Abstract
We contend that powerful group studies can be conducted using magnetoencephalography (MEG), which can provide useful insights into the approximate distribution of the neural activity detected with MEG without requiring magnetic resonance imaging (MRI) for each participant. Instead, a participant's MRI is approximated with one chosen as a best match on the basis of the scalp surface from a database of available MRIs. Because large inter‐individual variability in sulcal and gyral patterns is an inherent source of blurring in studies using grouped functional activity, the additional error introduced by this approximation procedure has little effect on the group results, and offers a sufficiently close approximation to that of the participants to yield a good indication of the true distribution of the grouped neural activity. T1‐weighted MRIs of 28 adults were acquired in a variety of MR systems. An artificial functional image was prepared for each person in which eight 5 × 5 × 5 mm regions of brain activation were simulated. Spatial normalisation was applied to each image using transformations calculated using SPM99 with (1) the participant's actual MRI, and (2) the best matched MRI substituted from those of the other 27 participants. The distribution of distances between the locations of points using real and substituted MRIs had a modal value of 6 mm with 90% of cases falling below 12.5 mm. The effects of this approach on real grouped SAM source imaging of MEG data in a verbal fluency task are also shown. The distribution of MEG activity in the estimated average response is very similar to that produced when using the real MRIs. Hum. Brain Mapping 20:142–147, 2003. © 2003 Wiley‐Liss, Inc.
Keywords: image co‐registration, synthetic aperture magnetometry, SAM, group functional imaging, brainswapping
INTRODUCTION
Currently the growth of our understanding of the link between brain and cognition is being driven by research in neuroimaging with positron emission tomography (PET), functional magnetic resonance imaging (fMRI), and electro‐ and magneto‐encephalography (EEG/MEG) providing new insights. All these functional neuroimaging methods rely on anatomical information from MRI, and clearly with fMRI the correspondence between anatomy and function is most directly made. With EEG/MEG researchers must bring their data into co‐registration with MRI before any conclusion can be made about anatomical sources of the observed functional data. All functional imaging methods are beset, however, by the problem of inter‐individual variability in the sulcal and gyral patterns of the brain [Mazziotta et al., 2001; Talairach and Tournoux, 1988; Thompson et al., 1996; Toga and Thompson, 2001]. Because of this spatial uncertainty, blurring of the functional images results when images are combined, and this puts all the methods on a more comparable basis when used to infer the distribution of brain activity from grouped functional imaging results.
In our investigations using whole head MEG recordings we have followed two lines of methodological development of relevance here. First, we have improved the co‐registration of the MEG results with the participants' MRIs such that we now attain registration errors typically of 2–4 mm in an individual subject [Singh et al., 1997]. This is clearly advantageous when using MEG to localise function in an individual such as required by pre‐surgical localisation of eloquent cortex. Second, we have shown that volumetric statistical images of changes in neural activity, obtained with synthetic aperture magnetometry (SAM) [Robinson and Vrba, 1999; Van Veen et al., 1997] can be combined and processed over a pool of subjects in the same manner as in PET and fMRI investigations [Singh et al., 2002]. Using this latter method, called here groupSAM, experiments on verbal fluency and biological motion perception have shown widespread regions of event related desynchronisation (ERD) and synchronisation (ERS) (Fig. 3a) that bear a striking resemblance to patterns of BOLD signal change [Singh et al., 2002]. GroupSAM requires that each participant have an MRI for co‐registration with the MEG data. This is then transformed to the MNI template brain using SPM99 [Friston et al., 1995] upon which the final group functional imaging results are displayed.
Figure 3.
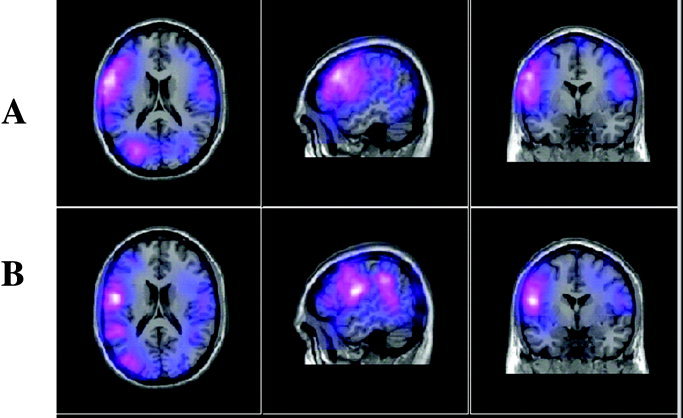
The results of averaging the MEG responses of six participants in a verbal fluency task using: (A) the participants MRIs [Singh et al., 2002] and (B) MRIs selected as the best match from a pool of 27 other MRIs.
As with all functional imaging techniques based on spatial normalisation, groupSAM suffers from the inherent variability of individual brains discussed above. In addition it also suffers from the resource limitations imposed by the requirement for MRIs of the participants. One of the most attractive features of MEG for cognitive experimentation is that the participant's experience of the experiment is favourable in comparison with both PET and fMRI/MRI. A typical recording with visual or auditory stimulation will require no longer than 15 min, with subject preparation time in the MEG system below 2 min. There are no alarming or distracting noises, nor any injections, and naive participants are generally happy to comply with the procedures. In requiring participants in an MEG experiment to also attend for an MRI scan these attractive features of the MEG methodology are lost and there are significant costs in terms of the additional time and procedures required. Furthermore one also has the problem of dropout of participants between the two sessions.
The contention of this study is that powerful group studies can be conducted using MEG, which can provide insight into the anatomical distribution of the brain activity subsuming cognitive functions without requiring an MRI for each participant in the study. Instead, for each participant in a study without an MRI one is substituted as a best match solely on the basis of the external head shape from a pool of available MRIs, and the MEG source reconstruction is done using the substituted MRI. This article concerns the errors that arise in making this approximation and does not relate to the MEG method used explicitly. The same approach could be used to combine participants' results in studies using other imaging modalities having similarly limited spatial resolution with respect to MRI, for example EEG. The small reduction in accuracy that results is acceptable, given the advantages in terms of resources and participant numbers that can be studied. We suggest that, as inter‐individual variability is large, little is lost in a group MEG study by approximating a participant's MRI using one substituted with a “best match” from a database of MRIs.
In this method the surface of the head is digitized first, and then this surface is matched to the scalp surfaces of an available pool of MRIs to find the closest matching MRI. If this pool is large then a very close match can be found. Our assumption is that the underlying brain structure of the participant will be a reasonable approximation to that in the substituted brain, at least as good as that provided by reference to the Talairach and Tournoux Atlas that refers to a single individual's brain. Other workers have advanced similar suggestions recently and argued for the utility of an approach using approximations of the brain volume. Carducci et al. [ 2001] used a method in which scalp locations on each participant were identified on the MNI averaged brain and the underlying brain location estimated by perpendicular projection from the surface of the MNI brain. They report that the mean difference in Talairach co‐ordinates between estimates using the real scalp and the averaged MNI scalp locations is <1 cm, and typically 4–6 mm. van't Ent et al. [ 2001] applied finite element analysis combined with principal components analysis to use the scalp surface to approximate the boundaries between the differently conductive partitions of the brain for EEG/MEG source reconstruction using realistically shaped head models. We examine the distribution of errors that arises in simulated functional MEG data when using substituted as opposed to actual brains, and show that typically there is <6 mm of additional error introduced. We also compare the results of a group MEG experiment on verbal fluency [Singh et al., 2002] with the results obtained using the same data processed with the cortical source estimation method.
MATERIALS AND METHODS
MRI and Simulation Functional Images
MRIs of 28 adults, 21 male and 7 female, were used in the study. T1‐weighted MRIs were acquired in a variety of MR systems, re‐sampled to 256 × 256 × 256 8‐bit images. A simulated functional image was prepared for each person (Fig. 1a). These were 3‐D images in which eight discrete regions representing local functional activity were placed at representative brain locations. The precise anatomical accuracy of placement of the regions is not as important as their purpose was simply to widely sample the distribution of the difference in locations with and without the MRI approximation being applied, and therefore operator placement accuracy is not an issue, and the initial location could have been chosen at random. The size of these small regions simulating functional activity was 5 × 5 × 5 mm, which is a typical voxel size used in MEG source reconstruction using SAM.
Figure 1.
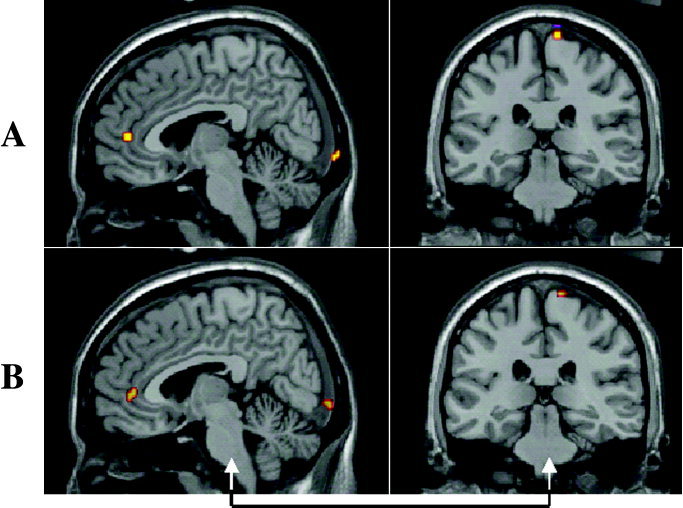
A: Sagittal and coronal sections of the results of spatial normalisation with the MNI template brain using a participant's own MRI with the simulated functional activations overlaid. B: Performing the spatial normalisation using an MRI with the best matching scalp surface extracted from a set of 27 MRIs.
Synthetic Data Used
The approximate locations of the eight representative brain regions used in the simulations were: (1) in the occipital pole at the end of the calcarine sulcus in the right hemisphere; (2) at the junction of the right calcarine sulcus and the parieto‐occipital sulcus near the midline; (3) on the lateral extremity of Heschl's gyrus in the left hemisphere (LHS); (4) on the superior surface of the medial termination of Heschl's gyrus in the left hemisphere; (5) at the most superior point of the posterior bank of the left central sulcus; (6) in the centre of the left insula cortex; (7) at the most anterior point of the cingulate cortex; and (8) in the left middle frontal gyrus. Figure 1a shows the location of a subset of these regions co‐registered with the MNI template for one participant. These locations have no significance other than to provide a range of locations throughout the brain to widely sample the distribution of errors in spatial location produced by using a substitute for a participants MRI. Hence, the activities represented by these small active regions were not derived from MEG SAM source modelling or dipole localisation algorithms.
Processing of SAM Images: Normal Method Using Participants' MRIs
To co‐register MEG data with a participant's MRI the external surface of the head is digitised with a Polhemus Isotrak 3‐D digitiser (Polhemus, Colchester, VT) and then a surface‐matching algorithm (ALIGN) [Kozinska et al., 1997] is used to calculate the transformation to apply to the MEG data to bring it into registration with the MRI. Using the co‐registered MRI a SAM image is then computed. For group analysis of SAM data the participant's MRI is first re‐sliced so that the MRI slices are in the same position and orientation as the SAM images. This re‐sliced MRI is then brought into registration with a template brain (Montreal Neurological Institute [MNI] template) using the non‐linear spatial normalisation functions in SPM99 [Friston et al., 1995]. The spatial normalisation was computed with non‐linear basis functions in a 7 × 8 × 7 mm volume for a maximum of 12 iterations. The computed normalisation transformation is then applied to the SAM images, thus generating a spatially normalised SAM image set. The results from multiple participants are then averaged or processed with other statistical procedures to reveal activated regions common to the group, which can then be visualised in co‐registration with the template MRI.
Processing of Artificial Functional Images Using Substituted MRIs
The artificial functional images were first processed as above, each using the participant's correct MRI. This resulted in a set of eight reference points in Talairach space for each participant, namely the locations of the centroids of the eight simulated regions of functional activity under spatial normalisation with the correct MRI. Then each participant's head shape was used to form the co‐registration surface for matching with the other 27 MRIs in the dataset. A full surface match on these 27 MRIs was carried out to identify the best substitute MRI. The surface was then re‐fitted after exclusion of outlying points, defined as those more than 2cm distance from the target MRI surface. In a real application computational efficiency could be improved when searching large sets of MRIs for substitutes by describing MRI surfaces in terms of their spherical harmonics [van't Ent et al., 2001]. After a substitute MRI was identified it was co‐registered with the MNI template as outlined above to produce an approximation to the true transformation. The resultant approximated synthetic functional images were produced by applying the transformation computed for the substitute MRI to the simulated functional images. The locations of the transformed synthetic data were again determined as the centroids of the activated regions, and the error introduced by the approximation was computed as the Euclidean distance between corresponding points in the transformed images produced using the real transformation and the approximated transformation.
RESULTS
Figure 1 shows for one participant the locations of some of the artificial functional activation regions displayed on the MNI template brain after spatial normalisation with SPM99 using the participant's actual MRI (Fig. 1a) and the best matched MRI obtained from those of the other 27 participants (Fig. 1b). The MRI slice locations are the same in both Figures 1a and 1b, chosen to best display the simulated activation regions under the transformation obtained with the participant's own MRI. Note that in Figure 1b, obtained with the best matching MRI, at each location activity remains visible on the slice although the peaks are displaced out of the slice plane. To estimate the localization error arising from the approximation procedure, the distances were computed between the centroids of the synthetic activity for each target source obtained using the correct and the approximate spatial normalisations for each participant. Figure 2 shows the histogram of these distances for the eight artificial activation regions in all 28 MRIs. The modal distance between the centroids of corresponding points is 6 mm with 90% of cases falling below 12.5 mm. To demonstrate the effects of the approximation procedure on the results of an actual SAM source imaging experiment we examined the responses of six people in a verbal fluency task [Singh et al., 2002] in which participants were asked to silently name as many words as they could when prompted with a visually displayed initial letter cue. The results of the experiment (Fig. 3a), produced using the participants' MRIs and the normal processing procedure (see Materials and Methods) show widespread event related desynchronisation, with left language related activation clearly evident in the frequency range shown here (15–25 Hz). Figure 3b shows the results derived from the best approximation of the participants' MRIs from a pool of 27 MRIs of other people. The distribution of MEG activity in the estimated average response is broadly similar to that produced using the real MRIs. Under both transformations cortical desynchronisation is observed in the left hemisphere, with distinct activations in the left frontal and temporal regions. Local differences arising from the approximation are apparent and reflect the same order of displacement illustrated in Figures 1a and 1b for a single point source. For example, a posterior tempero‐parietal source is more strongly represented in the sagittal view of Figure 3b because under the approximate transformation the functional activity is rendered more superficially than it is under the correct transformation. Figure 2 shows the typical extent of the local perturbations that are induced by the approximation.
Figure 2.
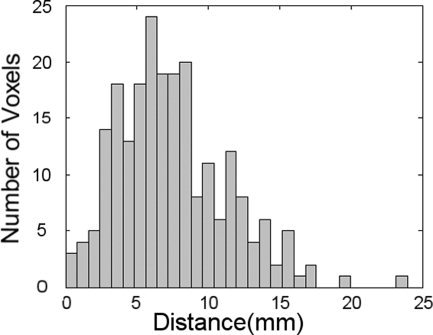
Histogram of the distribution of distances between the centroids of each of eight corresponding regions of simulated brain activation in each of two artificial functional images produced using 28 MRIs. For each MRI the synthetic activations were subject to two transformations to bring them into registration with the MNI template, the first using the participants MRI, and the second using the best matched head shape from the other 27 MRIs to approximate the correct transformation.
DISCUSSION
The results of substituting an MRI selected from a pool of 28 MRIs using the closest matching external head shape to estimate the distribution of ERS and ERD from MEG responses show that the results are broadly similar to those obtained when the real MRIs are used for the analysis (Fig. 3). The additional localisation error introduced by the procedure is of the same order as the grid used to reconstruct the MEG SAM source image (5 mm) and is similar to that reported by Carducci [ 2001]. As we apply MEG/MRI co‐registration methods in which millimetre accuracy in registration can be obtained, we suspect that the accuracy of the approximated group functional images approaches or even exceeds that in MEG studies using MRIs for each participant but which neglect a precise co‐registration methodology [Singh et al., 1997]. Note that in a very few cases more significant (>1.5 cm) errors resulted from applying the MRI estimation approach. This may be due to the limited surface matching accuracy possible with the relatively small (28) pool of MRIs used in this study. The use of a larger pool will no doubt reduce the incidence of large errors, and increase the accuracy of the procedure, but the question of the optimal pool size remains open and may be the subject of a future study.
The method advanced here is not exclusively linked to the SAM source imaging technique, but could be applied to any method yielding distributed cortical source information such as obtained using other beamformer methods [Sekihara et al., 1999], minimum current or minimum norm algorithms [Grave de Peralta‐Menendez and Gonzales‐Andino, 1998; Uutela et al., 1999]. The LORETA method of electrical source reconstruction (Pascual‐Marqui, et al., 1994) applies a similar strategy in that electrode locations defined in the 10–20 electrode placement system can be combined over a group of participants and displayed on a template MRI [Pizzagalli et al., 2000]. We also note that if the results of MEG source analysis using equivalent current dipole methods were expressed in terms of 3‐D Gaussian confidence volumes, calculated for example using a Monte‐Carlo method, these too could be combined across subjects in a similar manner. Furthermore, we anticipate that the method could be usefully applied to EEG‐derived source information. We do not suppose that the method is suitable for situations in which accurate anatomical information is required for a single participant, such as is needed for pre‐surgical evaluation for epilepsy, but in such cases patients' MRIs are invariably acquired. In all group functional imaging studies, including group fMRI studies, anatomical accuracy is limited by inter‐subject variation in cortical anatomy and the common co‐registration with a template brain. Furthermore we note that caution must be adopted in the application of source reconstruction methods to MEG data, such as SAM, because the accuracy of localisation depends heavily on factors such as the strength of the activity and its distance from the surface of the head. A recent study by Hillebrand and Barnes [ 2002] deals extensively with this issue.
We have shown that MEG can give information about the distribution of cortical activity in cognitive tasks, and in this context the approximation procedure advanced here can yield useful results about the approximate distribution of cortical activity where participants' MRIs are not available for an MEG study, perhaps because of ethical issues, or due to prohibitive costs in time and resources. The proposed method offers a reasonable and simply implemented alternative with which to investigate brain activity in a population of individuals. The method also suggests that the use of experimental designs in which large numbers of participants are required become practical. Improvements in the cost and availability of MRI technology may make strategies such as the one offered here redundant with time, but even so the viability of an MEG study could in any case be usefully assessed using the cortical approximation method before acquiring any MRIs. From the results of this study it seems reasonable to suppose that using the true MRIs in pooled MEG analysis will reveal only subtle differences not seen using approximated MRIs for each participant, and that much can be learned from MEG investigations using approximated anatomical information.
REFERENCES
- Carducci F, Babiloni C, Babiloni F, Cincotti F, Moretti D, Rizzuto M, Rossini PM (2001): Localization of cortical sites in subjects not having MRIs. Neuroimage 13: S91. [Google Scholar]
- Friston K, Holmes A, Worsley K, Poline J, Frith C, Frackowiak R (1995): Statistical parametric mapping in functional imaging: a general linear approach. Hum Brain Mapp 2: 189–210. [Google Scholar]
- Grave de Peralta‐Menendez R, Gonzales‐Andino SL (1998): A critical analysis of linear inverse solutions to the neuroelectromagnetic inverse problem. IEEE Trans Biomed Eng 45: 440–448. [DOI] [PubMed] [Google Scholar]
- Hillebrand A, Barnes GR (2002): A quantitative assessment of the sensitivity of whole‐head MEG to activity in the adult human cortex. Neuroimage 16: 638–650. [DOI] [PubMed] [Google Scholar]
- Kozinska D, Tretiak OJ, Nissanov J, Ozturk C (1997): Multidimensional alignment using the Euclidean distance transform. Graphical Models Image Process 59: 373–387. [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero‐Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B (2001): A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 356: 1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual‐Marqui RD, Michel CM, Lehmann D (1994): Low‐resolution electromagnetic tomography: a new method for localizing electrical activity in the brain. Int J Psychophysiol 18: 49‐65. [DOI] [PubMed] [Google Scholar]
- Pizzagalli D, Lehmann D, Koenig T, Regard M, Pascual‐Marqui RD (2000): Face‐elicited ERPs and affective attitude: brain electric microstate and tomography analyses. Clin Neurophysiol 111: 521–531. [DOI] [PubMed] [Google Scholar]
- Robinson SE, Vrba J (1999): Functional neuroimaging by synthetic aperture magnetometry (SAM) In: Recent advances in biomagnetism. Sendai: Tohoku University Press; p 302–305. [Google Scholar]
- Sekihara K, Poeppel D, Marantz A, Koizumi H, Miyashita Y (1999): MEG spatio‐temporal analysis using a covariance matrix calculated from nonaveraged multiple‐epoch data. IEEE Trans Biomed Eng 46: 515–521. [DOI] [PubMed] [Google Scholar]
- Singh KD, Barnes GR, Hillebrand A, Forde EME, Williams A (2002): Task‐related changes in cortical synchronisation are spatially coincident with the haemodynamic response. Neuroimage 16: 103–114. [DOI] [PubMed] [Google Scholar]
- Singh KD, Furlong P, Holliday I, Harding G (1997): Evaluation of MRI‐MEG/EEG co‐registration strategies using Monte‐Carlo simulation. Electroencephalogr Clin Neurophysiol 102: 81–85. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P (1988): Stereotactic coplanar atlas of the human brain. New York: Thieme Medical. [Google Scholar]
- Thompson PM, Schwartz C, Lin RT, Khan AA, Toga AW (1996): Three‐dimensional statistical analysis of sulcal variability in the human brain. J Neurosci 16: 4261–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Thompson PM (2001): Maps of the brain. Anat Rec 265: 37–53. [DOI] [PubMed] [Google Scholar]
- Uutela K, Hamalainen M, Somersalo E (1999): Visualization of magnetoencephalographic data using minimum current estimates. Neuroimage 10: 173–180. [DOI] [PubMed] [Google Scholar]
- van't Ent D, de Munck JC, Kaas AL (2001): A fast method to derive realistic BEM models for E/MEG source reconstruction. IEEE Trans Biomed Eng 48: 1434–1443. [DOI] [PubMed] [Google Scholar]
- van Veen BD, van Drongelen W, Yuchtman M, Suzuki A (1997): Localization of brain electrical activity via linearly constrained minimum variance spatial filtering. IEEE Trans Biomed Eng 44: 867–880. [DOI] [PubMed] [Google Scholar]


