Bacteria tolerate severe external stresses, including antibiotics, through a nonreplicative persistent (NRP) survival state, yet the proteins regulating this survival state are largely unknown. We show a specific universal stress protein (UspA616) controls the NRP state in Micrococcus luteus. Usps are widely conserved across bacteria, but their biological function(s) has remained elusive. UspA616 inactivation renders M. luteus susceptible to stress: bacteria die instead of adapting through the NRP state. UspA616 regulates malate synthase and isocitrate lyase, glyoxylate pathway enzymes important for chronic Mycobacterium tuberculosis infection. These data show that UspA616 regulates NRP stress survival in M. luteus and suggest a function for homologous proteins in other bacteria. Importantly, inhibitors of UspA616 and homologs may render NRP bacteria more susceptible to stresses, including current antibiotics.
KEYWORDS: universal stress protein, metabolism, stress response, glyoxylate shunt, proteomics, latency, antibiotic resistance, antibiotic tolerance, metabolic regulation
ABSTRACT
Bacteria have remarkable mechanisms to survive severe external stresses, and one of the most enigmatic is the nonreplicative persistent (NRP) state. Practically, NRP bacteria are difficult to treat, and so inhibiting the proteins underlying this survival state may render such bacteria more susceptible to external stresses, including antibiotics. Unfortunately, we know little about the proteins and mechanisms conferring survival through the NRP state. Here, we report that a universal stress protein (Usp) is a primary regulator of bacterial survival through the NRP state in Micrococcus luteus NCTC 2665, a biosafety level 1 (BSL1) mycobacterial relative. Usps are widely conserved, and bacteria, including Mycobacterium tuberculosis, Mycobacterium smegmatis, and Escherichia coli, have multiple paralogs with overlapping functions that have obscured their functional roles. A kanamycin resistance cassette inserted into the M. luteus universal stress protein A 616 gene (ΔuspA616::kan M. luteus) ablates the UspA616 protein and drastically impairs M. luteus survival under even short-term starvation (survival, 83% wild type versus 32% ΔuspA616::kan M. luteus) and hypoxia (survival, 96% wild type versus 48% ΔuspA616::kan M. luteus). We observed no detrimental UspA616 knockout phenotype in logarithmic growth. Proteomics demonstrated statistically significant log-phase upregulation of glyoxylate pathway enzymes isocitrate lyase and malate synthase in ΔuspA616::kan M. luteus. We note that these enzymes and the M. tuberculosis UspA616 homolog (Rv2623) are important in M. tuberculosis virulence and chronic infection, suggesting that Usps are important stress proteins across diverse bacterial species. We propose that UspA616 is a metabolic switch that controls survival by regulating the glyoxylate shunt.
IMPORTANCE Bacteria tolerate severe external stresses, including antibiotics, through a nonreplicative persistent (NRP) survival state, yet the proteins regulating this survival state are largely unknown. We show a specific universal stress protein (UspA616) controls the NRP state in Micrococcus luteus. Usps are widely conserved across bacteria, but their biological function(s) has remained elusive. UspA616 inactivation renders M. luteus susceptible to stress: bacteria die instead of adapting through the NRP state. UspA616 regulates malate synthase and isocitrate lyase, glyoxylate pathway enzymes important for chronic Mycobacterium tuberculosis infection. These data show that UspA616 regulates NRP stress survival in M. luteus and suggest a function for homologous proteins in other bacteria. Importantly, inhibitors of UspA616 and homologs may render NRP bacteria more susceptible to stresses, including current antibiotics.
INTRODUCTION
Bacterial survival mechanisms are critical components of antibiotic resistance, which is a recognized existential threat to populations worldwide (1–4). Resistance from antibiotic-inactivating genes or mutation is well known (5, 6), but bacteria can also resist antibiotics through a poorly understood nonreplicative persistent (NRP) state, which has been linked with viable-but-not-culturable (VBNC) and persister bacterial cell populations (7–9). Recent studies suggest VBNC and persistence are a continuum of bacterial survival states that coexist stochastically under stress (7, 10, 11). However, studies show that NRP bacteria exhibit an altered metabolism, making them difficult to treat (e.g., latent tuberculosis) (12, 13). Therefore, our research has focused on identifying and targeting the poorly understood proteins and mechanisms that underlie NRP survival to more effectively understand bacterial survival mechanisms and to combat difficult-to-treat bacterial infections.
An enigmatic family of conserved stress response proteins, denoted universal stress proteins (Usps), was discovered in Escherichia coli as a cytoplasmic 13.5-kDa protein upregulated under starvation, redox, toxic metals, pH, and antibiotic stresses (14–17). This protein family contains single-and two-domain variants and is distributed widely in Gram-positive and Gram-negative bacteria, Archaea, and even some invertebrates and plant species (18). Studies in NRP model bacteria Escherichia coli, Mycobacterium tuberculosis, and Mycobacterium smegmatis have failed to identify the clear biological function(s) of Usps, likely because of multiple functionally redundant paralogs (14, 16, 18–21). For example, Micrococcus luteus has only three universal stress protein (Usp) paralogs compared to six for E. coli, nine for M. tuberculosis, and 14 for M. smegmatis. Current thinking suggests Usps have a general protective role in stress responses.
Previously, we found a new link between NRP and a specific Usp (UspA616, WP_010079616.1, Mlut_01830) in Micrococcus luteus NCTC 2665, a Gram-positive biosafety level 1 (BSL1) actinobacterial relative of Mycobacteria, including M. tuberculosis and M. smegmatis (22). We hypothesized that a bacterial species with fewer paralogs might provide more defined Usp knockout phenotypes, thus linking function to this class of proteins (22). M. luteus is an ideal bacterial species to answer this question. M. luteus has a reproducible quiescent (nonproliferative) state reminiscent of VBNC and NRP states found in pathogenic Staphylococcus sp., Escherichia coli, Vibrio cholerae, and M. tuberculosis (8, 23–27). The small 2.5-Mb genome (compared to the M. tuberculosis genome at >4 Mb) suggests fewer M. luteus proteins and less potential protein level redundancy than in other NRP model bacteria (28). For example, M. luteus has only three UspA homologues (UspA184, WP_010080184.1, Mlut_19290; UspA712, Wp_101179712, Mlut_00830; and UspA616, WP_010079616.1, Mlut_01830). Importantly, our quantitative proteomics experiments identified only UspA616 as upregulated in quiescent M. luteus (22). This specific protein is a two-domain (tandem domains of class III/IV according to Tkaczuk et al. [29]) member of the Usp family and is conserved across diverse bacterial species at the structure and sequence levels (see Fig. S1A and B and S2 in the supplemental material) (18, 29). Knockout of UspA616 in a M. luteus competition assay resulted in a drastic loss of fitness upon starvation, supporting our hypothesis (30). UspA616 is the homolog of M. tuberculosis Rv2623, a Usp important for virulence and chronic infection in this pathogen in vivo (31). However, the association of Usps with dormancy and latency has never been reported (18, 19).
Here, we show that UspA616 has a critical role for bacterial survival beyond any general protective function. M. luteus without UspA616 loses NRP survival capacity directly because of cell death under both starvation and hypoxia. Quantitative proteomics identified that the glyoxylate shunt enzymes isocitrate lyase and malate synthase are upregulated in the logarithmic phase by loss of UspA616 protein. These data identify a novel biological function for this protein regulating a protein level pathway required for bacterial survival through regulation of the tricarboxylic acid (TCA) cycle and glyoxylate shunt proteins (Fig. 1) (32). We propose a new paradigm: UspA616 is a molecular switch that “turns on” stress-responsive bacterial survival programs at the protein level.
FIG 1.
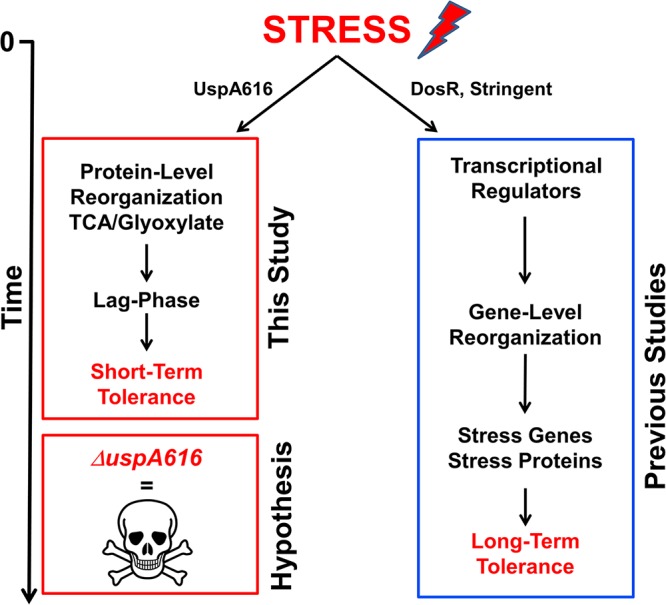
A global stress model and molecular model for induction of bacterial tolerance to stress. Stress induction initiates two integrated survival pathways at the transcriptional and protein levels. (Right) Transcriptional regulation through DosR, two-component stringent response, and other transcriptional regulators reorganizes bacterial gene expression to enhance stress survival but requires considerable time to upregulate appropriate stress genes (long-term tolerance). (Left) Protein level reorganization rapidly enhances short-term survival by switching metabolism from energy consumption to increased synthesis of survival proteins and molecular species, leading to a potential lag-phase adaptation to stress (short-term tolerance). We hypothesized that UspA616 protein is critical for this protein level reorganization and that inactivation of this protein results in cell death prior to acquiring longer-term stress tolerance, which was supported by the data presented in this article.
RESULTS
Generation and confirmation of uspA616 knockout in M. luteus.
The uspA616 gene (MLUT_RS12480, 1,323 bp) was inactivated by insertion of the pUCK4 kanamycin resistance gene internal to the uspA616 gene (2,587 bp) and transformation/homologous recombination as previously reported (33, 34). Transformed M. luteus cells were kanamycin resistant, and colony PCR analysis identified kanamycin insertion within the uspA616 gene (2,587 bp) (see Fig. S3 in the supplemental material). Loss of UspA616 protein was supported by mass spectrometry, which identified the UspA616 peptide EGVQALLEEVAGK in wild-type M. luteus but not in ΔuspA616::kan M. luteus [MH(+2) = 671.8643 atomic mass units (amu) monoisotopic] (Fig. S3 and supplemental spectra). Whole-genome sequencing identified a single kanamycin resistance cassette inserted only within the uspA616 gene sequence (Fig. S3). These data show complete and specific knockout of both the uspA616 gene and the resulting UspA616 protein.
Control for genetic manipulation in bacterial stress experiments.
We previously evaluated knockout of the crtE carotenoid gene in M. luteus for developing a colorimetric fitness assay (30). Genetic manipulations and the pUCK4 kanamycin resistance cassette were identical for insertional inactivation of the uspA616 and crtE genes, respectively. Fitness and survival deficits were not observed for crtE knockout but were severe for the uspA616 knockout. Therefore, ΔuspA616::kan M. luteus starvation and hypoxia phenotypes were attributed to insertional inactivation of the uspA616 gene and ablation of the protein, not to the pUCK4 kanamycin cassette or genetic manipulations.
ΔuspA616::kan M. luteus cannot survive nutrient starvation.
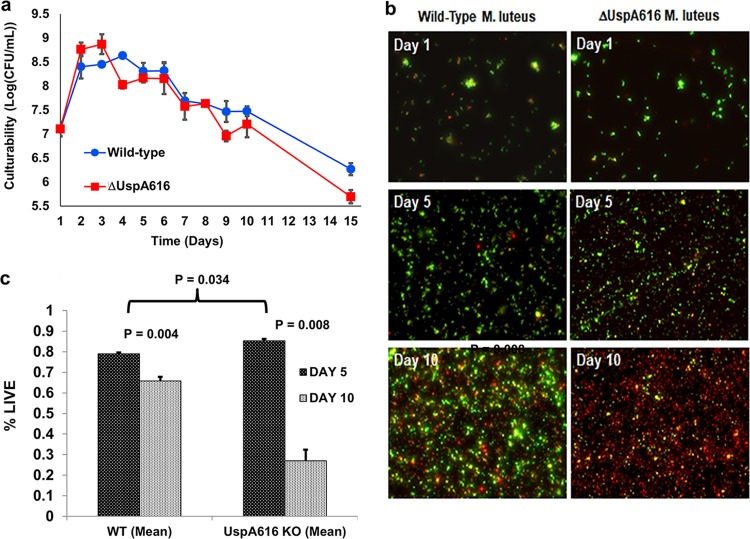
M. luteus grown on acetate minimal medium (AMM) grows in exponential phase (3 to 4 days) followed by transition to a NRP state as observed by loss of CFU. This quiescent state is initiated within hours of stationary phase and extends to ≥30 days, as previously observed (22, 35, 36). Wild-type and ΔuspA616::kan M. luteus strains were cultured in AMM to evaluate growth and starvation phases as previously described (22). Initial experiments on the ΔuspA616::kan M. luteus strain displayed an increase in logarithmic growth compared to that of wild-type bacteria followed by more significant decline as measured by optical density (data not shown). However, optical density does not measure cell culturability or viability. Therefore, subsequent experiments measured CFU culturability and fluorescence-based viability. CFU number supported an initial increase in culturability for ΔuspA616::kan M. luteus (Fig. 2a, days 1 to 3) followed by a more rapid decline (Fig. 2a, days 4 to 15). These data showed the same trends as previously observed for ΔuspA616::kan M. luteus, early logarithmic-phase growth enhancement followed by nearly zero survival fitness upon starvation after 4 to 6 days (30). This logarithmic-phase growth has important implications for the reported increased virulence phenotype in M. tuberculosis and considered in our discussion (31). The loss of culturability in ΔuspA616::kan M. luteus suggested increased cell death, which required LIVE/DEAD fluorescence measurements (green, live; red, dead). LIVE/DEAD fluorescence demonstrated high levels of viable bacteria (green cells) compared to that of the wild type in logarithmic phase (Fig. 2b, day 1) and in early phases of starvation (Fig. 2b, day 5). Quantitative analysis of these data also show enhanced logarithmic cell growth compared to that of wild-type M. luteus (Fig. 2c). This situation drastically changed after 10 days, where ΔuspA616::kan M. luteus bacteria were no longer viable by visual inspection and exhibited a 70% decrease in viability by quantitative measurements compared to <20% for wild-type M. luteus (Fig. 2b, day 10, and c).
FIG 2.
ΔuspA616::kan M. luteus cannot survive starvation. (a) Quiescence and viability of wild-type and ΔuspA616 M. luteus under starvation conditions in acetate minimal medium (AMM) was measured by CFU for 15 days. ΔuspA616::kan M. luteus demonstrated a significant advantage in logarithmic growth compared to that of wild-type M. luteus but reduced viability after onset of starvation at days 4 through day 15. (b) Fluorescence microscopy using BacLight (Thermo Scientific) differentiates viable (green) cells from cell death (red) based on cell membrane permeability. At day 1 and day 5, the numbers of viable cells are not observably different between the wild type and ΔuspA616::kan M. luteus, which correlates well with CFU data prior to onset of starvation. However, day 10 data show massive cell death for ΔuspA616 M. luteus, demonstrating that the viability loss observed in CFU numbers is directly attributable to a survival defect under starvation conditions. (c) Quantitation of the BacLight microscopy data using ImageJ (NIH) demonstrated clear statistical significance for cell death of ΔuspA616 M. luteus compared to that of wild-type bacteria. Wild-type M. luteus exhibited a modest loss from 78% viable cells at day 5 to 65% viable cells at day 10 (P = 0.004). In contrast, ΔuspA616 M. luteus exhibited 85% viability at day 5 followed by massive cell death by day 10 with <27% viability (P = 0.008). The small growth advantage observed in ΔuspA616::kan M. luteus in CFU viability numbers in panel a was confirmed by these quantitative data (P = 0.034). All error bars reported are standard errors of the means (SEMs).
These data demonstrate that ΔuspA616::kan M. luteus has a severe defect in survival under starvation and corroborate our published competition experiments (30). usp knockout experiments in E. coli or M. tuberculosis generated only modest in vitro phenotypes, but we observed robust in vitro phenotypes for ΔuspA616::kan M. luteus that correlated with the in vivo phenotype for Rv2623 knockout in M. tuberculosis (15, 21, 31). Therefore, the reduced redundancy in ΔuspA616::kan M. luteus simplified the observation of the importance of Usps for stress survival, supporting our hypothesis and suggesting that UspA616 homologs in other bacterial species may be important targets for study.
ΔuspA616 M. luteus cannot survive hypoxia.
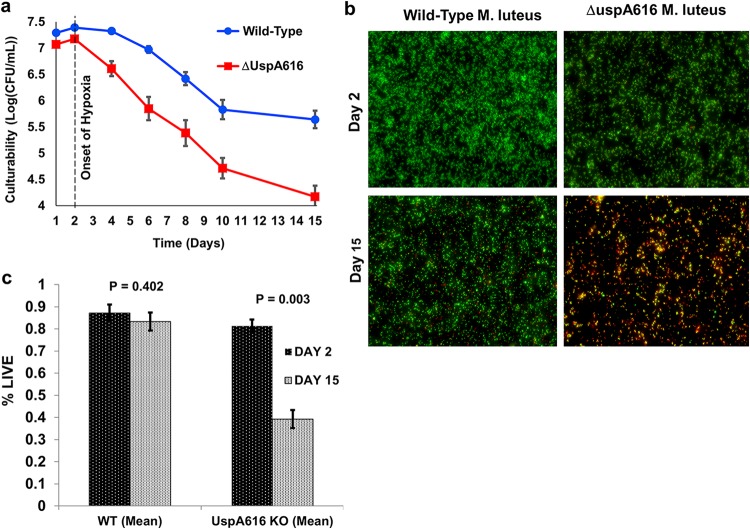
Wild-type and ΔuspA616::kan M. luteus strains were cultured in sealed tubes with limited air to rapidly deplete available oxygen (37). Cultures contained Luria broth (LB) buffered to pH 7.4 to prevent starvation or pH stress, thus isolating hypoxia for study. Oxygen depletion lower than 1% was monitored by methylene blue (1.5 μg/ml) (38). Initial inoculation levels of bacteria were modestly higher in the wild type than in ΔuspA616::kan M. luteus bacteria (Fig. 3a). However, logarithmic growth was essentially identical prior to hypoxia at approximately 48 h. After hypoxia, turbidity in ΔuspA616 M. luteus culture was reduced compared to that in wild-type bacteria, and CFU measurements showed a rapid decline in culturability (Fig. 3a). As shown for starvation, LIVE/DEAD fluorescence viability visualization and quantitative measurements demonstrated impaired ΔuspA616::kan M. luteus hypoxic survival (Fig. 3b and c). The fluorescence images in Fig. 3b for ΔuspA616::kan M. luteus at day 15 show high levels of yellow fluorescent cells that are likely in the process of cell death. These cells would exhibit both green and red fluorescence, leading to overestimation of the number of live cells in quantitative measurements (Fig. 3c).
FIG 3.
ΔuspA616::kan M. luteus has impaired hypoxic stress response. (a) CFU numbers under hypoxic conditions for ΔuspA616::kan M. luteus show reduced viability at all time points compared to that for wild-type M. luteus. Importantly, the CFU numbers for wild-type M. luteus began to stabilize by day 15, whereas ΔuspA616::kan M. luteus continued to rapidly lose viability. The dashed line at day 2 indicates the onset of hypoxia, as evident from loss of methylene blue coloration (see Materials and Methods). (b) Fluorescence microscopy using BacLight (Thermo Scientific) demonstrated viability differences between wild-type and ΔuspA616 M. luteus were attributable to differences in survival similar to those observed under starvation conditions. (c) Quantitation of the BacLight microscopy data for hypoxic cells demonstrated clear statistical significance for cell death of ΔuspA616::kan M. luteus compared to that of wild-type bacteria. Wild-type M. luteus demonstrated no significant loss in percentage of live cells from hypoxia (P = 0.402), while ΔuspA616 M. luteus demonstrated severe loss of living cells under these same conditions (P = 0.0003). The data here and from Fig. 2 demonstrate that ΔuspA616::kan M. luteus has impaired survival under starvation and hypoxia, both well-established stress models used for latency studies in M. tuberculosis. All error bars reported are SEMs.
These data show that UspA616 protein is important for survival under hypoxic conditions as well as under starvation, both primary stress models for M. tuberculosis latency that upregulate the DosR stress-responsive transcription system (39). Not surprisingly, Rv2623 in M. tuberculosis is a target of DosR-type transcription factors and is upregulated under starvation and hypoxia (40–43). M. luteus has a similar two-component system which has not been characterized in the stress responses to starvation and hypoxia.
UspA616 regulates survival through the glyoxylate shunt.
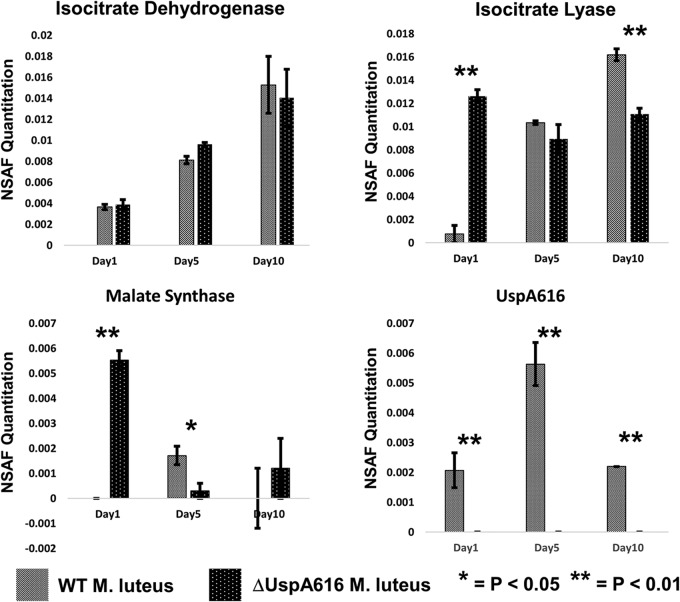
High levels of UspA616 were observed in the quiescent state by both accurate mass/retention time (AMT) and normalized spectral abundance factor (NSAF) data (Fig. S4A), which corroborated the severe deficits in starvation survival in ΔuspA616::kan M. luteus. We were able to differentiate logarithmic and quiescent M. luteus cultures using AMT and NSAF quantitation (Fig. S4B). Global proteomics experiments in wild-type and ΔuspA616::kan M. luteus identified important trends. Wild-type M. luteus showed increasing isocitrate dehydrogenase and isocitrate lyase levels as starvation progressed but also increased malate synthase only at day 5 (Fig. 4). As expected, we observed UspA616 at all time points. In ΔuspA616::kan M. luteus, we found wild-type isocitrate dehydrogenase levels but abnormally high levels of isocitrate lyase and malate synthase in early logarithmic phase (Fig. 4) (22).
FIG 4.
Normalized spectral abundance factor (NSAF) quantitation for isocitrate dehydrogenase, isocitrate lyase, malate synthase, and UspA616 proteins in wild-type and ΔuspA616::kan M. luteus. Wild-type and ΔuspA616::kan M. luteus strains were incubated in acetate minimal medium and sampled at day 1 (logarithmic growth), day 5 (transition into quiescence), and day 10 (fully quiescent) for proteomics analysis. Isocitrate dehydrogenase was progressively upregulated during transition to quiescence in both wild-type and UspA616 knockout bacterial strains. Isocitrate lyase exhibited progressive upregulation from day 1 through day 10 in wild-type M. luteus. However, this protein was unusually upregulated in early logarithmic phase in ΔuspA616::kan M. luteus. Malate synthase was observed at day 5 in wild-type M. luteus, but could not be reliably observed at day 1 or day 10. In ΔuspA616::kan M. luteus, malate synthase was highly upregulated in early logarithmic growth, and almost completely downregulated in the transition to quiescence. The upregulation of malate synthase and isocitrate lyase in wild-type M. luteus occurs at precisely the time required for starvation survival responses. UspA616 was observed to be modestly increased at day 5 compared to day 1 and day 10 in wild-type M. luteus. As expected, the UspA616 protein was never observed in ΔuspA616::kan M. luteus. All error bars reported are SEMs.
These data suggest a mechanism for rapid proliferation in logarithmic growth followed by cell death in ΔuspA616::kan M. luteus. Increased malate synthase may be a response to toxic levels of glyoxylate from high levels of isocitrate lyase in ΔuspA616::kan M. luteus. High levels of both isocitrate lyase and malate synthase in early logarithmic phase would enhance growth in AMM compared to that of wild-type bacteria (Fig. 2a and c) (30). Critically, isocitrate lyase was retained and malate synthase was lost after acetate was depleted at day 5. The loss of malate synthase during stress induction of the NRP state at day 5 in ΔuspA616::kan M. luteus (Fig. 4) may increase cell death upon starvation by glyoxylate toxicity, as previously observed in M. tuberculosis (44).
Molecular basis for starvation survival and quiescence in M. luteus.
Previously, we used normalized spectral abundance factor (NSAF) proteomics to measure protein quantities in logarithmic and quiescent M. luteus cultures (22). We expanded this analysis using targeted proteomics based on accurate mass/retention time (AMT) (45–47). AMT required only the peak area from accurate parent masses and retention time for effective quantitation. Of critical importance, the reduced complexity of the M. luteus proteome (∼2,200 open reading frames [ORFs]) enabled AMT to uniquely identify and quantitate the peak areas of multiple proteins under study in both logarithmic and quiescent M. luteus (see Table S1). Tandem mass spectrometry (MS/MS) fragmentation assignment confirmed accurate identification of these peptides (supplemental spectra).
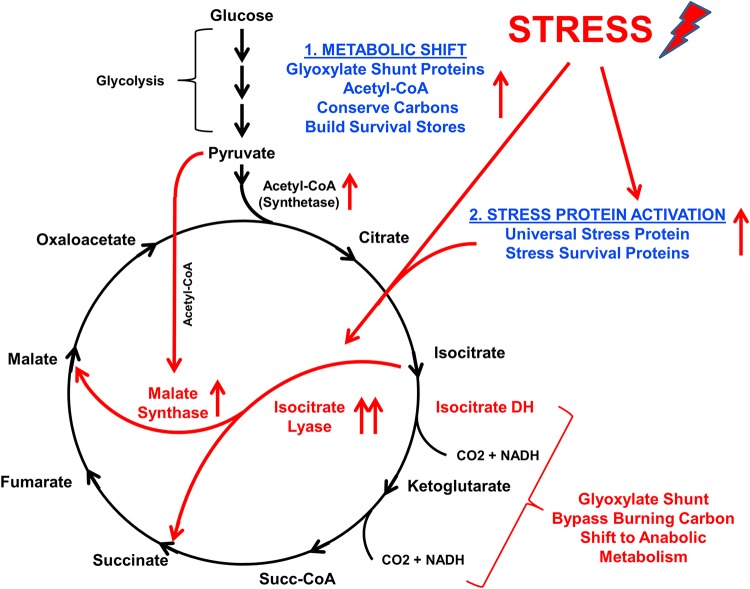
These quantitative data in logarithmic, quiescent, and ΔuspA616::kan M. luteus were mapped to the TCA cycle and glyoxylate shunt (Fig. 5). Starvation survival through the NRP state requires activation of two important biological processes: (i) glyoxylate shunt bypass of carbon degradation/oxidation steps in the TCA cycle and (ii) upregulation of stress survival proteins that control energy versus survival metabolism. These data suggest that ablation of the UspA616 protein alone is sufficient to prevent the NRP state and impair stress survival in M. luteus.
FIG 5.
A molecular model for induction of bacterial tolerance to stress through the glyoxylate shunt. Specific proteins upregulated in response to nutrient starvation are mapped to the TCA cycle and glyoxylate shunt. Starvation increases the glyoxylate shunt proteins isocitrate lyase and malate synthase, a metabolic shift away from energy production and carbon consumption (bypass of isocitrate dehydrogenase and α-ketoglutarate dehydrogenase steps in the TCA cycle). Increased levels of other TCA cycle proteins, including acetyl-CoA synthetase, suggest repurposing of carbon nutrient stores to anabolic synthesis of molecular species important for survival. Simultaneously, stress protein activation upregulates proteins, including universal stress proteins that critically switch bacterial metabolism into survival mode through regulation of the TCA cycle and glyoxylate shunt.
DISCUSSION
Nonreplicative persistent state.
Our initial experiments in Micrococcus luteus were focused on understanding this important bacterial survival mechanism in one of only few bacterial species with a well-defined and reproducible NRP state (22, 30). Persistence was first observed by failure of penicillin to sterilize Staphylococcus aureus cultures (25). The NRP state was first described by Wayne in 1976 in microaerophilic cultures of Mycobacterium tuberculosis (48). Since these descriptions, the NRP state has been proposed for diverse pathogenic and nonpathogenic bacterial species (10, 49). Phenotypic characteristics include the cessation of replication, reduction in metabolic activity (but not necessarily cessation), morphological changes, and capacity to survive potentially lethal external stresses, including starvation, hypoxia, host immunity, and antibiotics (48, 50). In addition to these NRP phenotypes, M. luteus also has changes to the entire protein complement of the NRP cell population (22). Diverse in vitro models have been used to model the NRP state, the best studied being hypoxia and starvation (23, 37, 51). However, it is unclear how these focused stress models replicate the complex physiological conditions that cause the NRP state in pathogenic and nonpathogenic bacteria in the environment (51). Implications for M. luteus experiments in evaluating these limitations are considered below.
Importance of the glyoxylate shunt and acetate in survival.
Upregulation of the glyoxylate shunt would bypass isocitrate dehydrogenase and oxidative degradation of carbon sources, rerouting metabolism to the synthesis of proteins and molecules, such as succinate, critical for survival (Fig. 5). This is consistent with an increased requirement for succinate during hypoxia in M. tuberculosis (52). Surprisingly, increased malate synthase commensurate with increased isocitrate lyase was not initially observed in starved wild-type M. luteus. Further experiments identified that malate synthase is temporally regulated to coincide with stress-responsive survival programs as described below.
Inactivation of uspA in E. coli produced diauxic growth and excretion of acetate into the medium via acetyl coenzyme A (acetyl-CoA) synthetase (15). Isocitrate lyase and malate synthase (acetate metabolism enzymes) were undetectable until complete depletion of glucose or gluconate (15). Wild-type M. luteus quiescence is achieved in AMM but not under rich medium conditions, and no diauxic growth was observed (22, 30). These observations suggest UspA616 may control survival responses via acetate metabolism through isocitrate lyase and malate synthase. This hypothesis was proposed by Nyström and Neidhardt for UspA in E. coli but was never substantiated (15). Acetate as a sole carbon source also replicates exposure to long-chain fatty acids (52). Historically, the glyoxylate pathway was an anaplerotic metabolic pathway important for adaptation to different carbon nutrients (32). Our data indicate that the glyoxylate pathway (and perhaps anaplerotic metabolism in general) provides a robust mechanism to regulate bacterial survival under extreme external stresses. Metabolic stresses (e.g., starvation, hypoxia, antibiotics, etc.) blocking any part of the metabolic pathway can be rerouted (bypassed) through ancillary pathways and preserve growth and survival.
Challenges in studying the NRP state.
The mechanisms for the nonreplicative persistent (NRP) state including antibiotic tolerance are poorly understood (7–9). However, NRP-type tolerance may contribute to acquisition of bona fide genetic resistance (53, 54). Systems biology studies in traditionally studied NRP bacterial systems have significant limitations from the complexity in bacterial genetic and protein responses (42, 43, 55, 56). Our data show that M. luteus has less redundancy at the genetic and protein levels, is amenable to targeted mechanistic studies, and provides new and complementary knowledge about the molecular basis for bacterial stress survival. This research links genes and proteins to stress phenotypes, establishing M. luteus as a valuable system for understanding starvation and hypoxic stress responses (22, 30).
Studying bacterial stress is challenging, because bacteria do not grow or behave consistently under stress. Experimental data and current literature indicate exceptional biological variability in bacterial responses under external stress. Additionally, statistical methods well-suited for Gaussian distributions and well-ordered and reproducible data sets may fail when confronted with such variability. For example, some ΔuspA616::kan M. luteus cultures exhibited significant cell death at day 5 (Fig. 3B), but in aggregate, this was not statistically significant in the quantitative data until day 10 (Fig. 3C). This increased variability in bacterial behavior may not reflect a failure in experimental procedures or statistical methods but rather may be a critical adaptive mechanism to enhance stress survival via a “hedge-betting” strategy (57, 58).
Our experiments suggest a combinatorial approach is needed to study such variation. Targeted and earlier global proteomics studies identified proteins upregulated in the quiescent state that were not readily observable in transcriptomic or metabolomics studies: UspA616 is upregulated in the quiescent state, and this protein regulates malate synthase and isocitrate lyase, enzymes known to play an important role in chronic infection in other bacterial species including M. tuberculosis. Therefore, a more complete picture of bacterial survival processes may require diverse and complementary experimental approaches applied across multiple bacterial species, including M. luteus, E. coli, M. smegmatis, and M. tuberculosis.
M. luteus as a model for tolerance mechanisms.
Studies in E. coli (a facultative anaerobe) identified lag-time adaptation under short-term severe stress (59). Unlike E. coli, M. luteus is an obligate aerobic bacterium similar to mycobacteria. Unlike in E. coli experiments, we observed no lag-time defect (i.e., time for initial growth induction) in ΔuspA616::kan M. luteus by any measurements (optical density at 600 nm [OD600], CFU, or fluorescence viability microscopy). This was despite obvious enhancement of logarithmic growth and culturability under initial conditions and survival deficits under starvation and hypoxia. Tolerance of one stress can confer tolerance of other stresses, suggesting conserved survival mechanisms (7, 9, 59). Combined, our protein level data and genetic and metabolic data, including those from the study by Fridman et al. (59), provide a satisfying, albeit more complex, hypothesis for tolerance (Fig. 1). Bacteria exposed to stress such as starvation, antibiotics, or pH rapidly shift glyoxylate shunt and TCA cycle regulation to enable short-term survival, leading to a metabolically active lag state with reduced proliferation (Fig. 1, left). Longer-term survival may be enhanced through stress-responsive transcription factors (i.e., DosR), which increase expression of critical stress response genes and proteins (Fig. 1, right) (42, 60). The purpose of DosR, and potentially other stress-responsive transcriptional systems, may be to upregulate the genes and proteins, including Usps that “execute” the stress survival program. The uspA616 gene and the homolog Rv2623 in M. tuberculosis appear to be regulated by such transcription factors. Therefore, bacterial tolerance is likely a consequence of reorganized stress metabolism that requires integration of both protein and genetic “programs” (Fig. 1).
M. luteus as a model for studying Usps in bacterial pathogenesis.
The study of the NRP state and latency is difficult, as most current knowledge on this survival state is limited to studies in M. tuberculosis and M. smegmatis, actinobacterial relatives of M. luteus. Therefore, it may be instructive to compare our data in M. luteus to established literature in these mycobacteria. Loebel et al.’s starvation model (23, 24) and Wayne et al.’s hypoxia model (37) are two established models for latency in M. tuberculosis, and ΔuspA616::kan M. luteus exhibits severe survival deficits under these same stress models. Our data explain multiple conflicting published observations through Usp control of the glyoxylate shunt enzymes isocitrate lyase and malate synthase.
In one study, inactivation of the M. tuberculosis Usp Rv2623 (the homolog of UspA616) generated hypervirulence in vivo, which was interpreted as a defect in achieving latent infection (31). Logarithmic (acute phase) ΔuspA616::kan M. luteus exhibited abnormally high levels of isocitrate lyase and malate synthase. If similar in Rv2623-deficient M. tuberculosis, this would enhance acute-phase bacterial growth through additional acetate/fatty acid metabolism during acute infection. Supporting this proposition, glyoxylate metabolism has been correlated with M. tuberculosis anaerobic adaptation, and malate synthase is critical for the detoxification of glyoxylate (44, 61). Multiple other metabolomics studies also point to isocitrate lyase and malate synthase in stress adaptation (52, 62, 63). As hypothesized in Fig. 1, ΔuspA616::kan M. luteus under longer-term starvation or hypoxia shows enhanced cell death (e.g., defect in survival is not an enhanced NRP state, but cell death). Therefore, hypervirulence from Rv2623 inactivation may be a hyperactive logarithmic phase phenotype, not a latency defect.
In a second study, inactivation of isocitrate lyase in M. tuberculosis attenuated chronic and persistent infection in immunocompetent animals without impairing acute infection (64), which apparently conflicts with the study described above. M. luteus quiescence does not occur under rich nutrient conditions but does in acetate medium, which may be considered a nutrient-poor carbon source (30). By extension, M. tuberculosis without isocitrate lyase might proliferate unimpeded while in a rich nutrient environment but not under nutrient-poor conditions correlated to latency in this pathogen (i.e., macrophage endosomes and hypoxic granulomas). The original papers on quiescence and reactivation in M. luteus never used glucose, gluconate, or rich nutrient conditions (22, 27, 35, 65). Acetate or other poor nutrient sources may signal for starvation and quiescence in M. luteus and perhaps latency in M. tuberculosis and other bacterial species. An important prediction from these data is that Rv2623-deficient M. tuberculosis may be hypersensitive to starvation, hypoxia, or antibiotics, which were stresses not apparently tested (31). Therefore, we suggest that Usp inhibition may sensitize pathogenic bacteria to external stresses, including antibiotics, by blocking tolerance inherent in the NRP state.
Potential functions and mechanisms for Usp-type proteins.
Biological and biochemical understanding of Usps has been severely impaired by (i) limitations in model bacterial systems exhibiting weak phenotypes from multiple functionally redundant proteins and (ii) limited molecular-based experimental approaches for studying these proteins. These limitations have relegated current ideas about Usp function to a “general protective role” in bacterial survival, and the mechanisms are largely unknown (18). Despite this situation, there are clues to functions and mechanisms for these proteins. Rv2623, the M. tuberculosis homolog of UspA616, appears to be regulated by the DosR stress-responsive transcription factor (60), and other usp genes may be responsive to different transcriptional regulators (66, 67). Nucleotide binding is variable among Usp-family members (19), but Rv2623 requires ATP-binding for biological activity (31). Diverse evidence suggests that protein dimerization (68), protein interactions (69), and phosphorylation/signaling (70, 71) may have importance for Usp biological functions. While these potential mechanisms are speculative at present, the data presented here clearly demonstrate that UspA616 is critical for bacterial survival and that M. luteus is a valuable and uniquely viable model capable of deciphering universal stress protein function. These data suggest a new paradigm for Usps as molecular switches that turn on bacterial survival under starvation and hypoxia.
MATERIALS AND METHODS
Reagents and materials.
Chemicals, buffers, and cell culture materials and reagents were purchased from Thermo Fisher Scientific, Sigma-Aldrich, or VWR and were of ACS reagent-grade certification or better. Chromatography and mass spectrometry solvents were purchased from Fisher Scientific and were Optima grade or better. The pGEM3ZF(+) plasmid was purchased from Promega Corporation (Madison, WI). The BacLight LIVE/DEAD bacterial viability kit for microscopy (L7007) was purchased from Thermo Fisher Scientific. Whole-genome sequencing materials were purchased from Qiagen and Illumina.
M. luteus growth conditions.
Micrococcus luteus NCTC 2665 was purchased from ATCC (Manassas, VA) and cryogenically stored long-term at −80°C in 20% glycerol. For all experiments, M. luteus colonies were obtained from LB agar plates inoculated from a glycerol stock and incubated at 30°C until colonies appeared. Starter cultures of M. luteus were inoculated from a single colony in LB medium and incubated at 30°C at 250 rpm on an orbital shaker with subculturing every 24 h using a 1:100 dilution in fresh medium. Culturing in defined acetate minimal media (AMM) was performed as previously described (22).
Transformation of M. luteus.
Transformation of M. luteus was performed as previously described (30) and based on the methods described by Angelov et al. (34) and Kloos and Schultes (33). Briefly, a single colony of M. luteus was grown aerobically overnight at 30°C in LB. From the starter culture, 500 μl of cells was washed in 1× phosphate-buffered saline (PBS) and inoculated in 50 ml of defined glutamate medium (DGM). Cells were grown aerobically at 30°C for 24 to 36 h and harvested by centrifugation. Harvested cells were chilled on ice, washed with 50 mM Tris-HCl (pH 7.4) and 100 mM CaCl2, and resuspended in 2 ml of 50 mM Tris-HCl (pH 7.4), 100 mM CaCl2, and 0.5% monosodium glutamate. The uspA616 gene (1,323 bp) sequence was amplified by Pfu polymerase using primers (forward) 5′-GGGCTCCGAGGCGTAGACTG-3′ and (reverse) 5′-GGTGGTGGAGGACGAGTAC-3′ and blunt-end cloned into pGEM-5ZF(+) at the EcoRV site. The kanamycin cassette from pUC4K was inserted into the uspA616 gene sequence via the unique BamHI site. One to ten micrograms of NdeI-linearized pGEM-5ZF(+) ΔuspA616::kan DNA (prepared as previously described) (30) was added to each aliquot of cells for 30 to 45 min at 30°C with orbital shaking at 250 rpm. After incubation, 1 ml of LB medium was added to the cells, and incubation continued for 10 to 16 h at 30°C with shaking. Cells were harvested by centrifugation for 5 min at 3,000 × g and resuspended in 100 μl of 1× PBS. Three milliliters of Top agar (0.7% agar) was melted at 45°C, and 100 μg/ml kanamycin and 100 μl of diluted cells were added, mixed, and poured onto the surface of a warm LB agar plate supplemented with kanamycin. Top agar was evenly dispersed until it solidified at room temperature, and plates were incubated at 30°C until colonies appeared.
Confirmation of knockout: colony PCR and whole-genome sequencing.
(i) Colony PCR. Colonies were picked and resuspended in 15 μl of 10 mM Tris-HCl, 0.1% SDS (pH 8). From this suspension, 5 μl was used to inoculate 5 ml of LB medium supplemented with kanamycin (100 μg/ml). The remaining 10 μl of resuspended colony was lysed by boiling at 90°C for 15 min, and lysed colony debris was harvested by centrifugation for 5 min at 16,000 × g. For a 25-μl PCR mixture, 0.5 μl of lysed colony supernatant was used as the template DNA.
(ii) Whole-genome sequencing. Genomic DNA was extracted by the University of Houston Seq-N-Edit Core using 1 ml of turbid ΔuspA616::kan M. luteus in LB medium. After homogenization with TissueLyser II (Qiagen), DNA was isolated with the PowerSoil Pro plus IRT miniprep kit automated on a QIAcube (Qiagen). Extracted genomic DNA (gDNA) was quantified by the Qubit 2.0 fluorometer (Thermo Fisher), and integrity was verified using a 4200 TapeStation (Agilent) genomic DNA assay. Sequencing libraries were prepared using the Nextera DNA Flex (Illumina) with 500 ng of gDNA input. After quantification, sequencing library size and purity were determined by the 4200 TapeStation D1000 assay. Libraries were sequenced on a MiSeq (Illumina) and assembled using SPAdes genome assembly algorithm.
(iii) Mass spectrometry. Procedures for accurate mass tag (AMT) and quantitative mass spectrometry to confirm ablation of UspA616 protein are described below.
Quantification of CFU.
Statistical calculations were performed within Excel or using the online tools from SciStatCalc (http://scistatcalc.blogspot.com). Images were scanned on an Epson Perfection 2580 photo scanner and processed using ImageJ (https://imagej.nih.gov/ij/index.html) to count colonies from .tif images in CFU calculations. CFU measurement images were counted using the “analyze particles” option in ImageJ using 16-bit image analysis on thresholded image data after applying the watershed binary. Calculated colony numbers were normalized for dilution, calculated to CFU per milliliter, and converted to log10 values. Calculations were manually checked, and statistics were calculated from a minimum of three replicate measurements (n = 3).
Bacterial viability quantification by fluorescence microscopy.
Bacterial viability was quantified using the Molecular Probes BacLight LIVE/DEAD bacterial viability kit for microscopy. Briefly, cells were harvested by centrifugation, washed with 500 μl of 0.85% sodium chloride, resuspended in a mixture of Syto 9 and propidium iodide in 0.85% sodium chloride, and incubated in the dark for 15 min. After incubation, 10 μl of stained cell suspension and a drop of mounting oil were added to the middle of a microscope slide and coverslipped. Samples were visualized by fluorescence microscopy using the EVO FL cell imaging system (Thermo Fisher) at 470 nm for Syto 9 and 531 nm for propidium iodide. Fluorescence images were merged, exported as .tiff files, and analyzed using ImageJ (https://imagej.nih.gov/ij/index.html) or FIJI (https://fiji.sc). Images were corrected for brightness and contrast, red, green, and blue (RGB) channels were split, and particle edges and thresholds were adjusted for best quality. Particles were then analyzed with set thresholds: 0 to infinite particle size and circularity between 0 and 1. The percentage of viable cells was calculated by dividing viable counts by the total counts from live and dead cell imaging.
Starvation stress in acetate minimal medium.
To begin starvation, 100 ml of AMM in 500-ml Erlenmeyer flasks was inoculated with 1 ml of M. luteus starter culture (see “M. luteus growth conditions” above). OD600 and culture viability using CFU and LIVE/DEAD fluorescence microscopy were monitored daily for 30 days. For mass spectrometry, 10 ml of cell suspension was removed at 1, 5, 10, and 25 days and harvested by centrifugation at 10,000 × g for 10 min at 4 °C, and cell pellets were processed for mass spectrometry or cryopreserved at −80 °C (n = 5 biological replicates).
Wayne model of hypoxia for oxygen limitation stress.
Thirty-four milliliters of LB medium (18-ml headspace ratio = 0.5), a flea stir bar, and 1.5 μg/ml methylene blue were placed in each 125-mm by 25-mm screw-cap test tube and autoclaved. ΔuspA616::kan M. luteus tubes were supplemented with 100 μg/ml of kanamycin. Sample sets were inoculated with 340 μl of wild-type or ΔuspA616::kan M. luteus starter cultures and sealed with a Suba-Seal silicone rubber septa wrapped with copper wire. Tubes were incubated at 30°C on a mechanical stir plate set to completely mix the culture without disturbing the water-air interface. Samples (n = 5 biological replicates) for OD600 growth, CFU measurements, and LIVE/DEAD imaging were taken on day 0 and every other day anaerobically with a syringe equipped with a beveled 10-cm metal needle flushed with nitrogen gas. For mass spectrometric analysis, 34-ml samples were taken just prior to hypoxia at day 2 and at day 15 (n = 5 biological replicates).
Proteomic analysis of M. luteus.
(i) M. luteus cell lysis and reduction/alkylation. The procedures described previously (22) were used with the following modifications. Cryopreserved cells (∼50 mg) were washed three times with 500 μl of cold 50 mM Tris-HCl (pH 7.4), harvested by centrifugation, and lysed by bead beating on a Qiagen TissueLyser II using lysis buffer (50 mM Tris-HCl, 4 M urea, pH 7.4, with 1× Halt protease inhibitor cocktail [Thermo Scientific]). Cell debris was removed by centrifugation for 10 min at 16,000 × g, and protein concentration was measured on a NanoDrop 2000c (Thermo Scientific) at 280 nm. Samples were reduced by 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) at 37°C for 30 min, alkylated by 10 mM iodoacetamide at room temperature in the dark, and stored at −80°C or subjected to quantitative protein chloroform-methanol precipitation as previously described (22, 72).
(ii) Trypsin digestion. Precipitated protein pellets were resuspended in 80 μl of 25 mM Tris-HCl (pH 7.4), and sequencing-grade trypsin (250 ng; Worthington Biochemicals) was added. The reaction mixture was incubated for 18 to 24 h at 37°C before quenching by addition of 10 μl of 10% formic acid. Samples were lyophilized and stored at −80°C or immediately used for mass spectrometry.
(iii) LC-MS/MS analysis using the Bruker MicrOTOF-QII. Proteomics experiments were performed on a Bruker Daltonics MicrOTOF-QII (Q-TOF) equipped with a CaptiveSpray nanospray source and an Agilent 1200 Nano high-pressure liquid chromatography (HPLC) system. Chromatography was with an in-house-packed 150-μm by 150-mm nanobore C18 reverse phase column with a gradient from 15% solvent A (water, 0.1% formic acid) to 90% solvent B (acetonitrile, 0.1% formic acid) over 45 or 75 min. Spray voltage was set to 1,400 V, dry gas to 2.0 liters/min, and capillary temperature to 150°C. All samples were analyzed either in data-dependent fragmentation (MS/MS) mode to accurately identify peptide fragmentation and retention times for normalized spectral abundance factor (NSAF) quantitation or targeted accurate mass tag (AMT) quantitation based on parent masses and retention time. Chromatographic runs and all mass spectra were internally calibrated using either Glu-fibrinopeptide (added as an internal standard) or an abundant peptide from EF-tu (endogenously present). All data analysis was performed using Compass Data Analysis software (Bruker Daltonics, version 4.0). Searches were performed in Open Mass Spectrometry Search Algorithm (OMSSA) against a custom M. luteus FASTA proteome database containing both standard and randomized decoy sequences (NCBI) (RefSeq/Bacteria/Micrococcus_luteus/all_assembly_versions/GCF_000023205.1_ASM2320v1). False discovery rate (FDR) was maintained at ∼1.0%. For AMT data analysis, the combination of accurate mass (±0.005 amu) and retention time within 0.2 min indicated a correct peptide assignment, but complete liquid chromatography tandem mass spectrometry (LC-MS/MS) experiments were performed after every 6 to 10 runs to confirm peptide identifications. Extracted ion chromatograms (EIC) from calculated accurate masses for target peptides were Gaussian smoothed and peak areas calculated using Bruker Daltonics Compass Data Analysis software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Amy Vollmer (Swarthmore College), James Sacchettini (Texas A&M University), James Oliver (UNC Charlotte), Steven Finkel (University of Southern California), and Dianne Newman (CalTech) for their constructive discussions, guidance, and unfailing support in this research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00497-19.
REFERENCES
- 1.Centers for Disease Control. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control, Atlanta, GA. [Google Scholar]
- 2.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson E, Kaur P. 2018. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol 9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim MY, Cho Y, Rho M. 2018. Diverse distribution of resistomes in the human and environmental microbiomes. Curr Genomics 19:701–711. doi: 10.2174/1389202919666180911130845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balaban NQ, Gerdes K, Lewis K, McKinney JD. 2013. A problem of persistence: still more questions than answers? Nat Rev Microbiol 11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 8.Ayrapetyan M, Williams TC, Oliver JD. 2015. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol 23:7–13. doi: 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 10.Ayrapetyan M, Williams T, Oliver JD. 2018. Relationship between the viable but nonculturable state and antibiotic persister cells. J Bacteriol 200:e00249-18. doi: 10.1128/JB.00249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J-S, Chowdhury N, Yamasaki R, Wood TK. 2018. Viable but non-culturable and persistence describe the same bacterial stress state. Environ Microbiol 20:2038–2048. doi: 10.1111/1462-2920.14075. [DOI] [PubMed] [Google Scholar]
- 12.Lillebaek T, Dirksen A, Baess I, Strunge B, Thomsen VØ, Andersen AB. 2002. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J Infect Dis 185:401–404. doi: 10.1086/338342. [DOI] [PubMed] [Google Scholar]
- 13.Cardona P-J. 2016. Reactivation or reinfection in adult tuberculosis: is that the question? Int J Mycobacteriol 5:400–407. doi: 10.1016/j.ijmyco.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 14.Nyström T, Neidhardt FC. 1992. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol Microbiol 6:3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 15.Nyström T, Neidhardt FC. 1993. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J Bacteriol 175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustavsson N, Diez A, Nyström T. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol Microbiol 43:107–117. doi: 10.1046/j.1365-2958.2002.02720.x. [DOI] [PubMed] [Google Scholar]
- 17.O’Connor A, McClean S. 2017. The role of universal stress proteins in bacterial infections. Curr Med Chem 24:3970–3979. doi: 10.2174/0929867324666170124145543. [DOI] [PubMed] [Google Scholar]
- 18.Vollmer AC, Bark SJ. 2018. Twenty-five years of investigating the universal stress protein: structure, function and applications, p 1–36. Advances in Applied Microbiology, 1st ed Academic Press, Inc., San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 19.Kvint K, Nachin L, Diez A, Nyström T. 2003. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol 6:140–145. doi: 10.1016/S1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 20.Nachin L, Nannmark U, Nyström T. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol 187:6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hingley-Wilson SM, Lougheed KEA, Ferguson K, Leiva S, Williams HD. 2010. Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis (Edinb) 90:236–244. doi: 10.1016/j.tube.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali S, Mitchell M, Havis S, Bodunrin A, Rangel J, Olson G, Widger WR, Bark SJ. 2017. A Proteomic signature of dormancy in the actinobacterium Micrococcus luteus. J Bacteriol 199:e00206-17. doi: 10.1128/JB.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loebel RO, Shorr E, Richardson HB. 1933. The influence of adverse conditions upon the respiratory metabolism and growth of human tubercle bacilli. J Bacteriol 26:167–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loebel RO, Shorr E, Richardson HB. 1933. The Influence of foodstuffs upon the respiratory metabolism and growth of human tubercle bacilli. J Bacteriol 26:139–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bigger J. 1944. Treatment of staphyloccal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 26.Kaprelyants AS, Kell DB. 1993. Dormancy in stationary-phase cultures of Micrococcus luteus: flow cytometric analysis of starvation and resuscitation. Appl Environ Microbiol 59:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaprelyants AS, Mukamolova GV, Davey HM, Kell DB. 1996. Quantitative analysis of the physiological heterogeneity within starved cultures of Micrococcus luteus by flow cytometry and cell sorting. Appl Environ Microbiol 62:1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young M, Artsatbanov V, Beller HR, Chandra G, Chater KF, Dover LG, Goh E-B, Kahan T, Kaprelyants AS, Kyrpides N, Lapidus A, Lowry SR, Lykidis A, Mahillon J, Markowitz V, Mavromatis K, Mukamolova GV, Oren A, Rokem JS, Smith MCM, Young DI, Greenblatt CL. 2010. Genome sequence of the Fleming strain of Micrococcus luteus, a simple free-living actinobacterium. J Bacteriol 192:841–860. doi: 10.1128/JB.01254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tkaczuk KL, A Shumilin I, Chruszcz M, Evdokimova E, Savchenko A, Minor W. 2013. Structural and functional insight into the universal stress protein family. Evol Appl 6:434–449. doi: 10.1111/eva.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havis S, Rangel J, Mali S, Bodunrin A, Housammy Z, Zimmerer R, Murphy J, Widger WR, Bark SJ. 2019. A color-based competition assay for studying bacterial stress responses in Micrococcus luteus. FEMS Microbiol Lett 366:fnz054. doi: 10.1093/femsle/fnz054. [DOI] [PubMed] [Google Scholar]
- 31.Drumm JE, Mi K, Bilder P, Sun M, Lim J, Bielefeldt-Ohmann H, Basaraba R, So M, Zhu G, Tufariello JM, Izzo AA, Orme IM, Almo SC, Leyh TS, Chan J. 2009. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-binding: requirement for establishing chronic persistent infection. PLoS Pathog 5:e1000460. doi: 10.1371/journal.ppat.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornberg HL. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kloos WE, Schultes LM. 1969. Transformation in Micrococcus lysodeikticus. J Gen Microbiol 55:307–317. doi: 10.1099/00221287-55-2-307. [DOI] [PubMed] [Google Scholar]
- 34.Angelov A, Bergen P, Nadler F, Hornburg P, Lichev A, Übelacker M, Pachl F, Kuster B, Liebl W. 2015. Novel Flp pilus biogenesis-dependent natural transformation. Front Microbiol 6:84. doi: 10.3389/fmicb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB. 1998. A bacterial cytokine. Proc Natl Acad Sci U S A 95:8916–8921. doi: 10.1073/pnas.95.15.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukamolova GV, Yanopolskaya ND, Kell DB, Kaprelyants AS. 1998. On resuscitation from the dormant state of Micrococcus luteus. Antonie Van Leeuwenhoek 73:237–243. doi: 10.1023/a:1000881918216. [DOI] [PubMed] [Google Scholar]
- 37.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64:2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raffia A, Fahim U. 2010. Photoredox reaction of methylene blue and lactose in alcoholic buffered solution. J Appl Chem Res 13:72–84. [Google Scholar]
- 39.Chao MC, Rubin EJ. 2010. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol 64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 40.Voskuil MI, Visconti KC, Schoolnik GK. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med 198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert OT, Ludwig C, Kogadeeva M, Zimmermann M, Rosenberger G, Gengenbacher M, Gillet LC, Collins BC, Röst HL, Kaufmann SHE, Sauer U, Aebersold R. 2015. Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18:96–108. doi: 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R, Pushparajan AR, Mundayoor S, Jaleel A, Kumar RA. 2015. Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol Cell Proteomics 14:2160–2176. doi: 10.1074/mcp.M115.051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puckett S, Trujillo C, Wang Z, Eoh H, Ioerger TR, Krieger I, Sacchettini J, Schnappinger D, Rhee KY, Ehrt S. 2017. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 114:E2225–E2232. doi: 10.1073/pnas.1617655114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 46.Zybailov BL, Florens L, Washburn MP. 2007. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst 3:354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 47.Zimmer JSD, Monroe ME, Qian W-J, Smith RD. 2006. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev 25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wayne LG. 1976. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am Rev Respir Dis 114:807–811. [DOI] [PubMed] [Google Scholar]
- 49.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 50.Fisher RA, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 51.Gibson SER, Harrison J, Cox J. 2018. Modelling a silent epidemic: a review of the in vitro models of latent tuberculosis. Pathogens 7:88. doi: 10.3390/pathogens7040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 55.Albrethsen J, Agner J, Piersma SR, Højrup P, Pham TV, Weldingh K, Jimenez CR, Andersen P, Rosenkrands I. 2013. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol Cell Proteomics 12:1180–1191. doi: 10.1074/mcp.M112.018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelkar DS, Kumar D, Kumar P, Balakrishnan L, Muthusamy B, Yadav AK, Shrivastava P, Marimuthu A, Anand S, Sundaram H, Kingsbury R, Harsha HC, Nair B, Prasad TSK, Chauhan DS, Katoch K, Katoch VM, Kumar P, Chaerkady R, Ramachandran S, Dash D, Pandey A. 2011. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics 10:M111.011627. doi: 10.1074/mcp.M111.011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versées W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 60.Boon C, Dick T. 2012. How Mycobacterium tuberculosis goes to sleep: the dormancy survival regulator DosR a decade later. Future Microbiol 7:513–518. doi: 10.2217/fmb.12.14. [DOI] [PubMed] [Google Scholar]
- 61.Wayne LG, Lin KY. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun 37:1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eoh H, Rhee KY. 2014. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc Natl Acad Sci U S A 111:4976–4981. doi: 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 64.McKinney JD, Höner zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 65.Shleeva M, Mukamolova GV, Young M, Williams HD, Kaprelyants AS. 2004. Formation of “non-culturable” cells of Mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology 150:1687–1697. doi: 10.1099/mic.0.26893-0. [DOI] [PubMed] [Google Scholar]
- 66.Munson GP, Holcomb LG, Alexander HL, Scott JR. 2002. In vitro identification of Rns-regulated genes. J Bacteriol 184:1196–1199. doi: 10.1128/jb.184.4.1196-1199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farewell A, Diez AA, DiRusso CC, Nyström T. 1996. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J Bacteriol 178:6443–6450. doi: 10.1128/jb.178.22.6443-6450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nachin L, Brive L, Persson K-C, Svensson P, Nyström T. 2008. Heterodimer formation within universal stress protein classes revealed by an in silico and experimental approach. J Mol Biol 380:340–350. doi: 10.1016/j.jmb.2008.04.074. [DOI] [PubMed] [Google Scholar]
- 69.Glass LN, Swapna G, Chavadi SS, Tufariello JM, Mi K, Drumm JE, Lam TT, Zhu G, Zhan C, Vilchéze C, Arcos J, Chen Y, Bi L, Mehta S, Porcelli SA, Almo SC, Yeh S-R, Jacobs WR, Torrelles JB, Chan J. 2017. Mycobacterium tuberculosis universal stress protein Rv2623 interacts with the putative ATP binding cassette (APC) transporter Rv1747 to regulate mycobacterial growth. PLoS Pathog 13:e1006515. doi: 10.1371/journal.ppat.1006515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Freestone P, Trinei M, Clarke SC, Nyström T, Norris V. 1998. Tyrosine phosphorylation in Escherichia coli. J Mol Biol 279:1045–1051. doi: 10.1006/jmbi.1998.1836. [DOI] [PubMed] [Google Scholar]
- 71.Heermann R, Weber A, Mayer B, Ott M, Hauser E, Gabriel G, Pirch T, Jung K. 2009. The universal stress protein UspC scaffolds the KdpD/KdpE signaling cascade of Escherichia coli under salt stress. J Mol Biol 386:134–148. doi: 10.1016/j.jmb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.