Abstract
Patients with bipolar disorder (BD) show higher frequency of obesity and type 2 diabetes (T2D), but the underlying genetic determinants and molecular pathways are not well studied. Using large publicly available datasets, we (1) conducted a gene-based analysis using MAGMA to identify genes associated with BD and body mass index (BMI) or T2D and investigated their functional enrichment; and (2) performed two meta-analyses between BD and BMI, as well as BD and T2D using Metasoft. Target druggability was assessed using the Drug Gene Interaction Database (DGIdb). We identified 518 and 390 genes significantly associated with BD and BMI or BD and T2D, respectively. A total of 52 and 12 genes, respectively, were significant after multiple testing correction. Pathway analyses conducted on nominally significant targets showed that genes associated with BD and BMI were enriched for the Neuronal cell body Gene Ontology (GO) term (p = 1.0E−04; false discovery rate (FDR) = 0.025) and different pathways, including the Signaling by Hedgehog pathway (p = 4.8E−05, FDR = 0.02), while genes associated with BD and T2D showed no specific enrichment. The meta-analysis between BD and BMI identified 64 relevant single nucleotide polymorphisms (SNPs). While the majority of these were located in intergenic regions or in a locus on chromosome 16 near and in the NPIPL1 and SH2B1 genes (best SNP: rs4788101, p = 2.1E−24), five were located in the ETV5 gene (best SNP: rs1516725, p = 1E−24), which was previously associated with both BD and obesity, and one in the RPGRIP1L gene (rs1477199, p = 5.7E−09), which was also included in the Signaling by Hedgehog pathway. The meta-analysis between BD and T2D identified six significant SNPs, three of which were located in ALAS1 (best SNP: rs352165, p = 3.4E−08). Thirteen SNPs associated with BD and BMI, and one with BD and T2D, were located in genes which are part of the druggable genome. Our results support the hypothesis of shared genetic determinants between BD and BMI and point to genes involved in Hedgehog signaling as promising targets.
Subject terms: Clinical genetics, Bipolar disorder
Introduction
Bipolar disorder (BD) is a chronic and disabling psychiatric illness, with a prevalence of 0.8–1.2% in the general population1, characterized by recurrent manic and depressive episodes. Being characterized by high rates of physical comorbidity and increased risk for suicide, BD is associated with decreased life expectancy and increased all-cause mortality, and therefore is a major socio-economic burden2.
Patients with BD show increased frequency of overweight and obesity (41% compared to 27% in general population in US)3,4. Additionally, patients with BD show a three-fold higher risk of type 2 diabetes (T2D)5,6 compared to the general population. These two conditions greatly contribute to a higher risk of cardiovascular diseases, which represent the leading cause for increased mortality in bipolar patients7. Indeed, obesity exerts a negative impact on the course of BD, as this comorbidity is associated with higher episode frequency8, rates of disability9, suicide attempts10, psychiatric and medical comorbidities11 as well as cognitive impairment12 and white matter abnormalities13,14. Conversely, a lower body mass index (BMI) has been associated with positive response to lithium treatment9, which represents the first line of treatment in BD, being effective in preventing relapses of both polarities15. These results suggest a potential interplay between BMI and molecular targets involved in the mechanism of action of this mood stabilizer. Although a link between BD and BMI has been established, the molecular mechanisms underlying this comorbidity have only recently started to be investigated and are still largely unknown16,17. Increasing our scarce knowledge of the factors predisposing patients with BD to develop metabolic comorbidities such as obesity and T2D would be of great importance, eventually allowing us to identify patients that could benefit from early monitoring and/or intervention.
Although lifestyle factors and adverse effects of pharmacotherapy play an important role in the increased susceptibility of BD patients to metabolic comorbidities5, a higher BMI has also been reported in adolescents and drug naïve patients18,19, thus suggesting the existence of common pathophysiological processes, as well as genetic/epigenetic links underlying these conditions5,20,21. Indeed, BD, obesity and T2D are multifactorial disorders that show a high heritability, estimated at 60–80% in BD22, 40–70% in obesity23,24 and 30–70% in T2D25. In a recent study, it was shown that Ets96B (the ETV5 homolog in Drosophila melanogaster) regulates a number of genes involved in neuroprotection and that its inhibition induces BD- and obesity-related phenotypes20. Moreover, single nucleotide polymorphisms (SNPs) located in the human ETV5 gene were associated with BD20. Furthermore, a recent review suggested that 24 genes previously associated with cardiometabolic phenotypes are also associated with mood disorders (major depressive disorder (MDD) and BD)26. An overview of studies previously investigating the role of genes potentially involved in both BD and obesity or T2D16,17,21,27–36 is reported in Table 1. The majority of these studies investigated variants already known to be associated with T2D or other metabolic phenotypes.
Table 1.
Overview of previous studies investigating molecular links between bipolar disorder and obesity or T2D in humans.
| Sample | Main results | Study |
|---|---|---|
| Post-mortem dlPFC samples from 268 patients (SCZ = 113, MD = 155)a and 191 HC; hippocampal samples from 219 patients (SCZ = 96, MD = 113)a and 169 HCa | Compared to HC, patients with mood disorders showed differential expression of GLP-1R and GLP-2R in the dlPFC and of GLP-1R in the hippocampus | Mansur et al.27 |
| Post-mortem dlPFC samples from 268 patients (SCZ = 113, MD = 155)a and 191 HC; hippocampal samples from 219 patients (SCZ = 96, MD = 113)a and 169 HC | Significant diagnosis by BMI interaction modulates expression of genes involved in the reeling pathway (RELN, CAMK2A, CAMK2N2 and GRIN2A) | Brietzke et al.17 |
| Post-mortem dlPFC samples from 321 patients (SCZ = 142, MDD = 99, BD = 80) and 209 HC; hippocampal samples from 196 patients (SCZ = 102, MDD = 52, BD = 42) and 180 HC | Changes in the expression of insulin receptor-related genes in the postmortem brain tissue of patients with mood and psychotic disorders mediate the expression of dopamine regulation-related genes | Mansur et al.16 |
| 100 patients with BD type II | Association between the TT genotype of the GNB3 C825T variant and lower BMI in patients treated with valproate | Chen et al.28 |
| 284 patients with psychosis (32 with BD) | The addition of genetic to clinical data does not improve prediction of BMI or BMI gain after 1 year | Harrison et al.29 |
| 662 patients with BD and 616 HC | Interactions between different TCF7L2 variants and BMI modulate susceptibility to BD | Cuellar-Barboza30 |
| 139 patients with BD and 137 HC | Association between the Met allele of the BDNF Val66Met variant and lower frequency of overweight and obesity | Morales-Marin et al.31 |
| 384 probands (SCZ = 129, SAD = 85, BD = 160), 413 non-affected relatives and 218 HC | Lack of association between a T2D PRS and proband or relative status; lack of association between a SCZ PRS and prevalence of diabetes | Padmanabhan et al.32 |
| 90 patients with BD, 76 with SCZ or SAD | Association between the Met66 allele of the BDNF Val66Met functional variant and weight gain in patients treated with atypical antipsychotics | Bonaccorso et al.33 |
| 81 patients with SCZ or BDa | Association between CYP2D6 phenotype and weight gain in patients treated with atypical antipsychotics | Nussbaum et al.34 |
| 388 BD patients and 1020 HC | Interaction between the TCF7L2 rs12772424 variant and BMI in modulating susceptibility to BD | Winham et al.21 |
| 930 patients with SCZ, 869 with BD and 876 HC | No association between BD and 32 SNPs previously associated with T2D | Kajio et al.35 |
| 96 patients with BD | Association between the T allele of the GNB3 C825T variant and lower BMI in patients treated with valproate | Chang et al.36 |
BD bipolar disorder, dlPFC dorsolateral prefrontal cortex, HC healthy controls, MD mood disorders, MDD major depressive disorder, PRS polygenic risk score, SAD schizoaffective disorder, SCZ schizophrenia
aThe number of mood disorder patients with a diagnosis of BD was not specified
Since the potential molecular mechanisms that could predispose BD patients to an increased susceptibility to obesity or T2D are largely unknown, studies not restricted to specific candidate genes or pathways, able to identify novel molecular targets and pathways commonly associated with BD and metabolic phenotypes, are urgently needed. Genetic variants suggested to be associated to BD or metabolic phenotypes by previous studies generally showed small effect sizes, as in the case of most complex phenotypes. To this regard, gene-based analysis is a powerful method to identify novel genes associated with a complex trait, as this method is able to globally evaluate the cumulative effect of multiple SNPs located in a gene. This approach gives the opportunity to build new knowledge upon genome-wide association studies (GWAS) that have already been produced with huge investments, thus accelerating the discovery of associations between genetic variation and complex traits. To our knowledge, no study has hitherto used this approach to investigate targets commonly associated with BD and metabolic phenotypes. In this sense, public databases of GWAS conducted on large samples of patients affected by BD or metabolic conditions and healthy controls represent an extraordinary opportunity to expand our knowledge on genes that might be implicated in these conditions.
In the present study, we took advantage of large publicly available GWAS datasets to conduct a gene-based analysis as well as cross-trait meta-analyses aiming at (1) identifying genes commonly associated with BD and BMI or BD and T2D, and (2) investigating if genes shared between these conditions are enriched for pathways previously implicated in BD pathophysiology or in the mechanism of action of medications used to treat BD.
Material and methods
Sample
The present study was conducted on the summary statistics of three large public GWAS datasets. Summary statistics include all SNPs that were analyzed, together with the calculated effect sizes. Evaluation of BD-associated genes was performed using the summary statistics from the largest GWAS conducted by the Psychiatric Genomics Consortium (PGC) Bipolar Disorder Working Group to date37. The dataset included GWAS results from logistic regression analyses on over 13.4 million autosomal SNPs on 20,352 BD cases according to DSM-IV, ICD-9 or ICD-10 criteria and 31,358 controls of European descent. Detailed characteristics of each included cohort have been described in the work from Stahl and coworkers37. Further, to identify genes associated with T2D, we used the DIAGRAM 1000G GWAS meta-analysis Stage 1 dataset, including 26,676 T2D cases and 132,532 Caucasian controls from 18 studies38. Criteria for T2D diagnosis differed among the studies and included self-reported diagnosis of diabetes by a physician, self-reported use of medication to treat diabetes, fasting glucose ≥ 7.0 mmol/l, or non-fasting glucose ≥ 11.1 mmol/l, hemoglobin A1c (HbA1c) > = 6.5%38. This dataset reported association summary statistics for over 12.06 million autosomal SNPs.
Finally, genes associated with BMI were investigated using the GWAS plus Metabochip meta-analysis dataset from the GIANT Consortium39. This dataset included summary association statistics of 322,154 subjects of European ancestry. Participants were recruited from 125 studies (82 with GWAS and 43 with results Metabochip results). In this dataset, over 2.47 million autosomal genotyped or imputed variants were tested for association with transformed BMI residuals in linear regressions assuming an additive genetic model39.
Gene-based analysis
For each of the three datasets, a gene-based analysis was performed with MAGMA40, using the FUMA platform41. MAGMA is a tool for gene analysis that estimates gene-based statistics taking into account the linkage disequilibrium (LD) of the included SNPs. LD was estimated using the European panel of the 1000 Genomes phase 3 data as reference42. Locations of protein-coding genes were defined as the regions from transcription start site to transcription stop site (default option in MAGMA). In case a dataset reported information on imputation quality or frequency of the minor allele (MAF), SNPs with imputation quality score < 0.3 or MAF < 0.01 were excluded. Additionally, in the case of the BD dataset, SNPs that were missing in more than 1% of the subjects were excluded. SNPs not represented in all three datasets as well as ambiguous SNPs (G/C or A/T) were also excluded. Three sets of genes significantly associated with BD, BMI and T2D were obtained. Multiple testing correction was performed according to the Benjamini and Hochberg (BH) procedure43 using the p.adjust function in R44. The hypergeometric test was used to assess over-representation of genes significantly associated with BD and either BMI or T2D after multiple testing correction. Analyses were conducted using R v. 3.6.1 (ref. 44).
Pathway analysis
Enrichment for non-redundant Gene Ontology (GO) terms and Reactome pathways among the genes commonly associated with BD and BMI, BD and T2D or all three phenotypes was assessed using the WebGestalt functional enrichment analysis tool (www.webgestalt.org/). In order to include the largest number of potentially relevant genes in the pathway analysis, we used the list of genes nominally associated with the traits of interests, with a p < 0.05, without applying a correction for multiple testing at this step. The enrichment analysis was performed using the hypergeometric test with default settings (minimum number of genes for a category: 5; multiple-testing adjustment: BH method). In order to obtain more information on the possible interactions between proteins coded by the genes found to be commonly associated with BD and BMI or T2D, a protein–protein interaction (PPI) analysis was conducted using STRING45. For this analysis, the interaction score was set to high confidence (score = 0.7) and all the active interaction sources supported by the tool were included (text mining, experiments, databases, co-expression, neighborhood, gene fusion and co-occurrence).
Meta-analysis and functional effects of SNPs associated with BD and BMI or T2D
In order to pinpoint specific SNPs associated with BD and BMI or T2D, we conducted two meta-analyses between BD and BMI, as well as BD and T2D using Metasoft46. This software provides effect estimates, heterogeneity estimates as well as a posterior probability that an effect exists in each study (m-value statistics > 0.9). The two meta-analyses were conducted with a conservative random-effect model using the same list of SNPs of the gene-based analysis as input. For both meta-analyses, we selected SNPs with a meta-analysis p < 5E−08, an m-value > 0.9 and a p < 0.05 in each original study. The putative functional role of the significant SNPs was evaluated using RegulomeDB47. Among SNPs predicted by RegulomeDB to likely affect binding of transcription factors (score < 3), the presence of significant expression quantitative trait loci (eQTL) in adipose tissue as well as in different brain regions was investigated using GTEx48. Finally, the presence of SNPs located in potentially druggable genes was investigated using the Drug Gene Interaction database (DGIdb)49.
Results
Gene-based analysis
A flow-chart of the analyses is reported in Supplementary Fig. 1. A total of 2,013,566 SNPs that passed quality control and were present in all datasets were used as input for the gene-based analysis. These SNPs allowed conducting a gene-based analysis for 17,455 genes. Gene-based analyses conducted with MAGMA identified 2700, 2144 and 1988 genes nominally associated with BD, BMI and T2D, respectively. Among these, a total of 579, 549 and 173 were significant after multiple testing correction with BH, respectively.
When comparing the three lists, 518 genes were found to be commonly associated with BD and BMI, 52 of which were significant after multiple testing correction (Supplementary Table 1). The number of genes associated with both phenotypes was 2.86-fold higher than expected based on results of the hypergeometric test (p = 9.4E−12, Supplementary Table 2). Regarding BD and T2D, 390 genes were associated with both phenotypes, 12 of which were significant after multiple testing correction (Supplementary Table 3). The number of overlapping genes was 2.09-fold higher than expected based on results of the hypergeometric test (p = 0.01, Supplementary Table 2). Similarly, genes associated with BD were enriched for targets associated with either BMI or T2D (p = 2.6E−12, Supplementary Table 2). Finally, 93 genes, three of which were significant after multiple testing correction, were associated with all investigated phenotypes (Supplementary Table 4). Genes associated with BD were not significantly enriched for targets associated with both BMI and T2D (p = 0.076, Supplementary Table 2).
Pathway analyses
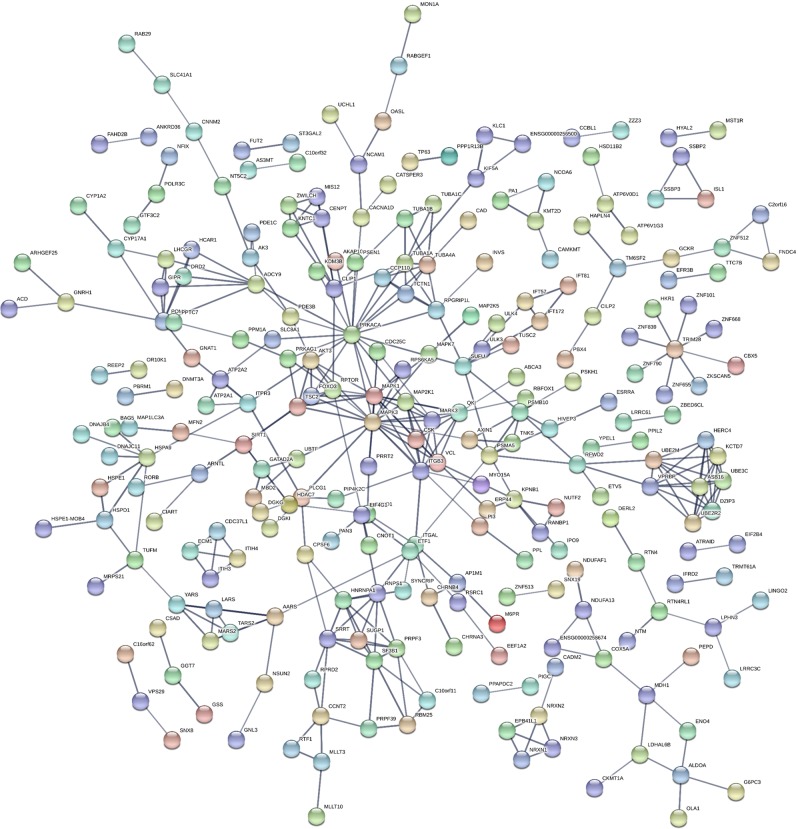
The analyses conducted using WebGestalt showed that the genes commonly associated with BD and BMI were enriched for one cellular-component GO term: Neuronal cell body (p = 0.0001; false discovery rate (FDR) = 0.025) (Table 2). Additionally, genes commonly associated with BD and BMI were enriched for 12 pathways (Supplementary Table 5). The weighted set cover method implemented by WebGestalt, which reduces redundancy of the gene sets, confirmed a significant association for four of these pathways: Hemostasis, Signaling by Hedgehog, L1CAM interactions and Signaling by BRAF and RAF fusions (Table 2), as well as for the GO term. We further explored the potential interactions between the proteins encoded by the 518 genes commonly associated with BD and BMI using STRING. This analysis showed that the network of proteins encoded by these genes presented a number of interactions greater than expected for a random set of proteins of similar size extracted from the genome (expected number of edges: 332; observed number of edges: 392; PPI enrichment p = 0.0007, Fig. 1), supporting the hypothesis that proteins encoded by genes commonly associated with BD and BMI could be biologically connected.
Table 2.
Weighted set of significantly enriched GO terms and pathways for genes commonly associated with bipolar disorder and BMI.
| Gene set size | Overlap | Enrichment ratio | p | FDR | Included genes | |
|---|---|---|---|---|---|---|
| GO terms | ||||||
| Neuronal cell body | 486 | 29 | 2.08 | 1.0E−04 | 0.025 | CAD, CHRNA3, CRTC1, CYP17A1, DDN, DGKI, DRD2, EEF1A2, FXR2, GNAT1, GNRH1, GRIK5, HPCA, ITPR3, KCNN2, KIF5A, KLC1, MAPK1, NRXN1, PARD3, PDE1C, PSEN1, RPTOR, RTN4, RTN4RL1, SLC1A3, SLC4A10, UCHL1, SHTN1 |
| Pathways | ||||||
| Hemostasis | 620 | 38 | 2.25 | 2.1E−06 | 0.004 | AK3, AKAP10, ALDOA, ATP2A1, ATP2A2, ATP2B2, CBX5, CDC37L1, CSK, DAGLA, DGKG, DGK1, ECM1, ITGAL, ITGB3, ITIH3, ITIH4, ITPR3, KIF5A, KLC1, MAPK1, MAPK3, MFN2, NEF2, OLA1, PDE3B, PLCG1, PPIL2, PRKACA, SH2B1, SLC7A6, SLC8A1, TUBA1A, TUBA1B, TUBA1C, TUBA4A, VCL, ZFPM2 |
| Signaling by Hedgehog | 147 | 14 | 3.50 | 4.8E−05 | 0.020 | ADCY9, DERL2, IFT172, IFT57, PRKACA, PSMA5, PSMB10, RPGRIP1L, SUFU, TUBA1A, TUBA1B, TUBA1C, TUBA4A, ULK3 |
| L1CAM interactions | 117 | 12 | 3.76 | 8.1E−05 | 0.020 | ITGB3, KCNQ2, MAP2K1, MAPK1, MAPK3, NCAM1, RPS6KA5, SHTN1, TUBA1A, TUBA1B, TUBA1C, TUBA4A |
| Signaling by BRAF and RAF fusions | 60 | 8 | 4.89 | 0.0002 | 0.030 | CSK, ITGB3, MAP2K1, MAPK1, MAPK3, MARK3, QKI, VCL |
Enriched GO terms and REACTOME pathways were obtained using the weighted set cover method implemented by WebGestalt, which reduces redundancy of the gene sets
BMI body mass index, FDR false discovery rate
Fig. 1. Predicted interactions between proteins encoded by genes commonly associated to bipolar disorder and BMI.
Output of the protein–protein interaction analysis conducted using STRING with genes associated with bipolar disorder and BMI as input. Each node represents all the proteins produced by a single protein-coding gene locus (splice isoforms are collapsed), while edges represent protein–protein associations. The interaction score was set to high confidence (score = 0.7) and all the active interaction sources supported by the tool were included (text mining, experiments, databases, co-expression, neighborhood, gene fusion and co-occurrence). The network of proteins encoded by genes commonly associated with bipolar disorder and BMI presents more interactions compared to the number expected for a random set of proteins of similar size extracted from the genome (number of nodes: 504, expected number of edges: 332, observed number of edges: 392, protein–protein interaction enrichment p = 0.0007).
No significantly enriched category of GO terms or pathways was observed for genes commonly associated with BD and T2D. However, the network of proteins encoded by these genes presented more interactions than what would be expected for a random set of proteins of similar size extracted from the genome (expected number of edges: 163; observed number of edges: 215; PPI enrichment p = 5.8E−05, Supplementary Fig. 2).
Genes associated with BD, BMI and T2D were not found to be significantly enriched for specific GO terms or pathways. Furthermore, the network of proteins encoded by these genes did not present a significantly higher number of interactions than what would be expected for a random set of proteins of similar size extracted from the genome (expected number of edges: 10; observed number of edges: 10; PPI enrichment p = 0.58).
Meta-analysis and functional effects of SNPs associated with BD and BMI or T2D
The meta-analyses with Metasoft were conducted on the 2,013,566 SNPs for which data were available in all three datasets, which were also used as input for the gene-based analysis. The meta-analysis between BD and BMI identified 64 significant SNPs relevant for both traits (m-value > 0.9) (Table 3).
Table 3.
Results of the meta-analysis showing SNPs significantly associated with bipolar disorder and BMI.
| Meta-analysis | Bipolar disorder | BMI | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr | Position (hg19) | Effect allele | Other allele | p | B | SE | OR | SE | p | B | SE | p | Direction | RegDB score |
| rs1516725 | ETV5 | 3 | 185824004 | T | C | 1.0E−24 | −0.046 | 0.004 | 0.94 | 0.02 | 0.002 | −0.05 | 0.005 | 1.9E−22 | −− | >=3 |
| rs3888190 | NPIPL1 | 16 | 28889486 | A | C | 1.1E−24 | 0.031 | 0.003 | 1.03 | 0.01 | 0.016 | 0.03 | 0.003 | 3.1E−23 | ++ | 1f |
| rs4788101 | NPIPL1-SH2B1 | 16 | 28867804 | T | C | 2.1E−24 | 0.031 | 0.003 | 1.03 | 0.01 | 0.015 | 0.03 | 0.003 | 4.8E−23 | ++ | >=3 |
| rs7498665 | NPIPL1-SH2B1 | 16 | 28883241 | A | G | 2.1E−24 | −0.031 | 0.003 | 0.99 | 1.14 | 0.109 | −0.03 | 0.003 | 9.2E−23 | −− | 1f |
| rs8061590 | NPIPL1-SH2B1 | 16 | 28883241 | A | G | 2.1E−24 | −0.031 | 0.003 | 0.97 | 0.01 | 0.023 | −0.03 | 0.003 | 4.3E−23 | −− | >= 3 |
| rs4234589 | ETV5 | 3 | 185818866 | A | G | 2.5E−24 | 0.046 | 0.004 | 1.07 | 0.02 | 0.002 | 0.04 | 0.005 | 3.8E−22 | ++ | >= 3 |
| rs8055982 | NPIPL1-SH2B1 | 16 | 28881202 | A | C | 2.8E−24 | −0.031 | 0.003 | 0.97 | 0.01 | 0.015 | −0.03 | 0.003 | 6.7E−23 | −− | 1f |
| rs4788102 | NPIPL1-SH2B1 | 16 | 28873398 | A | G | 2.8E−24 | 0.031 | 0.003 | 1.03 | 0.01 | 0.015 | 0.03 | 0.003 | 7.4E−23 | ++ | 1f |
| rs4788099 | NPIPL1-TUFM | 16 | 28855727 | A | G | 2.9E−24 | −0.031 | 0.003 | 0.97 | 0.01 | 0.010 | −0.03 | 0.003 | 1.1E−22 | −− | >= 3 |
| rs7359397 | NPIPL1-SH2B1 | 16 | 28885659 | T | C | 3.5E−24 | 0.031 | 0.003 | 1.03 | 0.01 | 0.020 | 0.03 | 0.003 | 7.4E−23 | ++ | 1f |
| rs12325113 | NPIPL1-ATXN2L | 16 | 28848668 | T | C | 5.2E−24 | −0.031 | 0.003 | 0.97 | 0.01 | 0.014 | −0.03 | 0.003 | 1.3E−22 | −− | >= 3 |
| rs12443881 | NPIPL1-ATXN2L | 16 | 28841777 | T | C | 6.2E−24 | 0.030 | 0.003 | 1.03 | 0.01 | 0.012 | 0.03 | 0.003 | 1.7E−22 | ++ | 1 f |
| rs6809651 | ETV5 | 3 | 185814641 | A | G | 1.1E−23 | −0.046 | 0.005 | 0.94 | 0.02 | 0.002 | −0.05 | 0.005 | 1.0E−21 | – | >= 3 |
| rs10513801 | ETV5 | 3 | 185822353 | T | G | 1.3E−23 | 0.046 | 0.005 | 1.07 | 0.02 | 0.001 | 0.04 | 0.005 | 1.1E−21 | + + | >= 3 |
| rs7187776 | NPIPL1-ATXN2L | 16 | 28857645 | A | G | 3.9E−23 | −0.030 | 0.003 | 0.97 | 0.01 | 0.023 | −0.03 | 0.003 | 1.1E−21 | −− | 1 f |
| rs8049439 | NPIPL1-ATXN2L | 16 | 28837515 | T | C | 1.0E−22 | −0.030 | 0.003 | 0.97 | 0.01 | 0.024 | −0.03 | 0.003 | 1.6E−21 | −− | 1b |
| rs7647305 | Intergenic | 3 | 185834290 | T | C | 2.2E−22 | −0.036 | 0.004 | 0.96 | 0.02 | 0.014 | −0.04 | 0.004 | 1.3E−20 | −− | >= 3 |
| rs2008514 | NPIPL1-AK125489 | 16 | 28825605 | A | G | 3.8E−22 | 0.030 | 0.003 | 1.03 | 0.01 | 0.023 | 0.03 | 0.003 | 1.7E−21 | ++ | >= 3 |
| rs8055138 | NPIPL1-ATP2A1 | 16 | 28891465 | T | C | 9.4E−21 | 0.031 | 0.003 | 1.03 | 0.01 | 0.016 | 0.03 | 0.003 | 1.3E−19 | ++ | 2b |
| rs151181 | NPIPL1-CLN3 | 16 | 28490517 | T | C | 3.0E−19 | −0.027 | 0.003 | 0.97 | 0.01 | 0.021 | −0.03 | 0.003 | 8.5E−18 | −− | >= 3 |
| rs4788084 | NPIPL1 | 16 | 28539848 | T | C | 6.5E−18 | 0.026 | 0.003 | 1.03 | 0.01 | 0.045 | 0.03 | 0.003 | 1.5E−17 | ++ | 1a |
| rs12446550 | NPIPL1 | 16 | 28543381 | A | G | 6.5E−18 | 0.026 | 0.003 | 1.03 | 0.01 | 0.045 | 0.03 | 0.003 | 1.5E−17 | ++ | >= 3 |
| rs7635103 | Intergenic | 3 | 185833759 | A | C | 7.8E−18 | 0.030 | 0.004 | 1.04 | 0.02 | 0.006 | 0.03 | 0.004 | 2.8E−16 | ++ | >= 3 |
| rs4788100 | NPIPL1-SH2B1 | 16 | 28864673 | T | C | 1.9E−15 | −0.029 | 0.004 | 0.96 | 0.01 | 0.010 | −0.03 | 0.004 | 5.2E−14 | −− | >= 3 |
| rs12325278 | NPIPL1 | 16 | 28848818 | A | G | 3.0E−15 | −0.029 | 0.004 | 0.97 | 0.01 | 0.010 | −0.03 | 0.004 | 7.8E−14 | −− | >= 3 |
| rs10968576 | LINGO2 | 9 | 28414339 | A | G | 3.4E−15 | −0.025 | 0.003 | 0.97 | 0.01 | 0.020 | −0.02 | 0.003 | 6.6E−14 | −− | >= 3 |
| rs7205323 | NPIPL1-SH2B1 | 16 | 28865892 | T | C | 3.9E−15 | 0.029 | 0.004 | 1.03 | 0.01 | 0.023 | 0.03 | 0.004 | 5.2E−14 | ++ | >= 3 |
| rs7187333 | NPIPL1-SH2B1 | 16 | 28865916 | A | G | 4.5E−15 | 0.029 | 0.004 | 1.03 | 0.01 | 0.028 | 0.03 | 0.004 | 5.2E−14 | ++ | >= 3 |
| rs2183824 | LINGO2 | 9 | 28412078 | T | C | 4.9E−15 | 0.025 | 0.003 | 1.03 | 0.01 | 0.024 | 0.02 | 0.003 | 8.3E−14 | ++ | >= 3 |
| rs8062405 | NPIPL1-ATXN2L | 16 | 28837906 | A | G | 5.0E−15 | −0.029 | 0.004 | 0.97 | 0.01 | 0.012 | −0.03 | 0.004 | 1.2E−13 | −− | >= 3 |
| rs2183825 | NPIPL1-ATXN2L | 16 | 28837906 | T | C | 9.4E−15 | −0.025 | 0.003 | 0.97 | 0.01 | 0.022 | −0.02 | 0.003 | 6.9E−14 | −− | >= 3 |
| rs16912921 | LINGO2 | 9 | 28413461 | A | C | 1.0E−14 | 0.025 | 0.003 | 1.03 | 0.01 | 0.026 | 0.02 | 0.003 | 9.6E−14 | ++ | >= 3 |
| rs12928404 | NPIPL1-ATXN2L | 16 | 28847246 | T | C | 2.9E−14 | −0.028 | 0.004 | 0.97 | 0.01 | 0.038 | −0.03 | 0.004 | 2.6E−13 | −− | 1b |
| rs4234588 | ETV5 | 3 | 185786406 | A | G | 5.4E−14 | 0.043 | 0.006 | 1.06 | 0.02 | 0.002 | 0.04 | 0.005 | 8.5E−18 | ++ | >= 3 |
| rs10968577 | LINGO2 | 9 | 28415512 | T | C | 4.1E−13 | 0.026 | 0.004 | 1.03 | 0.01 | 0.020 | 0.03 | 0.004 | 5.3E−12 | ++ | >= 3 |
| rs1620977 | NEGR1 | 1 | 72729142 | A | G | 8.7E−13 | 0.024 | 0.003 | 1.03 | 0.02 | 0.050 | 0.02 | 0.004 | 5.3E−12 | ++ | >= 3 |
| rs2968487 | Intergenic | 1 | 96887370 | T | C | 3.3E−12 | 0.025 | 0.004 | 1.03 | 0.01 | 0.025 | 0.02 | 0.004 | 2.8E−11 | ++ | >= 3 |
| rs587242 | Intergenic | 1 | 96886267 | A | G | 1.3E−11 | 0.025 | 0.004 | 1.04 | 0.02 | 0.015 | 0.02 | 0.004 | 2.1E−10 | ++ | >= 3 |
| rs2127162 | MAP2K5 | 15 | 68090824 | A | C | 4.7E−11 | −0.023 | 0.004 | 0.97 | 0.01 | 0.015 | −0.02 | 0.004 | 7.2E−10 | −− | >= 3 |
| rs1830868 | Intergenic | 1 | 96895906 | A | G | 5.9E−11 | 0.026 | 0.004 | 1.04 | 0.01 | 0.012 | 0.03 | 0.004 | 1.1E−09 | ++ | >= 3 |
| rs7543671 | Intergenic | 1 | 96892262 | T | C | 6.9E−11 | −0.026 | 0.004 | 0.96 | 0.01 | 0.016 | −0.03 | 0.004 | 1.1E−09 | −− | >= 3 |
| rs7543590 | Intergenic | 1 | 96892216 | T | C | 8.2E−11 | −0.026 | 0.004 | 0.96 | 0.01 | 0.016 | −0.02 | 0.004 | 1.3E−09 | −− | >= 3 |
| rs661573 | Intergenic | 1 | 96891371 | A | G | 1.1E−10 | 0.026 | 0.004 | 1.03 | 0.01 | 0.024 | 0.02 | 0.004 | 1.3E−09 | ++ | >= 3 |
| rs7551466 | Intergenic | 1 | 96892427 | T | G | 1.1E−10 | 0.025 | 0.004 | 1.04 | 0.01 | 0.016 | 0.02 | 0.004 | 1.7E−09 | ++ | >= 3 |
| rs1853738 | Intergenic | 1 | 96895992 | A | G | 1.2E−10 | −0.025 | 0.004 | 0.97 | 0.01 | 0.017 | −0.02 | 0.004 | 1.7E−09 | −− | >= 3 |
| rs12758621 | Intergenic | 1 | 96898421 | T | C | 1.2E−10 | 0.025 | 0.004 | 1.04 | 0.01 | 0.017 | 0.02 | 0.004 | 1.7E−09 | ++ | >= 3 |
| rs550739 | Intergenic | 1 | 96887109 | A | G | 1.2E−10 | 0.025 | 0.004 | 1.03 | 0.01 | 0.023 | 0.02 | 0.004 | 1.5E−09 | ++ | >= 3 |
| rs12030271 | Intergenic | 1 | 96892099 | A | G | 1.3E−10 | −0.025 | 0.004 | 0.96 | 0.01 | 0.016 | −0.02 | 0.004 | 2.0E−09 | −− | >= 3 |
| rs10489740 | Intergenic | 1 | 96919182 | A | G | 1.3E−10 | 0.025 | 0.004 | 1.04 | 0.01 | 0.012 | 0.02 | 0.004 | 2.3E−09 | ++ | >= 3 |
| rs722155 | Intergenic | 1 | 96906374 | A | G | 1.4E−10 | −0.025 | 0.004 | 0.97 | 0.01 | 0.017 | −0.02 | 0.004 | 2.0E−09 | −− | >= 3 |
| rs505066 | Intergenic | 1 | 96882671 | A | C | 3.8E−10 | 0.025 | 0.004 | 1.04 | 0.01 | 0.021 | 0.02 | 0.004 | 4.7E−09 | ++ | >= 3 |
| rs12739556 | Intergenic | 1 | 96977901 | T | C | 4.5E−10 | 0.025 | 0.004 | 1.04 | 0.01 | 0.018 | 0.02 | 0.004 | 6.3E−09 | ++ | >= 3 |
| rs12566597 | Intergenic | 1 | 96978626 | A | G | 6.9E−10 | 0.025 | 0.004 | 1.04 | 0.01 | 0.017 | 0.02 | 0.004 | 9.6E−09 | ++ | >= 3 |
| rs1412234 | LINGO2 | 9 | 28410683 | T | C | 8.5E−10 | −0.024 | 0.004 | 0.97 | 0.01 | 0.023 | −0.02 | 0.004 | 9.9E−09 | −− | >= 3 |
| rs4341405 | Intergenic | 1 | 96902589 | A | G | 9.0E−10 | −0.025 | 0.004 | 0.97 | 0.01 | 0.017 | −0.02 | 0.004 | 1.3E−08 | −− | >= 3 |
| rs4141973 | Intergenic | 1 | 50946521 | T | C | 1.6E−09 | 0.029 | 0.005 | 1.04 | 0.02 | 0.042 | 0.03 | 0.005 | 1.5E−08 | ++ | >= 3 |
| rs7541994 | Intergenic | 1 | 96984344 | A | G | 2.1E−09 | 0.024 | 0.004 | 1.03 | 0.02 | 0.025 | 0.02 | 0.004 | 2.2E−08 | ++ | >= 3 |
| rs4000303 | Intergenic | 1 | 96913765 | A | G | 4.8E−09 | −0.024 | 0.004 | 0.97 | 0.01 | 0.048 | −0.02 | 0.004 | 3.6E−08 | −− | >= 3 |
| rs1477199 | RPGRIP1L | 16 | 53712135 | A | G | 5.7E−09 | −0.025 | 0.004 | 0.96 | 0.02 | 0.035 | −0.02 | 0.004 | 4.6E−08 | −− | >= 3 |
| rs17203016 | Intergenic | 2 | 208255518 | A | G | 1.0E−08 | −0.022 | 0.004 | 0.96 | 0.02 | 0.033 | −0.02 | 0.004 | 8.1E−08 | −− | >= 3 |
| rs6671342 | Intergenic | 1 | 97000957 | T | G | 1.6E−08 | −0.023 | 0.004 | 0.97 | 0.02 | 0.031 | −0.02 | 0.004 | 1.4E−07 | −− | >= 3 |
| rs4729098 | DTX2P1-UPK3BP1-PMS2P11 | 7 | 76637070 | A | G | 2.9E−08 | −0.037 | 0.007 | 0.96 | 0.02 | 0.043 | −0.04 | 0.007 | 2.3E−07 | −− | >= 3 |
| rs12751763 | Intergenic | 1 | 96997655 | T | C | 3.6E−08 | 0.023 | 0.004 | 1.03 | 0.02 | 0.040 | 0.02 | 0.004 | 2.8E−07 | ++ | >= 3 |
| rs2814944 | Intergenic | 6 | 34552797 | A | G | 4.4E−08 | 0.023 | 0.004 | 1.04 | 0.02 | 0.050 | 0.02 | 0.004 | 2.2E−07 | ++ | >= 3 |
SNPs showing a RegulomeDB score < 3 are more likely to affect binding of transcription factors
BMI body mass index, Chr chromosome, OR odds ratio, RegDB RegulomeDB, SE standard error, SNP single nucleotide polymorphism
The majority of SNPs shared between BD and BMI were located in intergenic regions (n = 26) or in a locus in chromosome 16 spanning the NPIPL1, SH2B1, TUFM, ATP2A1, AK125489, CLN3 and ATXN2L genes (n = 24 SNPs; best SNP: rs3888190, p = 1.1E−24). Five SNPs were located in the ETV5 gene (best SNP: rs1516725, p = 1.0E−24), five in the LINGO2 gene (best SNP: rs2183824, p = 4.9E−15), while the remaining four SNPs were located in the NEGR1 (rs1620977, p = 8.7E−13), MAP2K5 (rs2127162, p = 4.7E−11), RPGRIP1L (rs1477199, p = 5.7E−09) and DTX2P1 genes (rs4729098, p = 2.9E−08). Interestingly, three of these genes (RPGRIP1L, SH2B1 and ATP2A1) were included in significantly enriched pathways identified by WebGestalt. Specifically, RPGRIP1L was included in the Signaling by Hedgehog pathway, while SH2B1 and ATP2A1 in the Hemostasis pathway (Table 2).
A total of 11 SNPs located in three genes were predicted to affect the binding of transcription factors by RegulomeDB (score < 3, Table 3). All SNPs for which RegulomeDB predicted a functional effect were also found to be significant eQTLs in adipose tissue and in different brain regions in GTEx (Supplementary Table 6). Finally, 13 SNPs were located in genes that are part of the druggable genome (LINGO2, NEGR1, MAP2K5, DTX2P1 and ATP2A1) according to DGIdb.
The meta-analysis between BD and T2D identified six SNPs relevant for both traits (Table 4), three of which were located in the ALAS1 gene (best SNP: rs352165, p = 3.4E−08), two in intergenic regions and one in the KCNG1 gene (rs6091248, p = 4.3E−08). The three SNPs rs61428, rs352162 (intergenic regions) and rs164640 (ALAS1) were predicted to affect the binding of transcription factors by RegulomeDB (Table 4) and were also found to be significant eQTLs in cerebellum (Supplementary Table 6). KCNG1 was found to be part of the druggable genome by DGIdb.
Table 4.
Results of the meta-analysis showing SNPs significantly associated with bipolar disorder and type 2 diabetes.
| Meta-analysis | Bipolar disorder | T2D | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr | Position (hg19) | Effect allele | Other allele | Beta | SE | p | OR | SE | p | B | SE | p | Direction | RegDB score |
| rs614288 | Intergenic | 3 | 52220203 | T | C | 0.05 | 0.01 | 2.2E−08 | 1.05 | 0.01 | 8.8E−05 | 0.05 | 0.01 | 8.9E−05 | ++ | 1f |
| rs352165 | ALAS1 | 3 | 52242902 | A | G | 0.05 | 0.01 | 2.6E−08 | 1.06 | 0.01 | 2.2E−05 | 0.04 | 0.01 | 3.2E−04 | ++ | >= 3 |
| rs164640 | ALAS1 | 3 | 52247314 | T | C | −0.05 | 0.01 | 3.3E−08 | 0.94 | 0.01 | 1.9E−05 | −0.04 | 0.01 | 3.5E−04 | −− | 1f |
| rs352162 | Intergenic | 3 | 52252969 | T | C | 0.05 | 0.01 | 3.3E−08 | 1.06 | 0.01 | 4.4E−06 | 0.04 | 0.01 | 3.4E−04 | ++ | 1d |
| rs352163 | ALAS1 | 3 | 52247110 | A | G | −0.05 | 0.01 | 3.5E−08 | 0.94 | 0.01 | 1.9E−05 | −0.04 | 0.01 | 4.0E−04 | −− | >= 3 |
| θrs6091248 | KCNG1 | 20 | 49630618 | A | G | 0.05 | 0.01 | 4.3E−08 | 1.06 | 0.01 | 1.7E−05 | 0.05 | 0.01 | 3.7E−04 | ++ | >= 3 |
SNPs showing a RegulomeDB score <3 are more likely to affect binding of transcription factors
Chr chromosome, OR odds ratio, RegDB RegulomeDB, SE standard error, SNP single nucleotide polymorphism
Finally, we found no SNP in common between the two meta-analyses (i.e. SNPs significant in both comparisons BD and BMI as well as BD and T2D)
Discussion
Our study supports the existence of shared genetic factors between BD and BMI. To the best of our knowledge, this is the first study to investigate genes commonly associated with BD, BMI and T2D using a gene-based approach and without a specific focus on single candidate genes and pathways previously suggested to be involved in BD. We showed that BD and BMI share a higher number of susceptibility genes compared to BD and T2D. Some of the genes we found to be associated with BD and either BMI or T2D (CACNA1D, ITIH4, NCAN, CRY2 and POMC) were previously reported by a recent systematic review to be associated with cardiometabolic phenotypes and mood disorders (MDD and BD)26. Besides validating these genes, we reported many novel targets and found that genes commonly associated with BD and BMI are enriched for the cellular-component Neuronal cell body GO term.
The pathway analysis from WebGestalt further supports the hypothesis that genes commonly associated with BD and BMI show a significant functional enrichment. Two of the pathways (Signaling by Hedgehog and Hedgehog “off” state) were related to Hedgehog signaling and the former was also included among the four pathways of the weighted set (i.e. the set including the most representative pathways that can cover all the genes from the enriched sets). The Hedgehog signaling pathway plays a crucial role in neural and limb development, cell growth, differentiation and survival50. The mechanisms underlying Hedgehog signaling are complex and not completely understood. This pathway takes the name from the Hedgehog (Hh) ligand, which was first identified in D. melanogaster51. This ligand has three counterparts in mammalians: sonic hedhgehog (SHH), Indian hedgehog (IHH) and desert hedgehog (DHH)52. Through a finely regulated switching between “off” and “on” states, the Hedgehog pathway modulates a signaling cascade that ultimately targets the Gli transcription factors. In the absence of the ligand (Hedgehog off state), the cytosolic Gli proteins are cleaved to a truncated form, translocate to the nucleus and repress gene expression. Conversely, in the presence of the Hedgehog ligand (Hedgehog on state), the Gli transcription factors are stabilized in their full-length form, which is able to stimulate gene expression50.
Recent evidence suggests that the Hedgehog transduction pathway shows a bi-directional connection with metabolism53. In fact, on one hand Hedgehog proteins are modified by fatty acids and cholesterol (these modifications being essential for their maturation and activity)53. On the other hand, this signaling system might play a role in adipocyte differentiation and regulation of energy homeostasis53. Although the role of the Hedgehog pathway in the pathophysiology of BD has not been systematically investigated, disruption of sonic hedgehog signaling associated with Ellis-van Creveld syndrome (a form of chondrodysplastic dwarfism) has been suggested to exert a protective role against BD54. Interestingly, in vitro treatment with lithium was found to modulate Hedgehog signaling in pancreatic adenocarcinoma cells via the inhibition of the GSK-3 serine/threonine kinase55. In this study, lithium exerted a biphasic effect, i.e. an initial increase of GLI1 cellular levels due to the inhibition of the ubiquitin-proteasome-mediated GLI degradation, followed by a downregulation of expression and activity of GLI1 after lithium treatment for 18 h55. These results suggest that lithium might interfere with Hedgehog signaling via modulation of the ubiquitin-proteasome degradation.
The meta-analysis approach also supported the hypothesis that genes involved in Hedgehog signaling might play an important role. In fact, among SNPs significantly associated with BD and BMI, rs1477199 was located in the RPGRIP1L gene. This gene encodes a protein localized at the transition zone of the primary cilium56 and is required for hypothalamic arcuate neuron development57. RPGRIP1L represents a particularly interesting target, as its expression and activity are regulated by intronic variants located in the FTO gene (which is strongly associated with obesity and T2D) through long-range regulation58. RPGRIP1L was recently associated with BD in a sample including 276 patients and 170 controls of Mexican origin59. Taken together, our findings suggest that complementary analytical approaches may provide converging evidence and should be used together to be able to identify genes in which multiple SNPs with small effect sizes might play an additive effect, as well as genes in which single SNPs might play a more relevant role. The two different approaches we used support the potential role of Hedgehog signaling in both BD and BMI.
Other relevant pathways shared between BD and BMI were revealed, such as “L1CAM interactions”, which has been implicated in neuritogenesis and neuroprotection60. Indeed, the neural cell adhesion molecule L1 plays a crucial role in nervous system development, being involved in neurite outgrowth, adhesion, axon guidance, myelination and synaptic plasticity61. Preclinical studies support the hypothesis that dysregulation of neuronal synaptic plasticity might mediate a potential role of L1 and other adhesion molecules in the pathogenesis of mood disorders62. Interestingly, state-dependent alterations of L1 peripheral messenger RNA (mRNA) levels in patients with BD have been suggested63. Specifically, L1 levels were increased in a sample of 13 BD patients in a current depressive state, but not in patients in a remissive state (n = 29)63.
Results from our meta-analysis confirmed the important role played by other genes previously suggested to be associated with BD and BMI. Specifically, we found five SNPs to be located in the ETV5 gene. Inhibition of the ETV5 homolog in D. melanogaster (Ets96B) induces BD- and obesity-related phenotypes20.
Our hypothesis that genetic variants might contribute to explain the increased comorbidity between BD and obesity is in contrast with findings from a recent study showing that the inclusion of genetic data in a model comprising clinical characteristics did not improve prediction of BMI or BMI gain after 1 year in a sample of 284 patients with psychosis29. However, in this study only 32 patients had a diagnosis of BD, suggesting the need to conduct further studies specifically including BD patients.
While our results support the existence of shared functional pathways between BD and BMI, we did not observe a functional enrichment for genes commonly associated with BD and T2D. However, proteins encoded by these genes showed more interactions compared to what would be expected for a random set or proteins of similar size extracted from the genome. The limited number of genetic targets we observed to be shared between BD and T2D is in accordance with previous studies showing a lack of association between T2D polygenic risk scores (PRS) and diagnosis of psychiatric disorders32,35. Although the aggregated effect of multiple T2D-associated variants might not play a relevant role in BD pathophysiology, the contribution of specific genes might still be important. In our meta-analysis, the largest number of SNPs commonly associated with BD and T2D was located in the ALAS1 gene, which might represent a promising target to further investigate. This gene catalyzes the rate-limiting step in heme (iron-protoporphyrin) biosynthesis, a process which has been shown to be bidirectionally connected with the regulation of the circadian clock64,65. Based on a large body of evidence supporting the existence of circadian disturbances in patients with BD66 as well as the important role of circadian genes in both BD and response to lithium67–70, it might be important for future studies to evaluate the role of genes interacting with circadian systems as in the case of ALAS1.
Our results have to be interpreted in light of some limitations. Firstly, we chose to select genes nominally associated with both BD and BMI or T2D in GWAS datasets without applying a correction for multiple testing at this step, in order to include the largest number of potentially relevant genes in the pathway analysis. Although this might have increased the risk to include less-relevant genes in the pathway analysis, this aspect was partly addressed by applying a FDR correction on pathways as well as searching for converging evidence from the meta-analysis approach to support the potential relevance of the identified pathways. Secondly, publicly available GWAS datasets do not generally include information on concomitant disturbances. Therefore, it was not possible to assess the prevalence of BD as well as other psychiatric comorbidities in the BMI and T2D datasets. As our analyses could not be adjusted for potential comorbidities, it is not possible to exclude that comorbidity with psychiatric disorders could play a role in explaining our findings. Thirdly, although GWAS summary statistics can be analyzed with a wide range of methods, they do not provide individual-level genotyping data, thus not allowing to conduct other analyses that can provide important insight such as PRS analysis. Finally, although we conducted our analyses on the largest and most recent BD dataset released by the PGC to date, the smaller number of participants included in this dataset compared to the BMI and T2D GWAS datasets might have limited our ability to identify shared genetics determinants between BD and the other traits. The main strengths of our study are the large sample size of the included datasets and the choice not to restrict our investigation to specific candidate genes or pathways to overcome limitations of previous studies focused on candidate targets.
It is important to note that our analyses were conducted on subjects of European ancestry. Further studies will be needed to assess the potential generalizability of these results to other populations. While our study was focused on BD, other psychiatric disorders such as MDD and schizophrenia show increased frequency of obesity71. A recent study investigating the role of the candidate gene CADM2, which encodes a synaptic cell adhesion molecule, reported that this gene might be associated with a wide range of psychological (neuroticism, mood instability and risk-taking) and metabolic traits, and that regulation of this gene in adipose tissue might mediate common biological mechanisms across phenotypes72. In accordance with these results, our gene-based analysis identified this gene as commonly associated with BD and BMI and this association survived multiple testing correction. Further studies will be needed to understand which of the targets we identified might be specific for BD and which genes or pathways might play a role in different psychiatric disorders.
To conclude, our results suggest the existence of shared genetic determinants between BD and BMI and support the relevance of genes implicated in Hedgehog signaling. Studies using other instruments such as PRS to further evaluate shared etiology between these traits, as well as to assess the aggregated effect of multiple variants with small effect sizes, are warranted. Future studies on independent samples for which information on comorbidities and potential confounding factors are available are needed to confirm our results and explore the potential role of these genes in the mechanism of action of mood stabilizers in patients with BD. Finally, based on the fact that some of the targets and pathways we reported to be associated with both BD and BMI have been implicated in the mechanism of action of lithium, it would be important to explore if at least part of the genetic determinants common to these traits might play a role in response to this mood stabilizer.
Supplementary information
Acknowledgements
C.P. is supported by a fellowship funded by Fondazione Umberto Veronesi. H.B.S. was supported by the Swedish Research Council and the Swedish Brain Research Foundation. The authors wish to thank Dr. Lyle Wiemerslage for advice and comments on the first version of manuscript.
Data availability
Datasets used in this article are publicly available and can be downloaded from the Psychiatric Genomics Consortium, the DIAGRAM Consortium and the GIANT consortium websites.
Code availability
Code is available upon request.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Claudia Pisanu, Phone: +390706754334, Email: claudia.pisanu@unica.it.
Helgi B. Schiöth, Email: Helgi.Schioth@neuro.uu.se
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-019-0652-x).
References
- 1.Merikangas KR, et al. Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Arch. Gen. Psychiatry. 2007;64:543–552. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes JF, Miles J, Walters K, King M, Osborn DP. A systematic review and meta-analysis of premature mortality in bipolar affective disorder. Acta Psychiatr. Scand. 2015;131:417–425. doi: 10.1111/acps.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vancampfort D, et al. Risk of metabolic syndrome and its components in people with schizophrenia and related psychotic disorders, bipolar disorder and major depressive disorder: a systematic review and meta-analysis. World Psychiatry. 2015;14:339–347. doi: 10.1002/wps.20252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein BI, et al. The burden of obesity among adults with bipolar disorder in the United States. Bipolar Disord. 2011;13:387–395. doi: 10.1111/j.1399-5618.2011.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calkin CV, Gardner DM, Ransom T, Alda M. The relationship between bipolar disorder and type 2 diabetes: more than just co-morbid disorders. Ann. Med. 2013;45:171–181. doi: 10.3109/07853890.2012.687835. [DOI] [PubMed] [Google Scholar]
- 6.Newcomer JW. Medical risk in patients with bipolar disorder and schizophrenia. J. Clin. Psychiatry. 2006;67:36–42. doi: 10.4088/JCP.1106e16. [DOI] [PubMed] [Google Scholar]
- 7.Osby U, Brandt L, Correia N, Ekbom A, Sparen P. Excess mortality in bipolar and unipolar disorder in Sweden. Arch. Gen. Psychiatry. 2001;58:844–850. doi: 10.1001/archpsyc.58.9.844. [DOI] [PubMed] [Google Scholar]
- 8.Fagiolini A, Kupfer DJ, Houck PR, Novick DM, Frank E. Obesity as a correlate of outcome in patients with bipolar I disorder. Am. J. Psychiatry. 2003;160:112–117. doi: 10.1176/appi.ajp.160.1.112. [DOI] [PubMed] [Google Scholar]
- 9.Calkin C, et al. Can body mass index help predict outcome in patients with bipolar disorder? Bipolar Disord. 2009;11:650–656. doi: 10.1111/j.1399-5618.2009.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein BI, Blanco C, He JP, Merikangas K. Correlates of overweight and obesity among adolescents with bipolar disorder in the National Comorbidity Survey-Adolescent Supplement (NCS-A) J. Am. Acad. Child Adolesc. Psychiatry. 2016;55:1020–1026. doi: 10.1016/j.jaac.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein BI, Liu SM, Schaffer A, Sala R, Blanco C. Obesity and the three-year longitudinal course of bipolar disorder. Bipolar Disord. 2013;15:284–293. doi: 10.1111/bdi.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIntyre RS, et al. Adverse effects of obesity on cognitive functions in individuals at ultra high risk for bipolar disorder: results from the global mood and brain science initiative. Bipolar Disord. 2017;19:128–134. doi: 10.1111/bdi.12491. [DOI] [PubMed] [Google Scholar]
- 13.Islam AH, Metcalfe AWS, MacIntosh BJ, Korczak DJ, Goldstein BI. Greater body mass index is associated with reduced frontal cortical volumes among adolescents with bipolar disorder. J. Psychiatry Neurosci. 2017;43:170041. doi: 10.1503/jpn.170041. [DOI] [PubMed] [Google Scholar]
- 14.Mazza E, et al. Body mass index associates with white matter microstructure in bipolar depression. Bipolar Disord. 2017;19:116–127. doi: 10.1111/bdi.12484. [DOI] [PubMed] [Google Scholar]
- 15.Alda M. Lithium in the treatment of bipolar disorder: pharmacology and pharmacogenetics. Mol. Psychiatry. 2015;20:661–670. doi: 10.1038/mp.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansur RB, et al. Expression of dopamine signaling genes in the post-mortem brain of individuals with mental illnesses is moderated by body mass index and mediated by insulin signaling genes. J. Psychiatr. Res. 2018;107:128–135. doi: 10.1016/j.jpsychires.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brietzke E, et al. The impact of body mass index in gene expression of reelin pathway mediators in individuals with schizophrenia and mood disorders: a post-mortem study. J. Psychiatr. Res. 2018;102:186–191. doi: 10.1016/j.jpsychires.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Maina G, Salvi V, Vitalucci A, D’Ambrosio V, Bogetto F. Prevalence and correlates of overweight in drug-naive patients with bipolar disorder. J. Affect. Disord. 2008;110:149–155. doi: 10.1016/j.jad.2007.12.233. [DOI] [PubMed] [Google Scholar]
- 19.Petry NM, Barry D, Pietrzak RH, Wagner JA. Overweight and obesity are associated with psychiatric disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Psychosom. Med. 2008;70:288–297. doi: 10.1097/PSY.0b013e3181651651. [DOI] [PubMed] [Google Scholar]
- 20.Williams MJ, et al. The drosophila ETV5 homologue Ets96B: molecular link between obesity and bipolar disorder. PLoS Genet. 2016;12:e1006104. doi: 10.1371/journal.pgen.1006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winham SJ, et al. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol. Psychiatry. 2014;19:1010–1016. doi: 10.1038/mp.2013.159. [DOI] [PubMed] [Google Scholar]
- 22.McGuffin P, et al. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 23.Turula M, Kaprio J, Rissanen A, Koskenvuo M. Body weight in the Finnish Twin Cohort. Diabetes Res. Clin. Pract. 1990;10:S33–S36. doi: 10.1016/0168-8227(90)90137-I. [DOI] [PubMed] [Google Scholar]
- 24.Willyard C. Heritability: the family roots of obesity. Nature. 2014;508:S58–S60. doi: 10.1038/508S58a. [DOI] [PubMed] [Google Scholar]
- 25.Almgren P, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54:2811–2819. doi: 10.1007/s00125-011-2267-5. [DOI] [PubMed] [Google Scholar]
- 26.Amare AT, Schubert KO, Klingler-Hoffmann M, Cohen-Woods S, Baune BT. The genetic overlap between mood disorders and cardiometabolic diseases: a systematic review of genome wide and candidate gene studies. Transl. Psychiatry. 2017;7:e1007. doi: 10.1038/tp.2016.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansur RB, et al. The effect of body mass index on glucagon-like peptide receptor gene expression in the post mortem brain from individuals with mood and psychotic disorders. Eur. Neuropsychopharmacol. 2019;29:137–146. doi: 10.1016/j.euroneuro.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen PS, et al. A longitudinal study of the association between the GNB3 C825T polymorphism and metabolic disturbance in bipolar II patients treated with valproate. Pharmacogenomics J. 2017;17:155–161. doi: 10.1038/tpj.2015.96. [DOI] [PubMed] [Google Scholar]
- 29.Harrison RNS, et al. Development of multivariable models to predict change in Body Mass Index within a clinical trial population of psychotic individuals. Sci. Rep. 2017;7:14738. doi: 10.1038/s41598-017-15137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuellar-Barboza AB, et al. Accumulating evidence for a role of TCF7L2 variants in bipolar disorder with elevated body mass index. Bipolar Disord. 2016;18:124–135. doi: 10.1111/bdi.12368. [DOI] [PubMed] [Google Scholar]
- 31.Morales-Marín ME, et al. Association between obesity and the brain-derived neurotrophic factor gene polymorphism Val66Met in individuals with bipolar disorder in Mexican population. Neuropsychiatr. Dis. Treat. 2016;12:1843–1848. doi: 10.2147/NDT.S104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padmanabhan JL, et al. Polygenic risk for type 2 diabetes mellitus among individuals with psychosis and their relatives. J. Psychiatr. Res. 2016;77:52–58. doi: 10.1016/j.jpsychires.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonaccorso S, et al. The brain-derived neurotrophic factor (BDNF) Val66Met polymorphism is associated with increased body mass index and insulin resistance measures in bipolar disorder and schizophrenia. Bipolar Disord. 2015;17:528–535. doi: 10.1111/bdi.12294. [DOI] [PubMed] [Google Scholar]
- 34.Nussbaum LA, et al. Molecular study of weight gain related to atypical antipsychotics: clinical implications of the CYP2D6 genotype. Rom. J. Morphol. Embryol. 2014;55:877–884. [PubMed] [Google Scholar]
- 35.Kajio Y, et al. Genetic association study between the detected risk variants based upon type II diabetes GWAS and psychotic disorders in the Japanese population. J. Hum. Genet. 2014;59:54–56. doi: 10.1038/jhg.2013.116. [DOI] [PubMed] [Google Scholar]
- 36.Chang HH, et al. C825T polymorphism of the GNB3 gene on valproate-related metabolic abnormalities in bipolar disorder patients. J. Clin. Psychopharmacol. 2010;30:512–517. doi: 10.1097/JCP.0b013e3181f03f50. [DOI] [PubMed] [Google Scholar]
- 37.Stahl EA, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott RA, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The 1000 Genome Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc.Ser. B.57, 289–300 (1995).
- 44.R. Core Team. R: A Language And Environment For Statistical Computing (Foundation for Statistical Computing, Vienna, Austria, 2018).
- 45.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffith M, et al. DGIdb: mining the druggable genome. Nat. Methods. 2013;10:1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niciu MJ, Ionescu DF, Mathews DC, Richards EM, Zarate CA., Jr. Second messenger/signal transduction pathways in major mood disorders: moving from membrane to mechanism of action, part II: bipolar disorder. CNS Spectr. 2013;18:242–251. doi: 10.1017/S1092852913000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson LJ, et al. New perspectives, opportunities, and challenges in exploring the human protein kinome. Cancer Res. 2018;78:15–29. doi: 10.1158/0008-5472.CAN-17-2291. [DOI] [PubMed] [Google Scholar]
- 52.Karege F, et al. Association of AKT1 gene variants and protein expression in both schizophrenia and bipolar disorder. Genes Brain Behav. 2010;9:503–511. doi: 10.1111/j.1601-183X.2010.00578.x. [DOI] [PubMed] [Google Scholar]
- 53.Teperino R, Aberger F, Esterbauer H, Riobo N, Pospisilik JA. Canonical and non-canonical Hedgehog signalling and the control of metabolism. Semin. Cell Dev. Biol. 2014;33:81–92. doi: 10.1016/j.semcdb.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ginns EI, et al. Disruption of sonic hedgehog signaling in Ellis-van Creveld dwarfism confers protection against bipolar affective disorder. Mol. Psychiatry. 2015;20:1212–1218. doi: 10.1038/mp.2014.118. [DOI] [PubMed] [Google Scholar]
- 55.Peng Z, et al. Lithium inhibits tumorigenic potential of PDA cells through targeting hedgehog-GLI signaling pathway. PLoS ONE. 2013;8:e61457. doi: 10.1371/journal.pone.0061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, et al. Ciliary gene RPGRIP1L is required for hypothalamic arcuate neuron development. JCI Insight. 2019;4:123337. doi: 10.1172/jci.insight.123337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rask-Andersen M, Almén MS, Schiöth HB. Scrutinizing the FTO locus: compelling evidence for a complex, long-range regulatory context. Hum. Genet. 2015;134:1183–1193. doi: 10.1007/s00439-015-1599-5. [DOI] [PubMed] [Google Scholar]
- 59.Saucedo-Uribe E, et al. Differential effects on neurodevelopment of FTO variants in obesity and bipolar disorder suggested by in silico prediction of functional impact: an analysis in Mexican population. Brain Behav. 2019;9:e01249. doi: 10.1002/brb3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loers G, Chen S, Grumet M, Schachner M. Signal transduction pathways implicated in neural recognition molecule L1 triggered neuroprotection and neuritogenesis. J. Neurochem. 2005;92:1463–1476. doi: 10.1111/j.1471-4159.2004.02983.x. [DOI] [PubMed] [Google Scholar]
- 61.Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- 62.Fransen E, et al. L1 knockout mice show dilated ventricles, vermis hypoplasia and impaired exploration patterns. Hum. Mol. Genet. 1998;7:999–1009. doi: 10.1093/hmg/7.6.999. [DOI] [PubMed] [Google Scholar]
- 63.Wakabayashi Y, et al. State-dependent changes in the expression levels of NCAM-140 and L1 in the peripheral blood cells of bipolar disorders, but not in the major depressive disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2008;32:1199–1205. doi: 10.1016/j.pnpbp.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 64.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 65.Simcox JA, et al. Dietary iron controls circadian hepatic glucose metabolism through heme synthesis. Diabetes. 2015;64:1108–1119. doi: 10.2337/db14-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steardo L, Jr., et al. Sleep disturbance in bipolar disorder: neuroglia and circadian rhythms. Front. Psychiatry. 2019;10:501. doi: 10.3389/fpsyt.2019.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCarthy MJ, et al. Chronotype and cellular circadian rhythms predict the clinical response to lithium maintenance treatment in patients with bipolar disorder. Neuropsychopharmacology. 2019;44:620–628. doi: 10.1038/s41386-018-0273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pisanu C, Heilbronner U, Squassina A. The role of pharmacogenomics in bipolar disorder: moving towards precision medicine. Mol. Diagn. Ther. 2018;22:409–420. doi: 10.1007/s40291-018-0335-y. [DOI] [PubMed] [Google Scholar]
- 69.Geoffroy PA, et al. Lithium response in bipolar disorders and core clock genes expression. World J. Biol. Psychiatry. 2018;19:619–632. doi: 10.1080/15622975.2017.1282174. [DOI] [PubMed] [Google Scholar]
- 70.Papadima EM, et al. Evidence towards RNA binding motif (RNP1, RRM) protein 3 (RBM3) as a potential biomarker of lithium response in bipolar disorder patients. J. Mol. Neurosci. 2017;62:304–308. doi: 10.1007/s12031-017-0938-5. [DOI] [PubMed] [Google Scholar]
- 71.Chao AM, Wadden TA, Berkowitz RI. Obesity in adolescents with psychiatric disorders. Curr. Psychiatry Rep. 2019;21:3. doi: 10.1007/s11920-019-0990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morris J, et al. Genetic variation in CADM2 as a link between psychological traits and obesity. Sci. Rep. 2019;9:7339. doi: 10.1038/s41598-019-43861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets used in this article are publicly available and can be downloaded from the Psychiatric Genomics Consortium, the DIAGRAM Consortium and the GIANT consortium websites.
Code is available upon request.