SUMMARY
Chronic liver disease due to alcohol use disorder contributes markedly to the global burden of disease and mortality1–3. Alcoholic hepatitis is a severe and life-threatening form of alcohol-associated liver disease. The gut microbiota promotes ethanol-induced liver disease in mice4, but little is known about microbial factors responsible for this process. We identified cytolysin, a two-subunit exotoxin secreted by Enterococcus faecalis (E. faecalis)5,6, to cause hepatocyte death and liver injury. Compared with controls, patients with alcoholic hepatitis have increased fecal numbers of E. faecalis. The presence of cytolysin-positive (cytolytic) E. faecalis correlated with liver disease severity and mortality in patients with alcoholic hepatitis. Using humanized mice colonized with bacteria from feces of patients with alcoholic hepatitis, we investigated the therapeutic effects of bacteriophages that target cytolytic E. faecalis. We found these phages to decrease cytolysin in the liver and abolish ethanol-induced liver disease in humanized mice. Our findings link cytolysin-positive E. faecalis with worse clinical outcomes and mortality in patients with alcoholic hepatitis. We show that bacteriophages can specifically target cytolytic E. faecalis, providing a method to precisely edit the intestinal microbiota. A prospective clinical trial with a larger cohort is required to validate human relevance of our findings and to test whether this new therapeutic approach is effective for patients with alcoholic hepatitis.
The most severe form of alcohol-related liver disease is alcoholic hepatitis; mortality ranges from 20% to 40% at 1–6 months, and as many as 75% of patients die within 90 days of a diagnosis of severe alcoholic hepatitis7–9. Therapy with corticosteroids is only marginally effective9. Early liver transplantation is the only curative therapy, but is offered only at select centers, to a limited group of patients10.
Alcohol-related liver disease can be transmitted via fecal microbiota4. We investigated microbes and microbial factors responsible for this transmissible phenotype and for progression of alcohol-related liver disease.
Cytolysin presence associates with increased mortality
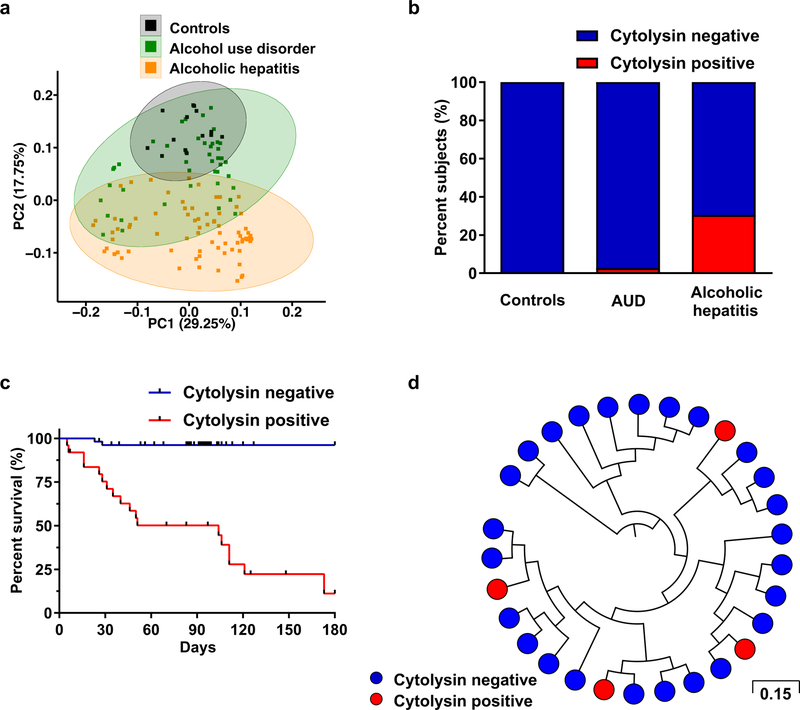
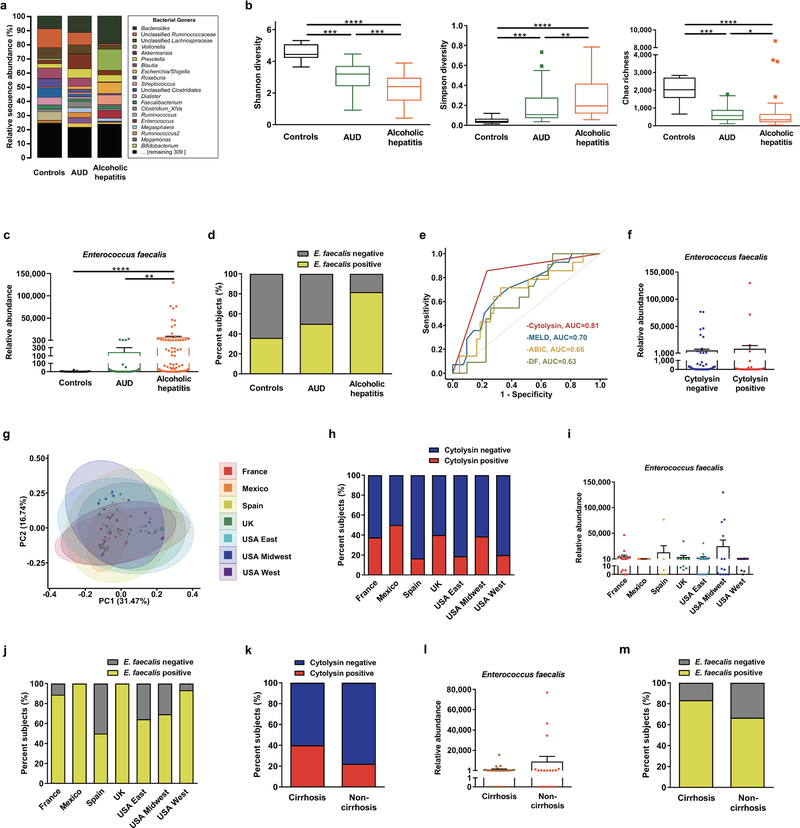
To determine whether chronic alcohol use and alcoholic hepatitis are associated with an altered composition of the fecal microbiota, 16S ribosomal RNA (rRNA) gene sequencing was performed. Differences in fecal microbiota composition were noted in patients with alcohol use disorder and alcoholic hepatitis, compared to subjects without alcohol use disorder (controls) (Fig. 1a; Extended Data Fig. 1a–1b; Supplementary Tables 1 and 2). One significant difference we observed was an increase in the proportion of Enterococcus spp. In patients with alcoholic hepatitis, 5.59% of fecal bacteria were Enterococcus spp, compared with almost none in controls (0.023%; 0.004% of all reads in the Human Microbiome Project) or patients with alcohol use disorder (0.024%). Fecal samples from patients with alcoholic hepatitis had about 2,700-fold more Enterococcus faecalis (E. faecalis) than samples from controls, measured by quantitative PCR (qPCR) (Extended Data Fig. 1c), consistent with 16S rRNA sequencing results. About 80% of alcoholic hepatitis patients are E. faecalis positive in feces (Extended Data Fig. 1d).
Figure 1. E. faecalis cytolysin associates with mortality in patients with alcoholic hepatitis.
(a) 16S rRNA sequencing of fecal samples from controls (n=14), patients with alcohol use disorder (AUD; n=43), or alcoholic hepatitis (n=75). Principal coordinate analysis (PCoA) based on Jaccard dissimilarity matrices was used to show ß-diversity among groups, at the genus level. Composition of fecal microbiota was significantly different between each group (P<0.01). (b) Percentage of subjects with fecal samples positive for cylLL and cylLS DNA sequences (cytolysin-positive), in controls (n=25), patients with AUD (n=38), or alcoholic hepatitis (n=82), assessed by qPCR. Statistically significant differences were detected between controls and alcoholic hepatitis patients (P<0.01), and between patients with alcohol use disorder and alcoholic hepatitis patients (P<0.001). (c) Kaplan-Meier curve of survival of patients with alcoholic hepatitis whose fecal samples were cytolysin-positive (n=25) or cytolysin-negative (n=54) (P<0.0001). (d) Core genome single nucleotide polymorphism (SNP) tree of E. faecalis strains isolated from patients with alcoholic hepatitis (n=93 from 24 patients), showing phylogenetic diversity of cytolysin-positive (red) E. faecalis. Genomically identical isolates from the same patient were combined and are shown as one single dot. Scale bar represents the nucleotide substitutions per SNP site. P values are determined by permutational multivariate analysis of variance (PERMANOVA) followed by false discovery rate (FDR) procedures (a), two-sided Fisher’s exact test followed by FDR procedures (b), or two-sided Log-rank (Mantel-Cox) test (c). The exact group size (n) and P values for each comparison are listed in Supplementary Table 10.
Colonization of mice with E. faecalis induces mild hepatic steatosis and exacerbates ethanol-induced liver disease11 by unclear mechanisms. Cytolysin is a bacterial exotoxin (bacteriocin) produced by E. faecalis12 that contains two post-translationally modified peptides, CylLL” and CylLS”, in its bioactive form6. The two peptides are encoded by two separate genes, cylLL and cylLS12. Cytolysin has lytic activity against not only Gram-positive bacteria but also eukaryotic cells13. We detected cylLL and cylLS genomic DNA (cytolysin-positive) in fecal samples from 30% of patients with alcoholic hepatitis; none of the fecal samples from controls and only one sample from a patient with alcohol use disorder was cytolysin-positive, detected by qPCR (Fig. 1b). Importantly, 89% of cytolysin-positive patients with alcoholic hepatitis died within 180 days after admission compared to only 3.8% of cytolysin-negative patients (P<0.0001) (Fig. 1c). Among those cytolysin-positive patients, 72.2% died due to liver failure (including liver failure related complications such as gastrointestinal bleeding) (Supplementary Table 2). Infection was not associated with 30-day, 90-day or 180-day mortality (P=0.403, 0.234 or 0.098) in patients with alcoholic hepatitis.
Univariate logistic and Cox regression of laboratory and clinical parameters associated detection of cytolysin-encoding genes in feces with international normalized ratio (INR), platelet count, model for end-stage liver disease (MELD) score, sodium MELD (MELDNa) score, age, serum bilirubin, INR, and serum creatinine (ABIC) score, and death (Supplementary Table 3). In the multivariate Cox analysis, detection of cytolysin-encoding genes in feces was still associated with 90-day (P=0.004) or 180-day mortality (P=0.001) (Supplementary Table 3), after we adjusted for the geographic origin of the patients; antibiotic treatment; platelet count; and creatinine, bilirubin, and INR as components of the MELD score. We found no serious multicollinearity between detection of fecal cytolysin and these cofactors (variance inflation factor (VIF) <1.6), indicating that cytolysin is an independent predictor of mortality in patients with alcoholic hepatitis. When we performed receiver operating characteristic (ROC) curve analysis for 90-day mortality, cytolysin had an AUC of 0.81, which was superior to other widely used predictors for mortality in clinical practice (Extended Data Fig. 1e). Based on our findings, we propose that the detection of cytolysin may be a prognostic factor for worse liver-related outcomes and death, and a stronger predictor of mortality than MELD, ABIC, and discriminant function (DF).
To determine phylogeny of E. faecalis in patients with alcoholic hepatitis, we performed targeted culturing from stool samples. Whole-genome sequencing of 93 E. faecalis isolates revealed broad phylogenetic diversity of cytolysin-positive E. faecalis from patients with alcoholic hepatitis (Fig. 1d), indicating that cytolysin production is a variable trait among E. faecalis isolates and that cytolysin is carried in mobile (genetic) elements, which include both chromosomally-encoded pathogenicity islands (PAI) and plasmids14. Detection of any other antimicrobial resistance genes or virulence genes in E. faecalis isolates did not correlate with disease severity or mortality in patients with alcoholic hepatitis (Supplementary Table 4).
The total amount of fecal E. faecalis or fecal E. faecalis positivity did not correlate with disease severity or mortality in patients with alcoholic hepatitis (Supplementary Tables 5 and 6). Cytolysin-positive and cytolysin-negative patients with alcoholic hepatitis had similar amounts of fecal E. faecalis (Extended Data Fig. 1f). Although there were differences in the composition of the gut microbiota in patients with alcoholic hepatitis from different geographic regions (Extended Data Fig. 1g), the proportion of cytolysin-positive patients, total amount of fecal E. faecalis, fecal E. faecalis positivity (Extended Data Fig. 1h–1j), treatment, and clinical outcomes (30 day and 90 day mortality) did not differ significantly among the regions/centers (Supplementary Table 7). In addition, cirrhosis was not associated with cytolysin-positivity, the total amount of fecal E. faecalis, or fecal E. faecalis positivity in alcoholic hepatitis patients (Extended Data Fig. 1k–1m; Supplementary Tables 4–6). These results confirm our findings that the presence of cytolysin producing E. faecalis, rather than the total amount or presence of E. faecalis, determines the severity of alcoholic hepatitis and mortality.
Cytolysin promotes ethanol-induced liver disease
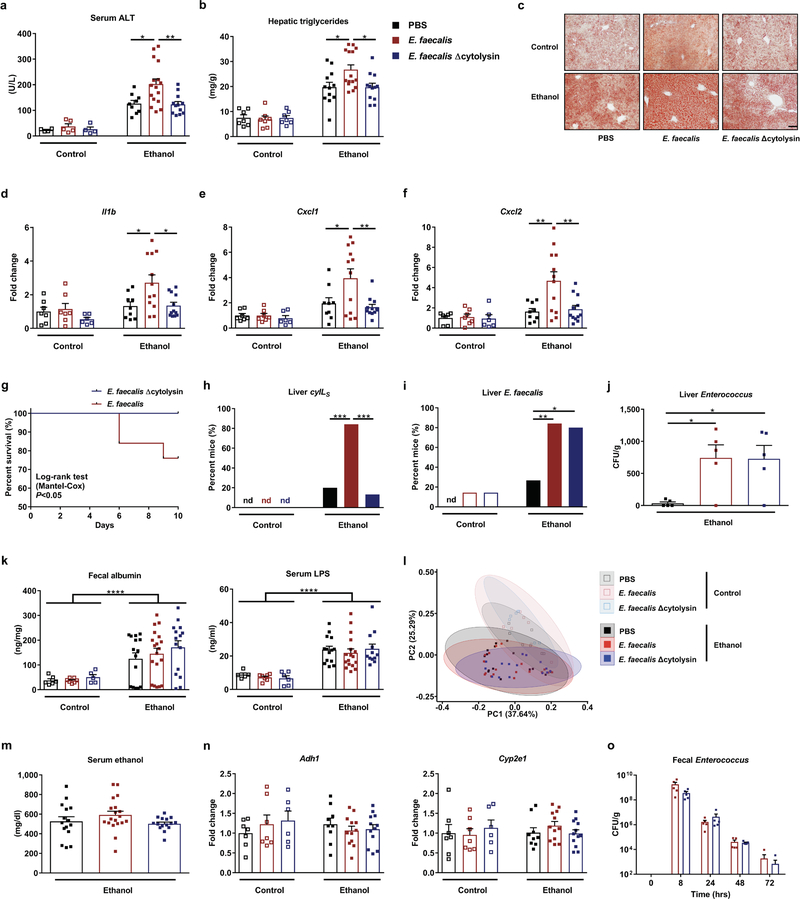
To determine whether cytolysin contributes to liver damage mediated by E. faecalis, we gavaged mice with a cytolytic E. faecalis strain (FA2–2(pAM714)) or a non-cytolytic E. faecalis strain (FA2–2(pAM771))5; the mice were then placed on a chronic–binge ethanol diet15. Compared to mice given phosphate-buffered saline (PBS), mice fed ethanol after they were gavaged with cytolytic E. faecalis developed more severe liver injury, indicated by higher level of alanine amino-transferase (ALT) (Extended Data Fig. 2a) and increased hepatic steatosis (Extended Data Fig. 2b and 2c). Mice fed ethanol after they were gavaged with cytolytic E. faecalis also had more liver inflammation with higher expression levels of mRNAs encoding inflammatory cytokines and chemokines (Il1b, Cxcl1, and Cxcl2) (Extended Data Fig. 2d–2f) compared with mice given PBS. Mice fed ethanol after they were gavaged with non-cytolytic E. faecalis had significantly less ethanol-induced liver injury, steatosis, and inflammation (Extended Data Fig. 2a–2f) and longer survival times (Extended Data Fig. 2g), compared with mice fed ethanol after they were administered cytolytic E. faecalis.
To explore the mechanism of cytolysin-associated liver damage, we measured cytolysin in the liver. CylLS was significantly increased in livers of mice given cytolytic E. faecalis but not mice that were not given E. faecalis or mice gavaged with non-cytolytic E. faecalis following chronic ethanol administration (Extended Data Fig. 2h). E. faecalis was detectable in the liver of mice given cytolytic and non-cytolytic E. faecalis and fed an ethanol diet, but not when fed an isocaloric (control) diet (Extended Data Fig. 2i), indicating that ethanol-induced changes in the gut barrier are necessary for translocation of cytolytic E. faecalis from the intestine to the liver. Livers of ethanol-fed mice given cytolytic and non-cytolytic E. faecalis had positive E. faecalis cultures (Extended Data Fig. 2j). We observed increased intestinal permeability in ethanol-fed mice compared with isocaloric diet-fed mice, but this was independent of gavaging cytolytic or non-cytolytic E. faecalis following chronic ethanol administration (Extended Data Fig. 2k), indicating that cytolysin does not affect intestinal barrier function.
Administration of cytolytic or non-cytolytic E. faecalis to mice did not significantly change the composition of the intestinal microbiota, based on 16S rRNA gene sequencing (Extended Data Fig. 2l). Cytolytic E. faecalis did not affect intestinal absorption or hepatic metabolism of ethanol, based on serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs (encoding the two primary enzymes that metabolize ethanol in the liver) (Extended Data Fig. 2m and 2n). These results indicate that E. faecalis that produce cytolysin promote ethanol-induced liver disease in mice.
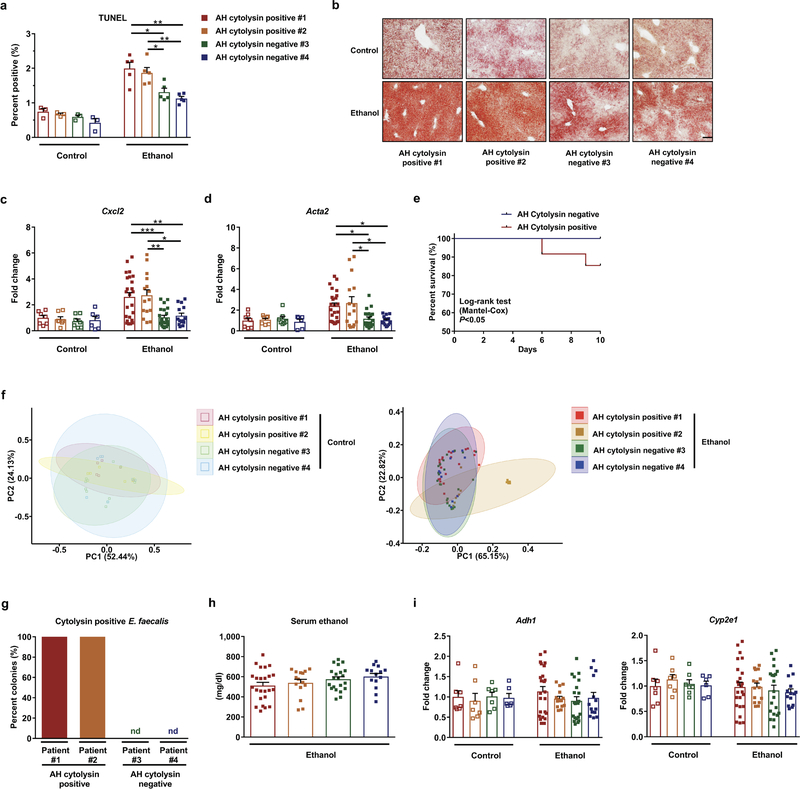
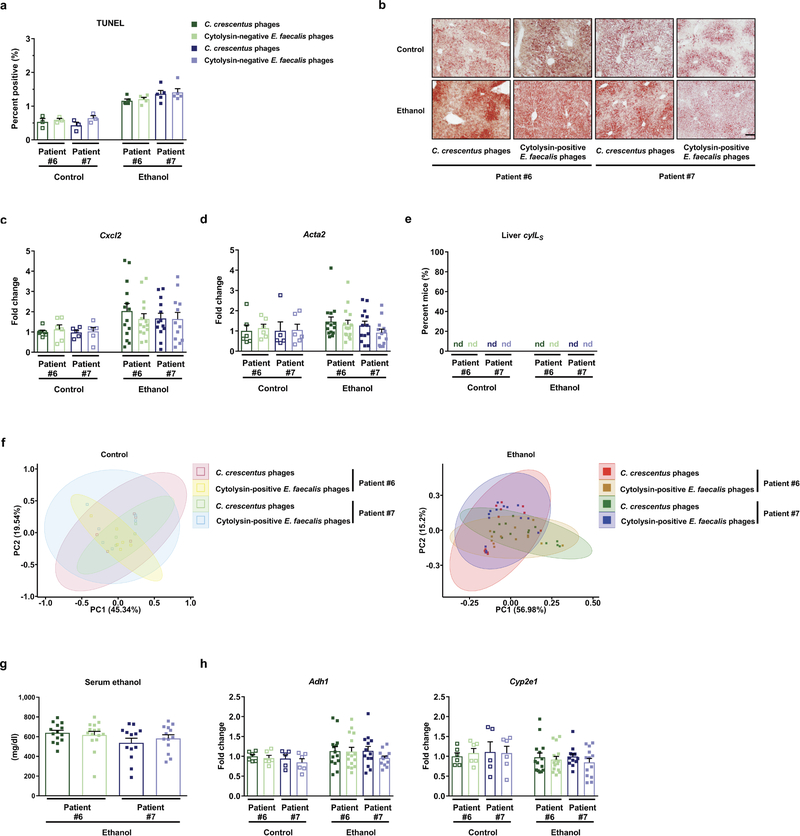
To extend our findings to humans, we colonized germ-free mice with feces from cytolysin-positive and -negative patients with alcoholic hepatitis (Supplementary Table 8). Consistent with our findings from mice colonized with cytolytic E. faecalis, gnotobiotic C57BL/6 mice colonized with feces from two different cytolysin-positive patients developed more severe ethanol-induced liver injury, steatosis, inflammation, and fibrosis than mice given feces from two different cytolysin-negative patients (Fig. 2a–2f; Extended Data Fig. 3a–3d). Transplantation of feces from cytolysin-positive patients reduced survival time of the mice (Extended Data Fig. 3e) and increased translocation of cytolytic E. faecalis to the liver following ethanol administration (Fig. 2g). Overall composition of the intestinal microbiota was not different between mice colonized with feces from cytolysin-positive or -negative alcoholic hepatitis donors following the control diet, based on 16S rRNA gene sequencing. Mice transplanted with feces from one cytolysin-positive alcoholic hepatitis patient (#2) showed a microbiota significantly different from the other mouse groups following ethanol administration (Extended Data Fig. 3f). Interestingly, non-cytolytic E. faecalis was not detected in stool samples from donors with cytolytic E. faecalis (Extended Data Fig. 3g). We did not observe differences in intestinal absorption or hepatic metabolism of ethanol between mice colonized with feces from cytolysin-positive vs cytolysin-negative patients (Extended Data Fig. 3h and 3i). Together, these results provide further evidence that cytolysin promotes ethanol-induced liver disease.
Figure 2. Transplantation of feces from cytolysin-positive patients with alcoholic hepatitis exacerbates ethanol-induced liver disease in gnotobiotic mice.
(a–g) C57BL/6 germfree mice were colonized with feces from two different cytolysin-positive and two different cytolysin-negative patients with alcoholic hepatitis and subjected to the chronic–binge feeding model. (a) Serum levels of ALT. (b) Hepatic triglyceride content. (c) Representative H&E-stained liver sections. (d–f) Hepatic levels of mRNAs encoding Il1b, Cxcl1, and Col1a1. (g) Proportions of mice positive for cytolysin in liver, measured by qPCR for cylLS. (h) LDH assay to measure cytotoxicity of hepatocytes isolated from mice fed oral isocaloric (control) diet (5 groups, left) or chronic–binge ethanol diet (5 groups, right) and incubated with vehicle, CylLS”, CylLL”, or both cytolysin subunits, at indicated concentrations, without (–) or with (+) ethanol (25mM) for 3 hours. Survival of hepatocytes was determined in 3 independent experiments. Scale bar=100 μm. Results are expressed as mean ± s.e.m (a, b, d–f, h). P values are determined by One-way ANOVA with Tukeýs post-hoc test (a, b, d–f), two-sided Fisher’s exact test followed by FDR procedures (g), or Two-way ANOVA with Tukeýs post-hoc test (h). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
To determine the mechanism by which cytolysin increases liver disease, we isolated hepatocytes from mice on ethanol or control diets and stimulated them with pure bioactive cytolysin peptides (CylLL” and CylLS”)6. Incubation of the primary mouse hepatocytes with two cytolysin subunits caused a dose-dependent increase in cell death compared to hepatocytes incubated with vehicle or with one subunit alone (Fig. 2h). Interestingly, when we isolated hepatocytes from ethanol-fed mice and then incubated those hepatocytes with ethanol, we did not observe increased levels of cytolysin-induced cell death compared to hepatocytes isolated from mice on the control diet, indicating that cytolysin-induced hepatocyte cell death was independent of ethanol. The cytotoxic effects of cytolysin are likely mediated by pore formation, resulting in cell lysis14.
Bacteriophage treatment attenuates alcohol-related liver disease in mice
To further demonstrate the potential causative role of cytolytic E. faecalis for the development of ethanol-induced steatohepatitis, we investigated the effects of treatment with bacteriophages. Bacteriophages (phages) are ubiquitous in bacteria-rich environment, including the gut16. E. faecalis phages that are highly-strain specific can be isolated17, potentially making direct editing of gut microbiota feasible. We have shown that Atp4aSl/Sl mice, which lack gastric acid, have overgrowth of intestinal enterococci, associated with increased susceptibility to alcohol-induced steatohepatitis11. Gavaging of wild-type mice with an E. faecalis strain isolated from Atp4aSl/Sl mice increased ethanol-induced steatohepatitis11. We found that this E. faecalis strain expressed cytolysin. We then isolated four distinct bacteriophages from sewage water. These bacteriophages lyse the cytolytic E. faecalis strain isolated from Atp4aSl/Sl mice. All four phages were podophages of the virulent Picovirinae group (Extended Data Fig. 4). Atp4aSl/Sl mice and their wild-type littermates were then placed on the chronic–binge ethanol diet and gavaged with the lytic bacteriophage cocktail (Extended Data Fig. 5). Bacteriophages directed against Caulobacter crescentus (C. crescentus), a bacterium that is present in fresh water lakes and streams18, but does not colonize humans or rodents19,20, were used as controls. Compared to Atp4aSl/Sl mice gavaged with control bacteriophage or vehicle, Atp4aSl/Sl mice gavaged with bacteriophages that target cytolytic E. faecalis had less liver injury, steatosis, and inflammation following chronic ethanol feeding (Extended Data Fig. 5a–5f). Administration of E. faecalis bacteriophage significantly reduced levels of cytolysin in the liver (Extended Data Fig. 5g) and fecal amounts of Enterococcus (Extended Data Fig. 5h). Bacteriophage administration did not affect the overall composition of the fecal microbiome, intestinal absorption or hepatic metabolism of ethanol (Extended Data Fig. 5i–5k).
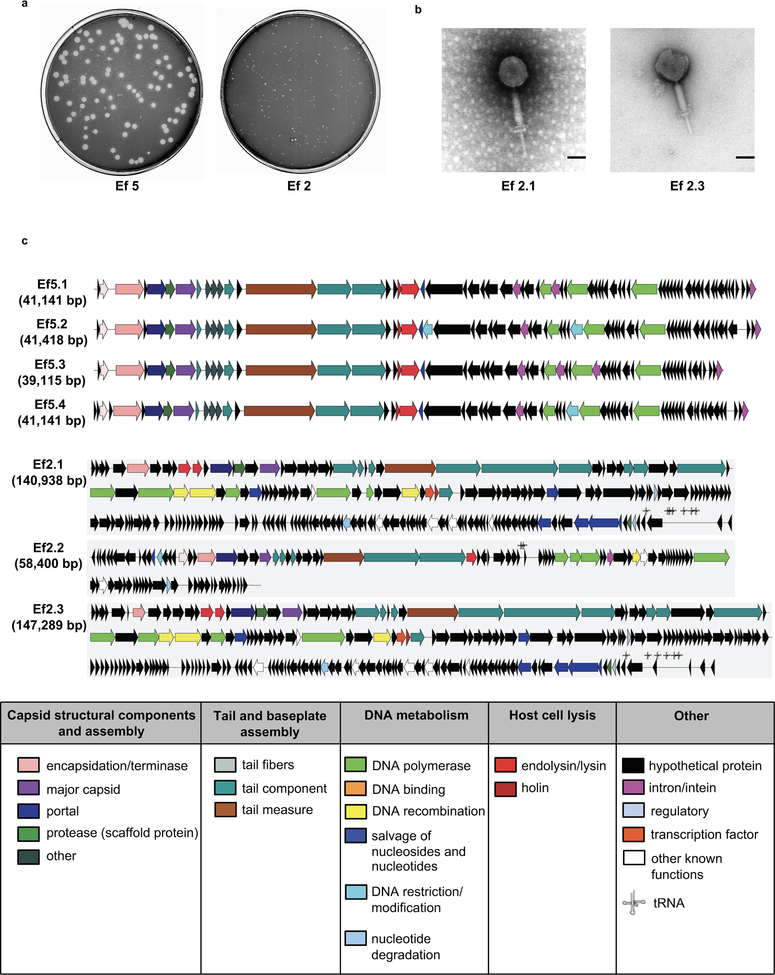
To develop a novel therapeutic approach to precisely edit the intestinal microbiota, cytolytic E. faecalis strains were cultured from fecal samples of patients with alcoholic hepatitis. We then isolated lytic bacteriophages from sewage water against these cytolytic E. faecalis strains and these phages had siphophage or myophage morphology (Fig. 3a; Extended Data Fig. 6). Gnotobiotic mice were colonized with feces from two different cytolysin-positive patients with alcoholic hepatitis (Supplementary Table 8) and given 3–4 different, but patient-specific lytic phages against cytolytic E. faecalis. The phages against cytolytic E. faecalis abolished ethanol-induced liver injury and steatosis, based on lower levels of ALT, percentage of TUNEL-positive hepatic cells, hepatic triglycerides and oil red O-staining (Fig. 3b–3d; Extended Data Fig. 7a–7b), decreased hepatic levels of Il1b, Cxcl1, Cxcl2, Col1a1 and Acta2 mRNAs, and reduced hepatic levels of cylLs, compared with mice given control phages (against C. crescentus) (Fig. 3e–3h; Extended Data Fig. 7c–7d). Treatment with phages against cytolytic E. faecalis also reduced fecal amounts of Enterococcus (Extended Data Fig. 7e) without affecting the overall composition of the gut microbiota (Extended Data Fig. 7f). Intestinal absorption of ethanol and hepatic metabolism were similar in all groups (Extended Data Fig. 7g–7h).
Figure 3. Phage therapy against cytolytic E. faecalis abolishes ethanol-induced liver disease in gnotobiotic mice.
(a) Transmission electron microscopy revealed that bacteriophages isolated were either siphophages (Ef5.1, Ef5.2, Ef5.3, Ef5.4 and Ef2.2) or myophages (Ef2.1 and Ef2.3). Scale bar=50 nm. (b–h) C57BL/6 germfree mice were colonized with feces from two different cytolysin-positive patients with alcoholic hepatitis (feces from 1 patient also used in Figure 2) and subjected to chronic–binge feeding model, gavaged with control phages against C. crescentus (1010 PFUs), or a cocktail of 3 or 4 different phages targeting cytolytic E. faecalis (1010 PFUs) 1 day before an ethanol binge. (b) Serum levels of ALT. (c) Hepatic triglyceride content. (d) Representative H&E-stained liver sections. Scale bar=100 μm. (e–g) Hepatic levels of mRNAs encoding Il1b, Cxcl1 and Col1a1. (h) Proportions of mice positive for cytolysin in liver, measured by qPCR for cylLS. Results are expressed as mean ± s.e.m (b, c, e–g). P values are determined by Two-way ANOVA with Tukeýs post-hoc test (b, c, e–g) or two-sided Fisher’s exact test followed by FDR procedures (h). All results are generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05, **P<0.01, *** P<0.001.
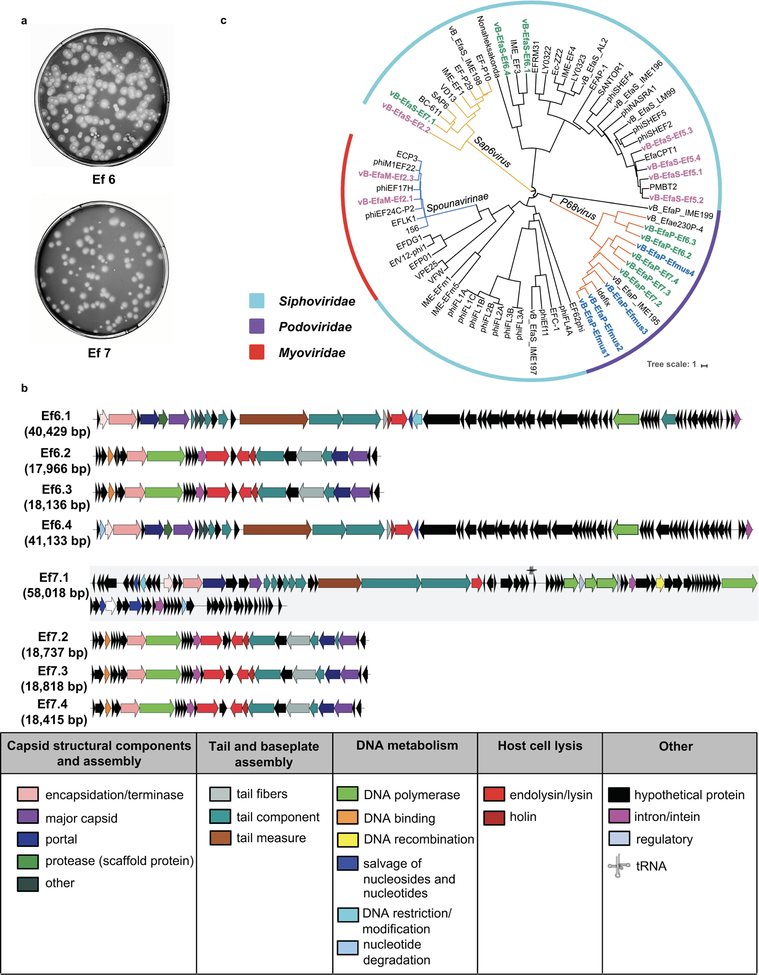
To demonstrate that the effect of bacteriophage treatment occurs via targeting of cytolysin-positive E. faecalis, rather than reduction in cytolysin-negative E. faecalis, we colonized gnotobiotic mice with feces from cytolysin-negative patients with alcoholic hepatitis (Supplementary Table 8). Bacteriophages against non-cytolytic E. faecalis from patients were isolated from sewage water, and they had siphophage or podophage morphology (Fig. 4a; Extended Data Fig. 8). These phages did not reduce features of ethanol-induced liver disease compared with control phages (Fig. 4b–4g; Extended Data Fig. 9a–9h), despite the reduction of fecal Enterococcus (Fig. 4h). Our findings indicate that lytic bacteriophage treatment can selectively attenuate ethanol-induced liver disease caused by cytolysin-positive E. faecalis in humanized mice.
Figure 4. Phages that target non-cytolytic E. faecalis do not reduce ethanol-induced liver disease in gnotobiotic mice.
(a) Transmission electron microscopy revealed that bacteriophages isolated were either podophages (Ef6.2, Ef6.3, Ef7.2, Ef7.3 and Ef7.4) or siphophages (Ef6.1, Ef6.4 and Ef7.1). Scale bar=50 nm. (b–h) C57BL/6 germfree mice were colonized with feces from two different cytolysin-negative patients with alcoholic hepatitis and subjected to chronic–binge feeding model, gavaged with control phages against C. crescentus (1010 PFUs), or a cocktail of 4 different phages targeting non-cytolytic E. faecalis (1010 PFUs) 1 day before an ethanol binge. (b) Serum levels of ALT. (c) Hepatic triglyceride content. (d) Representative H&E-stained liver sections. Scale bar=100 μm. (e–g) Hepatic levels of mRNAs encoding Il1b, Cxcl1, and Col1a1. (h) Fecal colony forming units (CFUs) of Enterococcus. Results are expressed as mean ± s.e.m (b, c, e–h). P values are determined by Two-way ANOVA with Tukeýs post-hoc test (b, c, e–h). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05.
Discussion
Phage-based therapies have been studied predominantly in patients with bacterial infections in the gastrointestinal tract21–23, urinary tract24,25 and other organ systems26–28. The results of these studies, while mixed in terms of efficacy, strongly suggest that phage treatment offers a safe alternative to antibiotics26,27. However, safety studies are required for complex populations such as patients with alcoholic hepatitis, since phages can induce a strong immune reaction29. Future work is required to determine if bacteriophages that target cytolytic E. faecalis might be used to treat patients with alcoholic hepatitis, a life-threatening disease with no effective treatment. Our data also suggest that cytolysin may be used as a predictive biomarker of severe alcoholic hepatitis; So an independent, prospective cohort is now needed to validate cytolysin as a biomarker and to extend the bacteriophage findings to human patients.
Eradication of this specific bacterial strain might produce better outcomes than current treatments. Remarkably, environmental sources can be used to easily isolate phages that target cytolysin-positive E. faecalis. We provide the first example of the efficacy of phage-based approaches in mice for a disease that is not considered a classic infectious disease. A clinical trial with a larger cohort is required to validate human relevance of our findings and to test this new therapeutic approach for patients with alcoholic hepatitis.
Methods
Patient cohorts.
Patient cohorts have been described30–32. We evaluated 26 subjects without alcohol use disorder (controls; social drinkers consuming less than 20 g/day), 44 patients with alcohol use disorder, and 88 patients with alcoholic hepatitis. Patients with alcohol use disorder fulfilling the DSM IV criteria33 of alcohol dependence and with active alcohol consumption (self-reported > 60 g/day) presented with various stages of liver disease (21% had advanced F3/4 fibrosis based on fibrosis-4 index (FIB-4); Supplementary Table 1). Patients with alcohol use disorder were recruited from an alcohol withdrawal unit in San Diego, USA and Brussels, Belgium where they followed a detoxification and rehabilitation program. At admission to the hospital, a complete medication and medical history is taken, and a complete physical examination is performed, including collection of biospecimens, basic demographic data, such as age, gender, weight and height, and self-reported daily alcohol consumption. Patients were actively drinking until the day of admission. Controls or patients with alcohol use disorder did not take antibiotics or immunosuppressive medication during the two months preceding enrollment. Other exclusion criteria were diabetes, inflammatory bowel disease, known liver disease of any other etiology, and clinically significant cardio-vascular, pulmonary or renal co-morbidities. Alcoholic hepatitis patients were enrolled from the InTeam Consortium (ClinicalTrials.gov identifier number: ) from centers in the USA, Mexico, United Kingdom, France and Spain. Inclusion criteria were active alcohol abuse (> 50 g/day for men and > 40 g/day for women) in the last 3 months, aspartate aminotransferase (AST) > alanine aminotransferase (ALT) and total bilirubin > 3 mg/dl in the past 3 months, liver biopsy and/or clinical picture consistent with alcoholic hepatitis. Exclusion criteria were autoimmune liver disease (ANA > 1/320), chronic viral hepatitis, hepatocellular carcinoma, complete portal vein thrombosis, extrahepatic terminal disease, pregnancy, and lack of signed informed consent. In all patients, the clinical picture was consistent with alcoholic hepatitis and in patients who underwent liver biopsy, the histology was in line with the diagnosis of alcoholic hepatitis. Liver biopsies were only done if clinically indicated as part of routine clinical care for diagnostic purposes of alcoholic hepatitis. Biospecimens were collected during their admission to the hospital. The median time of specimen collection was 4 days following admission to the hospital (range 0–24, n=82). For one patient who underwent liver transplantation, the transplantation date was considered as date of death. Patients were censored at the time point they were last seen alive. The baseline characteristics are shown in Supplementary Tables 1 and 2. Fecal 16S rRNA sequencing, Enterococcus culture and qPCR were performed. The MELD score, ABIC score and DF were calculated from all alcoholic hepatitis patients from whom respective laboratory values were available. The protocol was approved by the Ethics Committee of Hôpital Huriez (Lille, France), Universidad Autonoma de Nuevo Leon (Monterrey, México), Hospital Universitari Vall d’Hebron (Barcelona, Spain), King’s College London (London, UK), Yale University (New Haven, USA), University of North Carolina at Chapel Hill (Chapel Hill, USA), Weill Cornell Medical College (New York, USA), Columbia University (New York, USA), University of Wisconsin (Madison, USA), VA San Diego Healthcare System (San Diego, USA), University of California San Diego (La Jolla, USA) and Université Catholique de Louvain (Brussels, Belgium). Patients were enrolled after written informed consent was obtained from each patient.
Mice.
C57BL/6 mice were purchased from Charles River and used in Figure 2h and Extended Data Figure 2. C57BL/6 germ-free mice were bred at UCSD and used in Figure 2a–2g, Figure 3, Figure 4, Extended Data Figure 3, Extended Data Figure 7, and Extended Data Figure 9. Sublytic Atp4aSl/Sl mice on a C57BL/6 background have been described11,34 and heterozygous mice were used for breeding; sublytic Atp4aSl/Sl littermate mice and their wild-type littermates were used in Extended Data Figure 5.
Female and male mice (age, 9–12 weeks) were placed on a chronic–binge ethanol diet (NIAAA model) as described15. Mice were fed with Lieber-DeCarli diet and the caloric intake from ethanol was 0% on days 1–5 and 36% from day 6 until the end of the study period. At day 16, mice were gavaged with a single dose of ethanol (5 g/kg body weight) in the early morning and sacrificed 9 hours later. Pair-fed control mice received a diet with an isocaloric substitution of dextrose.
Stool samples from patients with alcoholic hepatitis (see Figure 1) were used for fecal transplantation in germ-free mice. Mice were gavaged with 100 μl of stool samples (1 g stool dissolved in 30 ml Luria-Bertani (LB) medium containing 15% glycerol under anaerobic conditions), starting at an age of 5–6 weeks and repeated two weeks later. Two weeks after the second gavage, mice were placed on the ethanol or control (isocaloric) diet.
In studies of the effects of cytolysin, 5 × 108 colony forming units (CFUs) of a cytolytic E. faecalis strain (FA2–2(pAM714)), a non-cytolytic E. faecalis strain (FA2–2(pAM771))5 (E. faecalis Δcytolysin) (kindly provided by Dr. Michael S. Gilmore), or PBS (vehicle control) were fed to mice by gavage every third day, starting from day 6 through day 15 of ethanol feeding (see above). Administration every third day was necessary, given that E. faecalis does not colonize mice11 (Extended Data Fig. 2o). To determine the effect of bacteriophage treatment, 1010 plaque forming units (PFUs) E. faecalis phages (or C. crescentus phage phiCbK as control)35 were gavaged to the mice one day before the ethanol binge (at day 16). All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of the University of California, San Diego.
Bacteriophages isolation and amplification.
E. faecalis strain from Atp4aSl/Sl mice feces was isolated before11 and was used to isolate phages Efmus1, Efmus2, Efmus3 and Efmus4. E. faecalis strains from human stool samples were isolated using methods described below and the corresponding phages were named as Ef with patient number plus a digit (Ef for E. faecalis, last digit for isolation order). All E. faecalis strains were grown statically in brain heart infusion (BHI) broth or on BHI agar at 37°C. C. crescentus phage phiCbK was purified as previously described35.
E. faecalis phages were isolated from untreated raw sewage water obtained from North City Water Reclamation Plant in San Diego, California. Fifty milliliter raw sewage water was centrifuged at 8,000 × g for 1 min at room temperature (RT) to pellet large particles. The supernatant was passed through a 0.45 μm and then a 0.2 μm syringe filter (Whatman, PES membrane). One hundred microliter of the clarified sewage was mixed with 100 μl overnight E. faecalis culture and then added to BHI broth top agar (0.5% agar) and poured over a BHI plate (1.5% agar). After overnight growth at 37°C, the resulting plaques were recovered using a sterile pipette tip in 500 μl PBS. Phages were replaqued on E. faecalis three more times to ensure that the phages were clonal isolates.
High-titer phage stocks were propagated by infecting 200 ml of exponentially growing E. faecalis at a multiplicity of infection (MOI) of 0.1 in BHI broth containing 10 mM MgSO4. Lysis was allowed to proceed for up to six hours at 37°C with shaking. The lysates were centrifuged at 10,000 × g for 20min at RT to remove the remaining bacterial cells and debris. Supernatant was then vacuum filtered through a 0.2 μm membrane filter and kept at 4°C until use.
Before mice were gavaged, 10–20 ml lysates were concentrated using Corning Spin-X UF Concentrators with 100,000-molecular-weight-cutoff (MWCO) to a volume of approximately 1 ml. Following concentration, the culture medium was replaced with PBS via diafiltration. The resulting lysate was further concentrated to a final volume of 0.5 ml and adjusted to the required PFUs.
Whole-genome sequencing for bacteriophages.
For all phages except Efmus4, 10 ml of lysates were treated with 10 μg/ml each of DNase and RNase at 37°C for 1 hour and phages were precipitated by adding 1M NaCl and 10% (w/v) polyethylene glycol 8000 (PEG 8000) and incubated at 4°C overnight. Precipitated phages were then pelleted by centrifugation at 10,000 × g for 10 min at 4°C and resuspended in 500 μl of resuspension buffer (5 mM MgSO4). Phage DNA was then extracted using Promega Wizard DNA Clean-up kit (Promega). Phage genomes were sequenced using a combination of Illumina and Oxford Nanopore Technologies (ONT) MinION platforms. Illumina sequencing libraries were prepared using the Nextera XT library kit with bead-based size selection prior to loading onto Illumina flow cells. Sequencing was performed with either Illumina MiSeq Reagent Kit v3 in 2 × 300-bp or NextSeq 500 Mid Output Kit in 2 × 150-bp paired-end formats. ONT MinION sequencing libraries were prepared using the Rapid Barcoding Kit (SQK-RBK004) and loaded onto MinION R9.4 flow cells. ONT reads were basecalled with Albacore v2.3.4 (ONT). The sequence reads were demultiplexed and adapters trimmed from ONT reads using Porechop v0.2.336. A hybrid Illumina-ONT de novo assembly was performed using the Unicycler v0.4.7 pipeline37. Subsequently, Pilon v1.2238 was used iteratively to polish the assemblies with Illumina reads until no additional corrections could be made.
For phage Efmus4, 109 PFUs of the phage was filtered sequentially using 0.45 μm and 0.2 μm filters (VWR) and purified on a cesium chloride (CsCl) density gradient39. One milliliter of the CsCl fraction was purified on Amicon YM-100 protein columns (Millipore) and treated with DNase I. DNA was isolated using a Qiagen UltraSens virus kit (Qiagen), amplified using GenomiPhi V2 (GE Healthcare), and fragmented to 200 to 400 bp using a Bioruptor (Diagenode). Libraries were created using the Ion Plus fragment library kit and sequenced using a 316 Chip on an Ion Torrent Personal Genome Machine (Life Technologies). Reads were trimmed according to modified Phred scores of 0.5 using CLC Genomics Workbench 4.9 (Cambridge), and the remaining reads were assembled using CLC Genomics Workbench 4.9 based on 98% identity with a minimum of 50% read overlap39. Reads were assembled into a single contig of 18,186 bp (20,118 × coverage).
Mapping of ONT reads to the hybrid assemblies was used to determine the orientation and terminal ends of linear phage genomes while reference genomes served as guides to orient circular phage genomes. Phage genome assemblies were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP)40,41.
Phage raw sequence reads and annotated genomes are available at NCBI under the following consecutive BioSample IDs (SAMN11089809– SAMN11089827). GenBank accession numbers include: Efmus1 (MK721195), Efmus2 (MK721197), Efmus3 (MK721185), Efmus4 (MK721193), Ef2.1 (MK693030), Ef2.2 (MK721189), Ef2.3 (MK721192), Ef5.1 (MK721199), Ef5.2 (MK721186), Ef5.3 (MK721200), Ef5.4 (MK721191), Ef6.1 (MK721187), Ef6.2 (MK721188), Ef6.3 (MK721196), Ef6.4 (MK721190), Ef7.1 (MK721194), Ef7.2 (MK721183), Ef7.3 (MK721184) and Ef7.4 (MK721198).
Genetic maps of bacteriophage genomes were generated by LinearDisplay.pl (https://github.com/JCVenterInstitute/LinearDisplay), a PERL script that uses Xfig (https://sourceforge.net/projects/mcj/) to render high-quality images. Preliminary annotation of genes was derived from the automated annotation and from Phage_Finder42, which uses curated HMMs and databases of core phage gene to annotate core gene functions. Annotation was then manually reviewed to assign the final colors.
Bacteriophage phylogenetic tree.
A phage whole genome phylogeny tree was generated from a pairwise distance matrix calculated with the MASH program, which approximates average nucleotide identity (ANI)43. First, a sketch file was created from all the 19 E. faecalis phage genomes isolated and sequenced in this study plus 54 Enterococcus phage genomes obtained from GenBank, with 5000 12mers generated per genome (mash sketch -k 12 -s 5000). The sketch file was then compared to all the initial phage genome sequences to generate the ANI matrix using the mash distance command using default settings. The GGRaSP R-package was used to calculate the UPMGA phylogeny from the ANI distance matrix, after redundant phage genomes (genomes ANI > 99.985) were removed using the GGRaSP R-package with a user defined cutoff of 0.015 (ggrasp.cluster (threshold = 0.015)). The resulting dendrogram was translated into newick format using the APE R package44, loaded into the iTOL tree viewer45, and annotated with taxonomic information and manually-entered clade identification.
Electron microscopy.
Bacteriophage morphology was examined by transmission electron microscopy of negatively stained grids, prepared using the valentine method46 with either 2% uranyl-acetate or 2% phosphotungstic acid, and examined at an acceleration voltage of 100KV in the JEOL 1200 EX Transmission Electron Microscope.
Bacterial DNA extraction and 16S rRNA sequencing.
DNA from human stool samples, mouse liver sections or bacterial culture was extracted as described before11, and DNA from mouse feces was extracted using QIAamp Fast DNA Stool kit (QIAGEN). 16S ribosomal RNA (rRNA) PCR was completed using Illumina adaptor and barcode ligated 16S primers targeting the V4 region of the 16S rRNA gene47,48. Amplicons were purified using the Qiaquick PCR purification kit (QIAGEN) using manufacturer’s specifications. Purified amplicons were then quantified via TECAN assay (Tecan, Switzerland), normalized, and pooled in preparation for 16S rRNA sequencing. Pooled library was quantified and checked for quality using Agilent 2100 Bioanalyzer (Agilent Technologies). Library was sequenced on Illumina MiSeq (Illumina) using V2 reagent chemistry, 500 cycles, 2 × 250bp format using manufacturer’s specifications. 16S sequence reads were processed and OTUs were determined using our MOTHUR-based 16S rDNA analysis workflow as described previously11,49. Raw 16S sequence reads can be found in the NCBI SRA associated with Bioproject PRJNA525701.
Real-time quantitative PCR.
Bacterial genomic DNA was extracted from human stool samples and mouse liver11. RNA was extracted from mouse liver and cDNAs were generated11. Primer sequences for mouse genes were obtained from the NIH qPrimerDepot. Primer sequences for E. faecalis 16S rRNA gene, E. faecalis cylLS and cylLL genes were described before50,51. All primers used in this study are listed in Supplementary Table 9. Mouse gene expression and amplification of bacterial genes were determined with Sybr Green (Bio-Rad Laboratories) using ABI StepOnePlus real-time PCR system. The qPCR value of mouse genes was normalized to 18S.
E. faecalis isolation and whole-genome sequencing.
To isolate E. faecalis strains from human subjects, 50–300 mg of human stool was resuspended in 500 μl PBS, serial dilutions were made, and 100 μl was placed on plates with selective medium, BBL Enterococcosel broth (Becton Dickinson). Enterococci colonies were identified by the production of dark brown or black color generated by hydrolysis of esculin to esculetin that reacts with ferric ammonium citrate. Each Enterococcus colony was then picked, and qPCR was performed to identify E. faecalis, using specific primers against the E. faecalis 16S rRNA gene50. For each subject, 1 – 6 E. faecalis colonies were analyzed and bacterial genomic DNA was then extracted as described in above section.
DNA sequencing was performed on the Illumina HiSeq Ten X generating paired-end reads (2 × 151bp). Bacterial genomes were assembled and annotated using the pipeline described previously52. Antimicrobial resistance and virulence genes including cytolysin (cyl) genes carried by E. faecalis isolates were identified by comparing individual genome assemblies against the CARD and VFDB databases using abricate v0.8.10 (https://github.com/tseemann/abricate), respectively53,54.
For the phylogeny of E. faecalis, the genome assemblies of the study isolates were annotated with Prokka55, and a pangenome estimated using Roary52. A 95% identity cut-off was used, and core genes were defined as those in 99% of isolates. A maximum likelihood tree of the SNPs in the core genes was created using RAxML56 and 100 bootstraps. The resulting tree was visualized using iTOL45. Genome sequence data of E. faecalis strains isolated in this study have been deposited in the European Nucleotide Archive (ENA) under the accession number PRJEB25007. Sequence reads are available at ENA under Run accession IDs ERR3200171-ERR3200263.
E. faecalis culture.
All E. faecalis strains were grown statically in brain heart infusion (BHI) broth or on BHI agar plate at 37°C. 50 μg/ml erythromycin was added when cytolytic and non-cytolytic E. faecalis strains were grown (Extended Data Figure 2).
Fecal Enterococcus level determination.
To determine fecal enterococci level in mice, 10–30 mg of mouse feces was resuspended into 500 μl PBS and serial dilutions were made. Five microliters of each dilution from each sample were spotted onto a plate with a selective medium, BBL Enterococcosel broth (Becton Dickinson) and the plates were then incubated at 37°C overnight. For Extended Data Fig. 2o, agar plates contained 50 μg/ml erythromycin. Enterococci colonies were identified by the production of dark brown or black color generated by hydrolysis of esculin to esculetin that reacts with ferric ammonium citrate. Colony numbers of each sample were then counted and CFUs were calculated.
Cytolysin expression and purification.
To purify bioactive CylLL” and CylLS”, an E. coli heterologous expression system was used. Briefly, either Hisx6-CylLL or Hisx6-CylLS were co-expressed with CylM (enzyme that performs dehydration and cyclization reactions on cytolysin) in E. coli to yield fully dehydrated and cyclized full-length peptides. The His-tag and leader peptide were then removed using recombinant CylA (27–412), the soluble domain of the native peptidase used in cytolysin maturation, to yield bioactive CylLL” or CylLS”. The resulting core peptides were further purified by reversed-phase HPLC.
The cylLL and cylLS genes were previously cloned into the MCSI of a pRSFDuet-1 backbone vector which contained the cylM gene in MCSII6. The cylA (27–412) gene was previously cloned into MCSI of a pRSFDuet-1 backbone vector57. E. coli BL21 Star™ (DE3) cells (50 μl) were transformed with 100 ng of either the cylLL_cylM:pRSFDuet, cylLS_cylM:pRSFDuet or cylA (27–412):pRSFDuet plasmids via KCM chemical transformation. The cells were plated on LB agar plates supplemented with kanamycin (50 μg/ml) and grown at 37°C overnight. One colony was picked to inoculate 15 ml of LB broth supplemented with kanamycin overnight at 37°C. The culture was used to inoculate 1.5 liters of terrific broth supplemented with kanamycin. Cultures were grown with shaking at 37°C to an OD600 of 0.8. The temperature of the incubator was lowered to 18°C and expression was induced with the addition of 0.3 mM final concentration of isopropyl β-D-thiogalactoside. The cultures were allowed to incubate at 18°C for 18 hours. The cells were collected by centrifugation at 5000 × g for 12 min. The cell paste was collected and frozen at −70°C.
For the purification of the protease CylA (27–412), the cell paste was thawed and resuspended in 50 ml LanP buffer (20 mM HEPES, 1 M NaCl, pH 7.5). The cell suspension was lysed by homogenization. The lysate was clarified by centrifugation at 13,000 × g for 45 min and filtered through a 0.45 μm centrifugal filter (Thermo Scientific). The clarified lysate was applied to a pre-equilibrated HisTrap HP 5 ml column (GE Healthcare) through a peristaltic pump. The loaded column was connected to an ÄKTA pure 25 M system. The protein was eluted by a linear gradient of LanP buffer to Elution Buffer (20 mM HEPES, 1 M NaCl, 500 mM imidazole, 10% glycerol, pH 7.5) over 30 min. The purest fractions, as determined by 4%–20% SDS-PAGE, were combined, concentrated to 1 mg/ml by Amicon Ultra Centrifugal Filters (30 kDa MWCO), and buffer exchanged into storage buffer (20mM HEPES, 300 mM KCl, 10% glycerol, pH 7.5) by PD-10 desalting column (GE Healthcare). Protein concentration was determined by absorbance at 280 nm.
For the purification of CylLL” and CylLS” peptides, the cell paste was thawed and resuspended in 50 ml of LanA Buffer B1 (6 M guanidine HCl, 20 mM NaH2PO4, 500 mM NaCl, 0.5 mM imidazole, pH 7.5). The cell suspension was lysed via sonication (2 sec pulse on, 5 sec pulse off, 7 min total pulse on time). The cell lysate was clarified by centrifugation at 13,000 × g for 45 min. The clarified cell lysate was filtered through a 0.45 μm centrifugal filter and applied via gravity flow to a pre-equilibrated, 2 ml bed volume of His60 Ni Superflow Resin (Clonetech). After the lysate had been applied, the resin was washed with 15 ml of LanA Buffer B2 (4 M guanidine HCl, 20 mM NaH2PO4, 500 mM NaCl, 30 mM imidazole, pH 7.5). The resin was washed again with 15 ml of LanA Wash Buffer (20 mM NaH2PO4, 500 mM NaCl, 30 mM imidazole, pH 7.5) to remove the guanidine HCl. The peptides were eluted with 10 ml of LanA Elution Buffer (20 mM NaH2PO4, 500 mM NaCl, 500 mM imidazole, pH 7.5). A 0.02 mg/ml final concentration of CylA (27–412) was added to the elution fraction and allowed to incubate at room temperature overnight to remove the leader peptide.
The digestion was quenched by adding 2% (v/v) final concentration of trifluoroacetic acid. The solution was centrifuged at 4,500 × g for 10 min and filtered through a 0.45 μm syringe filter (Thermo Scientific). The core peptides were purified by semi-preparative reverse phase HPLC using a Phenomenex Jupiter Proteo column (10 mm × 250 mm, 4 μm, 90 Å) connected to an Agilent 1260 Infinity II liquid chromatography system. The peptides were separated using a linear gradient of 3% (v/v) solvent B (acetonitrile + 0.1% trifluoroacetic acid) in solvent A (water + 0.1% trifluoroacetic acid). The fractions were spotted on a MALDI target plate by mixing 1 μl of sample with 1μl of a 25 mg/ml solution of Super-DHB (Sigma) in 80% acetonitrile/water + 0.1% trifluoroacetic acid. The fractions were analyzed by MALDI-TOF MS on a Bruker UltrafleXtreme MALDI-TOF/TOF operating in positive ionization, reflector mode.
Primary mouse hepatocytes.
Hepatocytes were isolated from C57BL/6 female mice fed the chronic–binge ethanol diet (NIAAA model)15. Livers were perfused in situ with 0.5 mM EGTA containing calcium-free salt solution and then perfused with a solution containing 0.02% (w/v) collagenase D (Roche Applied Science). Livers were then carefully minced and filtered using a 70 μm nylon cell strainer. Hepatocytes were centrifuged at 50 × g for 1 min after three times washing. Hepatocyte viability was assessed by Trypan Blue (Thermo Fisher Scientific). 1.5 × 105 hepatocytes were seeded on 12-well plates coated with rat collagen type I in DMEM-F12 (Thermo Fisher Scientific) with insulin-transferrin-selenium (1% v/v) (Thermo Fisher Scientific) and 40 ng/ml dexamethasone (MP Biomedicals) containing 10% (v/v) fetal bovine serum (FBS; Gemini Bio-Products) and antibiotics. After 4 hours, the culture was washed with DMEM-F12 media and changed to the same complemented media without FBS58. Then 16 hours later, hepatocytes were cultured with 0 or 25 mM ethanol and stimulated with 0, 200 or 400 nM CylLS” and/or CylLL” in the same culture medium without FBS. After 3 hours stimulation, hepatocyte cytotoxicity was assessed using Pierce LDH Cytotoxicity Detection Kit (Thermo Fisher Scientific).
Biochemical analysis.
Serum levels of ALT were determined using Infinity ALT kit (Thermo Scientific). Hepatic triglyceride levels were measured using Triglyceride Liquid Reagents kit (Pointe Scientific). Levels of serum LPS and fecal albumin were determined by ELISA kits (Lifeome Biolabs and Bethyl Labs, respectively). Serum levels of ethanol were measured using Ethanol Assay kit (BioVision).
Staining procedures.
Formalin-fixed tissue samples were embedded in paraffin and stained with hematoxylin-eosin. To determine lipid accumulation, liver sections were embedded in OCT compound. 8 μm frozen sections were then cut and stained with Oil Red O (Sigma-Aldrich). Representative pictures from each group of mice were shown in each figure. The terminal deoxynucleotide transferase-mediated dUTP nick-end labeling (TUNEL) assay was performed using an in situ cell death detection kit (Sigma-Aldrich). We randomly selected five high-power fields for counting TUNEL-positive cells and normalized numbers to total cells.
Data availability.
Raw 16S sequence reads can be found in the NCBI SRA associated with Bioproject PRJNA525701. Bacteriophage raw sequence reads and annotated genomes are available at NCBI under the following consecutive BioSample IDs (SAMN11089809 – SAMN11089827). Genome sequence data of E. faecalis strains isolated in this study were registered at ENA under Study PRJEB25007.
Code availability.
The PERL script for making the genetic maps of bacteriophage genomes can be found at https://github.com/JCVenterInstitute/LinearDisplay.
Statistical analysis.
Results are expressed as mean ± s.e.m. (except when stated otherwise). Univariate and multivariate Cox regression analysis was used to detect associations of cytolysin with overall mortality. The multivariate model was adjusted for geographic origin of the patients; antibiotic treatment; platelet count; and creatinine, bilirubin and INR as components of the MELD score. Univariate logistic regression analysis of laboratory and clinical parameters associated with the detection of cytolysin and E. faecalis was performed. Univariate linear regression analysis of laboratory and clinical parameters associated with the log-transformed total amount of fecal E. faecalis measured with qPCR was performed. To associate log-transformed total E. faecalis and E. faecalis positivity with mortality, univariate Cox regression was used. P values from Univariate and Multivariate Cox regression, Univariate logistic regression and Univariate linear regression were determined by Wald test. Multicollinearity was examined using the variance inflation factor (VIF). Kaplan-Meier curves were used to compare survival between cytolysin-positive and cytolysin-negative alcoholic hepatitis patients. Fecal E. faecalis, bacterial diversity and richness from controls and patients were compared using Kruskal-Wallis test with Dunn’s post-hoc test. Region/center specific clinical characteristics of alcoholic hepatitis patients were compared with Kruskal-Wallis test for continuous and Fisher’s exact test for categorical variables. Fecal E. faecalis in alcoholic hepatitis patients with or without cytolysin, and with or without cirrhosis, were compared with Mann-Whitney-Wilcoxon rank-sum test. Fecal E. faecalis in alcoholic hepatitis patients from different region/centers were compared with Kruskal-Wallis test. Percentage of subjects with fecal samples positive for E. faecalis and cytolysin was compared using Fisher’s exact test, followed by FDR procedures for multiple group comparisons. Jaccard dissimilarity matrices were used for principal coordinate analysis (PCoA) and P values were determined by permutational multivariate analysis of variance (PERMANOVA) followed by FDR procedures to correct for multiple comparisons.
For mouse and cell culture studies, significance of multiple groups was evaluated using one-way or two-way analysis of variance (ANOVA) with Tukey’s post-hoc test. Fisher’s exact test was used in the analysis of liver E. faecalis and cytolysin with FDR correction for multiple comparisons. Kaplan-Meier curves were used to compare survival between experimental mouse groups. PCoA based on Jaccard dissimilarity matrices was performed between experimental mouse groups and the P values were determined by PERMANOVA followed by FDR procedures to correct for multiple comparisons.
Exact P values for all comparisons, together with group size for each group, were listed in Supplementary Table 10. Statistical analyses were performed using R statistical software, R version 3.5.1, 2018 the R Foundation for Statistical Computing and GraphPad Prism v6.01. A P < 0.05 was considered to be statistically significant (adjusted for multiple comparison when performing multiple tests).
Extended Data
Extended Data Figure 1. Intestinal dysbiosis in patients with alcoholic hepatitis.
(a) 16S rRNA sequencing of fecal samples from controls (n=14), patients with alcohol use disorder (AUD; n=43), or alcoholic hepatitis (n=75). The graph demonstrates the relative abundance of sequence reads in each genus. (b) Bacterial diversity (Shannon-Index and Simpson-Index) and richness (Chao-Richness) was calculated in controls (n=14), patients with AUD (n=43), or alcoholic hepatitis (n=75). (c) E. faecalis in fecal samples from controls (n=25), patients with AUD (n=38), or alcoholic hepatitis (n=82), assessed by qPCR. (d) Percentage of fecal samples positive for E. faecalis in controls (n=25), patients with AUD (n=38), or alcoholic hepatitis (n=82), assessed by qPCR. E. faecalis was detected in feces from 80% of patients with alcoholic hepatitis vs 36% of controls (P<0.001). There was also a significant difference between patients with alcohol use disorder and alcoholic hepatitis (P<0.01). (e) ROC curves and AUC for the comparison of 90-day mortality and cytolysin positivity (red; n=57), MELD score (blue; n=56), ABIC score (yellow; n=57), and DF (green; n=42) in patients with alcoholic hepatitis. (f) E. faecalis in fecal samples from patients with alcoholic hepatitis whose fecal samples were cytolysin positive (n=25) or cytolysin negative (n=54), assessed by qPCR (P=0.8174). (g) 16S rRNA sequencing of fecal samples from patients with alcoholic hepatitis from different centers (France n=9; Mexico n=6; Spain n=5; UK n=11; USA East n=16; USA Midwest n=12; USA West n=16). Principal coordinate analysis (PCoA) based on Jaccard dissimilarity matrices was used to show ß-diversity among groups, at the genus level. Composition of fecal microbiota was significantly different (P<0.01). (h) Percentage of fecal samples positive for cylLL and cylLS DNA sequences (cytolysin-positive), in patients with alcoholic hepatitis from different centers (France n=16; Mexico n=6; Spain n=6; UK n=10; USA East n=16; USA Midwest n=13; USA West n=15), assessed by qPCR (P=0.6094). (i) E. faecalis in fecal samples from patients with alcoholic hepatitis from different centers, assessed by qPCR (P=0.5648). (j) Percentage of fecal samples positive for E. faecalis in patients with alcoholic hepatitis from different centers (France n=16; Mexico n=6; Spain n=6; UK n=10; USA East n=16; USA Midwest n=13; USA West n=15), assessed by qPCR (P=0.0529). (k) Percentage of subjects with fecal samples positive for cylLL and cylLS DNA sequences (cytolysin positive), in patients with alcoholic hepatitis and cirrhosis (n=30), or without cirrhosis (n=18), assessed by qPCR (P=0.3431). (l) E. faecalis in fecal samples from patients with alcoholic hepatitis and cirrhosis (n=30), or without cirrhosis (n=18), assessed by qPCR (P=0.5736). (m) Percentage of fecal samples positive for E. faecalis in alcoholic hepatitis patients with cirrhosis (n=30), or without cirrhosis (n=18), assessed by qPCR (P=0.2878). Results are expressed as as mean ± s.e.m (c, f, i, l). For the Box and Whisker plots in (b), the box extends from 25th to 75th percentiles, with the center line representing the median; for all three groups, the lower whiskers show the minimum values; for the “Controls” group (black), the higher whisker shows the maximum value; for the other two groups, the higher whiskers represent the 75th percentile plus 1.5 times inter-quartile distance (the distance between the 25th and 75th percentiles), all values greater than this are plotted as individual dots. P values are determined by Kruskal-Wallis test (i) with Dunn’s post-hoc test (b, c), two-sided Fisher’s exact test (h, j, k, m) followed by false discovery rate (FDR) procedures (d), two-sided Mann-Whitney-Wilcoxon rank-sum test (f, l), or permutational multivariate analysis of variance (PERMANOVA) (g). The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Figure 2. Cytolytic E. faecalis causes progression of ethanol-induced liver disease in mice.
(a–n) C57BL/6 mice were fed oral isocaloric (control) or chronic–binge ethanol diets and gavaged with vehicle (PBS), a cytolytic E. faecalis strain (FA2–2(pAM714)) (E. faecalis) (5×108 colony forming units (CFUs)), or a non-cytolytic E. faecalis strain (FA2–2(pAM771))5 (E. faecalis Δcytolysin) (5×108 CFUs) every third day. (a) Serum levels of ALT. (b) Hepatic triglyceride content. (c) Representative oil red O-stained liver sections. (d–f) Hepatic levels of mRNAs. (g) Kaplan-Meier curve of survival of mice on chronic–binge ethanol diets (day 0, start of ethanol feeding). Mice gavaged with PBS all survived and were not included in figure. A higher proportion of mice (n=15) gavaged with non-cytolytic E. faecalis survived than mice (n=25) gavaged with cytolytic E. faecalis. (h) Proportions of mice positive for cytolysin in liver, measured by qPCR for cylLS (the gene encoding cytolysin subunit CylLS”). (i) Proportions of mice positive for E. faecalis in liver, measured by qPCR. About 80% of mice colonized with cytolytic E. faecalis, as well as non-cytolytic E. faecalis, were positive for E. faecalis in their livers. (j) Liver CFUs of Enterococcus in mice on a chronic–binge ethanol diet. (k) Paracellular intestinal permeability was evaluated by measuring fecal albumin content and serum levels of lipopolysaccharide (LPS) by ELISAs. (l) Fecal samples were collected and 16S rRNA genes were sequenced. Principal coordinate analysis based on Jaccard dissimilarity matrices showed no significant differences among mice gavaged with PBS, cytolytic or non-cytolytic E. faecalis following the diets. Compared to control-diet fed mice, mice fed with ethanol diet had significant different fecal microbiomes after gavaging E. faecalis (P<0.05). (m and n) Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among these mice after ethanol feeding. (o) Mice were gavaged with cytolytic or non-cytolytic E. faecalis strains (carrying erythromycin resistance gene; 5×108 CFUs) at time 0, and feces was collected at 0, 8, 24, 48, and 72 hrs. Fecal CFUs of Enterococcus were determined by culturing fecal samples on BBL Enterococcosel Broth agar plate with 50 μg/ml erythromycin. At time 0 and 72, 5/5 and 4/5 mice, respectively, had no detectable erythromycin-resistant Enterococcus in feces. These points are not shown on the graph, but have been included in the calculation of mean ± s.e.m. Scale bar=100 μm. Results are expressed as mean ± s.e.m (a, b, d–f, j, k, m–o). P values among groups of mice fed with control diet or ethanol diet are determined by One-way ANOVA with Tukeýs post-hoc test (a, b, d–f, j, k, m, n), two-sided Log-rank (Mantel-Cox) test (g), two-sided Fisher’s exact test followed by FDR procedures (h and i), or PERMANOVA followed by FDR procedures (l). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. P values between control-diet fed mice and ethanol-diet fed mice are determined by Two-way ANOVA (k). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Figure 3. Transplantation of cytolysin-positive feces increases ethanol-induced liver disease in gnotobiotic mice.
(a–f, h, i) C57BL/6 germ-free mice were colonized with feces from two different cytolysin-positive and two different cytolysin-negative patients with alcoholic hepatitis, and then fed isocaloric (control) or chronic–binge ethanol diets. (a) Percentage of terminal deoxynucleotide transferase-mediated dUTP nick-end labeling (TUNEL) positive hepatic cells. (b) Representative oil red O-stained liver sections. (c and d) Hepatic levels of mRNAs encoding the inflammatory cytokine Cxcl2, and Acta2 (marker of activated hepatic stellate cells). (e) Kaplan-Meier curve of survival of mice on chronic–binge ethanol diets (day 0, start of ethanol feeding) gavaged with feces from cytolysin-positive (n=48 mice) or cytolysin-negative (n=32 mice) patients with alcoholic hepatitis. (f) Fecal samples were collected and 16S rRNA genes were sequenced. The graph shows principal coordinate analysis of fecal microbiomes. No significant difference was observed between mice colonized with feces from cytolysin positive or negative alcoholic hepatitis donors following the control diet. Mice transplanted with feces from cytolysin-positive alcoholic hepatitis patient #2 showed a significantly different microbiota than the other mouse groups following ethanol administration (P<0.01). (g) Percentage of cytolysin-positive E. faecalis in 4 patients with alcoholic hepatitis. Stool samples from the four patients were placed on plates with selective medium and Enterococcus colonies were identified by the production of dark brown or black color generated by hydrolysis of esculin to esculetin that reacts with ferric ammonium citrate. Enterococcus colonies were confirmed to be E. faecalis by qPCR. Cytolysin status of each E. faecalis colony was determined by qPCR. (h) Serum levels of ethanol were comparable among colonized mice after ethanol feeding. (i) Hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice on control or ethanol diets. Scale bar=100 μm. Results are expressed as mean ± s.e.m (a, c, d, h, i). P values are determined by One-way ANOVA with Tukeýs post-hoc test (a, c, d, h, i), two-sided Log-rank (Mantel-Cox) test (e), or PERMANOVA followed by FDR procedures (f). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05, **P<0.01, ***P<0.001.
Extended Data Figure 4. Isolation and amplification of bacteriophages against cytolytic E. faecalis isolated from mice.
(a) BHI agar plate showing bacteriophage plaque morphology. Bacteriophage cocktail (100 μl, 102–103 PFUs) was mixed with overnight grown E. faecalis culture (100 μl) and then added to BHI broth top agar (0.5% agar) and poured over a BHI plate (1.5% agar). After overnight growth at 37°C, images were captured on an Epson Perfection 4990 Photo scanner. (b) Simplified illustration of different bacteriophage morphology. Siphophages have long, flexible, noncontractile tails (left), myophages have contractile tails (middle), and podophages have short, noncontractile tails (right). (c) Transmission electron microscopy revealed that bacteriophages isolated were all podophages (Efmus1, Efmus2, Efmus3 and Efmus4). Phages specific to E. faecalis strain isolated from mouse feces were named as Efmus with a number (Ef for E. faecalis, mus for mouse, digit for isolation order). (d) Genetic map of phage genomes. The linear maps are based on nucleotide sequences of the phage genomes and predicted open reading frames. The name and length (bp) of each genome are indicated to the left of each phage map. Protein-coding sequences are colored based on functional role categories (see box). Scale bar=50 nm. All results were generated from at least three independent replicates.
Extended Data Figure 5. Phages reduce translocation of cytolysin to the liver and reduce ethanol-induced liver disease in Atp4aSl/Sl mice.
(a–k) Wild-type (WT) and their Atp4aSl/Sl littermates were fed oral isocaloric (control) or chronic–binge ethanol diets and gavaged with vehicle (PBS), control phages against C. crescentus (1010 plaque forming units (PFUs)), or a cocktail of 4 different phages targeting cytolytic E. faecalis (1010 PFUs) 1 day before ethanol binge. (a) Serum levels of ALT. (b) Hepatic triglyceride content. (c) Representative oil red O-stained liver sections. (d–f) Hepatic levels of mRNAs. (g) Proportions of mice positive for cytolysin in liver, measured by qPCR for cylLS. (h) Fecal colony forming units (CFUs) of Enterococcus. (i) Fecal samples were collected and 16S rRNA genes were sequenced. Principal coordinate analysis based on Jaccard dissimilarity matrices found no significant difference in fecal microbiota among mice given PBS, control phage, or phages targeting cytolytic E. faecalis in each group. (j and k) Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice after ethanol feeding. Scale bar=100 μm. Results are expressed as mean ± s.e.m (a, b, d–f, h, j, k). P values are determined by Two-way ANOVA with Tukeýs post-hoc test (a, b, d–f, h, j, k), two-sided Fisher’s exact test followed by FDR procedures (g), or PERMANOVA followed by FDR procedures (i). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Figure 6. Isolation and amplification of bacteriophages against cytolytic E. faecalis isolated from patients with alcoholic hepatitis.
(a) BHI agar plates showing bacteriophage plaque morphology. (b) Transmission electron microscopy graphs of myophages Ef2.1 and Ef2.3, stained with phosphotungstic acid showing contracted tails. (c) Genetic map of phage genomes. The linear maps are based on nucleotide sequences of the phage genomes and predicted open reading frames. The name and length (bp) of each genome are indicated to the left of each phage map. Protein-coding sequences are colored based on functional role categories (see box). Sequences encoding tRNA genes are indicated by a cloverleaf structure. Scale bar=50 nm. All results were generated from at least three independent replicates.
Extended Data Figure 7. Phages that target cytolytic E. faecalis reduce ethanol-induced liver disease in gnotobiotic mice.
(a–h) C57BL/6 germ-free mice were colonized with feces from two different cytolysin-positive patients with alcoholic hepatitis (feces from 1 patient is used in Figure 2). The mice were then fed oral isocaloric (control) or chronic–binge ethanol diets, and gavaged with control phages against C. crescentus (1010 plaque forming units (PFUs)), or a cocktail of 3 or 4 different phages targeting cytolytic E. faecalis (1010 PFUs) 1 day before an ethanol binge. (a) Percentage of TUNEL-positive hepatic cells. (b) Representative oil red O-stained liver sections. (c and d) Hepatic levels of mRNAs encoding the inflammatory cytokine Cxcl2, and Acta2 (marker of activated hepatic stellate cells). (e) Fecal colony forming units (CFUs) of Enterococcus. (f) Fecal samples were collected and 16S rRNA genes were sequenced. Principal coordinate analysis based on Jaccard dissimilarity matrices shows no significant differences in fecal microbiota of mice gavaged with control phage and phages targeting cytolytic E. faecalis in each group. (g and h) Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice after ethanol feeding. Scale bar=100 μm. Results are expressed as mean ± s.e.m (a, c–e, g, h). P values are determined by Two-way ANOVA with Tukeýs post-hoc test (a, c–e, g, h), or PERMANOVA followed by FDR procedures (f). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10. *P<0.05, ***P<0.001.
Extended Data Figure 8. Isolation and amplification of bacteriophages against non-cytolytic E. faecalis isolated from patients with alcoholic hepatitis.
(a) BHI agar plates showing bacteriophage plaque morphology. (b) Genetic map of phage genomes. The linear maps are based on nucleotide sequences of the phage genomes and predicted open reading frames. The name and length (bp) of each genome are indicated to the left of each phage map. Protein-coding sequences are colored based on functional role categories (see box). Sequences encoding tRNA genes are indicated by a cloverleaf structure. (c) Phylogenetic tree of Enterococcus bacteriophages. A whole-genome average nucleotide distance tree was constructed for 73 available Enterococcus phage genomes, 54 from GenBank (black letters), 19 from this study (4 phages against cytolysin-positive E. faecalis isolated from mice in blue letters, 7 phages against cytolysin-positive E. faecalis isolated from alcoholic hepatitis patients in pink letters, and 8 phages against cytolysin-negative E. faecalis isolated from alcoholic hepatitis patients in green letters) with Mash43 using a sketch size of s=5000 and k-mer size of k=12 and GGRaSP59 (see Methods). Colored branches denote specific phage genera: Sap6virus, P68virus and Spounavirinae. The scale bar represents percent average nucleotide divergence. All results were generated from at least three independent replicates.
Extended Data Figure 9. Phages that target non-cytolytic E. faecalis do not reduce ethanol-induced liver disease in gnotobiotic mice.
(a–h) C57BL/6 germfree mice were colonized with feces from two different cytolysin-negative patients with alcoholic hepatitis. Transplanted gnotobiotic mice were fed oral isocaloric (control) or chronic–binge ethanol diets and gavaged with control phages against C. crescentus (1010 plaque forming units (PFUs)), or a cocktail of 4 different phages targeting non-cytolytic E. faecalis (1010 PFUs) 1 day before an ethanol binge. (a) Percentage of TUNEL-positive hepatic cells. (b) Representative oil red O-stained liver sections. (c and d) Hepatic levels of mRNAs encoding the inflammatory cytokine Cxcl2, and Acta2 (marker of activated hepatic stellate cells). (e) Proportions of mice positive for cytolysin in liver, measured by qPCR for cylLS. (f) Fecal samples were collected and 16S rRNA genes were sequenced. Principal coordinate analysis based on Jaccard dissimilarity matrices found no significant difference in fecal microbiota among mice gavaged with control phages and phages targeting cytolytic E. faecalis in each group. (g and h) Serum levels of ethanol and hepatic levels of Adh1 and Cyp2e1 mRNAs did not differ significantly among colonized mice after ethanol feeding. Scale bar=100 μm. Results are expressed as mean ± s.e.m (a, c, d, g, h). P values are determined by Two-way ANOVA with Tukeýs post-hoc test (a, c, d, g, h), two-sided Fisher’s exact test followed by FDR procedures (e), or PERMANOVA followed by FDR procedures (f). All results were generated from at least three independent replicates. The exact group size (n) and P values for each comparison are listed in Supplementary Table 10.
Supplementary Material
Acknowledgements
S.L. was supported by a DFG fellowship (LA 4286/1–1). This study was supported in part by a Biocodex Microbiota Foundation Grant, NIH grants R01 AA24726, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.), the Wellcome Trust (WT098051) (to T.D.L.), NIH grant U01AA021908 (to R.B.) and services provided by P30 DK120515 and P50 AA011999. Y.S. is supported by a Wellcome Trust PhD Studentship. I.R.R was funded by National Institute of General Medical Sciences (NIGMS)–NIH Chemistry–Biology Interface Training Grant (T32-GM070421).
Abbreviations:
- ABIC
Age, serum bilirubin, INR, and serum creatinine score
- Acta2
smooth muscle actin alpha 2
- Adh1
alcohol dehydrogenase 1
- AH
alcoholic hepatitis
- ALT
alanine amino-transferase
- ANOVA
analysis of variance
- AUC
area under the curve
- AUD
alcohol use disorder
- BHI broth
brain heart infusion broth
- C. crescentus
Caulobacter crescentus
- CFUs
colony forming units
- CI
confidence interval
- Col1a1
collagen, type I, alpha 1
- Cxcl
chemokine (C-X-C motif) ligand
- Cyp2e1
cytochrome P450 family 2 subfamily E polypeptide 1
- DF
discriminant function
- E. coli
Escherichia coli
- E. faecalis
Enterococcus faecalis
- FDR
false discovery rate
- FIB-4
fibrosis-4 index
- HR
hazard ratio
- Il1b
interleukin 1 beta
- INR
international normalized ratio
- LB medium
Luria-Bertani medium
- LDH
lactate dehydrogenase
- LPS
lipopolysaccharide
- MELD
model for end-stage liver disease
- MELDNa
sodium MELD
- MOI
multiplicity of infection
- PBS
phosphate-buffered saline
- PCoA
principal coordinate analysis
- PERMANOVA
permutational multivariate analysis of variance
- PFUs
plaque forming units
- qPCR
quantitative PCR
- ROC
receiver operating characteristic
- rRNA
ribosomal RNA
- RT
room temperature
- SNP
single nucleotide polymorphism
- TUNEL
terminal deoxynucleotide transferase-mediated dUTP nick-end labeling
- VIF
variance inflation factor
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Competing interests
B.S. is consulting for Ferring Research Institute. However, there is no competing interest with regard to this study. All other authors declare no competing interests.
References
- 1.Lozano R et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee BP, Vittinghoff E, Dodge JL, Cullaro G & Terrault NA National trends and long-term outcomes of liver transplant for alcohol-associated liver disease in the United States. JAMA Intern Med 179, 340–348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm J et al. Burden of disease associated with alcohol use disorders in the United States. Alcohol Clin Exp Res 38, 1068–1077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llopis M et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ike Y, Clewell DB, Segarra RA & Gilmore MS Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J Bacteriol 172, 155–163 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang W & van der Donk WA The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat Chem Biol 9, 157–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maddrey WC et al. Corticosteroid therapy of alcoholic hepatitis. Gastroenterology 75, 193–199 (1978). [PubMed] [Google Scholar]
- 8.Dominguez M et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol 103, 2747–2756 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Thursz MR et al. Prednisolone or pentoxifylline for alcoholic hepatitis. N Engl J Med 372, 1619–1628 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Mathurin P & Lucey MR Management of alcoholic hepatitis. J Hepatol 56 Suppl 1, S39–S45 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Llorente C et al. Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat Commun 8, 837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmore MS et al. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J Bacteriol 176, 7335–7344 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox CR, Coburn PS & Gilmore MS Enterococcal cytolysin: a novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr Protein Pept Sci 6, 77–84 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Van Tyne D, Martin MJ & Gilmore MS Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins (Basel) 5, 895–911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertola A, Mathews S, Ki SH, Wang H & Gao B Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat Protoc 8, 627–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogilvie LA & Jones BV The human gut virome: a multifaceted majority. Front Microbiol 6, 918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee A et al. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect Immun 87, pii: e00085–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poindexter JS Biological properties and classification of the Caulobacter group. Bacteriol Rev 28, 231–295 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin J et al. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep 6, 29681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcuk LM et al. Clinical studies of the use of bacteriophage in the treatment of cholera. Bull World Health Organ 45, 77–83 (1971). [PMC free article] [PubMed] [Google Scholar]
- 22.Sarker SA et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine 4, 124–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalmasso M, Hill C & Ross RP Exploiting gut bacteriophages for human health. Trends Microbiol 22, 399–405 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Ujmajuridze A et al. Adapted bacteriophages for treating urinary tract infections. Front Microbiol 9, 1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khawaldeh A et al. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J Med Microbiol 60, 1697–1700 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Schooley RT et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61, pii: e00954–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dedrick RM et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25, 730–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fish R et al. Compassionate use of bacteriophage therapy for foot ulcer treatment as an effective step for moving toward clinical trials. Methods Mol Biol 1693, 159–170 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Gorski A et al. Phages and immunomodulation. Future Microbiol 12, 905–914 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Brandl K et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol 69, 396–405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao B et al. Serum and fecal oxylipins in patients with alcohol-related liver disease. Dig Dis Sci 64, 1878–1892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang S et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology, doi: 10.1002/hep.30832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ball SA, Tennen H, Poling JC, Kranzler HR & Rounsaville BJ Personality, temperament, and character dimensions and the DSM-IV personality disorders in substance abusers. J Abnorm Psychol 106, 545–553 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Krieg L et al. Mutation of the gastric hydrogen-potassium ATPase alpha subunit causes iron-deficiency anemia in mice. Blood 118, 6418–6425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill JJ et al. The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13, 542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wick RR, Judd LM, Gorrie CL & Holt KE Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genom 3, e000132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wick RR, Judd LM, Gorrie CL & Holt KE Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker BJ et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santiago-Rodriguez TM et al. Transcriptome analysis of bacteriophage communities in periodontal health and disease. BMC Genomics 16, 549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haft DH et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46, D851–D860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatusova T et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44, 6614–6624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fouts DE Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res 34, 5839–5851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ondov BD et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paradis E & Schliep K Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valentine RC, Shapiro BM & Stadtman ER Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7, 2143–2152 (1968). [DOI] [PubMed] [Google Scholar]
- 47.Haas BJ et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21, 494–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108 Suppl 1, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen P et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology 148, 203–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryu H et al. Development of quantitative PCR assays targeting the 16S rRNA genes of Enterococcus spp. and their application to the identification of Enterococcus species in environmental samples. Appl Environ Microbiol 79, 196–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas W, Shepard BD & Gilmore MS Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415, 84–87 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Page AJ et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom 2, e000083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33, D325–D328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia B et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45, D566–D573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seemann T Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).24642063 [Google Scholar]
- 56.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang W, Bobeica SC, Wang L & van der Donk WA CylA is a sequence-specific protease involved in toxin biosynthesis. J Ind Microbiol Biotechnol 46, 537–549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwaisako K et al. Protection from liver fibrosis by a peroxisome proliferator-activated receptor delta agonist. Proc Natl Acad Sci U S A 109, E1369–E1376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke TH, Brinkac LM, Sutton G & Fouts DE GGRaSP: a R-package for selecting representative genomes using Gaussian mixture models. Bioinformatics 34, 3032–3034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S sequence reads can be found in the NCBI SRA associated with Bioproject PRJNA525701. Bacteriophage raw sequence reads and annotated genomes are available at NCBI under the following consecutive BioSample IDs (SAMN11089809 – SAMN11089827). Genome sequence data of E. faecalis strains isolated in this study were registered at ENA under Study PRJEB25007.