Iron and manganese play important roles in how cell’s cope with oxygen stress. However, how these metals affect the ability of cells to replicate after oxidative challenges is not known. Here, we show that replication in Escherichia coli is inhibited following a challenge with hydrogen peroxide and requires manganese for the rapid recovery of DNA synthesis. The manganese-dependent recovery of DNA synthesis occurs independently of lesion repair and modestly improves survival, but it also increases the mutation rate in cells. The results imply that replication in E. coli is likely to utilize an iron-dependent enzyme(s) that becomes oxidized and inactivated during oxidative challenges. We propose that manganese remetallates these or alternative enzymes to allow genomic DNA replication to resume, although with reduced fidelity.
KEYWORDS: DNA repair, DNA replication, manganese, oxygen toxicity
ABSTRACT
Divalent metals such as iron and manganese play an important role in the cellular response to oxidative challenges and are required as cofactors by many enzymes. However, how these metals affect replication after oxidative challenge is not known. Here, we show that replication in Escherichia coli is inhibited following a challenge with hydrogen peroxide and requires manganese for the rapid recovery of DNA synthesis. We show that the manganese-dependent recovery of DNA synthesis occurs independent of lesion repair, modestly improves cell survival, and is associated with elevated rates of mutagenesis. The Mn-dependent mutagenesis involves both replicative and translesion polymerases and requires prior disruption by H2O2 to occur. Taking these findings together, we propose that replication in E. coli is likely to utilize an iron-dependent enzyme(s) that becomes oxidized and inactivated during oxidative challenges. The data suggest that manganese remetallates these or alternative enzymes to allow genomic DNA replication to resume, although with reduced fidelity.
IMPORTANCE Iron and manganese play important roles in how cell’s cope with oxygen stress. However, how these metals affect the ability of cells to replicate after oxidative challenges is not known. Here, we show that replication in Escherichia coli is inhibited following a challenge with hydrogen peroxide and requires manganese for the rapid recovery of DNA synthesis. The manganese-dependent recovery of DNA synthesis occurs independently of lesion repair and modestly improves survival, but it also increases the mutation rate in cells. The results imply that replication in E. coli is likely to utilize an iron-dependent enzyme(s) that becomes oxidized and inactivated during oxidative challenges. We propose that manganese remetallates these or alternative enzymes to allow genomic DNA replication to resume, although with reduced fidelity.
INTRODUCTION
Iron serves as a cofactor for enzymes involved in a broad variety of metabolic pathways but is easily oxidized to its inactive form (1). At the same time, excess or unbound intracellular iron is a prooxidant that is deleterious to cell viability when peroxides are present, generating hydroxyl radicals via Fenton chemistry that damage DNA, proteins, and lipids (2–7). In contrast, manganese is relatively stable in aerobic environments and can often, but not always, remetallate iron-dependent enzymes or homologous Mn-dependent enzymes to carry out the required catalytic function with a similar or modestly compromised efficiency (reviewed in references 8, 9, and 10). Thus, cells tightly regulate the intracellular concentrations of iron and manganese when growing in aerobic environments to optimize the metal-dependent enzymes required for metabolism and minimize the toxicity associated with metabolically activated oxygen species.
Escherichia coli contains several interdependent regulatory genes to ensure that metal concentrations allow for proper protein function and limit oxygen free radical damage (11–14). The ferric uptake regulator (Fur) controls the expression of more than 60 genes encoding proteins performing functions required for iron transport and storage (fecABCDE) and manganese import (mntH), proteins performing essential metabolic functions such as ribonucleotide reductase (nrdEF) and cytochrome bo3 ubiquinol oxidase (cyoABCDE), and proteins performing detoxification of metabolically activated oxygen species such as hydroperoxidase II (katE), iron-dependent superoxide dismutase (sodB), and others (11, 12, 14–18). Other regulators include the redox sensors OxyR and SoxRS, which respond to oxidative stress and regulate expression of genes that enhance survival under these conditions, including fur itself, peroxide-scavenging enzymes (katG products), DNA repair enzymes, and a proton-dependent manganese(II) import channel (MntH) (9, 10, 12, 19–25). Increased intracellular manganese pools also provide the optimal conditions for incorporation of the correct metal into several enzymes, including SodA, the manganese-dependent superoxide dismutase, while activating several other enzymes that specifically require manganese for activity, such as the manganese-dependent ribonucleotide reductase (NrdEF) (18, 26, 27). The in vivo metallation state of many enzymes, as well as how their metallation status and activity are altered during oxidative stress, remains an active area of investigation.
In addition to metabolically activated oxygen species, other environmental agents can interact with, alter, and damage DNA, including ionizing radiation, UV light, and a variety of chemical agents (28–35). Replication in the presence of DNA damage is thought to produce most of the mutagenesis, genomic rearrangements, and lethality that occur in all cells. Of the agents described above, how replication processes and recovers from UV-induced damage has been the most extensively characterized. Irradiation with short-wave UV light forms cyclobutane pyrimidine dimers and 6-4 photoproducts in DNA that block the progression of the replication fork (31, 36–38). After arrest, RecA and several RecF pathway proteins are required to process the replication fork such that the blocking lesion can be removed and replication can resume (38–44). Cells lacking RecA or any of several RecF pathway proteins are hypersensitive to UV-induced damage and fail to recover replication following disruption (38, 39, 42, 45). The time and ability to resume DNA synthesis following UV-induced damage also correlates with the time at which the UV-induced lesions are removed from the DNA (38, 41, 43, 44). In cells deficient in nucleotide excision repair, the UV-induced lesions are not removed, and the recovery of replication is severely impaired. These cultures exhibit elevated levels of mutagenesis, strand exchanges, and cell lethality (46–50).
The E. coli genome also encodes three translesion DNA polymerases, polymerase II (Pol II), Pol IV, and Pol V. Although mutations in these proteins do not affect the ability of DNA replication to recover after DNA damage or the time required, they do contribute to the overall mutagenesis that occurs in the presence of lesions (43, 44, 51–55). In the case of UV irradiation, DNA Pol V is responsible for nearly all the mutagenesis that occurs and contributes modestly to survival at high UV doses that exceed the repair capacity of the cell (43, 44, 56–59).
Less is known about how replication recovers following DNA damage induced by oxygen free radical species. Further, although both iron and manganese are known to regulate the cellular response after oxidative challenges, how these divalent metals affect replication under these conditions has not been examined. Here, we investigated the survival, recovery, and mutagenic phenotypes exhibited by E. coli exposed to 10 mM H2O2 when supplemented with 200 μM iron, manganese, or no metals. We found that the rapid recovery of DNA synthesis was independent of lesion repair but depends on the presence of manganese. The manganese-dependent recovery modestly improves survival and is associated with an increased mutation frequency that likely involves both replicative and translesion DNA polymerases.
RESULTS
Replication recovery is not dependent on lesion repair following oxidative challenge in minimal growth medium.
To begin to characterize how replication responds to and recovers from oxidative stress, we monitored the rate of DNA synthesis in E. coli treated with H2O2. Wild-type cultures, grown in a defined phosphate-buffered medium, were exposed to 10 mM H2O2 for 5 min before the cells were filtered to remove excess H2O2, resuspended in fresh medium, and allowed to recover. To monitor replication, duplicate aliquots of culture were pulsed for 2 min with [3H]thymidine at various times before and after H2O2 treatment, and the rate of replication (3H incorporation into DNA/2 min) was then determined for each time point. For the purposes of comparison and as a control, we also monitored the recovery of cells following UV irradiation, which has been well characterized in previous work (41, 43, 44). Consistent with previous observations, the rate of DNA synthesis was initially inhibited by >90% following UV irradiation with 50 J/m2 in wild-type cells. However, within 20 min the DNA synthesis began to resume, and that rate continued to increase over the remaining time course (Fig. 1A). In contrast, in H2O2-treated cultures, although replication was inhibited to a similar degree, DNA synthesis did not resume, and no recovery was observed during the 60-min time course (Fig. 1A).
FIG 1.
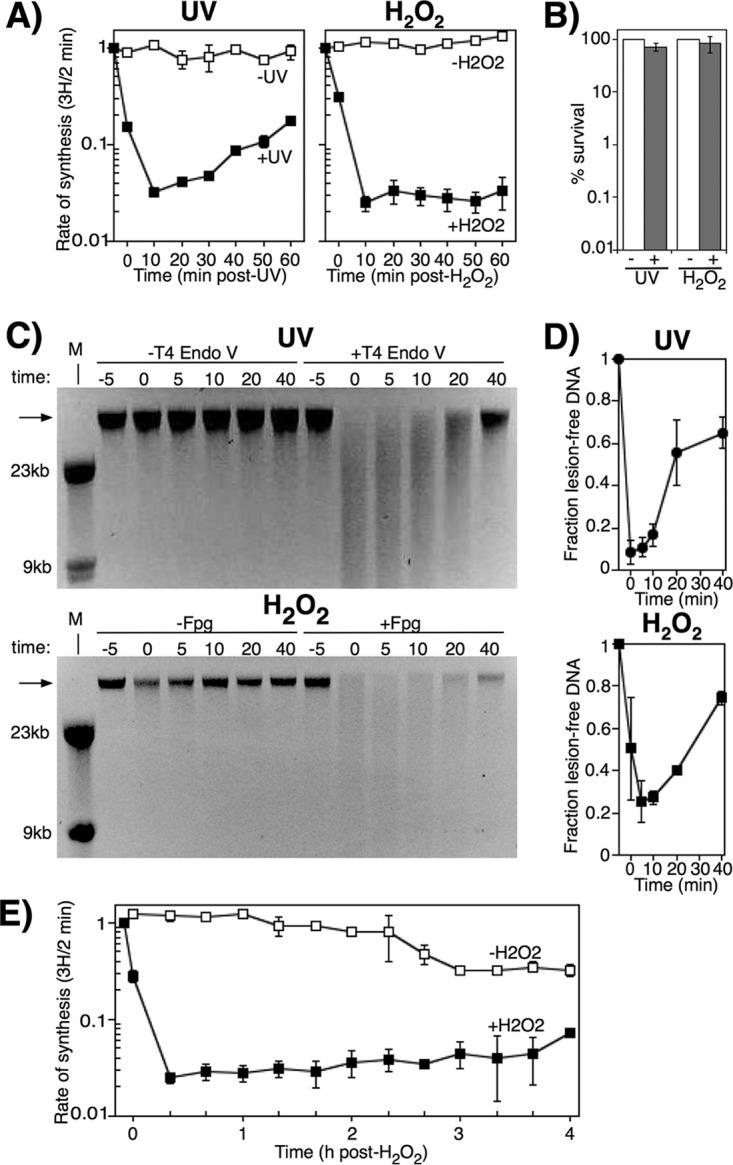
Lesion removal does not necessarily correlate with the onset of replication recovery following oxidative stress. (A) [3H]thymidine was added to cultures for 2 min at the indicated times following either UV irradiation or H2O2 treatment at time zero. The rate of DNA synthesis (3H/2 min) relative to the amount incorporated immediately prior to exposure are plotted for wild-type cells exposed to mock treatment (open symbols), 50 J/m2 UV (filled symbols), or 10 mM H2O2 (filled symbols). Graphs represent an average from at least two independent experiments. Error bars represent one standard error of the mean. (B) Wild-type cultures were either UV irradiated at 50 J/m2 or treated with 10 mM H2O2 for 5 min. The survival of wild-type cultures after UV irradiation or oxidative challenge is plotted. Bar graphs represent the averages from three independent experiments. Error bars represent one standard error of the mean. (C) Wild-type cultures were either UV irradiated at 50 J/m2 (top panel) or treated with 10 mM H2O2 for 5 min (bottom panel) and allowed to recover, and then genomic DNA was purified at the indicated times. For UV-irradiated samples, DNA was either treated with T4 endonuclease V (T4 Endo V) or no T4 Endo V for 1 h at 37°C and then analyzed on alkali agarose gels. For H2O2-treated samples, DNA was either treated with Fpg or no glycosylase for 1 h at 37°C and then analyzed on alkali agarose gels. A representative gel is shown for each treatment. Arrows indicate lesion-free DNA. (D) The fraction of lesion-free, high-molecular-weight (HMW) DNA in T4 Endo V-treated (top graph) or Fpg-treated (bottom graph) samples is plotted for each time point relative to mock-treated samples. Graphs represent the averages from at least two independent experiments. Error bars represent one standard error of the mean. (E) Cells were either exposed to 10 mM H2O2 for 5 min (filled symbols) or mock treated (open symbols) at time zero and then allowed to recover. At the indicated times, [3H]thymidine was added to cultures for 2 min. The amount of DNA synthesis/2 min (3H) is plotted. The graph represents an average from at least two independent experiments. Error bars represent one standard error of the mean.
The failure to recover DNA synthesis following H2O2 treatment was not due to elevated levels of lethality. Consistent with our previous work, under the conditions used in these assays, more than 90% of the cells survived both the H2O2 and the UV treatments that were used (Fig. 1B) (39, 41, 60).
The lack of recovery following H2O2 treatment was also not due to persisting oxidative lesions that prevented the resumption of DNA synthesis. To monitor the repair of both UV- and H2O2-induced lesions in cultures over time, cells were either UV irradiated or treated with H2O2 and allowed to recover as before. At various times during the recovery period, total genomic DNA was purified and treated with either T4 endonuclease V (T4 Endo V) or Fpg glycosylase/AP-lyase. These enzymes recognize and incise DNA at sites where the predominant lesions produced by UV irradiation and H2O2, cyclobutane pyrimidine dimers, and 8-oxoguanines occur, respectively (61–64). Genomic DNA samples from each time point were then electrophoresed in a denaturing alkali agarose gel, where the presence of UV- or H2O2-induced lesions was observed as the loss of high-molecular-weight DNA in the enzyme-treated samples. In the case of UV, a loss of high-molecular-weight DNA was observed immediately after UV irradiation in T4 Endo V-treated samples (Fig. 1C), demonstrating the presence of UV-induced DNA lesions. As the UV-induced lesions were repaired during the recovery period, the number of T4 Endo V-sensitive sites decreased, and high-molecular-weight DNA was restored to >60% by 40 min post-UV irradiation (Fig. 1D). The rate at which DNA synthesis resumed (Fig. 1A) correlated with the repair of the DNA damage (Fig. 1C and D), consistent with previous work (38, 41, 43, 44).
In H2O2-treated cultures, a similar induction of oxidative DNA lesions was observed at times immediately after treatment with H2O2. These lesions were removed, and high-molecular-weight fragments were restored at a rate that was similar to that observed for the UV-treated samples (Fig. 1C and D). However, despite the removal of the oxidative lesions, no resumption in DNA synthesis occurred (Fig. 1A).
To determine whether the timing of the recovery was simply delayed beyond 60 min in H2O2-treated cultures, we repeated the assay and extended the time course. We observed that the rate of synthesis remained suppressed for over 3 h, with a modest increase in DNA synthesis possibly beginning 4 h posttreatment (Fig. 1E). Taken together, the results demonstrate that replication is inhibited by H2O2 and fails to recover in defined minimal medium. The inability to recover DNA synthesis is not due to cell lethality or the presence of replication-blocking lesions in the genomic template.
Manganese promotes the recovery of DNA synthesis after treatment with H2O2.
The results presented above were unexpected, as we have previously observed that replication recovers when cultures are similarly treated with H2O2 in rich Luria-Bertani (LB) medium (60, 65). Therefore, we hypothesized that the lack of recovery after H2O2 treatment was due to a difference between using defined minimal medium and rich LB medium.
The cellular toxicity of H2O2 arises through the oxidation and reduction of iron in a disproportionation reaction that produces a hydroxyl radical species, which then reacts with and damages DNA, proteins, and lipids in the cell (2–6). The oxidation of iron also inactivates a range of Fe-dependent enzymes which play essential roles in a number of metabolic processes that are still being characterized (9, 10, 23, 66, 67). Manganese plays a large role in ameliorating the toxic effects of iron, since it does not react with H2O2 and can replace iron in many Fe-proteins to restore activity (9, 10, 23, 68–71). In addition, cells contain several genes devoted to limiting intracellular iron concentrations and upregulating manganese import during oxidative stress (23, 72–74). We noted that our rich medium was significantly enriched for both iron and manganese relative to our defined media (23, 68) (Table 1) and speculated that these metals may affect the ability to restore replication following H2O2 treatment. To examine this, cultures were grown in defined [14C]thymine medium, supplemented with 200 μM ferrous sulfate (FeSO4·7H2O) or 200 μM manganese (II) chloride (MnCl2) or without additional metals, and treated with 10 mM H2O2 for 5 min as before. At various times during the recovery period, aliquots from each culture were pulse-labeled for 2 min with [3H]thymidine before the DNA was precipitated and the amount of 14C and 3H incorporated was quantified. In this way, both replication fork speed (3H incorporation per 2 min) and overall DNA accumulation (14C incorporation) could be monitored over time. All experiments included a mock-treated control to ensure that any differences observed were due to H2O2 treatment and not an effect of thymine addition or differences in growth phase. As shown in Fig. 2, cultures in nonsupplemented medium or iron-containing medium did not resume DNA synthesis for the duration of the experiment. However, cultures supplemented with manganese began to recover DNA synthesis between 20 and 30 min after H2O2 was removed (Fig. 2). The total DNA accumulation correlated with the rate of DNA synthesis in each case, indicating that the newly made DNA was stable and not associated with degradation or DNA turnover. From these results, we infer that manganese promotes the recovery of DNA synthesis after acute exposure to H2O2.
TABLE 1.
Metal concentrations in minimal and rich media as measured by inductively coupled plasma mass spectrometry
| Medium | Metal concn |
|||||||
|---|---|---|---|---|---|---|---|---|
| Mg (mM) | Cr (μM) | Mn (μM) | Fe (μM) | Ni (μM) | Cu (μM) | Zn (mM) | Pb (μM) | |
| Minimal | 392.1 | 66.92 | 117.04 | 489.75 | 39.87 | 67.67 | 1.17 | 0.92 |
| Rich | 81.87 | 307.9 | 184.21 | 4,326.98 | 64.91 | 231.8 | 8.26 | 1.35 |
FIG 2.
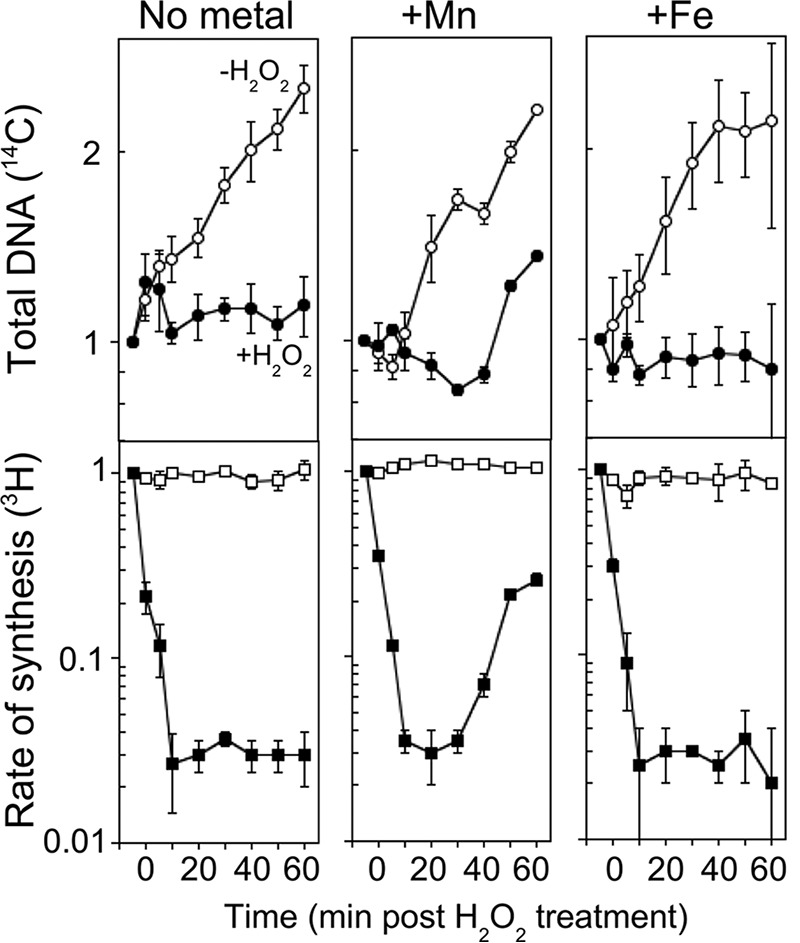
Manganese promotes replication recovery after H2O2 treatment. [3H]thymidine was added to [14C]thymine-prelabeled cultures for 2 min at the indicated times following treatment at time zero. The total DNA accumulation (14C) and rate of DNA synthesis (3H) relative to the amount incorporated immediately prior to exposure are plotted for wild-type cells grown in medium supplemented with manganese, iron, or no metal and exposed to 0 mM (open symbols) or 10 mM (filled symbols) H2O2. Graphs represent an average from at least three independent experiments. Error bars represent one standard error of the mean.
Manganese-dependent recovery increases survival and mutagenesis.
To determine how the ability to resume DNA synthesis affects survival in the presence of H2O2, cultures were grown in medium containing no metal supplements, 200 μM iron, or 200 μM manganese as before and then treated with 10 mM H2O2 and sampled at various times after exposure to determine the fraction of cells surviving to form colonies. When manganese was present in the growth medium, a modest increase in cell viability was observed compared to either no metal or iron supplementation, particularly with longer H2O2 exposure times (Fig. 3). Thus, the manganese-dependent recovery of DNA synthesis correlates with improved survival in H2O2-treated cultures. However, since manganese also affects several other enzymes and metabolic pathways, we do not rule out the possibility that other metabolic processes are contributing to the enhanced survival.
FIG 3.
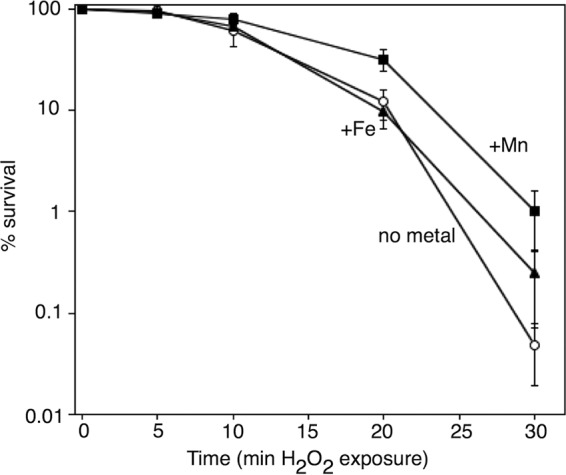
The presence of manganese increases survival of cells chronically exposed to H2O2. The survival of wild-type cells grown in medium containing manganese, iron, or no metal additive after exposure to 10 mM H2O2 is plotted after treatment for the indicated times. Graphs represent an average from five independent experiments. Error bars represent one standard error of the mean.
We also examined whether the manganese-promoted recovery of replication affected mutagenesis after oxidative challenge. To do this, we measured the frequency at which mutations conferring resistance to rifampin arose after H2O2 treatment. There are at least 69 base substitutions within the rpoB gene that confer rifampin resistance to cells, making this phenotype a robust system to monitor mutagenesis at multiple sites and in various sequence contexts (75). Replicating cultures containing no metal supplements, 200 μM iron, or 200 μM manganese were either mock treated or exposed to 10 mM H2O2 for 5 min and then allowed to recover overnight before the fraction of rifampin-resistant mutations per viable cells in each culture was determined. We found that a higher rate of mutations occurred after H2O2 treatment when manganese was present in the medium, relative to media containing either iron or no additional metals (Fig. 4A). In medium containing no metal supplements or in iron-supplemented medium, H2O2 treatment increased the frequency of rifampin-resistant mutations ∼3-fold. In comparison, in the presence of manganese, the frequency of rifampin-resistant mutations increased ∼9-fold. No significant increase in mutation frequency was caused by the addition of manganese alone, indicating that replication must first be inhibited by H2O2 before mutagenesis can occur. To determine whether manganese increased mutagenesis after other forms of DNA damage, we repeated the experiment treating cells with 27 J/m2 UV irradiation instead of H2O2. After UV treatment, the frequency of mutants increased ∼50-fold in unsupplemented medium and increased by a similar amount in the presence of either iron or manganese (Fig. 4B). Thus, although UV irradiation was a more potent mutagen overall, the presence of manganese did not significantly increase the frequency of mutants observed compared to cultures containing no additional metals or supplemented with iron, respectively.
FIG 4.
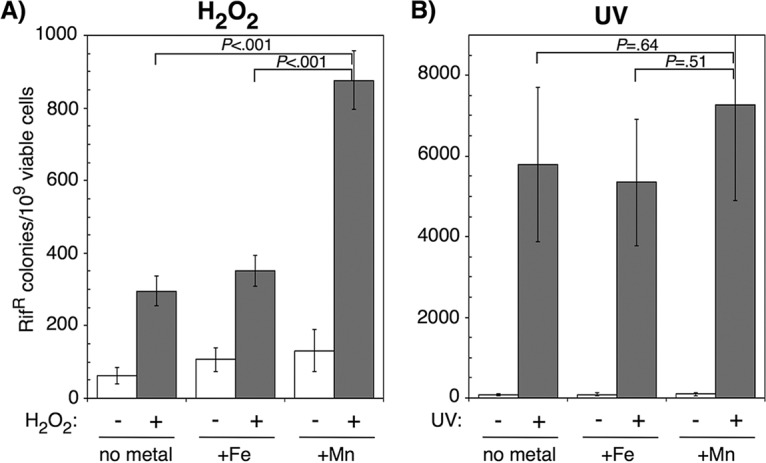
Manganese-dependent replication recovery after oxidative challenge is mutagenic. (A) Wild-type cultures grown in medium supplemented with manganese, iron, or no metals were treated with 10 mM H2O2 for 5 min and examined for the number of rifampin (Rif)-resistant colonies that appeared after overnight incubation. (B) Wild-type cultures grown in medium supplemented with manganese, iron or no metals were irradiated with 27 J/m2 UV and examined for the number of rifampin-resistant (Rifr) colonies that appeared after overnight incubation. The number of rifampin-resistant colonies per 109 surviving cells is plotted for each treatment. Graphs represent averages from six independent experiments. Error bars represent one standard error of the mean. P values represent the likelihood the observed difference could arise by chance, as determined by two-way analysis of variance (ANOVA).
Mutagenesis following many forms of DNA damage depends upon specialized polymerases that are capable of replicating across DNA lesions with lower fidelity (52, 55, 76). E. coli contains three such translesion DNA polymerases: Pol II (a polB gene product), Pol IV (a dinB gene product), and Pol V (a umuDC gene product) (52). We hypothesized that the mutagenic replication occurring after H2O2 may be due to a manganese-dependent translesion DNA polymerase. To address this, we examined how the absence of these polymerases affected mutagenesis in the presence or absence of manganese supplementation as before. We observed that the frequency of mutagenesis after H2O2 treatment was only partially reduced by the absence of polB, dinB, and umuDC compared to wild-type cells in the manganese medium (Fig. 5A). Further, no single translesion polymerase appeared to be responsible for partial reduction in manganese-dependent mutagenesis. Single polB, dinB, and umuDC mutants each displayed partially reduced mutation frequency when manganese-supplemented cultures were exposed to H2O2 (Fig. 5C). In comparison, when the translesion DNA polymerase mutants were UV irradiated with 27 J/m2, we observed a nearly complete lack of UV-induced mutagenesis irrespective of metal supplementation in the growth medium (Fig. 5B). Thus, the translesion DNA polymerases contribute to a portion of the manganese-dependent mutagenesis that occurs in the presence of oxidative damage. However, a significant portion of the mutagenesis seems likely to be due to the replicative and/or repair polymerases, and the overall magnitude of the mutations induced is relatively modest compared to UV-induced damage.
FIG 5.
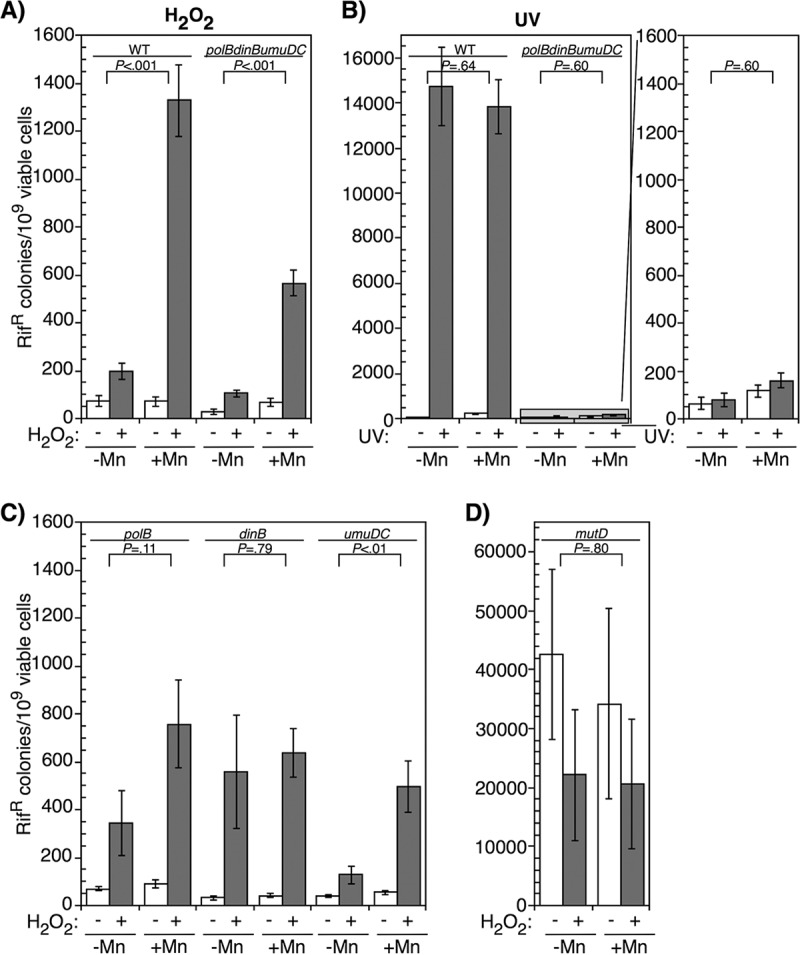
The absence of translesion DNA polymerases reduces, but does not completely inhibit, H2O2-induced mutagenesis in the presence of manganese. (A) Wild-type cultures and polB, dinB, and umuDC mutant cultures grown in medium supplemented with manganese or no metals were treated with 10 mM H2O2 for 5 min and examined for the number of rifampin-resistant (Rifr) colonies that appeared after overnight incubation. (B) Wild-type cultures and polB, dinB, and umuDC mutant cultures grown in medium supplemented with manganese or no metals were treated with 27 J/m2 UV and examined for the number of rifampin-resistant colonies that appeared after overnight incubation. (C) Cultures of polB, dinB, and umuDC mutants grown in medium supplemented with manganese or no metals were treated with 10 mM H2O2 for 5 min and examined for the number of rifampin-resistant colonies that appeared after overnight incubation. (D) mutD cultures were grown in the presence or absence of manganese and treated with 10 mM H2O2 for 5 min, then examined for the number of rifampin-resistant colonies after overnight incubation. The number of rifampin-resistant colonies per 109 surviving cells is plotted for each treatment. Graphs represent averages from six independent experiments. Error bars represent one standard error of the mean. P values represent the likelihood the observed difference could arise by chance, as determined by two-way ANOVA.
We also tried to assess the contribution of the replicative polymerase to manganese-dependent mutagenesis by examining a mutD5 mutant, which inactivates the proofreading exonuclease of Pol III and results in a high rate of spontaneous mutations. If the Mn-dependent mutations result from Pol III, we hypothesized that the mutation rate may increase after H2O2 treatment in the presence of manganese. However, contrary to this expectation, we observed the mutation rate decreased ∼2-fold in mutD5 after oxidative challenge irrespective of the presence of manganese (Fig. 5D). Although no manganese-dependent effect was observed, the basal mutation rate of mutD5 was 2 orders of magnitude higher than that seen after H2O2 exposure in any other strain we examined, making it unlikely that we would be able to detect any oxidative-stress-dependent mutagenesis occurring in this background. Nevertheless, given the manganese-dependent induction of mutations in wild-type cells and the partial reduction of mutations in the absence of translesion polymerases, we suggest that the reduced fidelity in the presence of manganese may generally apply to both replicative and translesion polymerases during recovery from H2O2 treatment.
Taken together, these results indicate that replication is sensitive to oxidative challenge and likely depends upon one or more oxygen-sensitive, iron-dependent enzymes. Manganese is required for the rapid resumption of DNA replication and presumably remetallates this or an alternative enzyme of similar function that allows replication to resume. The manganese-dependent recovery of replication modestly improves survival, but it is also associated with elevated levels of mutagenesis involving both translesion and replicative polymerases.
DISCUSSION
Ferrous iron (Fe2+) serves as a cofactor in a broad variety of mononuclear, heme, and iron-sulfur proteins. However, in the presence of H2O2, it can oxidize to its ferric (Fe3+) form and is stripped from these proteins (for a review, see reference 1). This has the dually detrimental effect of inactivating enzymes involved in multiple metabolic pathways and increasing the free intracellular iron concentration, which through Fenton chemistry produces free radicals that damage DNA, proteins, and lipids (2, 3, 5–7). The presence of intracellular manganese has been shown to correlate with increased resistance to oxidative DNA damage in a wide range of bacteria (68–71, 77, 78). Similarly, we observed a modest increase in the survival of E. coli when manganese is present in the media. In E. coli, manganese import is upregulated ∼10-fold in response to oxidative challenge (23, 79). The typical intracellular manganese concentration of cells grown in minimal medium is ∼15 μM, whereas manganese supplementation increases this concentration to ∼35 μM, and oxidative stress can further increase the concentration to ∼150 μM (23, 79). Whereas a manganese importer (mntH) mutant grows as well as wild-type cells under nonoxidizing growth conditions, manganese import is essential for viability under oxidizing or low-iron growth conditions (23). Manganese protection from H2O2 toxicity is thought to be conferred by remetallating inactivated, mononuclear iron enzymes, as well as through the activation of functionally redundant enzymes that specifically utilize manganese. In both cases, manganese acts to restore required enzymatic activities to essential metabolic pathways (9, 10, 23). We did not find any influence of iron supplementation on either survival or mutagenesis following oxidative challenge despite the potential for elevated free iron pools to induce DNA damage via Fenton chemistry. We speculate that the tight regulation of Fe2+ import by iron homeostasis genes is sufficient to repress iron uptake and limit additional iron-induced DNA damage.
A wide range of mononuclear iron enzymes involved in diverse cellular pathways have been characterized that can be remetallated by manganese to restore function, including those required for pyrimidine metabolism, amino acid catabolism, and carbohydrate metabolism (9, 10). In addition, several enzymes that specifically utilize manganese as a cofactor and are important in the response to oxidative stress or iron starvation have been identified. Many appear to have redundant activities with iron enzymes that are oxygen sensitive. These include the alternative manganese-dependent class Ib ribonucleotide reductase, NrdEF, the Mn-dependent superoxide dismutase, MnSOD, and the apurinic/apyrimidinic (AP) endonuclease IV (26, 27, 80–82). In this study, we show that the metabolic process of DNA replication likely requires oxygen-sensitive iron-enzymes that are inactivated by H2O2 but can be restored by the presence of manganese. Although we were unable to identify the specific enzyme(s) that allows replication to resume, it seems likely that they will fall into either the class of metabolic mononuclear iron enzymes or manganese-cofactored enzymes. At the time of writing, the ecocyc.org database listed approximately 90 proteins for which manganese serves as a cofactor (83). Several of these proteins function in nucleotide metabolism. These enzymes represent potential candidates which can be examined and include the alternative class Ib ribonucleotide reductase, NrdEF, that is required for cell replication under low-iron conditions (18).
Although the manganese-promoted recovery of replication increased survival after H2O2 treatment, it also compromised genomic stability as a higher frequency of mutations occurred in these cultures. We believe the increased mutagenesis most likely results from manganese reducing the overall fidelity of the DNA polymerases. A large number of studies have shown that the fidelity of multiple DNA polymerases is altered by the presence of manganese in vitro. The fidelity of E. coli DNA polymerase I, as well as human polymerases ι, Dpo4, and Primpol are all altered by manganese supplementation in a concentration-dependent manner (84–87). Further, the reduced fidelity of DNA polymerases in the presence of manganese has been frequently utilized for in vitro PCR-mediated mutational screens (88, 89). The observations we report here suggest that manganese also affects the fidelity of both translesion and replicative polymerases of E. coli in vivo.
The increased mutagenesis could also partially result from more frequent encounters of the replication machinery with oxidized base damage. Since replication resumes more rapidly when manganese is present, it is likely that replication would more frequently encounter lesions. Several polymerases have been shown to incorporate the wrong base on templates containing several common oxidized bases induced by H2O2, including 8-oxoguanine, thymine glycol, and 5-hydroxycytosine (90–94).
Following oxidative challenges, manganese has been shown to restore function to a range of metabolic pathways that utilize monofunctional iron enzymes (9, 10, 23). The results presented here extend these observations to the process of DNA replication. Following many forms of DNA damage, replication inhibition and recovery correlate with the presence and removal of blocking DNA lesions, respectively (36, 38, 41). Our results suggest that in the case of H2O2, the inhibition of replication may occur independent of blockage by DNA damage, and result from an inactivation of oxygen-sensitive iron enzymes. Eukaryotic DNA polymerases also contain iron-sulfur clusters that are required for function, raising the possibility that similar mechanisms operate in humans (95). These observations demonstrate that the mechanisms by which replication responds to and recovers from oxidative challenges are distinct from other agents that primarily target DNA. It will be important to consider these potential differences and broaden our understanding of the metallation state of proteins in order to understand how genomic stability is maintained during oxidative stress or acute oxidative challenges.
MATERIALS AND METHODS
Bacterial strains and plasmids.
SR108, a thyA36 deoC2 derivative of W3110, and CL646 (SR108 polB::Ω Sm-Sp dinB::Kanr umuDC595::cat) have been previously described (43, 96). CL716 (SR108 mutD5) was constructed by P1 transduction of mutD5 zaf-13::Tn10 from NR9458 (97) into SR108. pBR322 is a medium-copy-number, ColE1-based, 4.4-kb plasmid (Promega).
DNA synthesis and accumulation.
UV irradiation used a 15-W germicidal lamp (254 nm) at an incident dose of 0.9 J/m2/s. For experiments using UV irradiation, overnight cultures were diluted 1:100 and grown at 37°C in Davis medium supplemented with 0.4% glucose, 0.2% Casamino Acids, and 10 μg/ml thymine (DGCthy) to an optical density at 600 nm (OD600) of 0.25 to 0.35. At this time, half of the cells were mock irradiated, while the other half of the culture was irradiated with 50 J/m2.
For experiments using hydrogen peroxide as a DNA-damaging agent, overnight cultures were diluted 1:100 and grown at 37°C in DGCthy to an OD600 of precisely of 0.25 to 0.35. Where DNA accumulation was also monitored, cultures were grown in DGCthy supplemented with 0.1 μCi/ml [14C]thymine and, where indicated, either 200 μM ferrous sulfate (FeSO4⋅7H2O) or 200 μM manganese(II) chloride (MnCl2⋅4H2O) was added to the medium as well. At this time, half of the cells grown in each type of medium were mock treated, while the remaining culture was exposed to 10 mM H2O2 for 5 min at 37°C. After either mock or H2O2 treatment, cells were filtered on 0.45-μm membranes (Fisher) to remove excess H2O2 from the medium and resuspended in fresh DGCthy medium. As before, DGCthy medium was supplemented with 0.1 μCi/ml [14C]thymine in the case of DNA accumulation assays and, where indicated, the corresponding metal additive in which cells were cultured.
For both UV irradiation and H2O2 experiments, cultures were returned immediately to 37°C after treatment to allow recovery and continued growth. At the times indicated, duplicate 0.5-ml aliquots of culture were pulse-labeled with 0.5 μCi/ml [3H]thymidine for 2 min at 37°C. The cells were then lysed, and the DNA was precipitated in cold 5% trichloroacetic acid and filtered onto Millipore glass fiber filters. The amounts of 3H and 14C on each filter were determined by scintillation counting.
Metal concentration analysis.
Metal concentrations in minimal and rich media were determined by inductively coupled plasma mass spectrometry through Oregon Health Sciences University’s Elemental Analysis Core services.
H2O2 survival assays.
Fresh overnight cultures were diluted 1:100 in DGCthy medium supplemented with either iron or manganese or without metals as described above, grown at 37°C to an OD600 of 0.3, and then treated with 10 mM H2O2. At the times indicated, 0.1-ml aliquots of each culture were removed and serially diluted in 10-fold increments into DGCthy medium supplemented with the corresponding metal. Triplicate 10-μl aliquots of each dilution were then spotted onto LB plates supplemented with 10 μg/ml thymine (LBthy). Viable colonies were counted after overnight incubation at 37°C.
UV survival assays.
Fresh overnight cultures were diluted 1:100 in DGCthy medium, grown at 37°C to an OD600 of 0.3, and then treated with 0 or 50 J/m2 UV irradiation. Immediately after irradiation, 0.1-ml aliquots of each culture were removed and serially diluted in 10-fold increments into DGCthy medium. Triplicate 10-μl aliquots of each dilution were then spotted onto LBthy plates. Viable colonies were counted after overnight incubation at 37°C.
Lesion frequency.
For UV irradiation, fresh overnight cultures were diluted 1:100 and grown at 37°C in DGCthy medium to an OD600 of 0.3. At this time, cultures were irradiated with an incident dose of 50 J/m2 and then returned to 37°C to allow recovery. For H2O2 challenge, fresh overnight cultures were diluted 1:100, grown at 37°C in DGCthy medium to an OD600 of 0.3, and then treated with 10 mM H2O2 for 5 min at 37°C. Cells were filtered on 0.45-μm membranes to remove excess H2O2 from the medium, resuspended in fresh DGCthy medium, and returned to 37°C for the duration of the time course. At the times indicated, a 0.75-ml aliquot was transferred to an equal volume of 2× NET (200 mM NaCl, 20 mM Tris [pH 8.0], 40 mM EDTA [pH 8.0]). Cells were pelleted, resuspended in 0.14 ml of lysis buffer (1 mg/ml lysozyme and 0.5 mg/ml RNase A in 10 mM Tris [pH 8.0]–1 mM EDTA [pH 8.0]), and incubated for 30 min at 37°C. Then, 0.01-ml portions of 10-mg/ml proteinase K and 0.01-ml portions of 20% Sarkosyl were added to the samples, and incubation was continued for 30 min at 37°C. Samples were then extracted once with 4 volumes phenol-chloroform, followed by 2 volumes of chloroform, and dialyzed against 200 ml of 1 mM Tris (pH 8.0)–1 mM EDTA (pH 8.0) for 30 min using 47-mm Millipore (0.025-μm-pore) disks.
For UV-irradiated samples, 15 μl of each DNA sample was treated in reaction buffer (12.5 mM sodium phosphate [pH 6.8], 5 mM EDTA [pH 8.0], 50 mM NaCl, 0.5 mM dithiothreitol, 0.005% Triton X-100, 0.1 mg/ml bovine serum albumin) supplemented with either no enzyme or 2 U of T4 endonuclease V (T4 Endo V; Trevigen) for 1 h at 37°C. For H2O2-treated samples, 15 μl of each DNA sample was treated in reaction buffer (30 mM EDTA [pH 8.0], 22.5 mM NaCl, 5 mM Tris [pH 8.0]) supplemented with either no enzyme or 0.53 μM Fpg glycosylase for 1 h at 37°C. Enzyme preparations were titrated using purified undamaged genomic DNA as a template. The highest enzyme concentration that did not exhibit nonspecific activity on undamaged DNA was used. For the preparations in our lab, this corresponded to 2 U of T4 Endo V and 0.53 μM Fpg glycosylase. Treated samples were then electrophoresed on a 0.5% alkaline agarose gel in 30 mM NaOH–1 mM EDTA at 30 V for 16 h, stained, and visualized with ethidium bromide.
H2O2- and UV-induced mutagenesis.
Mutagenesis induced by either H2O2 or UV was measured by the appearance of rifampin-resistant colonies as a result of exposure to these agents. For experiments using H2O2 treatment, fresh overnight cultures were diluted 1:100 and grown at 37°C in DGCthy medium supplemented with iron, with manganese, or without metals to an OD600 of 0.3. At this time, cultures were divided equally and mock treated or exposed to 10 mM H2O2 for 5 min. After either mock or H2O2 treatment, the cells were filtered on 0.45-μm membranes to remove excess H2O2 from the medium and resuspended in fresh DGCthy medium supplemented with the corresponding metal additive in which they were cultured.
For experiments using UV irradiation, fresh overnight cultures were diluted 1:100 and grown at 37°C in DGCthy medium supplemented with iron, with manganese, or without metals to an OD600 of 0.3. At this time, cultures were divided equally and irradiated with an incident dose of 0 or 27 J/m2.
For both H2O2 treatment and UV irradiation, cultures were returned to 37°C after exposure to allow growth and recovery overnight. After overnight incubation, cultures were plated on LBthy plates containing 100 μg/ml rifampin to determine the mutagenic effect, as well as on LBthy plates to determine the number of viable colonies. Rifampin-resistant colonies and viable cells were counted after overnight incubation at 37°C.
ACKNOWLEDGMENT
This study was supported by National Science Foundation grant MCB1916625.
REFERENCES
- 1.Imlay JA. 2014. The mismetallation of enzymes during oxidative stress. J Biol Chem 289:28121–28128. doi: 10.1074/jbc.R114.588814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haber F, Weiss J. 1934. The catalytic decomposition of hydrogen peroxide by iron salts. Proc R Soc London Ser A 147:332–351. [Google Scholar]
- 3.Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 4.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 5.Tamarit J, Cabiscol E, Ros J. 1998. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem 273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- 6.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Leapman RD, Lai B, Ravel B, Li SM, Kemner KM, Fredrickson JK. 2007. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol 5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol 177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehres DG, Maguire ME. 2003. Emerging themes in manganese transport, biochemistry, and pathogenesis in bacteria. FEMS Microbiol Rev 27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 9.Anjem A, Imlay JA. 2012. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem 287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobota JM, Imlay JA. 2011. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci U S A 108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patzer SI, Hantke K. 2001. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J Bacteriol 183:4806–4813. doi: 10.1128/JB.183.16.4806-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J Bacteriol 184:3151–3158. doi: 10.1128/jb.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagg A, Neilands JB. 1987. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev 51:509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHugh JP, Rodríguez-Quinoñes F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli: a new mechanism for iron homeostasis. J Biol Chem 278:29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 15.Angerer A, Braun V. 1998. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch Microbiol 169:483–490. doi: 10.1007/s002030050600. [DOI] [PubMed] [Google Scholar]
- 16.Hassan HM, Sun HC. 1992. Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc Natl Acad Sci U S A 89:3217–3221. doi: 10.1073/pnas.89.8.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoerter JD, Arnold AA, Ward CS, Sauer M, Johnson S, Fleming T, Eisenstark A. 2005. Reduced hydroperoxidase (HPI and HPII) activity in the Δfur mutant contributes to increased sensitivity to UVA radiation in Escherichia coli. J Photochem Photobiol B 79:151–157. doi: 10.1016/j.jphotobiol.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Martin JE, Imlay JA. 2011. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol 80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng M, Doan B, Schneider TD, Storz G. 1999. OxyR and SoxRS regulation of fur. J Bacteriol 181:4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng M, Storz G. 2000. Redox sensing by prokaryotic transcription factors. Biochem Pharmacol 59:1–6. doi: 10.1016/S0006-2952(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 21.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol 13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 22.Aslund F, Zheng M, Beckwith J, Storz G. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci U S A 96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anjem A, Varghese S, Imlay JA. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol 72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J, Weiss B. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol 174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Demple B. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli: purification and interaction with DNA. J Biol Chem 269:18371–18377. [PubMed] [Google Scholar]
- 26.Hassan HM, Fridovich I. 1977. Physiological function of superoxide dismutase in glucose-limited chemostat cultures of Escherichia coli. J Bacteriol 130:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beyer WF, Fridovich I. 1991. In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J Biol Chem 266:303–308. [PubMed] [Google Scholar]
- 28.Hutchinson F. 1985. Chemical changes induced in DNA by ionizing radiation. Prog Nucleic Acids Res Mol Biol 32:115–154. doi: 10.1016/S0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- 29.Teoule R. 1987. Radiation-induced DNA damage and its repair. Int J Radiat Biol Relat Stud Phys Chem Med 51:573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- 30.Varghese AJ, Wang SY. 1967. Ultraviolet irradiation of DNA in vitro and in vivo produces a 3d thymine-derived product. Science 156:955–957. doi: 10.1126/science.156.3777.955. [DOI] [PubMed] [Google Scholar]
- 31.Varghese AJ. 1972. Photochemistry of nucleic acids and their constituents. Photophysiology:207–274. [PubMed] [Google Scholar]
- 32.Singer B, Kuśmierek JT. 1982. Chemical mutagenesis. Annu Rev Biochem 51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- 33.Fridovich I. 1978. The biology of oxygen radicals. Science 201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 34.Ames BN. 1983. Dietary carcinogens and anticarcinogens: oxygen radicals and degenerative diseases. Science 221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- 35.Cross CE, Halliwell B, Borish ET, Pryor WA, Ames BN, Saul RL, McCord JM, Harman D. 1987. Oxygen radicals and human disease. Ann Intern Med 107:526–545. doi: 10.7326/0003-4819-107-4-526. [DOI] [PubMed] [Google Scholar]
- 36.Setlow RB, Swenson PA, Carrier WL. 1963. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science 142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell DL, Nairn RS. 1989. The biology of the (6–4) photoproduct. Photochem Photobiol 49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 38.Courcelle J, Crowley DJ, Hanawalt PC. 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and RecF protein function. J Bacteriol 181:916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courcelle J, Carswell-Crumpton C, Hanawalt PC. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci U S A 94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Courcelle J, Hanawalt PC. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet 262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 41.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 42.Chow KH, Courcelle J. 2004. RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli. J Biol Chem 279:3492–3496. doi: 10.1074/jbc.M311012200. [DOI] [PubMed] [Google Scholar]
- 43.Courcelle CT, Belle JJ, Courcelle J. 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol 187:6953–6961. doi: 10.1128/JB.187.20.6953-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Courcelle CT, Chow KH, Casey A, Courcelle J. 2006. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci U S A 103:9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Courcelle J, Hanawalt PC. 2003. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 46.Courcelle J, Ganesan AK, Hanawalt PC. 2001. Therefore, what are recombination proteins there for? Bioessays 23:463–470. doi: 10.1002/bies.1065. [DOI] [PubMed] [Google Scholar]
- 47.Howard-Flanders P, Theriot L, Stedeford JB. 1969. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J Bacteriol 97:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rupp WD, Howard-Flanders P. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 49.Foucault F, Vaury C, Barakat A, Thibout D, Planchon P, Jaulin C, Praz F, Amor-Guéret M. 1997. Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom’s syndrome. Hum Mol Genet 6:1427–1434. doi: 10.1093/hmg/6.9.1427. [DOI] [PubMed] [Google Scholar]
- 50.Courcelle J, Hanawalt PC. 2001. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc Natl Acad Sci U S A 98:8196–8202. doi: 10.1073/pnas.121008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ona KR, Courcelle CT, Courcelle J. 2009. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J Bacteriol 191:4959–4965. doi: 10.1128/JB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. 2000. All three SOS-inducible DNA polymerases (Pol II, Pol IV, and Pol V) are involved in induced mutagenesis. EMBO J 19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khidhir MA, Casaregola S, Holland IB. 1985. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet 199:133–140. doi: 10.1007/bf00327522. [DOI] [PubMed] [Google Scholar]
- 54.Witkin EM, Roegner-Maniscalco V, Sweasy JB, McCall JO. 1987. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc Natl Acad Sci U S A 84:6805–6809. doi: 10.1073/pnas.84.19.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. 1999. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli Pol V. Proc Natl Acad Sci U S A 96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bagg A, Kenyon CJ, Walker GC. 1981. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A 78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato T, Shinoura Y. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet 156:121–131. doi: 10.1007/bf00283484. [DOI] [PubMed] [Google Scholar]
- 58.Steinborn G. 1978. Uvm mutants of Escherichia coli K-12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet 165:87–93. doi: 10.1007/bf00270380. [DOI] [PubMed] [Google Scholar]
- 59.Elledge SJ, Walker GC. 1983. Proteins required for ultraviolet light and chemical mutagenesis: identification of the products of the umuC locus of Escherichia coli. J Mol Biol 164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 60.Schalow BJ, Courcelle CT, Courcelle J. 2011. Escherichia coli Fpg glycosylase is nonrendundant and required for the rapid global repair of oxidized purine and pyrimidine damage in vivo. J Mol Biol 410:183–193. doi: 10.1016/j.jmb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell DL, Haipek CA, Clarkson JM. 1985. (6–4)Photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat Res 143:109–112. doi: 10.1016/S0165-7992(85)80018-X. [DOI] [PubMed] [Google Scholar]
- 62.Koehler DR, Courcelle J, Hanawalt PC. 1996. Kinetics of pyrimidine (6–4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol 178:1347–1350. doi: 10.1128/jb.178.5.1347-1350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 1991. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A 88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. 1998. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci U S A 95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nazaretyan SA, Savic N, Sadek M, Hackert BJ, Courcelle J, Courcelle CT. 2017. Replication rapidly recovers and continues in the presence of hydroxyurea in Escherichia coli. J Bacteriol 200:e00713-17. doi: 10.1128/JB.00713-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Djaman O, Outten FW, Imlay JA. 2004. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem 279:44590–44599. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 67.Jang S, Imlay JA. 2007. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem 282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, Wackett LP, Fredrickson JK, Ghosal D. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 69.Seib KL, Tseng HJ, McEwan AG, Apicella MA, Jennings MP. 2004. Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: distinctive systems for different lifestyles. J Infect Dis 190:136–147. doi: 10.1086/421299. [DOI] [PubMed] [Google Scholar]
- 70.Tseng HJ, McEwan AG, Paton JC, Jennings MP. 2002. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect Immun 70:1635–1639. doi: 10.1128/iai.70.3.1635-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol 36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 72.Yeo WS, Lee JH, Lee KC, Roe JH. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol 61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 73.Varghese S, Wu A, Park S, Imlay KR, Imlay JA. 2007. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol 64:822–830. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andrews SC. 2010. The Ferritin-like superfamily: evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim Biophys Acta 1800:691–705. doi: 10.1016/j.bbagen.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 75.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2:593–608. doi: 10.1016/S1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 76.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem 274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 77.Colomer-Winter C, Flores-Mireles AL, Baker SP, Frank KL, Lynch AJL, Hultgren SJ, Kitten T, Lemos JA. 2018. Manganese acquisition is essential for virulence of Enterococcus faecalis. PLoS Pathog 14:e1007102. doi: 10.1371/journal.ppat.1007102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. 2001. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol Microbiol 40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 79.Martin JE, Waters LS, Storz G, Imlay JA. 2015. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet 11:e1004977. doi: 10.1371/journal.pgen.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monje-Casas F, Jurado J, Prieto-Alamo MJ, Holmgren A, Pueyo C. 2001. Expression analysis of the nrdHIEF operon from Escherichia coli: conditions that trigger the transcript level in vivo. J Biol Chem 276:18031–18037. doi: 10.1074/jbc.M011728200. [DOI] [PubMed] [Google Scholar]
- 81.Levin JD, Shapiro R, Demple B. 1991. Metalloenzymes in DNA repair. Escherichia coli endonuclease IV and Saccharomyces cerevisiae Apn1. J Biol Chem 266:22893–22898. [PubMed] [Google Scholar]
- 82.Tsaneva IR, Weiss B. 1990. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol 172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karp PD, Weaver D, Paley S, Fulcher C, Kubo A, Kothari A, Krummenacker M, Subhraveti P, Weerasinghe D, Gama-Castro S, Huerta AM, Muñiz-Rascado L, Bonavides-Martinez C, Weiss V, Peralta-Gil M, Santos-Zavaleta A, Schröder I, Mackie A, Gunsalus R, Collado-Vides J, Keseler IM, Paulsen I. 2014. The EcoCyc Database. EcoSal Plus 6. doi: 10.1128/ecosalplus.ESP-0009-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beckman RA, Mildvan AS, Loeb LA. 1985. On the fidelity of DNA replication: manganese mutagenesis in vitro. Biochemistry 24:5810–5817. doi: 10.1021/bi00342a019. [DOI] [PubMed] [Google Scholar]
- 85.Frank EG, Woodgate R. 2007. Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J Biol Chem 282:24689–24696. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- 86.Tokarsky EJ, Wallenmeyer PC, Phi KK, Suo Z. 2017. Significant impact of divalent metal ions on the fidelity, sugar selectivity, and drug incorporation efficiency of human PrimPol. DNA Repair (Amst) 49:51–59. doi: 10.1016/j.dnarep.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vaisman A, Ling H, Woodgate R, Yang W. 2005. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J 24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kunkel TA, Loeb LA. 1979. On the fidelity of DNA replication. Effect of divalent metal ion activators and deoxyrionucleoside triphosphate pools on in vitro mutagenesis. J Biol Chem 254:5718–5725. [PubMed] [Google Scholar]
- 89.Goodman MF, Keener S, Guidotti S, Branscomb EW. 1983. On the enzymatic basis for mutagenesis by manganese. J Biol Chem 258:3469–3475. [PubMed] [Google Scholar]
- 90.Ide H, Kow YW, Wallace SS. 1985. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res 13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evans J, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace S. 1993. Thymine ring saturation and fragmentation products: lesion bypass, misinsertion, and implications for mutagenesis. Mutat Res 299:147–156. doi: 10.1016/0165-1218(93)90092-R. [DOI] [PubMed] [Google Scholar]
- 92.Purmal AA, Lampman GW, Bond JP, Hatahet Z, Wallace SS. 1998. Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J Biol Chem 273:10026–10035. doi: 10.1074/jbc.273.16.10026. [DOI] [PubMed] [Google Scholar]
- 93.Purmal AA, Lampman GW, Kow YW, Wallace SS. 1994. The sequence context-dependent mispairing of 5-hydroxycytosine and 5-hydroxyuridine in vitro. Ann N Y Acad Sci 726:361–363. doi: 10.1111/j.1749-6632.1994.tb52852.x. [DOI] [PubMed] [Google Scholar]
- 94.Blaisdell JO, Hatahet Z, Wallace SS. 1999. A novel role for Escherichia coli endonuclease VIII in prevention of spontaneous G→T transversions. J Bacteriol 181:6396–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. 2011. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol 8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mellon I, Spivak G, Hanawalt PC. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 97.Schaaper RM, Cornacchio R. 1992. An Escherichia coli dnaE mutation with suppressor activity toward mutator mutD5. J Bacteriol 174:1974–1982. doi: 10.1128/jb.174.6.1974-1982.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


