Abstract
Objective
The lack of highly sensitive and specific diagnostic biomarkers is a major contributor to the poor outcomes of patients with hepatocellular carcinoma (HCC). We sought to develop a non-invasive diagnostic approach using circulating cell-free DNA (cfDNA) for the early detection of HCC.
Design
Applying the 5hmC-Seal technique, we obtained genome-wide 5-hydroxymethylcytosines (5hmC) in cfDNA samples from 2554 Chinese subjects: 1204 patients with HCC, 392 patients with chronic hepatitis B virus infection (CHB) or liver cirrhosis (LC) and 958 healthy individuals and patients with benign liver lesions. A diagnostic model for early HCC was developed through case-control analyses using the elastic net regularisation for feature selection.
Results
The 5hmC-Seal data from patients with HCC showed a genome-wide distribution enriched with liver-derived enhancer marks. We developed a 32-gene diagnostic model that accurately distinguished early HCC (stage 0/A) based on the Barcelona Clinic Liver Cancer staging system from non-HCC (validation set: area under curve (AUC)=88.4%; (95% CI 85.8% to 91.1%)), showing superior performance over α-fetoprotein (AFP). Besides detecting patients with early stage or small tumours (eg, ≤2.0 cm) from non-HCC, the 5hmC model showed high capacity for distinguishing early HCC from high risk subjects with CHB or LC history (validation set: AUC=84.6%; (95% CI 80.6% to 88.7%)), also significantly outperforming AFP. Furthermore, the 5hmC diagnostic model appeared to be independent from potential confounders (eg, smoking/alcohol intake history).
Conclusion
We have developed and validated a non-invasive approach with clinical application potential for the early detection of HCC that are still surgically resectable in high risk individuals.
Keywords: hepatobiliary cancer, hepatocellular carcinoma, cancer
Significance of this study.
What is already known on this subject?
Early detection of hepatocellular carcinoma (HCC) improves clinical outcomes and patient survival.
There are currently no effective biomarkers for early detection of HCC, especially in the context of high risk individuals with a history of chronic hepatitis B virus infection (CHB) or liver cirrhosis.
The 5hmC-Seal technique has been demonstrated to be a sensitive and robust epigenomic tool for cancer biomarker discovery using circulating cell-free DNA (cfDNA).
What are the new findings?
Genome-wide 5hmC in cfDNA derived from patients with HCC reflected liver tissue and gene regulatory relevance.
5hmC markers identified in cfDNA showed high capability for distinguishing patients with early HCC from those with a history of CHB or liver cirrhosis, as well as from patients with benign liver lesions and healthy controls, showing superior performance over α-fetoprotein (AFP).
How might it impact on clinical practice in the foreseeable future?
The 5hmC-Seal technique is a non-invasive tool that can aid in early detection of HCC in high risk individuals with a history of CHB or liver cirrhosis. This technique could serve as a non-invasive screening tool for the general population in the future, as well.
Introduction
Despite having a multitude of therapeutic modalities available, patients with hepatocellular carcinoma (HCC) have undesirable outcomes, with 5-year overall survival rates of less than 50%. However, survival rates can reach 70% with surgical resection or transplantation in early stage patients, highlighting the tremendous clinical need for effective early diagnostic approaches.1 2 Epidemiologically, HCC represents the second most common cause of cancer deaths worldwide (~7 50 000 annual deaths) and is present with a particularly high frequency in East Asia, where major risk factors (eg, chronic hepatitis B virus infection (CHB) and liver cirrhosis (LC)) are endemic.3 Indeed, China accounts for more than 50% of new HCC cases and related mortality.3 Almost half of patients with HCC are diagnosed at an advanced stage, which often prevents the possibility of curative therapies. Challenges to early diagnosis of HCC include the absence of pathognomonic symptoms and the lack of sensitive and specific biomarkers, which contribute to poor clinical outcomes.
At present, α-fetoprotein (AFP) is the most common serological test used for screening and diagnosis of HCC, as well as for surveillance after treatment. However, there are serious limitations with AFP, such as low sensitivity,4 false-negatives (eg, a small HCC tumour with AFP under the detectable level) and false-positives owing to conditions such as pregnancy and certain gastrointestinal tumours.5 6 In addition, the unequivocal diagnosis of a nodule detected using ultrasonography remains clinically challenging with unsatisfactory diagnostic accuracy (eg, ~60%–80% sensitivity).7
Pathological analysis of tumour tissue is currently the ‘gold standard’ for clinical diagnosis of cancers. However, tissue pathology-based approaches suffer from high cost, limited tumour tissue accessibility during invasive procedures, and the fact that a tissue biopsy may not reflect intra-tumour heterogeneity,8 9 thus limiting its use in the early detection of HCC. There are now some appealing alternatives, including methods based on liquid biopsy. Early liquid biopsy assays targeted proteins or microRNAs. Though promising, these approaches did not offer satisfactory performance in terms of specificity and sensitivity suitable at the clinical scale yet.10 11 More recently, circulating cell-free DNA (cfDNA) in plasma, which carries genetic and epigenetic information from cells of origin,12 has been shown to indicate the presence of cancer.13 Though limited by sample size and/or technical restrictions, several recent studies have begun to show the promise of epigenetic markers in cfDNA for diagnosis and prognosis in human cancers, including HCC.14–17
In the human genome, 5-hydroxymethylcytosines (5hmC) are abundant epigenetic marks that are generated through oxidation of 5-methylcytosines by the ten-eleven translocation enzymes.18 The 5hmC modifications in promoters, gene bodies and gene regulatory elements (eg, enhancers) faithfully reflect gene expression activation in mammalian genomes, and thus can serve as ideal markers for specific gene/locus activation in chromatin.18 Recent studies have suggested that 5hmC modifications are related to cancer pathobiology, including the observed global reduction of 5hmC levels in various cancer types.19 Therefore, 5hmC has emerged to be a novel class of cancer epigenetic biomarkers with promise in precision medicine, considering its cancer and gene regulatory relevance, tissue specificity, and also importantly, the technical advances in enabling technologies for its application in convenient liquid biopsy.16 20–22
Specifically, we employed the 5hmC-Seal, a highly sensitive and selective chemical labelling-based sequencing technology developed and optimised by our team,21 23 to characterise genome-wide 5hmC profiles in cfDNA from 2554 Chinese subjects including patients with HCC, patients with high risk conditions including CHB and LC and controls comprised of patients with benign liver lesions and healthy individuals (figure 1). We developed and validated a 5hmC-based diagnostic model for distinguishing early HCC from high risk individuals with CHB/LC as well as from non-HCC subjects, and compared its performance with AFP.
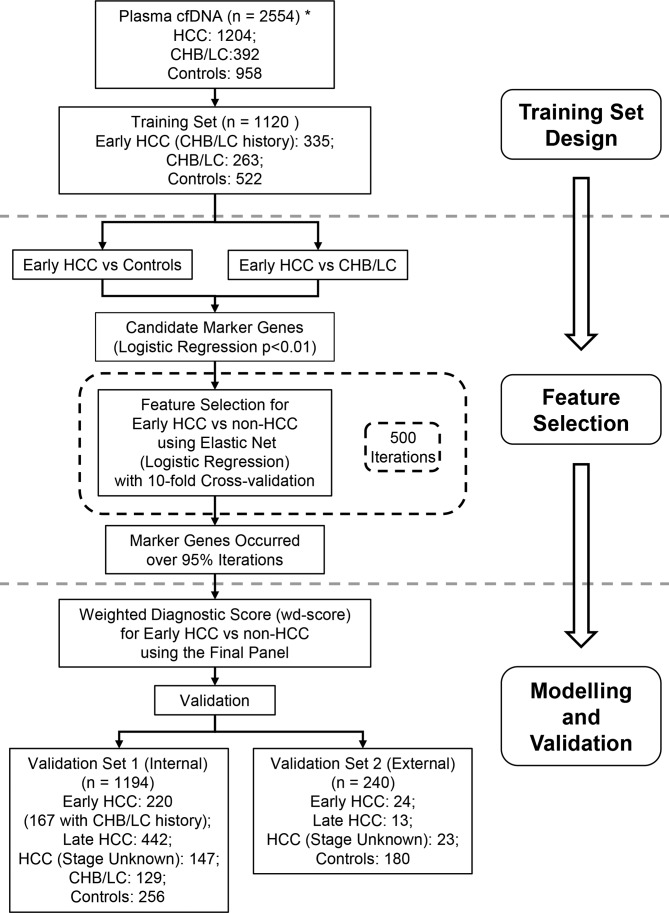
Figure 1.
Study design. The primary aim is to develop a 5hmC-based diagnostic model for early detection of HCC) using the genome-wide 5hmC-Seal profiles derived from plasma cfDNA. A two-step procedure is designed to identify a diagnostic model for early HCC (stage 0/A). The training set and the main validation set (‘validation set 1’) are comprised of HCC samples from Zhongshan Hospital of Fudan University and The Eastern Hepatobiliary Surgery Hospital, Shanghai, China. An independent set of HCC samples from other participating hospitals (‘validation set 2’) are used to evaluate external performance of the 5hmC diagnostic model for HCC. Due to sample size limitation, only controls and patients with HCC are available in the external validation set. *The total number of study subjects does not include the 20 samples that were removed due to technical reasons. Control: healthy individuals and patients with benign liver lesions. CHB, chronic hepatitis B virus infection; cfDNA, cell-free DNA; HCC, hepatocellular carcinoma; 5hmC, 5-hydroxymethylcytosines; LC, liver cirrhosis.
Materials and methods
Study subject
A total of 2554 out of 2574 prospectively enrolled adult subjects (≥18 years) were included in this study, including patients with HCC (n=1204), patients with a history of CHB or patients with LC (n=392), patients with benign liver lesions (n=388) and healthy individuals (n=570) (figure 1) from Zhongshan Hospital of Fudan University, The Eastern Hepatobiliary Surgery Hospital, and other participating institutions from July 2016 to November 2017 (see online supplementary methods and tables 1, 2). The patient population was socioeconomically diverse, with most patients coming from Shanghai, China and neighbouring provinces. Peripheral blood samples (5–10 mL/subject) were collected at the time of diagnosis and before any radical treatment, followed by plasma preparation and cfDNA extraction.21 We conducted central pathology review on all of the tumours from patients who underwent surgery. HCC stage was determined according to the Barcelona Clinic Liver Cancer (BCLC) staging system.24 Informed consent was obtained from all participants before the study.
Sample preparation, 5hmC-Seal profiling and data processing
Detailed information about preparation of cfDNA or genomic DNA (gDNA) samples, 5hmC-Seal library construction, sequencing and data processing has been reported by us previously.21 25 Briefly, the 5hmC-Seal profiling combines a series of routine sample preparation and sequencing steps with a unique pull-down step based on covalent chemistry. The raw 5hmC-Seal data were removed for adaptors and checked for quality, followed by aligning to the human genome reference (hg19).21 High quality alignments were then summarised by counting overlaps with gene bodies or other genomic features (eg, histone modification marks). The 5hmC-Seal count data were normalised using DESeq2,26 which performs the variance-stabilising transformation to correct for sequencing depth and library size. The raw and processed 5hmC-Seal data are available at the NCBI/Gene Expression Omnibus Database (GSE112679).
Batch design
The majority of patients with HCC (n=1144) were recruited from Zhongshan Hospital of Fudan University and The Eastern Hepatobiliary Surgery Hospital. We randomly assigned patients with early HCC (stage 0/A) with CHB or LC history from these two institutions into the training set (2/3 of samples) and a validation set (1/3 of samples) (ie, internal validation—‘validation set 1’, figure 1). To allow evaluation of HCC samples collected from other hospitals, we combined samples from Shanghai Public Health Clinic Center and The Tenth People’s Hospital of Shanghai into a separate validation set (ie, external validation—‘validation set 2’, figure 1).
Development of a weighted diagnostic score for early HCC
A two-step procedure was used to select optimal marker genes (ie, 5hmC modification summarized for gene bodies) for distinguishing early HCC from non-HCC subjects (ie, patients with CHB, LC, or controls) (figure 1). In the first step, a logistic regression model adjusted for age (>50 year vs ≤ 50 year) and gender was used to exclude the most unlikely marker genes to allow more efficient feature selection in the next step by selecting candidates that differed between early HCC and CHB/LC as well as between early HCC and controls (p value<0.01). In the second step, these candidates were subjected to further feature selection for distinguishing early HCC from non-HCC using the elastic net regularization on a multivariable logistic regression model, as implemented in the glmnet package (V.2.0-16).27 In order to select the best possible marker genes, the elastic net model was cross-validated for a grid of parameter values for α and λ (α range: 0.05–1 with 0.05 increment; λ range: 10-5–1 with logarithmically equal increment), where α controls for the relative proportion between the Ridge and Lasso penalty, and λ controls for the overall strength of penalty. This selection process was repeated 500 times, and a panel of 5hmC marker genes cross-validated in at least 95% iterations was retained for the final diagnostic model. A weighted diagnostic score (wd-score) was then calculated as the sum of the gene-wise product of logistic model coefficients and corresponding 5hmC marker value for each individual: , where is the coefficient from the final multivariable logistic model for the kth marker gene, and is the 5hmC level of the kth marker gene. The area under curve (AUC) and 95% CIs were generated to evaluate the model performance. The score cutoff that maximized the Youden’s index in the training set was used to estimate sensitivity and specificity. Linear regression models or Wilcoxon rank-sum tests were used to assess whether the wd-scores were independent from other clinical and demographic features, such as alanine aminotransferase (ALT), smoking history, alcohol intake and body mass index (BMI) whenever available. The DeLong, DeLong and Clarke-Pearson test was used to compare AUCs between the 5hmC-based diagnostic model and AFP.28 The multivariable logistic regression model was also performed to evaluate whether the 5hmC-based diagnostic model remained significant after controlling age, gender, BMI and AFP. All statistical analyses were performed with R Statistical Computing Environment (V.3.5.1).29
Results
Characterisation of the 5hmC-Seal data
A total of 2554 study subjects were included in the current report (figure 1, see online supplementary tables 1,2). The 5hmC-Seal libraries were sequenced to produce a median number of ~20.7 million reads in each cfDNA sample, with a median number of ~6.9 million unique reads mapped to the gene bodies. The 5hmC-Seal data showed high correlation for replicates from the same individual across a series of input DNA amount and different batches (mean Pearson’s r>0.99, see online supplementary figure 1A), demonstrating robustness of this technique and clinical feasibility with convenient amount of specimens (eg, <5 mL of plasma). Based on the genome-wide 5hmC data in cfDNA, we did not observe obvious subgroup stratification within each diagnosis group, indicating no systematic biases in the 5hmC-Seal profiling (see online supplementary figure 1B).
gutjnl-2019-318882supp004.pdf (162.6KB, pdf)
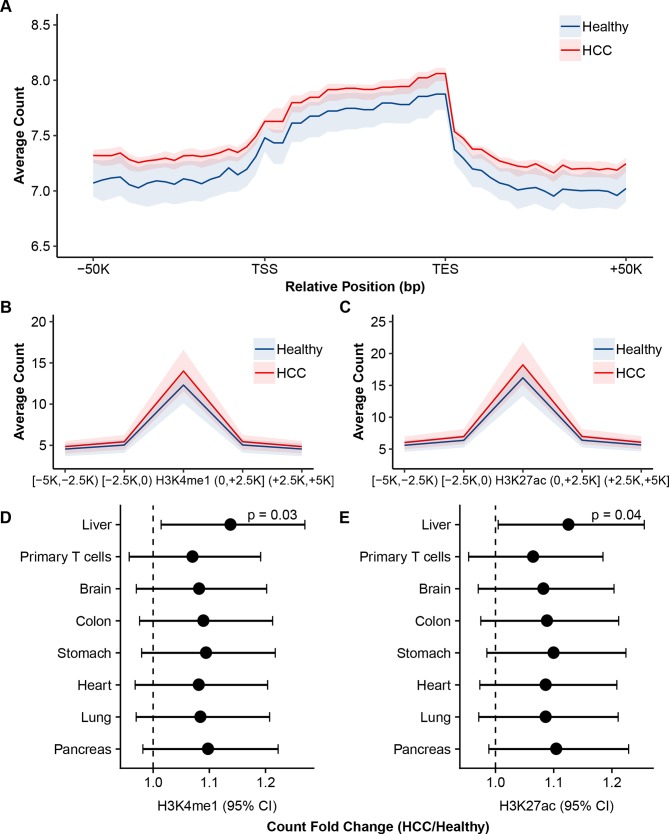
In a random set of cfDNA samples from 50 patients with HCC and 50 healthy individuals, the 5hmC profiles were found to be significantly enriched in the regions between transcription start sites and transcription end sites, and depleted in the flanking regions (figure 2A), consistent with our previous observations.21 To explore gene regulatory relevance of 5hmC in cfDNA, the 5hmC-Seal data were summarised for H3K4me1 and H3K27ac, which are histone modification marks for enhancers30–32 and with available annotations for various adult tissues from the Roadmap Epigenomics Project.33 Compared with healthy individuals, patients with HCC were enriched with liver-derived H3K4me1 and H3K27ac marks (figure 2B–E), indicating regulatory and tissue relevance of the profiled 5hmC in patient-derived cfDNA.
Figure 2.
Genomic distribution and regulatory relevance of 5hmC in cfDNA. The 5hmC-Seal data from a random set of 50 patients with HCC and 50 healthy individuals are shown. (A) The profiled 5hmC-Seal data in cfDNA are enriched in gene bodies and depleted in the flanking regions. (B, C) The profiled 5hmC-Seal data in cfDNA are enriched in liver-derived histone modification peaks and depleted in the flanking regions of (B) H3K4me1 and (C) H3K27ac. (D, E) The average fold changes of 5hmC-Seal read counts between HCC and healthy individuals are plotted against histone modification peaks derived from various adult tissues from the Roadmap Epigenomics Project for (D) H3K4me1 and (E) H3K27ac. In (A–C), the shaded area represents the first quantile to the third quantile. In (B–C), each mark at x-axis represents a region relative to the start or end positions of the histone modification peaks. In (D, E), p values of the two-sided t-tests for the ratios of two means were estimated for the fold changes between patients with HCC and healthy individuals, and are shown for the liver-derived peaks. The error bar represents the 95% CI for the fold change. cfDNA, cell-free DNA; HCC, hepatocellular carcinoma; 5hmC, 5-hydroxymethylcytosines; K: kilo base pair; TSS, transcription start site; TES, transcription end site.
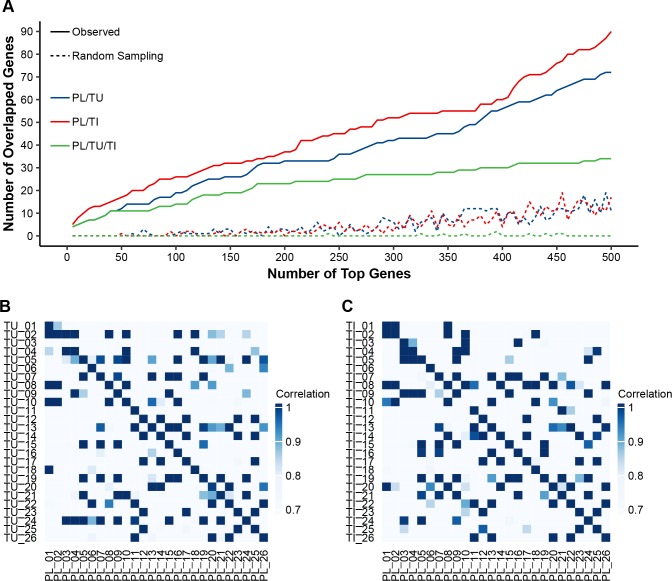
Furthermore, those genes with high variability in 5hmC modification across patient cfDNA samples coincided with genes showing high variability in tumours or adjacent tissues, as shown by a comparison in 26 sets of tumour, adjacent tissue and plasma cfDNA samples from the same individuals (figure 3A). For the top ranked genes in terms of variability in cfDNA, there was a higher within-subject correlation of cfDNA and tumour/adjacent tissue profiles than between-subject pairs (mean Pearson’s r 0.88 vs 0.73, Wilcoxon rank-sum test p<0.0001), providing further evidence that 5hmC in cfDNA was relevant to the tissue origin (figure 3B, C). Notably, the observation that only a portion of genes in tumours or adjacent tissues showed similarity with cfDNA21 is likely due to such reasons as different DNA degradation properties in cell-free circulation and heterogeneous cell origins.
Figure 3.
Tissue relevance of the 5hmC-Seal data in HCC patient-derived cfDNA. (A) For the most variable genes in cfDNA samples, the number of overlapped genes between cfDNA and tumours (TU: blue line) or adjacent tissues (TI: red line) is significantly higher than that from random sampling (eg, hypergeometric test p<0.0001 for the top 500 modified genes in cfDNA). The green line indicates the shared genes across plasma cfDNA, tumours and adjacent tissues. (B, C) Within-subject correlation is significantly higher (Wilcoxon rank-sum test p<0.0001) between plasma cfDNA and TU/TI genomic DNA (diagonal line, mean of Pearson’s r: 0.88) than that between different individuals (mean of Pearson’s r: 0.73), based on the top 30 most variable genes in cfDNA samples in terms of 5hmC modification. cfDNA, cell-free DNA; HCC, hepatocellular carcinoma; 5hmC, 5-hydroxymethylcytosines; PL, plasma cfDNA; TI, adjacent tissue; TU, tumour.
Development of a weighted diagnostic score for early HCC
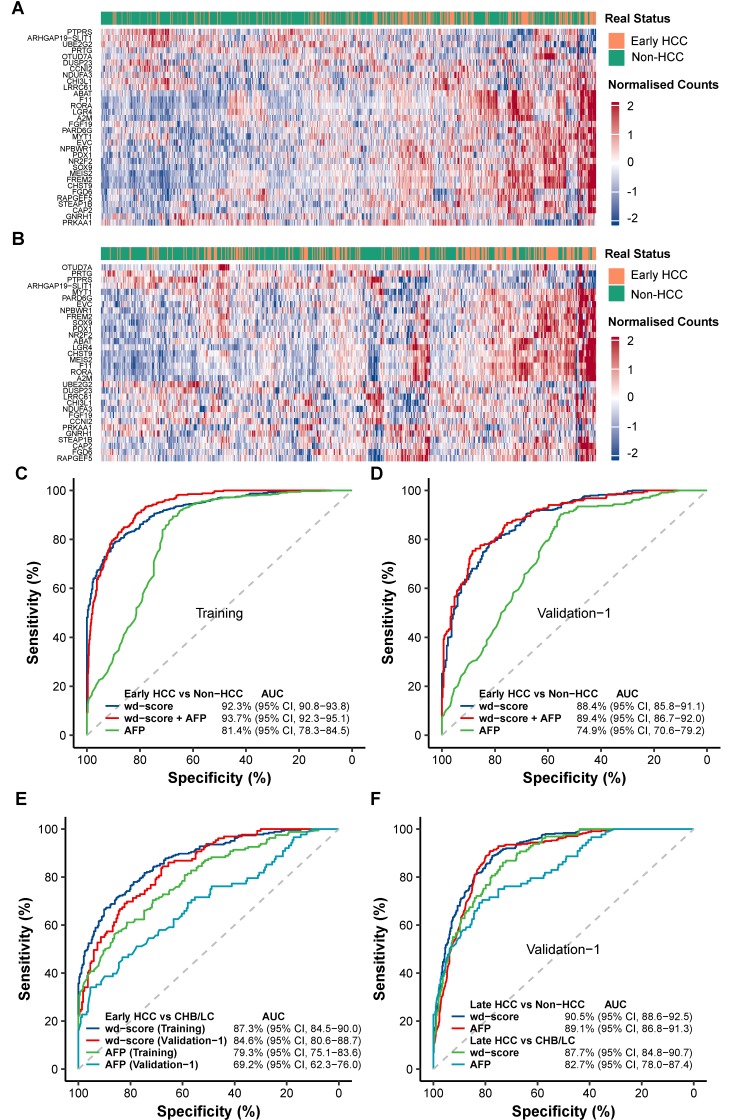
Our primary aim was to develop a convenient and integrated diagnostic model using the 5hmC profiles in cfDNA to distinguish patients with early (stage 0/A) HCC from high risk subjects with CHB/LC, as well as from all non-HCC subjects including both high risk individuals and controls. We first selected 917 candidate marker genes in the training set that showed evidence of differential 5hmC modification in cfDNA between early HCC and controls (see online supplementary table 3), as well as between early HCC and CHB/LC (see online supplementary table 4), at a less stringent cut-off (p-value<0.01, see online supplementary table 5) to balance inclusiveness and disease relevance. The application of elastic net regularisation approach on these candidates using logistic regression modelling identified a consistent panel of 32 marker genes (figure 4A, B, see online supplementary table 6).
Figure 4.
Development and validation of a 5hmC-based diagnostic model. (A, B) The 32 marker genes used to compute the wd-scores for early HCC (stage 0/A) detection are used to generate the heatmaps for (A) the training set and (B) validation set 1. (C, D) The performance of the wd-scores, AFP, or the combination of wd-scores and AFP in distinguishing early HCC from non-HCC subjects is shown for (C) the training set and (D) validation set 1. (E) The performance of the wd-scores or AFP in distinguishing early HCC from CHB/LC is shown for the training set and validation set 1. (F) The performance of the wd-scores or AFP in distinguishing late HCC (ie, advanced stage B/C) from non-HCC or CHB/LC subjects is shown for validation set 1. Non-HCC: CHB/LC and controls. AFP, α-fetoprotein; AUC, area under curve; CHB, chronic hepatitis B virus infection; HCC, hepatocellular carcinoma; 5hmC, 5-hydroxymethylcytosines; LC, liver cirrhosis; wd-scores, weighted diagnostic score.
gutjnl-2019-318882supp005.pdf (111.5KB, pdf)
gutjnl-2019-318882supp006.pdf (89.3KB, pdf)
gutjnl-2019-318882supp007.pdf (162.6KB, pdf)
gutjnl-2019-318882supp008.pdf (111.5KB, pdf)
The wd-scores computed based on these 32 marker genes showed excellent capacity for distinguishing patients with early HCC from non-HCC subjects in both the training set (AUC=92.3%; (95% CI 90.8% to 93.8%); sensitivity=89.6%; specificity=78.9%; score cut-off=27.9) and validation set 1 (AUC=88.4%; (95% CI 85.8% to 91.1%); sensitivity=82.7%; specificity=76.4%, figure 4C, D), as well as distinguishing patients with early HCC from control individuals (see online supplementary figure 2). This 5hmC-based model markedly outperformed the AFP-based model, which showed an AUC range of 74.9%–81.4% in the training set and validation set 1, in diagnostic accuracy for early HCC versus non-HCC (DeLong test p<10-10, figure 4C, D). Importantly, the 5hmC-based wd-scores demonstrated the capability of detecting those patients with early HCC that would be misclassified based on AFP alone. For example, for the 160 patients with early HCC that would be misclassified by AFP (cut-off=20 ng/mL) in the training set, the wd-scores achieved an AUC of 92.4% (see online supplementary figure 3). In addition, combining the wd-scores and AFP would further improve the diagnostic performance by ~1% in terms of AUC (figure 4C, D).
gutjnl-2019-318882supp009.pdf (89.3KB, pdf)
gutjnl-2019-318882supp010.pdf (162.6KB, pdf)
Individuals with a history of CHB are at 5–100 fold higher risk for developing HCC.34 CHB-related LC is one major risk factor for HCC in China, and LC frequently complicates HCC diagnoses. In the most challenging clinical setting of distinguishing early HCC and patients with CHB or LC, the 5hmC-based wd-scores significantly outperformed AFP-based detection model in the training set (AUC=87.3%; (95% CI 84.5% to 90.0%)) and validation set 1 (AUC=84.6%; (95% CI 80.6% to 88.7%), figure 4E). At a score cut-off of 27.9 the diagnostic model achieved 82.7% sensitivity and 67.4% specificity to separate early HCC (182 out of 220) and CHB/LC (87 out of 129) in validation set 1, compared with 44.8% sensitivity and 76.1% specificity for AFP (cut-off=20 ng/mL).
The 5hmC-based wd-scores also showed equivalent performance related to AFP in distinguishing late stage HCC (ie, advanced stage B/C) from non-HCC patients (AUC=90.5%; (95% CI 88.6% to 92.5%)) or from CHB/LC (AUC=87.7%; (95% CI 84.8% to 90.7%), figure 4F) in validation set 1. For a small set of patients with HCC (n=147) with unknown BCLC stage information, the wd-scores showed performance comparable to AFP in distinguishing them from non-HCC or from CHB/LC (see online supplementary figure 4). Notably, the 32-gene wd-scores could distinguish patients with HCC from non-HCC, regardless of the CHB or LC background in HCC, for example showing comparable performance for cirrhotic HCC and non-cirrhotic HCC (see online supplementary figure 5).
gutjnl-2019-318882supp011.pdf (111.5KB, pdf)
gutjnl-2019-318882supp012.pdf (89.3KB, pdf)
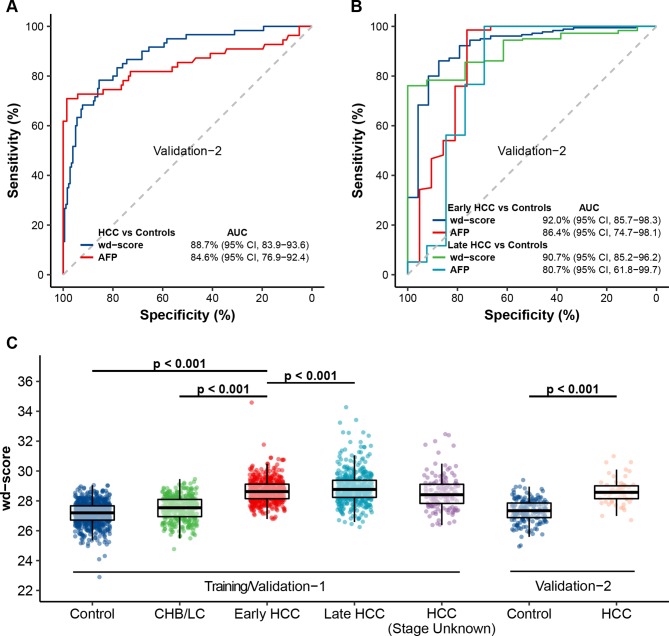
We then evaluated and confirmed the 5hmC-Seal approach in an external set of 60 patients with HCC (ie, validation set 2). Though the stage information was not available for all patients, the 5hmC-based diagnostic model still achieved high accuracy for distinguishing HCC from controls (AUC=88.7%; (95% CI 83.9% to 93.6%)), outperforming AFP (figure 5A, B).
Figure 5.
Further evaluation of the 5hmC-based diagnostic model. (A) The performance of the wd-scores or AFP in distinguishing HCC from controls is shown for the independent validation set 2. (B) For those patients with confirmed stages, the performance of the wd-scores or AFP in distinguishing early or late HCC from controls is shown for validation set 2. (C) The boxplots show the relationships between wd-scores and the clinical diagnosis across the training set and both validation sets. Control: healthy individuals and patients with benign liver lesions. AFP, α-fetoprotein; AUC, area under curve; CHB, chronic hepatitis B virus infection; HCC, hepatocellular carcinoma; 5hmC, 5-hydroxymethylcytosines; LC, liver cirrhosis; wd-score, weighted diagnostic score.
Diagnostic scores and additional clinical characteristics
Overall, the wd-scores showed an increasing trend from controls to HCC, noting the significantly higher scores in patients with early HCC than in subjects with CHB/LC or controls in the training set and validation set 1 (Wilcoxon rank-sum test p<0.001, figure 5C). The wd-scores also increased as BCLC stage advanced within patients with HCC with available stage information (n=997) (effect size=0.271, p-trend <0.001). In particular the wd-scores showed excellent detection power for stage 0 patients from non-HCC with 90.4% AUC in the training set and 87.1% AUC in validation set 1 (see online supplementary figure 6). Further, while we are fully aware that tumour size is not a highly meaningful indication of progression in HCC, the wd-scores accurately distinguished patients with different tumour sizes from non-HCC (see supplementary figure 7). Specifically, the wd-scores showed a strong detection performance by separating patients with HCC with small HCC (≤2.0 cm, n=220) from non-HCC (AUC=85.1%; (95% CI 80.8% to 89.5%)) as well as from CHB/LC (AUC=85.5%; (95% CI 81.8% to 89.3%), see online supplementary figure 8) in validation set 1.
gutjnl-2019-318882supp013.pdf (162.6KB, pdf)
gutjnl-2019-318882supp014.pdf (111.5KB, pdf)
gutjnl-2019-318882supp015.pdf (89.3KB, pdf)
Moreover, the wd-scores remained significantly associated with diagnosis under the multivariable logistic regression model (p<0.001 in both the training set and validation set 1) after controlling variables including age, gender and BMI for those early HCC and non-HCC with available data. In those patients with HCC with available demographic information, the wd-scores appeared to be independent from potential confounders: smoking history (p=0.78), alcohol intake history (p=0.07), or ALT level (p=0.12), under multivariable linear regression models adjusted for age, gender and BMI, though detailed relationships between these potential confounders and the diagnostic model will need to be confirmed in future studies or trials.
In addition, applying the HCC diagnostic model in a set of 89 Chinese patients with primary pancreatic ductal adenocarcinoma (PDAC)35 showed strong distinguishing capability of the wd-scores for HCC and PDAC, regardless of HCC stage, therefore suggesting potential cancer specificity of the HCC model (see online supplementary figure 9).
gutjnl-2019-318882supp016.pdf (162.6KB, pdf)
Functional relevance of the 5hmC-based diagnostic markers for early HCC
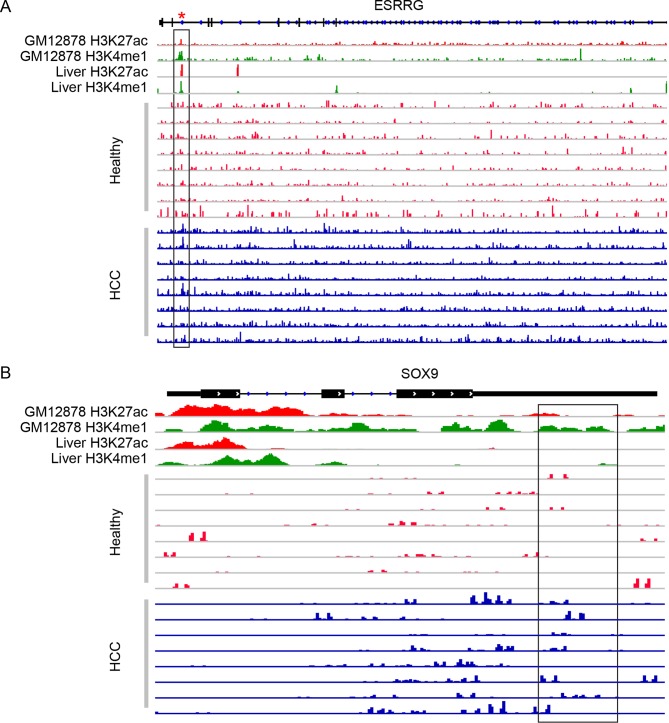
We sought to explore potential mechanisms underlying the marker genes by linking them with cis-regulatory elements. For the majority of the final marker genes, their 5hmC profiles in gene bodies were found be significantly associated with their 5hmC profiles in the combined, liver-derived H3K4me1 or H3K27ac peak regions or the predicted enhancers (see online supplementary table 6-7). Figure 6 shows ESRRG and SOX9 as two examples in a set of randomly selected patients with HCC and healthy individuals, noting the general overlapping pattern of 5hmC-Seal reads and H3K4me1/H3K27ac peaks based on the Roadmap Epigenomics Project33 or the Encyclopaedia of DNA Elements Project.31 For example, in ESRRG, a gene known to be implicated in the pathobiology of HCC,36 the gene-level 5hmC differential modification we observed between HCC and controls was found to coincide with the differential modification in the H3K4me1 and H3K27ac peak regions, thus offering a potential interpretation to the observed differential modification through histone modification marks.
Figure 6.
Read distribution in candidate marker genes co-localised with cis-regulatory elements. The histone modification marks (H3K4me1, H3K27ac) from the ENCODE Project (GM12878) or liver tissue-derived data from the Roadmap Epigenomics Project are shown together with the 5hmC-Seal sequencing reads in a random set of cfDNA samples from patients with HCC and healthy individuals. The boxed regions are examples where patients with HCC and healthy individuals show differences in read distribution overlapped with histone marks or predicted enhancers. The red asterisk represents a predicted enhancer region from the ENCODE Project. Genomic positions are based on the human genome reference (hg19). (A) ESRRG (Chromosome 1q41; boxed region: chr1:216 700 111–216 704 999); and (B) SOX9 (Chromosome 17q24.3; boxed region: chr17:70 121 229–70 122 119). cfDNA, cell-free DNA; HCC, hepatocellular carcinoma; ENCODE, Encyclopaedia of DNA Elements.
Further functional annotation analysis of the 917 candidate marker genes suggested an enrichment of various Kyoto Encyclopaedia of Genes and Genomes pathways involving metabolism processes (eg, carbon, amino acids), such as ‘glyoxylate and dicarboxylate metabolism’, ‘glycine, serine and threonine metabolism’, as well liver functions like ‘bile secretion’, and ‘complement and coagulation cascades’ (see online supplementary table 8). Interestingly, numerous candidate genes in these pathways have been implicated in the physiopathogenesis of HCC, hepatitis B virus infection, or fibrosis, for example, A2M, KNG1 of ‘complement and coagulation cascades’,37 38 and ALDH3A1 in tyrosine and phenylalanine metabolism.39 40
gutjnl-2019-318882supp018.pdf (111.5KB, pdf)
In addition, we explored gene expression relevance of the 5hmC marker genes in cfDNA utilising The Cancer Genome Atlas (TCGA)41 gene expression data on HCC tumours and normal liver tissues. Interestingly, the top ranked genes in terms of differential 5hmC modification between early HCC and controls in the training set were significantly enriched with those top ranked differentially expressed genes from TCGA (eg, hypergeometric test p<0.0001 for the top 200 modified genes in cfDNA, see online supplementary figure 10), thus suggesting gene expression relevance of the detected 5hmC markers in patient-derived cfDNA.
gutjnl-2019-318882supp019.pdf (89.3KB, pdf)
Discussion
We sought to develop clinically convenient, liquid biopsy-based biomarkers to help diagnose HCC from related liver diseases and controls using the 5hmC-Seal, a highly sensitive chemical labelling technique. Our primary analysis identified a 32-gene based 5hmC marker panel, which includes genes implicated in HCC, hepatitis B virus infection, or hepatic fibrosis (see online supplementary tables 6,8). A weighted model (wd-score) based on this panel demonstrated significantly improved performance over serum AFP testing alone for early HCC versus non-HCC (figure 4C, D) and, of the most significant clinical importance, patients with early HCC (stage 0/A) versus high risk individuals with CHB or LC (figure 4E), regardless of the CHB or LC background for HCC (see online supplementary figure 5). Notably, the wd-scores showed superior sensitivity over AFP by accurately detecting those HCC cases that would have failed to be detected by AFP testing alone (see online supplementary figure 3). In addition, the wd-scores demonstrated consistently high capacity for distinguishing patients with small tumours (≤2.0 cm) from non-HCC or CHB/LC subjects (see online supplementary figure 7-8). Taken together, our findings suggested that the 5hmC-Seal had the promise of becoming an integrated part of HCC management, from early detection of patients with HCC from high risk individuals through real-time post-treatment surveillance. We envision that patients diagnosed using the 5hmC markers could be put on more frequent monitoring by ultrasonography, CT, or MRI, thus improving patient survival in the long run. Furthermore, by combining the 5hmC-based model and AFP, a slightly improved detection accuracy could be achieved for distinguishing early HCC from non-HCC or from CHB/LC (figure 4C–F), indicating the potential benefit of integrating these two approaches in the clinic.
Technically, in contrast to other screening technologies, the 5hmC-Seal approach does not require pathology-related assumptions to inform probe targeting strategies. Because of its covalent chemical labelling nature which prevents sequencing biases, the 5hmC-Seal approach is not limited to any specific sequence context.23 Given the limited yield and highly fragmented (~160–320 bp) property of cfDNA, the 5hmC-Seal technique offers sensitivity that is critical for applications using liquid biopsy.21 The 5hmC-Seal technique requires only sequencing of the enriched 5hmC-contatining cfDNA or DNA fragments at low-coverage,21 22 thus offering cost-efficiency. The 5hmC-Seal data are genome-wide in nature, not limited to specific sites, thus avoiding the potential problem of missing specific sites that could be encountered in mutation-based analysis.42 Targeted sequencing of the marker panel in combination of covalent labelling could in theory further reduce cost in the future with further assay development to ensure robustness in clinical applications.
Regarding public health importance, notable utilities of the 5hmC markers in cfDNA for HCC include: (i) increased sensitivity over current ultrasonography regimens, and (ii) population-level screening of high risk individuals. Even though screening of patients with established liver cirrhosis with ultrasonography is recommended every 6 months for early diagnosis and improved overall survival, ultrasonography has only a sensitivity of 60%–80%.43 Our sensitive 5hmC markers in cfDNA for early HCC could fill an urgent need to identify early stage tumours amenable to curative treatments. Thus we offered a significantly improved tool for early detection of HCC, especially in high risk subjects with CHB/LC history, for which a confident diagnosis of nodules is almost impossible.44 Particularly, current diagnostic imaging techniques for HCC require a combination of equipment and infrastructural support which might not be readily available in developing regions where HCC burden is extremely high. Additionally, given its non-invasiveness and the tissue-specificity of 5hmC,21 22 the 5hmC-Seal approach may also serve as a convenient tool even for the population at large.
We acknowledge several limitations that could be addressed in future studies. First, although major clinical variables (eg, gender, age) have been controlled in our analyses, future independent validation studies will help address problems such as the potential selection bias for the validation sets or to confirm the implications of potential confounders (eg, alcohol intake history). Second, our study was in a Chinese patient population mostly with CHB/LC background, and therefore more validation will be necessary to demonstrate the generalisability of the results in prospective studies which will cover other populations, geographical regions, and disease risk factors, such as hepatitis C virus infection-related HCC. Finally, we used a case-control design in the current study to demonstrate the overall accuracy of the 5hmC-Seal and its ability to distinguish patients with early HCC from high risk non-HCC subjects. Future development phases, including retrospective longitudinal studies, and prospective screening studies, will help validate and establish the ultimate clinical utilities of this approach.45 Functional studies will also provide insights into the potential mechanism of the detected 5hmC marker genes in HCC pathogenesis.
In conclusion, we have developed and validated a novel non-invasive 5hmC-based diagnostic model for early HCC that are still surgically resectable, using the highly sensitive 5hmC-Seal assay. The 5hmC-Seal approach was demonstrated to be clinically useful, with the potential to aid in the existing diagnostic approaches and screening in high risk individuals. Our findings in this study of early HCC lay the foundation for developing a future pan-cancer, non-invasive screening tool as well.
gutjnl-2019-318882supp001.pdf (162.6KB, pdf)
gutjnl-2019-318882supp002.pdf (104.4KB, pdf)
gutjnl-2019-318882supp003.pdf (111.5KB, pdf)
gutjnl-2019-318882supp017.pdf (89.3KB, pdf)
Footnotes
JC, LC, ZZ and XZ contributed equally.
CH, HW, WZ and JF contributed equally.
Contributors: Conceptualisation: JF, WZ, HW and CH; Methodology: XL, JN and CH; Collection of samples and clinical data: JC, LC, XZ, WL, GS, YG, PG, YY, AK, LX, RD, YZ, XY, JW, TZ, DY, XH, HS, SQ, FS, CS, WZ and JZ; Formal data analysis: ZZ, CZ, XZ and WZ; Writing original draft: JC, LC, ZZ, XZ, XL, EKS, BCHC, CH, WZ and JF; Funding acquisition: GS, AK, JC, HW, and LC, CH, WZ, and JF.
Funding: National Institutes of Health (U01CA217078 and R01CA223662); Chinese State Key Project for Liver Cancer (2018ZX10732202-001); National Natural Science Foundation of China (81790633, 91729303, 81672860, 81572061, 81602513, 81472840, 81530077 and 81672825); The University of Chicago Ludwig Center; and The Howard Hughes Medical Institute.
Competing interests: The 5hmC-Seal technology was invented by CH and was licensed by Shanghai Epican Genetech Co. Ltd. for clinical applications in human diseases from the University of Chicago. XL is a co-founder of Shanghai Epican Genetech Co. Ltd. CH and WZ are shareholders of Shanghai Epican Genetech Co. Ltd. CH is a scientific founder of Accent Therapeutics, Inc. and a member of its scientific advisory board. All other authors report no potential conflicts of interest.
Ethics approval: The human research ethics committees at The Eastern Hepatobiliary Surgery Hospital, The Second Military Medical University, Shanghai, China; Zhongshan Hospital, Fudan University, Shanghai, China; The Tenth People’s Hospital of Shanghai, Tongji University, Shanghai, China.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not required.
References
- 1. Hasegawa K, Aoki T, Ishizawa T, et al. . Comparison of the therapeutic outcomes between surgical resection and percutaneous ablation for small hepatocellular carcinoma. Ann Surg Oncol 2014;21(Suppl 3):348–55. 10.1245/s10434-014-3585-x [DOI] [PubMed] [Google Scholar]
- 2. Ishizawa T, Hasegawa K, Aoki T, et al. . Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology 2008;134:1908–16. 10.1053/j.gastro.2008.02.091 [DOI] [PubMed] [Google Scholar]
- 3. Chen W, Zheng R, Baade PD, et al. . Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 4. Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann Intern Med 2003;139:46–50. 10.7326/0003-4819-139-1-200307010-00012 [DOI] [PubMed] [Google Scholar]
- 5. El-Bahrawy M. Alpha-fetoprotein-producing non-germ cell tumours of the female genital tract. Eur J Cancer 2010;46:1317–22. 10.1016/j.ejca.2010.01.028 [DOI] [PubMed] [Google Scholar]
- 6. Sterling RK, Wright EC, Morgan TR, et al. . Frequency of elevated hepatocellular carcinoma (HCC) biomarkers in patients with advanced hepatitis C. Am J Gastroenterol 2012;107:64–74. 10.1038/ajg.2011.312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hernaez R, Lazo M, Bonekamp S, et al. . Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology 2011;54:1082–90. 10.1002/hep.24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wan JCM, Massie C, Garcia-Corbacho J, et al. . Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 9. Bedard PL, Hansen AR, Ratain MJ, et al. . Tumour heterogeneity in the clinic. Nature 2013;501:355–64. 10.1038/nature12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwarzenbach H, Nishida N, Calin GA, et al. . Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014;11:145–56. 10.1038/nrclinonc.2014.5 [DOI] [PubMed] [Google Scholar]
- 11. Mao Y, Yang H, Xu H, et al. . Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut 2010;59:1687–93. 10.1136/gut.2010.214916 [DOI] [PubMed] [Google Scholar]
- 12. Sun K, Jiang P, Chan KC, et al. . Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015;112:E5503–12. 10.1073/pnas.1508736112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426–37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 14. Chan KC, Jiang P, Chan CW, et al. . Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761–8. 10.1073/pnas.1313995110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wen L, Li J, Guo H, et al. . Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell Res 2015;25:1250–64. 10.1038/cr.2015.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeng C, Stroup EK, Zhang Z, et al. . Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun 2019;39:12 10.1186/s40880-019-0356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai J, Chen L, Zhang Z, et al. . 5-Hydroxymethylcytosines from circulating cell-free DNA as diagnostic and prognostic markers for hepatocellular carcinoma. BioRxiv 2018. [Google Scholar]
- 18. Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 2011;13:7–13. 10.1038/nrg3080 [DOI] [PubMed] [Google Scholar]
- 19. Mariani CJ, Madzo J, Moen EL, et al. . Alterations of 5-hydroxymethylcytosine in human cancers. Cancers 2013;5:786–814. 10.3390/cancers5030786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao P, Lin S, Cai M, et al. . 5-Hydroxymethylcytosine profiling from genomic and cell-free DNA for colorectal cancers patients. J Cell Mol Med 2019;23:3530–7. 10.1111/jcmm.14252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W, Zhang X, Lu X, et al. . 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 2017;27:1243–57. 10.1038/cr.2017.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Song CX, Yin S, Ma L, et al. . 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 2017;27:1231–42. 10.1038/cr.2017.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song CX, Szulwach KE, Fu Y, et al. . Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 2011;29:68–72. 10.1038/nbt.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pons F, Varela M, Llovet JM. Staging systems in hepatocellular carcinoma. HPB 2005;7:35–41. 10.1080/13651820410024058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han D, Lu X, Shih AH, et al. . A highly sensitive and robust method for genome-wide 5hmC profiling of rare cell populations. Mol Cell 2016;63:711–9. 10.1016/j.molcel.2016.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–45. 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 29. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2013. [Google Scholar]
- 30. Chen H, Li C, Peng X, et al. . A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 2018;173:386–99. 10.1016/j.cell.2018.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Creyghton MP, Cheng AW, Welstead GG, et al. . Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010;107:21931–6. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kundaje A, Meuleman W, Ernst J, et al. . Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264–73. 10.1053/j.gastro.2011.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Z, You L, Lu X, et al. . Genome-wide 5-hydroxymethylcytosine mapping in circulating cell-free DNA Identifies robust diagnostic markers for pancreatic adenocarcinoma [abstract]. Proceedings of the 110th Annual Meeting of the AACR. Atlanta, USA, 2019:433. [Google Scholar]
- 36. Yuan B, Liang Y, Wang D, et al. . MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer Sci 2015;106:819–24. 10.1111/cas.12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang W, Zhang L, Guo Q, et al. . Identification of the pathogenic biomarkers for hepatocellular carcinoma based on RNA-seq analyses. Pathol Oncol Res 2019. 10.1007/s12253-019-00596-2 [DOI] [PubMed] [Google Scholar]
- 38. Kurokawa Y, Matoba R, Takemasa I, et al. . Molecular features of non-B, non-C hepatocellular carcinoma: a PCR-array gene expression profiling study. J Hepatol 2003;39:1004–12. 10.1016/S0168-8278(03)00473-2 [DOI] [PubMed] [Google Scholar]
- 39. Zhang X, Yang XR, Sun C, et al. . Promyelocytic leukemia protein induces arsenic trioxide resistance through regulation of aldehyde dehydrogenase 3 family member A1 in hepatocellular carcinoma. Cancer Lett 2015;366:112–22. 10.1016/j.canlet.2015.06.014 [DOI] [PubMed] [Google Scholar]
- 40. Calderaro J, Nault JC, Bioulac-Sage P, et al. . ALDH3A1 is overexpressed in a subset of hepatocellular carcinoma characterised by activation of the Wnt/ß-catenin pathway. Virchows Arch 2014;464:53–60. 10.1007/s00428-013-1515-0 [DOI] [PubMed] [Google Scholar]
- 41. Weinstein JN, Collisson EA, Mills GB, et al. . The cancer genome atlas pan-cancer analysis project. Nat Genet 2013;45:1113–20. 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Merker JD, Oxnard GR, Compton C, et al. . Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol 2018;36:1631–41. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 43. Singal A, Volk ML, Waljee A, et al. . Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol Ther 2009;30:37–47. 10.1111/j.1365-2036.2009.04014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology 2016;150:835–53. 10.1053/j.gastro.2015.12.041 [DOI] [PubMed] [Google Scholar]
- 45. Pepe MS, Etzioni R, Feng Z, et al. . Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001;93:1054–61. 10.1093/jnci/93.14.1054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-318882supp004.pdf (162.6KB, pdf)
gutjnl-2019-318882supp005.pdf (111.5KB, pdf)
gutjnl-2019-318882supp006.pdf (89.3KB, pdf)
gutjnl-2019-318882supp007.pdf (162.6KB, pdf)
gutjnl-2019-318882supp008.pdf (111.5KB, pdf)
gutjnl-2019-318882supp009.pdf (89.3KB, pdf)
gutjnl-2019-318882supp010.pdf (162.6KB, pdf)
gutjnl-2019-318882supp011.pdf (111.5KB, pdf)
gutjnl-2019-318882supp012.pdf (89.3KB, pdf)
gutjnl-2019-318882supp013.pdf (162.6KB, pdf)
gutjnl-2019-318882supp014.pdf (111.5KB, pdf)
gutjnl-2019-318882supp015.pdf (89.3KB, pdf)
gutjnl-2019-318882supp016.pdf (162.6KB, pdf)
gutjnl-2019-318882supp018.pdf (111.5KB, pdf)
gutjnl-2019-318882supp019.pdf (89.3KB, pdf)
gutjnl-2019-318882supp001.pdf (162.6KB, pdf)
gutjnl-2019-318882supp002.pdf (104.4KB, pdf)
gutjnl-2019-318882supp003.pdf (111.5KB, pdf)
gutjnl-2019-318882supp017.pdf (89.3KB, pdf)