Damage to DNA occurs continuously. DNA quality control mechanisms, such as DNA repair and replicative pathways, mitigate that damage and preserve the DNA sequence. Mutations are believed to arise when DNA lesions are not repaired appropriately, and they are thought to be an indicator of ineffectual DNA quality control. Yet, DNA sequencing of normal tissues reveals that considerable somatic mutagenesis is common. Mutagenesis appears to be the inevitable outcome of cellular wear and tear and is not necessarily cancer associated. Perhaps mutagenesis is not due to failings of DNA quality control mechanisms. Rather, such pathways may be naturally limited in activity, resulting in permissiveness to mutagenesis. We suggest that this is a prioritization of survival over genomic perfection, given that most DNA damage is inconsequential and thus, affordable.
Water and oxygen are highly mutagenic to DNA, but essential for life (1). Hydrolysis and oxidation continuously cause DNA modifications that could become permanently embedded in the genome, as mutation (1). In addition to endogenous sources of DNA damage, DNA is regularly exposed to environmental genotoxins such as ultraviolet (UV) radiation or tobacco smoke. DNA damage is thus inevitable. DNA quality control pathways survey and fix the genome to mitigate damage (2, 3). DNA damage, such as double-strand breaks, are poorly tolerated and if unfixed, can induce cell death (2, 3). Moreover, DNA damage checkpoints, which pause cell division to allow time for DNA repair, highlight that maintaining DNA integrity prior to cell division is vital for cell survival (4). Comprehensive DNA quality control pathways are therefore considered crucial to preserving genomic health.
The biologist Theodor Boveri hypothesized that aggregations of heritable material, later called chromosomes, were associated with cancer. His work lay the foundations for the somatic mutation theory of carcinogenesis, in which stepwise acquisition of mutations are believed to be central to the development of cancer (5). Through statistical analyses of inherited versus sporadic retinoblastoma, Alfred Knudson hypothesized that two “hits” to DNA (driver mutations) were necessary to cause cancer. This explained early, childhood onset of heritable retinoblastoma, because the first mutation was inherited. A raft of “driver” mutation discoveries soon followed. Cancers carry only a handful (less than 10 per tumor) of causally implicated driver mutations that occur in “cancer genes,” so-called because these genes are recurrently mutated across many tumors. This buttressed the somatic mutation theory and was instrumental to the development of targeted therapeutics.
Cancer cells also carry thousands of “passenger” mutations believed to be bystander events devoid of biological effects. Passenger mutations are thought to arise because of incompetencies of DNA quality control pathways to adequately fix DNA damage (6). Indeed, an early step in tumorigenesis has been proposed to be the induction of a “mutator phenotype” (6). Large-scale cancer genome sequencing efforts have revealed the extraordinary extent of mutagenesis such that extensive genetic changes are considered pathognomonic of cancer.
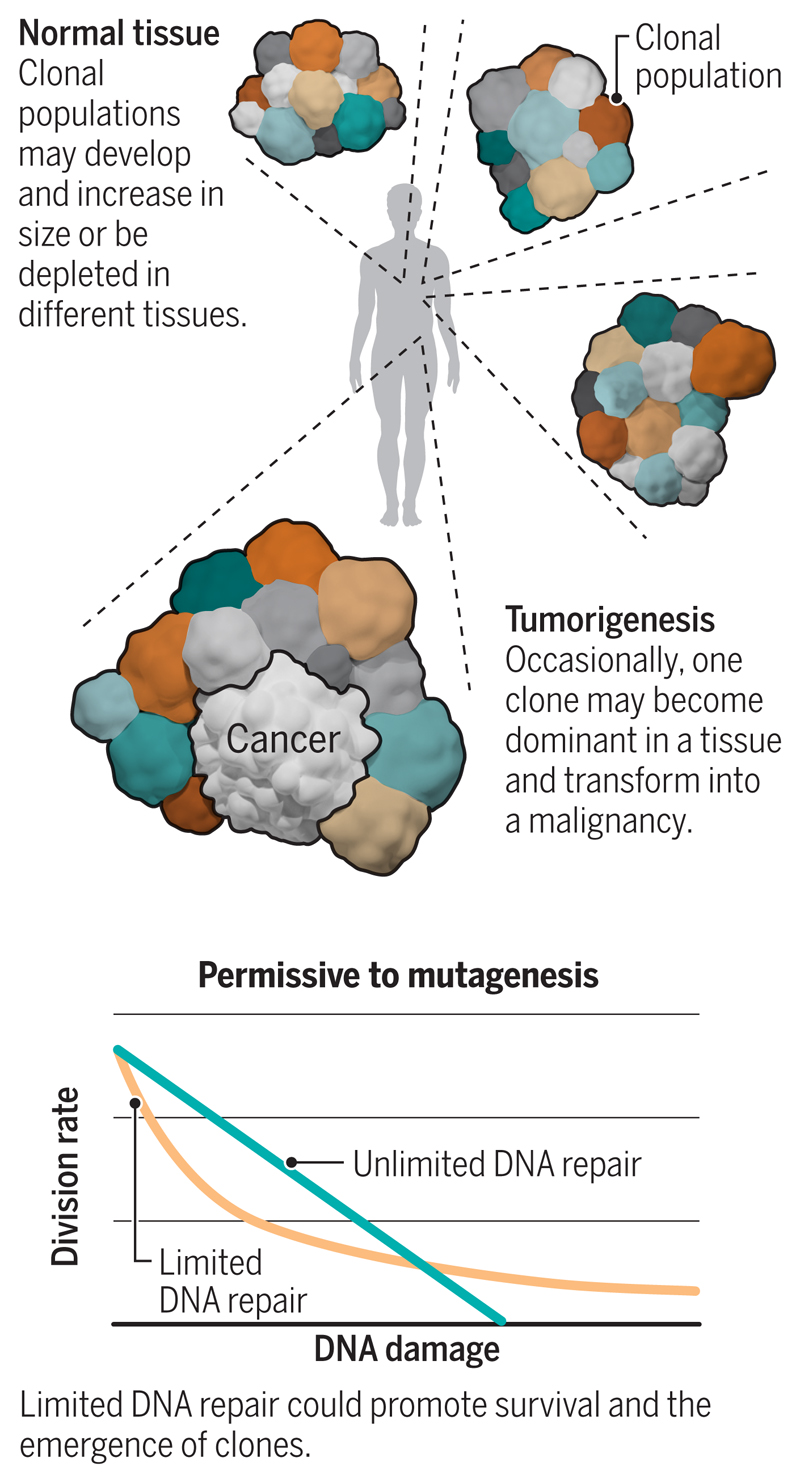
Recently, somatic mutations, including driver mutations and high numbers of passenger mutations, were found to occur in normal tissue with no sign of cancer, implying that mutagenesis could be extensive in normal cells, prior to malignant transformation (7). Notably, samples of redundant eyelid skin removed at blepharectomy were used for ultra-deep sequencing of 74 cancer genes, and characteristic mutations associated with skin cancer (e.g., melanoma) were found in these nonmalignant tissues. Qualitative mutational signatures (patterns) of UV radiation–induced damage were also appreciable, and the quantitative burden of mutagenesis in normal skin was comparable to that of skin cancers (8). Moreover, ultra-deep targeted sequencing of normal esophageal epithelium discerned hundreds of mutated clonal cell populations per square centimeter. The cancer gene NOTCH1 was one of the most commonly mutated genes in normal esophageal epithelium, with a frequency exceeding that observed in esophageal cancers (9, 10). In a comprehensive analysis of RNA sequencing data from >6000 samples of 29 healthy tissue types (11), cancer-associated mutations were found across tissues, levels of mutagenesis were associated with exposure to exogenous mutagens and tissue-specific proliferation rates, and macroscopic clonal expansions were also demonstrable. Hence, driver mutations, widespread mutagenesis, and even a mélange of competitive clones are all compatible with normal tissue and are not necessarily associated with cancer (see the figure).
Mutagenesis in normal tissue.
DNA is continuously subject to damage, which may become fixed as mutation if unrepaired by tolerant DNA quality control pathways. Most normal tissues are likely to have many clonal populations defined by the distinct set of mutations they harbor.
The high number of passenger mutations, equivalent to 1000 to 10,000 per genome, in normal cells (8, 12) raises questions regarding why DNA quality control mechanisms have failed to limit mutagenesis. Perhaps a somewhat counterintuitive perspective (13) can be considered: If DNA quality control pathways monitor and preserve DNA integrity too strictly, it could be detrimental to cellular survival. The repair of DNA lesions has a cost: It requires time and cellular resources. If every DNA lesion in a cell were repaired, avoiding mutations altogether, the cellular cost associated with performing that repair would have to increase in direct proportion to the amount of damage. In conditions of high DNA damage—through exposure to environmental mutagens, for example—DNA repair could be too costly for cellular survival. Instead, if restrictions or thresholds were placed on DNA quality control to perform some repair but not necessarily of all lesions, then DNA-repair–limited cells are more likely to survive than cells caught in comprehensive repair, which can lead to cell death (apoptosis). This implies that DNA quality control pathways are fully functional but naturally permissive of mutagenesis, even in normal cells.
Evidence to support this hypothesis comes from experiments in which induced pluripotent stem cells (iPSCs) were treated with an array of environmental genotoxins (12). These normal stem cells accumulated extensive passenger mutations showing distinct mutational signatures. Some mutational signatures showed transcriptional strand bias—whereby the non-transcribed DNA strand is more heavily mutated than the transcribed strand (12), the mark of transcription-coupled repair (TCR) activity. TCR fixes DNA damage on both strands, but preferentially on the transcribed strand, and its activity in normal cells was comparable to that in cancer cells. Accordingly, in iPSCs and cancer cells, the presence of strand bias indicates that TCR is competent and operative, but unable to mitigate all DNA damage, and is thus tolerant of mutagenesis.
Limited DNA quality control during circumstances of high damage has been suggested through mathematical modeling (13). The model shows that in a situation where individual DNA damaging events are rarely deleterious (that is, when most mutations are passengers) during periods of high damage, DNA-repair–unlimited cells incur high repair costs and become depleted owing to cell death. By contrast, DNA-repair–limited cells may continue with cell division and survive (see supplementary movies S1 and S2). This model could be a useful starting point to explore permissiveness to mutagenesis. Given that most (~98%) of the genome is intronic and intergenic and the majority of mutations that arise as a result of DNA damage are unlikely to affect protein function, the model might explain why normal, healthy cells show considerable amounts of mutagenesis: survival through tolerance of mutation, rather than compromised DNA quality control processes.
The tolerance of mutagenesis in normal cells can, however, lead to the occurrence of new drivers, but not necessarily cancer. For example, in the testes of older men, acquired mutations in genes encoding components of receptor tyrosine kinase–RAS–mitogen-activated protein kinase (MAPK) and fibroblast growth factor receptor (FGFR) signaling pathways, which are commonly associated with cancer, result in increased cellular proliferation rates but seldom cause cancer. Clonal populations of seminiferous tubules generate spermatozoa with these mutations. This explains the high spontaneous birth prevalence and strong paternal age-effect of disorders such as achondroplasia, Apert, and Costello syndromes—all associated with activating mutations in MAPK and FGFR signaling genes. A study of 276 testicular biopsies obtained from five older men (median age of 83) identified additional cancer gene variants, all known to activate RAS-MAPK signaling (14). The aging male germ line therefore accumulates deleterious cancer driver mutations that rarely (if ever) switch to malignancy.
Another example is segmental overgrowth syndromes caused by somatic mutations of the cancer gene, PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α). Although these syndromes are benign in terms of oncology, they can produce debilitating clinical phenotypes with unrelenting tissue growth (15). Referred to as PIK3CA-related overgrowth syndromes (PROS), affected people are mosaics, wherein discrete clonal populations exhibit mutations. The most frequent PIK3CA mutations in PROS (Glu542Lys, Glu545Lys, and His1047Arg) are identical to hotspot cancer driver mutations although other, non–cancer-associated PIK3CA mutations also occur in PROS. Notably, surrounding cells in affected tissues that are not PIK3CA-mutated are also overgrown (15). It has consequently been suggested that PIK3CA-mutated cells do not simply possess cell-autonomous characteristics, they may exert proliferation-enhancing properties on neighboring, non–PIK3CA-mutated cells.
Therefore, driver mutations alone are not sufficient to initiate cancer, although they can be disease-causing. Considering that an adult human has ~30 trillion cells, and only one cell develops into a cancer, human cells are remarkably robust at preventing cancer. Because the burden of mutagenesis and the frequencies of driver mutations can be high in normal tissues, mutations alone are possibly not sufficient to initiate cancer. Additional extrinsic factors, including cellular interactions and the microenvironment, seem to be required to create a selective environment that triggers a cell to become a dominant cancer clone. It is likely that the mutated genome of a cell contributes to the potential for malignant transformation, but it is not deterministic of it. This concept is not unfamiliar: Inherited mutations in DNA repair genes, such as BRCA1, BRCA2, and MLH1 (mutL homolog 1), cause familial cancer predisposition. They confer an increased lifetime risk of cancer, but cancer does not always arise. Additional factors seem to be required to tip the balance toward cancer development.
Mutagenesis remains important to explore. Identifying clinically relevant drivers and mutational signatures should provide meaningful directions for patient stratification and prognostication. The use of specific driver mutations is a favored method for patient stratification in clinical studies. However, a cancer gene could play disparate roles in tumors affecting different organs, and its functionality could evolve over the patient’s lifetime. Given the mutagenesis observed in normal tissues, perhaps it is time to reflect on whether this is the best approach for stratification. Similarly, although many mutational signatures have been identified, only a subset are clinically instructive—some may occur normally and not be causative of cancer. Others may be biomarkers of DNA repair deficiencies, which are potentially informative for therapy selection. It is imperative to identify mutations that are truly clinically meaningful, to improve cancer management effectively.
Supplementary Material
Acknowledgments
Thanks to M. Hall for valuable discussions. B.A.H. is supported by the Royal Society (grant no. UF130039). S.N.-Z. is supported by a Cancer Research UK (CRUK) Advanced Clinician Scientist Award (C60100/A23916) and the Josef Steiner Foundation. Both authors are supported by a Medical Research Council (MRC) Grant-in-Aid to the MRC Cancer unit.
References
- 1.Lindahl T. Nature. 1993;362:709. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Rich T, Allen RL, Wyllie AH. Nature. 2000;407:777. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. N Engl J Med. 2009;361:1475. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 4.Kastan MB, Bartek J. Nature. 2004;432:316. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, et al. N Engl J Med. 1988;319:525. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA. Cancer Res. 1991;51:3075. [PubMed] [Google Scholar]
- 7.Tomasetti C, Vogelstein B, Parmigiani G. Proc Natl Acad Sci USA. 2013;110:1999. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martincorena I, et al. Science. 2015;348:880. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martincorena I, et al. Science. 2018;362:911. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A, et al. Nature. 2019;565:312. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 11.Yizhak K, et al. Science. 2019;364 doi: 10.1126/science.aaw0726. eaaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kucab JE, et al. 2019;177:821. doi: 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breivik J, Gaudernack G. FEBS Lett. 2004;563:7. doi: 10.1016/S0014-5793(04)00282-0. [DOI] [PubMed] [Google Scholar]
- 14.Maher GJ, et al. Genome Res. 2018;28:1779. doi: 10.1101/gr.239186.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madsen RR, Vanhaesebroeck B, Semple RK. Trends Mol Med. 2018;24:856. doi: 10.1016/j.molmed.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.