Abstract
Immunosuppression increases the risk of cancers associated with viral infection1. In particular, squamous cell carcinoma (SCC) of the skin has a >100-fold increased risk in immunosuppressed patients and has been associated with beta human papillomavirus (β-HPV) infection2–4. Previous studies, however, have failed to establish a causative role for HPVs in driving skin cancer development. Herein, we provide an alternative explanation for this association by demonstrating that the T cell immunity against commensal papillomaviruses suppresses skin cancer in immunocompetent hosts. The loss of this immunity, rather than the oncogenic effect of HPVs, is the reason for the markedly increased risk of skin cancer in immunosuppressed patients. To investigate the impact of papillomavirus on carcinogen-driven skin cancer, we colonized several strains of immunocompetent mice with mouse papillomavirus type 1 (MmuPV1)5. Mice with natural anti-MmuPV1 immunity after colonization and acquired immunity due to T cell transfer from immune mice or MmuPV1 vaccination were protected against chemical- and ultraviolet (UV)-induced skin carcinogenesis in a CD8+ T cell-dependent manner. RNA and DNA in situ hybridizations for 25 commensal β-HPVs revealed a significant reduction in viral activity and load in human skin cancer compared to the adjacent normal skin, suggesting a strong immune selection against virus-positive malignant cells. Consistently, β-HPV E7 peptides activated CD8+ T cells from normal human skin. Our findings reveal a beneficial role for commensal viruses and establish the foundation for novel immune-based approaches to block skin cancer development by boosting immunity against the commensal HPVs present in all of our skin.
Keywords: Commensal papillomavirus, skin cancer, T cell immunity, β-HPV, MmuPV1, UV, DMBA, TPA, Virome
Cutaneous SCC is the second most common type of cancer with severe morbidity and mortality especially among the immunosuppressed patients including organ transplant recipients (OTRs)2. Although UV radiation is the main and preventable cause of skin cancer, the incidence of skin cancer in the United States has doubled from 1992 to 20122, highlighting the urgent need to develop novel approaches for skin cancer prevention and treatment. Considering that β-HPVs have been found in more than 80% of SCCs among OTRs, a potential viral cause of skin cancer has been proposed2–4. However, unlike high-risk α-HPVs, there are no predominant β-HPV types identified in skin cancers and the β-HPV genome is rarely integrated into the DNA of cancer cells or transcriptionally active6. Findings like these have led to a “hit-and-run” theory in which β-HPV facilitates the initiation of UV-driven skin cancer but is later lost during tumor maintenance4,7.
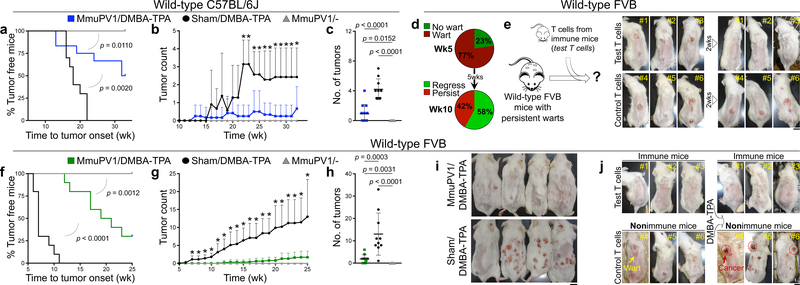
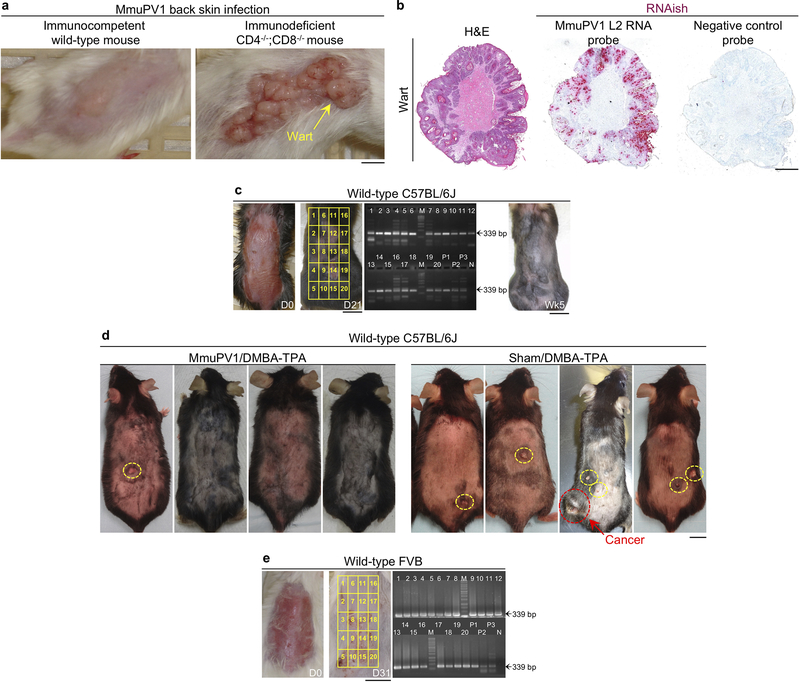
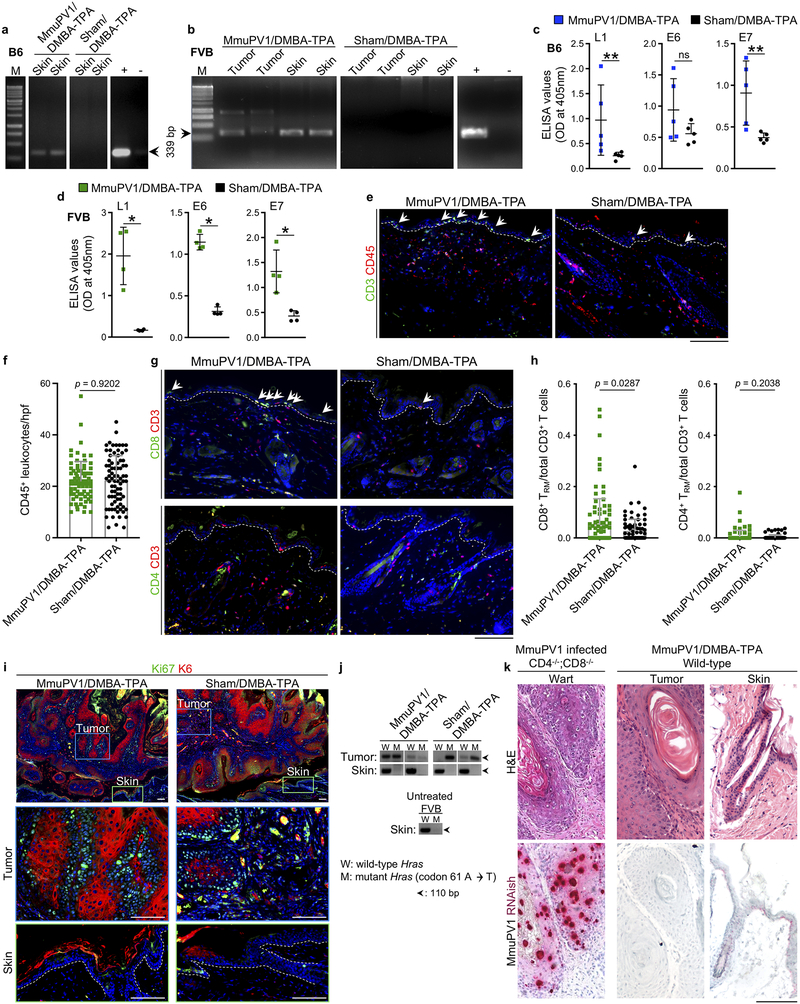
To investigate the role of papillomavirus in carcinogen-driven skin cancer, we utilized a novel murine MmuPV1 back skin infection system, which led to a confluent wart development in CD4−/−;CD8−/− mice but no skin lesions in immunocompetent, wild-type animals (Extended Data Fig. 1a–c). Two months after infection, MmuPV1- and sham-infected C57BL/6J mice were subjected to a chemical carcinogenesis protocol for 30 weeks. Surprisingly, MmuPV1-colonized mice showed a significant delay in skin tumor onset, developed significantly fewer tumors over time, and completed the study with significantly less tumor burden compared with sham-infected controls (Fig. 1a–c and Extended Data Fig. 1d). In the FVB strain, 23% of MmuPV1-infected wild-type mice showed complete immunity five weeks after infection (i.e., no skin warts, Fig. 1d and Extended Data Fig. 1e). At ten weeks post-infection, warts completely regressed in 58% of wart-bearing mice, which is indicative of antiviral adaptive immunity. Adoptive transfer of memory T cells from MmuPV1-immune mice into CD4−/−;CD8−/− animals led to fewer warts following MmuPV1 back skin infection compared to control T cell-deficient mice and CD4−/−;CD8−/− mice that received memory T cells from parvovirus vaccine-treated wild-type mice (Extended Data Fig. 2a, b). T cells from MmuPV1-immune mice also provided immunity (i.e., wart rejection) to wild-type FVB mice with persistent warts (Fig. 1e). MmuPV1-colonized immune FVB mice that received 7,12-dimethylbenz[a]anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA) for 20 weeks were protected against chemical carcinogenesis compared with sham-infected mice (Fig. 1f–i). Importantly, mice with acquired immunity after T cell transfer were also protected from chemical carcinogenesis (Fig. 1j). The MmuPV1-specificity of the transferred T cells from MmuPV1-immune mice was further substantiated by their inability to protect against the growth of uninfected SCC cells (Extended Data Fig. 2c). At the completion of the carcinogenesis studies, MmuPV1 viral DNA and anti-MmuPV1 antibodies were detectable in the normal skin and the blood of MmuPV1-colonized mice, respectively (Extended Data Fig. 3a–d). Despite no change in overall inflammatory cell infiltrates, MmuPV1-colonized skin had an increased ratio of epidermal CD8+ tissue-resident memory T (TRM) to total T cells (Extended Data Fig. 3e–h). DMBA-TPA-induced skin tumors in MmuPV1-colonized mice showed similar proliferative and mutational signatures to those in sham-infected animals and lacked MmuPV1 viral transcripts (Extended Data Fig. 3i–k).
Fig. 1: MmuPV1 skin colonization protects animals against chemical skin carcinogenesis.
a-c, Skin tumor outcomes for DMBA-TPA-treated MmuPV1-colonized wild-type C57BL/6J mice (MmuPV1/DMBA-TPA, n = 12), DMBA-TPA-treated sham-infected (Sham/DMBA-TPA, n = 10) and MmuPV1-colonized mice (MmuPV1/-, n = 10) determined by (a) tumor latency (Log-rank test), (b) tumor counts per mouse over time, and (c) tumor burden at the completion of the carcinogenesis protocol (*p < 0.05, two-tailed Mann-Whitney U test). d, The percentage of wild-type FVB mice with wart development on their back skin at five weeks and a subgroup with persistent warts at ten weeks post MmuPV1 infection. e, T cells from skin-draining lymph nodes of MmuPV1-colonized immune mice (test T cells) transferred to mice with persistent warts (n = 3). The changes in skin wart burden is documented at two weeks post adoptive T cell transfer. In control group (n = 3), T cells represent naïve T cells as found in the spleen of uninfected wild-type FVB mice. f-i, Skin tumor development in DMBA-TPA-treated MmuPV1-colonized wild-type FVB mice (MmuPV1/DMBA-TPA, n = 10) compared with DMBA-TPA-treated sham-infected (Sham/DMBA-TPA, n = 10) and MmuPV1-colonized mice (MmuPV1/-, n = 10). (f) Time to tumor onset (Log-rank test), (g) number of skin tumors over time, (h) tumor burden at the completion of the study, and (i) representative images of mice in DMBA-TPA-treated cohorts (*p < 0.05, two-tailed Mann-Whitney U test). j, Mice that rejected their skin warts after receiving T cells from MmuPV1-colonized immune mice (test T cells in “e”, n = 3) compared with wart-bearing nonimmune mice (n = 3) following DMBA-TPA treatment. Note that the nonimmune mice developed invasive skin cancers at 8 (mouse #4), 15 (mouse #5) and 20 (mouse #6) weeks post DMBA treatment (red circles). Mice were shaved to enable better visualization of the skin tumors. Data represent two independent sets of experiments with similar results. Scale bars: 1 cm, error bars represent the mean ± SD.
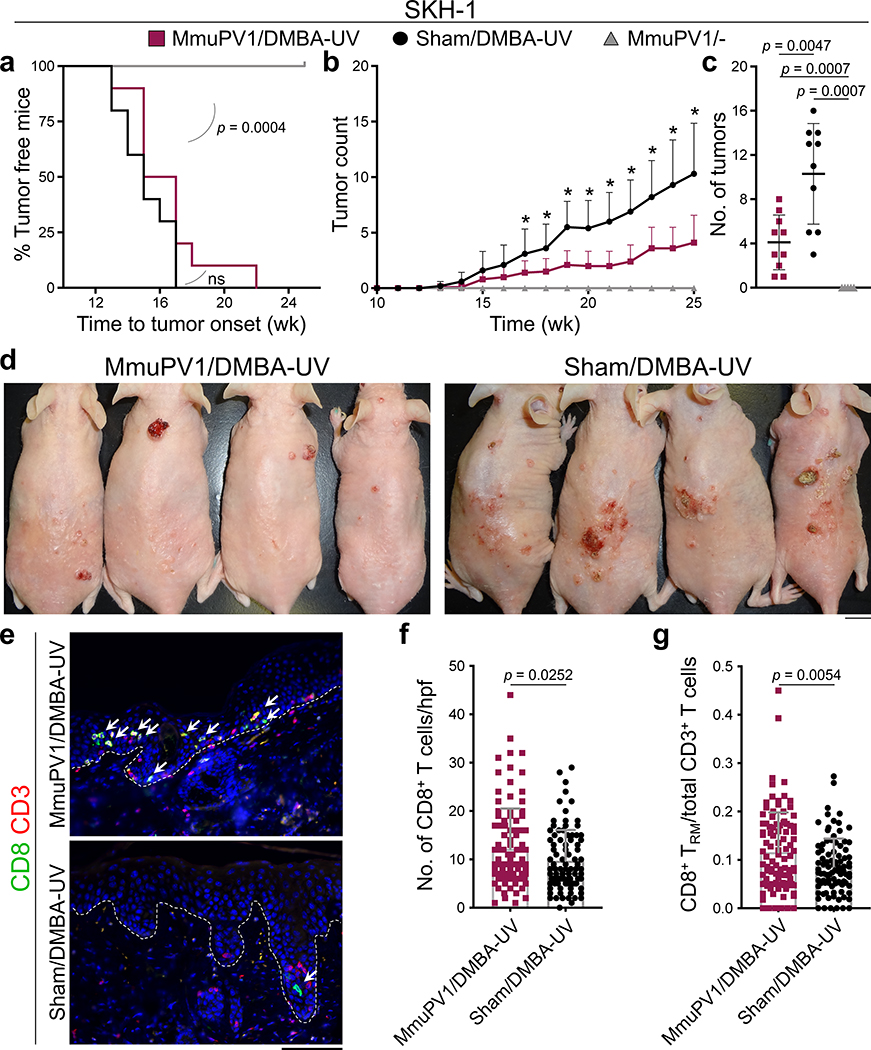
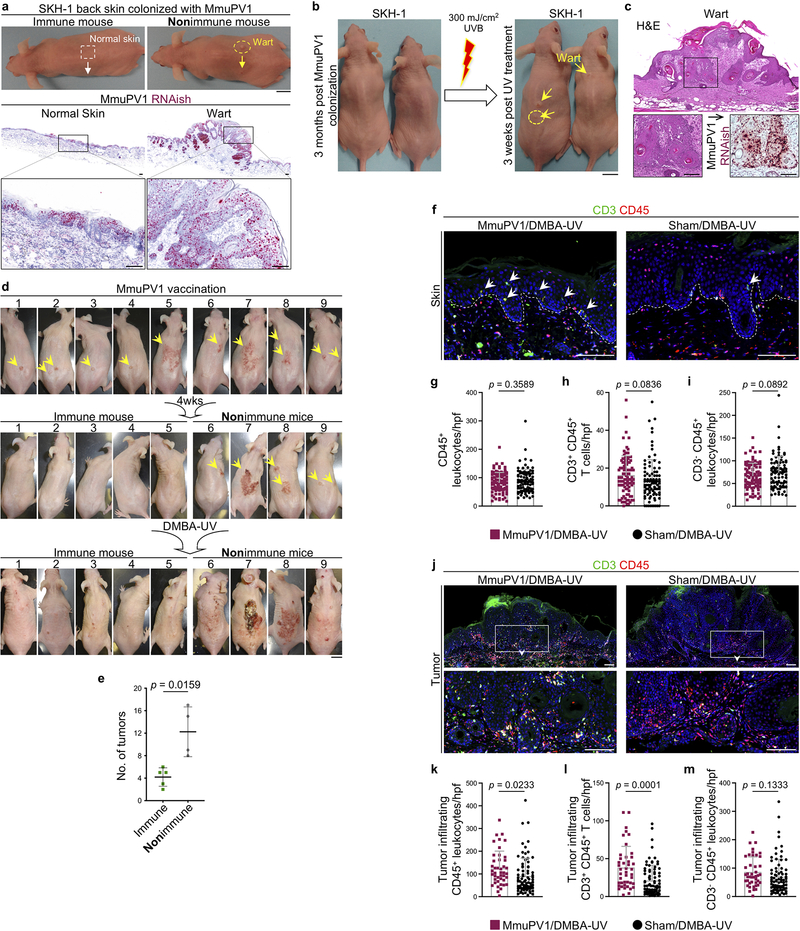
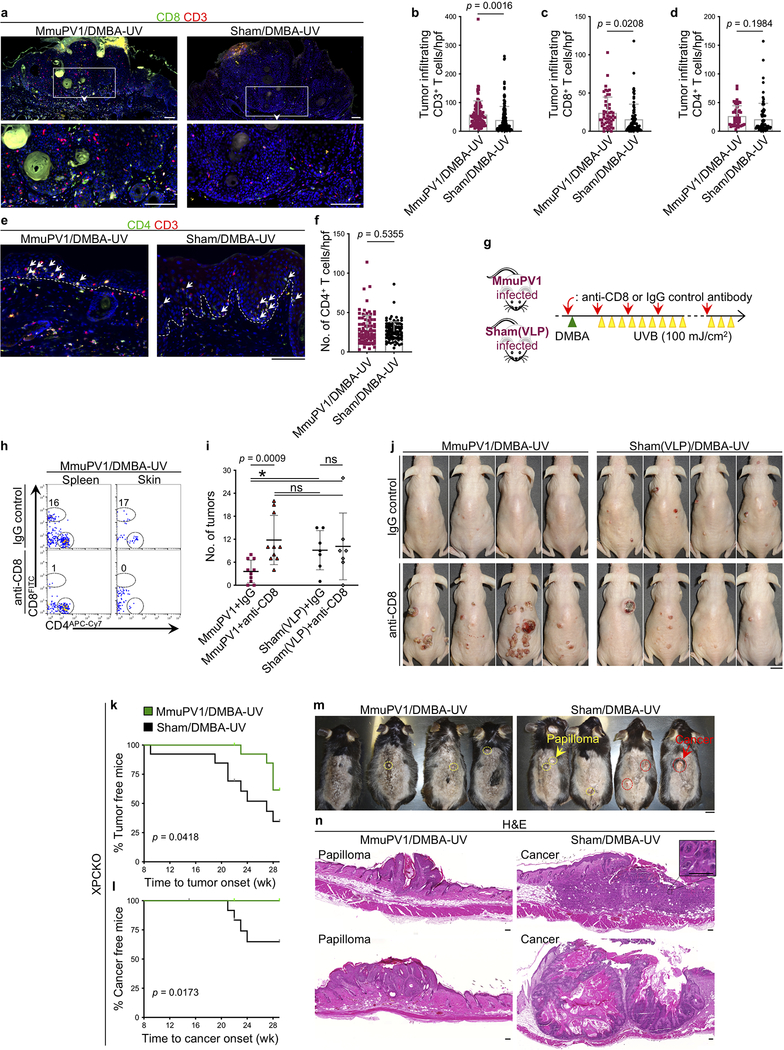
To study the impact of MmuPV1 on UV carcinogenesis, MmuPV1 back skin infection was performed on immunocompetent SKH-1 mice (Extended Data Fig. 4a). MmuPV1-infected immune mice8 that received a single immunosuppressive dose of UVB (300 mJ/cm2)9 at three months post-infection developed warts, indicating the long-term persistence of MmuPV1 skin colonization (Extended Data Fig. 4b, c). To avoid immunosuppressive UV exposure, MmuPV1- and sham-infected mice were treated with DMBA a week prior to receiving 100 mJ/cm2 UVB treatment triweekly for 25 weeks. MmuPV1-colonized SKH-1 mice developed significantly fewer tumors over time and had markedly less tumor burden at the completion of the study compared to the sham-infected controls (Fig. 2a–d). A small subset of SKH-1 mice with persistent warts two months after MmuPV1 back skin infection8 were vaccinated with MmuPV1 virus particles intraperitoneally three times over a two-week period. Four weeks later, five out of nine mice developed immunity against MmuPV1 as demonstrated by the rejection of their persistent warts (Extended Data Fig. 4d). The mice with acquired immunity against MmuPV1 developed markedly less skin tumors compared with the nonimmune mice at the completion of the UV carcinogenesis study (p = 0.0159, Extended Data Fig. 4d, e). A significant increase in the total number of CD8+ T cells and epidermal CD8+ TRM/total T cell ratio was detectable in the skin of MmuPV1-colonized mice compared with their sham-infected controls at the completion of UV carcinogenesis protocol (Fig. 2e–g). Furthermore, total T and CD8+ T cells were markedly increased in the skin tumors of MmuPV1-colonized mice (Extended Data Fig. 4f–m and 5a–c). Skin and tumor infiltrating CD3- CD45+ leukocytes and CD4+ T cells were not changed between the two groups (Extended Data Fig. 4f–m and 5d–f).
Fig. 2: MmuPV1 skin colonization protects immunocompetent SKH-1 mice against UV carcinogenesis.
a-c, SKH-1 mice with MmuPV1-colonized back skin and no warts (i.e., immune) were subjected to DMBA-UV carcinogenesis protocol. (a) Time to tumor onset (ns: not significant, Log-rank test), (b) tumor counts per mouse over time, and (c) the number of tumors for each mouse at the completion of the carcinogenesis protocol compared between the following groups: DMBA-UV-treated MmuPV1-colonized SKH-1 mice (MmuPV1/DMBA-UV, n = 10), DMBA-UV-treated sham-infected mice (Sham/DMBA-UV, n = 10) and MmuPV1-colonized mice (MmuPV1/-, n = 5). *p < 0.05, two-tailed Mann-Whitney U test. d, Representative images of mice in DMBA-UV treatment groups (scale bar: 1 cm). Note the resemblance of DMBA-UV-induced skin tumors to actinic keratosis and SCC in humans. e, Representative images of CD8+ T cells in the skin of MmuPV1/DMBA-UV and Sham/DMBA-UV mice at the completion of the carcinogenesis protocol. Arrows point to epidermal CD8+ T cells, dashed lines highlight the epidermal basement membrane, scale bar: 100 μm. f, CD8+ T cell infiltrates in MmuPV1/DMBA-UV (n = 10) and Sham/DMBA-UV (n = 9) skin quantified in ten randomly selected high power field (hpf) images per mouse and averaged across the mice in each group (two-tailed unpaired t-test). g, The ratio of epidermal CD8+ T cells (i.e., CD8+ TRM cells) over the total T cell count in each high power image calculated across MmuPV1/DMBA-UV (n = 10) and Sham/DMBA-UV (n = 9) groups (two-tailed unpaired t-test). Data represent two independent sets of experiments with similar results. Each dot represents one high power image, stained cells were counted blindly, error bars represent the mean ± SD.
To determine the role of CD8+ T cells in mediating the antitumor immunity induced by papillomavirus skin colonization, SKH-1 mice were infected with MmuPV1 or sham infected with MmuPV1 virus-like particles (sham(VLP)). MmuPV1- and sham(VLP)-infected mice underwent anti-CD8 antibody-based CD8+ T cell depletion together with UV carcinogenesis protocol (Extended Data Fig. 5g, h). Importantly, MmuPV1-colonized IgG control-treated SKH-1 mice developed markedly fewer tumors compared to CD8+ T cell-depleted MmuPV1-colonized mice, as well as both IgG and anti-CD8 antibody-treated sham(VLP)-infected control groups (Extended Data Fig. 5i, j). Consistent with our findings in other immunocompetent strains of mice, XPC−/− mice (XPCKO, deficient in repairing UV-induced DNA mutations10) colonized with MmuPV1 were protected from skin cancer compared with their sham-infected controls (Extended Data Fig. 5k–n).
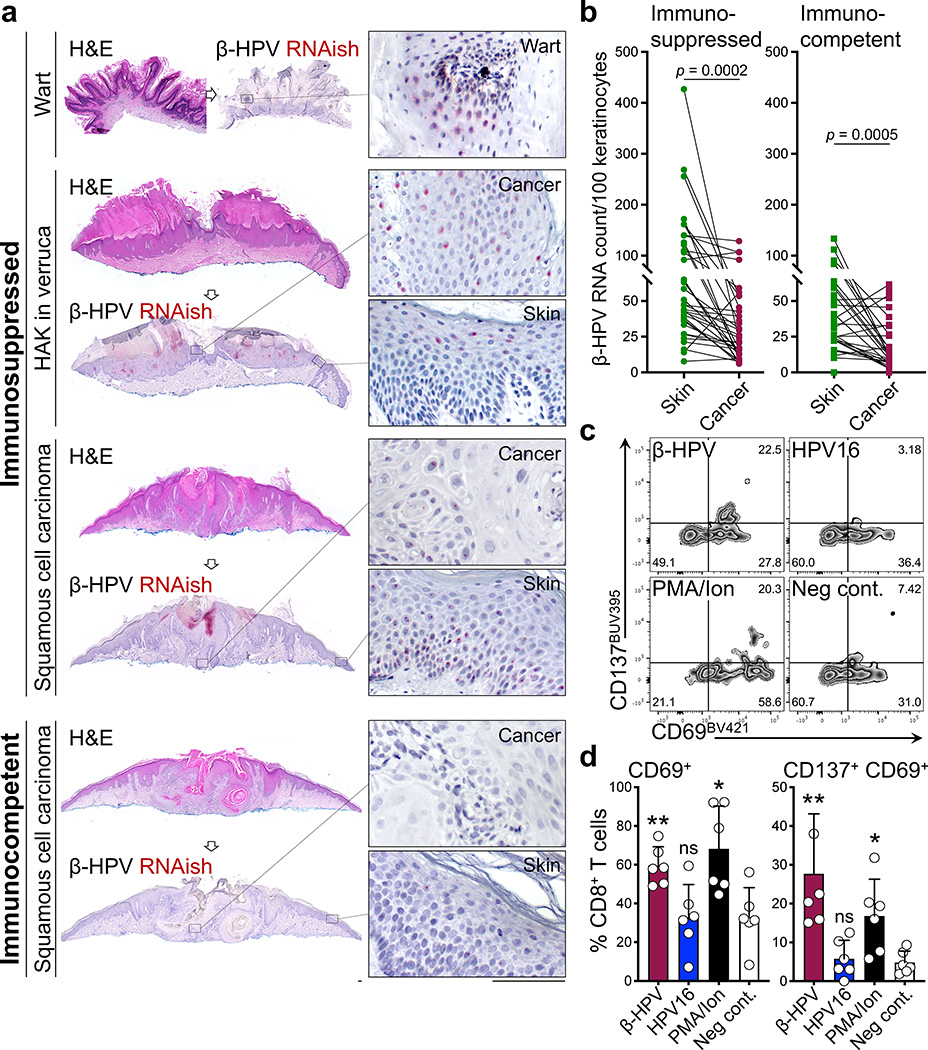
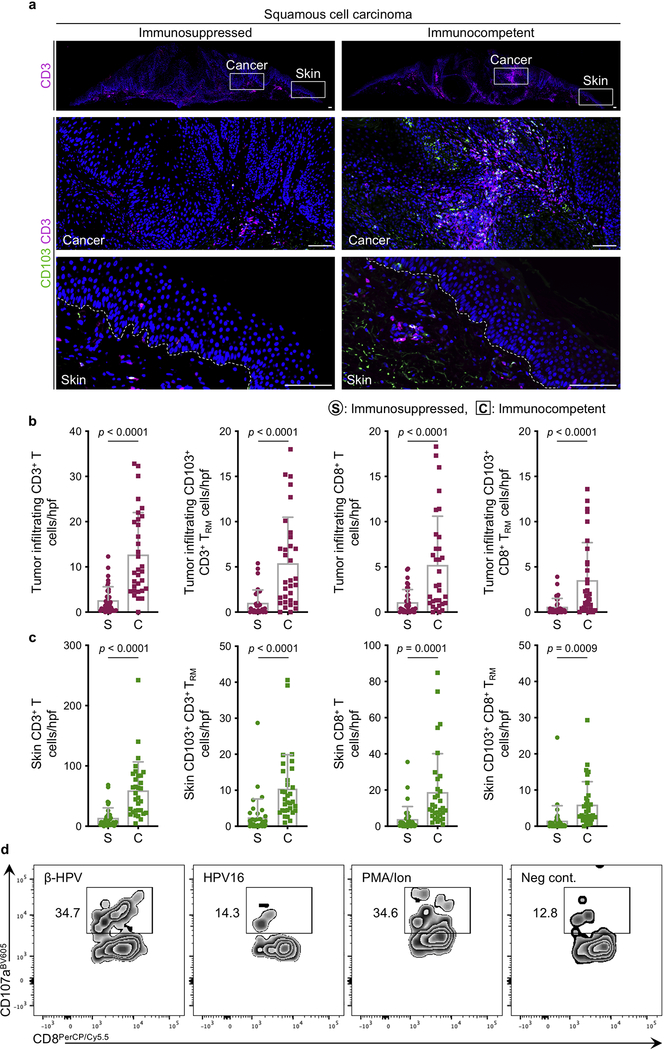
To determine whether β-HPVs plays a similarly protective role in the human skin, we utilized β-HPV RNA in situ hybridization (RNAish) to simultaneously detect the E6/7 transcripts of 25 β-HPV types in human tissue sections (Extended Data Fig. 6). In contrast to skin lesions from an immunosuppressed patient, β-HPV RNA expression was largely absent in the cancer cells of an SCC from an immunocompetent patient (Fig. 3a). β-HPV RNA expression was significantly reduced in cancer cells compared to adjacent normal skin keratinocytes among immunocompetent and immunosuppressed patients (Fig. 3b). Immunosuppressed patients’ skin lesions had significantly higher β-HPV viral transcripts compared to the skin lesions and normal facial skin samples from immunocompetent patients (Extended Data Fig. 7a–e). β-HPV DNAish probes for 25 β-HPV types (Extended Data Fig. 7f) detected higher viral load in an SCC from an immunosuppressed patient compared with an SCC from an immunocompetent patient (Extended Data Fig. 8a). β-HPV viral load was reduced in cancer cells compared to the adjacent normal skin of the immunosuppressed patient (Extended Data Fig. 8b) with this reduction being more pronounced in the lesions of immunocompetent patients (Extended Data Fig. 8c). The higher viral activity and load in the skin cancers of immunosuppressed patients correlated with significantly fewer tumor and skin infiltrating CD8+ T and CD103+ CD8+ TRM cells in their skin cancers compared with immunocompetent samples (Extended Data Fig. 9a–c). Importantly, β-ΗPV E7 peptides activated CD8+ T cells isolated from the normal facial skin of immunocompetent adults (Fig. 3c, d and Extended Data Fig. 9d). In contrast, high-risk HPV16 E7 peptides did not activate skin-derived CD8+ T cells (Fig. 3c, d and Extended Data Fig. 9d).
Fig. 3: A significant reduction in β-HPV activity from normal skin to skin cancer and the presence of β-HPV-specific CD8+ T cells in the normal human skin, pointing to a selective pressure by antiviral immunity against malignant cells with active HPV.
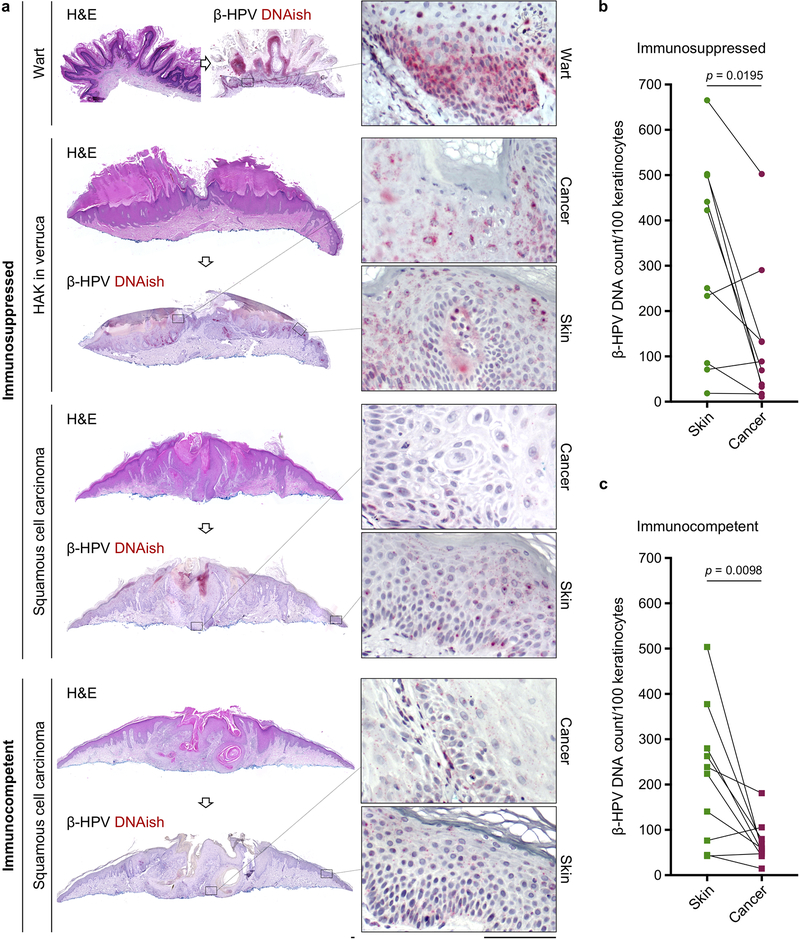
a, Representative β-HPV RNAish-stained (red dots) sections of SCC from immunosuppressed and immunocompetent patients. Wart sample serves as a positive control and exhibits the greatest amount of β-HPV activity. Hypertrophic actinic keratosis arising in association with a wart (HAK in verruca) is another example of a β-HPV-active lesion found on the skin of immunosuppressed patients. Insets highlight the representative areas of the cancer/wart and their adjacent normal skin (scale bars: 100 μm). b, β-HPV RNAish signals quantified in paired samples of skin cancer and its adjacent normal skin collected from immunosuppressed (n = 38) and immunocompetent patients (n = 32). Skin cancer characteristics are listed in Supplementary Table 2 (two-tailed paired t-test). c, d, Representative (c) flow plots and (d) quantification of CD69+ and CD137+ CD69+ CD8+ T cells isolated from human facial skin and used in a β-HPV peptide stimulation assay (n = 6 biological replicates in each treatment condition). T cells were isolated from eight facial skin samples (6 males and 2 females, average age: 75, age range: 60–89). Percentage of CD8+ T cells in each quadrant is listed on the flow plots. PMA/Ionomycin stimulation was used as a positive control. Details of peptide pool can be found in methods section and Supplementary Table 3. Error bars represent the mean + SD, RNAish signals were counted blindly, *p < 0.05, **p < 0.01, ns: not significant compared with negative control, two-tailed Mann-Whitney U test.
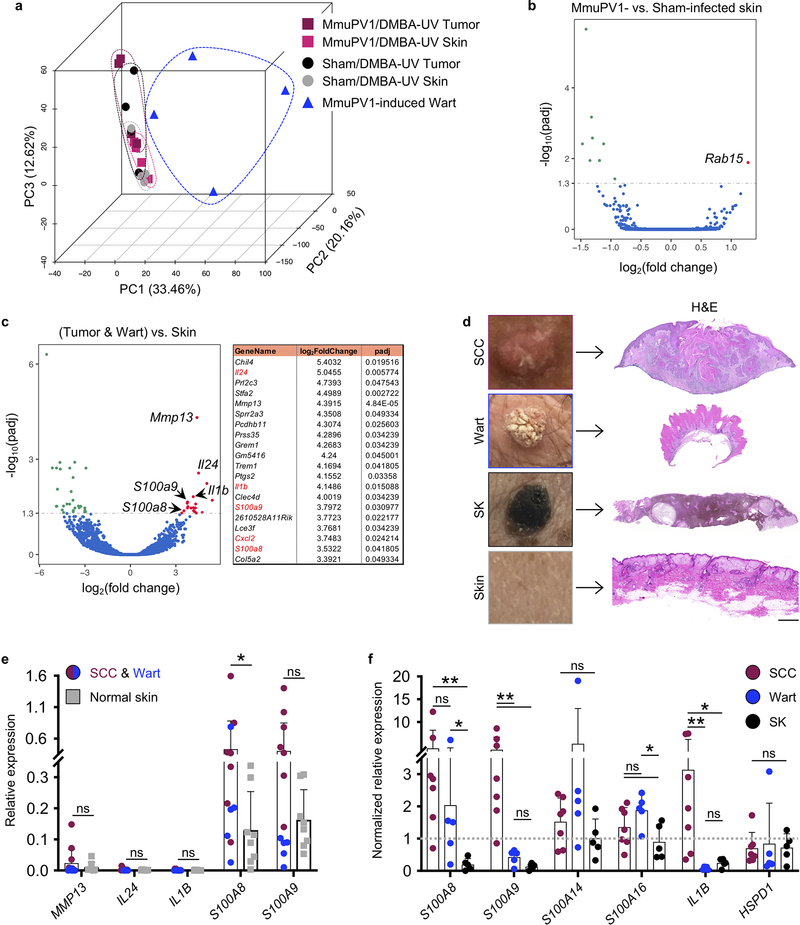
To identify the signals that lead to papillomavirus antigen presentation to T cells upon abnormal proliferation of keratinocytes, we performed RNA sequencing on skin warts, MmuPV1/DMBA-UV skin and tumors and Sham/DMBA-UV skin and tumors of SKH-1 mice (Extended Data Fig. 10a–c). Interestingly, among the 20 genes that were upregulated in both MmuPV1-induced warts and DMBA-UV-induced skin tumors (from both MmuPV1- and sham-infected groups) compared with skin (also from both groups), there were several immune related genes including damage-associated molecular pattern (DAMP) genes, S100a8 and S100a9 (Extended Data Fig. 10c). We confirmed the induction of S100 genes in human SCCs and warts compared with normal skin and seborrheic keratosis, a benign skin growth in which S100A8 and S100A9 genes were down-regulated compared with the normal skin (Extended Data Fig. 10d–f).
The findings presented herein reveal a previously unrecognized role for commensal HPVs in cancer development. Using a novel papillomavirus skin colonization model, we show that MmuPV1-colonized immunocompetent mice are protected against chemical- and UV-induced skin cancer in a CD8+ T cell dependent manner. Although specific-pathogen-free (SPF) mice may not fully reproduce the complex human skin microbiome, our findings strongly suggest that the antiviral adaptive immune responses define the role of papillomaviruses in skin carcinogenesis. Our discovery of β-HPV-specific CD8+ T cells in the normal human skin is indicative of an adaptive immunity that is primed against commensal HPVs in healthy adults at baseline. These skin-resident T cells can target keratinocytes with active virus during their abnormal proliferation to form a wart or a skin cancer. Accordingly, T cell-based vaccines against commensal HPVs may provide an innovative approach to boost this antiviral immunity in the skin and help prevent warts and skin cancers in high-risk populations. In addition, the augmentation of the anti-HPV immunity may contribute to high efficacy of immune checkpoint blockade therapy against SCC11. Considering the emerging diversity of the skin virome12, it is critical to characterize the skin-resident viral communities in immunocompetent and immunosuppressed individuals and determine how these viruses contribute to human health and disease.
Methods
Human Tissue Studies
Discarded de-identified human tissue samples were obtained through Mohs surgery clinics and pathology department at Massachusetts General Hospital. The skin lesions and normal skin samples were (a) processed for immune cell or RNA isolation or (b) obtained as formalin fixed paraffin embedded sections for histological assays.
Animal Studies
All mice were housed under pathogen-free conditions in the animal facilities at Massachusetts General Hospital and University of Louisville in compliance with animal care and all relevant ethical regulations. Six to ten week old female C57BL/6J (The Jackson Laboratory, Bar Harbor, ME, strain code: 000664), female FVB (Charles River, Wilmington, MA, strain code: 207), female SKH-1 Elite (Charles River Laboratories, strain code: 477), and male and female XPC−/− (The Jackson Laboratory, strain code: 010563) were used in the immunocompetent arms of this study. Female CD4−/−;CD8−/− mice in the FVB background were used as T cell deficient hosts (provided by Dr. David G. DeNardo; CD8−/−: The Jackson Laboratory, strain code: 032563). Age- and gender-matched groups of mice were used in all experiments. Wherever possible, animals were randomized into test versus control groups and power analysis was used to determine optimal number of animals in each group. In tumor studies, skin tumor onset and tumor counts were recorded from the time of DMBA treatment (week 0) and the maximum tumor diameter allowed was 2 cm. MmuPV1-infected mice were housed in a biocontainment unit in an animal facility at University of Louisville in accordance with animal care regulations.
Statistics and Reproducibility
Two-tailed Mann-Whitney U test was used for tumor counts and T cell activation assay. Two-tailed paired t-test was used for comparing RNAish and two-tailed Wilcoxon matched-pairs signed rank test for DNAish signal counts between skin cancers and their adjacent normal skin. Two-tailed unpaired t-test was used for immunostained cell counts, RNAish signal counts comparing skin lesions to normal human skin, and other continuous variables. Log-rank test was used as the test of significance for time to tumor onset outcomes. Two-tailed fisher’s exact test was used as the test of significance for skin cancer anatomical distribution outcomes. Pearson’s χ2 tests were used for other categorical variables. A P value less than 0.05 was considered significant. Bar graphs show mean ± standard deviation. Representative data were repeated in at least two independent sets of experiments with similar results.
Study approval
De-identified human tissue sample analysis was reviewed and approved by Massachusetts General Hospital IRB. Massachusetts General Hospital and University of Louisville IACUC approved the animal studies.
MmuPV1 purification
Muzzle warts of B6.Cg-Foxn1nu/Foxn1nu mice were homogenized via pulverization with a mortar and pestle in liquid nitrogen then homogenized with a tissue grinder (DWK Life Sciences, Millville, New Jersey, catalogue no. 885450–0023). Tissue was then subjected to three freeze-thaw cycles between liquid nitrogen and a 37˚C water bath. Tissue was then sonicated for two minutes (amplitude = 20, 10s pulse). Cesium Chloride (Sigma-Aldrich, St. Louis, MO, catalogue no. 289329) dissolved in phosphate-buffered saline (PBS) was added to the wart homogenate for a final density of 1.3623 g/mL, determined via refractometer (product discontinued). Tissue was ultracentrifuged overnight at 36,000 rpm and opaque bands at densities ranging from 1.27–1.31 g/mL were extracted. Extracted bands were dialyzed three times for eight hours using Slide-A-Lyzer cassette (VWR, catalogue no. PI66230) in three liters of PBS. The purity of the viral preparation was confirmed using SDS-PAGE gel electrophoresis.
MmuPV1 inoculation
MmuPV1 viral stock was purified from MmuPV1-induced muzzle warts of B6.Cg-Foxn1nu/Foxn1nu mice using the cesium chloride gradient method following a protocol described above13. Back skin of the wild-type, XPC−/− (XPCKO) and CD4−/−;CD8−/− mice was shaved with electric razor and waxed. Next, skin was scarified using a nail file x10–20 passages across the skin to generate microaberrations in the skin barrier, which was accompanied by skin erythema. Purified virus inoculum (20 μL) was pipetted onto scarified skin and spread homogenously. The same viral inoculum was used for all infected mice, which yielded confluent wart development on the back skin of T cell-deficient FVB mice. Sham-infected mice received 20 μL of sterile normal saline topically after skin aberration. Vaseline gauze (McKesson, San Francisco, CA, catalog no. 61–20056) was cut to fit the site of the injury and applied under a standard adhesive bandage. Meloxicam (0.5 mg/kg, Boehringer Ingelheim Vetmedica, St. Joseph, MO) was injected subcutaneously for pain relief and again the next day. Bandages were removed at 48 hours post inoculation and 200 μL of sterile normal saline was injected subcutaneously to any lethargic mice.
PCR Detection of MmuPV1 in mouse skin
To confirm skin colonization after MmuPV1 back skin infection and at the completion of carcinogenesis protocols, DNA was isolated from the skin biopsies using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany, catalog no. 69506). PCR amplification of MmuPV1 L1 gene was performed following previously described method (primers are listed in Supplementary Table 4b)13.
Wart development
For ten weeks following viral infection or sham infection, mice were monitored for the development of warts. As previously described14, mice with warts lasting greater than two months were considered to have “persistent” warts. We classified these mice as “nonimmune” and they were subjected to T cell transfer or MmuPV1 vaccination before entering the chemical- and UV-carcinogenesis studies. Mice that showed either no wart development or spontaneous wart rejection were classified as “immune” and entered into carcinogenesis studies. MmuPV1 vaccination in wart-bearing SKH-1 mice was performed by intraperitoneal injection of MmuPV1 virus inoculum in 200 μL of sterile PBS three times over two weeks.
T cell isolation and transfer
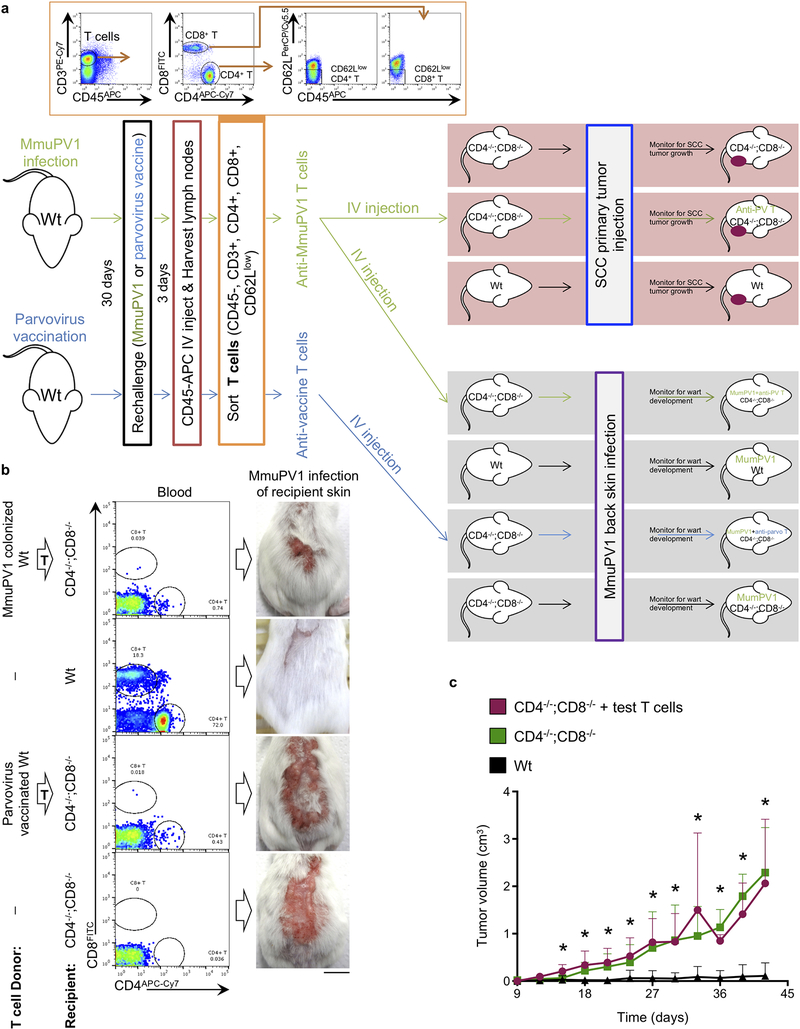
MumPV1-colonized FVB mice that never developed warts or exhibited spontaneous regression of warts by ten weeks following infection (immune mice) were used as T cell donors. A single cell suspension of CD4+ and CD8+ T cells from skin-draining lymph nodes was prepared using EasySep™ Mouse T Cell Isolation Kit (Stemcell Technologies, Vancouver, Canada, catalog no. 19851). To assess the MmuPV1-specific nature of T cells from MumPV1-colonized immune mice, we transferred their sorted CD4+ and CD8+ T cells from skin-draining lymph nodes into CD4−/−;CD8−/− recipients. Donor mice were injected intravenously with 2 μg CD45-APC (BioLegend, San Diego, CA, catalog no. 103112) three minutes prior to harvest to exclude any circulating T cells. At harvest, single cell suspensions of skin-draining lymph nodes were stained with CD3e-PE-Cy7 (Biolegend, catalog no. 100320), CD4-APC-Cy7 (Biolegend, catalog no. 100414), CD8α-FITC (Biolegend, catalog no. 100706), and CD62L-PerCP/Cy5.5 (Biolegend, catalog no. 104432, Supplementary Table 4a). Sorted CD45- CD3+ CD4+ CD62Llow and CD45- CD3+ CD8+ CD62Llow donor memory T cells15 were injected intravenously into CD4−/−;CD8−/− mice at 129,600 cells per mouse (6:1 CD4+:CD8+ ratio) in 200 μL sterile normal saline. As a control for MumPV1-specific T cells, a group of wild-type FVB mice were vaccinated against an unrelated virus (mouse parvovirus type 1) in parallel with MmuVP1-infected T cell donor mice in order to propagate a population of T cells that would not respond to MmuPV1. This group of T cell donors was vaccinated with a cocktail of 50 μg polyinosinic–polycytidylic acid (poly(I:C), Sigma-Aldrich, catalog no. P1530) combined with mouse parvovirus virus-like particles (VLPs) in 200 μL of sterile normal saline delivered via subcutaneous injection at four sites (50 μL per site per vaccination) on the back skin at 30 days and 3 days prior to T cell transfer. 200 μL of 5% Imiquimod (Sigma-Aldrich, catalog no. 1338313) dissolved in dimethyl sulfoxide (DMSO) and diluted in 100% EtOH (Sigma-Aldrich, catalog no. 276855) was applied topically following each vaccination. T cell recipients, T cell-deficient CD4−/−;CD8−/− and wild-type FVB mice, were infected with MmuPV1 two days after T cell transfer, including mice that received T cells from parvovirus vaccine plus a topical imiquimod-treated donors. Another subgroup of MmuPV1-T cell recipients including T cell-deficient CD4−/−;CD8−/− and wild-type mice received a DMBA-TPA-induced primary squamous cell carcinoma (SCC) cell injection into their right flank and monitored for tumor growth (Extended Data Fig. 2a). Mice were monitored closely for wart development in MmuPV1 infection cohorts and SCC growth in tumor cohorts for two months including pictures and tumor size measurements. To examine the presence/absence of T cells in the recipient mice, peripheral blood was collected from the mice three weeks following the T cell transfer. 2–3 drops of blood per mouse via submandibular vein extraction was collected in 10 mL of RBC Lysis Buffer (Biolegend, catalog no. 420301), stained with CD3e-PE-Cy7, CD4-APC-Cy7 and CD8α-FITC, and examined by flow cytometry. One million T cells in 200 μL sterile normal saline from skin-draining lymph nodes of MumPV1-colonized immune mice versus naïve T cells were injected intravenously into the tail vein of wart-bearing (nonimmune) wild-type FVB mice. The recipient mice were monitored for the resolution of their skin warts and their response to skin chemical carcinogenesis.
Chemical Carcinogenesis
Following infection and evidence of MmuPV1 immunity, C57BL/6J and FVB mice underwent a skin chemical carcinogenesis protocol16. All animals were shaved and seven days later received a single dose of 100 μg 7,12-dimethylbenz[a]anthracene (DMBA) (Sigma-Aldrich, catalog no. D3254) in 200 μL acetone on the back skin. One week later, treatments with 12-O-tetradecanoylphorbol-13-acetate (TPA) (Sigma-Aldrich, catalog no. P1585) dissolved in 200 μL acetone were initiated (three times per week for 30 weeks in C57BL/6J and two times per week for 20 weeks in FVB cohorts). Throughout the carcinogenesis protocol, tumors were counted every week and pictures were collected every other week. Final tumor burden was determined based on the total number of palpable skin lesions developed on the animals’ back skin.
UV Carcinogenesis
Following infection and evidence of MmuPV1 immunity, SKH-1 and XPCKO mice underwent UV skin carcinogenesis protocol. Mice received a single dose of 50 μg (SKH-1) or 100 μg (XPCKO) DMBA in 200 μL acetone on the back skin. One week later, mice received up to 25 weeks (SKH-1) or 30 weeks (XPCKO) of narrow-band ultraviolet B (UVB) (302–312 nm) three times weekly via UVP Black-Ray® Lamp UVB (VWR, Radnor, PA, catalog no. 36575–052), which was periodically calibrated using International Light IL 1400A Digital Lightmeter (International Light Technologies, Peabody, MA). Mice received 100 mJ/cm2 UVB at each UV treatment timepoint. This is considered a suberythemic dose for a fair-skinned individual of average tanning ability (Fitzpatrick skin types I-II), which approximates 25–50 minutes of sun exposure in Florida midday in the summer17,18. Throughout the carcinogenesis protocol, tumors were counted every week and pictures were collected every other week. Any palpable discrete lesion that was discontinuous/separate from other lesions was considered a tumor. Tumor counts were performed by a single individual to maintain consistency from week to week. Final tumor burden was determined based on the total number of palpable discrete skin lesions developed on the animals’ back skin after DMBA treatment. For the immunosuppressive UV dosing experiment, SKH-1 mice received 300 mJ/cm2 UVB on their back skin one time.
CD8+ T cell depletion
SKH-1 mice were infected with MmuPV1 or sham infected with MmuPV1 virus-like particles (VLPs: L1Met3019, 105 μg in 40 μL PBS per mouse) applied to their abraded back skin. Four weeks later, MmuPV1-infected immune mice and sham(VLP)-infected controls were started on anti-CD8 (Rat Anti-Mouse CD8⍺, YTS 169.4, BioXCell, West Lebanon, NH) or IgG (Rat Isotype Control, Sigma-Aldrich) antibody treatment at 750 μg in 200 μL sterile PBS (first dose) followed by 250 μg in 200 μL sterile PBS weekly intraperitoneal injections (Extended Data Fig. 5g and Supplementary Table 4a). One day after the first antibody treatment, mice underwent UV carcinogenesis protocol as described above.
Hras Mutation Specific PCR
Following the carcinogenesis protocol, DNA was extracted from tumors and skin of MmuPV1-, sham-infected, or untreated wild-type FVB mice using the DNeasy Blood & Tissue Kit (Qiagen, catalog no. 69506). Mutation specific primers were designed as previously described with the addition of a wild-type specific primer (primers listed in Supplementary Table 4b)20. PCR was performed using 500 ng of genomic DNA, 12.5 pmol of each primer, 2.5 μL of 10X Klentaq1 Reaction Buffer (DNA Polymerase Technology, St. Louis, MO, catalog no. RB20), 200 mM dNTPs (Bio Basic, Amherst, NY, catalog no. DD0056), 2.0% v/v DMSO, 1.25 Units of Klentaq-LA (DNA Polymerase Technology, catalog no. 110), and water to a final volume of 25 μL. Amplification was performed as described previously20. Briefly, DNA was denatured at 95˚C for five minutes then cycled 30 times through denaturation at 95˚C for one minute, hybridization at 55˚C for one minute, and extension at 72˚C for one minute. After cycling, extension was continued for five minutes at 72˚C. PCR products (110 bp) were analyzed on a 2% agarose gel (Genesee Scientific, San Diego, CA, catalog no. 20–102QD) and visualized with Ethidium Bromide.
Histology and immunofluorescence staining
Mice tissue samples were harvested and fixed in 4% paraformaldehyde (PFA, Sigma-Aldrich, catalog no. P6148) overnight at 4°C. Next, tissues were dehydrated in ethanol, processed, and paraffin embedded. 5 μm sections of paraffin-embedded tissues from mice and humans were cut, deparaffinized, and stained with hematoxylin and eosin (H&E). For immunofluorescence staining, rehydrated tissue sections were permeated with 1X PBS supplemented with 0.2% v/v Triton X-100 (Thermo Fisher Scientific, Waltham, MA, catalog no. BP151) for five minutes. Antigen retrieval was performed in Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA, catalog no. H-3300) using a Cuisinart pressure cooker for 20 minutes at high pressure. Slides were washed three times for three minutes each in 1X PBS supplemented with 0.1% v/v Tween 20 (Sigma-Aldrich, catalog no. P1379). Sections were blocked with 5% m/v bovine serum albumin (Fisher Scientific, Hampton, NH, catalog no. BP1600) and 5% v/v goat serum (Sigma-Aldrich, catalog no. G9023). The slides were stained overnight at 4˚C with primary antibodies (Supplementary Table 4a). The following day, slides were washed as above and incubated for two hours at room temperature with secondary antibodies conjugated to fluorochromes (Supplementary Table 4a). After washing as above, slides were incubated with 1:4000 4’,6-Diamidino-2-Phenylindole (DAPI, Invitrogen, Carlsbad, CA, catalog no. D3571) for five minutes at room temperature, then washed as above. Slides were mounted with Prolong Gold Antifade Reagent (Invitrogen, catalog no. P36930). Once stained, ten randomly selected images of the tissue at 200x magnification (i.e., high power field) were obtained for each section. Blinded manual counting of CD3+, CD4+, CD8+, CD103+, CD45+ cells was performed using the ZEN Blue ‘event’ tool (Zeiss, Oberkochen, Germany). Positive cells were determined by comparing fluorescent intensity to the background, which was minimized using ZEN. Further analyses were performed based on the number of double positive cells (e.g., CD3+ CD8+) and the number of T cell subtypes in the epidermal compartment over the total number of CD3+ T cells in each image.
Serology
Using methods described previously21, anti-MmuPV1-specific antibodies in mouse serum were detected via enzyme-linked immunosorbent assay (ELISA).
RNA and DNA in situ hybridization
RNAish and DNAish were performed on formalin fixed paraffin embedded (FFPE) human and mouse tissue sections using RNAscope® probes and protocols (Supplementary Table 1; DNA probes were generated using the sense strand of viral DNA at the same RNA probes binding sites; Advanced Cell Diagnostics, California, USA)22. We used the HybEZ™ Hybridization System to perform RNAscope® Assay hybridization and incubation steps. Briefly, 5 μm sections were baked in a dry oven for one hour at 60°C and immediately deparaffinized in xylene, followed by rehydration in an ethanol series. Epitope retrieval was performed by placing the slides in RNAscope® 1X Target Retrieval Reagent (Advanced Cell Diagnostics, catalog no. 322000) at 102°C for 15 minutes and then washed. Protease treatment was performed by adding RNAscope® Protease Plus (Advanced Cell Diagnostics, catalog no. 322331) to the section and incubated at 40°C for 30 minutes in a HybEZ™ Oven II (Advanced Cell Diagnostics, catalog no. 321720). After probe hybridization with target probes, preamplifier and amplifier, sections were stained with Fast RED reagent (RNAscope® 2.5 HD Detection Reagents – RED, Advanced Cell Diagnostics, catalog no. 322360). 50% Hematoxylin plus 0.02% ammonia water was used as a counterstain. Positive and negative probes were used in each assay to ensure proper controls. We used probes to an endogenous housekeeping gene peptidylprolyl isomerase B (PPIB, Advanced Cell Diagnostics, catalog no. 313901) and the bacterial gene dapB (Advanced Cell Diagnostics, catalog No. 310043) as positive and negative controls, respectively. We assessed RNAish and DNAish red signals under a standard bright field microscope at 400X magnification. Ten representative areas of skin cancer and normal skin from each slide were imaged at 400X magnification and positive RNAish/DNAish signals and keratinocyte nuclei were counted in each image in a blinded manner.
qRT-PCR
RNA samples were extracted from human tissues that were stored in Allprotect (Qiagen, catalog no. 76405) at 4°C and flash frozen samples stored at −80°C. A piece of tissue (~50–100 mg) was washed using sterile 1x PBS and placed into tube containing a 5 mm TissueLyser bead, then 600 μL of RNeasy Lysis Buffer (Buffer RLT, Qiagen, catalog no. 79216) and 2-mercaptoethanol (βΜΕ) was added to the sample-bead mixture. The tissue was homogenized for five minutes through mechanical manipulation. The liquid was transferred into a new tube where 1 mL of TRIzol was added. Using standard Thermo Fisher protocols for TRIzol, the solution was mixed and centrifuged at 4°C for ten minutes. The clear supernatant was collected and 0.2 mL of chloroform/ 1 mL of TRIzol was added. The mixture was centrifuged, and the clear supernatant was retrieved. For extraction of RNA, the Allprep DNA/RNA mini kit was used (Qiagen, catalog no. 80284). The clear supernatant was then added to the Allprep DNA spin column, the flow through was mixed with one volume of 70% ethanol. This solution was mixed and applied to the RNAeasy spin column where standard methods of purification and DNase digestion were followed. RNA was quantified using a nanodrop Spectrophotometer (Nano Drop Technologies, Wilmington, DE, catalog no. ND-1000) and 1 μg of RNA was used for reverse-transcriptase reaction using SuperScript III RT Kit (ThermoFisher, catalog no. 18080044). 1 μg of RNA was mixed with 0.25 mg/mL random primers, 10 mM dNTP mix and nuclease free H2O for a total of 13 μL. This sample was then incubated at 65°C for five minutes. A mix of diluted 1x first strand buffer, 0.1 M of dithiothreitol (DTT), 40 U/μl of RNaseOUT and 200 U of SuperScript III was added to the nucleotide mix. The sample was then incubated in a thermocycler. The program consisted of five minutes at 25°C, one hour at 50°C, and 15 minutes at 70°C. Following PCR, cDNA samples were diluted 1:9 using UltraPure™ DNase/RNase-Free Distilled Water. 3 μL of the 1:9 dilution was used in the total 10 μL qPCR reaction. For forward and reverse primers (Integrated DNA Technologies, Coralville, IA, Supplementary Table 4b), 0.5 μL of 10 μM concentration was used. 5 μL of SYBR® Green master mix was used along with 1 μL of UltraPure™ DNase/RNase-Free Distilled Water per reaction for Keratin 14 and β-HPV23 qPCRs. For other gene expression analyses, a premixed cocktail of primers and probes were added in addition to PrimeTime® Gene Expression Master Mix according to the manufacturer’s instructions (Integrated DNA Technologies). The qPCR was run on LightCycler 480 II (Roche, Basel, Switzerland, product no. 05015278001). qRT-PCR products were verified by electrophoresis on a 1% agarose gel at 120 V for 60 minutes. The relative gene expression analysis was performed in triplicate for each sample by comparing the test genes to GAPDH as the refence gene. Average relative gene expressions from the normal skin samples were used to normalize the relative gene expressions in SCCs, warts and seborrheic keratoses.
RNA sequencing and analysis
Total RNA was extracted from the warts of SKH-1 mice after MmuPV1 back skin infection and skin and tumors of SKH-1 mice following completion of the UV carcinogenesis protocol using the RNeasy Mini Kit (Qiagen, catalog no. 74104) according to the manufacturer’s instructions. A total of 2 μg RNA per sample was used for RNA sample preparations. RNA integrity was assessed with an Agilent Bioanalyzer 2100. Libraries were prepared by Novogene (Sacremento CA) using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, catalog no. E7770). Sequencing was performed by Novogene using the Illumina NovaSeq 6000 System. Reads were aligned to the mouse reference genome (mm10) using STAR. Differential expression analysis was performed by Novogene using the DESeq2 R package. Unsupervised clustering was performed and visualized as PCA and volcano plots. Original data are available in the NCBI Gene Expression Omnibus (accession number GSE128476).
Human T cell isolation and peptide stimulation
T cells were isolated from human skin as previously described24. Briefly, de-identified normal facial skin samples generated as part of Mohs surgery repair was obtained. Subcutaneous fat tissue was removed from the skin tissue, and the remaining tissue was minced. Small fragments of tissue were digested in RPMI 1640 including 1% DNase-I (Sigma-Aldrich) and 0.2% collagenase-I (Fisher Scientific) for two hours at 37°C. Then cells were passed through 40 μm cell strainer and incubated in RPMI 1640 supplemented with 20% v/v FBS, 1% v/v penicillin/streptomycin, 1% m/v glutamine, 0.00035% m/v 2-mercaptoethanol, and 50 U/mL human IL-2 recombinant (BioLegend). Human skin T cells were seeded in 96 well plate and treated with a pool of 5 β-HPV E7 peptides (HPV5/8/9/20/38, 5 μg/mL of each peptide, custom peptides, JPT, Berlin, Germany), pool of HPV16 E7 peptides (5 μg/mL of each peptide, PepMix™ HPV 16 (Protein E7), JPT, product code PM-HPV16-E7) or 50 ng/mL phorbol-12-myristate-13-acetate (PMA) plus 500 ng/mL ionomycin (Ion). Peptide pools were generated as 15-mers with 11 amino acid overlap across the length of E7 proteins. After 24 hours of peptide exposure, cells were collected and stained with antibodies to surface markers for T cell activation (Supplementary Table 4a) and examined by flow cytometry (BD LSRFortessa X-20). Flow data were analyzed using FlowJo software (Ashland, OR).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA sequencing data have been deposited to NCBI Gene Expression Omnibus (accession number GSE128476).
Extended Data
Extended Data Fig. 1: Back skin infection with MmuPV1 in wild-type and T cell deficient mice and the impact of MmuPV1 colonization on chemical carcinogenesis outcomes in wild-type C57BL/6J mice.
a, Wart burden in CD4−/−;CD8−/− mice (right) compared with the absence of warts in wild-type mice (left) following MmuPV1 infection of the back skin at ten weeks post-infection. Note the confluent pattern of wart development in the T cell deficient mouse. b, MmuPV1-induced wart in CD4−/−;CD8−/− mouse stained with hematoxylin and eosin (H&E) (left), MmuPV1 L2 RNAish (middle) and negative control RNAish probe (right; scale bar: 1 mm). c, Representative images of wild-type C57BL/6J mice back skin on the day of MmuPV1 infection and 21 days post-infection. MmuPV1 L1 PCR on 20 segments of the back skin. A typical wild-type C57BL/6J mouse five weeks post-infection, highlighting the absence of warts in 100% of the animals. d, Macroscopic images of wild-type C57BL/6J mice infected with MmuPV1 on their back skin or sham infected and treated with DMBA-TPA. Representative images of the mice are shown. Papillomas and invasive skin cancer are highlighted with yellow and red circles, respectively. e, Representative images of the back skin of wild-type FVB mice on the day of MmuPV1 infection and 31 days post-infection. Mice were shaved for visualization of the skin and skin tumors, scale bars: 1 cm, MmuPV1 L1 PCR bands are marked by arrows, PCR amplicon size: 339 bp, PCR primers, forward: GAGCTCTTTGTTACTGTTGTC, reverse: ATCCTCTCTTTCCTTGGGC, M: molecular-weight size marker, P1–3: positive controls, N: negative control.
Extended Data Fig. 2: T cells transferred from wild-type MmuPV1-colonized immune mice to T cell-deficient mice reduce the wart burden upon MmuPV1 infection but have no impact on the uninfected SCC growth.
a, Schematic of T cell transfer experiment. Flow sort gating strategy is provided in the inset showing the selection of memory T cells. T cell donor mice received CD45-APC (BioLegend) intravenously three minutes prior to harvest to label and exclude the circulating immune cells. Note that parvovirus vaccination control experiment was done in parallel with the MmuPV1 challenged mice. b, Representative images of the warts on the back skin of mice three weeks after MmuPV1 infection. Flow cytometry demonstrates the presence of CD4+ and CD8+ T cells in the peripheral blood of the recipient mice, indicating a successful adoptive T cell transfer (n = 4 per group, scale bar: 1cm). c, Growth of subcutaneously injected DMBA-TPA-induced primary SCC tumor cells in wild-type (Wt, n = 9), CD4−/−;CD8−/− (n = 5), and CD4−/−;CD8−/− mice that received T cells from MmuPV1-immune donors (test T cells, n = 4). Note that SCC primary tumor growth experiment was done in parallel with the MmuPV1 challenged mice. Error bars represent the mean + SD, *p < 0.05 compared with Wt group, two-tailed Mann-Whitney U test.
Extended Data Fig. 3: Evidence of MmuPV1 colonization and T cell homing into the epidermis of MmuPV1-infected mice are found at the completion of the chemical carcinogenesis protocol.
a, b, MmuPV1 L1 PCR on DNA isolated from the skin of wild-type (a) C57BL/6J (B6) and (b) FVB mice >6 months after the infection. MmuPV1 L1 PCR bands are highlighted by arrows, PCR amplicon size: 339 bp, M: molecular-weight size marker, +: positive control, -: negative control. c, d, Anti-MmuPV1 seroconversion in DMBA-TPA-treated cohorts of (c) C57BL/6J (n = 5 per group) and (d) FVB mice (n = 4 per group). *p < 0.05, **p < 0.01, ns: not significant, two-tailed Mann-Whitney U test. e, Representative images of CD3 and CD45-stained skin from MmuPV1-colonized FVB mice compared with their sham-infected controls at the completion of the chemical carcinogenesis protocol. Arrows point to T cells in the epidermis, dashed lines highlight the epidermal basement membrane. f, CD45+ leukocytes quantified in skin sections of MmuPV1/DMBA-TPA and Sham/DMBA-TPA FVB mice across ten random high power field (hpf) images of normal skin from each mouse and averaged across the mice in each group (two-tailed unpaired t-test). Each dot represents leukocyte count in one high power image, n = 8 for the number of mice in each group. g, h, Epidermal T cell homing in DMBA-TPA-treated MmuPV1-colonized skin compared with DMBA-TPA-treated sham-infected skin of wild-type FVB mice. (g) Representative images CD8/CD3- and CD4/CD3-stained skin sections. Arrows point to the epidermal CD8+ TRM cells, dashed lines highlight the epidermal basement membrane. (h) The ratios of epidermal CD8+ TRM and CD4+ TRM cells to total CD3+ T cells in the skin per hpf image (two-tailed unpaired t-test). T cells in up to ten random hpf images of the normal skin from each mouse were counted. Each dot represents one high power image. n = 10 for MmuPV1/DMBA-TPA and 9 for Sham/DMBA-TPA group. i, Representative skin tumors from MmuPV1/DMBA-TPA and Sham/DMBA-TPA wild-type FVB mice stained with keratin 6 (K6, a marker for epidermal hyperplasia) and Ki67 (a proliferation marker). Dashed lines highlight the epidermal basement membrane in the skin. j, PCR amplification of the wild-type (A) and mutant (T) region of Hras gene in DNA of MmuPV1/DMBA-TPA and Sham/DMBA-TPA tumors, skin, and an untreated skin from a wild-type FVB mouse (band size: 110 bp). Note that Hras codon 61 A to T mutation highlights DMBA-TPA induced skin tumors in MmuPV1/DMBA-TPA and Sham/DMBA-TPA wild-type FVB cohorts. k, Matched H&E and MmuPV1 RNAish images of a wart from an MumPV1-infected CD4−/−;CD8−/− mouse, a skin tumor and normal skin from MmuPV1-colonized DMBA-TPA-treated wild-type mouse. Note the dense and confluent RNAish signals in the wart from T cell deficient mouse. After the completion of DMBA-TPA treatment, positive MmuPV1 RNAish signals are detectable in the normal skin of a wild-type mouse. The skin tumor from the same mouse lacks MmuPV1 RNAish signal. Stained cells were counted blindly, error bars represent the mean ± SD, scale bars: 100 μm.
Extended Data Fig. 4: The immunization of MmuPV1-infected SKH-1 mice with MmuPV1 vaccine protects against UV carcinogenesis.
a, Representative images of SKH-1 mice with no evidence of disease following infection (immune) and mice with visible warts after back skin infection with MmuPV1 (nonimmune). MmuPV1 L2 RNAish of an immune mouse and a nonimmune mouse skin harvested three weeks after MmuPV1 infection to detect viral activity in the normal skin and the MmuPV1-driven wart. Insets highlight the active virus in the normal skin of the immune mouse and the wart of the nonimmune mouse. b, Macroscopic images of the SKH-1 mice three months after MmuPV1 back skin infection. SKH-1 mice with spontaneous immunity to the virus (no wart) treated one time with an immunosuppressive dose of UVB (300 mJ/cm2). Images of the mice three weeks after UV treatment are shown. Arrows point to the newly developed warts on the UV-treated skin. c, The histological images of H&E and MmuPV1 RNAish stained wart (yellow circle). Inset is magnified to highlight MmuPV1-induced cytopathic changes in the H&E image and confluent positive MmuPV1 RNAish signals in the wart. d, Macroscopic images of MmuPV1-infected SKH-1 mice that continued to have warts (yellow arrows) before MmuPV1 vaccination, four weeks after vaccination and at the completion of the UV carcinogenesis protocol. The nine wart-bearing mice were treated with MmuPV1 virus particles intraperitoneally three times over two weeks. Four weeks later, the mice underwent UV carcinogenesis protocol. Mice with acquired antiviral immunity (n = 5) are compared with nonimmune mice that have persistent warts (n = 4). e, Skin tumor burden in vaccinated immune (n = 5) and nonimmune mice (n = 4) treated with UV carcinogenesis protocol. In mice with confluent pattern of skin tumors, counts represent the individual lesions prior to their coalescence, error bars represent the mean ± SD, two-tailed Mann-Whitney U test. f, Representative images of CD3/CD45-stained skin from MmuPV1-colonized SKH-1 mice compared with their sham-infected controls at the completion of the UV carcinogenesis protocol. Arrows point to T cells in the epidermis. g-i, Skin infiltrating (g) total CD45+ leukocytes, (h) CD3+ CD45+ T cells and (i) CD3- CD45+ leukocytes quantified in CD3/CD45-stained skin sections of MmuPV1/DMBA-UV (n = 10) and Sham/DMBA-UV (n = 9) SKH-1 mice across ten random hpf images of each skin sample and averaged across the mice in each group. Each dot represents one high power image. Note the trend toward T cell increase and CD3- inflammatory cell decrease in MmuPV1/DMBA-UV skin compared with Sham/DMBA-UV control. Error bars represent the mean + SD, two-tailed unpaired t-test. j, Representative images of CD3/CD45-stained cells in the skin tumors of MmuPV1-colonized SKH-1 mice compared with their sham-infected controls at the completion of the UV carcinogenesis protocol. Insets are magnified to highlight the immune cells in the tumor parenchyma. k-m, Tumor infiltrating (k) total CD45+ leukocytes, (l) CD3+ CD45+ T cells and (m) CD3- CD45+ leukocytes quantified in CD3/CD45-stained sections of MmuPV1/DMBA-UV and Sham/DMBA-UV SKH-1 skin tumors across hpf images of each tumor and averaged across the mice in each group. 12 early skin tumors are included in each group, each dot represents one high power image. Error bars represent the mean + SD, two-tailed unpaired t-test, stained cells were counted blindly, dashed lines highlight the epidermal basement membrane. Scale bars, mouse: 1 cm, tissue: 100 μm.
Extended Data Fig. 5: CD8+ T cell immunity is required to protect MmuPV1-colonized mice from UV carcinogenesis and MmuPV1-colonization protects XPCKO mice from UV carcinogenesis.
a, Representative images of CD8+ T cells in the skin tumors of MmuPV1-colonized SKH-1 mice compared with their sham-infected controls at the completion of the UV carcinogenesis protocol. Insets are magnified to highlight T cells in the tumor parenchyma. b-d, Tumor infiltrating (b) CD3+, (c) CD8+ and (d) CD4+ T cells quantified in CD8/CD3- and CD4/CD3-stained tumor sections of MmuPV1/DMBA-UV and Sham/DMBA-UV SKH-1 mice across hpf images of each tumor and averaged across the mice in each group. 12 early skin tumors are included in each group, each dot represents one high power image. e, f, CD4+ T cell infiltrates in the MmuPV1-colonized and sham-infected SKH-1 skin shown by (e) representative images of the CD4/CD3-stained skin sections (arrows point to the epidermal CD4+ TRM cells) and (f) quantification of CD4+ T cells per high power image of the skin. Ten random hpf images of the skin from each mouse in each group are included. Each dot represents one high power image. n = 10 in MmuPV1/DMBA-UV and n = 9 in Sham/DMBA-UV group. g, Schematic diagram of anti-CD8/IgG antibody treatment combined with UV carcinogenesis protocol. Four weeks post-MmuPV1 or sham(VLP) infection, mice were started on anti-CD8 or IgG isotype control antibody treatment (red arrows). A day after first antibody treatment, the back skin of SKH-1 mice was treated with 50 μg of DMBA once (green triangle). Seven days later, mice were started on UVB (100 mJ/cm2) three times a week (yellow triangles). h, Flow analysis on spleen and skin of MmuPV1/DMBA-UV mice treated with anti-CD8 or IgG antibody to evaluate CD8+ T cell depletion efficiency at six weeks post-DMBA. Percentage of CD8+ T cells are listed on each plot. i, Skin tumor burden in MmuPV1-colonized mice treated with IgG control (MmuPV1+IgG, n = 10), anti-CD8 antibody (MmuPV1+anti-CD8, n = 10), sham(VLP)-infected mice treated with IgG control (Sham(VLP)+IgG, n = 7) and anti-CD8 antibody (Sham(VLP)+anti-CD8, n = 7) after DMBA-UV treatment (error bars represent the mean ± SD, *p < 0.05, ns: not significant, two-tailed Mann-Whitney U test). j, Representative images of mice in the four treatment groups. Due to the large skin tumors in MmuPV1-colonized CD8+ T cell-depleted mice, the UV carcinogenesis study was terminated at 18 weeks post-DMBA. k, l, XPCKO mice infected with MmuPV1 on their back skin (n = 15) or sham infected (n = 13), subjected to the UV carcinogenesis protocol and their skin tumor outcomes documented as latency to (k) first skin tumor development and (l) first invasive skin cancer development (Log-rank test). Note that all XPCKO mice in the study were immune to MmuPV1 (i.e., no wart development). m, Representative images of XPCKO mice at the completion of 30-week UV carcinogenesis protocol. Premalignant tumors (papillomas) and invasive skin cancers are highlighted with yellow and red circles, respectively. Mice were shaved for UV treatments and the visualization of the skin tumors. n, Representative H&E-stained histological images of a papilloma in MmuPV1/DMBA-UV and invasive skin cancer in Sham/DMBA-UV XPCKO mice. Inset shows the cellular atypia in the Sham/DMBA-UV skin cancer (scale bar: 50 μm). ns: not significant, two-tailed unpaired t-test, error bars represent the mean + SD, stained cells were counted blindly, dashed lines highlight the epidermal basement membrane, scale bars, mouse: 1 cm, tissue: 100 μm.
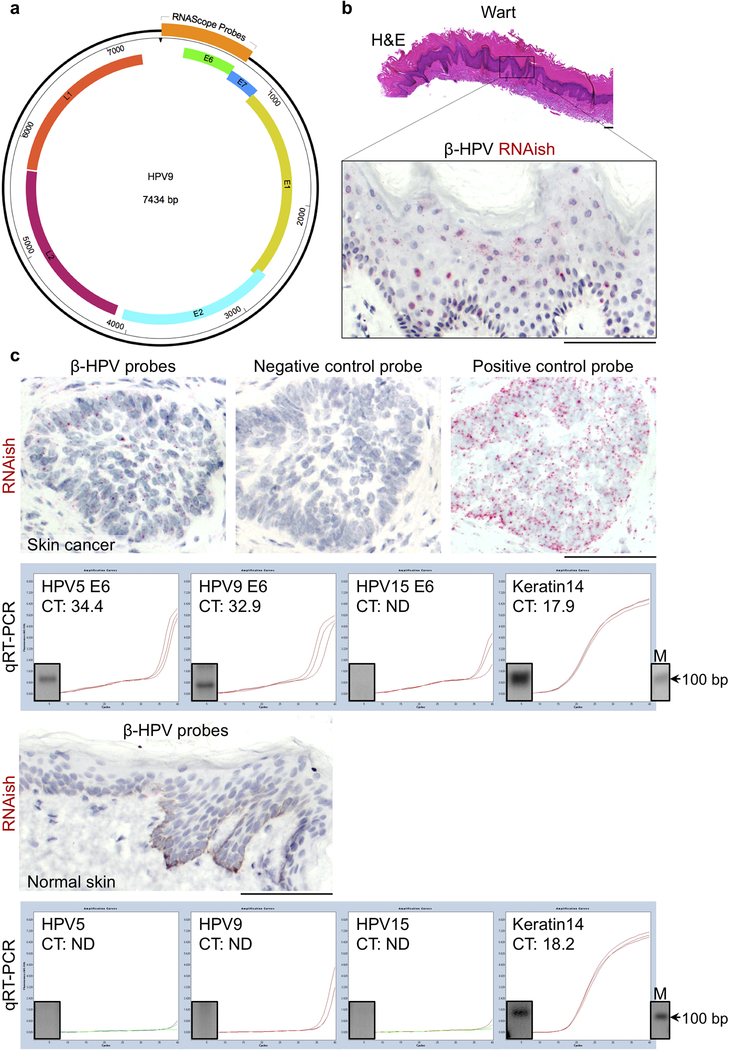
Extended Data Fig. 6: β-HPV RNAish is validated with a positive control wart and quantitative real time PCR (qRT-PCR) on RNAish positive and negative human samples.
a, β-HPV RNAish and DNAish probes binding site shown on HPV9 genome. The RNAish and DNAish probe against each β-HPV type was constituted of a pool of 20 double Z probes targeting a region of 1,000 bases (Advanced Cell Diagnostics, California, USA). b, H&E and RNAish staining of a wart from a 63-year-old immunosuppressed female. Note the abundance of positive signals (red dots) throughout the wart. c, β-HPV RNAish of a skin cancer from an 87-year-old immunosuppressed female including the positive and negative control probe stains. The detection of β-HPV by RNAish correlates with qRT-PCR positivity for HPV5 and 9 E6 protein transcripts in the same skin cancer. A sample of normal skin from an 18-year-old immunocompetent African American female stained with β-HPV RNAish probes. The lack of RNAish signal (red) in this sample correlates with undetectable HPV5, 9 or 15 E6 protein transcripts on qRT-PCR of the same sample. qRT-PCR products visualized using gel electrophoresis. PCR band size: HPV5 E6: 100 bp, HPV9 E6: 66 bp, HPV15 E6: 78 bp, Keratin 14: 109 bp, M: molecular-weight size marker, scale bars: 100 μm.
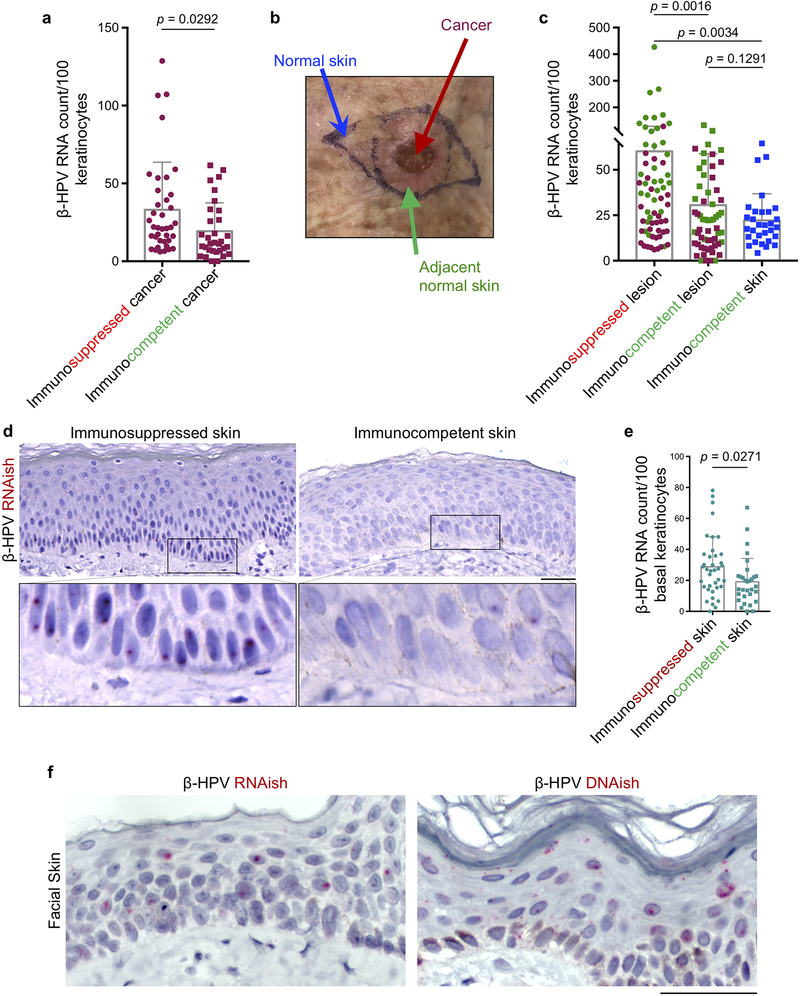
Extended Data Fig. 7: Immunosuppressed patients have greater β-HPV viral activity in their skin lesions compared to immunocompetent patients.
a, β-HPV RNAish signal counts in the skin cancer cells compared between immunosuppressed (n = 38) and immunocompetent (n = 32) patients. b, A clinical image of a skin cancer surgical site showing the skin cancer (red arrow), its adjacent normal skin (green arrow) and the normal skin away from cancer site (blue arrow). c, Quantification of β-HPV RNAish signals in high power images across the immunosuppressed lesions, immunocompetent lesions and normal facial skin away from a cancer site. Skin lesions include β-HPV RNAish signal counts from skin cancer (red dots) and the adjacent normal skin (green dots) images. 30 normal facial skin samples (blue dots) from immunocompetent patients are included (18 males and 12 females, average age: 71, range: 39–94). d, Representative low and high magnification images of β-HPV RNAish-stained normal skin samples from immunosuppressed and immunocompetent patients. Note the density and size of the apparent RNAish signals in basal layer keratinocytes of an immunosuppressed patient. e, The density of β-HPV RNAish signals in basal layer keratinocytes quantified across 38 immunosuppressed and 31 immunocompetent skin samples. f, β-HPV DNA in situ hybridization (DNAish) to detect β-HPV viral load in the skin. Compared to β-HPV RNAish that marks viral transcripts, β-HPV DNAish is a novel tool to detect viral load at a subcellular resolution in skin keratinocytes. Note the higher level of viral DNA signals compared with RNA, and the localization of the signals in the keratinocytes’ nucleus and cytoplasm. Error bars represent the mean + SD, ns: not significant, two-tailed unpaired t-test, scale bars: 50 μm.
Extended Data Fig. 8: β-HPV viral load is markedly reduced in skin cancer cells compared to their adjacent normal skin in immunocompetent patients.
a, Representative DNAish of a wart, hypertrophic actinic keratosis arising in association with a wart (HAK in verruca), and SCC in immunosuppressed patients and an SCC in an immunocompetent patient. b, c, Quantification of β-HPV DNAish signals in paired samples of skin cancer and the adjacent normal skin from (b) immunosuppressed patients (n = 10) and (c) immunocompetent patients (n = 10). Two-tailed Wilcoxon matched-pairs signed rank test, scale bars: 100 μm.
Extended Data Fig. 9: Significantly fewer T and TRM cells infiltrate the skin cancer and the adjacent normal skin in immunosuppressed versus immunocompetent patients.
a, Representative images of CD3/CD103-stained squamous cell carcinoma from immunosuppressed and immunocompetent patients (same cancers are shown for β-HPV RNAish and DNAish stains in Fig. 3a and Extended Data Fig. 8a). Insets are magnified to highlight CD103+ TRM cells in the cancer and adjacent normal skin, scale bars: 100 μm. b, c, CD3/CD8/CD103-stained skin cancer sections used to quantify (b) CD3+ T, CD103+ CD3+ TRM, CD8+ T and CD103+ CD8+ TRM cells infiltrating the skin cancer parenchyma (tumor infiltrating), and (c) CD3+ T, CD103+ CD3+ TRM, CD8+ T and CD103+ CD8+ TRM cells in the adjacent normal skin of immunosuppressed (S) versus immunocompetent (C) patients. Note that most T cells in the normal skin reside in the dermis. Stained cells were counted blindly in ten random hpf images of skin cancer and adjacent normal skin from each tissue specimen and averaged across the samples in each group, 37 immunosuppressed and 32 immunocompetent skin cancer samples are included (skin cancer characteristics are listed in Supplementary Table 2), each dot represents the average of the T cell counts in the high power images from each sample, error bars represent the mean + SD, two-tailed unpaired t-test. d, Cytotoxic T lymphocyte degranulation following exposure to β-HPV peptides. T cells isolated from normal facial skin of adults were exposed to β-HPV E7 peptides (far left), HPV16 E7 peptides (middle left), PMA/ionomycin (positive control) and media (negative control). Representative flow plots are shown. Percentage of CD107a+ CD8+ T cells are listed on each plot. Data represent two independent sets of experiments with similar results.
Extended Data Fig. 10: Damage Associated Molecular Pattern molecules are upregulated during wart and skin cancer development.
a, Principle Component Analysis (PCA) of gene expression profiles obtained from MmuPV1-induced warts (n = 4, blue triangles), MmuPV1 (n = 4, pink squares) or sham (n = 4, grey circles) infected skin, and MmuPV1 (n = 4, red squares) or sham (n = 4, black circles) infected tumors of SKH-1 mice. Note that DMBA-UV-induced skin tumors from MmuPV1-infected mice are indistinguishable from skin tumors from Sham-infected mice, while both have a very distinct transcriptional profiles compared with MmuPV1-driven warts. b, c, Volcano plots of differentially expressed genes in (b) MmuPV1- versus sham-infected skin (n = 4 per group) and (c) skin tumors and warts (n = 12) compared with MmuPV1- and sham-infected skin (n = 8). Differential expression analysis was performed by Novogene Co. (Sacramento, CA) using the DESeq2 R package. P values were calculated using DESeq2 R package (V2_1.6.3). The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR). The 20 genes that were up-regulated in skin tumors and warts compared with MmuPV1- and sham-infected skin are shown in a table. d-f, The immune gene expression analysis on human skin lesions based on the mouse RNAseq data. d, Representative macroscopic and H&E-stained histological images of SCC, wart, seborrheic keratosis (SK) and normal human skin (scale bar: 500 μm). e, The relative gene expression in SCCs (n = 7) and warts (n = 5) compared with normal skin (n = 8). f, The normalized relative gene expression in SCCs (n = 7), warts (n = 5) and SKs (n = 5) compared for several DAMP genes. Average relative gene expression in the normal skin was used for normalization. GAPDH is used as the refence gene, *p < 0.05, **p < 0.01, two-tailed Mann-Whitney U test, error bars represent the mean + SD.
Supplementary Material
Acknowledgements
We thank D. DeNardo for providing CD4−/−;CD8−/− mice. S.D. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is supported by a Cancer Research Institute CLIP Grant. T.H. was supported by Shiseido Co. J.D.S., J.L.M., T.H., D.T.H., P.A.B., K.H.N. and S.D. were supported by grants from the Burroughs Wellcome Fund, Sidney Kimmel Foundation, Cancer Research Institute and NIH (K08AR068619, DP5OD021353 and U01CA233097). H.W.N., A.B.J., J.J. and animal studies were supported by anonymous donations for Oral Cancer Research, the Elsa U. Pardee Foundation, the Helmsley Trust Foundation, and the James Graham Brown Cancer Center, University of Louisville.
Footnotes
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Grulich AE, van Leeuwen MT, Falster MO & Vajdic CM Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet 370, 59–67, doi: 10.1016/S0140-6736(07)61050-2 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Nehal KS & Bichakjian CK Update on Keratinocyte Carcinomas. The New England journal of medicine 379, 363–374, doi: 10.1056/NEJMra1708701 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Aldabagh B, Yu J & Arron ST Role of human papillomavirus in cutaneous squamous cell carcinoma: a meta-analysis. Journal of the American Academy of Dermatology 70, 621–629, doi: 10.1016/j.jaad.2014.01.857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chockalingam R, Downing C & Tyring SK Cutaneous Squamous Cell Carcinomas in Organ Transplant Recipients. Journal of clinical medicine 4, 1229–1239, doi: 10.3390/jcm4061229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingle A et al. Novel laboratory mouse papillomavirus (MusPV) infection. Veterinary pathology 48, 500–505, doi: 10.1177/0300985810377186 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Howley PM & Pfister HJ Beta genus papillomaviruses and skin cancer. Virology 479–480, 290–296, doi: 10.1016/j.virol.2015.02.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hufbauer M & Akgul B Molecular Mechanisms of Human Papillomavirus Induced Skin Carcinogenesis. Viruses 9, doi: 10.3390/v9070187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JW et al. Immunologic Control of Mus musculus Papillomavirus Type 1. PLoS Pathog 11, e1005243, doi: 10.1371/journal.ppat.1005243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uberoi A, Yoshida S, Frazer IH, Pitot HC & Lambert PF Role of Ultraviolet Radiation in Papillomavirus-Induced Disease. PLoS Pathog 12, e1005664, doi: 10.1371/journal.ppat.1005664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sands AT, Abuin A, Sanchez A, Conti CJ & Bradley A High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature 377, 162–165, doi: 10.1038/377162a0 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Migden MR et al. PD-1 Blockade with Cemiplimab in Advanced Cutaneous Squamous-Cell Carcinoma. The New England journal of medicine 379, 341–351, doi: 10.1056/NEJMoa1805131 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Tirosh O et al. Expanded skin virome in DOCK8-deficient patients. Nature medicine, doi: 10.1038/s41591-018-0211-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 13.Joh J et al. Molecular diagnosis of a laboratory mouse papillomavirus (MusPV). Exp Mol Pathol 93, 416–421, doi: 10.1016/j.yexmp.2012.07.001 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Jiang RT et al. Spontaneous and Vaccine-Induced Clearance of Mus Musculus Papillomavirus 1 Infection. Journal of virology 91, doi: 10.1128/JVI.00699-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen BJ, Cui X, Sempowski GD, Liu C & Chao NJ Transfer of allogeneic CD62L- memory T cells without graft-versus-host disease. Blood 103, 1534–1541, doi: 10.1182/blood-2003-08-2987 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Abel EL, Angel JM, Kiguchi K & DiGiovanni J Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nature protocols 4, 1350–1362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Orazio JA et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature 443, 340–344, doi: 10.1038/nature05098 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Corporation, T. P. a. M. Report on Carcinogens Background Document for Broad-Spectrum Ultraviolet (UV) Radiation and UVA, and UVB, and UVC. (Technology Planning and Management Corporation; 2000). [Google Scholar]
- 19.Joh J et al. MmuPV1 infection and tumor development of T cell-deficient mice is prevented by passively transferred hyperimmune sera from normal congenic mice immunized with MmuPV1 virus-like particles (VLPs). Exp Mol Pathol 100, 212–219, doi: 10.1016/j.yexmp.2016.01.003 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Nelson MA, Futscher BW, Kinsella T, Wymer J & Bowden GT Detection of mutant Ha-ras genes in chemically initiated mouse skin epidermis before the development of benign tumors. Proceedings of the National Academy of Sciences of the United States of America 89, 6398–6402 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joh J, Jenson AB, Ingle A, Sundberg JP & Ghim SJ Searching for the initiating site of the major capsid protein to generate virus-like particles for a novel laboratory mouse papillomavirus. Exp Mol Pathol 96, 155–161, doi: 10.1016/j.yexmp.2013.12.009 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics : JMD 14, 22–29, doi: 10.1016/j.jmoldx.2011.08.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang C et al. E6/E7 expression of human papillomavirus types in cutaneous squamous cell dysplasia and carcinoma in immunosuppressed organ transplant recipients. The British journal of dermatology 155, 129–136, doi: 10.1111/j.1365-2133.2006.07378.x (2006). [DOI] [PubMed] [Google Scholar]
- 24.Watanabe R et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Science translational medicine 7, 279ra239, doi: 10.1126/scitranslmed.3010302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. RNA sequencing data have been deposited to NCBI Gene Expression Omnibus (accession number GSE128476).