Abstract
PURPOSE
Adenocarcinoma is the most common histologic subtype of non–small-cell lung cancer, representing 40% of all diagnoses. Several biomarkers are currently used to determine patient eligibility for targeted treatments, including analysis of molecular alterations in EGFR and ALK, as well as programmed death-ligand 1 (PD-L1) protein expression. Epidemiologic data reporting the frequency of these biomarkers in Brazilian patients with lung adenocarcinoma (LUAD) are limited, and existing studies predominantly included patients from the southeast region of the country.
MATERIALS AND METHODS
The goal of this study was to investigate the frequency of somatic mutations in the EGFR, KRAS, NRAS, and BRAF genes, ALK, and PD-L1 expression in a series of Brazilian patients diagnosed with LUAD predominantly recruited from centers in southern Brazil. Molecular analysis of the EGFR, KRAS, NRAS, and BRAF genes was performed by next-generation sequencing using DNA extracted from tumor tissue. Immunohistochemistry was used to detect ALK and PD-L1 expression.
RESULTS
Analysis of 619 tumors identified KRAS mutations in 189 (30.2%), EGFR mutations in 120 (19.16%), and BRAF mutations in 19 (3%). Immunohistochemistry demonstrated ALK and PD-L1 expression in 4% and 35.1% of patients, respectively.
CONCLUSION
To our knowledge, this is the first study investigating the molecular epidemiology of patients with LUAD from southern Brazil and the largest assessing the frequency of multiple predictive biomarkers for this tumor in the country. The study also reveals a distinct mutation profile compared with data originating from other regions of Brazil.
INTRODUCTION
Lung cancer (LC) is the leading cause of cancer mortality worldwide and responsible for 1.7 million deaths every year.1 In Brazil, the National Cancer Institute estimated there would be 31,270 new patients with LC from 2018 to 2019, accounting for the second most common tumor type in the country. It is the leading cause of deaths among men, ahead of prostate cancer, and the second leading cause among women, only behind breast cancer. In southern Brazil in 2018, 5,350 and 3,110 new cases were estimated in men and women, respectively, which makes LC the third most frequent cancer in the region.2
Non–small-cell LC (NSCLC) accounts for approximately 85% of pulmonary neoplasm diagnoses.3,4 Effective treatments remain scarce, considering that the 5-year survival rate does not reach 20%, even in countries such as the United States.5 In Brazil, this number is even lower, estimated at 16%.6
The use of predictive biomarkers allows therapeutic decisions to be based on tumor molecular profile.7 For instance, certain somatic changes in the EGFR, ALK, ERBB2, and BRAF genes are substantial targets for tyrosine kinase inhibitors (TKIs).8 In addition, new treatments for NSCLC using immune checkpoint inhibitors have recently been approved.9 Its prescription depends on the expression of certain biomarkers on the tumor cell surface, such as the programmed death-ligand 1 (PD-L1) protein, a molecule in which the binding to its programmed death-1 receptor on T cells allows immune escape and tumor cell proliferation. The use of anti–programmed death-1/PD-L1 drugs blocks such binding and reactivates the patient's immune response.10
Although the molecular profile of predictive biomarkers in LUAD is already well documented in Europe, the United States, and some regions of Asia, there are few studies exploring these data in Latin America. In Brazil, only a few reports have been published since 2012, and all were essentially restricted to the southeast region. Thus, these data may not be representative of all regions in Brazil, given the differences in ancestry according to regions.11,12
On the basis of this information, the main goal of this study was to investigate the frequency of somatic alterations in EGFR, KRAS, NRAS, and BRAF genes by next-generation sequencing (NGS), as well as ALK and PD-L1 expression in a series of Brazilian patients diagnosed with LUAD. To our knowledge, this is the first study to include a large number of patients who were tested by a biomarker panel in southern Brazil. These results might be important for new public policies in the treatment of LUAD.
MATERIAL AND METHODS
Study Population
This was a retrospective study conducted by the Precision Medicine Program of the Hospital de Clínicas de Porto Alegre (HCPA) in Brazil, which enrolled a case series of patients with LUAD who underwent molecular testing from September 2016 to January 2019.
Samples from 619 individuals were obtained from different hospitals and clinics distributed in 22 centers located in the three states of the southern region of Brazil: Rio Grande do Sul (N = 516), Santa Catarina (N = 24), and Paraná (N = 74). The five remaining patients were obtained from Rio de Janeiro. All included patients had confirmed adenocarcinoma histology. The diagnostic slides and formalin‐fixed, paraffin‐embedded tissue blocks were retrieved and reviewed by pathologists with expertise in LC. This project was approved by the HCPA Research Ethics Committee (No. 18-0099) and registered under the Certificate of Presentation for Ethical Appreciation (No. 83557418.5.0000.5327).
Tumor Selection and DNA Extraction
For all patients, 10-µm thick sections representative of the tumor tissue were cut, and regions with a higher percentage of tumor cells were selected for DNA extraction. DNA from the tissue samples was extracted using the ReliaPrep FFPE gDNA Miniprep System (Promega, Madison, WI) according to the manufacturer's recommendations. After extraction, the DNA samples were quantified using the fluorescence method (Qubit 2.0 Fluorometer, Invitrogen, Carlsbad, CA), which provides an accurate DNA quantification.
Of the 799 NSCLC samples that were received by the Precision Medicine Program of the HCPA, 619 (77.4%) were considered suitable for NGS analysis on the basis of tumor cell content, DNA amount, and purity.
Molecular Analysis by NGS
Molecular analysis of the EGFR, KRAS, NRAS, and BRAF genes was performed with an NGS platform (Ion Torrent PGM, server version 5.0; ThermoFisher Scientific, Waltham, MA). We used an AmpliSeqTM customized panel (ThermoFisher Scientific) to identify mutations in the EGFR gene (exons 18 to 21), KRAS (exons 2 and 3), NRAS (exons 2 and 3), and BRAF (exons 11 and 15). Data were analyzed on the bioinformatics platform Ion Torrent Suite and Ion Reporter version 5.0 with a minimum depth of 500x. Sequences NM_005228.3 (EGFR), NM_0033360.3 (KRAS), MM_002524.3 (NRAS), and NM_004333.4 (BRAF) were used as references. The test was performed using research-use-only reagents with internal validation.
PD-L1 and ALK Analysis
For PD-L1 analysis, which included 202 patients, the immunohistochemistry (IHC) expression scores were based on the Interpretation Guide for Window PD-L1 (SP263) Assay Staining of NSCLC 2017 (Roche, Basel, Switzerland). The analysis considers the overall percentage of positive neoplastic cells in the sample of any intensity above the eventual observed background staining in the negative control slide showing a cytoplasmic membrane pattern (circumferential, discontinuous, or basolateral). A negative control slide with an antibody provided in the PD-L1 kit was used for all patients. A tissue sample with PD-L1 positivity was also used as a positive control. Positivity in immune cells was not considered for determining scores in the Window PD-L1 test.
Similarly, for the ALK assay, paraffin sections of the lesions from 350 patients were submitted to the IHC technique. Tissue sections were stained with the ALK antibody clone D5F3 (Roche), which is able to recognize ALK fusion proteins and EML4-ALK variant expression. The slides were analyzed using the Ventana BenchMark XT Automated System (Roche). All reactions had negative and positive controls on the blade itself.
Statistical Analysis
To verify any possible association of the predictive biomarkers with gender, age, and whether there was any statistically significant difference between our study and others performed in the Brazilian population, we performed χ2 tests. The results were considered statistically significant when the P was < .05.
RESULTS
NGS Analysis of EGFR, KRAS, NRAS, and BRAF Genes
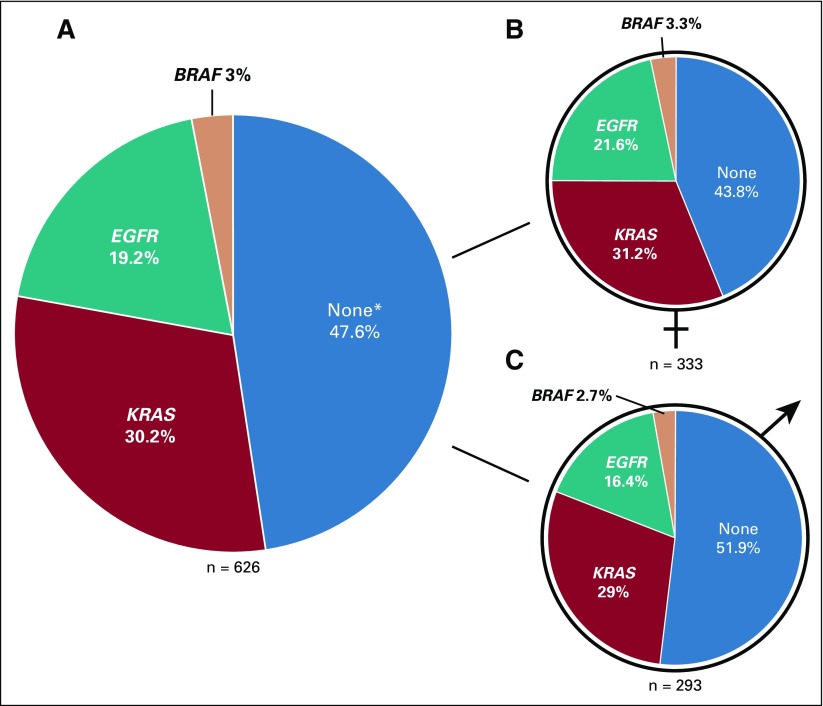
NGS results of 619 tumors revealed 189 patients (30.2%) with mutations in KRAS, 120 (19.16%) with mutations in EGFR, and 19 (3%) with mutations in BRAF. No NRAS mutations were identified, and seven patients harbored double mutations. In 298 patients (47.6%), we did not detect any alteration using our NGS panel (Fig 1).
FIG 1.
Frequency of somatic mutations in EGFR, KRAS, and BRAF in lung adenocarcinoma tumors from patients in southern Brazil. (A) Female and male patients, (B) only female patients, and (C) only male patients. (*) Not detected using our next-generation sequencing panel. It does not exclude the presence of alterations in other driver genes.
As expected, the frequency of EGFR mutations was 5.3% higher in women compared with men; however, this difference was not statistically significant (P = .096). In contrast, the frequency of patients with no mutations detected by our panel was higher in men, with a statistically significant difference (P = .044; Fig 1B and 1C; Data Supplement).
Among patients with KRAS mutations, p.(Gly12Cys) was the most frequent, accounting for 37% of the mutations found in this gene. This alteration was followed by p.(Gly12Val) and p.(Gly12Asp), found in 43 (22.75%) and 30 (15.9%) patients, respectively. Another 29 mutations were registered in exon 2 (15.3%). KRAS mutations in codon 61 of exon 3 were less frequently found in 17 patients (8.9%; Data Supplement).
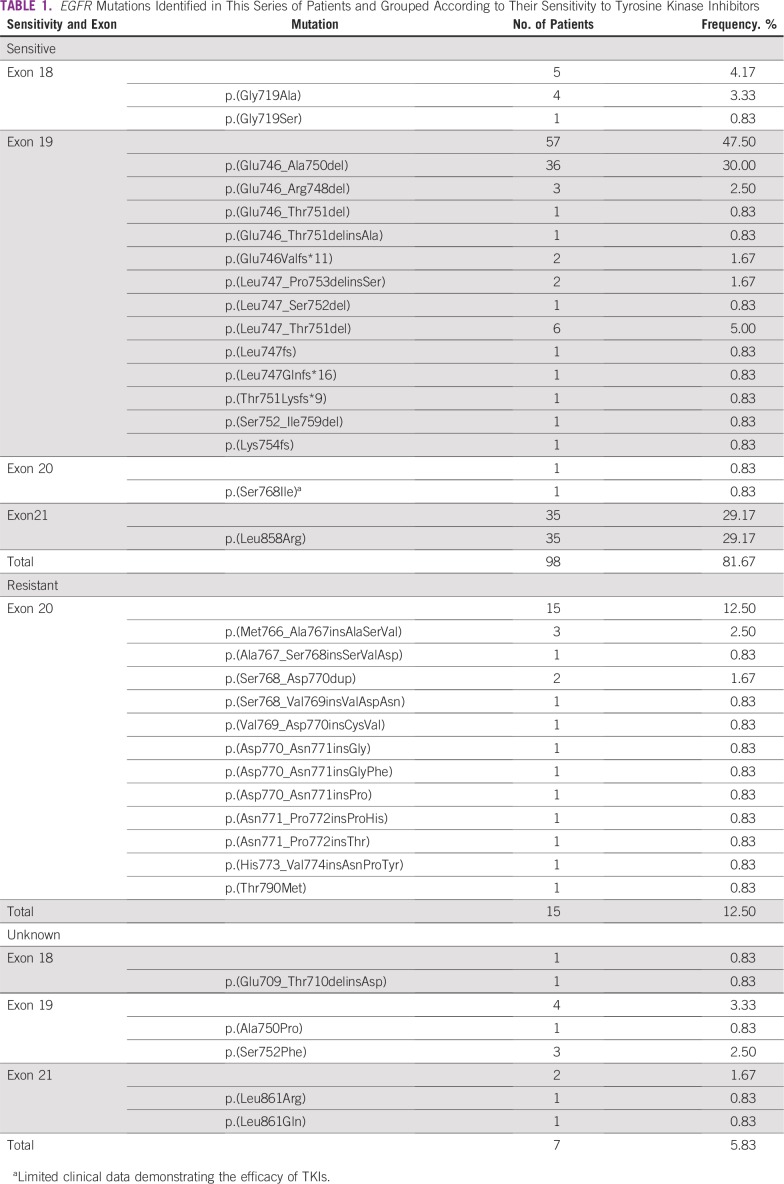
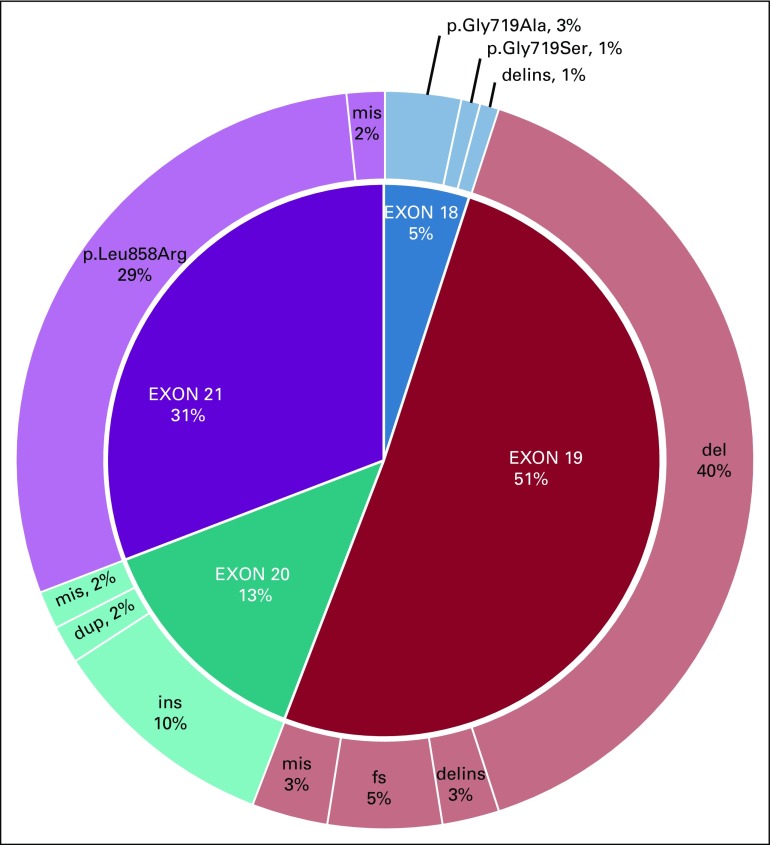
Regarding the effect of EGFR mutations on TKI response, the majority was classified as sensitive (n = 98; 81.67%). Most of the mutations were located in exon 19 (n = 57; 47.50%), including the common deletion p.(Glu746_Ala750del; n = 36; 30.00%), followed by the point mutation p.(Leu858Arg; n = 35; 29.17%) in exon 21. Resistance alterations were restricted to exon 20 (n = 15; 12.50%). We also reported seven (5.83%) missense mutations of unknown clinical significance, including four in exon 19 and two in exon 21, and one complex mutation p.(Glu709_Thr710delinsAsp) in exon 18. In exon 20, there were 12 (10%) in-frame insertions and two (1.7%) duplications. Interestingly, one patient with no progression of disease was diagnosed with the p.(Thr790Met) mutation at a low frequency (3.2% of 883 sequence reads), which might be a case of primary resistance to first- and second-generation TKIs (Table 1; Fig 2).
TABLE 1.
EGFR Mutations Identified in This Series of Patients and Grouped According to Their Sensitivity to Tyrosine Kinase Inhibitors
FIG 2.
EGFR mutation frequencies according to their location in the gene. Del, deletions; delins, insertions/deletions; dup, duplications; fs, frameshifts; ins, insertions; mis, missense mutations.
Finally, among the 19 BRAF mutations, 13 (68.42%) were the well-known p.(Val600Glu) in exon 15. Another four mutations were located in exon 15, and two were located in exon 11, which also belongs to the kinase domain of the BRAF protein.
Although positive results of all predictive biomarkers were more commonly found in elderly patients (≥ 60 years of age), only KRAS had a statistically significant difference (P = .044; Data Supplement). The mean age at diagnosis of patients with KRAS, EGFR, and BRAF alterations was 67.5, 66.6, and 61.0 years, respectively.
PD-L1 and ALK IHC Analysis
PD-L1 staining by IHC was positive in 71 of the 202 analyzed tumors (35.1%). Approximately one fifth of tumors (21.3%) had moderate expression, with positive staining noted between 1% and fewer than 50% of the cells. High expression of PD-L1, which is indicated by positive staining in 50% or more of the cells, was observed in 28 tumors (13.8%). ALK staining by IHC was positive in 14 of the 350 patients analyzed (4%), most of them identified in women (78.6%).
DISCUSSION
Although the prevalence of predictive biomarkers for molecular-targeted treatment in NSCLC has been established in several continents, including Asia, Europe, and North America, only a few studies have been conducted in Latin America. Furthermore, most of the studies characterizing samples from patients with LC in Brazil have recruited patients from the southeast, which limits the true understanding of the scenario in the entire country. In accordance with this assumption, in our study, we identified a particular frequency pattern of predictive biomarkers compared with that observed in other Brazilian regions.
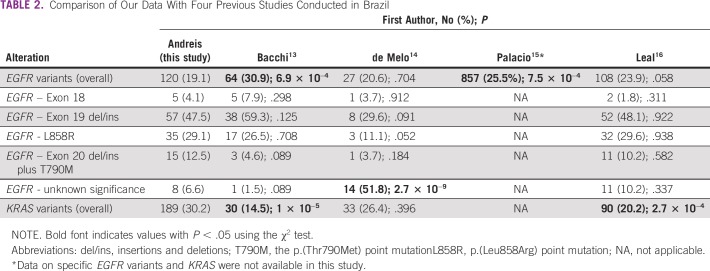
To our knowledge, the frequency of EGFR mutations (19.16%) was the lowest ever reported when considering those observed in four previous studies performed in Brazil (Table 2; Data Supplement).13-16 In agreement with this observation, it is expected that the proportion of EGFR-mutated LUAD would decrease as the proportion of European ancestry increases in a given population. Several studies conducted in Europe showed a frequency of EGFR mutation of approximately 10%.17-19
TABLE 2.
Comparison of Our Data With Four Previous Studies Conducted in Brazil
In line with these observations, the prevalence of European ancestry in southern Brazil has been estimated at 80% to 90%, the highest among all Brazilian regions. African and Amerindian populations have a lower but no less important contribution.11,12 Studies comparing the human leukocyte antigen allelic diversity in the region also confirmed a high similarity to Europeans and a significant difference from Asians or even Brazilian indigenous populations.20-23 Together, this information supports the reason why the frequency of EGFR mutations in southern Brazil is lower than other regions of the country, but still higher than in Europe.
Considering the work of Bacchi et al,13 which showed EGFR mutation in 30.9% of patients, it is important to note that the majority of the patients were from southeast and northeast Brazil. A recent and larger study performed in southeast Brazil also found a higher frequency of mutations in the gene (23.9%), but this difference was not statistically significant compared with our data (P = .058). Although European ancestry was predominant in this series of patients, a relatively high proportion of them had Asian ancestry (7.3%),13 a population in which the frequency of EGFR mutations may reach 60%.16,24 Although de Melo et al14 found a percentage of patients with EGFR-mutated disease similar to ours, more than half (51.8%) were variants of unknown therapeutic impact compared with only 6.6% in our study (P = 2.7 × 10−9). Finally, the largest published study including data from EGFR molecular testing in Brazil (N = 3,364) also identified a higher and statistically significant mutation frequency (25.5%; P = 7.5 × 10−4; Table 2; Data Supplement).15 Interestingly, a study conducted in Uruguay, a country geographically close to the southern region of Brazil, showed similar results regarding EGFR mutations (18.3%). The proportion of actionable alterations in exon 19 was similar (48.7% v 47.5%), but the missense substitution p.(Leu858Arg) in exon 21 was lower (22% v 29.17%; χ2,1.025; P = .311). Our proportion of resistance mutations in exon 20 was also slightly higher (8.5% v 12.5%; χ2; 0.646; P = .422).25
Previous reports showed a statistically significant association between female patients and a higher prevalence of EGFR mutations,14,16,17,26 which was not observed in our participants (Data Supplement). In addition, we reported six rare EGFR mutations of unknown therapeutic impact: p.(Glu709_Thr710delinsAsp), p.(Ala750Pro), p.(Ser752Phe), p.(Leu861Arg), and p.(Leu861Gln), and p.(Ser768Ile). The efficacy of TKIs on tumors harboring these mutations is not well established, but some case reports suggest a benefit from treatment with first-generation TKIs.27-32
We also found EGFR alterations co-occurring in the same tumor in four patients. Two had the p.(Gly719Ala) coexisting with the p.(Ser768Ile) and the p.(Leu861Gln). Two other patients were carriers of two sensitive mutations in exon 19—p.(Glu746Valfs*11) plus p.(Lys754fs) and p.(Leu747Glnfs*16) plus p.(Thr751Lysfs*9). Another three patients with coexisting resistance mutations were also reported. One had the EGFR alteration in exon 20 p.(Asp770_Asn771insGly) plus BRAF p.(Val600Glu), another had a double mutation in KRAS (p.Gly13Cys plus p.Gln61His), and one had the KRAS p.(Gly12Cys) plus the BRAF p.(Asp594Asn). Only p.(Gly719Ala) plus p.(Ser768Ile) was already reported in a Brazilian patient.14 Our data are in accordance with other studies conducted in Brazil, which revealed that all EGFR and KRAS alterations were mutually exclusive.13,14,16,33
Regarding KRAS alterations, some studies reported mutations in tumors diagnosed at more advanced ages,26,34 which were confirmed in our participants (Data Supplement). The general frequency of such alterations identified in our study is notable (30.2%) and significantly higher than that observed in two previous Brazilian cohorts (Table 2; Data Supplement).13,16 However, it is important to note that these studies used Sanger sequencing for the molecular analysis, a method that requires the presence of the mutant allele in at least 15% to 20% of tumor DNA. For comparison purposes, if we had excluded the positive patients with a mutant allele frequency below 15% and 20%, our KRAS mutation percentage would decrease from 30.2% to 18.21% and 15.49%, respectively. For EGFR mutations, the frequencies in these two scenarios also decreased, ranging from 14.85% to 13.57% when excluding samples with mutant allele frequencies less than 15% and less than 20%, respectively. These numbers are closer to the previous studies cited earlier and could indicate that our NGS panel is more sensitive in detecting these driver mutations in LUAD. In addition, actionable mutations would have been missed in our study if we used a cutoff of allele frequency at 15%, including seven sensitive deletions in exon 19, four p.(Leu858Arg) substitutions in exon 21 and five resistance mutations, four insertions in exon 20, and one p.(Thr790Met) point mutation.
We next focused on biomarkers with a lower mutation prevalence. To our knowledge, our study is the first to report BRAF mutations in Brazilian patients with LUAD, found in 3% of the participants. This prevalence is in agreement with results in non-Latin populations.33,35,36 The European Medicines Agency and US Food and Drug Administration recently approved the combined use of dabrafenib and trametinib for the treatment of patients with NSCLC harboring the BRAF p.(Val600Glu) mutation.37
In Brazil, the only study to assess ALK expression was performed in the northeast region.38 The authors found a much higher prevalence of ALK-positive tumors (13.3% compared with 4% in our study).38 A recent report that included samples from nine countries in Latin America found that the prevalence of ALK rearrangements ranged from 4.1% (Colombia) to 10.8% (Peru). Argentina and Uruguay, which are more geographically close to the southern region of Brazil, had a frequency of 5.4% and 4.4%,39 respectively, which is similar to our findings. Co-occurrence of ALK rearrangements with EGFR or KRAS mutations is rare, with only a few reports in the literature.26,40-46 We found two individuals with positive IHC for ALK protein who also harbored the KRAS variants p.(Gly12Val) and p.(Gly12Asp). Ulivi et al47 reported that patients with EML4-ALK translocations co-occurring with KRAS point mutations had decreased responsiveness to crizotinib.
Alves da Silva et al38 evaluated the PD-L1 expression in LC patients from the northeast region of Brazil and observed a similar proportion of positivity (40.5% v 35.1% in our study). The prevalence of tumors scoring from 1% to 50% or less and 50% or more was also similar between both studies (24% v 21.2% and 16.5% v 13.8%, respectively). These data reveal a much lower proportion of tumors with a staining score of 50% or more, due to a larger case series that had found high PD-L1 expression in 30.2% of the patients.48
Our study has some limitations. Because of its retrospective approach and the use of data from different private and public medical centers, we had to anonymize patients’ information, including clinical data on treatment response, survival, and smoking status. However, we believe that this fact does not compromise the relevance of our results since the association between biomarkers and these clinical aspects has been widely studied.
In summary, to our knowledge, this is the first and largest study assessing the frequency of multiple predictive biomarkers for LUAD in Brazil. In addition, it reveals a unique pattern of mutation frequencies in different genes compared with data originating from other regions of the country. The frequency of EGFR mutations is the lowest found in Brazilian patients, possibly reflecting a higher proportion of individuals with European ancestry.11,12 Our results also underscore the need to expand LUAD molecular testing in the Brazilian public health system, given that approximately 15% of patients with LUAD from the southern region would benefit from the use of TKIs. Araujo et al49 estimated that fewer than half of the patients have their tumors submitted for molecular testing in Brazil. This number can be even lower in public health care institutions.49 We expect that these data, together with results from other studies, will help to change this scenario and accelerate the implementation of new public policies for the treatment of LC in the country, on the basis of cost-effective analysis of our population.
ACKNOWLEDGMENT
We thank the undergraduate students Julia Tsao Schein, from the Medicine School of Universidade Federal do Rio Grande do Sul, and Guilherme Danielski Viola, from the Institute of Biosciences of Universidade Federal do Rio Grande do Sul, for their collaboration.
Footnotes
Presented in part at Next Frontiers to Cure Cancer, São Paulo, Brazil, May 10 to 12, 2018; and International Congress of Genetics, Foz do Iguaçu, Brazil, September 10 to 14, 2018.
Supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico and Research Incentive Fund of Hospital de Clínicas de Porto Alegre (Grant no. 2018-0099). PhD scholarship awarded to T.F.A. (Grant no. 140361/2018-9). All results presented here were performed by the Precision Medicine Program of Hospital de Clínicas de Porto Alegre, a diagnostic program that provides clinical services to AstraZeneca Brazil, Bristol-Myers Squibb, and Pfizer. Although the diagnostic tests were paid by these companies, none of them participated in or interfered with the design of the study, result analyses, and conclusions.
AUTHOR CONTRIBUTIONS
Conception and design: Tiago F. Andreis, Marina Siebert, Sandra Leistner-Segal, Vinícius Lorandi, Patricia Ashton-Prolla, Gabriel S. Macedo
Administrative support: Jane M. Ulbrich
Provision of study materials or patients: Sandra Leistner-Segal, Jane M. Ulbrich, Luis F.R. Rivero, Patricia Ashton-Prolla
Collection and assembly of data: Tiago F. Andreis, Bruno S. Correa, Fernanda De-Paris, Marina Siebert, Jane M. Ulbrich, Luis F.R. Rivero, Francine H. De Oliveira, Patricia Ashton-Prolla, Gabriel S. Macedo
Data analysis and interpretation: Tiago F. Andreis, Fernanda S. Vianna, Sandra Leistner-Segal, Jane M. Ulbrich, Luis F.R. Rivero, Vinícius Lorandi, Patricia Ashton-Prolla, Gabriel S. Macedo
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jgo/site/misc/authors.html.
Vinícius Lorandi
Travel, Accommodations, Expenses: Astellas Pharma, MSD Oncology, Novartis, AstraZeneca, Genentech
Patricia Ashton-Prolla
Research Funding: AstraZeneca Brazil (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brasil: Estimativa 2018: Incidência de Câncer no Brasil. Rio de Janeiro - RJ, Brazil, Ministério da Saúde, Instituto Nacional de Câncer José Alencar Gomes da Silva (INCA), Coordenação de Prevenção e Vigilância, 2017 pp. 128.
- 3.Lortet-Tieulent J, Soerjomataram I, Ferlay J, et al. International trends in lung cancer incidence by histological subtype: Adenocarcinoma stabilizing in men but still increasing in women. Lung Cancer. 2014;84:13–22. doi: 10.1016/j.lungcan.2014.01.009. https://www.inca.gov.br/sites/ufu.sti.inca.local/files//media/document//estimativa-incidencia-de-cancer-no-brasil-2018.pdf. [DOI] [PubMed] [Google Scholar]
- 4.Pirker R, Filipits M. Personalized treatment of advanced non-small-cell lung cancer in routine clinical practice. Cancer Metastasis Rev. 2016;35:141–150. doi: 10.1007/s10555-016-9612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. SEER: Cancer Stat Facts: Lung and Bronchus Cancer 5-year survival rates (2009-2015), in Surveillance E, and End Results (SEER) Program, National Cancer Institute, DCCPS, Surveillance Research Program (ed), 2019. https://seer.cancer.gov/statfacts/html/lungb.html.
- 6. Polato CPB, Bonfante MDS, Andrae IG, et al: Análise de sobrevida em pacientes com câncer de pulmão tratados no Sistema Único de Saúde no Brasil entre 2002 e 2003 [in Portuguese]. Cad Saude Colet 21:173-181, 2013. [Google Scholar]
- 7.Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21:2213–2220. doi: 10.1158/1078-0432.CCR-14-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 9.Rizvi NA, Mazières J, Planchard D, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): A phase 2, single-arm trial. Lancet Oncol. 2015;16:257–265. doi: 10.1016/S1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Linares A, Adhikari K, Acuña-Alonzo V, et al. Admixture in Latin America: Geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10:e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chacón-Duque JC, Adhikari K, Fuentes-Guajardo M, et al. Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat Commun. 2018;9:5388. doi: 10.1038/s41467-018-07748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacchi CE, Ciol H, Queiroga EM, et al. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics (São Paulo) 2012;67:419–424. doi: 10.6061/clinics/2012(05)03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Melo AC, Karen de Sá V, Sternberg C, et al. Mutational profile and new IASLC/ATS/ERS classification provide additional prognostic information about lung adenocarcinoma: A study of 125 patients from Brazil. Oncology. 2015;89:175–186. doi: 10.1159/000376552. [DOI] [PubMed] [Google Scholar]
- 15.Palacio S, Pontes L, Prado E, et al. EGFR mutation testing: Changing patterns of molecular testing in Brazil. Oncologist. 2019;24:e137–e141. doi: 10.1634/theoncologist.2018-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leal LF, de Paula FE, De Marchi P, et al. Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci Rep. 2019;9:3209. doi: 10.1038/s41598-019-39965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gahr S, Stoehr R, Geissinger E, et al. EGFR mutational status in a large series of Caucasian European NSCLC patients: Data from daily practice. Br J Cancer. 2013;109:1821–1828. doi: 10.1038/bjc.2013.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemée F, Bergoglio V, Fernandez-Vidal A, et al. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci USA. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits AJJ, Kummer JA, Hinrichs JWJ, et al. EGFR and KRAS mutations in lung carcinomas in the Dutch population: Increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell Oncol (Dordr) 2012;35:189–196. doi: 10.1007/s13402-012-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro MS, Issler HC, Gelmini GF, et al. High-resolution characterization of 12 classical and non-classical HLA loci in southern Brazilians. HLA. 2019;93:80–88. doi: 10.1111/tan.13488. [DOI] [PubMed] [Google Scholar]
- 21.Reis PG, Ambrosio-Albuquerque EP, Fabreti-Oliveira RA, et al. HLA-A, -B, -DRB1, -DQA1, and -DQB1 profile in a population from southern Brazil. HLA. 2018;92:298–303. doi: 10.1111/tan.13368. [DOI] [PubMed] [Google Scholar]
- 22.Boquett J, Schüler-Faccini L, Jobim LF, et al. Self-assessment of color categories and its relationship with HLA profiling in Brazilian bone marrow donors. Biol Blood Marrow Transplant. 2015;21:1140–1144. doi: 10.1016/j.bbmt.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31:184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berois N, Touya D, Ubillos L, et al. Prevalence of EGFR mutations in lung cancer in Uruguayan population. J Cancer Epidemiol. 2017;2017:6170290. doi: 10.1155/2017/6170290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee B, Lee T, Lee SH, et al. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget. 2016;7:23874–23884. doi: 10.18632/oncotarget.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JY, Yu CJ, Chang YC, et al. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 28.Wu SG, Chang YL, Lin JW, et al. Including total EGFR staining in scoring improves EGFR mutations detection by mutation-specific antibodies and EGFR TKIs response prediction. PLoS One. 2011;6:e23303. doi: 10.1371/journal.pone.0023303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ackerman A, Goldstein MA, Kobayashi S, et al. EGFR delE709_T710insD: A rare but potentially EGFR inhibitor responsive mutation in non-small-cell lung cancer. J Thorac Oncol. 2012;7:e19–e20. doi: 10.1097/JTO.0b013e3182635ab4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi Y, Togashi Y, Yatabe Y, et al. EGFR exon 18 mutations in lung cancer: Molecular predictors of augmented sensitivity to afatinib or neratinib as compared with first- or third-generation TKIs. Clin Cancer Res. 2015;21:5305–5313. doi: 10.1158/1078-0432.CCR-15-1046. [DOI] [PubMed] [Google Scholar]
- 31.Chiu CH, Yang CT, Shih JY, et al. Epidermal growth factor receptor tyrosine kinase inhibitor treatment response in advanced lung adenocarcinomas with G719X/L861Q/S768I mutations. J Thorac Oncol. 2015;10:793–799. doi: 10.1097/JTO.0000000000000504. [DOI] [PubMed] [Google Scholar]
- 32.Leventakos K, Kipp BR, Rumilla KM, et al. S768I mutation in EGFR in patients with lung cancer. J Thorac Oncol. 2016;11:1798–1801. doi: 10.1016/j.jtho.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carneiro JG, Couto PG, Bastos-Rodrigues L, et al. Spectrum of somatic EGFR, KRAS, BRAF, PTEN mutations and TTF-1 expression in Brazilian lung cancer patients. Genet Res. 2014;96:e002. doi: 10.1017/S0016672314000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dacic S, Shuai Y, Yousem S, et al. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol. 2010;23:159–168. doi: 10.1038/modpathol.2009.154. [DOI] [PubMed] [Google Scholar]
- 35.Brustugun OT, Khattak AM, Trømborg AK, et al. BRAF-mutations in non-small cell lung cancer. Lung Cancer. 2014;84:36–38. doi: 10.1016/j.lungcan.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 37.Khunger A, Khunger M, Velcheti V. Dabrafenib in combination with trametinib in the treatment of patients with BRAF V600-positive advanced or metastatic non-small cell lung cancer: Clinical evidence and experience. Ther Adv Respir Dis. 2018;12:1753466618767611. doi: 10.1177/1753466618767611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alves da Silva AV, Martins Neto F. The frequency of high PD-L1 expression is low in lung adenocarcinoma patients from northeast Brazil. Surg Exp Pathol. 2019;2:4. [Google Scholar]
- 39.Arrieta O, Cardona AF, Bramuglia G, et al. Molecular epidemiology of ALK rearrangements in advanced lung adenocarcinoma in Latin America. Oncology. 2019;96:207–216. doi: 10.1159/000493733. [DOI] [PubMed] [Google Scholar]
- 40.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo YW, Wu SG, Ho CC, et al. Good response to gefitinib in lung adenocarcinoma harboring coexisting EML4-ALK fusion gene and EGFR mutation. J Thorac Oncol. 2010;5:2039–2040. doi: 10.1097/JTO.0b013e3181f43274. [DOI] [PubMed] [Google Scholar]
- 43.Rossing HH, Grauslund M, Urbanska EM, et al. Concomitant occurrence of EGFR (epidermal growth factor receptor) and KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog) mutations in an ALK (anaplastic lymphoma kinase)-positive lung adenocarcinoma patient with acquired resistance to crizotinib: A case report. BMC Res Notes. 2013;6:489. doi: 10.1186/1756-0500-6-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jürgens J, Engel-Riedel W, Prickartz A, et al. Combined point mutation in KRAS or EGFR genes and EML4-ALK translocation in lung cancer patients. Future Oncol. 2014;10:529–532. doi: 10.2217/fon.13.194. [DOI] [PubMed] [Google Scholar]
- 45.Yang JJ, Zhang XC, Su J, et al. Lung cancers with concomitant EGFR mutations and ALK rearrangements: Diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res. 2014;20:1383–1392. doi: 10.1158/1078-0432.CCR-13-0699. [DOI] [PubMed] [Google Scholar]
- 46.Lee JK, Kim TM, Koh Y, et al. Differential sensitivities to tyrosine kinase inhibitors in NSCLC harboring EGFR mutation and ALK translocation. Lung Cancer. 2012;77:460–463. doi: 10.1016/j.lungcan.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Ulivi P, Chiadini E, Dazzi C, et al. Nonsquamous, non-small-cell lung cancer patients who carry a double mutation of EGFR, EML4-ALK or KRAS: Frequency, clinical-pathological characteristics, and response to therapy. Clin Lung Cancer. 2016;17:384–390. doi: 10.1016/j.cllc.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 49.Araujo LH, Baldotto C, Castro G, Jr, et al. Lung cancer in Brazil. J Bras Pneumol. 2018;44:55–64. doi: 10.1590/S1806-37562017000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]