Graphical abstract
Abbreviations: DSHP, Dataset of single helix-mediated protein-protein interactions; EGF, Epidermal growth factor; EGFR, EGF receptor; MD, molecular dynamics; PDB, Protein data bank; PME, Particle Mesh Ewald; PPI, Protein-protein interactions; RMSD, Root mean square deviation; TNF, Tumor necrosis factor
Keywords: Protein design, Protein-protein interaction, Helix design, Peptide design
Abstract
Protein-protein interactions (PPIs) play a key role in numerous biological processes. Many efforts have been undertaken to develop PPI modulators for therapeutic applications; however, to date, most of the peptide binders designed to target PPIs are derived from native binding helices or using the native helix binding site, which has limited the applications of protein-protein interface binding peptide design. Here, we developed a general computational algorithm, HPer (Helix Positioner), that locates single-helix binding sites at protein-protein interfaces based on the structure of protein targets. HPer performed well on known single-helix-mediated PPIs and recaptured the key interactions and hot-spot residues of native helical binders. We also screened non-helical-mediated PPIs in the PDBbind database and identified 17 PPIs that were suitable for helical peptide binding, and the helical binding sites in these PPIs were also predicted for designing novel peptide ligands. The L2 domain of EGFR, which was the top ranked, was selected as an example to show the protocol and results of designing novel helical peptide ligands on the searched binding site. The binding stability of the designed sequences were further investigated using molecular dynamics simulations.
1. Introduction
Protein-protein interactions (PPIs) play a key role in numerous biological processes, and many of PPIs are therapeutic targets. Developing PPI modulators for therapeutic applications has therefore attracted considerable attention. PPI modulators comprise three main classes: proteins, mini proteins/peptides, and small molecules; however, the flat surface and relatively large area of PPIs makes developing small molecules targeting PPIs challenging [1]. On the other hand, protein therapeutics (particularly antibodies) that regulate PPIs are hindered by limited drug delivery systems, a short half-life, and immunogenicity. Compared to small molecules and antibodies, mini proteins or peptides are promising due to their high binding specificity, good design-ability, and lower immunogenicity [2].
The most abundant secondary protein structure in nature is the α-helix, which plays important roles in both protein folding and executing functions, such as protein-protein association, and α-helices mediate many PPIs [3], [4]. Additionally, α-helices are good templates for modification or mimicry. Several strategies have been used to develop helix mimics, including helix stabilization, helical foldamers, and helical surface mimetics [5]. To date, most of the de novo designed protein binders use helical structures to bind at the interfaces. For example, several successful de novo designs of protein binders from Baker’s group use helices as interface motifs, including protein binders targeting hemagglutinin [6], interleukin-2 and interleukin-5 [7], and botulinum neurotoxin B [8]. Our group has successfully designed single-helix peptides targeting tumor necrosis factor alpha (TNFα) [9]. Thus, there is enormous potential for designing helical binders at protein-protein interfaces as PPI modulators.
Although many PPIs are mediated by helices, they can also contain other common recognition motifs, such as loops [10] or β-strands [11]. Designing protein-protein interface binders based only on the structures of native binders limits design approaches, and the same protein binding site can often bind different structural motifs. For example, Kuhlman et al.’s redesigned the C-terminal of the RGS14 GoLoco motif, which has a coil structure, into a helical structure and found it bound to Gαi1 at the original binding site [12]. In our previous work designing TNF-binding peptides, we identified a helical binding site on TNF surface that was originally used to bind the β-structured TNF receptor. We successfully designed helical peptides that bind to the identified site and inhibit the cellular activity of TNFα [9], [13].
Considering the advantages of the peptide modulators mentioned above, there is a need to identify more protein-protein interfaces that are suitable for single-helix peptide binding. In the present study, we developed a computational method that can locate single-helix binding sites at protein-protein interfaces based on the structure of the protein targets. To reveal the interacting profiles of a single α-helix peptide ligand, we first collected a dataset of single-helix-mediated PPIs (protein-peptide or protein-protein interactions mediated by only one helix in one of the two partners) and analyzed their structural features and properties. Guided by these binding features, we then developed a general computational algorithm, HPer (Helix Positioner) that locates single-helix binding sites at protein-protein interfaces (https://github.com/proteincraft/HPer). HPer recaptured most of the known helix binding sites in the dataset and predicted potential helix binding sites in PPIs in the PDBbind database, which are not naturally mediated by a helix. These identified helix binding sites may be very helpful for developing novel PPI modulators, including helical peptides, helix containing mini proteins, or helical mimetics. We selected the L2 domain of EGFR from the identified targets, and computational designed novel helical peptide ligands. The molecular dynamic (MD) simulation results showed that the designed complex structures were stable.
2. Methods and materials
2.1. Construction of the dataset of single helix-mediated PPIs
We constructed a unique Dataset of Single-Helix-mediated PPIs (DSHP) based on the protein-protein complex structures from the PDBbind database 2014 [14]. The interfacial residues in the complex structures were defined using Rosetta InterfaceAnalyzer with a 5 Å distance cutoff. Secondary structure types of interfacial residues were assigned using DSSP [15]. For highly helical interfaces (whose helical content is larger than 80%), the continuity of helical residues was checked by residue number to ensure the protein-protein interfaces were mediated by a single α-helix.
The selected complexes were then clustered with a single linkage clustering method based on the target protein sequences, using a similarity cutoff of 95%. In each of the clusters, the complex structure with highest resolution was selected. All of the resulting protein-protein complex structures were further validated by visual inspection. 30 single-helix-mediated PPIs were collected in the current version of the DSHP dataset (Table S1).
2.2. Computational design of helical peptide binders using RosettaScripts
RosettaScripts [16] was used to design, filter, and evaluate helical peptides sequences at the surface of the PPIs. The backbones of helical peptide ligands were first remodeled using “LoopModeler”. Subsequently, “FlxbbDesign” was used to design sequences of helical peptides. Before filtering and evaluation of the designed complex models, we used “FlexPepDock” in refining mode to optimize the binding pose of the designed peptides. If the binding pose of a helical peptide was significantly changed, the sequence was optimized again. The helical sequences were allowed to be mutated to 19 types of amino acids (proline excluded). The helical peptide with N, C- terminus excluded was defined as the loop region for “LoopModeler”. The resulting complex structures were evaluated using “InterfaceAnalyzer”, and structure models with lowest binding energy and good packing quality were selected for further computational analysis. Scripts for the above design protocol are provided in the supplementary material.
2.3. Molecular dynamics simulations
All of the energy minimizations and MD simulations were performed with Gromacs5.1.4 using Amber ff99SB-ildn force field and TIP3P water model. The water box of a target-helix complex was first minimized for 10,000 steps with all protein atoms restrained. A 200 ps NVT and a 200 ps NPT MD simulations were performed for equilibration. Then, a 100 ns NPT production simulation was performed for the dynamic study of the complex structures. The simulations were carried out at T = 300 K and P = 1.015 bars using a 2-fs time step. The V-rescale thermostat with a coupling constant τT = 0.1 ps, and the Parrinello-Rahman barostat with a coupling constant τp = 2.0 ps were used. The P-LINCS algorithm was used to restrain all bond lengths to their equilibration values. The van der Waals interaction cutoff was set to 14 Å, and the long-range electrostatic interaction was calculated using the Particle Mesh Ewald (PME) method. Analysis of MD simulation trajectories were performed using VMD [17] and Gromacs [18] built-in modules.
3. Results and discussion
3.1. Analysis of naturally occurring single helix-mediated PPIs
To reveal the interacting profiles of single α-helix ligands, we collected 30 single-helix-mediated protein-protein complex structures from the PBDbind database (Table S1) and performed statistical analysis.
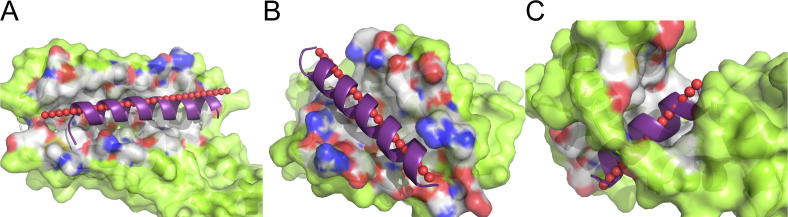
Generally, the recognition between a single helix and its protein target can be divided into three categories, with hot-spot residues distributed on one face, two faces, or three faces [5]. All of these three situations are present in the DSHP dataset (Fig. 1). The target protein structures are diverse based on the CATH [19] assignments. The target protein of 16 cases adopted all-helical structures, including alpha horseshoe, up-down bundle, and orthogonal bundle, and in half of these cases, the single-helix ligands bound to the groove formed by orthogonal bundles (e.g., Bcl-2 family proteins) (Table S1). The other 14 proteins were α/β or mixed α + β structures.
Fig. 1.
Typical cases in the DSHP dataset with single helix in three different binding modes, the one face (A), two face (B), and three face (C) modes. The corresponding PDB codes are (A) 1YDI, (B) 2WH6, and (C) 2BE6. Interacting profiles (white, blue, and red surfaces indicates interfacial carbon, nitrogen, and oxygen atoms, respectively) between helical ligands (purple cartoons) and the target proteins (green surfaces) were extracted from the complex structures. Predicted binding sites (red spheres in line) aligned well with the corresponding original helices. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
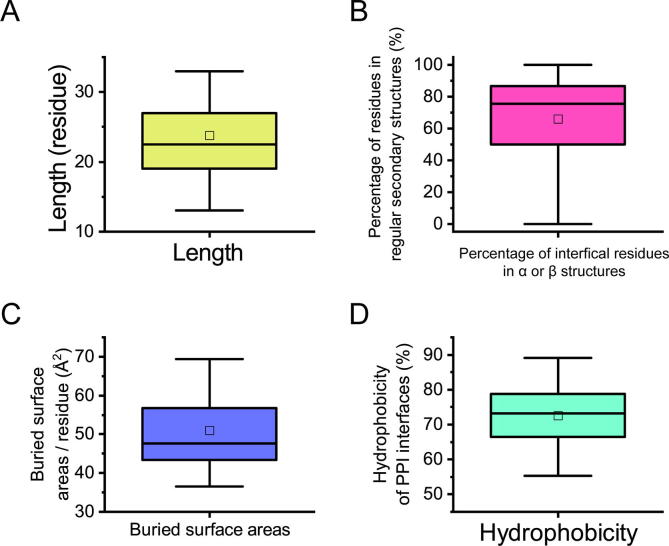
The length of the helices ranged from 15 to 33 residues, with an average of 24 residues (Fig. 2A), which was longer than the average length of participating helix (not solely helix) PPIs, which was 14 residues [3]. This indicates that, for PPIs mediated only by a single helix, longer helices are needed to maintain stability. We calculated the buried surface area per residue, the percentage of interfacial target protein residues in a regular secondary structure (α-helix or β-strand), and the percentage of hydrophobic buried surface area on the target protein side. In DSHP, the average buried surface area per residue of the helix binder ranged between 36.5 Å2 and 86 Å2 (Fig. 2B). The percentage of interfacial residues in a regular secondary structure on the receptor fell between 0 and 100%, with a 66% average value and a 27.9% standard deviation (Fig. 2C). This average value exceeds the corresponding value of general PPIs with diversified interacting structures [20]. The percentage of hydrophobic buried surface areas on the target protein side of the PPIs mediated by a single helix showed an average of 72.5% and standard deviation of 8.9% (Fig. 2D), which is higher than that from Regan’s study using 113 heterodimeric complexes with an average hydrophobicity of approximately 65% [21].
Fig. 2.
Distribution of interacting profiles of structures in a dataset of single-helix-mediated protein-protein interactions in the DSHP dataset. (A) Length of the α-helical ligands in DSHP. (B) Percentage of interfacial residues in regular secondary structures (α and β). (C) Buried surface area per residue of the target proteins covered by their helical binding ligands. (D) The percentage of hydrophobic buried surface areas on the target protein side in DSHP. Box plots indicate the quartile of the distributions. The lower edge of the box represents the 25th percentile; the upper edge indicates the 75th percentile. The line and the square within the box denote the median and mean values, respectively. The two whiskers indicate the maximum and minimum values.
3.2. Computational algorithm for locating helical peptide binding sites at protein-protein interfaces
Our algorithm searched for suitable positions for helical peptide binding at protein-protein interfaces with good shape complementarity, which is regarded as the dominant factor for PPIs [22], [23]. A set of atoms in the target protein that would be blocked in order to modulate the PPIs were defined as the targeted atoms and used as the input of our algorithm. In this work, all of the target proteins were in a known complex structure, and their atoms within 5 Å of the binding partner were defined as interfacial and used as the targeted atoms. However, our algorithm was not limited to be used on proteins with complex structures. Without complex structures, the targeted atoms may be obtained from other experimental or computational evidences.
Three steps were taken to explore the best binding sites of a single helix on a protein-protein interface:
-
(1)
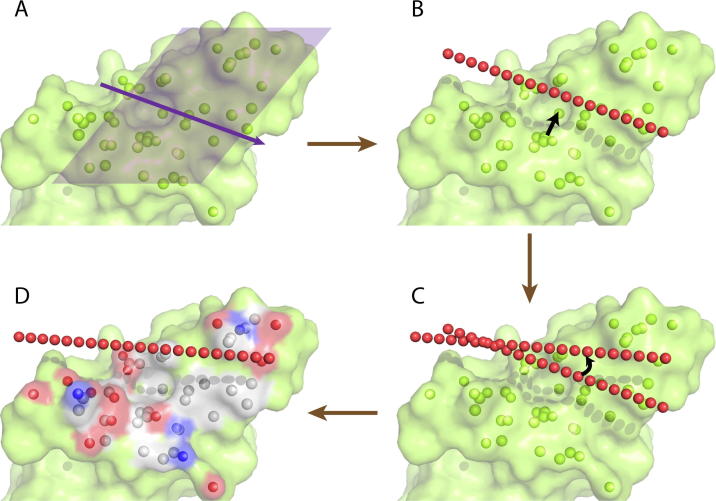
Definition of the principal axis and principal plane of the targeted atoms. The principal axis of the targeted atoms was a straight line passing through the center of mass of the targeted atoms, and the direction was represented as a normal vector. A set of 2000 normal vectors were generated presenting all possible directions in the 3D rotation space using quasiuniformly distributed points on a half unit sphere (Fig. S1) [24]. The neighboring two vectors had an angle smaller than 7° (Fig. S2). For each of the directions, the distance of every targeted atom to the axis was calculated and the distances were summed up. The axis with minimum distance summation was defined as the principal axis of the targeted atoms. Subsequently, a plane crossing the principal axis was rotated from 0 to 360° with a 6° interval, and generated 30 planes. The plane with minimum summation of targeted atom distances was selected and defined as the principal plane (Fig. 3A).
-
(2)
Placement of the initial helix binding position. The distance between the helical axis and the side chain atoms of the helix ranged from 3.25 to 9 Å (Fig. S3) and the average value was ∼5 Å. As there is a 4 Å nearest helix-target interaction distance, we placed the initial position of the detecting axis vector 9 Å above the principal plane and parallel to the principal axis (Fig. 3B).
-
(3)
Search for the best binding site around the initial binding position. We then sampled the position of the helix axis by uniformly translating and rotating around the center of the initial binding position (Fig. 3C). In the process of translation, the center of the axis was moved along a 3D grid, and each of the three dimensions ranged from −7.5 to +7.5 Å with an interval of 0.3 Å. After the center was moved to a new position, all possible directions from the quasiuniformly distributed set of normal vectors were sampled. In most cases, with the sampled helix axes, the corresponding helix peptides were estimated to be able to cover the whole targeting surface.
Fig. 3.
Illustration of the HPer algorithm for helix binding position detection. (A) Definition of the principal axis and the principal plane of targeted atoms. (B) The helix axis is placed to mimic helix binding. (C) Rotation and translation of the helix axis for searching for positions with the best fitness score. (D) Interacting profile (red: negatively charged atoms and surface; blue: positively charged atoms and surface, white: hydrophobic atoms and surface) analysis around the potential helix binding site (red spheres). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The sampled binding poses of the axis were first filtered by the distance (<4 Å) between the axis and target protein to eliminate clashes. The remaining binding poses were then evaluated using a fitness score, which represented the geometry complementarity between the helix and the interacting surface. The score contains two terms: fraction (F) of the interfacial targeted atoms, which has a 7 to 13 Å distance to the helix axis, and cosine of the angle (θ) between the helix axis and the principal axis of the targeted atoms (Eq. (1)). Higher fitness score means the helix has a larger probability interacting with more targeted atoms and more parallel to the principal plane.
| (1) |
In addition to geometry complementarity, the stability of a PPI may also depends on hydrophobic interactions, electrostatic interactions, and hydrogen bonding interactions [25], as well as on the composition of amino acid residues in the ligand helix and the types of secondary structures of targeted residues. Thus, we estimated the interface properties of the target protein interacting with the helix axis, including hydrophobicity, buried area, and secondary structure composition. Interfacial residues were defined as residues with a distance of 7–13 Å to the helix axis. The secondary structure types of these interacting residues were defined using DSSP [15], and the polar or hydrophobic buried surface areas were calculated by summation of the solvent accessible surface area of the interacting polar or hydrophobic atoms given by NACCESS [26] of the interacting atoms (Fig. 3D).
3.3. HPer (Helix positioner) identifies known single helix binding sites
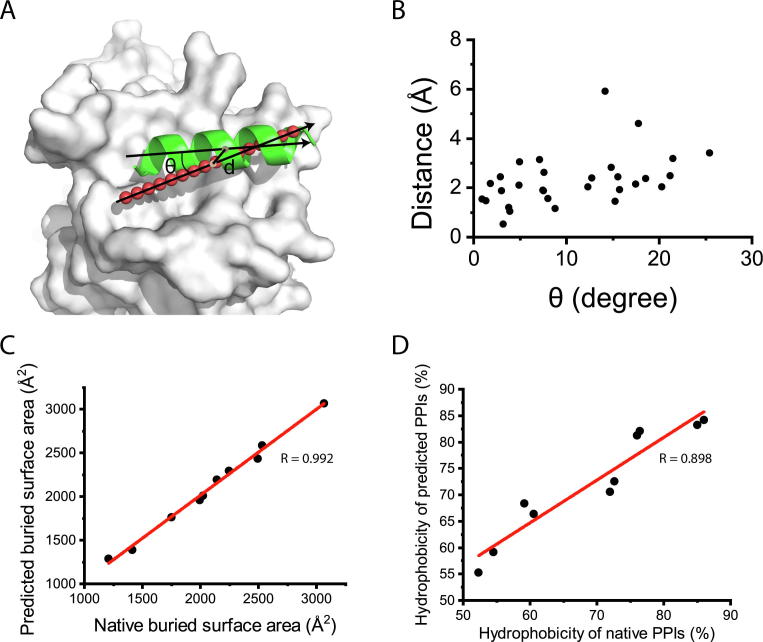
We tested HPer on the DSHP dataset to see if HPer could identify the position of the α-helix peptide ligands. The best position for a helical peptide binding on the corresponding protein-protein interfaces was predicted and compared with the position of the native helical binder. We then defined two parameters to measure the offset of a predicted helix binding position to the position of the native helix binder. The first was the distance between the center of the predicted helix axis and the center of the native helix binder. The second was the angle between the predicted axis and the axis of the native helix binder (Fig. 4A). As shown in Fig. 4B, the offsets of these two parameters in 10 of 30 cases in the DSHP dataset were less than 2 Å and 10°, respectively. For 25 out of the 30 cases, the offsets were less than 4 Å and 20°. The inaccuracies of the remaining five cases came from the bending of the helices, which are common in long helices, but HPer currently uses the straight axis representation for a single helix.
Fig. 4.
Recapturing helical peptide positions in the DSHP dataset. (A) The distance (d) between the center and the angle (θ) between the axis are used to evaluate the offsets between the predicted helix binding site and corresponding native helix position. (B) Distribution of the two offsets of all the cases in DSHP. (C) Correlation between predicted buried surface area using HPer and the buried surface area value calculated from native complex structure. (D) Correlation between the predicted and the real hydrophobicity of the interfacial surface of target proteins.
Overall, HPer correctly identified helix binding positions of all three binding modes present in the DSHP dataset with good accuracy (Fig. 1). Meanwhile, for the ten cases that were predicted with high accuracy, the predicted interacting profiles given by HPer showed good correlation with the corresponding values calculated using the real complex structure (Fig. 4C and D). These results suggest that HPer is capable of locating helix binding sites on protein-protein interfaces, and at the same time gives good prediction on interacting profiles for the potential α-helix ligand binding, which is important for de novo single helix binder design.
3.4. Recovery of key interactions and hot-spot residues in native helix-mediated PPIs
To validate the capability of designing helical binders using the binding sites predicted by HPer, we redesigned helical binders for 8 PPIs in the DSHP dataset, of which the length were less than 30 residues and the binding positions have been accurately predicted by HPer, with a distance offset d < 2 Å and an angle offset θ < 10°. Structures of poly-alanine in helical conformation were first generated using standard α-helix backbone parameters (ϕ = −60°, ψ = −42°) and were placed at the predicted binding sites with the N-terminal to C-terminal in two different directions along the axis. The two helical poly-alanines in opposite N-C directions were then rotated along the helical axis by 60° five times. Therefore, the resulting 12 helical poly-alanine peptides were generated in various orientations as the starting structures for sequence design.
For each input binding pose, the backbones were remodeled and the sequences were designed using RosettaScripts [16]. In total, 2400 sequences were generated for each of the eight cases. The designed sequences were then filtered using the binding energy and packing score given by RosettaInterfaceAnalyzer. For each case, the lowest binding energy sequence with the packing score larger than 0.6 was selected as the designed result.
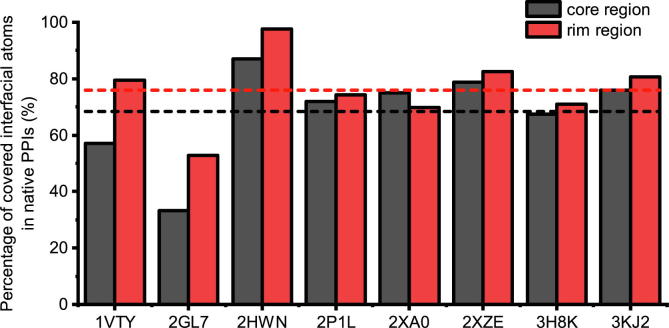
According to previous studies on structural properties of PPIs, the interfaces can be dissected into core region (fully buried areas) and rim regions [27]. The interfaces we designed were compared with native ones. In seven of the eight cases, approximately 68% of the core regions and 76% of the rim regions of native PPIs were covered by our designed helices (Fig. 5). The interface of 2GL7 consists of a crooked groove, and the straight designed helical peptide only docked to a portion of the groove (Fig. S4B), which caused the low coverage of native interfaces in this case.
Fig. 5.
Recovery of key interactions of core regions and rim regions in designed models of the eight helix-mediated protein-protein interactions.
Hot-spot residues are a small set of interfacial residues that contribute the most to binding. Several computational protein design strategies depend on hot-spot residues, such as grafting [28] and anchor-based design [29]. Thus, we further identified native hot-spot residues for the eight cases using SpotOn method [30], [31] and compared the results with those in the designed models. In six of the eight cases (2HWN, 2P1L, 2XA0, 2XZE, 3H8K, and 3KJ2), the hot-spot residues were successfully recovered with average sequence identity and similarity of 35.0% and 85.0% (Table 1) at corresponding sub-pockets (Fig. S4). For the other two cases (1VTY and 2GL7), the backbone of the designed peptides were more seriously deviated from the native positions, which result in the worse reproducing native hot-spot residues.
Table 1.
Redesign of native helix-mediated protein-protein interactions. Binding energy and packing quality of designed models were calculated using Rosetta Scripts.
| PDB_ID | Hot-spot residues Native/Designeda |
Binding energy (REUb) | Packing quality |
|---|---|---|---|
| 1VTY | Y437/W; W440/W; I441/V | −47.2 | 0.604 |
| 2GL7 | L366/Null; I369/I; L373/Null | −15.0 | 0.610 |
| 2HWN | I8/I; I12/L; V16/A | −42.3 | 0.684 |
| 2P1L | L112/I; L116/L; F123/L | −57.7 | 0.634 |
| 2XA0 | L59/E; L63/I; R64/R, I66/A; L70/L; M74/V | −61.2 | 0.608 |
| 2XZE | L210/L; M213/L; L217/W | −48.7 | 0.638 |
| 3H8K | L82/F; L89/V | −49.6 | 0.736 |
| 3KJ2 | I6/L; I13/I; E17/Q | −56.0 | 0.632 |
The corresponding designed residue type to each native hot-spot residue is given after the slash.
REU: Rosetta Energy Unit.
3.5. Prediction of novel helical peptide binding sites
HPer was developed for the purpose of extending PPI druggable spaces and providing starting structures for novel helical peptide binder design. We subsequently screened the 1592 structures of protein-protein complexes from the PDBbind 2014 for non-helical containing protein-protein interfaces suitable for helical peptide binding.
The complex structures were firstly screened according to the following steps. 21,017 complex structures containing pairwise chains were extracted from multiple chain structures. Then, the complex structures with the interfacial helical content of the ligand protein higher than 30% were discarded, and the 13,547 non-helix containing protein-protein interfaces were maintained. Secondary structures of interfacial residues were assigned using DSSP and interfacial residues were defined using Rosetta InterfaceAnalyzer with a distance cutoff of 5 Å.
We used HPer to search for potential helical peptide binding sites around the interface of the remaining structures. Because stable helical peptide ligands are assumed to have at least 4 turns [32] and α-helix contains ∼3.6 residues per turn, we defined the minimum length of the predicted helical peptide ligand to be 15 residues. The minimum value of the 4 interface properties from the statistical analysis of the DSHP dataset were defined as the criteria to determine whether the site was suitable for helical peptide binding, including scorefitness > 0.7, buried surface areas/residue > 35 Å2, percent of hydrophobicity > 0.5, and α + β proportion > 0.27 (outliers excluded).
The 144 structures satisfying the above criteria were then clustered by similarity of sequences with a cutoff of 90%. Finally, 17 interfaces with the best fitness score from each cluster were selected as representative cases suitable for single helical peptide binding (Table 2, Fig. S5, https://github.com/proteincraft/HPer). Some of these PPIs are drug design targets of great interest, including interleukin-17, bone morphogenetic protein receptor type I receptor, epidermal growth factor receptor (EGFR), proprotein convertase subtilisin/kexin type 9, interleukin-2, and nerve growth factor.
Table 2.
Non-helical containing protein-protein interactions predicted suitable for helical peptide binding.
| PDB_ID | Length (AA) | pα+β | Scorefitnees | hydrophobicity | BSA/AA (Å2) | Annotation | CATH classification |
|---|---|---|---|---|---|---|---|
| 3EO1 | 22 | 0.45 | 1 | 0.80 | 41.9 | TGF-beta | Sandwich |
| 3EOB | 29 | 0.42 | 1 | 0.70 | 46.5 | LFA-1 alpha L | Sandwich |
| 3JVF | 45 | 0.38 | 0.729 | 0.66 | 48.2 | IL-17 receptor | Ribbon |
| 3K2U | 30 | 0.38 | 1 | 0.67 | 46.3 | HGFA | Beta Barrel |
| 3NFP | 32 | 0.38 | 1 | 0.54 | 48.1 | IL-2 receptor | Sandwich |
| 3NH7 | 24 | 0.43 | 0.82 | 0.51 | 53.0 | BMP type I receptor | Ribbon |
| 3W9E | 33 | 0.35 | 0.743 | 0.63 | 50.5 | Antibody Fab heavy chain | Sandwich |
| 4DN4 | 16 | 0.38 | 0.76 | 0.60 | 51.9 | C-C motif chemokine 2 | Sandwich |
| 4FAO | 24 | 0.4 | 1 | 0.69 | 54.0 | Activin receptor type-2B | Ribbon |
| 4KRP | 28 | 0.44 | 0.78 | 0.72 | 68.1 | EGFR | Alpha-Beta Horseshoe |
| 4KVN | 25 | 0.47 | 0.71 | 0.56 | 47.1 | Hemagglutinin | Alpha-Beta Complex |
| 4OV6 | 31 | 0.32 | 0.83 | 0.68 | 45.3 | PCSK9 | 2-Layer Sandwich |
| 2ERJ | 33 | 0.46 | 1 | 0.69 | 46.5 | IL-2 receptor | Ribbon |
| 2IFG | 42 | 0.36 | 0.84 | 0.55 | 44.6 | Nerve growth factor | Alpha-Beta Horseshoe |
| 2JIX | 28 | 0.45 | 1 | 0.62 | 39.4 | ERYTHROPOIETIN RECEPTOR | Sandwich |
| 2RA3 | 27 | 0.32 | 0.72 | 0.63 | 56.2 | BPTI | Beta Barrel |
| 2VXS | 19 | 0.40 | 1 | 0.56 | 50.2 | IL-17 | Ribbon |
3.6. A case study: de novo design of helical peptides binding to the EGFR L2 domain
Among the selected cases in Table 2, the interface of the EGFR L2 domain binding with a nanobody (PDB ID: 4KRP) showed the highest fitness score (0.78) and good predicted properties for the binding with helical peptide binders (Table 1). We used the EGFR-EGF interface as an example to test the feasibility of designing helical binders at the predicted binding site. EGFR is an important therapeutic target associated with many cancers and other diseases [33]. Currently, there are two types of EGFR inhibitors: antibodies targeting the extracellular domain [34] and small molecule inhibitors of the tyrosine kinase domain [35]. Although anti-EGFR monoclonal antibodies have advantages over anti-EGFR tyrosine kinase inhibitors, including their high efficiency, high specificity, and low toxicity, the antibodies suffer from limited drug delivery routes and immunogenicity [36]. Thus, there is an urgent need to develop novel EGFR peptide inhibitors with high specificity and better pharmaceutical properties.
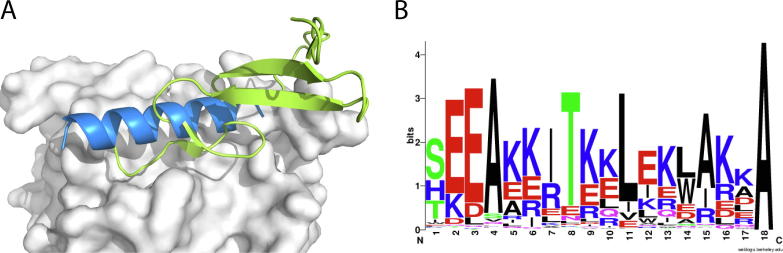
The extracellular portion of EGFR consists of four domains, L1, CR1, L2, and CR2. Downstream signaling of EGFR is activated by the dimerization of EGFRs after EGF binds to EGFR’s L1 and L2 domain [33]. The helix binding site we detected on the EGFR L2 domain was used for binding with EGF (Fig. 5A). The EGFR L2 domain comprises six turns of a β helix [37]. The helix binding site we predicted lies on the flat surface formed by the five β strands. The loops at the end of the β sheets present a groove shape, which is suitable for helix binding. The Cα atoms of the predicted helix were superimposed well with some of the interfacial Cα atoms of EGF (Fig. 6A).
Fig. 6.
Computational design of novel helical peptide binding with EGFR L2 domain. (A) Structure model of EGFR L2 domain (white) in complex with TGFalpha (green, PDB ID: 1MOX). The predicted helix binding site (blue) identified by HPer. (B) Weblogo plot of designed results. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We then designed sequences for the helical peptide binding to the EGFR L2 domain at the identified binding site using RosettaScripts. Among the 24,000 sequences we designed (Fig. 6B), we selected the sequence (EGFR-Pep1) with the lowest binding energy (−40.35 REU) and very good packing quality (0.704). EGFR-Pep1 formed hydrophobic interactions with the two hydrophobic pockets on the surface of EGFR L2 domain, which are the pocket formed by residues L325, L348, and V350, and the pocket formed by residues F412, V417, and I438. There were also two hydrogen bonds formed on their interface, including T10-Q384 and E9-S418. The side chain oxygen of Q384 on EGFR is the only buried polar atom without hydrogen bonding. In addition, electrostatic interactions were designed around the core region of the EGFR interface, including R353, D355, and K465 (Fig. S6).
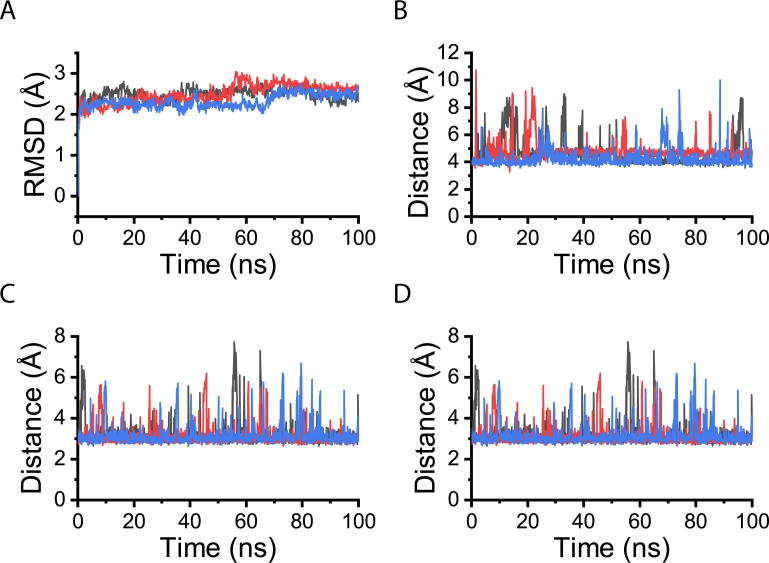
MD simulations were then carried out to test the binding stability of EGFR-Pep1 to the EGFR L2 domain. The designed complex model of EGFR-Pep1 and the EGFR L2 domain was taken as the initial structure. For the three trajectories of 100 ns, the system showed convergence in the first several nanoseconds of the simulation and the root mean square deviation (RMSD) of the Cα atoms during the simulation was maintained as less than 3 Å (Fig. 7A). This indicates the designed model might be stable.
Fig. 7.
Molecular dynamics simulations of a designed helical peptide in complex with the EGFR L2 domain. (A) Root mean square deviation (RMSD) of the complex Cα atoms as a function of simulation time. (B) Distance between the carboxyl of Glu5 and guanidine group of Arg353 as a function of simulation time. (C) Distance between the hydroxyl of Thr10 and amide of Gln384 as a function of simulation time. (D) Distance between the carboxyl of Glu9 and hydroxyl of Ser418 as a function of simulation time. Results from three independent trajectories are shown in different colors.
The designed salt-bridges between EGFR-Pep1 Glu5 and EGFR Arg353 were maintained, and the distance was within 5 Å in the simulations (Fig. 7B). The designed hydrogen bonds at the interface Thr10-Gln384 (Fig. 7C) and Glu9-Ser418 (Fig. 7D) were also maintained, and the corresponding heavy atom distance were around 3 Å in the simulations. Buried surface areas were slightly fluctuated around 800 Å2, and most of the interfacial contacts in the complex model were also not changed. (Fig. S7).
The results from MD simulations showed that the hydrogen bonds, salt bridges, and hydrophobic interactions we have designed between EGFR-Pep1 and EGFR were stable during the simulation. The novel helical binder EGFR-Pep1 might has the ability to inhibit EGF-EGFR interaction.
4. Conclusions
We have developed a computational method HPer for searching for and evaluating potential α-helical peptide binding sites to modulate PPIs. In contrast with other programs such as Peptiderive [38] or PepComposer [39], which can be used to design linear peptide binders to a given protein surface, HPer focuses on searching for helix binding sites on the interfaces of the targeted PPIs. The predicted binding sites provided the initial backbone positions for the subsequent sequence de novo design of the helical peptide binders. With the cases we collected in the DSHP dataset, we have extracted the structural and property features of single α-helix-mediated protein-protein complex structures, which were used as guidelines for ranking the designed helical peptide ligands.
We demonstrated that HPer recaptured the positions of α-helix ligands and predicted interacting profiles in the DSHP dataset. Using the predicted positions, we carried out sequence design for eight PPIs in the DSHP dataset and recovered native hot-spot residues in most cases. We further predicted potential helical peptide binding sites in the PPIs from the PDBbind database and identified 17 preferable helical peptide binding sites in non-helix-mediated PPIs. Many of the 17 examples of PPIs are important therapeutic targets that can be explored further in future studies. We used EGFR as case study and designed a novel helical peptide, EGFR-Pep1, for the EGFR-EGF interface, which might have the potential as a novel EGFR inhibitor.
In conclusion, HPer performed well in searching for helical peptide binding sites for PPI targets and provided good initial structural models for novel helical peptides and their mimetic design. The DSHP dataset we have compiled should be useful for testing methodologies of computational protein-protein interaction design and for understanding principles of helical peptide and protein recognition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported in part by the Ministry of Science and Technology of the People’s Republic of China (2015CB910300, 2016YFA0502303) and the National Natural Science Foundation of China (21633001, 8200905085). The computational work was carried out on the High Performance Computing Platform of Peking-Tshinghua Center for Life Sciences at Peking University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.11.001.
Contributor Information
Changsheng Zhang, Email: changshengzhang@pku.edu.cn.
Luhua Lai, Email: lhlai@pku.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Availability
The source code, executives, and examples are available at https://github.com/proteincraft/HPer.
References
- 1.Jin L.Y., Wang W.R., Fang G.W. Targeting protein-protein interaction by small molecules. Annu Rev Pharmacol Toxicol. 2014;54(54):435–456. doi: 10.1146/annurev-pharmtox-011613-140028. [DOI] [PubMed] [Google Scholar]
- 2.Bruzzoni-Giovanelli H. Interfering peptides targeting protein-protein interactions: the next generation of drugs? Drug Discovery Today. 2018;23(2):272–285. doi: 10.1016/j.drudis.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Jochim A.L., Arora P.S. Assessment of helical interfaces in protein-protein interactions. Mol BioSyst. 2009;5(9):924–926. doi: 10.1039/b903202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guharoy M., Chakrabarti P. Secondary structure based analysis and classification of biological interfaces: identification of binding motifs in protein-protein interactions. Bioinformatics. 2007;23(15):1909–1918. doi: 10.1093/bioinformatics/btm274. [DOI] [PubMed] [Google Scholar]
- 5.Bullock B.N., Jochim A.L., Arora P.S. Assessing helical protein interfaces for inhibitor design. J Am Chem Soc. 2011;133(36):14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleishman S.J. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332(6031):816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva D.A. De novo design of potent and selective mimics of IL-2 and IL-15. Nature. 2019;565(7738):186–191. doi: 10.1038/s41586-018-0830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevalier A. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550(7674):74–79. doi: 10.1038/nature23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C.S. Computational design of helical peptides targeting TNF alpha. Angew Chem-Int Ed. 2013;52(42):11059–11062. doi: 10.1002/anie.201305963. [DOI] [PubMed] [Google Scholar]
- 10.Siegert T.R. Analysis of loops that mediate protein-protein interactions and translation into submicromolar inhibitors. J Am Chem Soc. 2016;138(39):12876–12884. doi: 10.1021/jacs.6b05656. [DOI] [PubMed] [Google Scholar]
- 11.Watkins A.M., Arora P.S. Anatomy of beta-strands at protein-protein interfaces. ACS Chem Biol. 2014;9(8):1747–1754. doi: 10.1021/cb500241y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sammond D.W. Computational design of the sequence and structure of a protein-binding peptide. J Am Chem Soc. 2011;133(12):4190–4192. doi: 10.1021/ja110296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W. Computational design and optimization of novel d-peptide TNFalpha inhibitors. FEBS Lett. 2019;593(12):1292–1302. doi: 10.1002/1873-3468.13444. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z.H. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 2015;31(3):405–412. doi: 10.1093/bioinformatics/btu626. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch W., Sander C. Dictionary of protein secondary structure - pattern-recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 16.Fleishman S.J. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph Model. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 18.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013;29(7):845–54. [DOI] [PMC free article] [PubMed]
- 19.Dawson N.L. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45(D1):D289–D295. doi: 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansari S, Helms V. Statistical analysis of predominantly transient protein-protein interfaces. Proteins 2005;61(2):344–55. [DOI] [PubMed]
- 21.Chen J.M., Sawyer N., Regan L. Protein-protein interactions: general trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22(4):510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuchiya Y., Kinoshita K., Nakamura H. Analyses of homo-oligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng Des Sel. 2006;19(9):421–429. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda D., Gray J.J. Shape complementarity and hydrogen bond preferences in protein-protein interfaces: implications for antibody modeling and protein-protein docking. Bioinformatics. 2016;32(16):2451–2456. doi: 10.1093/bioinformatics/btw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindemann S.R., Yershova A., LaValle S.M. Incremental grid sampling strategies in robotics. Algorithmic Found Rob VI. 2005;17:313–328. [Google Scholar]
- 25.Veselovsky A.V. Protein-protein interactions: mechanisms and modification by drugs. J Mol Recognit. 2002;15(6):405–422. doi: 10.1002/jmr.597. [DOI] [PubMed] [Google Scholar]
- 26.Hubbard SJ, Thornton JM. NACCESS. Computer Program, Department of Biochemistry and Molecular Biology, University College London, 1993;2(1).
- 27.Janin J., Bahadur R.P., Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41(2):133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 28.Liu S. Nonnatural protein-protein interaction-pair design by key residues grafting. Proc Natl Acad Sci U S A. 2007;104(13):5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis S.M., Kuhlman B.A. Anchored design of protein-protein interfaces. PLoS ONE. 2011;6(6) doi: 10.1371/journal.pone.0020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melo R. A machine learning approach for hot-spot detection at protein-protein interfaces. Int J Mol Sci. 2016;17(8) doi: 10.3390/ijms17081215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreira I.S. SpotOn: high accuracy identification of protein-protein interface hot-spots. Sci Rep. 2017:7. doi: 10.1038/s41598-017-08321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning M.C., Illangasekare M., Woody R.W. Circular-dichroism studies of distorted alpha-helices, twisted beta-sheets, and beta-turns. Biophys Chem. 1988;31(1–2):77–86. doi: 10.1016/0301-4622(88)80011-5. [DOI] [PubMed] [Google Scholar]
- 33.Ward C.W. The insulin and EGF receptor structures: new insights into ligand-induced receptor activation. Trends Biochem Sci. 2007;32(3):129–137. doi: 10.1016/j.tibs.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Martinelli E. Anti-epidermal growth factor receptor monoclonal antibodies in cancer therapy. Clin Exp Immunol. 2009;158(1):1–9. doi: 10.1111/j.1365-2249.2009.03992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciardello F., Tortora G. EGFR antagonists in cancer treatment (vol 358, pg 1160, 2008) N Engl J Med. 2009;360(15):1579. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 36.Seshacharyulu P. Targeting the EGFR signaling pathway in cancer therapy. Exp Opin Ther Targets. 2012;16(1):15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrett T.P. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110(6):763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 38.Sedan Y. Peptiderive server: derive peptide inhibitors from protein-protein interactions. Nucleic Acids Res. 2016;44(W1):W536–W541. doi: 10.1093/nar/gkw385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obarska-Kosinska A., et al. PepComposer: computational design of peptides binding to a given protein surface. Nucleic Acids Res 2016;44(W1):W522–28. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code, executives, and examples are available at https://github.com/proteincraft/HPer.