Abstract
Objectives
Intraductal papillary mucinous neoplasms (IPMNs) are pancreatic cysts that can progress to invasive pancreatic cancer. Associations between oncogenesis and oral microbiome alterations have been reported. This study aims to investigate a potential intracystic pancreatic microbiome in a pancreatic cystic neoplasm (PCN) surgery patient cohort.
Design
Paired cyst fluid and plasma were collected at pancreatic surgery from patients with suspected PCN (n=105). Quantitative and qualitative assessment of bacterial DNA by qPCR, PacBio sequencing (n=35), and interleukin (IL)-1β quantification was performed. The data were correlated to diagnosis, lesion severity and clinical and laboratory profile, including proton-pump inhibitor (PPI) usage and history of invasive endoscopy procedures.
Results
Intracystic bacterial 16S DNA copy number and IL-1β protein quantity were significantly higher in IPMN with high-grade dysplasia and IPMN with cancer compared with non-IPMN PCNs. Despite high interpersonal variation of intracystic microbiota composition, bacterial network and linear discriminant analysis effect size analyses demonstrated co-occurrence and enrichment of oral bacterial taxa including Fusobacterium nucleatum and Granulicatella adiacens in cyst fluid from IPMN with high-grade dysplasia. The elevated intracystic bacterial DNA is associated with, but not limited to, prior exposure to invasive endoscopic procedures, and is independent from use of PPI and antibiotics.
Conclusions
Collectively, these findings warrant further investigation into the role of oral bacteria in cystic precursors to pancreatic cancer and have added values on the aetiopathology as well as the management of pancreatic cysts.
Keywords: pancreatic tumours, bacterial translocation, inflammation, pancreatic surgery, endoscopic procedure
Significance of this study.
What is already known on this subject?
Intraductal papillary mucinous neoplasm (IPMN) is a pancreatic cystic tumour that displays various degrees of cellular dysplasia up to cancer.
Alterations in oral microbiome are associated with increased risk for the development of pancreatic cancer.
The microbiome in cancer tissue may enhance oncogenesis through local immune modulation.
What are the new findings?
Elevated pancreatic intracystic bacterial DNA and interleukin-1β correlate in synergy with IPMN and neoplastic grade.
Microbiome of pancreas cyst fluid display co-occurrence and enrichment of oral bacterial species.
Elevated intracystic bacteria load in pancreas is associated but not limited to prior exposure to invasive endoscopic procedures, and is independent from use of proton-pump inhibitor and antibiotics.
How might it impact on clinical practice in the foreseeable future?
Intracystic bacterial DNA testing may have added values to guide the management of IPMN.
Reducing the pancreatic inflammatory microbiome may represent a potential therapeutic strategy for patients with IPMN.
Introduction
Pancreatic cystic neoplasms (PCN) are increasingly detected in the general population. They comprise a clinically challenging entity with potential to progress to invasive cancer.1 2 Among PCNs, intraductal papillary mucinous neoplasm (IPMN) are most common.3–5 IPMNs are pancreatic cysts of epithelial origin, characterised by malignant transformation from low-grade dysplasia (LGD) to high-grade dysplasia (HGD), and further to invasive carcinoma, following adenoma-carcinoma sequence.5 6 IPMN can be an early detection marker of pancreatic cancer, but since the majority of suspected IPMNs are benign lesions or LGD, the treatment strategy is either conservative or surgical resection of the pancreas. Unfortunately, the preoperative prediction accuracy of PCN types, in particular the IPMN neoplastic grades, is still suboptimal, and the final diagnosis is usually revealed only after operation.1 5 7–9
Much progress has been made to find molecular markers to support PCN differentiation diagnostics. Patients at higher risk of malignant progression10 11 often show typical cyst features, for example, worrisome features, abnormal cyst fluid cytology, serum carcinoembryogenic antigen levels, inflammatory cytokines or mutations in genes such as KRAS or GRAS.1 These findings have advanced the understanding of the oncogenic mechanisms of malignant transformation of PCNs, which is of importance for disease management and cancer prevention.
Novel findings suggest that cancerous pancreas tissues can have increased bacterial abundance compared with normal pancreas.12 This has a clinical impact as bacteria like Fusobacterium species in cancerous tissues correlate with poor pancreatic cancer prognosis13 and intratumoural bacteria can metabolise the anticancer drug gemcitabine, thereby reducing therapeutic effects.14 Using endoscopic ultrasound fine-needle aspirated (EUS-FNA) pancreas cyst fluids, a retrospective study recently showed that cyst fluids from PCNs harbour a unique microbiome that is independent of cyst type, clinical or biochemical parameters.15 However, the EUS-FNA procedure collects a GI specimen through the mouth and oesophagus, posing a risk for sample contamination by oral and gut microbiota.
As an alternative to the EUS-FNA approach, we present here a prospective study based on cyst fluid and plasma samples collected on pancreatic resection for studying the pancreatic microbiome. A total of 105 PCN patients undergoing pancreatic resection with subsequent histologically validated diagnosis were included.
Methods
Ethical considerations
All study participants gave written informed consent prior to sample collection.
Study population and sample collection
Cyst fluid and peripheral blood liquid biopsies were collected at the day of surgery from 105 patients undergoing surgical pancreatectomy for suspected pancreatic cystic lesions based on preoperative diagnosis at the Karolinska University Hospital, Stockholm, Sweden. Clinical and laboratory data were extracted from electronic journals by clinical doctors. Directly after surgery, cyst fluid was aspirated from the resected pancreas and immediately stored at −80°C until further processing. Fasting venous blood was collected in K2 EDTA tubes (BD Vacutainer) and Ficoll Paque PLUS (GE Life Sciences) density gradient centrifugation was performed according to manufacturers’ instructions to isolate the plasma fraction, which was frozen at −80°C until further analysis.
DNA extraction and microbiota analysis
Microbial DNA was isolated from cyst fluid and plasma using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research, Irvine, California, USA) in a biological flow cabinet. DNA was isolated from formalin-fixed paraffin embedded (FFPE) pancreas tissue slices using the AllPrep DNA/RNA FFPE Kit (Qiagen, Sollentuna, Sweden). Total bacterial 16S DNA gene copy number was quantified using TaqMan qPCR as previously described.16 Cyst fluid microbial composition and diversity was assessed by PacBio Single Molecule, real-time full-length 16S rRNA gene sequencing (GATC Biotech, Konstanz, Germany). All DNA samples were coded before sending to GATC without information that could reveal the sample classification.
Interleukin-1β and lipopolysaccharide quantification
The Human interleukin (IL)-1β/IL-1F2 ELISA DuoSet (R&D Systems) was used to quantify plasma and cyst fluid IL-1β levels. The PyroGene Recombinant Factor C Endotoxin Detection Assay (Lonza, Falun, Sweden) was used to quantify bacterial lipopolysaccharide (LPS) in cyst fluid.
A detailed description of subjects, histopathological diagnosis, assays, microbiota analysis and statistical methods can be found in the online supplementary material.
gutjnl-2018-317458supp001.pdf (105.6KB, pdf)
Results
Patient cohort overview and diagnose-based group comparisons
A total of 105 patients diagnosed with PCN undergoing pancreatectomy were studied. The final pancreas diagnoses on the resected tissue specimens indicated non-IPMN cysts (n=21) comprising mucinous cystic neoplasm (MCN) and serous cystic neoplasm (SCN), IPMN cysts (n=57) or IPMN cysts with invasive cancer (hereafter referred to as Cancer; n=27). This gives a preoperative diagnosis accuracy of 52.4%, 94.7% and 51.9% for non-IPMN, IPMN and Cancer, respectively, based on comparison of preoperative clinical and radiology findings and postoperative histologically confirmed diagnosis (table 1). The non-IPMN group is on average younger and represented by more females than the IPMN and Cancer groups. Serum S-Ca 19–9, haemoglobin A1c, albumin and bilirubin levels were also higher in Cancer compared with non-IPMN.
Table 1.
Patient cohort clinical characteristics
| Parameters | All subjects (n=105) |
Non-IPMN* (n=21) |
IPMN (n=57) |
Cancer (n=27) |
| Gender (F:M) | 59:46 | 20:1†‡ | 29:28† | 10:17‡ |
| Age (years) median (range) | 69 (30–84) | 47†‡ (30–77) | 71† (47–84) | 71‡ (46–83) |
| BMI (kg/m2) median (range) | 25.2 (18.1–40.4) | 26.4 (20.4–37.1) | 26.4 (18.1–40.4) | 24.5 (20.2–30.2) |
| Smoking (%) | 12.2 | 23.5 | 7.3 | 15.4 |
| Alcohol (%) | 34.8 | 29.4 | 32.7 | 42.3 |
| Diabetes (%) | 19.1 | 9.5 | 17.5 | 29.6 |
| Proton-pump inhibitor use (%) | 14.3 | 14.3 | 15.8 | 11.1 |
| Preoperative diagnostic accuracy (%) | n.a. | 52.4† | 94.7† | 51.9 |
| S-Ca 19–9 (kE/L) median (range) Above normal§ (%) |
12.0 (<1.0–4840) 33.3 |
9.9‡ (<1.0–62)
23.8‡ |
11.0 (<1.0–115) 19.3 |
94.0‡ (<1.0–4840)
70.4‡ |
| Haemoglobin A1c (mmol/mol) median (range) | 40.0 (15.1–80) | 35.0‡ (30–80) | 40.0 (28–72) | 42.0‡ (15.1–67) |
| Amylase (µkat/L) median (range) | 0.37 (<0.13–11.6) | 0.38 (0.19–1.96) | 0.39 (<0.13–4.18) | 0.27 (<0.13–11.6) |
| Albumin (g/L) median (range) | 36 (16–42) | 38‡ (33–42) | 37 (16–42) | 33.5‡ (19–38) |
| Bilirubin (μmol/L) median (range) | 6 (<3–315) | 5‡ (<3–18) | 6 (<3–315) | 14‡ (4–277) |
| C reactive protein (mg/L) median (range) | 2 (1–130) | 2 (1–13) | 1 (1–49) | 4 (1–130) |
| Invasive endoscopic procedure (%) mean frequency (range) | 38.1 0.62 (0–7) |
14.3‡
0.14 (0–1)‡ |
29.8 0.51 (0–7) |
74.1‡
1.2 (0–4)‡ |
| Unicystic/multicystic (%) | 52.0/48.0 | 80.0/20.0 | 36.8/63.2 | 64.0/26.0 |
Pairwise comparisons between the diagnosis groups were made using Fisher’s exact test (qualitative parameters) and Kruskal-Wallis test with Dunn’s multiple comparisons correction (quantitative variables).
Bold indicates statistical significance (p <0.05) in pairwise comparison.
*Non-IPMN group comprises MCN (n=9) and SCN (n=12).
†Indicates ‘non-IPMN cyst’ vs ‘IPMN’ p<0.05 (two-tailed).
‡Indicates ‘non-IPMN cyst’ vs ‘Cancer’ p<0.05 (two-tailed).
§Normal S-Ca 19–9<34 kE/L.
n.a., not applicable.
The intracystic bacterial DNA quantity is associated with cystic lesion severity
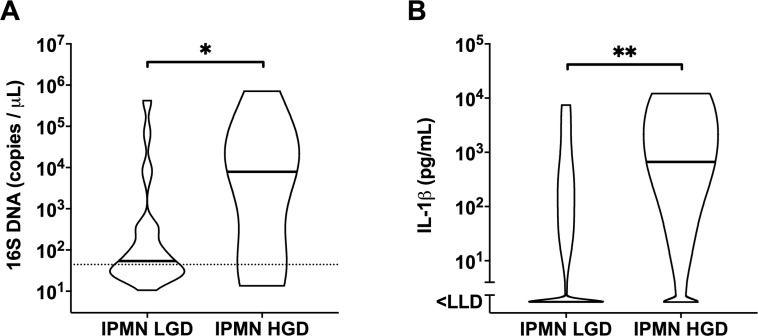
Paired cyst fluid and plasma were tested for the bacterial 16S gene copy number. Compared with non-IPMN, significantly higher bacterial DNA copies were found in the cyst fluid of IPMN (10.1-fold higher geometric mean, p=0.0042) and Cancer (11.4-fold higher geometric mean, p=0.0008), with a p-for-trend=0.0067. Likewise, the proportion of bacterial DNA-positive cyst samples increased stepwise (χ2 test, p=0.0033) with cyst lesion severity—from 33.0% in non-IPMN to 59.6% in IPMN and 81.5% in Cancer (figure 1A). Similar increases were seen for the inflammatory marker IL-1β in cyst fluid when comparing non-IPMN against IPMN (p=0.029), and Cancer (p=0.0024) (figure 1B). The elevated amount of intracystic bacterial DNA is accompanied by higher IL-1β levels in cyst fluid as shown by a positive Spearman’s correlation (r=0.51, p<0.0001) (figure 1C). This appeared to be a local rather than systemic effect, as both 16S DNA and IL-1β were found to be very low in plasma in all groups (figure 1D–F), which is in line with the low circulatory C reactive protein levels found in these groups (table 1). In addition, the quantity of bacterial LPS is elevated in IPMN (p=0.049) and Cancer (p=0.013) compared with non-IPMN, and positively correlated to 16S DNA (r=0.58, p=0.0004) and IL-1β (r=0.44, p=0.0096) in cyst fluid samples (see online supplementary figure 1). Next, the IPMN group was further stratified into HGD or LGD according to the histopathology evaluation. We found that cyst fluid from IPMN HGD harboured significantly higher 16S DNA copies (17.7-fold higher geometric mean, p=0.02) as well as IL-1β concentration (103.2-fold increase, p=0.0032) than that of IPMN LGD (figure 2A, B). Again, this effect was noted only in cyst fluid, and not in paired patient plasma.
Figure 1.
Intracystic quantities of (A) bacterial 16S DNA and (B) IL-1β in ’non-IPMN' (n=21), ’IPMN' (n=57), ’IPMN Cancer' (n=27) patients, and in paired plasma (D, E) of ’non-IPMN' (n=10), ’IPMN' (n=15), ’IPMN Cancer' (n=7) patients. Dotted line indicates assay cut-off value. Geometric mean for 16S DNA and mean for IL-1β are indicated per each group. Spearman’s correlation analysis on 16S DNA and IL-1β quantities in (C) cyst fluid and (F) plasma, displaying non-linear fit curve (solid line) and 95% CI (dashed line). Statistically significant differences were determined using Kruskal-Wallis test with Dunn’s multiple comparison test with *p<0.05, **p<0.01, ***p<0.001. IL, interleukin; IPMN, intraductal papillary mucinous neoplasm; LLD, lower limit of detection; n.d., not determined.
Figure 2.
Violin plot showing the distribution with median of (A) total bacterial 16S DNA in ’IPMN LGD' (n=45) and ’IPMN HGD' (n=14) and (B) IL-1β in ’IPMN LGD' (n=35) and ’IPMN HGD' (n=10) cyst fluid samples, determined by TaqMan qPCR and ELISA, respectively. Statistical comparisons between groups were made using Mann-Whitney U test with *p<0.05 and **p<0.01. Dotted line indicates assay cut-off value. HGD, high-grade dysplasia; IL, interleukin; IPMN, intraductal papillary mucinous neoplasm; LGD, low-grade dysplasia; LLD, lower limit of detection.
Compositional analysis revealed a highly diverse PCN microbiome
The finding of the elevated bacterial DNA quantity in IPMN and Cancer compared with non-IPMN prompted us to investigate the composition of the bacterial DNA-positive cyst fluid samples. We asked whether the microbial composition might differ depending on the type of PCN and/or progression of disease. Full-length bacterial 16S rRNA genes were sequenced using PacBio technology. Due to DNA quantity and quality thresholds applied before and after construction of full-length 16S rRNA gene libraries, cyst fluid samples from 35 patients classified as IPMN LGD (n=14) or IPMN HGD (n=8) and Cancer (n=14) were subjected to microbiome analysis. We found that these three groups showed no significant differences for Chao1 richness, Shannon diversity or inverted Simpson’s indices at operational taxonomic unit (OTU) level (figure 3A–C). At phylum level, IPMN LGD was found to be rather homogeneously dominated by Proteobacteria, while IPMN HGD and Cancer were generally more diverse with samples dominated by either Firmicutes or Proteobacteria (figure 3D). Relative abundance at genus level showed interpersonal variation in microbial composition, with no clear distinctive profile based on diagnosis group (figure 3E).
Figure 3.
Box plot displaying (A) Chao1 richness (white), (B) Shannon entropy (light grey) and (C) inverted Simpson’s (dark grey) indices per diagnose group at operational taxonomic unit (OTU) level. Bar plots showing relative abundance of OTUs per sample at (D) phylum level and (E) genus level. HGD, high-grade dysplasia; IPMN, intraductal papillary mucinous neoplasm; LGD, low-grade dysplasia.
Network analysis shows co-abundance of oral pathogens
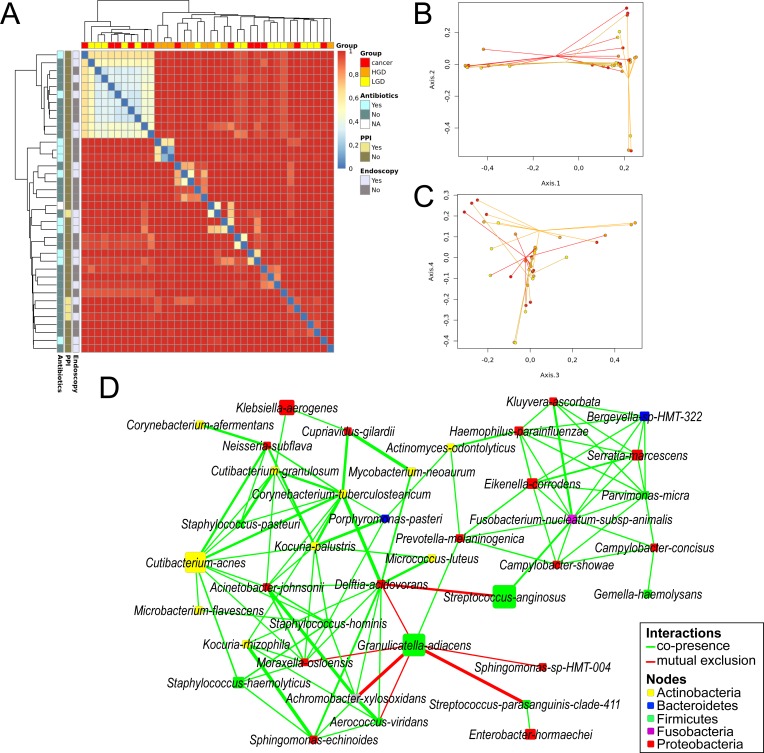
We noticed interpersonal variation in microbial communities of the cyst fluid even within the same diagnosis groups, as visualised by a clustered heat map displaying Bray-Curtis dissimilarity at OTU level between individual samples. This dissimilarity was independent of factors such as recent antibiotic medication, proton-pump inhibitor (PPI) use and previous invasive endoscopic procedures (Figure 4A). The bacterial composition could not separate samples entirely according to their corresponding diagnosis, although overall IPMN HGD appeared to stand out, as shown in principal coordinates analysis (PCoA) plots based on OTU relative abundance showing the first four dimensions (figure 4B, C). Indeed, non-parametric analysis of similarities analysis confirmed that the composition of IPMN HGD is unlike the other two groups (p=0.002 on 999 permutations).
Figure 4.
Beta diversity between samples visualised by (A) clustered heatmap showing Bray-Curtis dissimilarity at OTU level for each sample, characterised horizontally by diagnose group and vertically by previous endoscopic procedures, PPI use and antibiotic medication <1 month prior to surgery. (B) PCoA spider plots displaying dimensions 1 and 2 and (C) dimensions 3 and 4. Samples are coloured according to final diagnose (yellow=IPMN LGD, orange=IPMN HGD, red=IPMN Cancer). (D) Bacterial co-occurrence and antioccurrence visualised in a network, with nodes representing bacterial species (coloured according to phylum, size corresponds to mean relative abundance) and edges representing interactions (green=co-occurrence, red=mutual exclusion) supported by >3 methods at statistical significance of q<0.01. HGD, high-grade dysplasia; IPMN, intraductal papillary mucinous neoplasm; LGD, low-grade dysplasia; PPI, proton-pump inhibitor.
Stringent network analysis using five different metrics for similarity or dissimilarity was employed to gain deeper insight into the correlative bacterial populations at species level. Despite the apparent dissimilarity of microbial composition between all samples, the resulting network comprising 38 species show significant co-occurrence and antioccurrence (supported by ≥4 methods, q<0.01). The oral pathogen Fusobacterium nucleatum co-occurred with 10 other species, including Serratia marcescens, Parvimonas micra, Prevotella melaninogenica, Haemophilus parahaemolyticus, Streptococcus anginosus, Bergeyella sp. HMT322, Kluyvera ascorbata, Eikenella corrodens, Campylobacter concisus and Campylobacter showae, many of which are known oral species. Notably, Granulicatella adiacens co-occurred with another oral bacteria P. melaninogenica, while showing mutual exclusion with Streptococcus parasanguinis, Achromobacter xylosoxidans, Aerococcus viridans, Moraxella osloensis, Delftia acidovorans and Sphingomonas sp. HMT004. The network also contains other oral species, such as Actinomyces odontolyticus, Porphyromonas pasteri and Gemella haemolysans (figure 4D).
Identification of bacterial biomarkers associated with disease severity
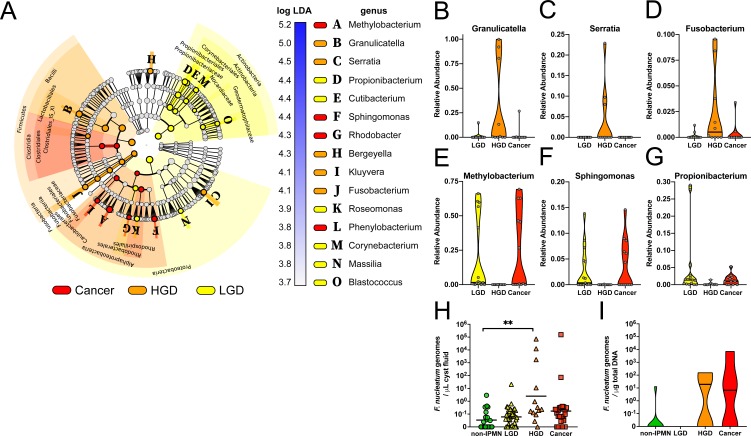
Linear discriminant analysis effect size analysis17 identified 15 bacterial genera that were differentially abundant between the three diagnosis groups. IPMN HGD was highly enriched for the genera Granulicatella, Serratia and Fusobacterium, but low on Methylobacterium, Sphingomonas and Propionibacterium. The latter three appeared more abundant in LGD and Cancer (figure 5A–G).
Figure 5.
(A) Cladogram representing linear discriminant analysis effect size results which identified 15 features that are differentially abundant between the three diagnose classes (red=Cancer, orange=IPMN HGD, yellow=IPMN LGD). (B–G) Violin plot showing relative abundance distribution and median of selected differentially abundant bacterial genera per diagnose group. Validation of Fusobacterium nucleatum genome quantification by TaqMan qPCR in (H) cyst fluid and (I) DNA extracted from 25 formalin-fixed PCN tissue samples (non-IPMN n=9, IPMN LGD n=6, IPMN HGD n=4, Cancer n=6). Statistically significant differences were determined using Kruskal-Wallis test with Dunn’s multiple comparison test **p<0.01. HGD, high-grade dysplasia; IPMN, intraductal papillary mucinous neoplasm; LGD, low-grade dysplasia; PCN, pancreatic cystic neoplasm.
Because F. nucleatum is reported to associate with oncogenesis,13 we validated the presence of F. nucleatum by a targeted qPCR assay on PCN cyst fluid samples (n=99), and confirmed the HGD group contained a highly increased F. nucleatum genome copy number in the cyst fluid (figure 5H). As F. nucleatum may also reside intracellularly, FFPE samples from a subset of 25 patients with PCN (see online supplementary table 1), validated to have >40% tumour tissue content, were also tested using qPCR. This confirmed F. nucleatum detection predominantly in IPMN HGD and Cancer lesion tissues (figure 5I).
gutjnl-2018-317458supp002.pdf (903.6KB, pdf)
Correlation of intracystic bacterial DNA quantity and composition with clinical factors
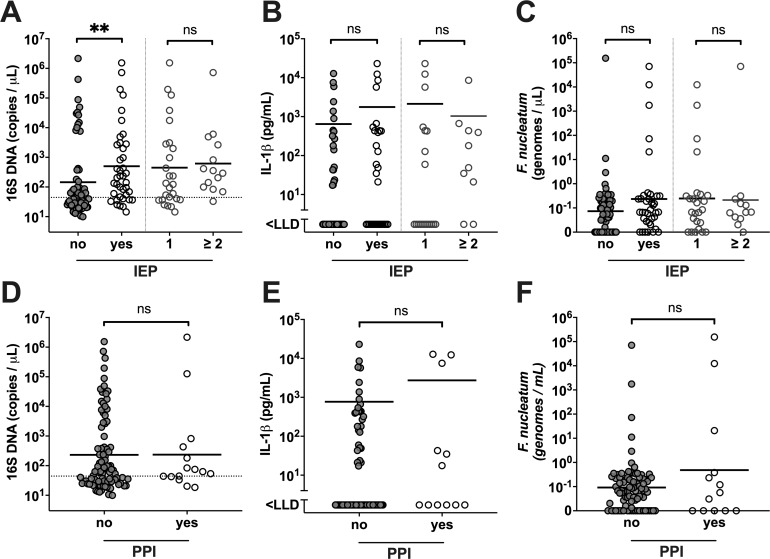
Next, we examined the possible clinical factors that may affect bacterial DNA quantity in pancreatic cyst fluid. Correlation analysis was performed against documented antibiotic and PPI usage, or prior invasive endoscopic procedures (IEP) including EUS-FNA, endoscopic retrograde cholangiopancreatography (ERCP) and percutaneous transhepatic cholangiography (PTC), which may be potential routes of bacterial ‘seeding’ of the pancreatic ducts. Our result indicated that patients who had a history of IEP prior to surgery also had significantly elevated intracystic 16S gene copies (p=0.0017) (figure 6A), although this appeared independent from the number of IEP, F. nucleatum genome copies or IL-1β concentration (figure 6B, C). Furthermore, when we stratified patients according to their history of IEP, the IPMN HGD and Cancer patients that never had prior IEP also had significantly higher intracystic bacterial 16S DNA (p=0.0268) compared with the SCN benign tumour group, similar to that observed for all patients in the current cohort (see online supplementary figure 2A, B). This is supported by a comparable positive correlation between 16S DNA and IL-1β in patients with or without history of IEP procedures (see online supplementary figure 2C, D). Use of PPI, which has been linked to alterations in gut microbiota composition and increased risk of bacterial translocation and enteric infections,18 19 had no significant impact on the intracystic bacterial 16S DNA, IL-1β or F. nucleatum quantity (figure 6D–F). Neither the use of PPI or antibiotics, nor prior IEP associated with the intracystic microbial diversity (figure 4A), and no difference in alpha-diversity was observed between groups with or without IEP (not shown). Analysis using multivariate analysis by linear models (MaAsLin) showed no statistically significant associations between relative abundance of bacterial genera and clinical parameters, including history of IEP, use of PPI and use of antibiotics <1 month prior to surgery (not shown).
Figure 6.
Patient stratification based on previous pancreatic invasive procedures (IEP), classified as ’no' (n=65) and ’yes' (n=40), comprising those with '1' IEP (n=26) and '≥2' (n=14), or use of proton-pump inhibitors (PPI), classified as ’no' (n=90) and ’yes' (n=15). Panels display intracystic (A) 16S DNA quantities, (B) interleukin (IL)-1β quantity and (C) Fusobacterium nucleatum genome copies, with the geometric mean value indicated per group stratified on IEP, and intracystic (D) 16S DNA quantities, (E) IL-1β quantity and (F) F. nucleatum genome copies, with the geometric mean value indicated per group stratified on PPI use. Statistically significant differences between ’no' and ’yes' groups, and '1' and '≥2' groups, respectively, were determined using Mann-Whitney U test, with **p<0.01. LLD, lower limit of detection; ns, not significant.
Discussion
In this prospective evaluation of pancreatic cyst fluid aspirated from surgically removed PCNs, we showed that intracystic bacterial DNA and IL-1β concentrations were significantly elevated in groups classified as IPMN and IPMN with invasive Cancer, as compared with other PCN types. Specifically, bacterial DNA and IL-1β concentrations were significantly positively correlated intracystically but not in paired patient plasma, and were found to be highest among patients diagnosed with IPMN HGD. Of note, the non-IPMNs, which comprised SCN (benign) and MCN pancreas tumours, were low on bacterial DNA and IL-1β. This is of interest as in contrast to IPMN, they do not communicate with the pancreatic ductal system, which drains and communicates with the gut. Our finding is in agreement with a previous study on pancreatic cyst fluid cytokines showing that IL-1β levels could discriminate between low-risk (IPMN LGD) and high-risk (IPMN HGD and Cancer) lesions.20 Release of the proinflammatory cytokine IL-β is known to be restricted to monocytes responding to pathogen-associated molecular patterns (PAMPs). Microbial nucleic acids and cell-wall components are an important group of IL-1β-stimulatory PAMPs which include microbial genomes, unmethylated CpG DNA motifs, LPS and peptidoglycan of Gram-positive/Gram-negative bacteria.21 The high intracystic IL-1β concentrations detected in IPMN HGD and Cancer groups, and their positive correlations with the bacterial DNA and LPS levels, therefore indicate a role for the pancreatic cyst microbiome in modulation of the local tumour environment. This in turn suggests an inflammatory potential of the pancreatic microbiome that is not related to systemic infections, as suggested by the paired blood plasma data.
Interestingly, the elevated intracystic bacterial DNA quantity appeared to associate with prior IEP when the entire cohort was examined. Because a selection bias might have occurred as patients with IPMN with more advanced or malignant lesions are likely to have undergone more invasive endoscopic procedures, we stratified our cohort by previous IEPs. We found that patients in IPMN and Cancer groups who never underwent any invasive procedure of the pancreas in fact also show a higher 16S DNA load and IL-1β load in their pancreatic cyst fluid. Hence, current study cannot rule out that IEP (ERCP, PTC or EUS-FNA) pose a possible iatrogenic route of bacterial translocation into the pancreas. But, the fact that no clear difference could be observed in the intracystic bacterial DNA quantity and composition from patients with and without IEP, further supports that other routes of bacterial translocation may exist.
Our network analysis showed that the intracystic pancreas microbiome harboured co-occurring bacterial species that are known to inhabit the human oral cavity, including G. adiacens, F. nucleatum, P. micra, E. corrodens, H. parahaemolyticus, A. odontolyticus, P. melaninogenica and Campylobacter ssp.22 Several of these species are found in biofilms that cause periodontitis and gingivitis and are also connected with increased risk for systemic disease.23 The presence of bacteria in PCN and pancreatic cancer (PC) tissue has been demonstrated before.12 14 15 Although it was recently shown that an abundant pancreatic microbiome may drive disease progression in PC by inducing intratumoural immune suppression,12 this is the first time bacterial DNA quantity was found to correlate with the neoplastic grade severity of IPMN. Transition from low-risk IPMN LGD to high-risk IPMN HGD with increased bacterial presence or an altered microbial community and subsequent immune signalling has not been reported to our knowledge and deserves to be further investigated. The present finding that high abundance of F. nucleatum was seen in both cyst fluid and tissues of HGD is in line with F. nucleatum being recognised as an onco-bacterium24 and more research should focus on elucidating its early involvement in tumourogenesis. Beyond this point, the pancreatic tumour microenvironment is known to be possess hypoxic and immunosuppressive properties, supporting the preferential growth of anaerobic bacteria, such as F. nucleatum, as reported in colorectal cancer.25 26
Oral pathogens and inflammation (periodontitis) have been linked to increased risk for PC, mainly through association studies with pancreatic cancer incidence and salivary microbiota composition or seroprevalence of Porphyromonas gingivalis antibodies.27–30 Specifically, oral carriage of P. gingivalis, and Aggregatibacter actinomycetemcomitans, as well as circulating antibodies against P. gingivalis were associated with a higher risk of PC. A recent meta-analysis concluded that there is a general positive association between periodontitis and PC, even after adjusting for common risk factors.31 In addition, gut microbiota dysbiosis, bacterial translocation and inflammation have been linked with several pancreatic disorders, including PC.32 Increased levels of salivary G. adiacens were shown to be associated with pancreatitis and PC compared with healthy individuals.33 In line with these findings, the intracystic microbiome we describe comprises several co-occurring bacterial species, such as F. nucleatum, C. showae, P. melaninogenica, G. adiacens and Haemophilus parainfluenzae, which have previously been linked with other human malignancies.34–36 Possible explanations to why the pancreatic cyst microbiome comprises oral bacterial species deserve to be investigated, including functional similarities shared by pancreas and salivary glands,37 possibly creating a biological environment attractive to similar types of microorganisms.
Our study demonstrates that distinct bacterial communities can exist in cyst fluid from IPMN with LGD, HGD and IPMN Cancer. While these disease groups show markedly different microbial compositions at the phylum level, at lower taxonomic levels high interpersonal variation in community structure becomes clear. This is in contrast to a previously reported pancreatic ‘core microbiome’ across different types of PCN, including MCN and SCN, by Li et al,15 although similar bacterial taxa were detected in our samples despite our different technological approach. Notwithstanding high interpersonal variation and our relatively small sample size, our results show enrichment of Fusobacteria, Granulicatella and Serratia in IPMN HGD, and warrant further investigation into the potential role of these oral bacteria in the progression of cyst precursors to PC.
A strength of this study is that postresection aspirated cyst fluid was collected in combination with histologically confirmed diagnosis, limiting the risk of sample contamination through endoscopic probing while maximising diagnostic accuracy. Furthermore, well-controlled and standardised procedures were applied for liquid biopsy collection and storage. A limitation in our current study is that only one cyst was sampled per patient, while IPMN can be of unifocal or multifocal nature with independently arising cysts that may contain different grades of dysplasia and varying malignant potential.2 38 39 As histological examination considers the whole resection specimen, the actual sampled cyst may not be fully representative for the multifocal type of IPMN. Preliminary data from our laboratory suggest a similar microbial profile in fluid taken from separate pancreatic locations in the same patient (n=2; unpublished observation), however future studies of microbiome compositional variation in the pancreatic ductal system should be considered. In addition, bacterial DNA quantity and quality obtained from cyst fluid and plasma were generally low, and therefore susceptible to environmental contaminations in assays such as qPCR and amplicon sequencing-based microbial composition analyses.40 To address this concern, we applied stringent DNA quantity and quality filters while maintaining sufficient sample numbers to allow for statistical analyses, as well as using appropriate negative controls that were analysed in parallel. Nevertheless, for an absolute optimal correlation between relative abundance of specific bacterial taxa and disease parameters, larger sample size and samples taken from multiple sites are needed to prove if different disease-specific intracystic microbiomes exist. Based on our present microbiota data, we estimate a sample size of >70 per patient group would be needed to reach a power of 0.8.
Our result may have an impact on clinical practice in the foreseeable future. First, additional means to reduce systemic dissemination of oral pathobionts in patients diagnosed with IPMN should be recommended, for instance, invasive endoscopy could be performed with minimal oral bacteria carryover, by infection control using local oral antiseptics and/or regular oral care. Second, employing salivary biomarker detection or serology29 33 of the bacteria identified in current study could pave ways to IPMN discrimination from other cystic lesions of the pancreas to reduce unnecessary surgeries. This is of particular importance and may balance current problems with overtreatment of IPMN and the same time prevent potential durable changes of pancreatic microbiome that could contribute to cell transformation and future PC resistance to chemotherapy.14
Furthermore, pancreatic cystic microbiome testing could have added values in guiding the management of IPMN, especially during follow-up, as IPMN recurrence is possible 5–10 years after resection. Further investigation is needed to assess whether high bacterial loads and inflammation in the postoperative pancreas are associated with increased risk of IPMN recurrence. This may lead to further improvement of the disease-free survival after resection, which at present for LGD is 5.4%–10%; 52 months, and for HGD/cancer in patients with IPMN is >50%; 29 months.1 Of note, DNA testing (16S and/or targeted oral bacteria multiplex) is very rapid, that is, 2–3 hours, compared with traditional histopathology which takes weeks, and may better support an effective and timely decision.
However, there are important considerations if microbiome testing is to become fully applicable to clinical practice. First, stringent laboratory quality control measures to prevent environmental contamination need be in place to ensure sequencing accuracy and data reliability. In addition, a pancreatic microbiome database and/or multicentre study which includes a substantial number of well-described patients may allow the detection of a ‘core’ pathobiont group hiding behind variations in the current cohort.27 29 30
Much work remains to be done in order to better understand the disease progression in various PCNs.1 The findings presented in this study might not directly lead to clinical applicability using the intracystic microbiota as a ‘predictive tool for malignancy’, but they might change the way clinicians think about the largely ignored role of microbes in PCN development and progression. Experimental (intervention) studies will be important to elucidate whether bacteria identified here have a driving role in IPMN progression, and if so, whether an antibiotic treatment in patients affected by IPMNs will be an important option to reduce the risk of progression to malignancy in the near future. The impact of pancreatic invasive endoscopic procedures as a potential iatrogenic route of bacterial translocation deserves to be further investigated with regard to tumour recurrence and progression.
Acknowledgments
The authors would like to thank all patients for making this study possible. The authors would also like to thank Sam Ghazi at Karolinska University Hospital for technical support and consultation.
Footnotes
Contributors: Study design: MDC, MSC, RV, RAG. Sample collection and processing: MDC, AH, HD, RAG, KH, ZA, CFM. Clinical data collection and interpretation: ZA, RV, AH, CFM, RAG. Laboratory work: RAG, HD, HA, KH, LL. Bioinformatic analysis and statistics: LWH, RAG. Manuscript preparation: RAG, MSC, RV, KH. Approval of final draft submission: RAG, AH, HA, LL, HD, KH, LWH, ZA, RV, CFM, MDC, MSC.
Funding: This work was supported by the Swedish Cancer Society (CAN2016/731 to MSC) (CAN 2014/634, CAN 2015/621, CAN 2017/409, CAN 2017/419 to MDC). Ruth och Richard Julins funds, Karolinska Institutet funds, KI KID funding, and SOF KI/SLL för odontologisk forskning (to MSC). The Stockholm County Council (ALF SLL20150113) (to MDC).
Competing interests: None declared.
Ethics approval: This study follows the Helsinki convention and good clinical practice and was approved by the Regional Ethics Committee at Stockholm (Dnr. 2015/1580-31/1).
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Not required.
References
- 1. European Study Group on Cystic Tumours of the Pancreas. European evidence-based guidelines on pancreatic cystic neoplasms. Gut 2018;67:789–804. 10.1136/gutjnl-2018-316027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chandwani R, Allen PJ. Cystic Neoplasms of the Pancreas. Annu Rev Med 2016;67:45–57. 10.1146/annurev-med-051914-022011 [DOI] [PubMed] [Google Scholar]
- 3. Valsangkar NP, Morales-Oyarvide V, Thayer SP, et al. . 851 resected cystic tumors of the pancreas: a 33-year experience at the Massachusetts General Hospital. Surgery 2012;152:S4–12. 10.1016/j.surg.2012.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferrone CR, Correa-Gallego C, Warshaw AL, et al. . Current trends in pancreatic cystic neoplasms. Arch Surg 2009;144:448–54. 10.1001/archsurg.2009.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aronsson L, Andersson R, Ansari D. Intraductal papillary mucinous neoplasm of the pancreas - epidemiology, risk factors, diagnosis, and management. Scand J Gastroenterol 2017;52:803–15. 10.1080/00365521.2017.1318948 [DOI] [PubMed] [Google Scholar]
- 6. Machado NO, Al Qadhi H, Al Wahibi K. Intraductal Papillary Mucinous Neoplasm of Pancreas. N Am J Med Sci 2015;7:160–75. 10.4103/1947-2714.157477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Del Chiaro M, Segersvärd R, Pozzi Mucelli R, et al. . Comparison of preoperative conference-based diagnosis with histology of cystic tumors of the pancreas. Ann Surg Oncol 2014;21:1539–44. 10.1245/s10434-013-3465-9 [DOI] [PubMed] [Google Scholar]
- 8. Tanaka M, Fernández-del Castillo C, Kamisawa T, et al. . Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 2017;17:738–53. 10.1016/j.pan.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 9. Crippa S, Pergolini I, Rubini C, et al. . Risk of misdiagnosis and overtreatment in patients with main pancreatic duct dilatation and suspected combined/main-duct intraductal papillary mucinous neoplasms. Surgery 2016;159:1041–9. 10.1016/j.surg.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 10. Singhi AD, McGrath K, Brand RE, et al. . Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2018;67:2131–41. 10.1136/gutjnl-2016-313586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Springer S, Wang Y, Dal Molin M, et al. . A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015;149:1501–10. 10.1053/j.gastro.2015.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pushalkar S, Hundeyin M, Daley D, et al. . The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8:403–16. 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mitsuhashi K, Nosho K, Sukawa Y, et al. . Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015;6:7209–20. 10.18632/oncotarget.3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geller LT, Barzily-Rokni M, Danino T, et al. . Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357:1156–60. 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S, Fuhler GM, Bn N, et al. . Pancreatic cyst fluid harbors a unique microbiome. Microbiome 2017;5:147 10.1186/s40168-017-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang S, Lin S, Kelen GD, et al. . Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J Clin Microbiol 2002;40:3449–54. 10.1128/JCM.40.9.3449-3454.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Segata N, Izard J, Waldron L, et al. . Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imhann F, Bonder MJ, Vich Vila A, et al. . Proton pump inhibitors affect the gut microbiome. Gut 2016;65:740–8. 10.1136/gutjnl-2015-310376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reveles KR, Ryan CN, Chan L, et al. . Proton pump inhibitor use associated with changes in gut microbiota composition. Gut 2018;67:1369–70. 10.1136/gutjnl-2017-315306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maker AV, Katabi N, Qin LX, et al. . Cyst fluid interleukin-1beta (IL1beta) levels predict the risk of carcinoma in intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res 2011;17:1502–8. 10.1158/1078-0432.CCR-10-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hemmi H, Takeuchi O, Kawai T, et al. . A Toll-like receptor recognizes bacterial DNA. Nature 2000;408:740–745. 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- 22. Zaura E, Keijser BJ, Huse SM, et al. . Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol 2009;9:259 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jia G, Zhi A, Lai PFH, Pfh L, et al. . The oral microbiota - a mechanistic role for systemic diseases. Br Dent J 2018;224:447–55. 10.1038/sj.bdj.2018.217 [DOI] [PubMed] [Google Scholar]
- 24. Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol 2019;17:156-166 10.1038/s41579-018-0129-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abed J, Emgård JE, Zamir G, et al. . Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016;20:215–25. 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gur C, Ibrahim Y, Isaacson B, et al. . Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015;42:344–55. 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fan X, Alekseyenko AV, Wu J, et al. . Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67:120–7. 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Öğrendik M. Periodontal Pathogens in the Etiology of Pancreatic Cancer. Gastrointest Tumors 2017;3:125–7. 10.1159/000452708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Michaud DS, Izard J, Wilhelm-Benartzi CS, et al. . Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013;62:1764–70. 10.1136/gutjnl-2012-303006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres PJ, Fletcher EM, Gibbons SM, et al. . Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ 2015;3:e1373 10.7717/peerj.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maisonneuve P, Amar S, Lowenfels AB, et al. . Periodontal disease, edentulism, and pancreatic cancer: a meta-analysis. Ann Oncol 2017;28:985–95. 10.1093/annonc/mdx019 [DOI] [PubMed] [Google Scholar]
- 32. Pagliari D, Saviano A, Newton EE, et al. . Gut Microbiota-Immune System Crosstalk and Pancreatic Disorders. Mediators Inflamm 2018;2018:1–13. 10.1155/2018/7946431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Farrell JJ, Zhang L, Zhou H, et al. . Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012;61:582–8. 10.1136/gutjnl-2011-300784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amer A, Galvin S, Healy CM, et al. . The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species. Front Microbiol 2017;8:2391 10.3389/fmicb.2017.02391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Warren RL, Freeman DJ, Pleasance S, et al. . Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 2013;1:16 10.1186/2049-2618-1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao H, Chu M, Huang Z, et al. . Variations in oral microbiota associated with oral cancer. Sci Rep 2017;7:11773 10.1038/s41598-017-11779-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kamisawa T, Tu Y, Egawa N, et al. . Salivary gland involvement in chronic pancreatitis of various etiologies. Am J Gastroenterol 2003;98:323–6. 10.1111/j.1572-0241.2003.07230.x [DOI] [PubMed] [Google Scholar]
- 38. Raman SP, Kawamoto S, Blackford A, et al. . Histopathologic findings of multifocal pancreatic intraductal papillary mucinous neoplasms on CT. AJR Am J Roentgenol 2013;200:563–9. 10.2214/AJR.12.8924 [DOI] [PubMed] [Google Scholar]
- 39. Felsenstein M, Noë M, Masica DL, et al. . IPMNs with co-occurring invasive cancers: neighbours but not always relatives. Gut 2018;67:1652–62. 10.1136/gutjnl-2017-315062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salter SJ, Cox MJ, Turek EM, et al. . Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12:87 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2018-317458supp001.pdf (105.6KB, pdf)
gutjnl-2018-317458supp002.pdf (903.6KB, pdf)