Abstract
The mammalian circadian clock has evolved as an adaptation to the 24-hour light/dark cycle on earth. Maintaining cellular activities in synchrony with the activities of the organism (such as eating and sleeping) helps different tissue and organ systems coordinate and optimize their performance. The full extent of the mechanisms by which cells maintain the clock are still under investigation, but involve a core set of clock genes that regulate large networks of gene transcription both by direct transcriptional activation/repression as well as the recruitment of proteins that modify chromatin states more broadly.
Keywords: mammalian, circadian clock, transcription, clock genes
Introduction
The ~24-hour rotation of the Earth has been a major evolutionary force on the development of intrinsic circadian clocks in most species (Pittendrigh, 1993). In plants that require light for energy production, it is obvious why there is a metabolic link to the day/night cycle (Greenham and McClung, 2015); however, animals have also adapted behavioral changes corresponding with light and temperature cycles to respond to and anticipate energetic demands (Bass and Takahashi, 2010). In addition to rhythmicity in sleep/activity cycles, in mammals there are also many other examples of ~24-hour physiological rhythms, including body temperature fluctuations (Buhr et al., 2010), circulating hormone levels (Lightman, 2016), and metabolism (Green et al., 2008).
An important (and defining) aspect of circadian rhythms is that they persist in the absence of external cues (Pittendrigh and Daan, 1976), yet external cues are important for synchronizing or entraining rhythms. It was originally thought that most circadian rhythms were entrained by light (Pittendrigh, 1960); however light is merely one of the many cues that can entrain the circadian clock. Moreover, while research on the mammalian circadian clock was long focused on the suprachiasmatic nucleus of the hypothalamus (SCN) as the central pacemaker (Welsh et al., 2010, Hastings et al., 2018), we now know that the circadian clock itself is actually an intrinsic property of cells in many different tissues (Yoo et al., 2004), with the SCN serving to synchronize “peripheral” clocks (Albrecht, 2012, Mohawk et al., 2012). The specific machinery underlying circadian clocks differ from organism to organism, but at the cellular level they depend on the transcription of sets of core clock genes, underscoring the evolutionary conservation of the core clock mechanism across species (Dunlap, 1999, Bell-Pedersen et al., 2005).
This review will describe what the field has learned about mammalian clock genes and the regulation of circadian gene transcription across tissues, with a focus on how these circadian genes influence metabolic pathways. While it is by no means exhaustive, it provides an overview of aspects of the circadian clock that the Takahashi lab continues investigating, and highlights unresolved questions that remain of great interest to the circadian field.
Cast of Characters: The Mammalian Circadian Clock
The first clock gene, period, was discovered through investigations of Drosophila mutants with abnormal behavioral cycles (Konopka and Benzer, 1971, Smith and Konopka, 1981, Reddy et al., 1984). These important studies laid the foundation for understanding the molecular basis of the clock, as the per gene was found to exhibit a circadian rhythm and the PER protein, itself, was found to regulate per gene expression (Hardin et al., 1990). Extending the studies in Drosophila, the first mammalian core clock gene, Clock, was discovered in a forward genetics screen for mice with abnormal circadian behavioral patterns (Vitaterna et al., 1994, King et al., 1997). The CLOCK protein In mice has features in common with Drosophila PER, including a PAS domain (for Per, ARNT, and Sim). However, CLOCK and its binding partner, BMAL1 (Gekakis et al., 1998), also have bHLH domains that allow them to bind DNA directly to regulatory elements (E-boxes) on rhythmic genes to influence their transcription.
The major targets of CLOCK/BMAL1 include other core clock genes that encode the mammalian Period ortholog (Per1, Per2, and Per3) (Shearman et al., 1997) and CRYPTOCHROME (Cry1 and Cry2) (Kume et al., 1999) repressor proteins. These negative regulators heterodimerize then translocate into the nucleus where they repress their own gene transcription by interacting directly with CLOCK/BMAL1 (Michael et al., 2017, Rosensweig et al., 2018). In addition to this direct transcriptional feedback, the mRNA expression of Per1/2/3 and Cry1/2 is also regulated by various mechanisms (Kojima et al., 2011, Lim and Allada, 2013). The degradation of PER and CRY proteins is also regulated by the serine/threonine kinases, casein kinase δ (CK1δ) and CK1ϵ (Gallego and Virshup, 2007, Narasimamurthy et al., 2018), the F-box proteins, FBXL3 and FBXL21 (Hirano et al., 2013, Yoo et al., 2013), and other proteins (Reischl et al., 2007). Once negative transcriptional feedback and post-transcriptional and post-translational regulation of PER and CRY is sufficient to decrease PER/CRY protein levels in the nucleus, repression is relieved and CLOCK/BMAL1 start a new cycle of Per/Cry gene transcription (Takahashi, 2017).
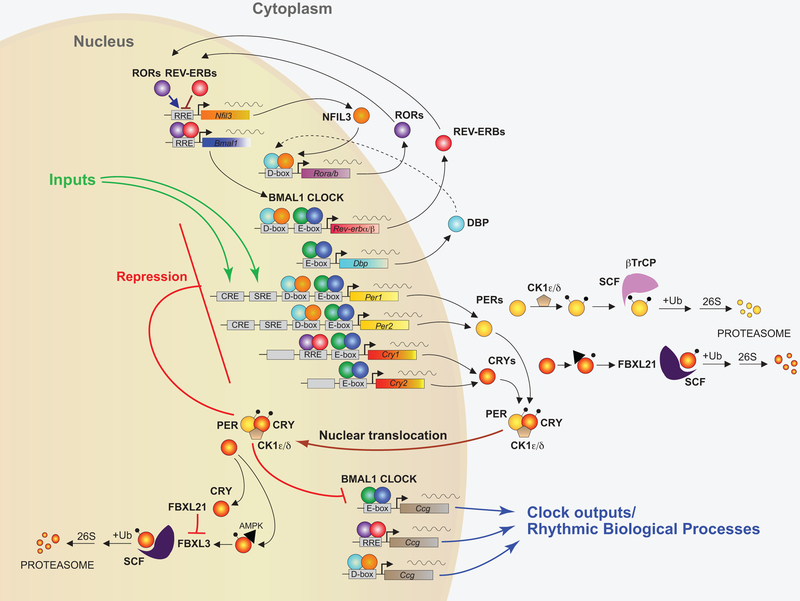
Since the initial discovery of these core mammalian clock genes, several additional genes and feedback loops have been uncovered, increasing the complexity of the mammalian circadian clock gene network (FIGURE 1). In the second major transcriptional loop, CLOCK/BMAL1 activate transcription of genes for the nuclear receptors REV-ERBα and REV-ERBβ (Preitner et al., 2002). These proteins compete with the retinoic acid-related orphan receptors, RORα, RORβ, and RORγ for binding sites (ROR-binding elements) on the BMAL1 gene, providing both positive (ROR) and negative (REV-ERB) regulation of transcription (Sato et al., 2004), and, as will be discussed later, they make an important link between the circadian clock and metabolism (Zhang et al., 2015). A third feedback loop involves the D-box binding protein (DBP) and the nuclear factor, interleukin-3 regulated protein (NFIL3, also known as E4BP4) which are regulated by CLOCK/BMAL1 (Ripperger and Schibler, 2006) and CRY1 (Stratmann et al., 2010), and bind to D-box elements on circadian promoters, including RORα and RORβ (Ueda et al., 2005). Together, these feedback loops that make up the “molecular clock” are governed by transcriptional (Takahashi, 2017), post-transcriptional (Kojima and Green, 2015), and post-translational (Gallego and Virshup, 2007) regulatory mechanisms that are sufficient to maintain circadian rhythms; however, external cues are still important for synchronizing rhythms of cells within and across tissues (Golombek and Rosenstein, 2010).
Figure 1: Core components of the mammalian circadian clock.
In the core feedback loop, the transcription factors BMAL1 (green circles) and CLOCK (blue circles) bind to E-box domains on gene promoters, including the genes for Per1 and Per2 (yellow) and Cry1 and Cry2 (red/yellow). PERs (yellow circles) and CRYs (red/yellow circles) dimerize and translocate to the nucleus after binding with casein kinase δ (CK1δ) or CK1ϵ, where they repress their own transcription. The stability of PER and CRY is regulated both in the cytoplasm and within the nucleus by several proteins, including FBXL21 and FBXL3. In a second feedback loop, CLOCK and BMAL1 also regulate the transcription of genes for the nuclear receptors REV-ERBα and REV-ERBβ (red circles), which compete with the retinoic acid-related orphan receptors, RORα, RORβ, and RORγ (purple circles) for binding to RRE elements on the BMAL1 gene promoter, providing both positive (ROR) and negative (REV-ERB) regulation of BMAL1 transcription. A third feedback loop is mediated by CLOCK/BMAL1-mediated transcription of the gene Dbp (light blue) and the ROR/REV-ERB-mediated transcription of Nfil3 (orange). DBP (light blue circles) and NFIL3 (orange circles) dimerize and bind to D-box elements on the promoters of many of the core clock genes, providing additional layers of regulation. In addition, CLOCK/BMAL1, ROR/REV-ERB, and DBP/NFIL3 regulate the transcription of many other clock output genes (Figure modified from (Takahashi, 2017)).
Circadian Rhythms Throughout the Body
Early studies of mammalian circadian rhythms suggested that the brain responds to light cues to regulate sleep/wake cycles and daily behavioral and neuroendocrine rhythms. The discovery of axons projecting from the retina to the suprachiasmatic nucleus of the hypothalamus (SCN) (Moore and Lenn, 1972) gave insights into the central pathway mediating these effects, and subsequent lesioning and transplantation studies established the SCN as a master regulator of circadian rhythms (Moore and Eichler, 1972, Stephan and Zucker, 1972, Ralph et al., 1990). It wasn’t until several years later that a specialized population of retinal ganglion cells (intrinsically photosensitive “ipRGCs”) containing the photopigment, melanopsin, were discovered (Provencio et al., 2000, Berson et al., 2002, Hattar et al., 2002). The light information transmitted by ipRGCs via the retinohypothalamic tract is sufficient to set the phase of the SCN (Guler et al., 2008), which responds by generating circadian patterns of action potentials (Hastings et al., 2018).
The SCN consists of a heterogeneous cluster of approximately 10,000 neurons in the ventral hypothalamus (Welsh et al., 2010). There are two main subdivisions of SCN neurons, defined based on their expression of the neuropeptides arginine vasopressin (AVP) and vasoactive intestinal peptide (VIP). However, almost all SCN neurons express the inhibitory neurotransmitter GABA (Okamura et al., 1989), and there are several other neuropeptides that are expressed across the SCN, increasing the complexity of these subregions (Abrahamson and Moore, 2001, Hastings et al., 2018). The VIP-expressing neurons in the ventrolateral “core” of the SCN receive synaptic inputs from ipRGCs, and the response of these neurons is thought to be important for maintaining synchrony within the SCN. However, there is recent evidence to suggest that astrocytes within the SCN are also important for maintaining synchrony (Brancaccio et al., 2017, Brancaccio et al., 2019), so the story is likely not so simple.
The response of SCN neurons to incoming light signals is well-characterized and involves the activation of NMDA receptors on SCN neurons, calcium activation of CAMKII, and the activation of gene transcription (Golombek and Rosenstein, 2010). Of the core clock components, the Per1 and Per2 genes are particularly responsive to photic entrainment and have been used as a molecular marker for circadian oscillations (Yamazaki et al., 2000, Yoo et al., 2004). In addition, the expression of many other genes in the SCN, including immediate early genes are induced by light (Porterfield et al., 2007).
Aside from the SCN, the circadian clock is also intrinsic to cells in many other tissues (Mohawk et al., 2012). This has been shown in isolated cells (Balsalobre et al., 1998, Yagita et al., 2001, Welsh et al., 2004), in rodent models generated to visualize PER gene expression rhythms from various tissues (Yamazaki et al., 2000, Yoo et al., 2004) and in studies of rhythmic gene expression across different tissues (Panda et al., 2002, Storch et al., 2002, Zhang et al., 2014). As will be discussed in more detail later, these other tissues respond to the signals coordinated by the SCN, but can also entrain to signals other than light (Damiola et al., 2000, Stokkan et al., 2001, Vollmers et al., 2009). Therefore, although the SCN serves to synchronize these “peripheral” clocks, SCN inputs are not required for maintaining circadian timing in these other tissues (Albrecht, 2012, Mohawk et al., 2012, Hastings et al., 2018).
Mechanisms of Circadian Transcriptional Regulation
The main output of the core circadian clock includes genes regulated by CLOCK/BMAL1 (Takahashi, 2017), REVERBs/RORs (Ueda et al., 2002), and DBP (Ripperger and Schibler, 2006), but there is also evidence that PER1 and CRY1/2 regulate expression of genes outside of the core regulatory feedback loop (Lamia et al., 2011). Within the past decade, there has been great interest in understanding how circadian transcription factors drive the rhythmic expression of a variety of genes across different tissues (Koike et al., 2012, Menet et al., 2012). CLOCK and BMAL1 form a much larger complex with histone modifying enzymes (Katada and Sassone-Corsi, 2010) and transcriptional coactivators such as Sirtuin 1 and CBP/p300 (Nakahata et al., 2008, Lee et al., 2010) to open chromatin and promote gene expression (Menet et al., 2014), along with the rhythmic recruitment of RNA Polymerase (Takahashi, 2017).
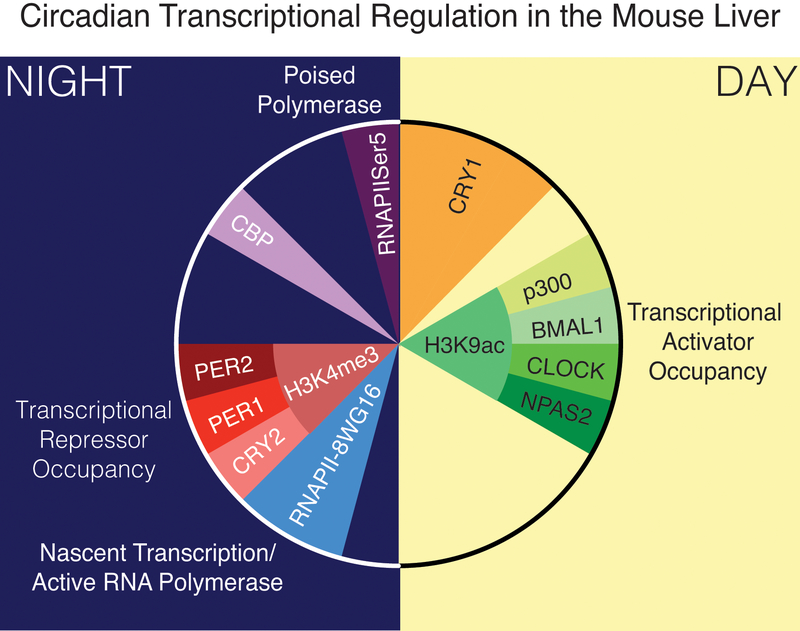
Taking a single gene (Dbp) as an example, one can observe the core clock transcription factors binding to the gene promoter at varying times throughout the day, as well as cyclic changes in chromatin state (Ripperger and Schibler, 2006, Koike et al., 2012). When one starts to look genome-wide, it becomes apparent that there are large-scale changes in transcription and chromosomal organization mediated by the circadian clock (Figure 2) (Koike et al., 2012, Le Martelot et al., 2012, Menet et al., 2012, Vollmers et al., 2012, Takahashi, 2017). While some of these chromosomal changes are associated with promoters or enhancers, recent studies have shown that long-range chromatin interactions also show rhythmic changes (Xu et al., 2016, Kim et al., 2018, Mermet et al., 2018, Yeung and Naef, 2018, Pacheco-Bernal et al., 2019). The field is still just beginning to understand what factors mediate these large changes in topology, but this is an exciting area in circadian research, particularly since chromatin interactions may underlie tissue-specific regulation of gene expression (Abruzzi et al., 2011, Yeung et al., 2018). Moreover, although most of the genome-wide studies have been performed using liver tissues, the latest evidence suggests that BMAL1 binding is highly variable across tissues and depends upon other tissue-specific transcription factors (Perelis et al., 2015, Beytebiere et al., 2019). Since some of the core clock transcription factors are also differentially expressed in specific tissues, it is likely that there are even more complex interactions of the circadian clock with tissue-specific factors.
Figure 2: 24-hour depiction of genome-wide circadian transcriptional regulation in the mouse liver.
Peak occupancy of transcriptional activators at gene promoters occurs in the middle of the day and corresponds with a peak in H3K9acetylation. Peak transcription occurs shortly after nightfall, as indicated by activated RNA polymerase binding. Transcriptional repressor occupancy peaks shortly thereafter, and corresponds with a peak in Hk34 tri-methylation. Additional transcription factors and co-factors, such as CRY1 and CBP appear to occupy promoters at different times, and poised RNA polymerase occupancy peaks just at the end of the 24-hour cycle (Based on data from (Koike et al., 2012) and figure from (Takahashi, 2017)).
Circadian Control of Metabolism
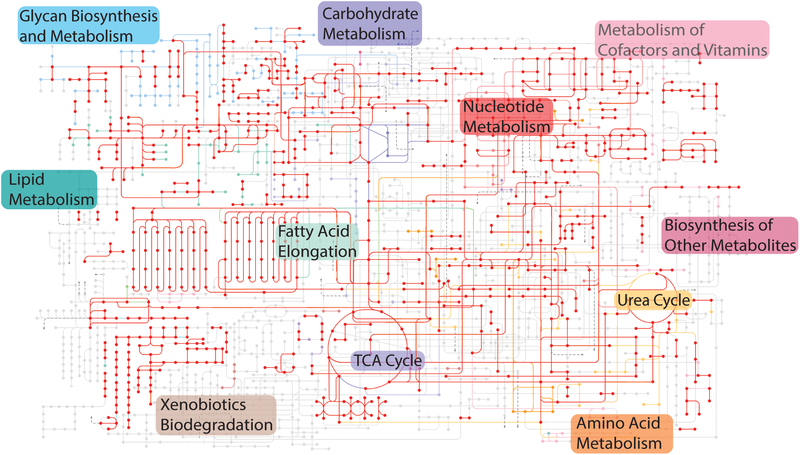
As major outputs of the circadian clock, CLOCK/BMAL1 regulate the transcription of thousands of genes (Koike et al., 2012, Menet et al., 2014, Beytebiere et al., 2019). In the liver there are about 3000 genes that display circadian rhythms in BMAL1 occupancy (Koike et al., 2012), and many of these genes are involved in regulating cellular metabolic pathways (Figure 3). CLOCK/BMAL1 regulation of whole-body metabolism has been noted previously, both in genomics studies and also in Clock and Bmal1 mutant mice, which display obesity and features of metabolic syndrome, such as altered glucose homeostasis, as well as disrupted skeletal muscle metabolism (Turek et al., 2005, Marcheva et al., 2010, Perelis et al., 2015, Harfmann et al., 2016). In addition, the transcriptional targets of REV-ERBs also connect the core clock machinery with metabolic pathways by regulating glucose and fatty acid metabolism (Cho et al., 2012, Delezie et al., 2012, Zhang et al., 2015), and Cry1 expression is associated with a reduction in gluconeogenesis in the liver (Zhang et al., 2010). Post-transcriptional regulation of core clock components also influences circadian gene expression in the liver (Wang et al., 2018), and many recent studies have shown that large-scale changes in the circadian epigenome (Sun et al., 2011, Vollmers et al., 2012, Masri et al., 2013) accompany the changes in metabolic gene expression. There are also several examples of genes involved in metabolic pathways that directly impact the expression of core clock genes (Asher et al., 2008, Lamia et al., 2009, Eckel-Mahan et al., 2013, Furlan et al., 2019). Thus, the core circadian clock is intimately linked to metabolism at the molecular level (Bass and Lazar, 2016, Challet, 2019).
Figure 3: BMAL1 regulation of metabolism.
Overlay of BMAL1 target genes (indicated in red) on diverse metabolic pathways in the liver. BMAL1 occupancy data are from a previously published ChIP-seq dataset (Koike et al., 2012). The original metabolic pathway is from a KEGG analysis (used with permission) and has been simplified to show major nodes (Kanehisa and Goto, 2000, Kanehisa et al., 2017). In KEGG, nodes indicate enzymes and lines indicate connections in metabolic pathways, with colors indicating pathways serving similar functions. The red dots and lines indicate BMAL1 interactions with genes involved in these pathways.
However, links between the circadian clock and metabolism were first made in studies showing that the liver circadian clock could be entrained by feeding time independently from the central SCN clock (Damiola et al., 2000, Stokkan et al., 2001). Since that time, many different feeding paradigms have shown that circadian gene expression in peripheral tissues is altered when the type or timing of food intake is manipulated (Vollmers et al., 2009, Eckel-Mahan et al., 2013, Mukherji et al., 2015). The circadian clock within several peripheral tissues, including the liver, skeletal muscle, and pancreas, is sensitive to hormonal signals and glucose levels (Saini et al., 2013, Dyar et al., 2014, Perelis et al., 2015, Schibler et al., 2015, Harfmann et al., 2016, Ikeda et al., 2018, Crosby et al., 2019), providing mechanisms through which food entrainment could occur. Interestingly, recent evidence suggests that, at least in the liver, the cell-autonomous circadian clock also depends on light synchronization and that food intake per se has large effects on transcription independent of effects on circadian gene expression (Atger et al., 2015, Greenwell et al., 2019, Koronowski et al., 2019). Thus, the simplistic view of liver clock being entrained by food, while the SCN is entrained by light, does not give sufficient credit to the complexity of the peripheral clock (Albrecht, 2012, Izumo et al., 2014).
What is clear, is that synchronization of central and peripheral clocks is important for overall health (Di Francesco et al., 2018, Dyar et al., 2018, Challet, 2019), and one of the most salient factors for this circadian synchronization appears to be the timing of food intake (Barclay et al., 2012, Hatori et al., 2012, Chaix et al., 2019). Notably, while caloric restriction paradigms have been successful in improving overall health and extending lifespan (Weindruch et al., 1986), these paradigms inadvertently impose temporal restriction of food intake. This was shown recently by my laboratory in experiments using automatic feeder cages that allowed us to regulate food intake as well as record activity of hundreds of mice simultaneously. We found that, under caloric restriction, mice consolidate their feeding to a 2-hour time interval, thus, self-imposing a time-restricted feeding pattern (Acosta-Rodriguez et al., 2017). These findings strongly suggest that for optimal metabolic performance, the timing of food intake must align with other circadian rhythms (i.e. activity, hormone secretion, temperature fluctuations).
Conclusions/Perspectives
While this review has only been able to touch on some of the highlights of work on circadian transcriptional regulation, the data show that the circadian regulation of gene expression is pervasive and extends far beyond CLOCK/BMAL1 occupancy on gene promoters, including RNA polymerase recruitment, the modulation of chromatin states, chromatin architecture, and nuclear localization. In addition, clock genes and the pathways they regulate are undoubtedly embedded in metabolic pathways as shown both by the effects of the timing of food intake as well as the intrinsic links to metabolic gene networks. Thus, multi-faceted levels of regulation of circadian gene transcription allow the organism to anticipate metabolic demands and optimize energy utilization by consolidating gene expression to certain times of day. Interestingly, emerging results from my laboratory suggest that the circadian regulation of the timing of metabolic events may be critical for maintaining health and extending lifespan. Using circadian gene transcription as a window into the overall synchrony of an organism, we hope to continue to learn about additional factors involved in the circadian regulation of transcription, which will no doubt give us perspective on the underlying basis for many human diseases.
DECLARATION OF INTERESTS AND FUNDING
The authors have no conflicts of interests to declare. JST is supported by the NIH (R01 NS106657) and is an Investigator in the Howard Hughes Medical Institute.
REFERENCES
- ABRAHAMSON EE & MOORE RY 2001. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res, 916, 172–91. [DOI] [PubMed] [Google Scholar]
- ABRUZZI KC, RODRIGUEZ J, MENET JS, DESROCHERS J, ZADINA A, LUO W, TKACHEV S & ROSBASH M 2011. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev, 25, 2374–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACOSTA-RODRIGUEZ VA, DE GROOT MHM, RIJO-FERREIRA F, GREEN CB & TAKAHASHI JS 2017. Mice under Caloric Restriction Self-Impose a Temporal Restriction of Food Intake as Revealed by an Automated Feeder System. Cell Metab, 26, 267–277 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALBRECHT U 2012. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron, 74, 246–60. [DOI] [PubMed] [Google Scholar]
- ASHER G, GATFIELD D, STRATMANN M, REINKE H, DIBNER C, KREPPEL F, MOSTOSLAVSKY R, ALT FW & SCHIBLER U 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell, 134, 317–28. [DOI] [PubMed] [Google Scholar]
- ATGER F, GOBET C, MARQUIS J, MARTIN E, WANG J, WEGER B, LEFEBVRE G, DESCOMBES P, NAEF F & GACHON F 2015. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci U S A, 112, E6579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALSALOBRE A, DAMIOLA F & SCHIBLER U 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell, 93, 929–37. [DOI] [PubMed] [Google Scholar]
- BARCLAY JL, HUSSE J, BODE B, NAUJOKAT N, MEYER-KOVAC J, SCHMID SM, LEHNERT H & OSTER H 2012. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One, 7, e37150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BASS J & LAZAR MA 2016. Circadian time signatures of fitness and disease. Science, 354, 994–999. [DOI] [PubMed] [Google Scholar]
- BASS J & TAKAHASHI JS 2010. Circadian integration of metabolism and energetics. Science, 330, 1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELL-PEDERSEN D, CASSONE VM, EARNEST DJ, GOLDEN SS, HARDIN PE, THOMAS TL & ZORAN MJ 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet, 6, 544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERSON DM, DUNN FA & TAKAO M 2002. Phototransduction by retinal ganglion cells that set the circadian clock. Science, 295, 1070–3. [DOI] [PubMed] [Google Scholar]
- BEYTEBIERE JR, TROTT AJ, GREENWELL BJ, OSBORNE CA, VITET H, SPENCE J, YOO SH, CHEN Z, TAKAHASHI JS, GHAFFARI N, et al. 2019. Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev, 33, 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANCACCIO M, EDWARDS MD, PATTON AP, SMYLLIE NJ, CHESHAM JE, MAYWOOD ES & HASTINGS MH 2019. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science, 363, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANCACCIO M, PATTON AP, CHESHAM JE, MAYWOOD ES & HASTINGS MH 2017. Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron, 93, 1420–1435 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUHR ED, YOO SH & TAKAHASHI JS 2010. Temperature as a universal resetting cue for mammalian circadian oscillators. Science, 330, 379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAIX A, LIN T, LE HD, CHANG MW & PANDA S 2019. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab, 29, 303–319 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHALLET E 2019. The circadian regulation of food intake. Nat Rev Endocrinol, 15, 393–405. [DOI] [PubMed] [Google Scholar]
- CHO H, ZHAO X, HATORI M, YU RT, BARISH GD, LAM MT, CHONG LW, DITACCHIO L, ATKINS AR, GLASS CK, et al. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature, 485, 123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROSBY P, HAMNETT R, PUTKER M, HOYLE NP, REED M, KARAM CJ, MAYWOOD ES, STANGHERLIN A, CHESHAM JE, HAYTER EA, et al. 2019. Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell, 177, 896–909 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAMIOLA F, LE MINH N, PREITNER N, KORNMANN B, FLEURY-OLELA F & SCHIBLER U 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev, 14, 2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DELEZIE J, DUMONT S, DARDENTE H, OUDART H, GRECHEZ-CASSIAU A, KLOSEN P, TEBOUL M, DELAUNAY F, PEVET P & CHALLET E 2012. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. FASEB J, 26, 3321–35. [DOI] [PubMed] [Google Scholar]
- DI FRANCESCO A, DI GERMANIO C, BERNIER M & DE CABO R 2018. A time to fast. Science, 362, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUNLAP JC 1999. Molecular bases for circadian clocks. Cell, 96, 271–90. [DOI] [PubMed] [Google Scholar]
- DYAR KA, CICILIOT S, WRIGHT LE, BIENSO RS, TAGLIAZUCCHI GM, PATEL VR, FORCATO M, PAZ MI, GUDIKSEN A, SOLAGNA F, et al. 2014. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab, 3, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DYAR KA, LUTTER D, ARTATI A, CEGLIA NJ, LIU Y, ARMENTA D, JASTROCH M, SCHNEIDER S, DE MATEO S, CERVANTES M, et al. 2018. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell, 174, 1571–1585 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECKEL-MAHAN KL, PATEL VR, DE MATEO S, OROZCO-SOLIS R, CEGLIA NJ, SAHAR S, DILAG-PENILLA SA, DYAR KA, BALDI P & SASSONE-CORSI P 2013. Reprogramming of the circadian clock by nutritional challenge. Cell, 155, 1464–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FURLAN A, JACQUIER M, WOLLER A, HELIOT L, DUEZ H, STAELS B & LEFRANC M 2019. Mathematical models converge on PGC1alpha as the key metabolic integrator of SIRT1 and AMPK regulation of the circadian clock. Proc Natl Acad Sci U S A, 116, 13171–13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALLEGO M & VIRSHUP DM 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol, 8, 139–48. [DOI] [PubMed] [Google Scholar]
- GEKAKIS N, STAKNIS D, NGUYEN HB, DAVIS FC, WILSBACHER LD, KING DP, TAKAHASHI JS & WEITZ CJ 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science, 280, 1564–9. [DOI] [PubMed] [Google Scholar]
- GOLOMBEK DA & ROSENSTEIN RE 2010. Physiology of circadian entrainment. Physiol Rev, 90, 1063–102. [DOI] [PubMed] [Google Scholar]
- GREEN CB, TAKAHASHI JS & BASS J 2008. The meter of metabolism. Cell, 134, 728–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREENHAM K & MCCLUNG CR 2015. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet, 16, 598–610. [DOI] [PubMed] [Google Scholar]
- GREENWELL BJ, TROTT AJ, BEYTEBIERE JR, PAO S, BOSLEY A, BEACH E, FINEGAN P, HERNANDEZ C & MENET JS 2019. Rhythmic Food Intake Drives Rhythmic Gene Expression More Potently than the Hepatic Circadian Clock in Mice. Cell Rep, 27, 649–657 e5. [DOI] [PubMed] [Google Scholar]
- GULER AD, ECKER JL, LALL GS, HAQ S, ALTIMUS CM, LIAO HW, BARNARD AR, CAHILL H, BADEA TC, ZHAO H, et al. 2008. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature, 453, 102–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARDIN PE, HALL JC & ROSBASH M 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature, 343, 536–40. [DOI] [PubMed] [Google Scholar]
- HARFMANN BD, SCHRODER EA, KACHMAN MT, HODGE BA, ZHANG X & ESSER KA 2016. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle, 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HASTINGS MH, MAYWOOD ES & BRANCACCIO M 2018. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci, 19, 453–469. [DOI] [PubMed] [Google Scholar]
- HATORI M, VOLLMERS C, ZARRINPAR A, DITACCHIO L, BUSHONG EA, GILL S, LEBLANC M, CHAIX A, JOENS M, FITZPATRICK JA, et al. 2012. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab, 15, 848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HATTAR S, LIAO HW, TAKAO M, BERSON DM & YAU KW 2002. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science, 295, 1065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRANO A, YUMIMOTO K, TSUNEMATSU R, MATSUMOTO M, OYAMA M, KOZUKA-HATA H, NAKAGAWA T, LANJAKORNSIRIPAN D, NAKAYAMA KI & FUKADA Y 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell, 152, 1106–18. [DOI] [PubMed] [Google Scholar]
- IKEDA Y, KAMAGATA M, HIRAO M, YASUDA S, IWAMI S, SASAKI H, TSUBOSAKA M, HATTORI Y, TODOH A, TAMURA K, et al. 2018. Glucagon and/or IGF-1 Production Regulates Resetting of the Liver Circadian Clock in Response to a Protein or Amino Acid-only Diet. EBioMedicine, 28, 210–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IZUMO M, PEJCHAL M, SCHOOK AC, LANGE RP, WALISSER JA, SATO TR, WANG X, BRADFIELD CA & TAKAHASHI JS 2014. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. Elife, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANEHISA M, FURUMICHI M, TANABE M, SATO Y & MORISHIMA K 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res, 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANEHISA M & GOTO S 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATADA S & SASSONE-CORSI P 2010. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol, 17, 1414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM YH, MARHON SA, ZHANG Y, STEGER DJ, WON KJ & LAZAR MA 2018. Rev-erbalpha dynamically modulates chromatin looping to control circadian gene transcription. Science, 359, 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING DP, ZHAO Y, SANGORAM AM, WILSBACHER LD, TANAKA M, ANTOCH MP, STEEVES TD, VITATERNA MH, KORNHAUSER JM, LOWREY PL, et al. 1997. Positional cloning of the mouse circadian clock gene. Cell, 89, 641–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOIKE N, YOO SH, HUANG HC, KUMAR V, LEE C, KIM TK & TAKAHASHI JS 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science, 338, 349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOJIMA S & GREEN CB 2015. Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry, 54, 124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOJIMA S, SHINGLE DL & GREEN CB 2011. Post-transcriptional control of circadian rhythms. J Cell Sci, 124, 311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONOPKA RJ & BENZER S 1971. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A, 68, 2112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KORONOWSKI KB, KINOUCHI K, WELZ PS, SMITH JG, ZINNA VM, SHI J, SAMAD M, CHEN S, MAGNAN CN, KINCHEN JM, et al. 2019. Defining the Independence of the Liver Circadian Clock. Cell, 177, 1448–1462 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUME K, ZYLKA MJ, SRIRAM S, SHEARMAN LP, WEAVER DR, JIN X, MAYWOOD ES, HASTINGS MH & REPPERT SM 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- LAMIA KA, PAPP SJ, YU RT, BARISH GD, UHLENHAUT NH, JONKER JW, DOWNES M & EVANS RM 2011. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature, 480, 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMIA KA, SACHDEVA UM, DITACCHIO L, WILLIAMS EC, ALVAREZ JG, EGAN DF, VASQUEZ DS, JUGUILON H, PANDA S, SHAW RJ, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science, 326, 437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LE MARTELOT G, CANELLA D, SYMUL L, MIGLIAVACCA E, GILARDI F, LIECHTI R, MARTIN O, HARSHMAN K, DELORENZI M, DESVERGNE B, et al. 2012. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol, 10, e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE Y, LEE J, KWON I, NAKAJIMA Y, OHMIYA Y, SON GH, LEE KH & KIM K 2010. Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J Cell Sci, 123, 3547–57. [DOI] [PubMed] [Google Scholar]
- LIGHTMAN S 2016. Rhythms Within Rhythms: The Importance of Oscillations for Glucocorticoid Hormones In: SASSONE-CORSI P & CHRISTEN Y (eds.) A Time for Metabolism and Hormones. Cham (CH). [PubMed] [Google Scholar]
- LIM C & ALLADA R 2013. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci, 16, 1544–50. [DOI] [PubMed] [Google Scholar]
- MARCHEVA B, RAMSEY KM, BUHR ED, KOBAYASHI Y, SU H, KO CH, IVANOVA G, OMURA C, MO S, VITATERNA MH, et al. 2010. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature, 466, 627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASRI S, PATEL VR, ECKEL-MAHAN KL, PELEG S, FORNE I, LADURNER AG, BALDI P, IMHOF A & SASSONE-CORSI P 2013. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci U S A, 110, 3339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENET JS, PESCATORE S & ROSBASH M 2014. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev, 28, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENET JS, RODRIGUEZ J, ABRUZZI KC & ROSBASH M 2012. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife, 1, e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MERMET J, YEUNG J, HURNI C, MAUVOISIN D, GUSTAFSON K, JOUFFE C, NICOLAS D, EMMENEGGER Y, GOBET C, FRANKEN P, et al. 2018. Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev, 32, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHAEL AK, FRIBOURGH JL, CHELLIAH Y, SANDATE CR, HURA GL, SCHNEIDMAN-DUHOVNY D, TRIPATHI SM, TAKAHASHI JS & PARTCH CL 2017. Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1. Proc Natl Acad Sci U S A, 114, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOHAWK JA, GREEN CB & TAKAHASHI JS 2012. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci, 35, 445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOORE RY & EICHLER VB 1972. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res, 42, 201–6. [DOI] [PubMed] [Google Scholar]
- MOORE RY & LENN NJ 1972. A retinohypothalamic projection in the rat. J Comp Neurol, 146, 1–14. [DOI] [PubMed] [Google Scholar]
- MUKHERJI A, KOBIITA A, DAMARA M, MISRA N, MEZIANE H, CHAMPY MF & CHAMBON P 2015. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc Natl Acad Sci U S A, 112, E6691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKAHATA Y, KALUZOVA M, GRIMALDI B, SAHAR S, HIRAYAMA J, CHEN D, GUARENTE LP & SASSONE-CORSI P 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell, 134, 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NARASIMAMURTHY R, HUNT SR, LU Y, FUSTIN JM, OKAMURA H, PARTCH CL, FORGER DB, KIM JK & VIRSHUP DM 2018. CK1delta/epsilon protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci U S A, 115, 5986–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKAMURA H, BEROD A, JULIEN JF, GEFFARD M, KITAHAMA K, MALLET J & BOBILLIER P 1989. Demonstration of GABAergic cell bodies in the suprachiasmatic nucleus: in situ hybridization of glutamic acid decarboxylase (GAD) mRNA and immunocytochemistry of GAD and GABA. Neurosci Lett, 102, 131–6. [DOI] [PubMed] [Google Scholar]
- PACHECO-BERNAL I, BECERRIL-PEREZ F & AGUILAR-ARNAL L 2019. Circadian rhythms in the three-dimensional genome: implications of chromatin interactions for cyclic transcription. Clin Epigenetics, 11, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PANDA S, ANTOCH MP, MILLER BH, SU AI, SCHOOK AB, STRAUME M, SCHULTZ PG, KAY SA, TAKAHASHI JS & HOGENESCH JB 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell, 109, 307–20. [DOI] [PubMed] [Google Scholar]
- PERELIS M, MARCHEVA B, RAMSEY KM, SCHIPMA MJ, HUTCHISON AL, TAGUCHI A, PEEK CB, HONG H, HUANG W, OMURA C, et al. 2015. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science, 350, aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PITTENDRIGH CS 1960. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol, 25, 159–84. [DOI] [PubMed] [Google Scholar]
- PITTENDRIGH CS 1993. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol, 55, 16–54. [DOI] [PubMed] [Google Scholar]
- PITTENDRIGH CS & DAAN S 1976. A Functional Analysis of Circadian Pacemakers in Nocturnal Rodents. J Comp Physiol, 106, 223–252. [Google Scholar]
- PORTERFIELD VM, PIONTKIVSKA H & MINTZ EM 2007. Identification of novel light-induced genes in the suprachiasmatic nucleus. BMC Neurosci, 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PREITNER N, DAMIOLA F, LOPEZ-MOLINA L, ZAKANY J, DUBOULE D, ALBRECHT U & SCHIBLER U 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell, 110, 251–60. [DOI] [PubMed] [Google Scholar]
- PROVENCIO I, RODRIGUEZ IR, JIANG G, HAYES WP, MOREIRA EF & ROLLAG MD 2000. A novel human opsin in the inner retina. J Neurosci, 20, 600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RALPH MR, FOSTER RG, DAVIS FC & MENAKER M 1990. Transplanted suprachiasmatic nucleus determines circadian period. Science, 247, 975–8. [DOI] [PubMed] [Google Scholar]
- REDDY P, ZEHRING WA, WHEELER DA, PIRROTTA V, HADFIELD C, HALL JC & ROSBASH M 1984. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell, 38, 701–10. [DOI] [PubMed] [Google Scholar]
- REISCHL S, VANSELOW K, WESTERMARK PO, THIERFELDER N, MAIER B, HERZEL H & KRAMER A 2007. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms, 22, 375–86. [DOI] [PubMed] [Google Scholar]
- RIPPERGER JA & SCHIBLER U 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet, 38, 369–74. [DOI] [PubMed] [Google Scholar]
- ROSENSWEIG C, REYNOLDS KA, GAO P, LAOTHAMATAS I, SHAN Y, RANGANATHAN R, TAKAHASHI JS & GREEN CB 2018. An evolutionary hotspot defines functional differences between CRYPTOCHROMES. Nat Commun, 9, 1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAINI C, LIANI A, CURIE T, GOS P, KREPPEL F, EMMENEGGER Y, BONACINA L, WOLF JP, POGET YA, FRANKEN P, et al. 2013. Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev, 27, 1526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATO TK, PANDA S, MIRAGLIA LJ, REYES TM, RUDIC RD, MCNAMARA P, NAIK KA, FITZGERALD GA, KAY SA & HOGENESCH JB 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron, 43, 527–37. [DOI] [PubMed] [Google Scholar]
- SCHIBLER U, GOTIC I, SAINI C, GOS P, CURIE T, EMMENEGGER Y, SINTUREL F, GOSSELIN P, GERBER A, FLEURY-OLELA F, et al. 2015. Clock-Talk: Interactions between Central and Peripheral Circadian Oscillators in Mammals. Cold Spring Harb Symp Quant Biol, 80, 223–32. [DOI] [PubMed] [Google Scholar]
- SHEARMAN LP, ZYLKA MJ, WEAVER DR, KOLAKOWSKI LF JR. & REPPERT SM 1997. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron, 19, 1261–9. [DOI] [PubMed] [Google Scholar]
- SMITH RF & KONOPKA RJ 1981. Circadian clock phenotypes of chromosome aberrations with a breakpoint at the per locus. Mol Gen Genet, 183, 243–51. [DOI] [PubMed] [Google Scholar]
- STEPHAN FK & ZUCKER I 1972. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A, 69, 1583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOKKAN KA, YAMAZAKI S, TEI H, SAKAKI Y & MENAKER M 2001. Entrainment of the circadian clock in the liver by feeding. Science, 291, 490–3. [DOI] [PubMed] [Google Scholar]
- STORCH KF, LIPAN O, LEYKIN I, VISWANATHAN N, DAVIS FC, WONG WH & WEITZ CJ 2002. Extensive and divergent circadian gene expression in liver and heart. Nature, 417, 78–83. [DOI] [PubMed] [Google Scholar]
- STRATMANN M, STADLER F, TAMANINI F, VAN DER HORST GT & RIPPERGER JA 2010. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev, 24, 1317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN Z, FENG D, EVERETT LJ, BUGGE A & LAZAR MA 2011. Circadian epigenomic remodeling and hepatic lipogenesis: lessons from HDAC3. Cold Spring Harb Symp Quant Biol, 76, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAHASHI JS 2017. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet, 18, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUREK FW, JOSHU C, KOHSAKA A, LIN E, IVANOVA G, MCDEARMON E, LAPOSKY A, LOSEE-OLSON S, EASTON A, JENSEN DR, et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science, 308, 1043–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UEDA HR, CHEN W, ADACHI A, WAKAMATSU H, HAYASHI S, TAKASUGI T, NAGANO M, NAKAHAMA K, SUZUKI Y, SUGANO S, et al. 2002. A transcription factor response element for gene expression during circadian night. Nature, 418, 534–9. [DOI] [PubMed] [Google Scholar]
- UEDA HR, HAYASHI S, CHEN W, SANO M, MACHIDA M, SHIGEYOSHI Y, IINO M & HASHIMOTO S 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet, 37, 187–92. [DOI] [PubMed] [Google Scholar]
- VITATERNA MH, KING DP, CHANG AM, KORNHAUSER JM, LOWREY PL, MCDONALD JD, DOVE WF, PINTO LH, TUREK FW & TAKAHASHI JS 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science, 264, 719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOLLMERS C, GILL S, DITACCHIO L, PULIVARTHY SR, LE HD & PANDA S 2009. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A, 106, 21453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOLLMERS C, SCHMITZ RJ, NATHANSON J, YEO G, ECKER JR & PANDA S 2012. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab, 16, 833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J, SYMUL L, YEUNG J, GOBET C, SOBEL J, LUCK S, WESTERMARK PO, MOLINA N & NAEF F 2018. Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc Natl Acad Sci U S A, 115, E1916–E1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEINDRUCH R, WALFORD RL, FLIGIEL S & GUTHRIE D 1986. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr, 116, 641–54. [DOI] [PubMed] [Google Scholar]
- WELSH DK, TAKAHASHI JS & KAY SA 2010. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol, 72, 551–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WELSH DK, YOO SH, LIU AC, TAKAHASHI JS & KAY SA 2004. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol, 14, 2289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU Y, GUO W, LI P, ZHANG Y, ZHAO M, FAN Z, ZHAO Z & YAN J 2016. Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression. PLoS Genet, 12, e1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAGITA K, TAMANINI F, VAN DER HORST GT & OKAMURA H 2001. Molecular mechanisms of the biological clock in cultured fibroblasts. Science, 292, 278–81. [DOI] [PubMed] [Google Scholar]
- YAMAZAKI S, NUMANO R, ABE M, HIDA A, TAKAHASHI R, UEDA M, BLOCK GD, SAKAKI Y, MENAKER M & TEI H 2000. Resetting central and peripheral circadian oscillators in transgenic rats. Science, 288, 682–5. [DOI] [PubMed] [Google Scholar]
- YEUNG J, MERMET J, JOUFFE C, MARQUIS J, CHARPAGNE A, GACHON F & NAEF F 2018. Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res, 28, 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YEUNG J & NAEF F 2018. Rhythms of the Genome: Circadian Dynamics from Chromatin Topology, Tissue-Specific Gene Expression, to Behavior. Trends Genet, 34, 915–926. [DOI] [PubMed] [Google Scholar]
- YOO SH, MOHAWK JA, SIEPKA SM, SHAN Y, HUH SK, HONG HK, KORNBLUM I, KUMAR V, KOIKE N, XU M, et al. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell, 152, 1091–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOO SH, YAMAZAKI S, LOWREY PL, SHIMOMURA K, KO CH, BUHR ED, SIEPKA SM, HONG HK, OH WJ, YOO OJ, et al. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A, 101, 5339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG EE, LIU Y, DENTIN R, PONGSAWAKUL PY, LIU AC, HIROTA T, NUSINOW DA, SUN X, LANDAIS S, KODAMA Y, et al. 2010. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med, 16, 1152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG R, LAHENS NF, BALLANCE HI, HUGHES ME & HOGENESCH JB 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A, 111, 16219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, FANG B, EMMETT MJ, DAMLE M, SUN Z, FENG D, ARMOUR SM, REMSBERG JR, JAGER J, SOCCIO RE, et al. 2015. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science, 348, 1488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]