Abstract
Enzymatic dehalogenation is an important and well-studied biological process in both the detoxification and catabolism of small molecules, many of which are anthropogenic in origin. However, dedicated dehalogenation reactions that replace a halogen atom with a hydrogen are rare in the biosynthesis of natural products. In fact, the debrominase Bmp8 is the only known example. It catalyzes the reductive debromination of the coral settlement cue and the potential human toxin 2,3,4,5-tetrabromopyrrole as part of the biosynthesis of the antibiotic pentabromopseudilin. Using a combination of protein crystallography, mutagenesis, and computational modeling, we propose a catalytic mechanism for Bmp8 that is reminiscent of that catalyzed by human deiodinases in the maintenance of thyroid hormones. The identification of the key catalytic residues enabled us to recognize divergent functional homologues of Bmp8. Characterization of one of these homologues demonstrated its debromination activity even though it is found in a completely distinct genomic context. This observation suggests that additional enzymes outside those associated with the tetrabromopyrrole biosynthetic pathway may be able to alter the lifetime of this compound in the environment.
Graphical Abstract
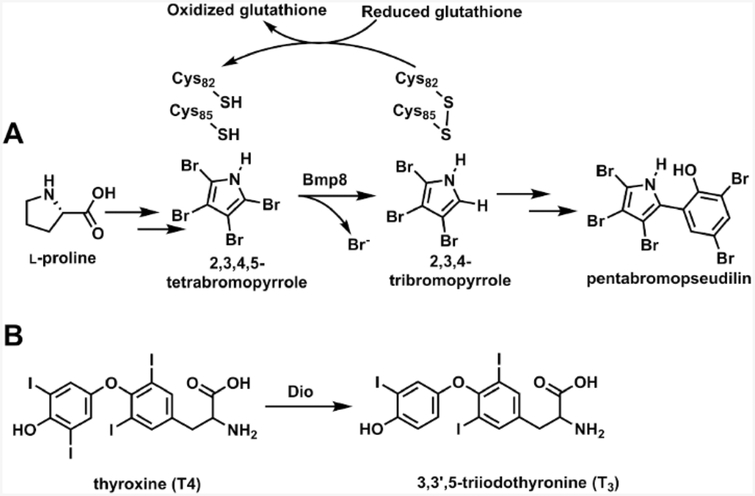
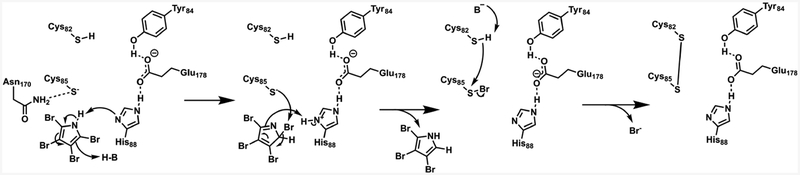
Halogenation reactions are common chemical transformations in the biosynthesis of natural products and are often critical for bioactivity.1 Five distinct families of halogenases typically install chlorine or bromine atoms, with a few examples of fluorination and iodination present as well. While diverse enzymatic dehalogenation reactions have been well studied in the context of small molecule detoxification or catabolism,2–4 dedicated dehalogenation in the biosynthesis of natural products is an extremely rare transformation. The only characterized example is found in the production of brominated pyrrole containing secondary metabolites such as the antibiotic pentabromopseudilin (Scheme 1).5,6 In these biosynthetic pathways, proline is converted to 2,3,4,5-tetrabromopyrrole,6,7 which itself has coral settlement promoting activity.8 Tetrabromopyrrole must then be debrominated to generate 2,3,4-tribromopyrrole before further modification yields the final product. The reductive debromination is catalyzed enzymatically by Bmp8 using a proposed pair of catalytic cysteines (Scheme 1A).5 This contrasts with several examples of cryptic dehalogenation reactions, where a halogen is rather used to activate a carbon for subsequent chemistry.9–12 While dedicated dehalogenation is rare in natural product biosynthesis, a parallel strategy is found in the maintenance of thyroid hormones in one class of human deiodinases (Scheme 1B).13 Similar to Bmp8, these enzymes employ a catalytic selenocysteine–cysteine pair to remove iodines from thyroid hormones which alters their bioactivity. Deiodinases (Dios) have been a focus of extensive research because their dysfunction in humans can lead to hyper- and hypothyroidism.13,14
Scheme 1. (A) Biosynthesis of Pentabromopseudilin; (B) Deiodination of Human Thyroid Hormonea.
a The reaction in (A) proceeds through the debromination activity of Bmp8. The reaction in (B) is catalyzed by deiodinases (Dios).
Not only does Bmp8 catalyze an unusual biosynthetic reaction, but also it acts as the important branch point between the coral settlement cue tetrabromopyrrole and downstream brominated products. In addition to biosynthetic production by marine bacteria,6 tetrabromopyrrole is also generated as an anthropogenic byproduct of salt water disinfection.15 It has been shown to exhibit developmental toxicity15 and is also implicated in neurotoxicity,16 making it a human health concern. The importance of Bmp8 in degrading this significant small molecule prompted us to explore the mechanism of Bmp8 by structural, biochemical, and computational methods to identify motifs that would enable bioinformatic identification of other tetrabromopyrrole debrominases. Furthermore, we sought to evaluate if the apparent similarities in catalysis between the unrelated Bmp8 and human Dio enzymes extend to a conserved dehalogenation strategy.
MATERIALS AND METHODS
PCR and Cloning.
PCRs were completed using PrimeStar or PrimeStar MAX polymerases according to the manufacturer’s instructions (Takara). Primers are listed in Table S1. PCR products were purified and cloned into a pET28 vector using the NEBuilder HiFi assembly kit (NEB) generating a N-terminal His6 construct. The pET28 vector used for cloning was linearized by PCR amplification. The Bmp8 variants were generated using mutagenic primers and assembled using the NEBuilder HiFi assembly kit. All Gibson Assembly reactions were transformed into DH5α Escherichia coli cells, and constructs were confirmed by Sanger sequencing (Genewiz).
Protein Expression and Purification.
Bmp8 was expressed and purified as previously described.17 Briefly, the pET28 Bmp8 and pCDFDuet Bmp1-TE plasmids were cotransformed into E. coli BL-21 cells. Five milliliters of an overnight LB culture was added to 1 L of sterile TB media. The flask was shaken in an incubator at 200 rpm at 37 °C until the OD600 reached ~0.8. The incubator was cooled to 18 °C, and the flasks were shaken for an additional 1 h, after which 0.3 mM IPTG was added. The cells remained shaking for 18 h and were harvested by centrifugation. The pellets were resuspending in 25 mL of 500 mM NaCl, 20 mM Tris, pH 8.0, and 10% glycerol. Cells were lysed by sonication with a Qsonica 6 mm tip at 40% amplitude for 12 cycles of 15 s on and 45 s off. The lysate was centrifuged at 15 000g to remove cellular debris. The cleared lysate was loaded at 2 mL/min onto a 5 mL HisTrap column (GE Healthcare Life Sciences) that was pre-equilibrated with buffer A (1 M NaCl, 20 mM Tris, pH 8.0, and 30 mM imidazole). After loading, the column was washed at 2 mL/min with 40 mL of buffer A to remove weakly bound proteins. Bmp8 was eluted using a linear gradient of 0 to 100% buffer B (1 M NaCl, 20 mM Tris, pH 8.0, and 250 mM imidazole) over 40 mL with 5 mL fractions collected. Pure fractions, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were combined, and 60 units of thrombin was added. The protein was dialyzed overnight against 50 mM KCl, 20 mM Tris, pH 8.9, and 2 mM dithiothreitol (DTT). After confirming complete cleavage of the N-terminal His6 tag by SDS-PAGE, the protein was loaded onto a 5 mL strong anion exchange Q FF column (GE Healthcare Life Sciences) that was pre-equilibrated with 95% buffer C (20 mM Tris, pH 8.9, and 3 mM DTT) and 5% buffer D (20 mM Tris, pH 8.9, 1 M KCl, and 3 mM DTT). The column was washed with 10 mL of 5% buffer D, and Bmp8 was eluted using an 80 mL linear gradient starting with 5% buffer D and ending with 70% buffer D with 3 mL fractions collected. Pure fractions, as determined by SDS-PAGE, were combined and concentrated. The concentrated Bmp8 was loaded onto a HiLoad 16/60 Superdex 75 prep grade column (GE Healthcare Life Sciences) pre-equilibrated with Bmp8 storage buffer (20 mM Tris, pH 8.0, 300 mM KCl, and 10% glycerol). Pure fractions were collected, concentrated, and frozen. PLCMD was purified in a similar manner except the Q FF anion exchange column purification was not used.
Crystallization and Data Processing.
Bmp8 was screened for initial crystal hits using commercial sparse screen matrix kits with hanging drop vapor diffusion. Optimized crystals were obtained using 1 μL of 7 mg/mL Bmp8 with 1 mM tris(2-carboxyethyl)phosphine (TCEP) and 1 μL of mother liquor (2.3 M ammonium sulfate and 0.1 M Bis-tris propane, pH 9.0) over a 150 μL well of mother liquor at 6 °C. Immediately prior to vitrification in LN2, crystals were briefly soaked in mother liquor supplemented with 30% glucose. Phases were obtained by single-wavelength anomalous dispersion (SAD) using crystals obtained with 2.3 M ammonium sulfate and 0.1 M HEPES, pH 8.5, that were soaked in 1 mM methylmercury chloride for 3 h prior to vitrification. The cocrystal structure of Bmp8 and tribromopyrrole was obtained with Bmp8 C82A crystals grown with the same conditions as WT Bmp8. Crystals were then soaked in mother liquor containing 1 mM tetrabromopyrrole for 3 h prior to vitrification. All data was collected at the Advanced Light Source Macromolecular Crystallography beamlines (8.2.1 and 8.2.2). A wavelength of 1.00 Å was used for the APO and methylmercury chloride-soaked crystals. The tetrabromopyrrole crystal data was collected at 0.91 Å. All data was indexed and scaled using autoPROC.18 AutoSol from the PHENIX software suite19 was used to obtain initial phases from the anomalous diffraction data. The Bmp8 structure was built using a combination of Phenix AutoBuild and Buccaneer.20 Structures were refined and manually adjusted by iteratively using phenix.refine and COOT.21 All refinement statistics are found in Table S2.
Gel Filtration Size Determination.
A 16/60 Superdex 75 prep grade column (GE Healthcare Life Sciences) was pre-equilibrated with buffer containing 20 mM Tris, pH 8.0, 300 mM KCl, and 10% glycerol. One vial of BioRad Gel Filtration standard (#1511901) was resuspended in 500 μL of H2O. All 500 μL was injected onto the column and run at a flow rate of 1 mL/min. Separately, 500 μL of Bmp8 was injected on the column and run in the same way. The standards were plotted, and the molecular weight of Bmp8 was estimated (Figure S1).
Activity Assays.
Bmp8 activity assays were completed as previously described.17 Briefly, 20 μM Bmp8 and 500 μM glutathione were mixed in a buffer containing 50 mM KCl and 20 mM Tris, pH 8.0. The 400 μL reaction was initiated with 100 μM substrate and quenched with 800 μL of ethyl acetate after a 30 min incubation at 28 °C. The samples were thoroughly vortexed to extract the remaining substrate and products from the reaction into the ethyl acetate layer. Seven hundred microliters of the ethyl acetate layer were removed and evaporated to dryness using a miVac DNA system (SP Scientific) system. The residue was resuspended in 80 μL of a 50% methanol/50% H2O mixture and subsequently analyzed by HPLC or LCMS. Assays with tetrachloropyrrole were completed in a similar manner with the exception of 2-chloro-3,4,5-tribromopyrrole. In this assay, 25 μM Bmp8 and 50 μM substrate were used in a reaction buffer of 50 mM KCl, 20 mM Tris, pH 8.0, and 10% glycerol.
HPLC Quantification.
The activity assay samples were analyzed using an Agilent 1200 series HPLC with a Phenomenex Luna 5 μm C18(2), 100 Å, 150 × 4.6 mm column. The following method was used: 20 min at 52% solvent B, gradient of 52% to 95% solvent B over 1 min, 95% solvent B for 3 min, gradient of 95% solvent B to 52% solvent B over 1 min, and 2 min at 52% solvent B. Solvent A is HPLC grade water + 0.1% trifluoroacetic acid (TFA), and solvent B is HPLC grade acetonitrile + 0.1% TFA. Absorbance was monitored at 220 nm, and the tribromopyrrole product peak was integrated and normalized to the amount produced by WT Bmp8.
LCMS Analysis.
To analyze samples by LCMS, 10 μL of an activity assay was injected onto a Phenomenex Luna 5 μm C18(2), 100 Å, 150 × 4.6 mm column and analyzed by an Agilent 1260 HPLC + Agilent 6530 Accurate Mass Q-TOF mass spectrometer using the following method: 5 min at 10% solvent B (0.5 mL/min), 10–70% solvent B over 10 min (0.5 mL/min), 70–80% over 10 min (0.5 mL/min), 80–100% solvent B over 0.5 min (0.75 mL/min), 2 min at 100% solvent B (0.75 mL/min), 100–10% solvent B over 0.5 min (0.75 mL/min), and 3 min at 10% solvent B (0.75 mL/min). Solvent A is H2O + 0.1% formic acid, and solvent B is acetonitrile + 0.1% formic acid.
Computational Methods.
All computations were carried out with the GAUSSIAN 0922 series of programs. All graphics on optimized structures were generated with CYLview.23 Reactants, transition states, intermediates, and products were optimized using B3LYP functional24–26 with Grimme’s D3 dispersion correction27 and 6–31G(d,p) basis set for all atoms except bromine and iodine, which used Lanl2dz basis set. Vibrational frequencies were computed to determine if the optimized structures are minima or saddle points on the potential energy surface corresponding to ground state and transition state geometries, respectively. Single point energies were subsequently obtained using B3LYP functional with Grimme’s D3 dispersion correction and 6–311+G(d,p) basis set for all atoms except bromine and iodine, which used SDD basis set. The implicit solvent model, CPCM,28–32 with dielectric constant of 4 was used to mimic the hydrophobic pocket of an enzyme. The association enthalpies of halogen bonded complexes were calculated according to eq 1.
| (1) |
RESULTS AND DISCUSSION
Bmp8 Crystal Structure.
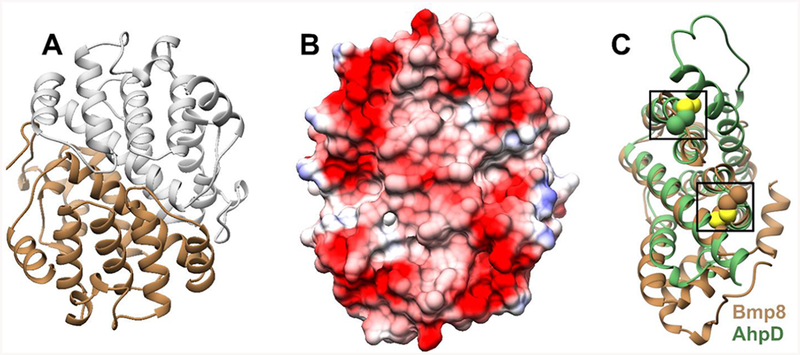
Previous mutagenesis and labeling work suggested a Bmp8 mechanism wherein a catalytic cysteine attacks tetrabromopyrrole to form a transiently modified cysteine that is resolved by a second cysteine to form tribromopyrrole and a disulfide bond (Scheme 1A).5 To evaluate and explore the details of this proposal, we began examining the mechanism of Bmp8 by crystallizing and solving the structure to a resolution of 2.3 Å. Bmp8 forms a dimer in the asymmetric unit with each monomer composed exclusively of α-helices (Figure 1A). These monomers come together to form a highly charged flat surface (Figure 1B). To be solubly produced in E. coli, Bmp8 must be coexpressed with the thioesterase domain of Bmp1 even though Bmp1 does not copurify with Bmp8.5,17 This charged surface is a possible location for the protein–protein interaction.
Figure 1.
(A) Bmp8 forms a dimer in the asymmetric unit. (B) The dimeric Bmp8 structure forms a highly acidic surface. (C) Bmp8 monomer (brown) aligned with CMD family protein AhpD (green). Locations of the catalytic cysteines are indicated.
The Bmp8 fold is completely distinct from the functionally similar Dio, which adopts a thioredoxin fold.33 Instead, analysis of Bmp8 with the DALI server34 revealed it to be most closely related to the carboxymuconolactone decarboxylase (CMD) family of proteins, PFAM PF02627. While the fold is conserved, CMDs are known to catalyze a variety of reactions. Canonical CMDs are decarboxylases and are found in degradation pathways of phenols and benzoates.35 Conversely, AhpD is a CMD family member36,37 that is primarily involved in protein disulfide bond reduction and is particularly important in the antioxidant pathway of Microbacterium tuberculosis.36,38 Notably, AhpD can also catalyze reduction of alkylhydroperoxides at a low rate.38 Structural alignment of Bmp8 with AhpD showed strong structural conservation with minimal sequence similarity (PDB: 1knc, r.m.s.d. of 2.4 Å over 133 aligned Cα, 16% identity). AhpD and other CMDs are typically found in a ring shaped trimeric or hexameric complex, but gel filtration experiments instead suggest that Bmp8 is most likely dimer in solution (Figure S1). Curiously, while the overall monomeric Bmp8 structure aligns well to AhpD, the putative cysteine containing active sites are located in entirely different regions of the proteins (Figure 1C).
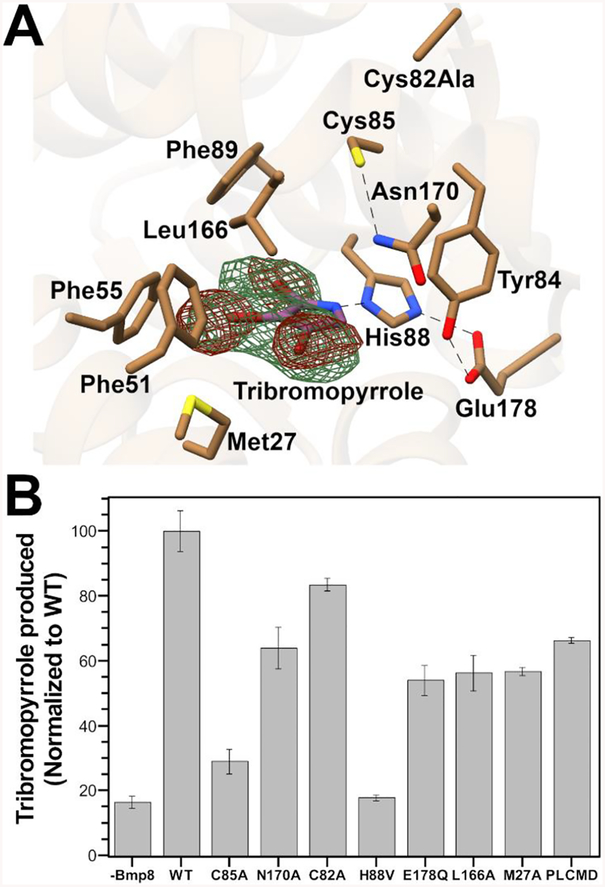
To better understand the enzymatic mechanism and basis for substrate binding, we attempted to obtain a cocrystal structure of the Bmp8 Cys82Ala variant with the tetrabromopyrrole substrate. However, by soaking the Bmp8 Cys82Ala crystals with tetrabromopyrrole, we were able solve a 3.2 Å structure that revealed electron density within the active site more consistent with the product, tribromopyrrole (Figures 2A and S2A). This assignment was further supported by analysis of anomalous diffraction maps that showed the presence of only three potential bromines within the active site. The observed tribromopyrrole may have formed by the non-enzymatic debromination of tetrabromopyrrole by residual TCEP in the crystallization buffer, which we previously showed is possible,17 or slow catalysis by the enzyme. Due to the low resolution and globular nature of the missing density in the Fo−Fc map (Figure S2A), the bromines of tribromopyrrole were fit into the anomalous densities. The C2 bromine appears to have weaker electron density than the other two bromines. This observation may indicate that some 3,4-dibromopyrrole is also present in the structure, as previously observed in Bmp8 activity assays.5
Figure 2.
(A) Active site of Bmp8 C82A with tribromopyrrole bound. Fourier (Fo−Fc) and anomalous maps calculated with tribromopyrrole removed prior to one round of refinement, contoured to 4 σ (green mesh) and 3 σ (red mesh), respectively. (B) HPLC-based activity assay with the amount of tribromopyrrole produced normalized to WT production.
When compared to the Apo WT Bmp8, there does not appear to be any conformational changes in the cocrystal structure (Figure S2B). The tribromopyrrole itself is located within a hydrophobic pocket composed of Met27, Phe51, Phe55, Phe89, and Leu166 (Figure 2A). The abundance of phenylalanines suggests that halogen–π interactions could be assisting in substrate binding. In particular, Phe55 appears well oriented for this interaction. Elsewhere in the active site, the nitrogen of the pyrrole is within hydrogen bonding distance (3.3 Å) of the histidine residue in the His88–Glu178–Tyr84 hydrogen bonded network (Figure S2C). This combination of a hydrophobic pocket and hydrogen bond locks the planar tribromopyrrole in an orientation perpendicular to the proposed catalytic Cys85 (Figure 2A).
Activity Assays and Mutagenesis.
On the basis of the tribromopyrrole Bmp8 cocrystal structure, we designed a series of mutants to test the importance of both the candidate catalytic residues and hydrophobic pocket (Figure 2B). While the native in vivo reductant is unknown, activity assays were completed in vitro using glutathione as a chemical reductant to generate a catalytic system.17 It should be noted that tetrabromopyrrole is not completely stable in this system and can be spontaneously debrominated by glutathione. The Cys85Ala mutation nearly abolished turnover, consistent with its proposed role as the catalytic nucleophile that enables debromination. For catalysis, Cys85 needs to be in the thiolate form. Examination of the active site identified a nearby Asn170 that could be responsible for stabilization of the Cys85 thiolate (Figure 2A). This hypothesis is supported by the diminishment of activity in the Bmp8 Asn170Ala variant. Mutation of the other member of the cysteine pair, Cys82, to alanine only slightly decreases product formation. This aligns with Cys82’s proposed role of resolving the modified Cys85 and generating the inactive disulfide form of the enzyme. If the Cys82 is absent, the Cys85 adduct instead spontaneously oxidizes to sulfinic acid and can be reduced by chemical reductants,5 making Cys82 unnecessary in this in vitro assay.
The importance of the His88, Glu178, and Tyr84 hydrogen bonding network was also explored. Mutation of His88 to Val abolished turnover and indicates that this residue is critical for activity. The Bmp8 Glu178Gln variant also possessed diminished turnover but was prone to degradation (Figures 2B and S3). Mutation of the final member of the hydrogen bonding network, Tyr84, resulted in insoluble protein. Together, these results demonstrate the importance of the His88, Glu178, and Tyr84 hydrogen bonding network in both catalysis and stability of the enzyme. Additionally, the significance of the hydrophobic binding pocket was explored. Expansion of the hydrophobic pocket with Leu166Ala and Met27Ala variants resulted in a decrease in the amount of tribromopyrrole formed, while the Phe55Ala mutation resulted in insoluble protein.
Mechanistic Proposal Based on Crystal Structure and Computation.
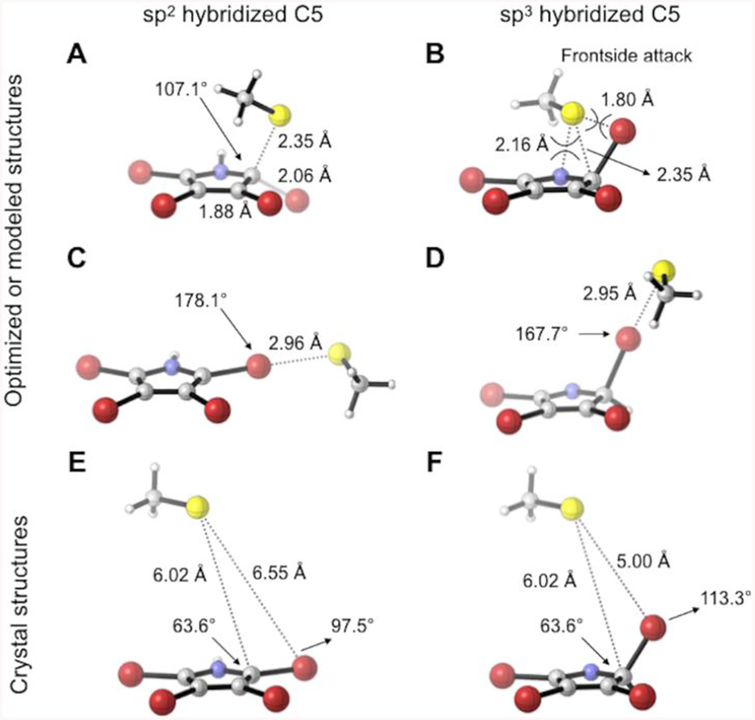
The cocrystal structure of Bmp8 and tribromopyrrole also enabled us to begin evaluating different mechanistic routes of debromination. Mutagenesis and the crystal structure strongly suggest that Cys85 is the nucleophile of the reaction, but multiple electrophilic targets can be proposed (Figure 3). The Cys85 could attack C5 in either the planar sp2-hybridization form or a tetrahedral sp3-hybridized form that is generated by isomerization of N1 to the imine or protonation of C5 to the iminium (2H-pyrrole). Both of these mechanisms would promote loss of the bromide anion (Figure 3A,B). Alternatively, direct attack of the bromine atom by Cys85 could occur (Figure 3C,D). Because of the low resolution of the structure and presence of only the product in the active site, we decided to complement the crystallographic data with computational analysis to better explore these possible mechanisms. We began by first manually docking tetrabromopyrrole into the Bmp8 active site by orienting it in the same location as tribromopyrrole (Figure 3E,F). This was followed by computational evaluation of each mechanistic hypothesis. Examination of the direct debromination by nucleophilic aromatic substitution (SNAr) at the C5 position showed that this reaction pathway involves a low energy barrier (6.0 kcal/mol) at an optimal C–S distance of 2.35 Å (Figure 3A). However, the crystal structure of the Bmp8 active site shows that the substrate is 6 Å away from Cys85 and likely too far away to achieve the transition state involving the cysteine attack of C5. Reaction with C5 in the sp3-hybridized form is also unlikely as a backside attack by thiolate is not possible in this orientation and frontside SN2 is disfavored (Figure 3B).
Figure 3.
Computationally optimized binding modes for the proposed Bmp8 transition states compared to the modeled location within the crystal structure. Cysteine has been truncated to thiolate for computational efficiency and because only the side chain is involved in the Br attack. (A) Attack on sp2-hydridized C5 (SNAr); (B) attack on sp3-hybridized C5 (SN2); (C) attack on C5sp2-Br (direct attack mechanism); (D) attack on C5sp3-Br (isomerization-based mechanism); (E, F) modeled substrate within the crystal structure.
If tetrabromopyrrole remains in the planar sp2-hybridized C5 form, the bromine atom can be removed by direct attack of thiolate through a halogen bond (Figure 3C). The halogen bond is a highly directional, strong intermolecular interaction between a Lewis-acidic halogen atom and a Lewis base.39 The strength of a halogen bond is comparable to that of a hydrogen bond and is increasingly recognized as a potential tool in the fields of self-assembly, crystal engineering, liquid crystals, materials science, biochemistry, and medicinal chemistry.39 The optimized complex between tetrabromopyrrole and thiolate is at a S–Br distance of 2.96 Å at a C–Br–S angle of nearly 180°. The modeled position of tetrabromopyrrole in the active site is more than 3.5 Å further away than the most favorable distance of Cys85 (Figure 3A). Furthermore, the angle of attack suggested by the crystal structure differs from the ideal angle by more than 80°. Together, these results indicate that a major active site rearrangement or different mode of tetrabromopyrrole binding would be required to mimic this transition state. Not only does geometry disfavor a direct attack mechanism, but also the energetics are unfavorable. Attempts to locate a transition state for the attack by Cys85 were unsuccessful, and the debrominated product complex is very thermodynamically unfavorable (Figure S4).
The formation of a sp3-hybridized carbon could position the bromine atom within 5 Å of Cys85, a closer distance for attack (Figure 3D). The formation of this intermediate would be facilitated by isomerization or protonation involving His88, suggesting possible roles for this residue in catalysis. His88 has increased basicity due to the hydrogen bonding network with Glu178 and Tyr84. A similar scenario is well established in serine proteases where a carboxylate modulates the pKa of histidine to deprotonate a serine residue. Likewise, His88 may be involved in deprotonating the pyrrole nitrogen during the isomerization step even though it has a relatively high pKa of ~17. Alternatively, the nitrogen could instead remain protonated and form an iminium intermediate that is stabilized by His88 (Figure S5).
Indeed, the computational results also favor the isomerization mechanism suggested by the crystal structure. While the cocrystal structure is not high resolution, the observed distance and angle of attack of the modeled sp3-hybridized 2H-pyrrole intermediate are better aligned with the computational results of an isomerization mechanism than any of the others proposed (Figure 3D). Moreover, the energetics of this isomerization mechanism are more favorable than the direct attack mechanism, as the reaction forms a thermodynamically favored debrominated product (Figure S4).
Collectively, the crystallography, mutagenesis, and computational work suggest a mechanism where tetrabromopyrrole binds in the hydrophobic pocket of Bmp8 (Scheme 2). His88 isomerizes or deprotonates the bromopyrrole to form a tetrahedral C5 intermediate (int1). The Cys85 thiolate, stabilized in the active site by Asn170, then attacks the bromine and forms the tribromopyrrole product upon rearomatization and reprotonation of N1. The brominated Cys85 can be directly attacked by the Cys85 thiolate to liberate bromide and form a disulfide bond. Alternatively, the brominated Cys85 could be spontaneously oxidized to release bromide and subsequently form the disulfide bond (Figure S5). Reduction of the disulfide bond by either a small molecule or disulfide reductase would complete the catalytic cycle.
Scheme 2. Proposed Bmp8 Mechanisma.
a Alternative mechanisms are proposed in Figure S5.
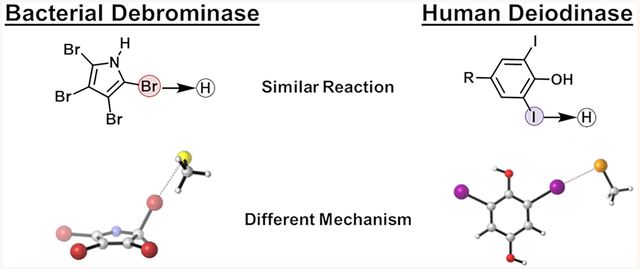
Mechanistic Comparison to Dio.
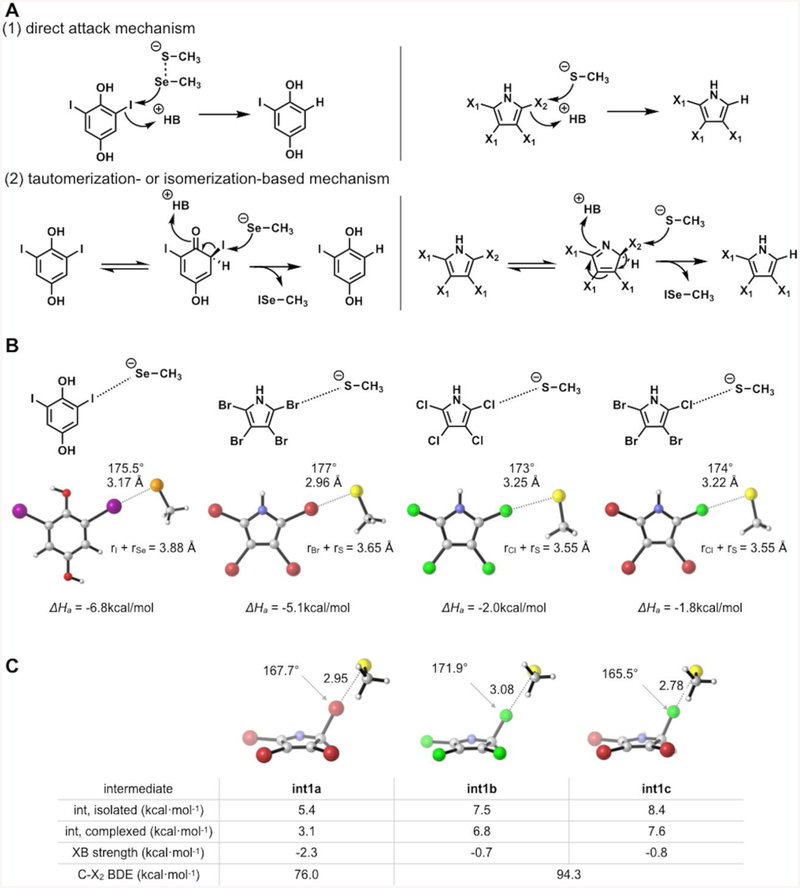
While the overall dehalogenation is similar to human Dio, some mechanistic details in our Bmp8 proposal differ from previous work on Dios.40,41 It had previously been hypothesized that the selenol could attack on the iodine center of the keto form of thyroxine, similar to Bmp8 (Figure 4A, mechanism 2).42 However, Dios can utilize a variety of substrates and remove iodine at different locations within a single compound. This lead to the isomerization mechanism to be disregarded because other substrates incapable of keto–enol isomerization, such as iodinated methoxybenzoate, could be converted by certain Dio enzymes.43–45 Instead, the current hypothesis is that the selenocysteine–cysteine pair in the Dio active site directly removes iodine atoms from a sp2-hybridized carbon through a strong halogen bond (Se–I bond) assisted by a chalcogen bond (S–Se) (Figure 4A, mechanism 1). The experimental and theoretical investigations suggest that two chalcogen atoms in proximity to each other are necessary for a complete cleavage of the C–I bond.41 Simple selenols, which lack any additional thiol or selenol groups, do not exhibit deiodinase activity as the strength of halogen bond is not sufficient to cleave the C–I bond.
Figure 4.
(A) Possible mechanisms of dehalogenation by Dio (left) and Bmp8 (right). For tetrabromopyrrole, X1 = Br and X2 = Br. For tetrachloropyrrole, X1 = Cl and X2 = Cl. For 2-chloro-3,4,5-tribromopyrrole, X1 = Br and X2 = Cl. (B) Computed halogen bonding strengths for 2,6-diiodobenzene-1,4-diol-selenolate adduct (Dio) and tetrahalopyrrole-thiolate adducts (Bmp8). (C) Computed energies and halogen bond strengths of tetrahalo-2H-pyrrole intermediates and thiolate. Halogen atoms are colored as follows: purple is I, red is Br, and green is Cl.
To further explore this mechanistic difference between Dio and Bmp8, we completed calculations with the thyroxine model compound 2,6-diiodobenzene-1,4-diol and a selenolate adduct (Figure 4B). Our results indicate that the Se–I halogen bond in the Dio model system is more energetically favorable than the S–Br halogen bond that forms in Bmp8 (−6.8 and −5.1 kcal/mol, respectively). This may result from both smaller sigma holes for Br and Cl compared to I39,46–48 and decreased nucleophilicity of thiolate compared to selenol.49 The energy difference likely enables direct loss of iodine in Dio, while Bmp8 evolved an isomerization step to overcome the thermodynamic limitation of the S–Br bond.
Bmp8 Activity with Alternative Substrates.
We sought to explore the limits of Bmp8’s ability to utilize alternative substrates and catalyze dechlorination reactions. When incubated with 2,3,4,5-tetrachloropyrrole, Bmp8 is able to form trichloropyrrole but at a much lower level than the debromination activity (Figure S6). We also tested specificity with the mixed halogenated substrate 2-chloro-3,4,5-tribromopyrrole and found that Bmp8 was selective for only the debromination reaction and did not catalyze dechlorination (Figure S7). Although the rate-determining step of the mechanism is unknown, this difference in reactivity is correlated with three factors: the intrinsic stabilities of tetrahalo-2H-pyrrole intermediates, strengths of their complexation with thiolate, and the bond energies of C–X2 (Figure 4C). Calculated results show that the brominated 2H-pyrrole intermediate (int1a) is intrinsically more stable than the chlorinated pyrroles (int1b and int1c). This may be related to the pyrrole isomerization step. In addition, halogen bond strengths between 2H-pyrrole intermediates and thiolate as well as C–X2 bond dissociation energies are greatest in the brominated 2H-pyrrole. If the second step of the mechanism (rearomatization) is the rate-determining step, then these correlations can explain the observed reaction rates.
Bmp8 Functional Homologue Identification.
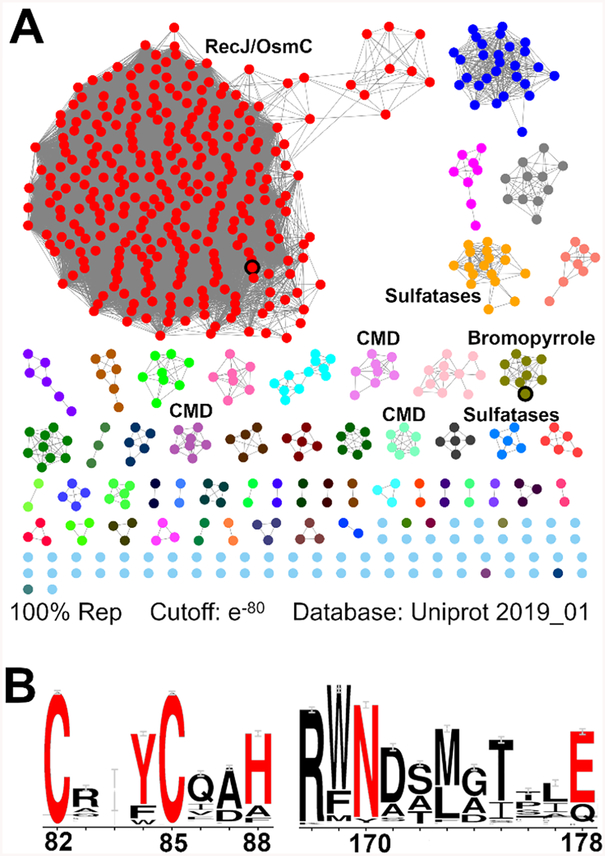
With a mechanism proposed and key residues identified, we aimed to examine the abundance and distribution of debrominase homologues. BlastP searches of both nonredundant and environmental sample databases identified over 1000 Bmp8 homologues with similarity scores of <e−5, most of which were isolated from marine sources. To analyze their predicted functions, we used the Bmp8 amino acid sequence to create a sequence similarity network50 and a genome neighborhood network51 (Figure 5A). Bmp8 itself is located within a small cluster of seven homologues that are all from Pseudoalteromonas and found within similar gene clusters, suggesting that they have the same enzymatic activity (Figures 5A (bromopyrrole cluster) and S8). Other Bmp8 homologues are located within clusters that have enzymes consistent with aromatic amino acid degradation, suggesting they are canonical members of the carboxymuconolactone decarboxylase super-family family (Figures 5A (CMD clusters) and S8). Notably, the major cluster, which is composed of 307 unique sequences, does not contain a consistent genomic context. Instead, homologues within this cluster appear to be often located near nucleases such as RecJ52 and the oxidative stress protein OsmC53 (Figure S8). Curiously, Pseudoalteromonas luteoviolacea 2ta16 contained two Bmp8 homologues, one within a homologous bromopyrrole gene cluster and the other within the RecJ/OsmC cluster context, which we named PLCMD. Bmp8 showed high sequence identity to PLCMD (32%), and all catalytic residues from Bmp8 are conserved (Figure S9). In fact, alignment of 1000 Bmp8 homologues generated conserved sequence motifs that are found throughout different isofunctional groups and correspond with key Bmp8 catalytic residues (Figure 5B). To determine if PLCMD is capable of debromination activity, we expressed and evaluated it in vitro. Surprisingly, PLCMD was able to catalyze debromination, albeit at a slower rate than Bmp8 (Figure 2B). This observation demonstrated that debromination activity can be found in the distantly related Bmp8 homologues even though they are likely responsible for orthogonal reactions in vivo.
Figure 5.
(A) Sequence similarity network generated from the Bmp8 amino acid sequence. Some clusters have been labeled with colocalized enzymes or putative functions. Bmp8 and PLCMD are indicated with a black outline in the bromopyrrole and RecJ/OsmC clusters, respectively. (B) Weblogo analysis of 1000 Bmp8 homologues. Residues in red were mutated, and the resulting Bmp8 variant was tested for activity (Figure 2B). Residues are numbered according to the Bmp8 sequence.
CONCLUSIONS
The high concentration of halogens within the ocean has facilitated the evolution of a rich source of halogenated natural products. While a wide variety of halogenases have been described and characterized, Bmp8 represents an unusual example of dedicated dehalogenation present in the bio-synthetic pathway of a natural product. This activity combined with the seeming similarity to human Dio prompted us to explore its structure and mechanism. Our data demonstrate that Bmp8 has adapted the conserved CMD fold to catalyze a dehalogenation reaction. By combining our structural results with computational calculations, we propose a reductive dehalogenation mechanism that first proceeds through an isomerization step before attack of the bromine atom by the Cys85 thiolate. The formation of the sp3-hybridized carbon is critical because it moves the bromine into closer contact with the nucleophilic Cys85 thiolate but also is necessary because the S–Br halogen bond is not strong enough to enable cleavage of the planar C–Br bond. This contrasts with human Dio that employs a catalytic selenocysteine. This unusual amino acid can form a much stronger Se–I bond and directly deiodinate human thyroid hormones. While the overall reaction between Bmp8 and Dio is similar, slight alterations in the mechanism are required to compensate for the differences in halogen bond energies. This mechanistic understanding of Bmp8 has allowed for bioinformatic identification of functional homologues and suggests that conserved enzymes in a variety of genomic contexts have the capacity to dehalogenate tetrabromopyrrole and alter its lifetime in the environment. This is significant as altering the persistence of tetrabromopyrrole in the environment has the potential to affect the settlement of new coral in the ocean. Together, our crystallographic, mutagenesis, and modeling work has offered new insights into the unusual chemistry of a biosynthetic debrominase.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Agarwal (Georgia Tech) for initial crystallographic screening of Bmp8, J. Noel and G. Louie (Salk Institute) for providing beamtime, and the staff of the ALS at beamlines 8.2.1 and 8.2.2 (Berkeley, California) for assistance with data collection.
Funding
This work was supported by the U.S. National Institutes of Health (R01-ES030316 to B.S.M.), the National Science Foundation (OCE-1837116 to B.S.M. and CHE-1764328 to K.N.H.), the Life Science Research Foundation through a Simons Foundation Fellowship to J.R.C., and NIH Marine Biotechnology Training Grant Predoctoral Fellowship (T32-GM067550) to T.N.P.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00318.
Experimental details, primers, crystallographic refinement statistics, energies from geometry optimization and single point calculations, gel filtration, maps of co-crystals and Bmp8 structural alignment, SDS-PAGE, calculated energetics, alternative Bmp8 mechanisms, activity assays, gene clusters, sequence alignment, and computational details (PDF)
Accession Codes
The accession codes are as follows: Bmp8, ADZ93287.1 (NCBI); PLCMD, ESP93448.1 (NCBI); APO Bmp8, 6OHI (PDB); Bmp8 C82A Tribromopyrrole Complex, 6OHJ (PDB).
The authors declare no competing financial interest.
REFERENCES
- (1).Agarwal V, Miles ZD, Winter JM, Eustáquio AS, El Gamal AA, and Moore BS (2017) Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev 117, 5619–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Fincker M, and Spormann AM (2017) Biochemistry of catabolic reductive dehalogenation. Annu. Rev. Biochem 86, 357–86. [DOI] [PubMed] [Google Scholar]
- (3).Mohn WW, and Tiedje JM (1992) Microbial reductive dehalogenation. Microbiol. Rev 56, 482–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fetzner S, and Lingens F (1994) Bacterial dehalogenases: Biochemistry, genetics, and biotechnological applications. Microbiol. Rev 58, 641–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).El Gamal A, Agarwal V, Rahman I, and Moore BS (2016) Enzymatic reductive dehalogenation controls the biosynthesis of marine bacterial pyrroles. J. Am. Chem. Soc 138, 13167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Agarwal V, El Gamal AA, Yamanaka K, Poth D, Kersten RD, Schorn M, Allen EE, and Moore BS (2014) Biosynthesis of polybrominated aromatic organic compounds by marine bacteria. Nat. Chem. Biol 10, 640–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).El Gamal A, Agarwal V, Diethelm S, Rahman I, Schorn MA, Sneed JM, Louie GV, Whalen KE, Mincer TJ, Noel JP, Paul VJ, and Moore BS (2016) Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair. Proc. Natl. Acad. Sci. U. S. A 113, 3797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sneed JM, Sharp KH, Ritchie KB, and Paul VJ (2014) The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc. R. Soc. London, Ser. B 281, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Marchand JA, Neugebauer ME, Ing MC, Lin C-I, Pelton JG, and Chang MCY (2019) Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 567, 420–4. [DOI] [PubMed] [Google Scholar]
- (10).Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, and Walsh CT (2005) Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature 436, 1191–4. [DOI] [PubMed] [Google Scholar]
- (11).Yamanaka K, Ryan KS, Gulder TAM, Hughes CC, and Moore BS (2012) Flavoenzyme-catalyzed atropo-selective N, C-bipyrrole homocoupling in marinopyrrole biosynthesis. J. Am. Chem. Soc 134, 12434–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hkansson K, Wipf P, Smith JL, Gerwick WH, and Sherman DH (2009) Metamorphic enzyme assembly in polyketide diversification. Nature 459, 731–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeöld A, and Bianco AC (2008) Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr. Rev 29, 898–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Maia AL, Goemann IM, Meyer ELS, and Wajner SM (2011) Deiodinases: The balance of thyroid hormone: Type 1 iodothyronine deiodinase in human physiology and disease. J. Endocrinol 209, 283–97. [DOI] [PubMed] [Google Scholar]
- (15).Yang M, and Zhang X (2014) Halopyrroles: A new group of highly toxic disinfection byproducts formed in chlorinated saline wastewater. Environ. Sci. Technol 48, 11846–52. [DOI] [PubMed] [Google Scholar]
- (16).Zheng J, McKinnie SMK, El Gamal A, Feng W, Dong Y, Agarwal V, Fenical W, Kumar A, Cao Z, Moore BS, and Pessah IN (2018) Organohalogens naturally biosynthesized in marine environments and produced as disinfection byproducts alter sarco/endoplasmic reticulum Ca2+ dynamics. Environ. Sci. Technol 52, 5469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chekan JR, and Moore BS (2018) Preparation and characterization of tetrabromopyrrole debrominase from marine proteobacteria. Methods Enzymol. 605, 253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, and Bricogne G (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr., Sect. D: Biol. Crystallogr 67, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Cowtan K (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr., Sect. D: Biol. Crystallogr 62, 1002–11. [DOI] [PubMed] [Google Scholar]
- (21).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, and Sonnenb DJ (2013) Gaussian 09, revision D.01, Gaussian Inc., Wallingford, CT. [Google Scholar]
- (23).Legault CY (2009) CYLview, Universitéde Sherbrooke. [Google Scholar]
- (24).Stephens PJ, Devlin FJ, Chabalowski CF, and Frisch MJ (1994) Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem 98, 11623–27. [Google Scholar]
- (25).Lee C, Yang W, and Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B: Condens. Matter Mater. Phys 37, 785–9. [DOI] [PubMed] [Google Scholar]
- (26).Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys 98, 5648–52. [Google Scholar]
- (27).Grimme S, Antony J, Ehrlich S, and Krieg H (2010) A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys 132, 154104. [DOI] [PubMed] [Google Scholar]
- (28).Takano Y, and Houk KN (2005) Benchmarking the conductor-like polarizable continuum model (CPCM) for aqueous solvation free energies of neutral and ionic organic molecules. J. Chem. Theory Comput 1, 70–7. [DOI] [PubMed] [Google Scholar]
- (29).Klamt A, and Schüürmann G (1993) COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc., Perkin Trans 2 2, 799–805. [Google Scholar]
- (30).Andzelm J, Kölmel C, and Klamt A (1995) Incorporation of solvent effects into density functional calculations of molecular energies and geometries. J. Chem. Phys 103, 9312–20. [Google Scholar]
- (31).Barone V, and Cossi M (1998) Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. A 102, 1995–2001. [Google Scholar]
- (32).Cossi M, Rega N, Scalmani G, and Barone V (2003) Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem 24, 669–81. [DOI] [PubMed] [Google Scholar]
- (33).Schweizer U, Schlicker C, Braun D, Köhrle J, and Steegborn C (2014) Crystal structure of mammalian selenocysteine-dependent iodothyronine deiodinase suggests a peroxiredoxin-like catalytic mechanism. Proc. Natl. Acad. Sci. U. S. A 111, 10526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Holm L, and Rosenström P (2010) Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ornston LN, and Stanier RY (1966) The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida.J. Biol. Chem 241, 3776–3786. [PubMed] [Google Scholar]
- (36).Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, and Nathan C (2002) Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295, 1073–7. [DOI] [PubMed] [Google Scholar]
- (37).Nunn CM, Djordjevic S, Hillas PJ, Nishida CR, and Ortiz De Montellano PR (2002) The crystal structure of Mycobacterium tuberculosis alkylhydroperoxidase AhpD, a potential target for antitubercular drug design. J. Biol. Chem 277, 20033–40. [DOI] [PubMed] [Google Scholar]
- (38).Hillas PJ, Soto Del Alba F, Oyarzabal J, Wilks A, and Ortiz De Montellano PR (2000) The AhpC and AhpD antioxidant defense system of Mycobacterium tuberculosis. J. Biol. Chem 275, 18801–18809. [DOI] [PubMed] [Google Scholar]
- (39).Cavallo G, Metrangolo P, Milani R, Pilati T, Priimagi A, Resnati G, and Terraneo G (2016) The halogen bond. Chem. Rev 116, 2478–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bayse CA, and Rafferty ER (2010) Is halogen bonding the basis for iodothyronine deiodinase activity? Inorg. Chem 49, 5365–7. [DOI] [PubMed] [Google Scholar]
- (41).Manna D, and Mugesh G (2012) Regioselective deiodination of thyroxine by iodothyronine deiodinase mimics: An unusual mechanistic pathway involving cooperative chalcogen and halogen bonding. J. Am. Chem. Soc 134, 4269–79. [DOI] [PubMed] [Google Scholar]
- (42).Goto K, Sonoda D, Shimada K, Sase S, and Kawashima T (2010) Modeling of the 5–deiodination of thyroxine by iodothyronine deiodinase: Chemical corroboration of a selenenyl iodide intermediate. Angew. Chem., Int. Ed 49, 545–7. [DOI] [PubMed] [Google Scholar]
- (43).Zefirov NS, and Makhon’kov DI (1982) X-Philic Reactions. Chem. Rev 82, 615–24. [Google Scholar]
- (44).Kunishima M, Friedman JE, and Rokita SE (1999) Transition-state stabilization by a mammalian reductive dehalogenase.J. Am. Chem. Soc 121, 4722–3. [Google Scholar]
- (45).Vasil’ev AA, and Engman L (1998) Iodothyronine deiodinase mimics. Deiodination of o, o’-diiodophenols by selenium and tellurium reagents. J. Org. Chem 63, 3911–3917. [Google Scholar]
- (46).Clark T, Hennemann M, Murray JS, and Politzer P (2007) Halogen bonding: The σ-hole. J. Mol. Model 13, 291–6. [DOI] [PubMed] [Google Scholar]
- (47).Amico V, Meille SV, Corradi E, Messina MT, and Resnati G (1998) Perfluorocarbon-hydrocarbon self-assembling. 1D infinite chain formation driven by nitrogen···iodine interactions. J. Am. Chem. Soc 120, 8261–2. [Google Scholar]
- (48).Metrangolo P, Panzeri W, Recupero F, and Resnati G (2002) Perfluorocarbon-hydrocarbon self-assembly: Part 16. 19F NMR study of the halogen bonding between halo-perfluorocarbons and heteroatom containing hydrocarbons. J. Fluorine Chem 114, 27–33. [Google Scholar]
- (49).Reich HJ, and Hondal RJ (2016) Why nature chose selenium. ACS Chem. Biol 11, 821–41. [DOI] [PubMed] [Google Scholar]
- (50).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta, Proteins Proteomics 1854, 1019–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zallot R, Oberg NO, and Gerlt JA (2018) ‘Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol 47, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Lovett ST, and Kolodner RD (1989) Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 86, 2627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Gutierrez C, and Devedjian JC (1991) Osmotic induction of gene osmC expression in Escherichia coli K12. J. Mol. Biol 220, 959–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.