster JL, Fox PT. (1995): A probabilistic atlas of the human brain: Theory and rational for its development. Neuroimage 2: 89–101. [DOI] [PubMed] [Google Scholar]
- Ono M, Kubik S, Abernathey CD. (1990): Atlas of the Cerebral Sulci. New York: Theime Medical Publishers. [Google Scholar]
- Pelizzari CA, Chen GTY, Spelbring DR, Weichselbaum RR, Chen CT. (1989): Accurate three‐dimensional registration of CT, PET, and/or MRI images of the brain. J Comput Assist Tomogr 13: 20–26. [DOI] [PubMed] [Google Scholar]
- Press GA, Murakami J, Courchesne E, Berthoy D, Grafe M, Wiley CA, Hesselink JR. (1989): The cerebellum in sagittal plane–Anatomic‐MR correlation: The cerebellar hemispheres. Am J of Neurol Res 10: 667–676. [DOI] [PubMed] [Google Scholar]
- Press GA, Murakami J, Courchesne E, Grafe M, Hesslink JR. (1990): The cerebellum: 3. Anatomic‐MR correlation in the coronal plane. Am J Neurol Res 11: 41–50. [PMC free article] [PubMed] [Google Scholar]
- Roland PE, Graufelds CJ, Wahlin J, Ingelman L, Andersson M, Ledberg A, Pederson J, Akerman S, Dabringhaus A, Zilles K. (1994): Human brain atlas: For high‐resolution functional and anatomical mapping. Hum Brain Mapp 1: 173–184. [DOI] [PubMed] [Google Scholar]
- Roland PE, Geyer S, Amunts K, Schormann T, Schleicher A, Malikovic A, Zilles K. (1997): Cytoarchitectural maps of the human brain in standard anatomical space. Human Brain Mapping 5: 222–227. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Doyon J, McDonald D, Holmes C, Lavoie K, Hurwitz AS, Kabani N, Toga A, Evans A, Petrides M. (1999): Three‐dimensional MRI atlas of the human cerebellum in proportional stereotaxic space. NeuroImage 10: 233–260. [DOI] [PubMed] [Google Scholar]
- Sobel DF, Gallen CC, Schwartz BJ, et al. (1993): Locating the central sulcus: comparison of MR anatomical and magnetoencephalographic functional methods. AJNR 14: 915–925. [PMC free article] [PubMed] [Google Scholar]
- Steinmetz H, Furst G, Freund HJ. (1989): Cerebral Cortical Localization: Application and validation of the proportional grid system in MR imaging. J Comput Assist Tomogr 13: 10–19. [PubMed] [Google Scholar]
- Stevens WR. (1990): UNIX Network Programming. Englewood Cliffs, NJ: Prentice Hall. [Google Scholar]
- Strother SC, Anderson JR, Xu XL, Liow JS, Bonar DC, Rottenberg DA. (1994): Quantitative comparisons of image registration techniques based on high‐resolution MRI of the brain. J Comput Asst Tomogr 18: 954–962. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. (1988): Co‐Planar Stereotaxic Atlas of the Human Brain. New York: Thieme Medical Publishers. [Google Scholar]
- Watson JDG, Meyers R, Frackowiak RSJ, Hajnal JV, Woods RP, Mazziotta JC, Shipp S, Zeki S. (1993): Area V5 of the human brain: Evidence from a combined study using positron emission tomography and magnetic resonance imaging. Cereb Cortex 3: 79–94. [DOI] [PubMed] [Google Scholar]
- Woods RP, Mazziotta JC, Cherry SR. (1993): MRI–PET registration with automated algorithm. J Comput Assist Tomogr 17: 536–546. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. (1998): Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Asst Tomogr 22: 139–152. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Watson JDG, Sicotte NL, Mazziotta JC. (1998a): Automated image registration: II. Intersubject validation of linear and nonlinear models. J Comput Asst Tomogr 22: 153–165. [DOI] [PubMed] [Google Scholar]
- Zilles K, Schleicher A, Langemann C, Amunts K, Morosan P, Palomero‐Gallagher N, Schormann T, Mohlberg H, Burgel U, Steinmetz H, Schlaug G, Roland PE. (1997): Quantitative analysis of sulci in the human cerebral cortex: Development, regional heterogeneity, gender difference, asymmetry, intersubject variability and cortical architecture. Hum Brain Mapp 5: 218–221. [DOI] [PubMed] [Google Scholar]


