Visual Abstract
Key Words: endothelial cells, gene expression, diabetes mellitus, endothelial cell dysfunction
Abbreviations and Acronyms: BSA, bovine serum albumin; ddCt, delta-delta cycle threshold; EC, endothelial cell; EDTA, ethylenediamine tetra-acetic acid; FACS, fluorescence activated cell sorting; FDR, false discovery rate; GSEA, gene set enrichment analysis; HUVEC, human umbilical vein endothelial cell; IV, intravenous; PBS, phosphate buffered saline; qPCR, quantitative polymerase chain reaction; Seq, sequencing; T2DM, type 2 diabetes mellitus; TGFβ, transforming growth factor beta; VEGF, vascular endothelial growth factor; VUMC, Vanderbilt University Medical Center; WBC, white blood cell
Highlights
-
•
Endothelial cells can be harvested directly from humans, rapidly sorted and subjected to RNA-sequencing to study global gene expression.
-
•
In endothelial cells isolated from patients with type 2 diabetes mellitus, pathways involved in TGF-β and Cyclin-D1 signaling were positively enriched while androgen signaling and oxidative phosphorylation were negatively enriched compared to healthy individuals.
-
•
Patient-derived endothelial cells can be used to discover and validate disease-associated pathways.
Summary
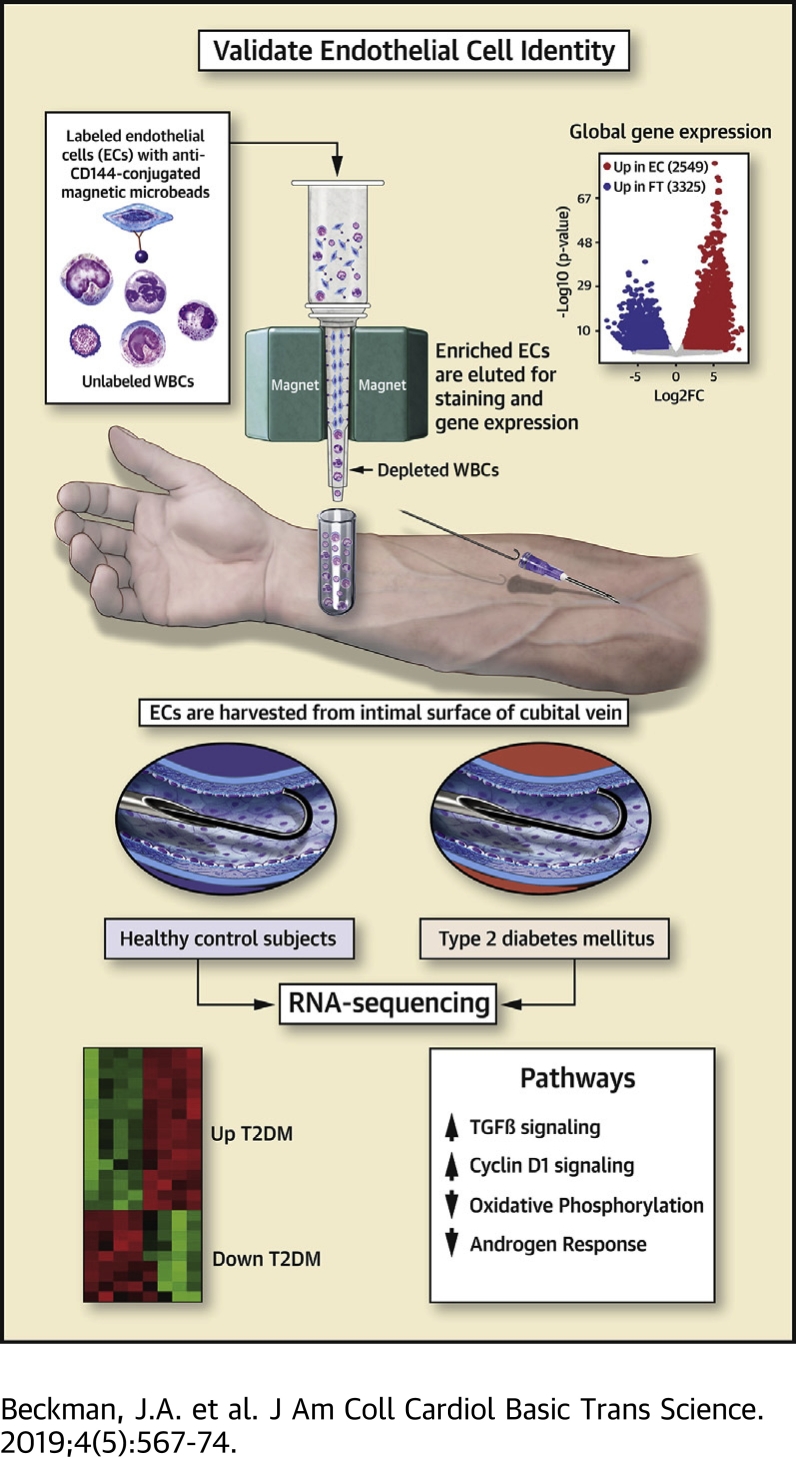
In this study low-input RNA-sequencing was used to annotate the molecular identity of endothelial cells isolated and immunopurified with CD144 microbeads. Using this technique, comparative gene expression profiling from healthy subjects and patients with type 2 diabetes mellitus identified both known and novel pathways linked with EC dysfunction. Modeling of diabetes by treating cultured ECs with high glucose identified shared changes in gene expression in diabetic cells. Overall, the data demonstrate how purified ECs from patients can be used to generate new hypotheses about mechanisms of human vascular disease.
The vascular endothelium—a single-cell thick layer situated at the interface of flowing blood and the vessel wall—plays a central role in cardiovascular homeostasis by regulating blood pressure, thrombosis, leukocyte trafficking, and metabolism (1). The ability to culture endothelial cells (ECs), including human umbilical vein ECs (HUVECs), in vitro revolutionized vascular biology by providing a renewable source of cells to study human EC structure and function. These model systems have generated key insights into EC biology involved in the pathophysiology of cardiovascular disease. However, the culture of ECs outside the context of the blood vessel alters function and cell differentiation in fundamental ways. For example, the nonphysiological static microenvironment of cell culture affects biochemical signaling and gene expression in ECs (2). Thus, acquiring ECs directly from patient vessels may facilitate the discovery of new mechanisms of EC dysfunction that arise from genetic mutations, chronic disease states, or drug toxicities, all of which are known to modulate vascular disease risk in vivo in humans. In addition, the inability to study patient-specific effects of drugs on EC signaling and gene regulation in vivo remains a significant obstacle for successful translation of potential vascular disease treatments. Accordingly, complementary methods for studying EC biology more directly in humans are needed.
Multiple groups have reported using wire biopsy of ECs from peripheral vein or artery to study the activity of specific signaling pathways implicated in EC dysfunction 3, 4, 5. These targeted approaches identify ECs on the basis of staining for known EC markers. Other blood cells, especially leukocytes, contaminate these preparations, thereby complicating analysis using conventional microscopy. To overcome these issues, fluorescence-activated cell sorting (FACS) (6) or magnetic microbeads (7) have been used to enrich for ECs during the procedure. However, FACS is time intensive and can damage fragile cells. In addition, the molecular identity of cells retrieved during biopsy and purification has not been determined using unbiased analysis. These issues raise questions about the utility of the EC biopsy technique in discovery-based investigation of vascular disease in humans.
In this study, we used a positive selection step with anti-CD144 microbeads to enrich for ECs rapidly and deplete non-ECs (i.e., circulating leukocytes). We then annotated the molecular identity of both selected and nonselected fractions of cells using unbiased, low-input RNA sequencing. Differential gene expression identified 2,549 up-regulated transcripts in the CD144-selected cell fraction. Gene ontology analysis of these transcripts revealed highly significant enrichment for pathways related to EC development and function. Furthermore, expression of classic EC markers clustered perfectly with selected cells, whereas leukocyte markers clustered in nonselected cells. We then used this approach to uncover novel gene expression pathways that are differentially regulated in ECs from patients with type 2 diabetes mellitus (T2DM) compared with healthy control patients. Comparison of genes induced by high glucose in HUVEC, a model of diabetic hyperglycemia, revealed a partial, but statistically significant overlap with genes identified in T2DM ECs. Collectively, these results demonstrate that immunopurification of cells isolated with wire biopsy strongly enriches for ECs, and comparative transcriptomic analysis can be used in conjunction with established EC culture models to discover clinically relevant disease-related pathways for future investigation.
Methods
EC harvest and enrichment
The protocol was approved by the Vanderbilt University Medical Center Human Research Committee, and all subjects provided written informed consent. A total of 5 healthy subjects (n = 3 men, 2 women, self-identified Caucasian = 4; self-identified African American = 1, average hemoglobin A1C = 5.1 ± 0.3) and 5 patients with diabetes (n = 4 men, 1 woman; self-identified Caucasian = 5, average hemoglobin A1C = 8.2 ± 1.2) were recruited. A 20-gauge intravenous (IV) catheter was inserted into a patient’s cubital vein under sterile conditions. ECs were gently scraped from the intimal surface of the cubital vein with a J-wire (Arrow International, Reading, Pennsylvania). The wire and cells were centrifuged (400 x g for 7 min at room temperature with no brake) in dissociation buffer (phosphate-buffered saline [PBS], 2 mM ethylenediamine tetra-acetic acid [EDTA], heparin 0.1 mg/ml, pH 7.4). After centrifugation, dissociation buffer was aspirated ,and cells were resuspended in 80 μl of labeling buffer (PBS supplemented with 0.5% bovine serum albumin [BSA], 2mM EDTA, pH 7.2) along with 20 μl of CD144 microbeads (Cat# 130-097-857 Miltenyi Biotec, Birgisch Gladbach, Germany). Cells were incubated for 15 min at 4°C. After incubation, cells were sorted through a magnetic column in a magnetic field (QuadraMACS, Miltenyi). Cells were washed 3 times with labeling buffer. Labeled ECs were recovered by gravity flow with 4 ml of cold labeling buffer after removing the column from the magnet.
Cell fixation and staining
Cells were fixed and stained as previously described (5). Cells were plated on poly-L-lysine-coated chamber slides. Slides were centrifuged at 400 revolutions/min for 5 s with no break, then rotated 180° for a second spin. Buffer was aspirated, and cells were stained with 4% paraformaldehyde for 10 min at room temperature. After second wash, slides were air dried then frozen at −80°C. For staining, slides were rehydrated with PBS/glycine 50 mM then incubated with anti-Von Willebrand factor antibody (Dako, Clone F8/86, Carpinteria, California) at 1:300 for 1 h at 37°C followed by secondary goat anti-mouse antibody conjugated to Alexa Fluor 488 dye (Invitrogen, Thermo Fisher Scientific, Waltham, Massachusetts) at 1:800 for 45 min at 37°C. Cells were mounted with ProLong Gold Antifade with DAPI (Invitrogen) and imaged with an immunofluorescence microscope (OIympus IX81, Olympus, Shinjuku, Tokyo, Japan).
RNA isolation and sequencing
For RNA isolation, isolated cells were quickly pelleted by centrifugation and immediately placed in RNA lysis buffer per protocol (Qiagen, RNeasy Plus Micro, Venlo, the Netherlands). Isolated RNA was quantified and sequencing libraries generated using low-input, mRNA sequencing DNA preparation kit per company protocol (Illumina, San Diego, California). Paired End-75 sequencing was performed on an Illumina HiSeq3000. Reads were trimmed to remove adapter sequences using Cutadapt v1.16 (8) and aligned to the GENCODE GRCm38.p5/human b37 genome using STAR v2.5.3a (9). GENCODE vM12/Ensembl v75 gene annotations were provided to STAR to improve the accuracy of mapping. Quality control on both raw reads and adaptor-trimmed reads was performed using FastQC. FeatureCounts v1.15.2 (10) was used to count the number of mapped reads to each gene. Differentially expressed, protein-coding genes were detected by DESeq2 (v1.18.1) (11). Heatmap3 was used for cluster analysis and visualization (12). Genome ontology analysis was performed on differentially expressed genes using the ToppGene suite. Gene set enrichment analysis (GSEA) was performed using the GSEA package (13). The gene set for the EC-restricted gene list (n = 151) was obtained from a previously published curated dataset (14).
EC culture
Pooled HUVEC were purchased from Lonza (Lonza Inc, Walkersville, Maryland) and maintained in growth media (ATCC VEGF bullet kit) on 0.1% gelatin-coated plates. Cells for gene expression were used at passage 3 or less. For glucose stimulation, media was supplemented with additional glucose to a final concentration of 25 mM. Cells were treated for 24 h with high glucose (25 mM) vs. standard glucose (5 mM). After treatment, cells were washed, trypsinized, counted on a hemocytometer, and 1,000 cells were used for RNA sequencing to mimic the low cell counts from the human EC harvests. Separate kinetic experiments compared high glucose and mannitol with a final concentration of 25 mM (n = 4). For the mannitol experiment, RNA was extracted by column purification after 6-h, 24-h, or 36-h treatment, then 100 ng was reverse transcribed (iScript, BioRad, Philadelphia, Pennsylvania), and real time-qPCR was performed using standard protocol (2-step amplification, Bio-Rad CFX96 cycler). All qPCR data are normalized to internal control gene 36B4. Primers available upon request.
Statistics
For RNA-sequencing data, differentially expressed protein-coding genes were identified with DESeq2 using false discovery rate (FDR)-adjusted p value <0.05 and absolute fold change >2.0 as the threshold for statistical significance (v1.18.1) (11). All the differential expression data are presented as volcano plots (Figures 1B and 2A). For GSEA, each individual gene set was tested against the entire experimental gene list, using the “stat” result generated by the DESeq2 analysis. The enrichment plots for GSEA and the FDR adjusted p values were generated using the GSEA package, as described in the original publication, without modification (Figures 1C and 2B) (13). For gene ontology analysis, we used the Bonferroni method to adjust for multiple hypothesis testing because it was the most stringent method in the ToppGene Suite (Figure 1D). To generate the heatmap comparing EC and FT samples, the gene-expression data were transformed using variance-stabilizing transformation in DESeq2 (Figure 1E). Samples were then clustered by Pearson correlation using the specified leukocyte and EC genes. In HUVEC stimulated with mannitol or glucose, the heatmap displays row normalized mean fold change compared with Time 0 (Supplemental Figure 1). Mean fold change from real-time qPCR data was calculated using the delta-delta cycle threshold (ddCt) method. We used 2-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test to determine significant changes in expression in mannitol- or glucose-treated HUVEC vs. Time 0 cells. The threshold for statistical significance was a p value <0.05. For comparison of fold changes in HUVEC vs. T2DM ECs gene expression, a standard box plot was drawn using ggplot2. The plot shows the minimum, first quartile, median, third quartile, and maximum values (Figure 2E). The p value was determined by Wilcoxon rank sum test.
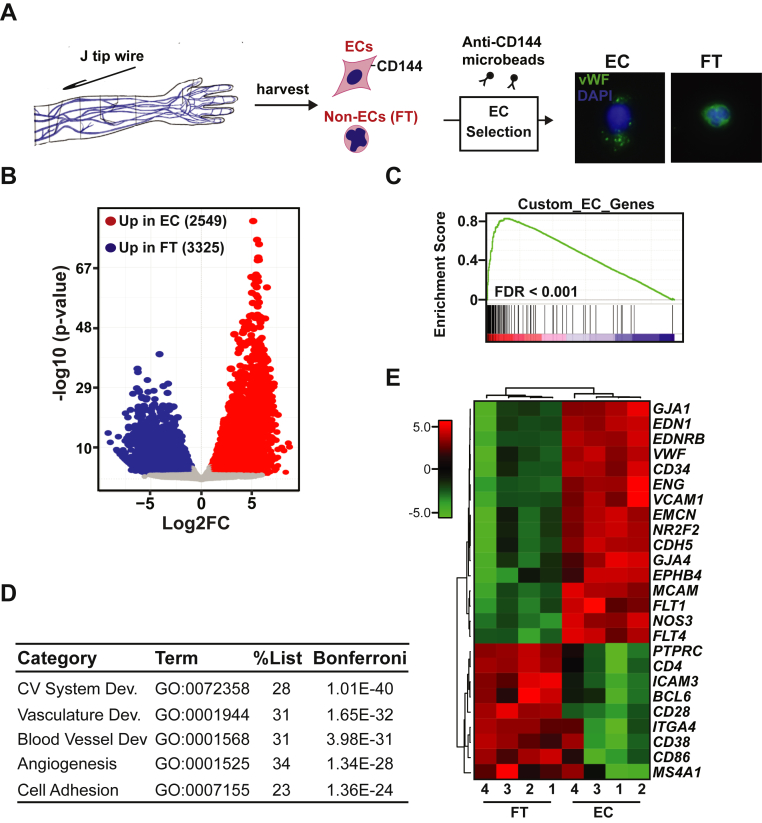
Figure 1.
Ex Vivo CD144-Selection Enriches Vascular ECs in Patient-Derived Samples
(A) Schematic of endothelial cell (EC) isolation procedure, selection of ECs using anti-CD144 magnetic microbeads and photomicrographs of immunofluorescence for von Willebrand factor (VWF) (green) in CD144 selected ECs and flow through cells (FT = nonselected). Nuclei were counterstained with DAPI (blue). Results shown at 40x magnification. (B) Volcano plot showing Log2 fold change (Log2FC) expression vs. log10 (p value) of differentially expressed genes in CD144-selected cells vs. FT. For significance cutoff, false discovery rate (FDR) adjusted p value <0.05 and 2-fold change was used. (C) Gene set enrichment plot showing positive enrichment of genes expressed in CD144-selected cells in a curated EC gene set. (D) Table showing results of gene ontology analysis (ToppGene suite) of differentially expressed genes from CD144-selected cells vs. FT cells. Bonferroni adjusted p value is shown. All statistically significantly upregulated genes in CD144 selected cells vs. FT cells from Figure 1B were included in the analysis using a cutoff of 2-fold; FDR <0.05. GO: Biological Processes are listed. (E) Heat map showing expression data of specific EC-restricted and leukocyte-restricted marker genes in EC and FT samples. Samples were clustered using Pearson correlation. For each EC and FT patient-derived sample, pairwise analysis was performed. Numbers below the heat map indicate paired samples.
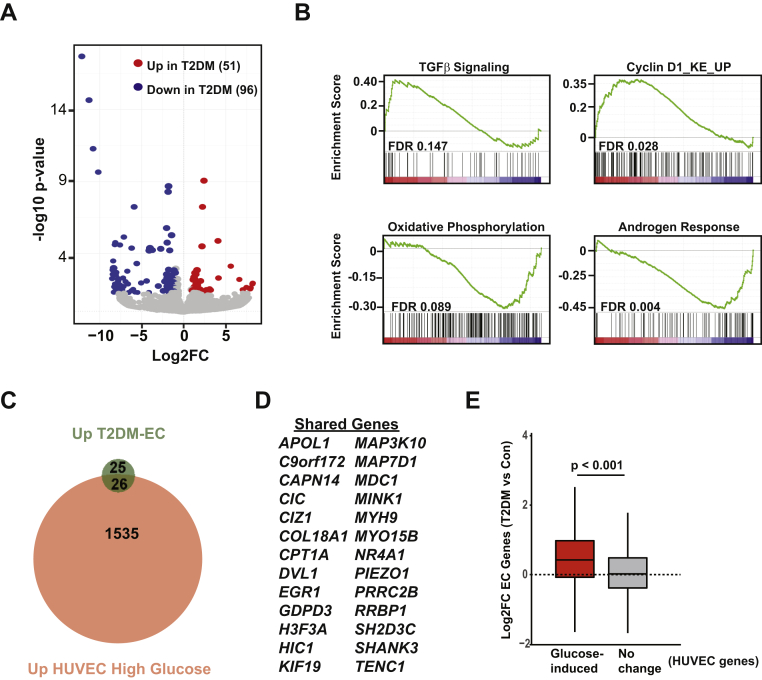
Figure 2.
Comparative Transcriptomics Identifies Changes in Pathways of Proliferation, Metabolism, and Androgen Signaling in ECs From Patients With T2DM
(A) Volcano plot showing Log2 fold change (Log2FC) in expression of protein coding genes vs. log10 (p value) in T2DM (endothelial cells) ECs compared with EC-Con (Same RNA-seq data from Figure 1). False discovery rate (FDR adjusted p value cutoff <0.05. (B) Gene set enrichment plots in T2DM ECs vs. EC-Con. Genes were ranked using the “stat” parameter generated in the DESeq2 pipeline. For each plot, FDR is included. (C) Venn diagram showing overlap in genes induced in T2DM-ECs (green) and HUVEC treated with 25 mM glucose for 24 h (red). (D) List of shared genes from panel C. (E) Boxplot showing median Log2FC (T2DM ECs vs. EC-Con) of all HUVEC genes induced by high glucose (2-fold, FDR <0.05) or HUVEC genes that did not change with glucose stimulation. For E, p value was determined by Wilcoxon rank sum test.
Results
Cells collected via venous cannulation and wire abrasion were immediately labeled with anti-CD144-conjugated magnetic microbeads, which recognize the EC surface marker VE-Cadherin (aka CD144). This method reproducibly recovers 1,000 ECs from 1 arm of a human donor within 45 min. CD144- selected cells stained positive for the EC protein Von Willebrand factor and possess characteristic nuclear morphology that distinguishes them from non-ECs, as previously described (Figure 1A) (5). To evaluate the molecular identity of these cells more comprehensively, we performed unbiased gene-expression profiling by paired-end RNA sequencing of EC (n = 4, EC-Con) and flow-through fractions (n = 4, FT-Con) from healthy control subjects. One pair of EC-FT was left out of this analysis because of low quality in the FT fraction. Pairwise analysis of differential expression identified 2,549 genes more highly expressed in the CD144-selected cell fraction and 3,325 genes in the FT fraction (Figure 1B) (adjusted p value <0.05; log2 fold change ≥1). We next used a curated list of genes previously identified as highly restricted to ECs to perform gene set enrichment analysis on the RNA-seq data from isolated cells (14). This approach identified striking positive enrichment for EC genes in the human CD144-selected cells (Figure 1C). To address the molecular identity of the EC fraction in an orthogonal manner, we evaluated the gene ontology of differentially up-regulated genes in CD144-selected cells. Pathways related to the cardiovascular system, blood vessel development, angiogenesis, and cellular adhesion were strongly over-represented, revealing excellent enrichment for ECs and depletion of white blood cells (WBCs) in these cell fractions (Figure 1D). Finally, we extracted expression data for well-known EC-specific and WBC-specific genes and detected a clear clustering of gene expression by cell fraction (Figure 1E). Overall, these unbiased data demonstrate that CD144 selection of extracted cells enriches for ECs, as defined by global gene-expression patterns and known markers of EC identity.
We next exploited this EC-enrichment method to examine how T2DM alters global gene expression by comparing transcriptomic profiles of ECs isolated from patients with T2DM (n = 4) with data from healthy controls (n = 5, EC-Con) (15). In T2DM ECs, 51 genes were significantly up-regulated and 101 genes significantly down-regulated compared with EC-Con (Figure 2A). GSEA identified that pathways of metabolism and growth, including TGFβ and oxidative phosphorylation, were positively and negatively enriched in T2DM ECs, respectively (Figure 2B). Significant positive enrichment occurred in Cyclin D1 signaling, a pathway that has not yet been implicated in vascular disease in T2DM. Notably, androgen signaling was negatively enriched in T2DM ECs, consistent with emerging links between androgen deficiency, T2DM, and cardiovascular disease (16). Finally, we modeled the hyperglycemia associated with T2DM in vitro by treating HUVEC with high glucose (25 mM, 24 h). Although many more genes are modulated by glucose in vitro, of the 51 genes induced in T2DM, a significant number of genes (p < 4.32e-13 for probability of overlap) were shared in glucose-stimulated HUVEC (Figures 2C and 2D). Independent validation of these targets in a separate experiment of HUVEC treated with high glucose or mannitol (as an osmolarity control) identified up-regulation of a subset of genes (Supplemental Figure 1). Based on these results, we considered the possibility that the trends in gene up-regulation may be similar in T2DM ECs and glucose-treated HUVEC. For this analysis, we derived 2 gene lists from the HUVEC RNA-seq dataset: glucose-induced genes and genes that did not change expression. We then compared the composite fold change of these 2 gene lists in the T2DM EC RNA-seq dataset. With this approach, we detected a significant global shift in expression of glucose-induced genes in T2DM ECs (Figure 2E).
Discussion
Vascular ECs play a central role in cardiovascular homeostasis and disease. Tools to study ECs in an unbiased manner directly in humans are limited but are critical to improve discovery-based research in human vascular biology. We address this limitation by developing a reproducible method for isolating, enriching, and then profiling global gene-expression programs in ECs from human subjects. Previous reports demonstrate rapid dedifferentiation of ECs and macrophages following their removal from resident tissue microenvironments 17, 18. The method presented here enables the study of context-dependent gene regulation in human ECs without supervening effects of cell culture. As a result, ECs isolated from patients can be used to broaden our insights into molecular mechanisms governing blood-vessel function in humans.
Previous work with freshly isolated ECs studied differences in candidate signaling pathways in ECs using targeted immunofluorescence 3, 5, 19, 20. A key goal of our study was to demonstrate that global differences in gene expression between 2 different patient groups could be measured with isolated ECs. We chose to study patients with T2DM, given the strong links between T2DM, EC dysfunction, and vascular disease (21). Notably, androgen signaling was negatively enriched in T2DM ECs, providing validation for our experimental system, given established associations between androgen deprivation—both physiologic and cancer treatment-induced—with metabolic and cardiovascular disease 16, 22, 23. This is also the first study to implicate impaired androgen signaling in ex vivo diabetic ECs. Down-regulation of oxidative phosphorylation observed here in isolated ECs is consistent with known changes in mitochondrial and EC function provoked by hyperglycemia and insulin resistance (24). In contrast to the androgen pathway, the role of cyclin D1 signaling in diabetic vascular disease is unknown and merits further study. Cyclin D2, a closely related homologue involved in cell-cycle control, is induced by glucose in rat ECs and promotes EC proliferation (25). Connections between cyclin D1 and EC dysfunction in diabetes mellitus may be particularly relevant now, given ongoing trials of new cancer therapies that inhibit the cyclin D1 axis. These therapies could be deployed to disrupt cyclin D1 signaling in ECs and test whether this intervention affects EC function under diabetic states in humans (26). Finally, the positive enrichment for TGF-β signaling may be relevant to the anticorrelation of diabetes mellitus and aneurysm disease that has been described, given that loss of function mutations in the TGF-β pathway are causally linked to genetic aneurysm syndromes such as Loeys-Dietz syndrome 27, 28.
Despite known differences in homeostatic gene-expression programs of cultured ECs vs. ECs immediately after isolation from organ depots, we detected a partially shared gene regulatory response between glucose-treated HUVECs and T2DM ECs (17). In addition, some genes were also up-regulated by mannitol in HUVEC, suggesting hyperosmolarity itself controls EC gene expression in vitro and may be relevant to gene regulation in vivo in patients with diabetes mellitus. Notably, this experiment compares stress responses in vitro and in vivo, unlike previous work examining baseline transcriptional profiles in ECs. Although the number of patients studied was small, the data presented herein suggest that stress signaling can lead to some convergences in gene expression in vitro and in vivo.
Study limitations
We cannot exclude the possibility that our CD144 cell-sorting method might bias these results if EC differentiation state and CD144 expression is down-regulated during progression of diabetes, as has been shown in other contexts (29). Orthogonal methods, such as multimarker sorting and single-cell RNA-sequencing could be used to overcome these potential limitations. Another limitation of this study is the relatively small number of patients included in this first study of diabetes. In the future, increasing sample size will improve statistical power to discern subtle changes in gene expression between disease or treatment groups.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Endothelial function is an important clinical feature of diabetes mellitus. However, tools to study endothelial molecular biology directly in patients are limited. The current study provides validation of a method to study gene regulation in endothelial cells from humans. This tool can be used to probe how endothelial function changes in systemic diseases including diabetes mellitus.
TRANSLATIONAL OUTLOOK: Patient-derived endothelial cells can be used to discover new biological pathways in systemic diseases that feature vascular dysfunction.
Conclusions
These results illustrate how a multimodal experimental platform that couples data from primary human samples with data from established in vitro model systems could prioritize pathways for further exploration as mediators of EC dysfunction in human disease. We anticipate that this approach, which couples immunopurification with unbiased gene expression, can be applied to study mechanisms of chronic cardiovascular diseases in humans as well as vascular effects and toxicities of drugs in clinical use or preclinical development.
Footnotes
This work was supported by grants HL131977 (to Dr. Beckman) and the Vanderbilt-Ingram Cancer Center Young Ambassador Award (to Dr. Brown). All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
Appendix
References
- 1.Gimbrone M.A., Jr., Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagel T., Resnick N., Dewey C.F., Jr., Gimbrone M.A., Jr. Vascular endothelial cells respond to spatial gradients in fluid shear stress by enhanced activation of transcription factors. Arterioscler Thromb Vasc Biol. 1999;19:1825–1834. doi: 10.1161/01.atv.19.8.1825. [DOI] [PubMed] [Google Scholar]
- 3.Colombo P.C., Ashton A.W., Celaj S. Biopsy coupled to quantitative immunofluorescence: a new method to study the human vascular endothelium. J Appl Physiol. 2002;92:1331–1338. doi: 10.1152/japplphysiol.00680.2001. [DOI] [PubMed] [Google Scholar]
- 4.Donato A.J., Eskurza I., Silver A.E. Direct evidence of endothelial oxidative stress with aging in humans: relation to impaired endothelium-dependent dilation and upregulation of nuclear factor-kappaB. Circ Res. 2007;100:1659–1666. doi: 10.1161/01.RES.0000269183.13937.e8. [DOI] [PubMed] [Google Scholar]
- 5.Tabit C.E., Shenouda S.M., Holbrook M. Protein kinase C-beta contributes to impaired endothelial insulin signaling in humans with diabetes mellitus. Circulation. 2013;127:86–95. doi: 10.1161/CIRCULATIONAHA.112.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waldo S.W., Brenner D.A., McCabe J.M. A novel minimally-invasive method to sample human endothelial cells for molecular profiling. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emin M., Wang G., Castagna F. Increased internalization of complement inhibitor CD59 may contribute to endothelial inflammation in obstructive sleep apnea. Sci Transl Med. 2016;8:320. doi: 10.1126/scitranslmed.aad0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 9.Dobin A., Davis C.A., Schlesinger F. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 11.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao S., Guo Y., Sheng Q., Shyr Y. Advanced heat map and clustering analysis using heatmap3. BioMed Res Int. 2014;2014:986048. doi: 10.1155/2014/986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhasin M., Yuan L., Keskin D.B., Otu H.H., Libermann T.A., Oettgen P. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genom. 2010;11:342. doi: 10.1186/1471-2164-11-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luscher T.F., Creager M.A., Beckman J.A., Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: Part II. Circulation. 2003;108:1655–1661. doi: 10.1161/01.CIR.0000089189.70578.E2. [DOI] [PubMed] [Google Scholar]
- 16.Rovira-Llopis S., Banuls C., de Maranon A.M. Low testosterone levels are related to oxidative stress, mitochondrial dysfunction and altered subclinical atherosclerotic markers in type 2 diabetic male patients. Free Radic Biol Med. 2017;108:155–162. doi: 10.1016/j.freeradbiomed.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Lacorre D.A., Baekkevold E.S., Garrido I. Plasticity of endothelial cells: rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood. 2004;103:4164–4172. doi: 10.1182/blood-2003-10-3537. [DOI] [PubMed] [Google Scholar]
- 18.Gosselin D., Skola D., Coufal N.G. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356 doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng L., Stern D.M., Pile-Spellman J. Human endothelium: endovascular biopsy and molecular analysis. Radiology. 1999;212:655–664. doi: 10.1148/radiology.212.3.r99au28655. [DOI] [PubMed] [Google Scholar]
- 20.Silver A.E., Christou D.D., Donato A.J. Protein expression in vascular endothelial cells obtained from human peripheral arteries and veins. J Vasc Res. 2010;47:1–8. doi: 10.1159/000231715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beckman J.A., Paneni F., Cosentino F., Creager M.A. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part II. Eur Heart J. 2013;34:2444–2452. doi: 10.1093/eurheartj/eht142. [DOI] [PubMed] [Google Scholar]
- 22.Bhatia N., Santos M., Jones L.W. Cardiovascular effects of androgen deprivation therapy for the treatment of prostate cancer: ABCDE steps to reduce cardiovascular disease in patients with prostate cancer. Circulation. 2016;133:537–541. doi: 10.1161/CIRCULATIONAHA.115.012519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarro G., Xu W., Jacobson D.A. Extranuclear actions of the androgen receptor enhance glucose-stimulated insulin secretion in the male. Cell Metab. 2016;23:837–851. doi: 10.1016/j.cmet.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X., Luo Y.X., Chen H.Z., Liu D.P. Mitochondria, endothelial cell function, and vascular diseases. Front Physiol. 2014;5:175. doi: 10.3389/fphys.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X.X., Liu Y.M., Li Y.J. High glucose concentration induces endothelial cell proliferation by regulating cyclin-D2-related miR-98. J Cell Mol Med. 2016;20:1159–1169. doi: 10.1111/jcmm.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner N.C., Ro J., Andre F. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 27.Raffort J., Lareyre F., Clement M., Hassen-Khodja R., Chinetti G., Mallat Z. Diabetes and aortic aneurysm: current state of the art. Cardiovasc Res. 2018;114:1702–1713. doi: 10.1093/cvr/cvy174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeys B.L., Schwarze U., Holm T. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 29.Widyantoro B., Emoto N., Nakayama K. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation. 2010;121:2407–2418. doi: 10.1161/CIRCULATIONAHA.110.938217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.