Abstract
Neurodegenerative disorders progress across the brain in characteristic spatio-temporal patterns. A better understanding of the factors underlying the specific cell and tissue vulnerability responsible for such patterns could help identify the molecular origins of these conditions. To investigate these factors, based on the observation that neurodegenerative disorders are closely associated with the presence of aberrant protein deposits, we made the hypothesis that the vulnerability of cells and tissues is associated to the overall levels of supersaturated proteins, which are those most metastable against aggregation. By analyzing single-cell transcriptomic and subcellular proteomics data on healthy brains of ages much younger than those typical of disease onset, we found that the most supersaturated proteins are enriched in cells and tissues that succumb first to neurodegeneration. Then, by focusing the analysis on a metastable subproteome specific to Alzheimer's disease, we show that it is possible to recapitulate the pattern of disease progression using data from healthy brains. We found that this metastable subproteome is significantly enriched for synaptic processes and mitochondrial energy metabolism, thus rendering the synaptic environment dangerous for aggregation. The present identification of protein supersaturation as a signature of cell and tissue vulnerability in neurodegenerative disorders could facilitate the search for effective treatments by providing clearer points of intervention.
Keywords: Biophysics, Neuroscience, Alzheimer's disease, Protein supersaturation, Neuronal vulnerability, Protein homeostasis, Neurodegenerative diseases
Biophysics; Neuroscience; Alzheimer's disease; Protein supersaturation; Neuronal vulnerability; Protein homeostasis; Neurodegenerative diseases
1. Introduction
Alzheimer's disease (AD); the primary cause of dementia, affects over 50 million people worldwide (De Strooper and Karran, 2016; Holtzman et al., 2011; Knowles et al., 2014; Selkoe and Hardy, 2016). The nature and the origins of this disease are still not known with certainty, as its onset and progression have been associated with a variety of factors, including mitochondrial dysfunction, disruption of the endoplasmic reticulum and membrane trafficking, disturbances in protein folding and clearance, and the activation of the inflammatory response (De Strooper and Karran, 2016; Holtzman et al., 2011; Knowles et al., 2014; Selkoe and Hardy, 2016). Despite this complexity, at the histopathological level AD is clearly associated with a characteristic pattern of progression across the brain, known as Braak staging (Bero et al., 2011; Braak and Braak, 1991; Mattson and Magnus, 2006; Miller et al., 2013). AD is hallmarked by aberrant deposits, known as amyloid plaques and neurofibrillary tangles, which are primarily composed of the amyloid beta peptide (Aβ) and the tau protein, respectively (De Strooper and Karran, 2016; Holtzman et al., 2011; Knowles et al., 2014; Selkoe and Hardy, 2016). The formation of these deposits has been implicated with the cellular dysregulation of protein homeostasis within affected tissues (Balch et al., 2008; Brehme et al., 2014; Gidalevitz et al., 2006; Hartl et al., 2011; Labbadia and Morimoto, 2015).
The specific staging of the disease and the presence of protein aggregates are characteristic not just of AD, but also of a wider range of neurodegenerative disorders, including Parkinson's disease (PD), frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (Balch et al., 2008; Brehme et al., 2014; Eisenberg and Jucker, 2012; Hartl et al., 2011; Knowles et al., 2014; Labbadia and Morimoto, 2015). Therefore, by investigating the relationship between tissue vulnerability and the phenomenon of protein aggregation one could hope to obtain insights into the molecular origins of these diseases. We have recently shown that tissues vulnerable to AD can be characterised by the expression levels of a set of proteins comprising those that co-aggregate to form plaques and tangles, and those that modulate the aggregation of Aβ and tau (Freer et al., 2016; Fu et al., 2019). Together, the expression levels of these proteins recapitulate the specific pattern of disease progression in AD (Freer et al., 2016).
Given that these results implicate dysregulation of protein aggregation in tissue vulnerability to AD, here we ask whether there are other proteins that place a burden on the protein homeostasis system. We focused attention on supersaturated proteins: proteins that are poorly soluble at the concentrations at which they are expressed. We posited that the presence of a specific subset of supersaturated proteins may impact the capacity of the protein homeostasis system to control aggregation, thus favoring the creation of an environment vulnerable to disease-specific aggregation. To identify this set of proteins, we began by investigating the role played by supersaturation in defining an environment vulnerable to the inception of neurodegenerative disease. In alignment with a role in vulnerability to aggregation, highly supersaturated proteins were found to be enriched in plaques and tangles, and in the aggregates found in ageing worms (Ciryam et al., 2013, 2015).
By exploiting recent advances in single-cell transcriptomics (Darmanis et al., 2015) and in subcellular proteomics (Wilhelm et al., 2014), the results that we report here indicate that cell and tissue vulnerability to neurodegeneration is associated with the elevated expression of a common set of highly supersaturated proteins. We further show that in AD, Braak staging is associated with the elevated expression of a set of metastable subproteome specific to Alzheimer's, which is composed of proteins that are supersaturated and downregulated in the disease. To explain why brain tissues, and neurons in particular, are so vulnerable to protein aggregation, we show that the metastable proteins are closely involved in synaptic processes, resulting in a vulnerability of the synaptic environment to aberrant protein aggregation.
2. Results
2.1. Superaturated proteins are enriched in cells and tissues vulnerable to neurodegenerative processes
In order to investigate the link between protein aggregation and neurodegenerative disorders, we first asked whether the vulnerability to neurodegenerative processes can be associated with a sustained burden on the protein homeostasis system (Balch et al., 2008; Brehme et al., 2014; Hartl et al., 2011; Knowles et al., 2014), resulting from a presence above average of aggregation-prone proteins (Gidalevitz et al., 2006; Labbadia and Morimoto, 2015; Olzscha et al., 2011; Vendruscolo, 2012; Walther et al., 2015).
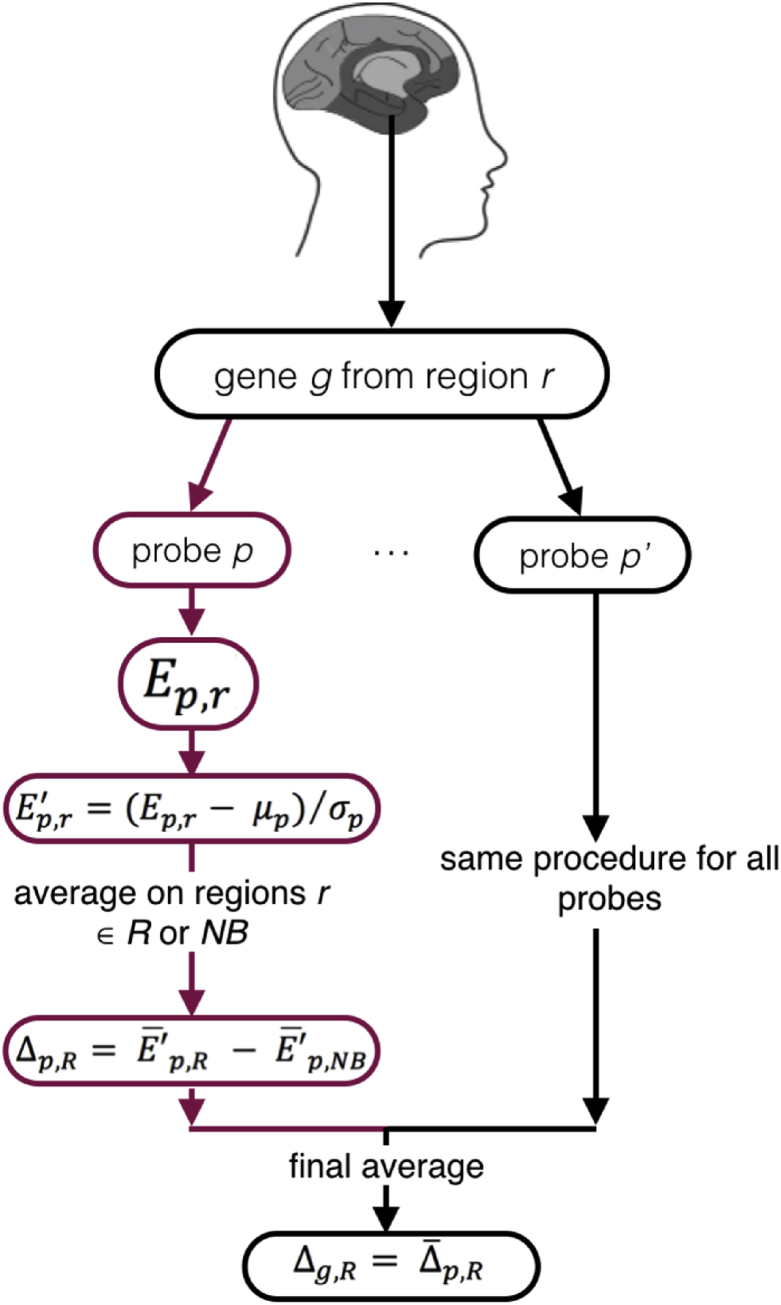
Following these considerations, we hypothesised that supersaturated proteins, which are expressed beyond their solubility limits, may have a role in tissue vulnerability to aberrant aggregation in neurodegenerative processes. We thus sought to establish whether such proteins may be found at elevated levels in those brain tissues most vulnerable to neurodegenerative processes in samples obtained from healthy brains much before the typical onset of disease. To address this question we utilised data from the Allen Brain Atlas and the Δ score method (Fig. 1) to quantify differential expression (Freer et al., 2016). The Δ scores represent the expression levels in vulnerable tissues relative to those in resistant tissues (see Experimental Procedures and Table S1 for a list of disease-specific vulnerable regions).
Fig. 1.
Schematic illustration of the calculation process to obtain the Δ scores. Multiple probes are used to measure the expression of each gene. For each probe p for a gene g expression is measured in each region r in the brain. Expression values for each probe are normalised across all tissues by subtracting the average value of the expression and by dividing the result by the standard deviation of the expression (Eq. 1). This step is carried out to enable the combination of expression readings from different probes. The Δ score of a probe (Δp) is calculated by subtracting the mean expression in non-Braak regions (NB) from the mean expression in a regional subset (R) of interest (Eq. 2). Finally, a Δ score for a gene (Δg) is calculated by averaging Δ scores from the corresponding probes (Eq. 5). We verified over a total of 18,918 genes that two probes measuring the same gene were highly correlated (coefficient of correlation R ≈ 0.7).
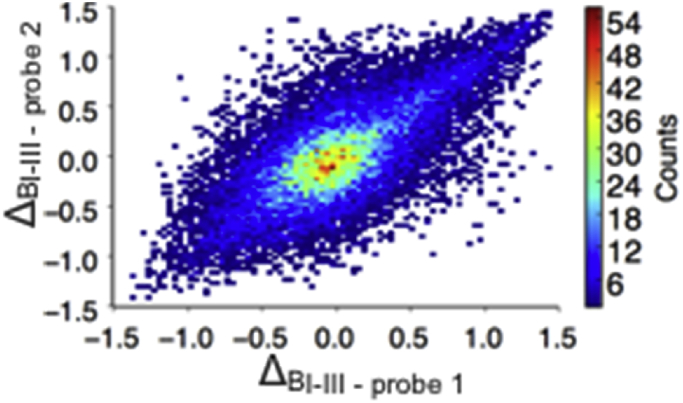
For this analysis we used mRNA expression data to estimate protein levels because, despite recent advances (Uhlén et al., 2015), systematic data concerning protein levels across different brain regions at high level of granularity are not yet available. Although it is well known that at the level of individual genes the correlation between mRNA expression levels and protein abundance levels is typically weak, here we are interested in averages over groups of genes, for which the correlation with the corresponding groups of proteins can be much stronger (Fig. 2). The Allen Brain Atlas is an online repository of microarray data for six normal human brains of ages 24–56 (Hawrylycz et al., 2012). Over 500 measurements are taken for each individual, with 93% of known genes represented by at least two probes. To evaluate the extent of noise in this dataset, we calculated the correlation between gene differential expression measured using different probes, as quantified by the Δ score (see Experimental Procedures), finding a good agreement between Δ scores for two probes measuring the same gene, with a Pearson coefficient of correlation of 0.7 (Fig. 3). To estimate the supersaturation levels, we used the measured expression levels of the genes relative to a computationally-derived aggregation propensity score as previously (Ciryam et al., 2013). For the analysis that follows we refer to supersaturated proteins as those in the top 5% of the distribution of supersaturation scores (Table S2).
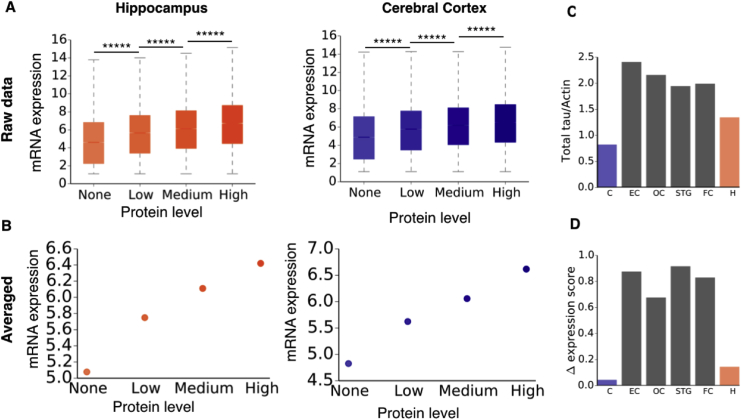
Fig. 2.
Analysis of the average correlation between mRNA and protein levels. (A) Correlation between mRNA and protein levels for two representative regions of the brain (Uhlén et al., 2015). (B) Correlation between mRNA and protein levels, averaged data. Error bars are smaller than the data point symbols. The correlation coefficients between mRNA levels and protein levels for hippocampal and cerebral cortex tissues are 0.98 (p = 0.02) in both cases. Protein expression levels are derived from immunohistochemical data scored according to staining intensity (negative, weak, moderate or strong), and fraction of stained cells (low (<25%), medium (25–75%) or high (>75%)). Each combination of intensity and fractions is converted into a protein expression level score as follows: negative – not detected; weak/low – not detected; weak/medium or weak/high – low; moderate/low – low; moderate/medium or moderate/high – medium; strong/low – medium, strong/medium or strong/high – high. (C) Measured levels of total tau protein (Čaušević et al., 2010), normalised to actin levels. The expression score for each region is relative to non-Braak tissues. C: cerebellum; EC: entorhinal cortex; OC: occipital cortex; STG: superior temporal gyrus; FC: frontal cortex and H: hippocampus. (D) Relative mRNA levels for the MAPT gene, for corresponding brain regions. The correlation coefficient between relative MAPT mRNA levels and relative tau protein levels is 0.9 (p = 0.01). Boxes represent the first and third quartiles of the distribution, whiskers the 1.5 inter-quartile range, and notches are standard errors on the median calculated with 100 bootstrap cycles. *****p < 0.00001, calculated with a Mann-Whitney U test with Benjamini-Hochberg multiple hypothesis testing correction (Benjamini and Hochberg, 1995). Protein expression scoring in (A) and (B) are based is based on immunohistochemical data manually scored with regard to staining intensity (negative, weak, moderate or strong), and fraction of stained cells (Uhlén et al., 2015).
Fig. 3.
Test of the level of noise in the Allen Brain Atlas dataset. The correlation between Δ scores for two probes measuring the same gene was tested, resulting in a coefficient of correlation of 0.7.
We first investigated the role played by these supersaturated proteins in defining the tissues most vulnerable to neurodegenerative processes, for example certain regions of the hippocampus in AD (Table S1). By carrying out a Δ score analysis (Table S3), we found that in those tissues most vulnerable to AD, ALS, FTD, and PD (Table S1), the top 5% supersaturated proteins (Table S2) are on average a significantly more expressed than in resistant tissues (Fig. 4).
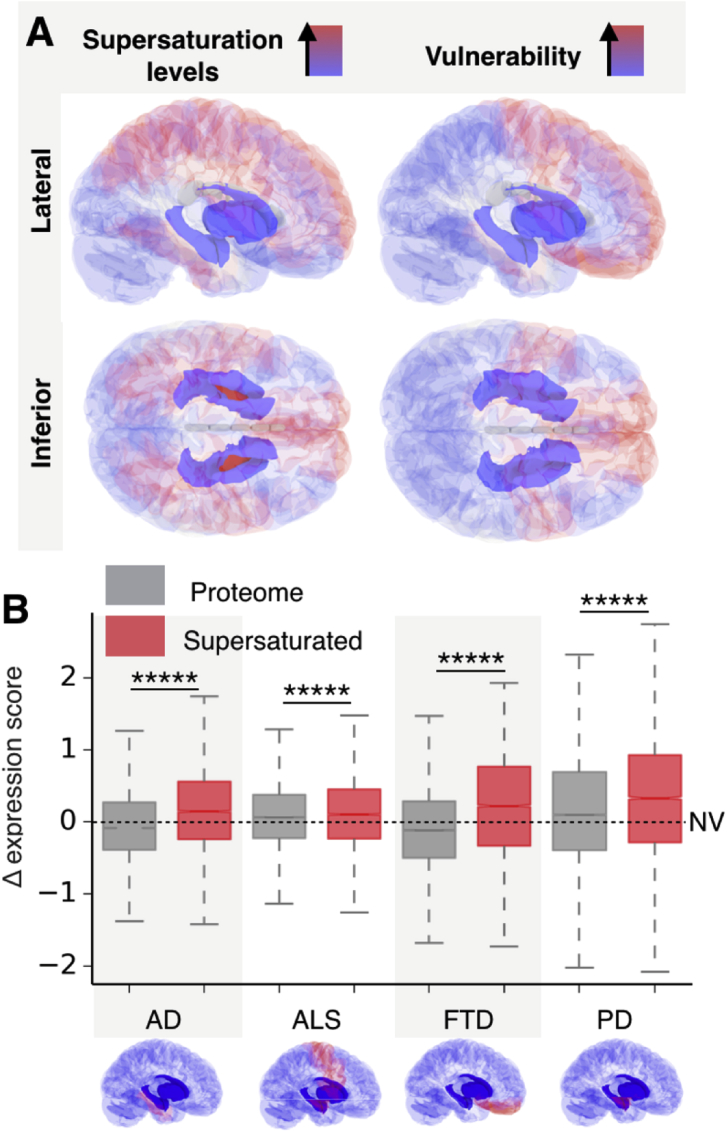
Fig. 4.
In healthy brains, supersaturated proteins are highly expressed in tissues and cells vulnerable to neurodegenerative processes. (A) Red regions in the brain on the left indicate an elevated expression of supersaturated proteins relative to the proteome, while blue regions indicate an average expression below that of the proteome as a whole. Red regions in the brain on the right indicate the tissues most vulnerable to neurodegenerative processes associated with the early stages of AD, PD, FTD and ALS (see panel B and Table S1). (B) Box plot of Δ scores of the top 5% of supersaturated proteins (red) compared to the whole proteome (grey) calculated for AD, PD, ALS and FTD (x-axis). For each disease, Δ scores represent expression in vulnerable tissues, relative to expression in resistant tissues (see Experimental Procedures). The Δ scores reported for PD, ALS and FTD are calculated with the same procedure described in Experimental Procedures for AD, using the disease-specific vulnerable regions defined in Table S1. *****p < 0.00001; the statistical significance of the differences between the distributions of supersaturated proteins and that of the proteome was calculated with a t-test. In addition, a one tailed t-test was used to test that the distribution of relative expression scores for supersaturated proteins is greater than zero. This test yielded a p value of less than 0.00001 for tissues vulnerable to AD, PD, ALS and FTD. The dashed line at Δ = 0 (NV) indicates non-vulnerable tissues.
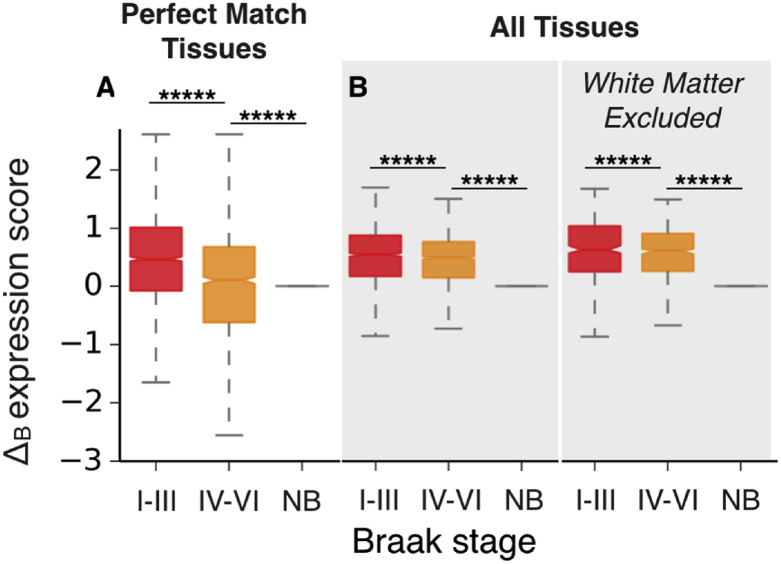
Overall, we found a broad agreement between the regional vulnerability to neurodegenerative processes (Table S1) and the tissue-specific expression levels of the genes encoding for supersaturated proteins (Fig. 4A). The underlying data plotted for individual diseases highlight that high differential expression is observed for the supersaturated proteins, but not for the proteome as a whole (Fig. 4B). In the case of AD, we further verified that these results are not significantly affected by the imperfect mapping between Braak regions and Allen Brain Atlas regions (see Experimental Procedures) and by an under-representation of white matter tissue (see Experimental Procedures) in the datasets that we used (Fig. 5).
Fig. 5.
Distributions of ΔB scores for tissues of consecutive Braak stages. (A) Boxplots of the ΔB scores for the metastable subproteome calculated for ‘perfect match’ tissues (see Table S8) progressively affected by AD (x-axis). (B) Same data as for panel A, but for all tissues. A comparison of ΔB score distributions is presented with white matter data included (left) and excluded from analysis (right). For each boxplot, the significance of the difference between ΔB distributions for Braak I-III and Braak IV-VI datasets, and for Braak IV-VI and non- Braak (NB) datasets was calculated with a Mann-Whitney U test and Benjamini-Hochberg multiple hypothesis testing correction (Benjamini and Hochberg, 1995) *****p < 0.00001.
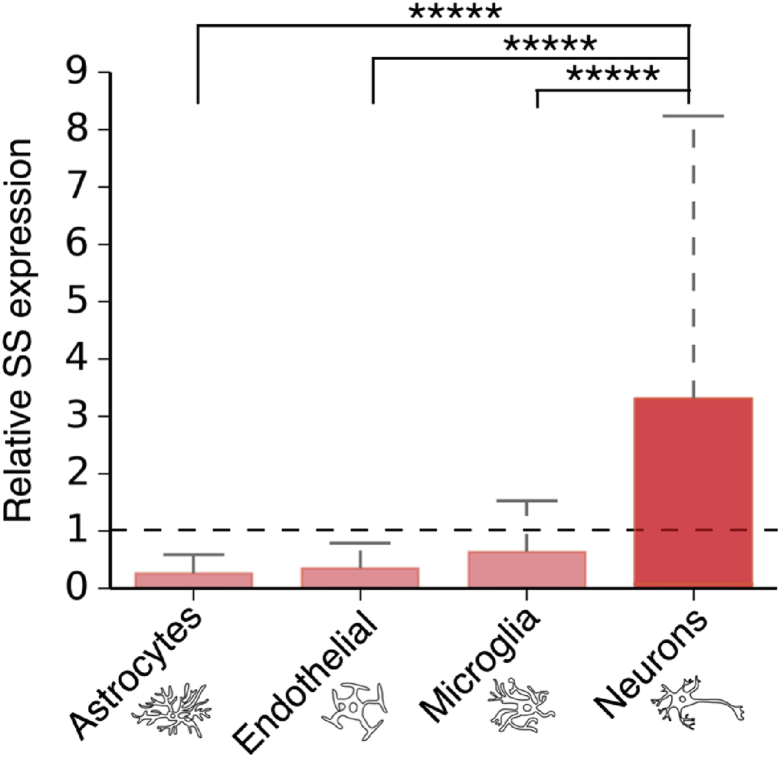
The levels of expression of supersaturated proteins are not only closely linked to vulnerability at the tissue level, but also at the cell-type level. From an analysis of single cell human RNAseq data (see Experimental Procedures), we found that neurons have levels of supersaturated proteins that are significantly more elevated than those observed in other brain cell types (Fig. 6). Thus, building on previous reports that supersaturated proteins are enriched in the characteristic aggregates observed in neurodegenerative diseases (Ciryam et al., 2015), these results demonstrate that supersaturated proteins preferentially localise to tissues and cell types most vulnerable in disease.
Fig. 6.
Relative expression of supersaturated proteins in different cell types. Expression analysis was carried out on human single cell RNAseq data (Darmanis et al., 2015). For further details on analysis method see Experimental Procedures. The p-value describes the significance of expression in neurons, relative to that in other brain cell types plotted, calculated with the Mann-Whitney U test with Benjamini-Hochberg multiple hypothesis testing correction (Benjamini and Hochberg, 1995) *****p < 0.0005.
2.2. A metastable subproteome specific to AD predicts tissue and cell vulnerability to AD
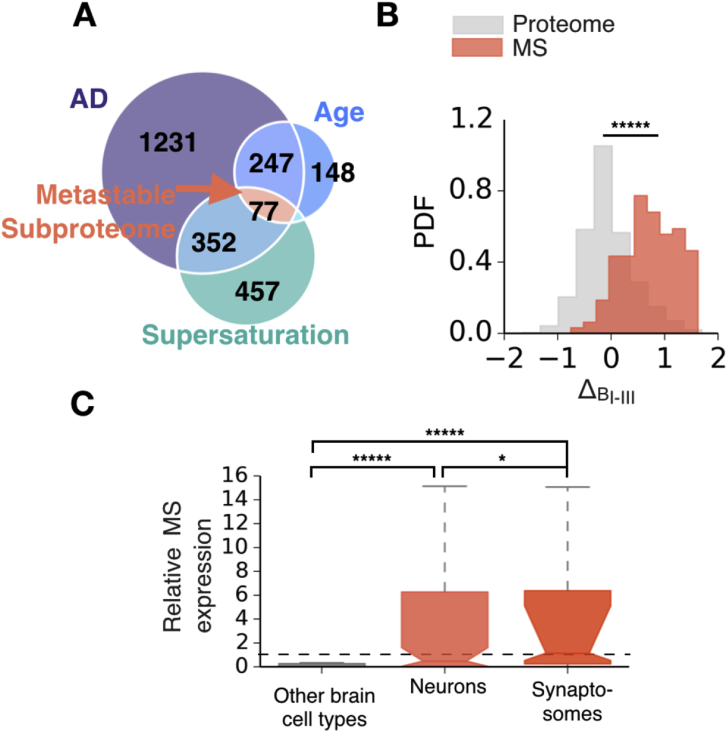
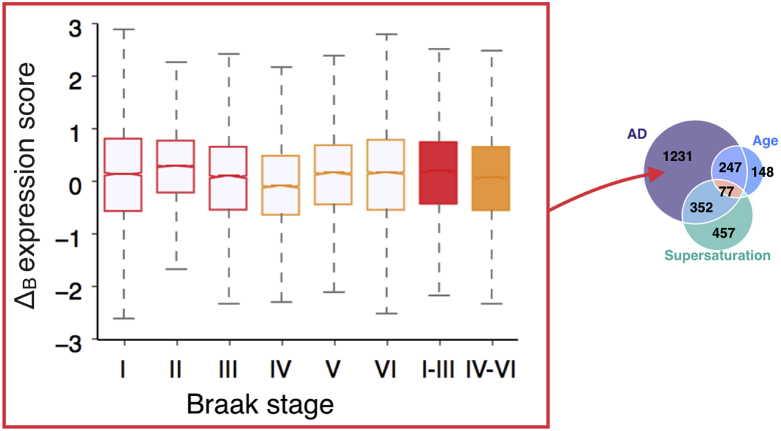
Having found an association between the regional expression levels of the most supersaturated proteins and tissue vulnerability to neurodegenerative processes, we next sought to assess the role of a metastable subproteome specific to AD (Ciryam et al., 2016). We predicted the existence of a subproteome metastable against aggregation, with potential to create an environment conducive to protein aggregation characteristic to AD. We defined this metastable subproteome as the intersection between highly supersaturated proteins, proteins downregulated in ageing, and proteins downregulated in disease (Ciryam et al., 2016); it contains 77 proteins (Fig. 7A and Table S2). The progression of AD across brain regions was determined using the Braak staging (Braak and Braak, 1991), which relies on the histopathological examination of the distribution of tangles in the brain during subsequent stages of AD, which is known to correlate with neuronal atrophy (Whitwell et al., 2008).
Fig. 7.
The expression of a metastable subproteome specific to AD is elevated in cell types and tissues vulnerable to AD. (A) The AD-specific metastable subproteome is defined as the intersection of three sets of proteins; those downregulated with age, downregulated in AD, and highly supersaturated (B) Comparison of the distributions of the Δ scores of genes comprising the metastable subproteome (MS, red) and the whole proteome (grey) (C) Expression of the metastable subproteome relative to the expression of the proteome in brain cell types and synaptosomes. ‘Other cell types’ include astrocytes, brain-derived endothelial cells, and microglia. For further details on analysis method see Experimental Procedures. *p < 0.01, *****p < 0.00001, Mann Whitney U-test (Benjamini and Hochberg, 1995).
Using the Δ score analysis, we found that this metastable subproteome has significantly elevated expression in the tissues most vulnerable to AD (tissues of Braak stages I-III) with respect to the other tissues (Fig. 7B and Tables S4 and S5). These results extend those that we previously reported, where we found that the specific set of proteins found in plaques and tangles in AD brains are also supersaturated (Ciryam et al., 2013). We established that these ‘co-aggregators’ had a significantly elevated expression in early AD tissues, however they could not predict differential vulnerability between tissues (Freer et al., 2016).
We next sought to use relative metastable subproteome expression to assess vulnerability to AD for different cell types. By calculating the relative expression of the metastable subproteome for different brain cells, utilising human single cell RNAseq data (Darmanis et al., 2015), we found that neurons have significantly higher levels of metastable-subproteome proteins than astrocytes, microglia and brain-derived endothelial cells (Fig. 7C).
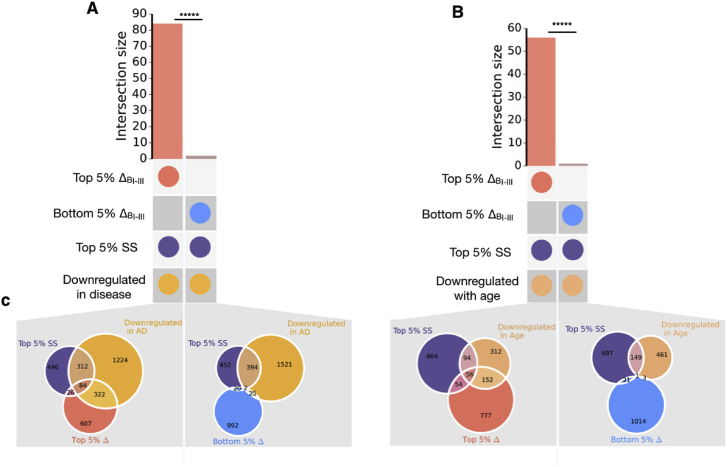
To test the robustness of our definition of metastable subproteome, we considered a range of alternative definitions corresponding to different overlaps between highly supersaturated proteins and proteins corresponding to genes with high relative expression in Braak I-III regions. We confirmed that a significant number of genes that intersect the top 5% of these two subsets, are also downregulated in AD and upon ageing. In contrast, significantly fewer of those genes in the intersection between the bottom 5% of ΔBI-III scores and the supersaturated proteins are downregulated in ageing and disease (Fig. 8 and Table S2). These results are fully consistent with those discussed above, and suggest that by analysing two properties that can be measured in normal brains (supersaturation and expression levels in early Braak regions), one can make predictions about tissue vulnerability to AD.
Fig. 8.
Test of the robustness of the results against the definition of the metastable subproteome. (a) Difference in overlap size between proteins downregulated in disease and supersaturated, and proteome members with top and bottom 5% ΔBI-III scores. (b) Difference in overlap size between proteins downregulated in ageing and supersaturated, and proteome members with top and bottom 5% ΔBI-III scores. (c) Sizes of all intersections related to panels (a) and (b). *****p < 0.0001, Fisher exact test performed for panels (a) and (b).
Taken together, the results that we have presented in this section indicate that tissue and cell vulnerability to AD is associated with the elevated expression of a metastable subproteome specific to AD. These results are consistent with previous single-nucleus RNA-seq analysis demonstrating that this metastable subproteome has significantly elevated expression in a neuronal subset with high relative vulnerability to degeneration in AD (Fu et al., 2019).
2.3. The synaptic environment is highly vulnerable to protein aggregation
To investigate vulnerability to neurodegenerative processes at a level of granularity more precise than tissue classification, we next focused on the expression signatures of cell types and subcellular localisations. Exploiting recent advances in proteomics, which enabled the systematic quantification of protein levels in synaptosomes (Wilhelm et al., 2014), we investigated whether our findings can also help rationalize why neuronal synapses succumb particularly early in neurodegerative diseases (Bero et al., 2011; De Strooper and Karran, 2016; Muratore et al., 2017).
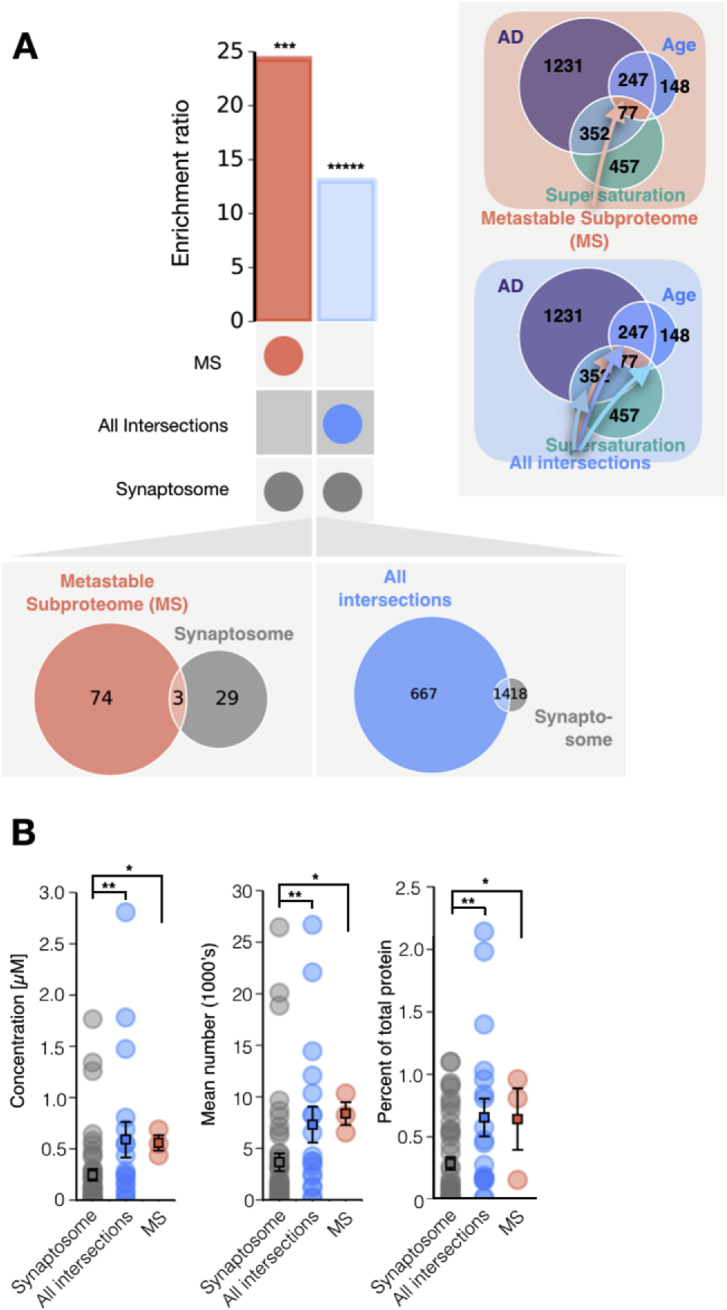
We found that both supersaturated proteins and the metastable subproteome have elevated expression in neurons relative to other cell types (Figs. 2C and 6), and the metastable subproteome is significantly enriched in the synaptosome relative to whole neurons (Fig. 7C). Robustness tests confirm the enrichment of synaptosomal proteins in the metastable subproteome, relative to the proteome as a whole (Fig. 9A). To further investigate these results, we identified a subset of proteins that we predicted to be more metastable than the proteome as a whole, but less aggregation prone than the metastable subproteome. We termed the members of this subset ‘intersectional proteins’. Intersectional proteins are defined as those that belong to at least two groups among supersaturated, downregulated in AD, and downregulated in ageing, but are not part of the metastable subproteome. We found that intersectional proteins have a significantly elevated enrichment for synapsosomal localisation, but with an enrichment ratio smaller than that of the metastable proteome (Fig. 9A).
Fig. 9.
Metastable subproteome proteins are significantly more abundant at the synapse than intersectional proteins and, more generally, the synaptosome. (A) Enrichment of the metastable subproteome (MS, left), and intersectional proteins (right), with respect to the most abundant synaptosomal proteins. Intersectional proteins are defined as those that belong to at least two groups among supersaturated, downregulated in AD, and downregulated in ageing, but are not part of the metastable subproteome. (B) Concentration, copy number, and percentage for the metastable subproteome (red), intersectional proteins (blue), and all proteins measured in a synaptic buton (grey). ***p < 0.01 and *****p < 0.0001, Fisher exact test performed for panel A; *p < 0.05, **p < 0.01 Mann-Whitney U test performed for panel B (Benjamini and Hochberg, 1995).
Having established an enriched presence of metastable proteins in the synaptosome set, we next sought to understand the abundance of metastable subproteome components in the synapse, by analysing mass-spectrometry data from isolated synaptic derivatives (Wilhelm et al., 2014). We consistently found that the metastable subproteome is highly abundant in the synaptosome for all metrics tested (Fig. 9B).
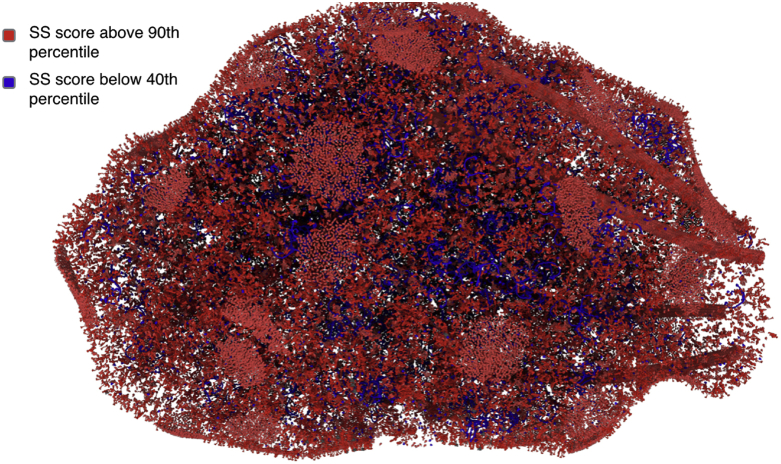
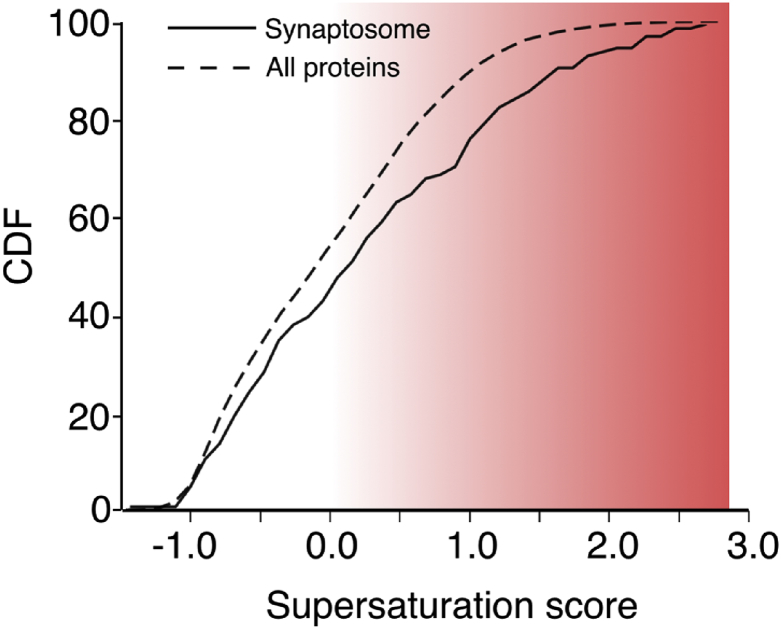
The prevalence of supersaturated proteins in the synaptic environment is particularly well illustrated by comparing the supersaturation of the synaptosome relative to the proteome (Figs. 10 and 11), and the abundance and localization of the synaptic proteins that are most supersaturated, including synaptotagmin, snap25, AP180 and tangle co-aggregator Hsc70 (Wang et al., 2005) (Fig. 10 and Movie S1). It is remarkable that supersaturated proteins are enriched at synapses relative to whole neurons, given that early synaptic degeneration has been observed in AD (Coleman et al., 2004; Selkoe, 2002).
Fig. 10.
Section a synaptic buton illustrating the high protein supersaturation levels in this environment. A supersaturation score quantifies the expression of a protein relative to its solubility limit (Ciryam et al., 2013). Proteins in red have supersaturation scores above the 90th percentile for the proteome as a whole. Proteins in blue have scores below the 40th percentile. Details on the proteomic contents of the synaptic buton are taken from a previous study (Wilhelm et al., 2014). Details on the figure configuration are given in Experimental Procedures.
Fig. 11.
The synaptic environment is enriched in aggregation-prone proteins. Cumulative distribution function (CDF) of supersaturation scores of the synaptosome and the proteome. The statistical significance of the difference between the mean values of the distributions is evaluated using a t-test, p = 1 × 10−18. Details on the proteomic contents of the synaptic buton are taken from a previous study (Wilhelm et al., 2014).
Overall, the results that we presented in this section indicate that the vulnerability of specific cell types and subcellular localisations to AD is correlated with the elevated expression at neuronal synapses of a common set of highly supersaturated proteins, which places the environment at risk to protein aggregation.
2.4. The metastable subproteome specific to AD is primarily involved in synaptic processes
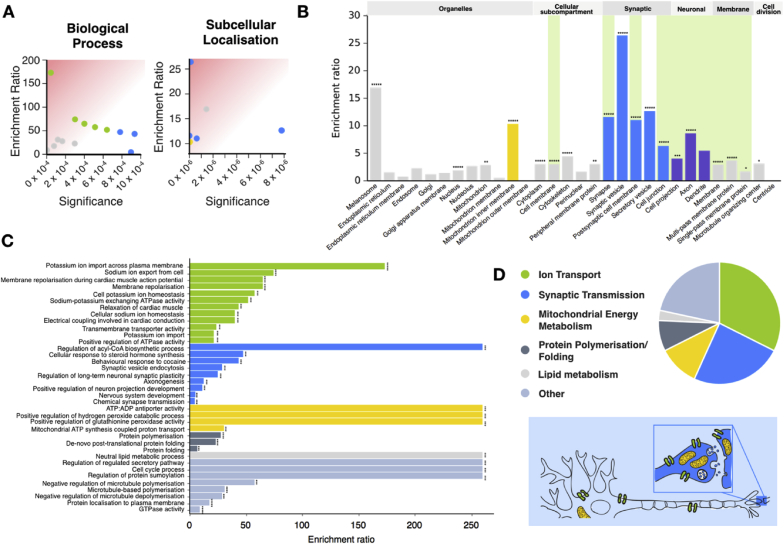
We next sought to understand the physiological role of the metastable subproteome specific to AD. Using enrichment analysis of biological processes (see Experimental Procedures), we found that the metastable subproteome plays a key role in biochemical pathways associated with synaptic functions, including ion transport, and synaptic transmission, as well as more generally with mitochondrial energy metabolism (Fig. 12 and Tables S6 and S7). Enrichment ratios were calculated as the ratio between observed and expected values for each category (see Experimental Procedures). Then, using enrichment analysis of subcellular localisations we found that the metastable subproteome is particularly highly expressed at synapses and in proximity of lipid membranes of neurons (Fig. 12 and Tables S6 and S7). These results complement those reported in Fig. 10, as they show that the synaptic environment is enriched in aggregation-prone proteins due to its physiological functions.
Fig. 12.
Synaptic processes and mitochondrial energy metabolism are enriched in metastable proteins. (A) Metastable subproteome enrichment ratio plotted against enrichment significance (p-value) for biological processes (left) and subcellular localisations (right). Point colours indicate the categories of the different processes, as in the legend of panel D (see also Tables S6–S7). (B) Metastable subproteome enrichment in specific subcellular localisations, categorized as in (A). Green shading coincides with subcellular localisations relevant to ion transport. (C) Enrichment ratio for the most significantly enriched biological processes (D) Chart showing the relative number of significantly enriched biological process categories, organized into the broad classifications indicated. *p < 0.05, **p < 0.01, ***p < 0.01, ****p < 0.001 and *****p < 0.0001 one tailed Fischer exact test and Bonferroni multiple hypothesis correction (Benjamini and Hochberg, 1995).
The results in this section thus indicate that the metastable proteins at risk of aggregation in AD are particularly involved in synaptic processes, which provides insights into the molecular origins of synaptic loss in AD. We also note that although the contributions of mitochondria to neurodegeneration has long been recognised, the precise mechanism of pathogenesis remains unclear (Gillingwater and Wishart, 2013; Wishart et al., 2006). Our results indicate that elevated levels of mitochondrial proteins can contribute to the metastability of a protein environment, conferring synaptic vulnerability independent to mitochondrial function.
2.5. Metastable subproteome levels in healthy brain tissues predict the progression of AD
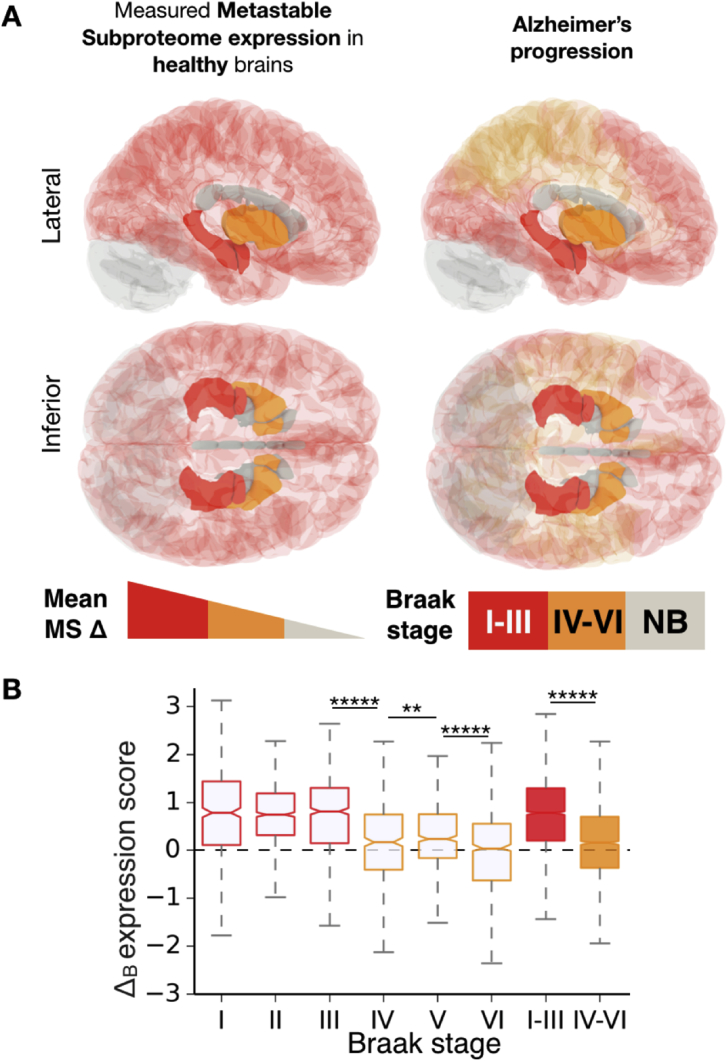
Finally, we sought to assess whether expression levels of the metastable subproteome in normal brains, decades before ages typical of disease onset, could predict the progression of AD. We posited that this relationship would be observed due to a similar mechanism to that we previously reported for the expression levels of components of the proteostasis system that regulates the amyloid beta peptide and tau (Freer et al., 2016). Using the Δ score analysis, we found that the most vulnerable tissues to AD (i.e. the early Braak tissues) have significantly elevated relative expression levels of the proteins in the metastable subproteome (Fig. 13). In addition, we found that the relative expression is lower in tissues with progressively more AD resistance, which succumb at later stages of the disease (late Braak tissues) (Fig. 13B and Table S8).
Fig. 13.
The metastable subproteome expression in healthy tissues recapitulates the progression of AD. (A) In the left panels, regions are coloured by the mean Δ score for metastable subproteome expression. In the right panels, regions are coloured according to Braak stage (see legend). (B) Box plot with Δ scores for the metastable subproteome (MS) in ‘perfect match’ regions affected at the different Braak stages (x-axis). Perfect match tissues have an optimal topological correspondence between their Braak and Allen Brain Atlas perimeters. **p < 0.01 and *****p < 0.00001 p-values for B, calculated with Mann-Whitney U test with Benjamini-Hochberg multiple hypothesis testing correction (Benjamini and Hochberg, 1995). The dashed horizontal line represents the Δ score of non-Braak regions, which is zero by definition.
It is possible that progressive neuronal death during AD progression could partly account for the trend in Fig. 13B, as the metastable subproteome, which is highly expressed within neurons, is derived from genes downregulated in AD (Ciryam et al., 2016). To control for this possibility, we calculated the Δ scores across progressive Braak stage tissues for genes downregulated in AD, but not supersaturated, nor downregulated with age (Fig. 14). We found no significant trend across the Braak stages, indicating that measurements of downregulation of gene expression during AD are not markedly affected by neuronal atrophy in vulnerable tissues.
Fig. 14.
Distributions of ΔB scores for tissues assigned to consecutive Braak stages, for a subset of genes downregulated in AD. Boxplots of the ΔB scores for genes downregulated in AD, but not supersaturated nor downregulated in ageing. Boxplots are calculated for tissues progressively affected by AD (x-axis). For each boxplot, the significance of the difference with the prior ΔB distribution for non-Braak (NB) tissues was calculated with a Mann-Whitney U test and Benjamini-Hochberg multiple hypothesis testing correction (Benjamini and Hochberg, 1995).
Overall, the results described in this section indicate that the levels of proteins in the metastable subproteome recapitulate the typical progression of AD across the brain described by the Braak staging. These results build on previous work which demonstrates that levels of proteins known to co-aggregate in Alzheimer's and their associated molecular chaperones predict AD progression in normal brains (Freer et al., 2016). Together, these studies collectively support a key role for protein supersaturation within the neuronal environment in determining vulnerability to the onset of AD.
3. Conclusions
We have reported evidence that protein supersaturation underlies vulnerability to aberrant aggregation in neurodegenerative processes, and shown that supersaturated proteins are expressed at elevated levels in the cell and tissues that first succumb to neurodegenerative diseases. We have then defined a metastable subset of proteins specific to AD and shown that these proteins are involved in synaptic processes and mitochondrial energy metabolism.
These results reveal that the synaptic environment is conducive to protein aggregation, and offer a method of predicting the progression of AD through the brain. By contributing to the identification of the molecular origins of cell and tissue vulnerability to neurodegenerative diseases, these results could provide clearer points of intervention related to protein supersaturation to develop increasingly effective therapeutic strategies.
3.1. Experimental Procedures
3.1.1. Data sources
A full list of data sources for the results reported in this work is provided in Table S9.
3.1.2. Allen Brain Atlas
We analyzed data from six normal human brains from individuals aged 24–57 years at the time of death. Samples were taken from more than 500 regions for each hemisphere, and more than 19,700 genes were studied with multiple probes. Microarray data were downloaded from the Allen Brain Atlas (Hawrylycz et al., 2012) on 19 December 2014. UniProt data (UniProt Consortium, 2014) for subcellular localization and biological process gene ontology assignments were downloaded from www.uniprot.org/downloads on 21 May 2015. Protein IDs were converted between UniProt and Entrez ID (used by the Allen Brain Atlas) using the UniProt ID mapping service. With this procedure, expression data were assigned to about 90% of the human reference proteome.
3.1.3. Braak staging
At progressive clinical stages of AD, conserved patterns of deposition of neurofibrillary tangles in brain tissue are observed, with increasingly large areas of the brain affected with advancing stages (Braak and Braak, 1991). In the Braak staging of AD (Braak and Braak, 1991), tissues are classified according to when, in the progression of AD, neurofibrillary tangles appear in neuron. This method is employed because the formation of neurofibrillary tangles is a pathological hallmark of AD, and correlates well with cell atrophy (Whitwell et al., 2008). In Braak stages I and II, the involvement of neurofibrillary tangles is confined primarily to the transentorhinal region of the brain. In stages III and IV, limbic regions are also affected, with all regions of the hippocampus exhibiting AD pathology. In stages V and VI, there is extensive neocortical involvement. Even at late stages of AD, some regions of the brain, notably the cerebellum, remain unaffected; we classified these regions as ‘non-Braak’ (NB). In the original paper describing the Braak staging (Braak and Braak, 1991), disease stages were discussed sequentially, with the regions affected noted at each stage, in addition to the severity of the pathology in these regions.
3.1.4. Mapping between the Braak staging and the Allen Brain Atlas
To assign the brain regions from the Allen Brain Atlas to a corresponding Braak stage, a rubric was developed. First, regions mentioned in the original paper (Braak and Braak, 1991), which we refer to as ‘Braak staging’ regions, were assigned to the earliest Braak stage that they are associated with. Next, these regions were matched to the regions characterized in the Allen Brain Atlas. When a region in the Allen Brain Atlas was larger than a Braak staging region, the whole of the Allen Brain Atlas region was allocated to the corresponding Braak stage. For instance, although only two isocortical layers were affected in Braak stage III, all isocortical tissues were assigned to Braak stage III because isocortical expression data were not distinguished by the layer they came from in the Allen Brain Atlas parcellation. For this reason, in the present investigation of the relationship between Braak staging and gene expression signatures, ‘perfect-match’ regions provide the most accurate data. Perfect-match tissues have a high correspondence between their Braak and Allen Brain Atlas perimeters. Assignments from Braak to Allen Brain Atlas regions are listed in Table S4. The same method was used to map tissues most vulnerable to AD, PD, FTD and ALS to the Allen Brain Atlas. These assignments are listed in Table S1.
Of the two main types of tissue in the brain, white matter consists mostly of glial cells and myelinated axons, whereas gray matter has a more diverse composition, including neuronal cell bodies, dendrites, myelinated and unmyelinated axons, glial cells, synapses, and capillaries. Thus, because neurofibrillary tangles are not found in AD in the axon hillock or initial axon segment, one would not expect to see them in white matter in AD (Fink, 2000). However, the volume of white matter does shrink in some regions during the progression of AD, where affected neurons have their cell bodies in gray matter and their axons in corresponding white matter. This fact implies that the appearance of neurofibrillary tangles, and thus the Braak staging, may not be ideal for describing vulnerability to AD in white matter tissues. However, the effect of this caveat on our study is limited because only 2 of the more than 500 regions studied in the Allen Brain Atlas include white matter. In Fig. 5b, the impact of white matter on observed results is tested and shown to be non-significant.
3.1.5. The Δ score of the expression of a gene
Because the expression of a given gene in the Allen Brain Atlas is measured by multiple probes (Hawrylycz et al., 2012), we first normalized the expression reading Ep,r for each probe p in each region r in the Allen Brain Atlas as
| (1) |
where μp and σp are the average and standard deviation of Ep,r across all regions, respectively. To quantify the variability of gene expression between tissues, we defined a Δ score (Figure S1) for a given probe p over a brain region R (which is typically made up of several Allen Brain Atlas regions) as
| (2) |
where
| (3) |
is the average of Ep,r for the non-Braak regions, that is, those regions that do not map onto any Braak staging regions (Braak and Braak, 1991), and
| (4) |
represents the average of Ep,r over the set of Allen Brain Atlas regions mapped onto brain region R under scrutiny (for example, a Braak stage). Then, Δ scores for different probes measuring the same gene were averaged to give a gene Δ score
| (5) |
where the average is over the Np(g) probes p(g) used to measure a gene g. The full list of Δ scores for the genes analysed in this work is given in Tables S3, S5 and S8. A comparison of Δ scores calculated for perfect-match tissues is given in Table S8.
3.1.6. Definition of a subset of highly supersaturated proteins
The supersaturation scores for about 20,000 proteins were calculated by quantifying measured tissue expression relative to a computationally-derived solubility score (Ciryam et al., 2013). The proteins making up the supersaturated subset used in this study are those proteins within the top 5% of supersaturation scores (Table S2). The classification of supersaturated protein that we have used is therefore based on an estimated score, and that proteome-wide measurements will be needed in the future to measure these levels experimentally.
3.1.7. Definition of a vulnerability landscape of the brain to neurodegeneration
Regions described as the most common location for initial onset of four prevalent neurodegenerative diseases associated with aberrant aggregation, were collected and together define the most vulnerable tissues. Table S1 lists these regions for each disease (AD, ALS, FTD, and PD), and associated references.
3.1.8. Definition of a set of genes downregulated in AD and ageing
Genes downregulated upon ageing and AD were calculated previously by including all the genes significantly downregulated in these cases (Ciryam et al., 2016). Corrections are made to exclude the influence of each factor on the other, further details of calculation techniques can be found in the original paper (Ciryam et al., 2013).
3.1.9. Relative gene expression for cell types
Data were obtained from a previous mRNA sequencing study of human brain tissue (Hawrylycz et al., 2012). To evaluate the vulnerability of different brain cell types, the relative expression was calculated for each gene within each cell type as
| (6) |
where Eg,c is the expression for each gene g in a given cell type c, and μg,c is the mean expression of that gene in a given cell type c.
3.1.10. Enrichment analysis
To evaluate enrichment, we created an ‘enrichment ratio’, which describes the ratio between observed and expected proteins in the metastable subproteome, for an assigned property under study
| (7) |
where MSx is the number of metastable subproteome members assigned to subcellular localisation or biological process x, MS is the total number of proteins in the metastable subproteome, Px is the number of human proteome members assigned to subcellular localisation or biological process x, and P is total the number of proteins in the human proteome as a whole. We evaluated the significance of our results using a one-tailed Fisher exact test and Bonferroni multiple hypothesis correction. The background dataset was downloaded from uniprot.org, with all reviewed human proteins included (http://www.uniprot.org/downloads). This dataset included subcellular localisation and biological process gene ontology assignments for each protein. Protein IDs were converted between UniProt and Entrez ID using the UniProt ID mapping service. The gene ontology (GO) project provides a controlled vocabulary describing the biological processes a protein is involved in. Data available from the UniProt website lists selected terms derived from the GO project.
3.1.11. Evaluation of statistical significance
To assess the differences in the distributions of Δ scores between various data sets, we used the nonparametric Wilcoxon/Mann-Whitney U test (Mann and Whitney, 1947), or a two-tailed t test (Fisher, 1925), as specified in the figure captions. Because of the high number of data and hypotheses tested in this study, we adjusted the P values to reduce the false discovery rate (FDR). Specifically, for Figs. 2, 5, 6, 7, 9, 12, 13, and 14, we used the Benjamini-Hochberg multiple hypothesis testing correction to control the FDR (Benjamini and Hochberg, 1995) because this method allows the cost paid for the control of multiplicity to be kept relatively low. More generally, from the analysis of the relationship between FDR, sensitivity, and study sample size, it is known that microarray studies can be susceptible to large FDR, which, besides measurement variability, is primarily determined by the proportion of truly differentially expressed genes, the magnitude of the true differences, and sample size (Pawitan et al., 2005; Storey and Tibshirani, 2003). Because our work relies on 3700 microarray studies (up to 900 samples from six brains), the FDR rate analysis was performed on a relatively large sample size, allowing for rather sensitive detection of truly differentially expressed genes. We further increased the statistical significance of the results and avoided a high false-negative rate by calculating the significance of the difference of Δ score distributions for groups of genes. In comparison to calculating the significance of the differences of Δ scores of individual genes, this approach greatly reduced the number of hypotheses in our study. These tests were performed using the SciPy and rpy2 modules for Python.
3.1.10. Shading of cortical and subcortical brain structures on three-dimensional representation
Figs. 4 and 13A were created using a set of three-dimensional meshes of a human brain, which were constructed from 12 volumes acquired using magnetic resonance imaging (Causevic et al., 2010; Klein and Tourville, 2012; Wilhelm et al., 2014). Images were colored using the computer graphics software Blender.
3.1.12. Creation of a three-dimensional representation of the synaptosome
Data on the proteomic contents of the synaptosome, and their relative location and abundance in this environment, were taken from (Wilhelm et al., 2014). The graphical model of the average presynaptic terminal was constructed using custom-written plug-ins and scripts in the 3D software Autodesk Maya (Autodesk Inc., San Rafael, CA). Information on protein structures was obtained from the Uniprot database. If available, protein data base (PDB) coordinates were used to determine the structure of a protein. In case no PDB data for an entire protein or parts of the protein existed, structural information from the following prediction servers was used to create the 3D structure manually: secondary structure (http://bioinf.cs.ucl.ac.uk/psipred/), alignment (http://web.expasy.org/sim/), coiled coil (http://toolkit.tuebingen.mpg.de/pcoils), disorder (http://mbs.cbrc.jp/poodle/poodle-s.html; http://mbs.cbrc.jp/poodle/poodle-w.html; http://mbs.cbrc.jp/poodle/poodle-l.html), transmembrane (http://www.ch.embnet.org/software/TMPRED_form.html), glycosylation (http://www.glycosciences.de/modeling/glyprot/php/main.php), domain search (http://smart.embl-heidelberg.de/index2.cgi), homologue proteins (http://web.expasy.org/blast/).
Declarations
Author contribution statement
Michele Vendruscolo, Rosie Freer, Pietro Sormanni, Prajwal Ciryam, Burkhard Rammner, Silvio Rizzoli, Christopher Dobson: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. Burkhard Rammner and Silvio Rizzoli were supported by the Deutsche Forschungsgemeinschaft (SFB1286/A3).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank J. Freer for his technical assistance in the production of Figs. 4 and 13A.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Video of a synaptic buton illustrating the high protein supersaturation levels in this environment (see also Fig. 10).
References
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995:289–300. [Google Scholar]
- Bero A.W., Yan P., Roh J.H., Cirrito J.R., Stewart F.R., Raichle M.E., Lee J.-M., Holtzman D.M. Neuronal activity regulates the regional vulnerability to amyloid-beta deposition. Nat. Neurosci. 2011;14:750–756. doi: 10.1038/nn.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brehme M., Voisine C., Rolland T., Wachi S., Soper J.H., Zhu Y., Orton K., Villella A., Garza D., Vidal M. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014;9:1135–1150. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causevic M., Farooq U., Lovestone S., Killick R. Beta-Amyloid precursor protein and tau protein levels are differently regulated in human cerebellum compared to brain regions vulnerable to Alzheimer's type neurodegeneration. Neurosci. Lett. 2010;485:162–166. doi: 10.1016/j.neulet.2010.08.088. [DOI] [PubMed] [Google Scholar]
- Čaušević M., Farooq U., Lovestone S., Killick R. β-Amyloid precursor protein and tau protein levels are differently regulated in human cerebellum compared to brain regions vulnerable to Alzheimer's type neurodegeneration. Neurosci. Lett. 2010;485:162–166. doi: 10.1016/j.neulet.2010.08.088. [DOI] [PubMed] [Google Scholar]
- Ciryam P., Kundra R., Freer R., Morimoto R.I., Dobson C.M., Vendruscolo M. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc. Natl. Acad. Sci. 2016;113:4753–4758. doi: 10.1073/pnas.1516604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Kundra R., Morimoto R.I., Dobson C.M., Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P., Tartaglia G.G., Morimoto R.I., Dobson C.M., Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman P., Federoff H., Kurlan R. A focus on the synapse for neuroprotection in Alzheimer disease and other dementias. Neurology. 2004;63:1155–1162. doi: 10.1212/01.wnl.0000140626.48118.0a. [DOI] [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Gephart M.G.H., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B., Karran E. The cellular phase of Alzheimer’s disease. Cell. 2016;164:603–615. doi: 10.1016/j.cell.2015.12.056. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. Academic Press; 2000. Encyclopedia of Stress, Three-Volume Set, Vol 1. [Google Scholar]
- Fisher R.A. Genesis Publishing Pvt Ltd; 1925. Statistical Methods for Research Workers. [Google Scholar]
- Freer R., Sormanni P., Vecchi G., Ciryam P., Dobson C.M., Vendruscolo M. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer’s disease. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Possenti A., Freer R., Nakano Y., Villegas N.C.H., Tang M., Cauhy P.V., Lassus B.A., Chen S., Fowler S.L. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 2019;22:47. doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gillingwater T., Wishart T. Mechanisms underlying synaptic vulnerability and degeneration in neurodegenerative disease. Neuropathol. Appl. Neurobiol. 2013;39:320–334. doi: 10.1111/nan.12014. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D.M., Morris J.C., Goate A.M. Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A., Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front. Neurosci. 2012;6:171. doi: 10.3389/fnins.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T.P., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947:50–60. [Google Scholar]
- Mattson M.P., Magnus T. Ageing and neuronal vulnerability. Nat. Rev. Neurosci. 2006;7:278. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.A., Woltjer R.L., Goodenbour J.M., Horvath S., Geschwind D.H. Genes and pathways underlying regional and cell type changes in Alzheimer’s disease. Genome Med. 2013;5:48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore C.R., Zhou C., Liao M., Fernandez M.A., Taylor W.M., Lagomarsino V.N., Pearse R.V., II, Rice H.C., Negri J.M., He A. Cell-type dependent Alzheimer's disease phenotypes: probing the biology of selective neuronal vulnerability. Stem Cell Rep. 2017;9:1868–1884. doi: 10.1016/j.stemcr.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H., Schermann S.M., Woerner A.C., Pinkert S., Hecht M.H., Tartaglia G.G., Vendruscolo M., Hayer-Hartl M., Hartl F.U., Vabulas R.M. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Pawitan Y., Michiels S., Koscielny S., Gusnanto A., Ploner A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005;21:3017–3024. doi: 10.1093/bioinformatics/bti448. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J. Alzheimer's disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2014:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M. Proteome folding and aggregation. Curr. Opin. Struct. Biol. 2012;22:138–143. doi: 10.1016/j.sbi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Walther D.M., Kasturi P., Zheng M., Pinkert S., Vecchi G., Ciryam P., Morimoto R.I., Dobson C.M., Vendruscolo M., Mann M. Widespread proteome remodeling and aggregation in aging C. elegans. Cell. 2015;161:919–932. doi: 10.1016/j.cell.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Woltjer R.L., Cimino P., Pan C., Montine K.S., Zhang J., Montine T.J. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19:869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- Whitwell J.L., Josephs K.A., Murray M.E., Kantarci K., Przybelski S., Weigand S., Vemuri P., Senjem M., Parisi J.E., Knopman D.S. MRI correlates of neurofibrillary tangle pathology at autopsy A voxel-based morphometry study. Neurology. 2008;71:743–749. doi: 10.1212/01.wnl.0000324924.91351.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B.G., Mandad S., Truckenbrodt S., Kröhnert K., Schäfer C., Rammner B., Koo S.J., Claßen G.A., Krauss M., Haucke V. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- Wishart T.M., Parson S.H., Gillingwater T.H. Synaptic vulnerability in neurodegenerative disease. J. Neuropathol. Exp. Neurol. 2006;65:733–739. doi: 10.1097/01.jnen.0000228202.35163.c4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video of a synaptic buton illustrating the high protein supersaturation levels in this environment (see also Fig. 10).