Abstract
One strategy to enhance tumor retention of imaging agents or anti-cancer drugs is rational design of probes that undergo a tumor-specific enzymatic reaction which prevents them from being pumped out of the cell. Here, the anticancer agent olsalazine (Olsa) was conjugated to the cell-penetrating peptide RVRR. Taking advantage of a biologically compatible condensation reaction, single Olsa-RVRR molecules were self-assembled into large intracellular nanoparticles (Olsa-NPs) by the tumor-associated enzyme furin. Both Olsa-RVRR and Olsa-NPs could be readily detected with chemical exchange saturation transfer magnetic resonance imaging (CEST MRI) by virtue of exchangeable olsalazine hydroxyl protons. In vivo studies using HCT116 and LoVo murine xenografts showed that the OlsaCEST signal and anti-tumor therapeutic effect were 6.5-fold and 5.2-fold increased, respectively, compared to olsalazine without RVRR, with an excellent “theranostic correlation” (R2=0.97) between the imaging signal and therapeutic response (normalized tumor size). This furin-targeted MRI-detectable platform has potential for imaging tumor aggressiveness, drug accumulation, and therapeutic response.
Unidirectional energy-dependent drug efflux mediated by cellular membrane proteins results in the failure of many anti-cancer chemotherapeutic agents1. Several strategies have been developed to overcome this multidrug resistance (MDR), including the inhibition of MDR efflux pumps with P-glycoprotein specific peptides or antibodies2, multi-target drug discovery3, and encapsulation of drugs inside synthetic nanocarriers4. Overall, successful use of nanomedicines has been rather disappointing, with less than 1% of the injected dose reaching the intracellular targets upon systemic administration5. Furthermore, most nanomedicines are deposited in the liver and spleen through non-specific uptake, leading to side toxicity concerns.
Intracellular self-assembly of small-molecule drugs into nanostructures is one effective strategy to increase the concentration of drugs locally and prolong their exposure time6,7. Such nanoparticles can be expected to increase the local concentration of anti-tumor agents, increasing sensitivity while minimizing non-target toxicity. The potential benefits of tumor-specific intracellular assembly prompted us to develop an enzyme-responsive theranostic platform for tumor imaging and therapy. To this end, we have chosen furin as enzyme, a type of proprotein convertase upregulated in multiple malignancies. Aside of being a potential therapeutic target8, the enzyme may serve as a biomarker to predict tumor progression9, with the degree of expression correlating to 5-year survival rates10. As a small molecule drug, we chose olsalazine (Olsa), a DNA methylation inhibitor11 that acts as a potential broad-spectrum anticancer agent. Importantly, belonging to the group of salicylates, the hydroxyl proton on olsalazine provides a distinct contrast on chemical exchange saturation transfer magnetic resonance imaging (CEST MRI).
CEST MRI is an imaging technique that can detect non-labeled, native molecules indirectly by manipulating the water proton signal through selective saturation of exchangeable protons12, 13. Among injectable diamagnetic CEST (diaCEST) agents, salicylic acid and its derivatives such as olsalazine provide labile protons with large chemical shift differences with water (8-12 ppm)14, 15, enabling highly specific detection of these molecules with minimal interference from common endogenous hydroxyl, amine, and amide groups. CEST MRI has now seen clinical translation for imaging dilute biomarkers associated with the microenvironment16, 17 as well as administered diaCEST contrast agents18. Compared with traditional MRI contrast enhancement, i.e. manipulating the T1 and T2 relaxation times through the use of metallic contrast agents, CEST MRI has several distinct advantages. First, the signal can be switched “on” and “off” through suitable RF irradiation, enabling simultaneous detection of multiple agents with different exchangeable protons, each resonating at a specific CEST frequency14. Second, the diamagnetic compound of interest does not need to be labeled and hence shows unaltered pharmacokinetic and safety profiles, which is important in (theranostic) drug development. Recently, CEST MRI has been applied to detect glucose19, glutamate20, glycogen21, glycosaminoglycan22, lactate23, mucin24, and certain enzymes25.
Typically, a concentration of labile protons in the millimolar range is needed for the detection of diaCEST agents. To enhance the sensitivity and signal-to-noise ratio, various strategies have been developed to enhance the sensitivity of molecular detection using polymers26, dendrimers14, liposomes27, nanofibers28, and nanoparticles29, aiming to accumulate sufficient labile protons in vivo for detecting contrast. An alternative strategy may be to employ molecular self-assembly of nanoscale/microscopic structures30,31, which has not been explored yet for designing CEST MRI-detectable contrast agents.
In this study, we rationally designed the single small molecular probe Olsa-RVRR as a CEST imaging agent with anti-tumor drug activity. Conjugation of the positively charged cell-penetrating peptide RVRR to Olsa serves two functions: it induces penetration of the agent across the membrane and it is a substrate for cleavage by the enzyme furin. After Olsa-RVRR enters furin-expressing cells, the peptide is cleaved by furin near the Golgi complex32, initiating a biocompatible click condensation reaction between the GSH-induced 1,2-aminothiol group (D-cysteine) and the cyano group of the 2-cyanobenzothiazole (CBT) motif, initiating the formation of clusters of aggregated olsalazine nanoparticles (Olsa-NPs) that enhance CEST signal. This reaction is also a biomimetic regeneration pathway of luciferin in fireflies33, 34. In this study, we hypothesized that the catalyzed intracellular formation of Olsa-NPs may not only enhance the intracellular concentration of olsalazine, but also prolong its retention time, leading to an amplified, localized OlsaCEST signal as well prolonged drug exposure.
Self-assembly of Olsa-RVRR in solution.
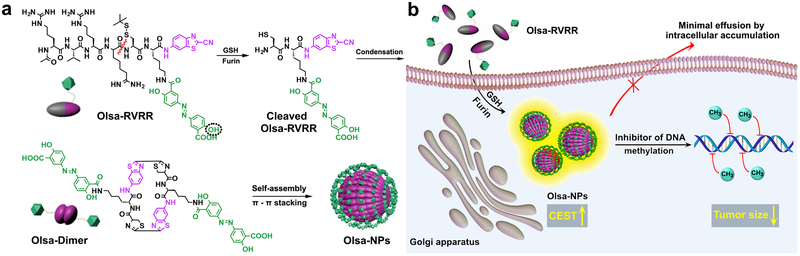
The intracellular self-assembly of Olsa-NPs was achieved through a sequence of reactions as outlined in Figure 1. The detailed synthetic procedures and characterization of Olsa-RVRR are shown in Supplementary Scheme 1 and Supplementary Figs. 1 and 2. Furin-controlled condensation of Olsa-RVRR was confirmed by combining high performance liquid chromatography (HPLC) and high-resolution matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (HR-MALDI-TOF MS).
Fig. 1. Schematic illustration for the formation of Olsa-NPs by furin-mediated intracellular reduction and condensation of Olsa-RVRR, resulting in enhanced CEST signal and tumor treatment efficacy.
(a) Self-assembly of Olsa-RVRR into Olsa-NPs through a series of steps. Red line indicates the site of furin cleavage, and the dotted circled hydroxyl group indicates the exchangeable hydroxyl proton that provides OlsaCEST signal at 9.8 ppm from the water frequency. (b) After Olsa-RVRR enters the cytoplasm of furin-overexpressing cells (HCT116 cells in this study), it undergoes cleavage of the peptide by activated furin near the Golgi complex, where the reduction of disulfide by glutathione (GSH) generates cleaved Olsa-RVRR. Amphiphilic oligomers (mostly dimers) are then formed from the click reaction between two cleaved Olsa-RVRR molecules, followed by self-assembly into Olsa-NPs as a result of intermolecular π-π stacking. The intracellular accumulation of Olsa-NPs then serves as a reservoir of olsalazine molecule enhancing CEST contrast and inhibiting DNA methylation for tumor therapy.
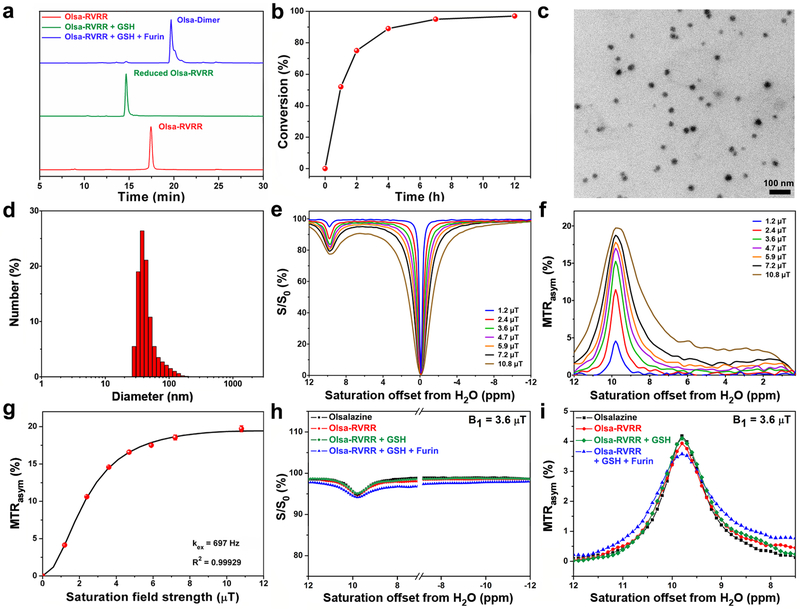
After treatment of 25 μM Olsa-RVRR with 250 μM GSH for 2 h at 37 °C at pH=7.4, the disulfide bond of Olsa-RVRR was reduced, producing a new compound (reduced Olsa-RVRR) as identified by the appearance of the HPLC peak at 14.7 min (Fig. 2a). After co-incubation of Olsa-RVRR with 250 μM GSH and 0.5 nmol U−1 furin for 12 h at 37 °C, the reduced Olsa-RVRR was gradually cleaved by furin to yield an active intermediate (cleaved Olsa-RVRR in Fig. 1a), which instantly condensed with each other to yield Olsa-Dimers (retention time at 19.6 min seen in Fig. 2a). Both the reduced Olsa-RVRR and Olsa-Dimer peaks from HPLC were collected and further characterized by HR-MALDI-TOF/MS (Supplementary Figs. 3 and 4). Olsa-RVRR was converted to Olsa-Dimer with a half-life less than 1 h (Fig. 2b), indicating efficient furin cleavage. Eventually, the amphiphilic Olsa-Dimer self-assembled to form Olsa-NPs due to noncovalent π-π stacking interaction5. Immediately after sonication, carbon-coated copper grids were dipped into the above reaction solutions, freeze-dried, and observed with transmission electron microscopy (TEM). TEM images revealed that, after furin treatment, Olsa-RVRR formed nanoaggregates with an average diameter of 22.89 ± 3.83 nm (Fig. 2c). In contrast, the Olsa-RVRR solution without furin did not produce any nanoparticles (Supplementary Fig. 5a). Dynamic light scattering (DLS) measurements showed that the generated Olsa-NPs had an average hydrodynamic diameter of 47.93 ± 10.45 nm (Fig. 2d).
Fig. 2. Physicochemical characterization of Olsa-RVRR and Olsa-NPs in solution.
(a) HPLC chromatogram of 25 μM Olsa-RVRR (red), 25 μM Olsa-RVRR + 250 μM GSH incubated for 2 h (green), and 25 μM Olsa-RVRR + 250 μM GSH + 0.5 nmol U−1 furin incubated for 12 h (blue). (b) The enzymatic conversion of Olsa-RVRR (25 μM) into Olsa-Dimers in the presence of 250 μM GSH and 0.5 nmol U−1 furin as function of time. (c) TEM image and (d) DLS size distribution of Olsa-NPs after incubation of 25 μM Olsa-RVRR + 250 μM GSH + 0.5 nmol U−1 furin for 12 h in furin buffer. (e) Z-spectra and (f) MTRasym values of 10 mM olsalazine for different saturation powers. (g) QUESP plot for 10 mM olsalazine. (h) Z-spectra and (i) MTRasym spectra of 2.5 mM olsalazine and 5 mM Olsa-RVRR before and after addition of 10 mM GSH and 2 nmol U−1 furin. B0=11.7 T, B1=3.6 μT, tsat=4 s, T=37 °C. All subpanels reflect representative data from in vitro experiments repeated three times.
Two control compounds were synthesized to validate that the self-assembly Olsa-NPs was indeed catalyzed by furin. The first control (CTR1) is an isomer of Olsa-RVRR but with the scrambled peptide sequence RKRCRV in lieu of the furin substrate RVRR. This substrate cannot be cleaved by furin to expose the 1,2-aminothiol group (Supplementary Scheme 2 and Supplementary Figs. 6 and 7). The second control (CTR2) is an olsalazine-conjugated KVRVRR sequence that is short of the CBT motif (Supplementary Scheme 3 and Supplementary Figs. 8 and 9). Thus, neither CTR1 nor CTR2 are substrates able to initiate a condensation reaction in the presence of GSH and furin, and the formation of nanoparticles would therefore not occur. To test this hypothesis, 25 μM CTR1 and CTR2 were incubated with 250 μM GSH for 2 h at 37 °C and pH=7.4, reducing the disulfide bond of both CTR1 and CTR2 (Supplementary Figs. 10-12). However, after co-incubation of CTR1 and CTR2 with 250 μM GSH and 0.5 nmol U−1 furin for 12 h at 37 °C, the reduced CTR1 could not be further cleaved, and reduced CTR2 was cleaved to cleaved CTR2 without condensation (Supplementary Figs. 10 and 13). TEM images demonstrated that no nanoparticles were formed when incubating CTR1 or CTR2 with GSH and furin (Supplementary Fig. 5c and Fig. 5d).
CEST properties of Olsalazine.
Measured Z-spectra (Fig. 2e) and MTRasym spectra (Fig. 2f) demonstrated a well-defined CEST peak at ~9.8 ppm from the water proton signal. The MTRasym increased as the saturation field strength B1 was raised from 1.2 to 10.8 μT. B1=3.6 μT was chosen for all further experiments based on the intensity of the OlsaCEST signal and scanner hardware settings for the in vivo studies. For a concentration range of 0.625 mM to 10 mM, MTRasym values at different olsalazine concentrations showed a linear correlation where MTRasym (%) = 0.03 + 1.51 × Colsalazine (mM) (Supplementary Fig. 14). The proton exchange rate (kex) of olsalazine at pH 7.4 was estimated to be 697 Hz by using the QUESP method35 (Fig. 2g), much smaller than the chemical shift difference value (9.8 ppm × 500 Hz/ppm = 4.9 kHz), indicating a slow exchange rate on the NMR time scale. The OlsaCEST signal was found to be stable between pH=6.5-8.0 (Supplementary Fig. 15).
We next compared the efficacy of generating CEST signal between Olsa-RVRR and Olsa-NPs (Figs. 2h,i). The OlsaCEST signal for 5 mM Olsa-RVRR was found to be similar to 2.5 mM olsalazine, attributable to the decrease of the number of exchangeable -OH proton by half for Olsa-RVRR compared to olsalazine. After furin-induced condensation and self-assembly to Olsa-NPs, as demonstrated by DLS measurements (Supplementary Fig. 16), the spectra broadened slightly, yet the intensity of OlsaCEST signal did not change significantly, indicating that most exchangeable protons remained unchanged in their interaction with water protons. Hence, modification of olsalazine and its self-assembly process does not affect the OlsaCEST signal.
In vitro cell experiments.
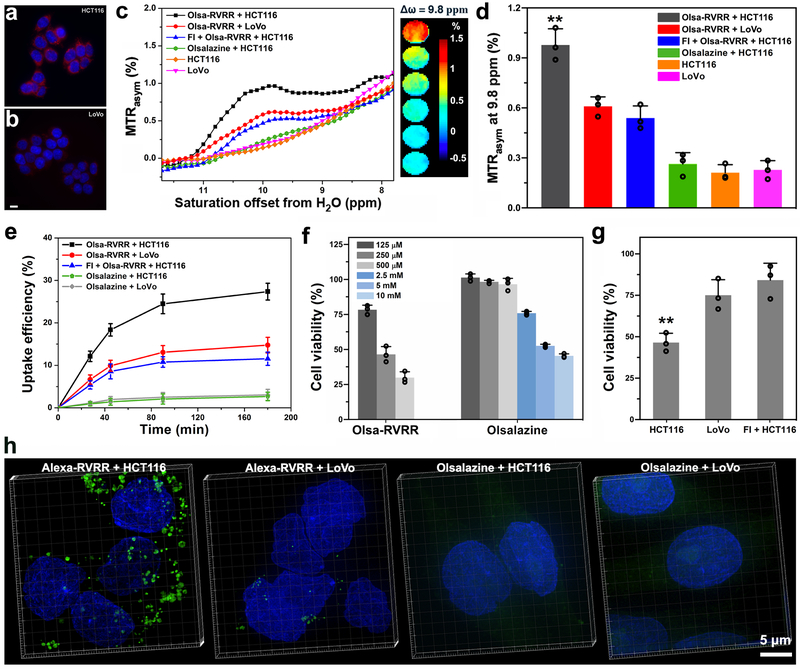
In order to assess the specificity of furin-mediated enhancement of the MRI CEST signal and accompanying cellular toxicity, the furin-overexpressing human colon carcinoma cell line HCT116 and the furin-lowexpressing human colon carcinoma cell line LoVo were tested side-by-side. Differences in furin expression were verified by immunofluorescent staining and western blotting (Figs. 3a,b and Supplementary Fig. 17). There was no significant difference between the intracellular GSH content of HCT116 and LoVo cells (Supplementary Fig. 18). After incubation with 500 μM Olsa-RVRR for 3 h, cells were washed and collected for CEST MRI measurements. As a result of higher furin expression, HCT116 cells exhibited a 1.8-fold larger OlsaCEST signal compared to LoVo cells, resulting from the higher accumulation of olsalazine (Figs. 3c,d). When HCT116 cells were co-incubated with Olsa-RVRR and the furin inhibitor II hexa-D-arginine amide (FI), a 45% reduction in OlsaCEST signal was observed, providing evidence that the CEST signal is enhanced by the furin-catalyzed intracellular assembly of Olsa-RVRR. To further prove that cellular penetration and intracellular nanoaggregation is essential for generating CEST contrast, HCT116 cells were incubated with 500 μM free olsazaline (without RVRR) for 3 h. The OlsaCEST signal was much lower compared to Olsa-RVRR, with no significant difference between free olsalazine-incubated cells and non-incubated cells (p>0.05).
Fig. 3. In vitro cell studies.
(a,b) Validation of furin expression in HCT116 (a) and LoVo cells (b) using anti-furin immunofluorescent staining (red). Nuclei are counterstained with DAPI (blue). Scale bar=10 μm. (c) MTRasym spectra and OlsaCEST signal (from top to bottom) of HCT116 cells + 500 μM Olsa-RVRR, LoVo cells + 500 μM Olsa-RVRR, HCT116 cells + 100 μM FI + 500 μM Olsa-RVRR, HCT116 cells + 500 μM olsalazine, and non-incubated HCT116 and LoVo cells. (d) OlsaCEST signal measured at 9.8 ppm corresponding to Fig. 3c. Data are shown as mean±SD; n=3 independent experiments; one-way ANOVA, followed by Dunnett's post-hoc test; ***P<0.001 vs. all other groups. (e) Time-dependent cellular uptake of the different substrates. Data are shown as mean±SD (n=3 independent experiments). (f) Cell viability of HCT116 cells incubated for 48 h with Olsa-RVRR and olsalazine as a function of substrate concentration. Data are shown as mean±SD (n=3 independent experiments). (g) Cell viability of HCT116 cells, LoVo cells, and FI-pretreated HCT116 cells incubated with 250 μM Olsa-RVRR for 48 h. Data are shown as mean±SD; n=3 independent experiments; two-tailed Student’s t-test; **P<0.01 vs. all other groups. (h) 3D-SIM super-resolution fluorescence images of HCT116 and LoVo cells incubated with 8 μM Alexa-RVRR or 8 μM Alexa 488 for 3 h. Green fluorescence represent furin-mediated, self-assembled Alexa 488 nanoparticles. Cell nuclei were counterstained with DAPI (blue). All subpanels reflect representative data from in vitro experiments repeated three times.
HPLC was then used to confirm furin-controlled condensation of Olsa-RVRR in cells. After incubation of HCT116 cells with Olsa-RVRR at 37 °C for 3 h, the cell lysate was analyzed by HPLC. The chromatogram showed the peak of the Olsa-Dimer as expected (Supplementary Fig. 19). To further characterize furin-mediated accumulation of Olsa-RVRR, we measured cell uptake kinetics by HPLC after incubation of HCT116 cells and LoVo cells with Olsa-RVRR, Olsa-RVRR with FI, and free olsalazine without RVRR. Both cell lines showed a faster absorption of agents at early times, with much higher cell uptakes for Olsa-RVRR than olsalazine (Fig. 3e). At 3 h, the absorption of Olsa-RVRR inside HCT116 cells reached 27%, nearly twofold of that for LoVo cells (15%). For HCT116 cells co-incubated with Olsa-RVRR and FI, the uptake efficiency decreased to 12%. To test our hypothesis that the higher cell content of olsalazine using Olsa-RVRR would be accompanied by a higher drug toxicity, we measured cell viability at 48 h post-incubation (Fig. 3f). A marked drug toxicity was observed for Olsa-RVRR at 125 μM, while for olsalazine no reduced cell viability could be observed up to 500 μM, and the dose needed to be increased 20-fold (2.5 mM) to obtain a comparable toxic effect. For Olsa-RVRR, cytotoxicity increased further with prolonged incubation times (Supplementary Fig. 20), as a result of nanoaggregation and the slow efflux rate of Olsa-NPs but not olsalazine in HCT116 cells (Supplementary Fig. 21). In addition, the toxicity of Olsa-RVRR towards different cells was also compared (Fig. 3g). For 250 μM Olsa-RVRR, the cell viability of HCT116 cells (46%) was significantly lower than those of LoVo cells (75%) and FI-pretreated HCT116 cells (78%) (Fig. 3g).
To further demonstrate that accumulation of olsalazine and enhanced CEST signal resulted from a condensation-induced self-assembly, 500 μM of the control compounds CTR1 or CTR2 were incubated with HCT116 cells for 3 h, and cells were then washed and collected for CEST MRI measurements (Supplementary Fig. 22). As neither CTR1 or CTR2 is able to undergo a condensation reaction to form nanoparticles, their CEST signal were 4.4-fold and 5.2-fold less compared to Olsa-RVRR (after subtracting the values from non-treated cells), respectively. However, due to the abundance of the positively charged amino acid Arg, CTR1 and CTR2 exhibited a 2.8-fold and 2.3-fold larger OlsaCEST signal compared to free olsalazine, respectively. When compared to 250 μM Olsa-RVRR, a much lower drug toxicity was observed for CTR1 (80.9%) or CTR2 (84.7%) at the same concentration (Supplementary Fig. 23), due to the lower concentration of olsalazine in CTR1 or CTR2 incubated cells.
To evaluate the mechanism of cell death induced by Olsa-RVRR, a DNA methyltransferase (DNMT) activity/inhibition assay kit was used to detect the inhibition of DNMT for Olsa-RVRR, olsalazine, CTR1, or CTR2 (Supplementary Fig. 24). The DNMT activity in Olsa-RVRR treated cells was decreased by nearly 50%, while no significant DNMT inhibition could be observed for olsalazine, CTR1, or CTR2 incubated cells (***P<0.001). These results are further proof that the observed DNA methylation inhibition is dependent on a higher drug concentration and prolonged exposure time resulting from the self-assembly of Olsa-NPs.
Having studied both furin high- and low-expressing tumor cells, we decided to also include a furin-deficient normal human colon cell line, i.e., CCD-18Co. Amongst the three cell lines HCT116, LoVo, and CCD-18Co, the lowest furin expression occurred in CCD-18Co cells accompanied by the lowest OlsaCEST signal after incubation with 500 μM Olsa-RVRR for 3h (Supplementary Fig. 17 and Fig. 25). The OlsaCEST signal was 5.0-fold and 2.1-fold lower compared to HCT116 and LoVo cells, respectively (after background signal correction of non-treated cells). After treatment of 250 μM Olsa-RVRR for 48h, lowest drug toxicity (86.4%) was observed for CCD-18Co cells (Supplementary Fig. 26), resulting from a lack of intracellular furin-mediated condensation and self-assembly of Olsa-NPs. After incubation of HCT116, LoVo, and CCD-18Co cells with 100 μM Olsa-RVRR at 37 °C for 12 h, the cell lysates were analyzed by HPLC (Supplementary Fig. 27). The HPLC peak area under the curve (AUC) values of Olsa-RVRR, reduced Olsa-RVRR, and Olsa-Dimer were calculated for each cell line. The percentage of Olsa-Dimer in HCT116, LoVo, and CCD-18Co cells was 88%, 52%, and 15%, respectively, in agreement with their furin expression levels. The total AUC value for all three peaks was about 1.9-fold larger for HCT116 compared to the LoVo cell lysate, and 3.7-fold larger than the CCD-18Co cell lysate, again resulting from the self-assembly of Olsa-NPs.
To get more visual insight into the intracellular self-assembly of Olsa-NPs, we synthesized the analogue Ac-Arg-Val-Arg-Arg-Cys(StBu)-Lys(Alexa 488)-2-cyanobenzothiazole (Alexa-RVRR, Supplementary Scheme 4 and Figs. 28-30), which replaced the olsalazine motif on Olsa-RVRR with the fluorophore Alexa 488. Incubation of HCT116 cells with a Golgi marker for 24 h followed by co-staining of the cells with Alexa-RVRR (8 μM) for 3 h revealed a good overlap of red fluorescence (from the Golgi marker) and green fluorescence (from the condensation product), confirming that intracellular condensation occurred at or near Golgi apparatus in cells where furin was produced. However, for LoVo cells having a lower level of furin expression, the green fluorescence was much weaker (Supplementary Fig. 31). To better observe the formation of nanoparticles in cells, after incubation of HCT116 cells or LoVo cells with 8 μM Alexa-RVRR for 3 h, three-dimensional structured illumination microscopy (3D-SIM) super-resolution fluorescence images were acquired to clearly show the clustering of green fluorescent nanoparticles in HCT116 cells, but much less inside LoVo cells (Fig. 3h and Supplementary Movies 1 and 2). In contrast, both cells showed very weak green fluorescence after incubation with 8 μM Alexa 488 alone (Fig. 3h and Supplementary Movies 3 and 4). As can be seen in the high-magnification 3D-SIM image of Alexa-RVRR-incubated HCT116 cells in Fig. 3h (Supplementary Fig. 32), the fluorescent dots were composed of many individual smaller fluorescent nanoparticles.
To get a better structural insight into the intracellular transformation of Olsa-RVRR into Olsa-NPs, we performed Raman imaging of live HCT116 cells after incubation with olsalazine or Olsa-RVRR (Supplementary Fig. 33). Generation of Raman spectral maps with a high signal-to-noise ratio requires long acquisition times – amounting to a few hours for a single cell. Therefore, to image the intracellular agents more accurately in a steady state, Raman imaging was also performed on fixed cells (Supplementary Fig. 34). Raman images generated from the 1168 cm−1 mode (characteristic of Olsa-NPs) confirmed the presence of Olsa-NPs in Olsa-RVRR-incubated HCT116 cells. No Olsa-NPs could be seen in free olsalazine-incubated HCT116 cells. Furthermore, the Raman images obtained from Olsa-RVRR-incubated HCT116 cells showed a higher Olsa-NP content compared to LoVo cells.
In vivo animal studies.
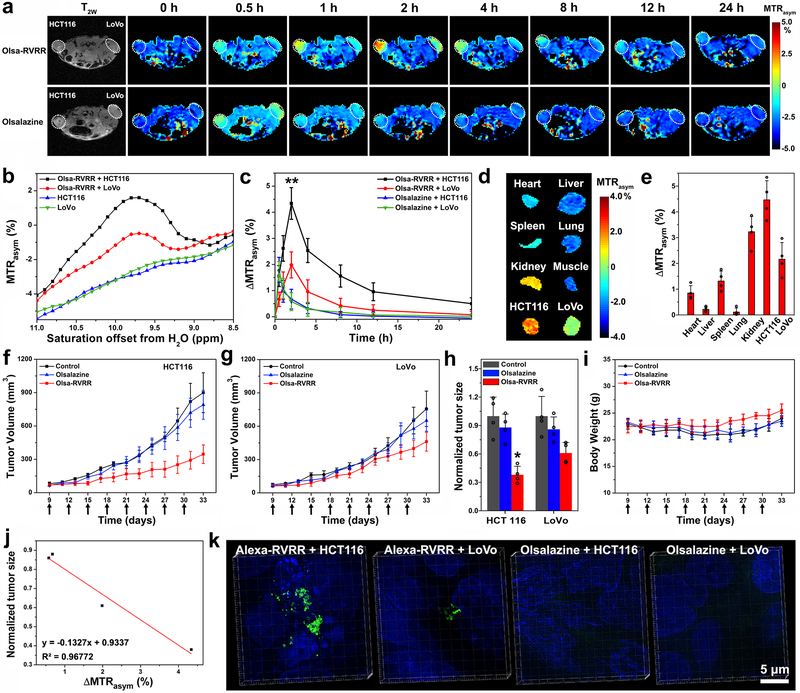
NU/J nude mice were subcutaneously (s.c.) injected with 1×106 HCT116 cells in the left and 1×106 LoVo cells in the right flank. When the tumor reached a volume of 100-200 mm3, 0.2 mmol/kg bw (278 mg/kg) Olsa-RVRR or 0.2 mmol/kg bw (69 mg/kg) olsalazine was injected intravenously (i.v.). Serial CEST MRI was obtained over 24 h to determine the time course of substrate uptake within the tumors (Figs. 4a-c). The OlsaCEST contrast was higher for HCT116 compared to LoVo for all time points. For HCT116, a maximum OlsaCEST signal increase of 4.3% was observed at 2 h post-injection (p.i.) of Olsa-RVRR, which was two-fold higher than that for the LoVo tumor (2.0%) at this time point, suggesting the accumulation of the olsalazine motif via the formation of Olsa-NPs in HCT116 tumors. In contrast to LoVo, OlsaCEST signal could still be detected in the HCT116 tumor at 24 h p.i. For the olsalazine group, no significant signal differences were detected between the tumors at any timepoint. Even with the two-fold higher number of exchangeable protons on olsalazine compared to Olsa-RVRR, the maximum intensity observed at 0.5 h p.i. was only ~1.8%, followed by a gradual decrease at 8 h p.i., after which the signal returned to background level. In order to assess the overall body biodistribution of the two substrates, mice were sacrificed at 2 h p.i. for Olsa-RVRR or 0.5 h p.i. for olsalazine, organs were collected, and conducted ex vivo CEST imaging. Mice injected with Olsa-RVRR showed the highest OlsaCEST signal in HCT116 tumor among the organs, followed by the kidney (Figs. 4d,e). For mice injected with olsalazine, both tumor sites showed comparable CEST signals (Supplementary Fig. 35). These ex vivo data are consistent with the in vivo imaging results.
Fig. 4. In vivo theranostic studies.
(a) Dynamic T2-weighted and OlsaCEST serial MRI of tumor-bearing mice after i.v. injection of 0.2 mmol/kg Olsa-RVRR or olsalazine (left: HCT116; right: LoVo). (b) MTRasym spectra of HCT116 and LoVo tumors before and 2 h after injection of Olsa-RVRR. (c) Time course of OlsaCEST signal for tumors after background correction by the subtraction of the MTRasym value at 0 h. Data are shown as mean±SD for n=4 mice; one-way ANOVA, followed by Dunnett's post-hoc test; ***P<0.001 vs. all other groups. (d) Ex vivo OlsaCEST images of various organs from tumor-bearing mice 2 h after i.v. injection of 0.2 mmol/kg Olsa-RVRR. (e) Organ distribution of OlsaCEST signal relative to muscle signal used as reference. Data are shown as mean±SD for n=4 mice. (f, g) Anti-tumor effects of olsalazine and Olsa-RVRR for HCT116 (f) and LoVo (g) tumors. Arrows indicate time points of repeated drug administration (QD3×8) after tumor cell injection. Data are shown as mean±SD (n=4 mice). (h) Relative tumor sizes at day 33 normalized to PBS group (set at 1.0). Data are shown as mean±SD for n= 4 mice; two-tailed Student’s t-test; *P=0.0179 vs. Olsa-RVRR group, **P<0.01 vs. control and olsalazine groups. (i) Measured mouse body weight over time time. Data are shown as mean±SD for n=4 mice. (j) Correlation between tumor OlsaCEST signal (HCT116 and LoVo) at 2h (Fig. 4c) and normalized tumor size (HCT116 and LoVo) at day 33 (Fig. 4h, n=4 mice). (k) 3D-SIM images of HCT116 and LoVo tumors after i.v. injection of 50 nmol Alexa-RVRR or Alexa 488. Green fluorescence represents self-assembled Alexa 488 nanoparticles. Cell nuclei were counterstained with DAPI (blue). The experiments in a-b, and d were repeated independently four times with similar results. The experiments in k were repeated independently three times with similar results.
We then studied the anti-tumor treatment effect. When the tumor volumes reached 50-100 mm3, mice were randomly divided into three groups (n=4 each). 0.1 mmol/kg bw 139 mg/kg of Olsa-RVRR, 0.1 mmol/kg bw (35 mg/kg) olsalazine, and PBS were individually injected into each group by i.v. (Q3D×8). The Olsa-RVRR substrate showed a more effective inhibition of tumor growth compared to olsalazine for both the HCT116 and LoVo tumor, resulting from the enhanced intracellular tumor retention of olsalazine, in agreement with the above CEST MRI results (Figs. 4f-h). Compared to the PBS control group set at 100%, at day 33 the average volume of the HCT116 and LoVo tumor reduced to ~38% and 61%, respectively, for the Olsa-RVRR-treated group, and to ~88% and 86% for the olsalazine-treated group. Direct evidence of effective treatment effect showing individual tumor sizes at day 33 is shown in Supplementary Fig. 36. All mice did not show any significant differences in body weight, in support of a tumor-specific toxicity (Fig. 4i). When we compared the therapeutic outcome to the diagnostic imaging data at 2 h p.i., an excellent “theranostic correlation” (R2=0.97) was found between the magnitude of the OlsaCEST signal and therapeutic response (normalized tumor size) (Fig. 4j).
Finally, we used 3D-SIM to prove the existence of self-assembled nanoparticles within tumor cells in vivo. Instead of using Olsa-RVRR, 50 nmol Alexa-RVRR or 50 nmol Alexa 488 was injected i.v., and tumors were resected at 2 h post-injection and processed for 3D-SIM imaging. A much higher quantity of green fluorescent nanoparticles was visible inside Alexa-RVRR-treated HCT116 tumors compared to LoVo tumors, while a very week green fluorescence was detected for animals injected with Alexa 488 only (Fig. 4k and Supplementary movies 5-8).
Based on a furin-mediated CBT-Cys click reaction followed by intracellular nanoparticle self-assembly, we successfully developed the olsalazine derivative Olsa-RVRR for furin-targeted cancer therapy and furin-specific CEST MRI. The MRI results showed that, compared to free olsalazine, Olsa-RVRR displayed a 15-fold and 6.5-fold increase of CEST signal for HCT116 cancer cells in vitro and in vivo, respectively. As a consequence of the enhanced intracellular retention and prolonged drug exposure time, Olsa-RVRR showed a 20-fold and 5.2-fold increased cytotoxicity for HCT116 cancer cells in vitro and in vivo, respectively. 3D-SIM images confirmed in situ formation of nanoparticles inside HCT116 cells in vitro and in vivo. The majority of injected Olsa-RVRR was observed within the tumor and kidney, suggesting renal excretion, in agreement with other studies36, 37.
Olsalazine gained Food and Drug Administration approval almost three decades ago as an anti-inflammatory drug for treatment of inflammatory bowel disease and ulcerative colitis. More recently, it was positively screened as a potential broad-spectrum anticancer agent, and identified as a DNA methylation inhibitor11. Olsalazine has shown efficacy to inhibit the development of colorectal cancer in pre-clinical models38-40 and tumor patients41. Being a therapeutic drug with inherent CEST MRI contrast, no additional labeling molecules are needed for detection that can potentially affect drug distribution and that would also require additional approval by the FDA. While MRI produces high spatial resolution images with excellent soft tissue contrast, inherent CEST contrast has been exploited for several compounds, including a series of anti-tumor drugs42. Until now, only a few enzyme-responsive CEST MRI agents have been studied in vivo25, 43-45, with only one pertaining to the study of intracellular protease43. Yet, few of these have CEST signal at the large 9.8 ppm frequency shift from water that is present in olsalazine and other salicylates, where the detection has almost no interference from endogenous background signals in vivo. Unlike the CEST signal for salicylic acid, where the proton exchange rate (ksw) is strongly dependent on pH15, the OlsaCEST signal is stable within pH values from 6.5 to 8.0. This is important, as solid tumors are characterized by an acidic microenvironment with different types of tumors exhibiting distinct extracellular pH values (pHe 6.5-7.2)46, 47, while having a higher intracellular pH (pHi) of about 7.4-7.648. The pH independence of the OlsaCEST signal would enable the use of olsalazine in different furin-expressing tumor models without contaminating signal changes associated with endogenous tumor-derived pH changes. The enhancement of acquired OlsaCEST signal for furin-overexpressed HCT116 tumor xenograft is about 4.3%, which is significantly larger compared to other CEST-detectable compounds, i.e, 1% for dextran26, 2% for underglycosylated mucin-124, 1% for the reporter probe 5-methyl-5,6-dihydrothymidine49, and 2%-3% for injectable hydrogels28, 50.
As for possible clinical translation, an Olsa-RVRR dosage extrapolation from our mouse studies to humans can be calculated based on the FDA body surface area normalization method using the following formula51: Human-equivalent dose (HED, mg/kg) = 139 mg/kg × (3/37) = 11.3 mg/kg. Besides colon cancer, Olsa-RVRR may be used in theranostic studies of other forms of cancer known to overexpress furin, including breast52, lung53, ovary10, head and neck55, and brain tumors55. Beyond furin, a myriad of other protein-based enzymes with aberrant activity or expression are currently used as biomarkers for early disease detection and monitoring treatment response. We hope that the current study may inspire further attempts to rationally design enzyme-specific substrates that can be condensed by a simple click reaction to form intracellular nanoassemblies and used as a theranostic platform for imaging cancer therapy.
Methods
In vitro CEST MRI of Olsalazine solutions.
Olsalazine was dissolved in 10 mM PBS, pH=7.4 at concentrations from 0.625 to 10 mM, and titrated using 3M HCl or NaOH to specific pH values of 5.0, 5.5, 6.0, 6.5, 7.0, 7.5 and 8.0. Sample phantoms were made by placing solutions into 1 mm glass capillaries assembled in a customized plastic holder for CEST MRI. Unless otherwise mentioned, samples were kept at 37 °C. MRI measurements were performed on a Bruker 11.7 T vertical bore MR scanner with a 20-mm birdcage transmit/receive coil. For CEST MRI, a modified rapid acquisition with relaxation enhancement (RARE) pulse sequence was used with the following parameters24: Repetition time/echo time (TR/TE)=6,000/5 ms, RARE factor=32, number of averages (NA)=2, slice thickness=2 mm, field of view (FOV)=14×17 mm, matrix size=64×64, resolution=0.22×0.27 mm, B1=3.6 μT, and tsat=4, with saturation offset frequencies from −12 to 12 ppm (with 0.3 ppm increments) with the water resonance set at 0 ppm. The total acquisition time was 32 min and 48 sec. B0 inhomogeneity was corrected using the water saturation shift referencing method. CEST signal was quantified by MTRasym = Ssat(−Δω)/S0 −Ssat(Δω)/S0, where Ssat(−Δω), Ssat(Δω), and S0 represents the water signal with a saturation frequency offset at −Δω, Δω, and without saturation, respectively.
In vitro cell studies.
Furin-expression on HCT116 and LoVo cells was visualized by anti-furin immunofluorescence staining, using overnight incubation with rabbit anti-furin antibody ab183495 (Abcam, 1:100) and anti-rabbit IgG (H&L) secondary antibody (Life Technologies, 1:500) for 1 h as the second step. For immunoblotting, cell lysates from cultured cells were prepared in modified RIPA buffer. Proteins were separated by SDS-PAGE, blotted onto nitrocellulose membranes, and probed with the primary antibody rabbit anti-furin antibody ab183495 (Abcam, 1:1000) and a HRP-conjugated secondary antibody (GE Healthcare, 1:2000), or with HRP-conjugated beta Actin monoclonal antibody (Thermo Fisher, 1:2000). The chemiluminescent signal was detected using ECL Plus (GE Healthcare). 1×107 furin-overexpressing HCT116, furin-low expressing LoVo human colon cancer cells, or furin-deficient CCD-18Co human colon fibroblast cells were incubated with 500 μM Olsa-RVRR, free olsalazine, CTR1, or CTR2 without RVRR for 3 h, then washed three times and resuspended in 200 μL, 10 mM PBS. After mixing homogeneously with 200 μL 1% (w/v) agarose to prevent sedimentation, cells were collected in 5 mm NMR tubes and subjected to CEST MRI immediately. A sequence similar to the above was used, except for the following parameters: TR/TE=6,000/3.8 ms, RARE factor=12, NA=1, slice thickness=6 mm, FOV=16×16 mm, matrix size=96×64, resolution=0.17×0.25 mm, B1=3.6 μT, and tsat=4 s, with saturation offset frequencies from −12 to −6.75 pm and 6.75 to 12 ppm (with 0.25 ppm increments and the water resonance set at 0 ppm). The total acquisition time was 18 min.
GSH detection.
2×104 HCT116, LoVo, or CDD-18Co cells were cultured in 96-well clear bottom black plates for 24 h. Background readings were obtained using three wells containing growth media without cells. A live cell GSH staining solution was added to each well according to the instructions of a commercial cellular glutathione detection assay kit (Cell Signaling Technology, 13859). Absorption spectra were obtained 1 hour after the start of incubation using a SpectraMax 340PC microplate reader (Marshall Scientific) using excitation/emission frequencies of 380/460 nm.
Cell uptake, efflux, and viability studies.
5×106 HCT116 and LoVo cells were incubated with 100 μM Olsa-RVRR (pretreated with FI or not) or 100 μM olsalazine. After 0, 30, 60, 90 or 120 min, the concentration of Olsa-RVRR or olsalazine in culture medium was analyzed with HPLC to determine the amount of cellular uptake. Cell uptake efficiency was calculated by dividing the uptake amount with the added amount. To study cellular efflux, HCT116 cells were incubated with 500 μM Olsa-RVRR or free olsalazine for 3 h, washed three times and then cultured in fresh McCoy’s 5A (modified) medium. After 0.5, 1, 2, 4, 8, or 12 h, the cell culture medium was analyzed with HPLC to determine the efflux of Olsa-RVRR or olsalazine, as manifested by the total amount of compounds containing the olsalazine motif (which has a unique absorption peak around 360 nm) that was pumped out. Untreated McCoy’s 5A (modified) medium was included for background correction. For cell viability testing, the total numbers of HCT116, LoVo, or CDD-18Co cells were counted after staining with acridine orange/propidium iodide staining (F23001, Logos Biosystems), using the “Fluorescence Cell Counting” mode on a Luna automated fluorescence cell counter (L20001, Logos Biosystems). Cells were either incubated Olsa-RVRR (pretreated with or without FI), olsalazine, CTR1 or CTR2 at different concentrations. Cell viability was calculated as a percentage of control (untreated cells) set at 100%. Data were expressed as the means ± SD from three independent experiments.
DNMT activity detection.
After incubation of 5×106 HCT116 cells with or without 500 μM Olsa-RVRR, olsalazine, CTR1, or CTR2 at 37 °C for 3 h, cells were washed three times and then cultured in fresh McCoy’s 5A (modified) medium. Forty-eight hours later, the nuclear proteins for each group were extracted using a nuclear extraction kit (Epigentek). After measuring the protein concentration using a Bradford protein assay, a DNA methyltransferase activity/inhibition assay kit (Epigentek) was used to detect DNMT activity. The protein content in each well was 8 μg (except for blank ones).
HPLC analysis.
After incubation of 5×106 HCT116 cells with 100 μM Olsa-RVRR in serum-free culture medium at 37 °C for 3 h, cells were washed three times with PBS, harvested and centrifuged for 5 min at 200xg. Cells were resuspended in 500 μL PBS, pulse-sonicated on ice at 40% power for 10 min with a 3 s pulse and 5 s rest, and centrifuged at 21,000xg for 5 min to collect the lysate supernatant, which was analyzed with HPLC. For cell lysate of 12 h, after incubation of 2×106 HCT116, LoVo, or CCD-18Co cells with 100 μM Olsa-RVRR at 37 °C for 12 h, cells were lysed with the aforementioned method and analyzed with HPLC.
Live cell imaging of self-assembled nanoparticles.
5×104 HCT116 and LoVo cells were first incubated with 5 μL Golgi marker (Golgi-RFP, BacMam 2.0, Invitrogen) for 24 h and washed with PBS three times before further incubation with Alexa-RVRR (8 μM) for 3 h in culture medium. After several times of washing with PBS, live cells were observed in PBS using a Zeiss AX10 fluorescence microscope.
3D-SIM super-resolution microscopy and image analysis.
After incubation with 8 μM Alexa-RVRR or Alexa 488 for 3 h, HCT116 and LoVo cells were washed with PBS three times, fixed with 4% paraformaldehyde for 10 min, and washed with PBS again three times, mounted onto glass microscope slides and cover-slipped with DAPI-containing mounting medium. To prepare ex vivo tumor samples for 3D-SIM, 50 nmol of Alexa-RVRR or Alexa 488 was injected i.v. into 6-8 weeks old female NU/J nude mouse (Jackson laboratories) xenografted with 1×106 HCT116 and LoVo tumor cells. All animal experiments were approved by the Johns Hopkins Animal Care and Use Committee. Two hours later, mice were euthanized and the tumors were resected. Tumors were fixed in 10% buffered formalin overnight and then cryoprotected in 30% w/v sucrose overnight at 4 °C. Tumors were frozen in optical cutting temperature (OCT) medium and cut into 10 μm slices. After mounting on slides coated with poly-L-lysine, OCT medium was removed by washing with PBS three times, DAPI-containing mounting medium was applied, and the coverslips were sealed with clear nail polish. 3D-SIM images of both cells and tumor samples were acquired using a DeltaVision OMX V3 imaging system (Applied Precision, GE) with simultaneous excitation at 405 nm for DAPI and 488 nm for Alexa 488. 3D-SIM images were further processed with Imaris software (Bitplane, Inc.).
Raman imaging.
1×105 HCT116 or LoVo cells were cultured in a 6-well plate with quartz slides for 24 h. After incubation with 100 μM Olsa-RVRR or olsalazine for 3 h at 37 °C, cells were washed three times with PBS. Live cell imaging was performed in McCoy’s 5A (modified) medium with a 60x water immersion objective. For fixed cell imaging, cells were fixed with 4% paraformaldehyde for 10 min, washed three times with PBS, and the slides were taken out for imaging. Raman measurements and imaging were performed using a Horiba Xplora Plus confocal Raman microscope with a motorized sample stage. A 785 nm laser and a 100x objective was used for sample excitation. Typical accumulation time used to acquire spectra was 30s. Spectral smoothening was done using a five-point fast Fourier transform (FFT) algorithm. The Raman images were acquired using LabSpec 6 (Horiba) software and the final image processing was done using ImageJ.
In vivo CEST MRI and drug efficacy studies.
All animal surgeries and protocols were carried out in compliance with all relevant ethical regulations, as approved by the Johns Hopkins Animal Care and Use Committee (ACUC). NU/J nude mice (female, 6-8 weeks) were subcutaneously injected with 1×106 HCT116 cells in the left flank and 1×106 LoVo cells in the right flank. Once the tumor reached a volume of 100-200 mm3, mice were i.v. injected with 0.2 mmol/kg Olsa-RVRR or 0.2 mmol/kg olsalazine (both in PBS with 1% DMSO). In vivo CEST MRI was performed at different time points using a 11.7 T Bruker horizontal bore scanner. A modified RARE pulse sequence was used with the following parameters: TR/TE=5,000/3.7 ms, RARE factor=23, NA=2, slice thickness=1 mm, FOV=32×32 mm, matrix size=64×64, resolution=0.5×0.5 mm, B1=3.6 μT, and tsat=4 s, with saturation offset frequencies from −12.6 to −7.4 pm and 7.4 to 12.6 ppm (with 0.2 ppm increments and the water resonance set at 0 ppm). The total acquisition time was 9 minutes and 20 seconds. For treatment response studies, once the tumor volume reached a volume of 50-100 mm3, mice were randomly divided into three groups (n=4 for each), and i.v. injected with 0.1 mmol/kg Olsa-RVRR, 0.1 mmol/kg olsalazine, or PBS only (with all solutions containing 1% DMSO) one time every 3 days. Tumors were measured using a digital caliper, and the tumor volume (V) expressed as mm3 was calculated by the formula: V = (smallest dimension)2 × largest dimension × 0.5.
Ex vivo CEST MRI of organs.
Two hours after i.v. injection of 0.2 mmol/kg Olsa-RVRR or 30 min after i.v. injection of 0.2 mmol/kg olsalazine, tumor-bearing mice were sacrificed, and the major organs were removed and placed in PBS in a cutted 48-well plate sealed with parafilm. Blank wells and spaces between wells were filled with Fomblin LC08, a perfluoroether devoid of proton signal. OlsaCEST MRI was acquired immediately using a 11.7 T Bruker horizontal bore scanner. A modified RARE pulse sequence was used with the following parameters: TR/TE=5,000/3.7 ms, RARE factor=23, NA=2, slice thickness=2 mm, FOV=29×29 mm, matrix size=64×64, resolution=0.45×0.45 mm, B1=3.6 μT, and tsat=4 s, with saturation offset frequencies from −12.6 to −7.4 pm and 7.4 to 12.6 ppm (with 0.2 ppm increments and the water resonance set at 0 ppm). The total acquisition time was 9 minutes and 20 seconds.
Statistics and reproducibility.
Data are expressed as means ± SD and analyzed with a one-way ANOVA or type 2, two-tailed Student’s t test where appropriate with the following significance values: *P<0.05, **P<0.01, and ***P<0.001. Sample sizes were chosen to ensure adequate power (>80%, p=0.05) to detect predicted effect sizes, which were estimated on the basis of either preliminary data or previous experiences with similar experiments. The number of times each experiment was independently repeated is listed in the figure captions. All statistical calculations were performed using Microsoft Excel or GraphPad Prism 8.0.0. Investigators were blinded to group allocation during experiments. Investigators processing the MRI data and performing tumor measurements were blinded to the treatment groups.
Data availability
The experimental data supporting the findings of this study are available in the main text or in the supplementary materials. Additional data are available from the corresponding author upon reasonable request.
Code availability
Custom-written MATLAB scripts including codes for correcting B0 inhomogeneity and image post-processing are available at our website http://godzilla.kennedykrieger.org/CEST/.
Supplementary Material
Acknowledgements
We thank Dr. Jiadi Xu, Dr. Zheng Han, Dr. Congxiao Wang, and Dr. Shaowei Bo for experimental assistance. This project was supported by the Pearl and Yueh-Heng Yang Foundation and NIH P41 EB024495.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper
References
- 1.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C & Gottesman MM Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5, 219–234 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Mechetner EB & Roninson IB Efficient inhibition of P-glycoprotein-mediated multidrug resistance with a monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 89, 5824–5828 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsay RR, Popovic-Nikolic MR, Nikolic K, Uliassi E & Bolognesi ML A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med 7,3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco E, Shen H & Ferrari M Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33, 941–951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm S et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014 (2016). [Google Scholar]
- 6.Yuan Y et al. Intracellular self-assembly of taxol nanoparticles for overcoming multidrug resistance. Angew. Chem. Int. Ed. 54, 9700–9704 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Du W, Hu X, Wei W & Liang G Intracellular peptide self-assembly: A biomimetic approach for in situ nanodrug preparation. Bioconjug. Chem. 29, 826–837 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Seidah NG & Prat A The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 11, 367–383 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Jaaks P & Bernasconi M The proprotein convertase furin in tumour progression. Int. J. Cancer 141, 654–663 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Page RE et al. Increased expression of the pro-protein convertase furin predicts decreased survival in ovarian cancer. Cell Oncol. 29, 289–299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendez-Lucio O, Tran J, Medina-Franco JL, Meurice N & Muller M Toward drug repurposing in epigenetics: Olsalazine as a hypomethylating compound active in a cellular context. ChemMedChem 9, 560–565 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Ward KM, Aletras AH & Balaban RS A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J. Magn. Reson. 143, 79–87 (2000). [DOI] [PubMed] [Google Scholar]
- 13.McMahon MT, Gilad AA, Bulte JWM & van Zijl PCM Chemical exchange saturation transfer imaging: advances and applications, Edn. 1st (Pan Stanford Publishing, Singapore; 2017). [DOI] [PubMed] [Google Scholar]
- 14.Lesniak WG et al. Salicylic acid conjugated dendrimers are a tunable, high performance CEST MRI NanoPlatform. Nano Lett. 16, 2248–2253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X et al. Salicylic acid and analogues as diaCEST MRI contrast agents with highly shifted exchangeable proton frequencies. Angew. Chem. Int. Ed. 52, 8116–8119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou JY et al. Three-Dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J. Magn. Reson. Imaging 38, 1119–1128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones KM, Pollard AC & Pagel MD Clinical applications of chemical exchange saturation transfer (CEST) MRI. J. Magn. Reson. Imaging 47, 11–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X et al. Dynamic glucose-enhanced (DGE) MRI: Translation to human scanning and first results in glioma patients. Tomography 1, 105–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker-Samuel S et al. In vivo imaging of glucose uptake and metabolism in tumors. Nat. Med. 19, 1067–1072 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai K et al. Magnetic resonance imaging of glutamate. Nat. Med. 18, 302–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Zijl PCM, Jones CK, Ren J, Malloy CR & Sherry AD MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST). Proc. Natl. Acad. Sci. U. S. A. 104, 4359–4364 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling W, Regatte RR, Navon G & Jerschow A Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST). Proc. Natl. Acad. Sci. U. S. A. 105, 2266–2270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBrosse C et al. Lactate chemical exchange saturation transfer (LATEST) imaging in vivo a biomarker for LDH activity. Sci. Rep. 6, 19517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song X et al. Label-free in vivo molecular imaging of underglycosylated mucin-1 expression in tumour cells. Nat. Commun. 6, 6719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoo B et al. Detection of in vivo enzyme activity with catalyCEST MRI. Magn. Reson. Med. 71, 1221–1230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu GS et al. A dextran-based probe for the targeted magnetic resonance imaging of tumours expressing prostate-specific membrane antigen. Nat. Biomed. Eng. 1, 977–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrauto G et al. Enzyme-Responsive LipoCEST agents: Assessment of MMP-2 activity by measuring the intra-liposomal water (1) H NMR shift. Angew. Chem. Int. Ed. 56, 12170–12173 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Lock LL et al. One-Component supramolecular filament hydrogels as theranostic label-free magnetic resonance imaging agents. ACS Nano 11, 797–805 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castelli DD, Terreno E, Longo D & Aime S Nanoparticle-based chemical exchange saturation transfer (CEST) agents. NMR Biomed. 26, 839–849 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Liang G & Xu B Enzymatic hydrogelation of small molecules. Acc. Chem. Res. 41, 315–326 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Tu Y et al. Mimicking the cell: Bio-Inspired functions of supramolecular assemblies. Chem. Rev. 116, 2023–2078 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Thomas G Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Bio. 3, 753–766 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren H et al. A biocompatible condensation reaction for the labeling of terminal cysteine residues on proteins. Angew. Chem. Int. Ed. 48, 9658–9662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang G, Ren H & Rao J A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat. Chem. 2, 54–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon MT et al. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly-L-lysine and a starburst dendrimer. Magn. Reson. Med. 55, 836–847 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye D et al. Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat. Chem. 6, 519–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y et al. Enzyme-Controlled intracellular self-assembly of (18)F nanoparticles for enhanced microPET imaging of tumor. Theranostics 5, 1058–1067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown WA et al. 5-aminosalicyclic acid and olsalazine inhibit tumor growth in a rodent model of colorectal cancer. Digest. Dis. Sci. 45, 1578–1584 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Eaden J Review article: the data supporting a role for aminosalicylates in the chemoprevention of colorectal cancer in patients with inflammatory bowel disease. Aliment. Pharm. Therap. 18, 15–21 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Herfarth H The role of chemoprevention of colorectal cancer with 5-aminosalicylates in ulcerative colitis. Digest. Dis 30, 55–59 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Velayos FS, Terdiman JP & Walsh JM Effect of 5-aminosalicylate use on colorectal cancer and dysplasia risk: A systematic review and metaanalysis of observational studies. Am. J. Gastroenterol. 100, 1345–1353 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Li Y et al. CEST theranostics: label-free MR imaging of anticancer drugs. Oncotarget 7, 6369–6378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haris M et al. In vivo magnetic resonance imaging of tumor protease activity. Sci. Rep. 4, 6081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinharay S et al. Noninvasive detection of enzyme activity in tumor models of human ovarian cancer using catalyCEST MRI. Magn. Reson. Med. 77, 2005–2014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinharay S, Randtke EA, Howison CM, Ignatenko NA & Pagel MD Detection of enzyme activity and inhibition during studies in solution, in vitro and in vivo with catalyCEST MRI. Mol. Imaging Biol. 20, 240–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb BA, Chimenti M, Jacobson MP & Barber DL Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Yu MX et al. Interactions of renal-clearable gold nanoparticles with tumor microenvironments: Vasculature and acidity effects. Angew, Chem. Int. Ed. 56, 4314–4319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White KA, Kisor K & Barber DL Intracellular pH dynamics and charge-changing somatic mutations in cancer. Cancer Metastasis Rev. (2019). doi: 10.1007/s10555-019-09791-8. [DOI] [PubMed] [Google Scholar]
- 49.Bar-Shir A, Liu GS, Greenberg MM, Bulte JWM & Gilad AA Synthesis of a probe for monitoring HSV1-tk reporter gene expression using chemical exchange saturation transfer MRI. Nat. Protoc. 8, 2380–2391 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang YJ et al. Label-free imaging of gelatin-containing hydrogel scaffolds. Biomaterials 42, 144–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickham KS et al. Single-Dose primaquine in a preclinical model of glucose-6-phosphate dehydrogenase deficiency: Implications for use in malaria transmission-blocking programs. Antimicrob. Agents Chemother. 60, 5906–5913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng M et al. Pro-protein convertase gene expression in human breast cancer. Int. J. Cancer 71, 966–971 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Mbikay M, Sirois F, Yao J, Seidah NG & Chretien M Comparative analysis of expression of the proprotein convertases furin, PACE4, PC1 and PC2 in human lung tumours. Br. J. Cancer 75, 1509–1514 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bassi DE et al. Elevated furin expression in aggressive human head and neck tumors and tumor cell lines. Mol. Carcinog. 31, 224–232 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Mercapide J et al. Inhibition of furin-mediated processing results in suppression of astrocytoma cell growth and invasiveness. Clin. Cancer Res. 8, 1740–1746 (2002). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental data supporting the findings of this study are available in the main text or in the supplementary materials. Additional data are available from the corresponding author upon reasonable request.