Abstract
Aims
A 25-base pair deletion in the cardiac myosin binding protein-C (cMyBP-C) gene (MYBPC3), proposed to skip exon 33, modifies the C10 domain (cMyBP-CΔC10mut) and is associated with hypertrophic cardiomyopathy (HCM) and heart failure, affecting approximately 100 million South Asians. However, the molecular mechanisms underlying the pathogenicity of cMyBP-CΔC10mutin vivo are unknown. We hypothesized that expression of cMyBP-CΔC10mut exerts a poison polypeptide effect leading to improper assembly of cardiac sarcomeres and the development of HCM.
Methods and results
To determine whether expression of cMyBP-CΔC10mut is sufficient to cause HCM and contractile dysfunction in vivo, we generated transgenic (TG) mice having cardiac-specific protein expression of cMyBP-CΔC10mut at approximately half the level of endogenous cMyBP-C. At 12 weeks of age, significant hypertrophy was observed in TG mice expressing cMyBP-CΔC10mut (heart weight/body weight ratio: 4.43 ± 0.11 mg/g non-transgenic (NTG) vs. 5.34 ± 0.25 mg/g cMyBP-CΔC10mut, P < 0.05). Furthermore, haematoxylin and eosin, Masson’s trichrome staining, as well as second-harmonic generation imaging revealed the presence of significant fibrosis and a greater relative nuclear area in cMyBP-CΔC10mut hearts compared with NTG controls. M-mode echocardiography analysis revealed hypercontractile hearts (EF: 53.4%±2.9% NTG vs. 66.4% ± 4.7% cMyBP-CΔC10mut; P < 0.05) and early diastolic dysfunction (E/E′: 28.7 ± 3.7 NTG vs. 46.3 ± 8.4 cMyBP-CΔC10mut; P < 0.05), indicating the presence of an HCM phenotype. To assess whether these changes manifested at the myofilament level, contractile function of single skinned cardiomyocytes was measured. Preserved maximum force generation and increased Ca2+-sensitivity of force generation were observed in cardiomyocytes from cMyBP-CΔC10mut mice compared with NTG controls (EC50: 3.6 ± 0.02 µM NTG vs. 2.90 ± 0.01 µM cMyBP-CΔC10mut; P < 0.0001).
Conclusion
Expression of cMyBP-C protein with a modified C10 domain is sufficient to cause contractile dysfunction and HCM in vivo.
Keywords: Cardiac myosin binding protein-C, Diastolic dysfunction, Fibrosis, Transgenic mouse model, South Asian population
1. Introduction
Hypertrophic cardiomyopathy (HCM) is a genetic heart disease affecting one in 200–500 people.1 HCM is clinically defined by left ventricular (LV) hypertrophy (mainly affecting the septal wall), diastolic dysfunction, mitral valve regurgitation, abnormal coronary microcirculatory function, and electrocardiographic abnormalities.2 The aetiology of HCM stems from mutations in genes encoding cardiac contractile proteins, in particular mutations in MYBPC3, encoding cardiac myosin binding protein-C (cMyBP-C), as the most common origin of disease pathogenesis.3 Evidence suggests that a subset of these mutations can cause an increase of toxic mutant and truncated proteins that leads to proteotoxicity in the heart owing to either specific interactions of the mutant protein or general misfolding that can cause cellular dysfunction and contribute to the development of HCM.4–7 In cases of mutations that cause misfolded protein aggregates, it follows that targeting proteotoxicity would be a reasonable therapeutic approach to counteract the resultant proteopathy6,8–10 by removing the offending truncated sarcomeric proteins.6,11–13 However, the aetiology that would explain why specific MYBPC3 mutant proteins lead to disease is likely variable and remains poorly understood.
The first MYBPC3 mutations associated with HCM were reported in 1995.14,15 Since then, more than 350 mutations have been published for this gene,16 with population-specific founder effects.17–19 cMyBP-C is a key sarcomeric thick-filament protein that interacts with both myosin and actin filaments to regulate sarcomere structure and function.20 Importantly, 70% of all MYBPC3 mutations include premature stop codons or frame shifts that encode proteins lacking key myosin binding residues in the C-terminus, which prevents protein incorporation into the sarcomere.21,22 The pathogenic mechanism of MYBPC3 mutations could stem from either an insufficient amount of gene product and, hence, reduced sarcomeric cMyBP-C, i.e. haploinsufficiency, direct pathogenic effects of the mutants on normal myofilament function, i.e. a poison polypeptide effect, or an accumulation of misfolded proteins, i.e. proteotoxicity.23–29 Studying the specific pathogenic mechanism for specific MYBPC3 mutations is confounded by an incomplete penetrance and variable onset of these mutations, which can be exacerbated by other physical or genetic modifiers,26,30 suggesting that several factors may be involved in the prognosis of HCM. Therefore, it is important to determine the pathogenicity and underlying molecular mechanism(s) of each mutation in the context of HCM as an essential first step in identifying targets for therapies directed towards sarcomeric cardiomyopathies.31,32
Previously, we discovered a specific variant in MYBPC3, a 25-base pair deletion that leads to the loss of the splicing branch point in intron 32.19,21,33,34 This variant affects approximately 100 million South Asians and is associated with HCM and heart failure (HF).19,35,36 This mutation is thought to result in the skipping of exon 33 with a corresponding replacement of 62 amino acids with 55 non-native amino acids at the C10 domain of the C-terminus of cMyBP-C (cMyBP-CΔC10mut).21 The mutation was found to be associated with the development of HCM and HF.19 Individuals carrying this variant are at high risk of developing HCM, but its functional and molecular effects are largely unknown.19 Adenoviral expression of cMyBP-CΔC10mut in adult rat cardiomyocytes showed contractile dysfunction,37 but whether expression of cMyBP-CΔC10mut alone is enough to cause an HCM phenotype in vivo remains elusive. Similar MYBPC3 mutations have been shown to be quickly degraded; however, we have recently identified an additional mutation, D389V, which occurs in a subpopulation of cMyBP-CΔC10mut carriers.38 This D389V variant occurs on the same allele as the cMyBP-CΔC10mut and is strongly associated with a HCM phenotype, suggesting that this protein is present in appreciable levels to cause dysfunction.
Therefore, to study the pathogenic potential of the cMyBP-CΔC10mut mutation in vivo, we generated transgenic (TG) mice expressing cardiac-specific cMyBP-CΔC10mut to define the link between cMyBP-CΔC10mut expression and HCM. We found that cMyBP-CΔC10mut did not incorporate into the C-zone of the sarcomere, but rather localized to the cytosol and Z-line. This evidence supports the poison-polypeptide hypothesis, in which an increase of toxic mutant and truncated proteins leads to a cascade of events beginning with improper incorporation of cMyBP-CΔC10mut, then contractile dysfunction, and finally the induction of pro-hypertrophic signalling and cardiac remodelling. This study details the progressive pathogenicity of these events, thus providing mechanistic insight into the in vivo pathogenicity of the most common mutation associated with the development of HCM.
2. Methods
An expanded Methods section can be found in the Supplementary material online.
2.1 Cardiac-specific cMyBP-CΔC10mut transgenic mouse model
The previously generated mouse cMyBP-C wild-type cDNA (∼3.8 kb) (Accession # NM_008653) was subjected to removal of exon 33, including 62 residues, and 55 non-native residues were introduced to generate the cMyBP-CΔC10mut protein.37 The sequence encoding the myc epitope tag (10 amino acids, EQKLISEEDL, Roche) was incorporated into the primer such that the tag was placed after the initiator Met residue. The anti-Myc antibody was used to distinguish the cMyBP-CΔC10mut protein from endogenous cMyBP-C. After the correct sequence was confirmed, the entire cDNA fragment was ligated into the mouse Myh6 promoter vector at the unique SalI site. The final construct was then resequenced to ensure the correct orientation. Next, the construct (∼10 kb) was digested free of the vector sequence (∼2.8 kb) with NotI, purified from agarose gels, and used to generate multiple TG lines (C57BL/6 strain). We obtained seven founder lines that were identified by PCR using primers corresponding to the α-MyHC 5’-untranslated (UTR) region with an internal control. An initial screening was performed on TG expression levels with a cMyc antibody (Data not shown), and the line with the highest levels of transgene expression was found and then selected for further analysis. All animal experiments were approved by the Institutional Animal Care and Use Committee at Loyola University Chicago and the University of Cincinnati and followed the policies described in the Guide for the Use and Care of Laboratory Animals published by the National Institutes of Health. Carbon dioxide (CO2) inhalation was used for euthanasia via the chamber method. This method employs a top-opening chamber into which the animal(s) was/were introduced. After the animal(s) was/were placed in the chamber, a slow flow of CO2 was initiated for a few minutes to slowly establish a high concentration at the bottom of the chamber. After breathing had stopped and the animal(s) were unconscious, euthanasia was completed by thoracotomy and removal of the heart for processing.
2.2 Mouse echocardiography
To assess cardiac function in non-transgenic (NTG) and cMyBP-CΔC10mut animals at 3 months of age, non-invasive M-mode echocardiography was performed using a VisualSonics Vevo 2100 imaging system (FUJIFILM VisualSonics Inc.) with an MS-550D 22–55 MHz transducer, as described previously.39 Under isoflurane anaesthesia delivered via inhalation (5% for knockdown and 1.5% for sedation during the duration of the procedure), the hearts were monitored to assess function and morphology.
2.2 Histopathology
To assess gross morphology, cardiac sections were taken from NTG and cMyBP-CΔC10mut hearts and stained with haematoxylin and eosin (H&E) or Masson’s trichrome (MT), as described previously.39
2.3 mRNA isolation, qPCR, and RNA-seq analysis
Gene expression changes between NTG and cMyBP-CΔC10mut hearts were evaluated as described previously.39
2.4 cMyBP-CΔC10mut protein localization analysis
To determine the localization of the cMyBP-CΔC10mut protein in the cardiac sarcomere of ΔC10 mice, cardiomyocytes were isolated from cMyBP-CΔC10mut hearts, as well as NTG hearts, for comparison. Following their isolation, cultured cardiomyocytes were fixed, permeabilized, and incubated with antibodies to detect endogenous cMyBP-C, myc-tagged cMyBP-CΔC10mut protein, and α-actinin.37 The immunofluorescence-labelled cardiomyocytes from NTG and cMyBP-CΔC10mut hearts were imaged using a Leica TCS SP5 microscope.
2.5 Second-harmonic generation imaging
Second-harmonic generation (SHG) imaging of NTG and cMyBP-CΔC10mut hearts was performed and quantified as we previously described.40
2.6 Cardiac muscle organization analysis
To examine whole heart morphology, hearts from NTG and cMyBP-CΔC10mut animals were processed and imaged as described previously.41
2.7 Western blot analysis
The level of endogenous cMyBP-C and cMyBP-CΔC10mut was evaluated by western blot analysis using both cMyBP-C (polyclonal) and anti-Myc antibody tag (Roche) antibodies in whole heart and myofilament isolations. Following our previously described method, we used a calibration curve of both cMyBP-C and Myc antibodies to perform semi-quantitative analysis.37 In addition to total protein levels, cMyBP-C phosphorylation was determined by using site-specific phospho-antibodies, as described previously.21
2.8 Force-calcium relationship in permeabilized cardiomyocytes
Isometric force measurements on permeabilized myocytes were performed, as described before.42
2.9 Electron microscopy
Hearts were dissected and then fixed with glutaraldehyde/paraformaldehyde. For sectioning, fixed muscles were post-fixed with 1% OsO4 in 0.1 M sodium cacodylate buffer for 1 h, dehydrated in graded alcohol and then embedded in Epon. Ultrathin sections were cut with a diamond knife on a Leica UC7 Ultramicrotome. A-band length, M-line width, and Z-line width were measured using ImageJ.43 The averaged data were analysed for statistical significance in GraphPad Prism using a one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test.
2.10 Statistical analyses
Tension-pCa relationships were fit with a modified Hill equation.44–46 The data are presented as mean ± SEM. Normal distribution of data was tested with the Shapiro–Wilk test. If data were normally distributed, a Student’s t-test was used to determine significant differences between groups. The Mann–Whitney U test was used for non-normal distribution of data. Statistically significant differences were considered as P < 0.05.
3. Results
3.1 TG expression of cMyBP-CΔC10mut causes HCM in mice
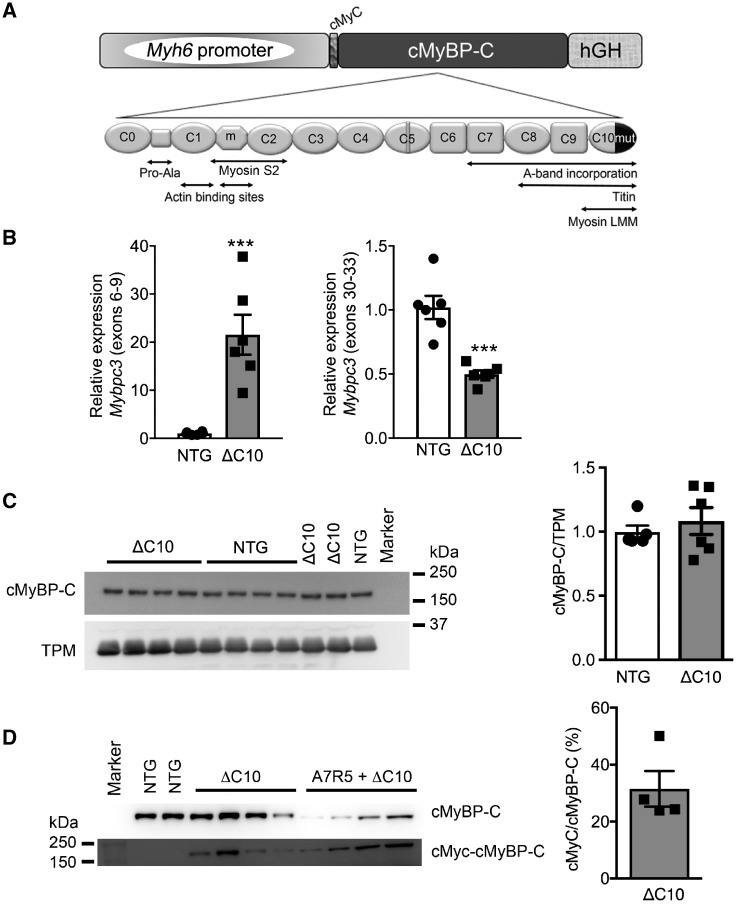
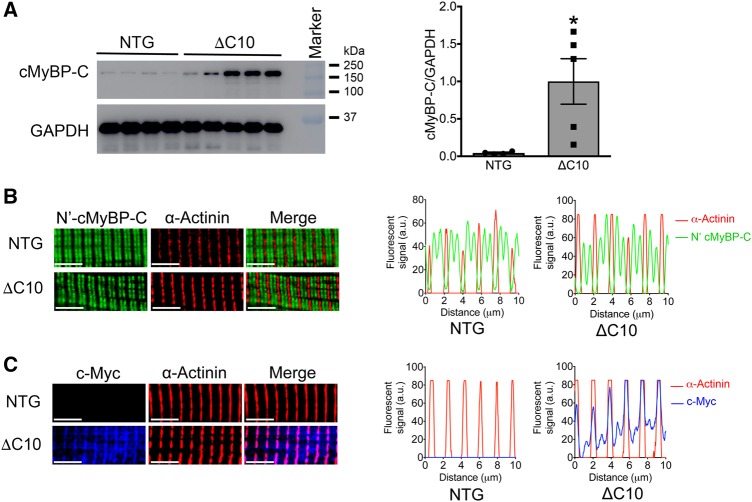
Data gained from previous in vitro experiments showed that cMyBP-CΔC10mut could not properly incorporate in adult rat cardiomyocytes which resulted in decreased sarcomere contractility.37 However, it is unknown whether abnormal localization of cMyBP-CΔC10mut or its overexpression can cause HCM in vivo. A knock-in model of this mutation was considered unsuitable because mouse intron 32 is entirely different from that of human, and ablation of exon 33 would result in a different stop codon when compared with human. Therefore, a cardiac-specific TG mouse model expressing cMyBP-CΔC10mut was established (Figure 1A) to determine the physiological changes that result from the presence of cMyBP-CΔC10mut and to define the functional defects underlying HCM. High levels of transgene expression were seen at the mRNA level (Figure 1B, panel left), as primers recognizing both endogenous and TG Mybpc3 showed 19.6 ± 1.3-fold higher expression in cMyBP-CΔC10mut mice than NTG littermates, while primers recognizing only endogenous Mybpc3 showed a slightly lower expression in cMyBP-CΔC10mut compared with NTG (Figure 1B, panel right). Analysis at the protein level revealed no significant change in total cMyBP-C levels (Figure 1C) or myofilament cMyBP-C levels (Supplementary material online, Figure S1A–C). cMyBP-C is heavily regulated by phosphorylation; therefore, the phosphorylation levels of the four most common sites were evaluated, but these were found to be unchanged (Supplementary material online, Figure S2). Additionally, cardiac troponin I (cTnI) phosphorylation was unchanged between the groups (Supplementary material online, Figure S1A, B, and D). Most HCM-associated MYBPC3 mutations lead to premature stop codons, resulting in mRNA degradation by nonsense-mediated decay (NMD) and/or quick degradation of the truncated protein.11,47 To establish if this is true for cMyBP-CΔC10mut, the percentage of cMyBP-C derived from the c-Myc-tagged transgene was determined. In cMyBP-CΔC10mut mice, 31.6 ± 6.2% of cMyBP-C was derived from the transgene (i.e. c-Myc tagged, Figure 1D). These data show that the cMyBP-CΔC10mut protein can be translated and remain stable enough for detection. Thus, any pathological effect elicited by cMyBP-CΔC10mut could be attributed to a poison polypeptide effect, or, in the alternative, through dysregulation of RNA or protein degradation pathways. This model is, therefore, suitable for studying the pathophysiological consequences of cMyBP-CΔC10mut.
Figure 1.
Cardiac-specific TG expression of cMyBP-CΔC10mut. (A) Schematic diagram of the TG construct expressing the cMyBP-CΔC10mut (ΔC10) under the control of α-myosin heavy chain promoter. (B) mRNA gene expression levels of MYBPC3 using primers targeting endogenous cMyBP-C and TG cMyBP-CΔC10mut (left) or endogenous cMyBP-C only (right) from NTG vs. cMyBP-CΔC10mut (n = 6). Quantification of cMyBP-CΔC10mut expression relative to total cMyBP-C. (C) Western blot analysis showing total cMyBP-C expression in NTG and cMyBP-CΔC10mut hearts (left) and their quantification normalized to the internal loading control α-tropomyosin (TPM) (5 NTG and 6 ΔC10). (D) Western blot analysis depicting total cMyBP-C and the c-Myc-tagged cMyBP-CΔC10mut in NTG and cMyBP-CΔC10mut mice, as well as adenovirus-expressed cMyBP-CΔC10mut in A7R5 rat aortic smooth muscle cells (left) (n = 4 ΔC10). ***P < 0.001.
3.2 Augmented fibrosis is evident in cMyBP-CΔC10mut hearts
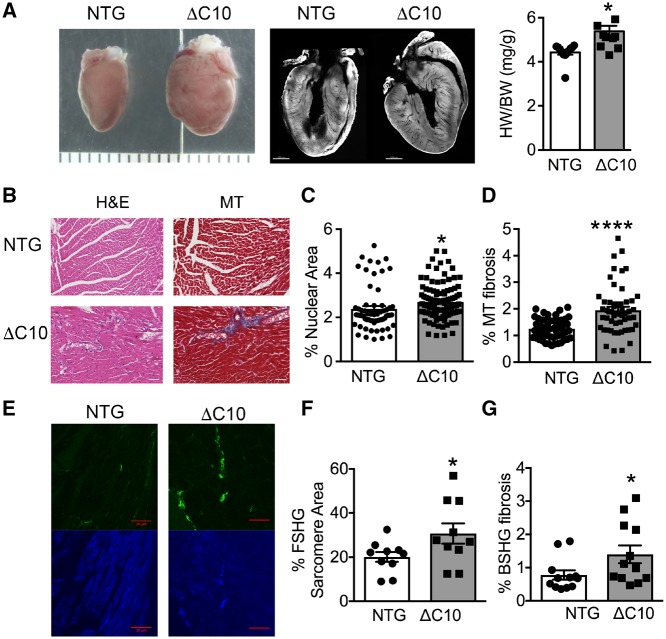
We next asked whether cMyBP-CΔC10mut expression alone would be sufficient to cause HCM in mice. Indeed, cMyBP-CΔC10mut expression resulted in increased heart size and weight compared with NTG mice (Figure 2A). A characteristic of HCM is myocardial fibrosis.48 Thus, histopathological analyses using myocardial sections from cMyBP-CΔC10mut and NTG hearts was performed. Percent nuclear area, an indicator of cellularity, was significantly elevated in cMyBP-CΔC10mut hearts (Figure 2B and C). Similarly, significant fibrosis was noticed in cMyBP-CΔC10mut myocardial sections, as shown by increased Masson’s trichrome staining (Figure 2B and D). This was confirmed with SHG imaging (collagen deposition, Figure 2E and G). SHG was also used to quantify sarcomere area (Figure 2E and F) and revealed a significant increase in sarcomere area in cMyBP-CΔC10mut myocardial sections compared with those from NTG mice. Together, these data demonstrate that myocardial fibrosis accompanies cardiac remodelling in cMyBP-CΔC10mut mice, further implicating that cMyBP-CΔC10mut expression leads to the macroscopic changes seen in HCM.
Figure 2.
Myocardial fibrosis is elevated in cMyBP-CΔC10mut hearts. (A) Gross morphology, cross section of whole isolated hearts and heart/body weight (HW/BW) ratio (mg/g). Scale marks are separated by 1000 μm in left panel and scale bar = 1000 μm in middle panel. (B) Representative H&E- and Masson trichrome (MT)-stained myocardial sections from cMyBP-CΔC10mut and NTG hearts. Scale bar = 50 μm. (C) Quantification of percent nuclear area and (D) percent MT fibrosis obtained from measurements of H&E- and MT-labelled myocardial sections from cMyBP-CΔC10mut and NTG hearts (n = 3 NTG hearts, 62 sections and 3 cMyBP-CΔC10mut hearts, 106 sections for panel C and n = 3 NTG hearts, 53 sections and 3 cMyBP-CΔC10mut hearts, 52 sections for D). (E) Representative SHG-imaged myocardial sections from cMyBP-CΔC10mut and NTG hearts. The green channel is backward SHG (BSHG) predominantly depicting collagen fibres, and the blue channel is forward-directed SHG (FSHG) showing the sarcomere pattern. Scale bar = 20 μm. Quantification of (F) percent FSHG sarcomere area and (G) percent BSHG fibrosis obtained from SHG-imaged myocardial sections from cMyBP-CΔC10mut and NTG hearts (n = 3 NTG hearts, 10 sections and 3 cMyBP-CΔC10mut hearts, 10 sections for F and n = 3 NTG hearts, 12 sections and 3 cMyBP-CΔC10mut hearts, 13 sections for G). *P < 0.05, ****P < 0.0001.
3.3 cMyBP-CΔC10mut mice show cardiac hypertrophy and diastolic dysfunction
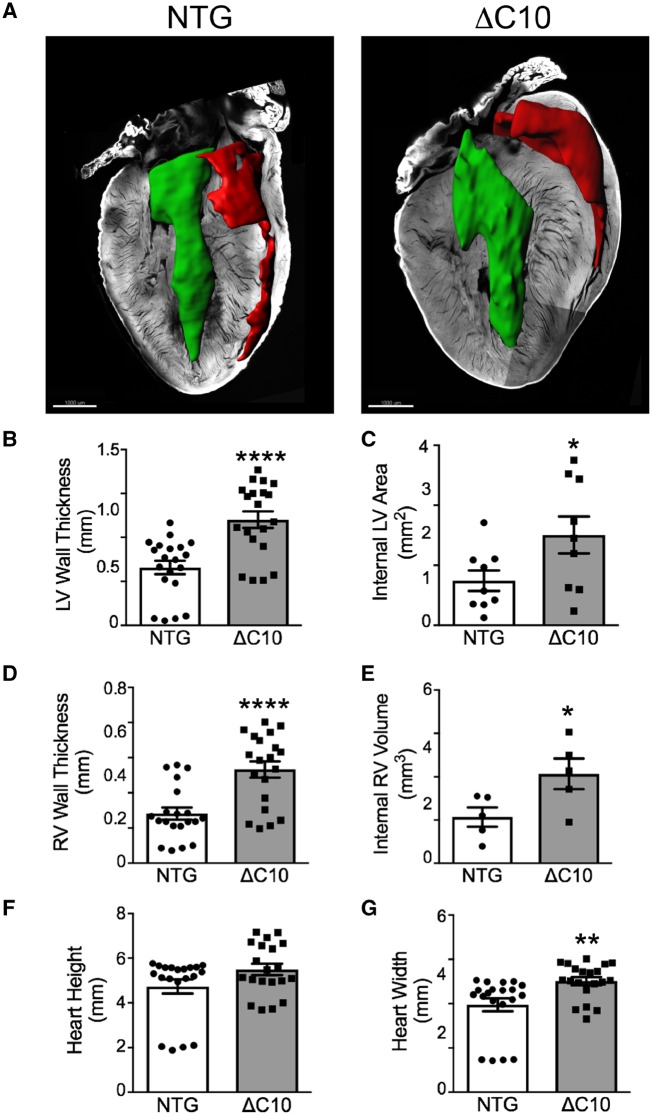
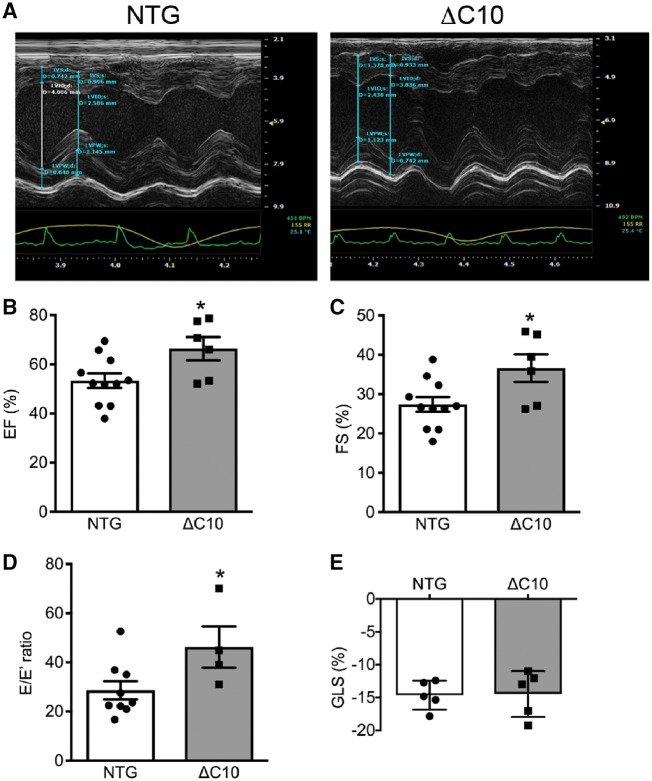
cMyBP-CΔC10mut protein expression caused hypertrophy in 3-month-old animals (Figures 2 and 3A). In line with this finding, morphometric measurements revealed a significant elevation in LV wall diameter and area (Figure 3B and C), right ventricular wall diameter and volume (Figure 3D and E), and heart width (Figure 3G) in cMyBP-CΔC10mut hearts compared with NTG controls. No significant difference in heart height was observed between cMyBP-CΔC10mut and NTG mice (Figure 3F). To evaluate cardiac function in cMyBP-CΔC10mut hearts compared with NTG controls, short-axis M-mode echocardiography was performed (Figure 4A). In keeping with an early HCM phenotype, systolic function was elevated (Figure 4B and C), while diastolic function, as determined by blood and tissue Doppler measurements, was impaired (Figure 4D) in cMyBP-CΔC10mut hearts compared with NTG controls. Evaluation of percent global longitudinal strain (GLS) in cMyBP-CΔC10mut hearts compared with NTG controls revealed no difference (Figure 4E). No other significant changes in cardiac function or morphology were seen by echocardiography (Table 1). cMyBP-CΔC10mut expression caused diastolic, but not systolic dysfunction, a pattern that is typical of HCM. Together, these data indicate that expression of cMyBP-CΔC10mut is sufficient to cause cardiac hypertrophy.
Figure 3.
cMyBP-CΔC10mut hearts display myocardial hypertrophy. (A) Confocal microscopy of myocardial sections from NTG and cMyBP-CΔC10mut hearts with cardiac chambers artificially filled with blood. Scale bar = 1000 μm. Morphometric measurements of confocal myocardial sections from NTG and cMyBP-CΔC10mut hearts quantifying (B) LV wall thickness, (C) Internal LV area, (D) RV wall thickness, (E) internal RV volume, (F) heart height, and (G) heart width (n = 5 NTG, 5 ΔC10). *P < 0.05, **P < 0.01, ****P < 0.0001.
Figure 4.
Diastolic dysfunction in cMyBP-CΔC10mut mice. (A) M-mode echocardiographic tracings showing LV hypertrophy in cMyBP-CΔC10mut hearts. Echocardiographic analysis of (B) ejection fraction (EF, %), (C) fractional shortening (FS, %), (D) E/E′ ratio and (E) global longitudinal strain analysis (GLS, %) (n = 6) *P < 0.05.
Table 1.
Echocardiographic data and cardiomyocyte force-Ca2+ data of NTG and cMyBP-CΔC10mut TG mice
| NTG | cMyBP-CΔC10mut | P-value | n value (NTG vs. cMyBP-CΔC10mut) | |
|---|---|---|---|---|
| HW/BW (mg/g) | 4.43 ± 0.11 | 5.38 ± 0.24 | 0.02* | 12 vs. 11 |
| IVSTd (mm) | 0.77 ± 0.05 | 0.82 ± 0.04 | 0.53 | 11 vs. 7 |
| IVSTs (mm) | 1.18 ± 0.05 | 1.23 ± 0.08 | 0.54 | 11 vs. 7 |
| LVPWd (mm) | 0.80 ± 0.05 | 0.77 ± 0.02 | 0.98 | 11 vs. 7 |
| LVPWs (mm) | 1.06 ± 0.08 | 1.14 ± 0.07 | 0.47 | 11 vs. 7 |
| LVIDd (mm) | 3.98 ± 0.17 | 3.61 ± 0.17 | 0.16 | 11 vs. 7 |
| LVIDs (mm) | 2.90 ± 0.16 | 2.39 ± 0.18 | 0.05 | 11 vs. 7 |
| EF (%) | 53.4 ± 2.9 | 66.4 ± 4.7 | 0.03* | 11 vs. 7 |
| FS (%) | 27.4 ± 1.9 | 36.6 ± 3.5 | 0.02* | 11 vs. 7 |
| GLS (%) | −13.0 ± −1.0 | −13 ± −1.1 | 0.98 | 5 vs. 5 |
| E/A | 1.66 ± 0.09 | 1.64 ± 0.10 | 0.88 | 6 vs. 6 |
| E/E′ | 28.7 ± 3.7 | 46.3 ± 8.4 | 0.04* | 6 vs. 6 |
| F max (kN/m2) | 19.96 ± 2.2 | 24.62 ± 3.4 | 0.36 | 5 vs. 10 |
| EC50 (µM) | 3.58 ± 0.33 | 2.74 ± 0.13 | 0.018* | 5 vs. 10 |
Summary of the heart weight (HW) to body weight (BW) ratio, echocardiographic findings, and functional measurements of cardiomyocytes from cMyBP-CΔC10mut compared with NTG hearts. P-values were calculated by two-tailed unpaired Student’s t-tests. *P < 0.05.
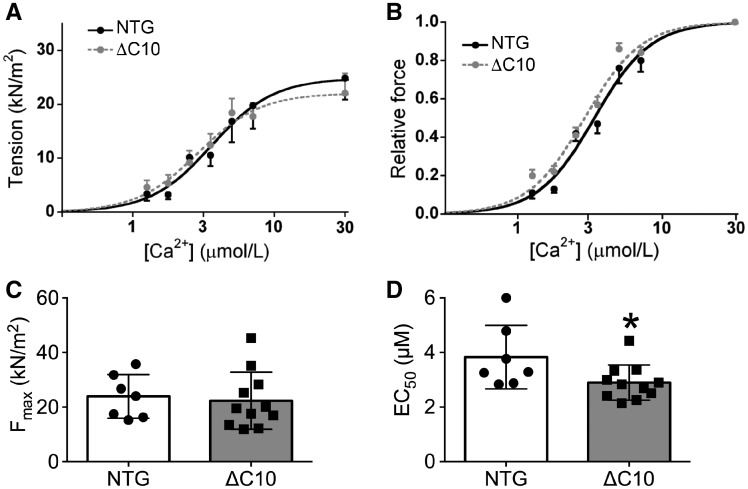
3.4 cMyBP-CΔC10mut myofilaments exhibit increased Ca2+-sensitivity
We next evaluated whether the diastolic dysfunction that was observed through echocardiography (Figure 4D) also occurred at the myofilament level. Maximal force-generating capacity of permeabilized cardiomyocytes did not change between cMyBP-CΔC10mut and NTG mice (Figure 5A and C), while an increased Ca2+-sensitivity of force development was observed in cMyBP-CΔC10mut cardiomyocytes (Figure 5B and D). When cardiomyocyte ultrastructure was studied by electron microscopy, no changes were apparent in thick filament length or arrangement, or in M-line or Z-line structure and dimensions (Supplementary material online, Figure S3). In vivo and in vitro contractile analyses indicated altered relaxation in cMyBP-CΔC10mut hearts owing to changes in myofilament Ca2+-sensitivity.
Figure 5.
Increased myofilament Ca2+-sensitivity of force generation in cMyBP-CΔC10mut skinned myocytes. Functional measurements of cardiomyocytes isolated from NTG and cMyBP-CΔC10mut hearts showing (A) force-Ca2+, (B) relative force curves, (C) maximal force, and (D) EC50 values (n = 5 NTG, 10 cMyBP-CΔC10mut). *P < 0.042.
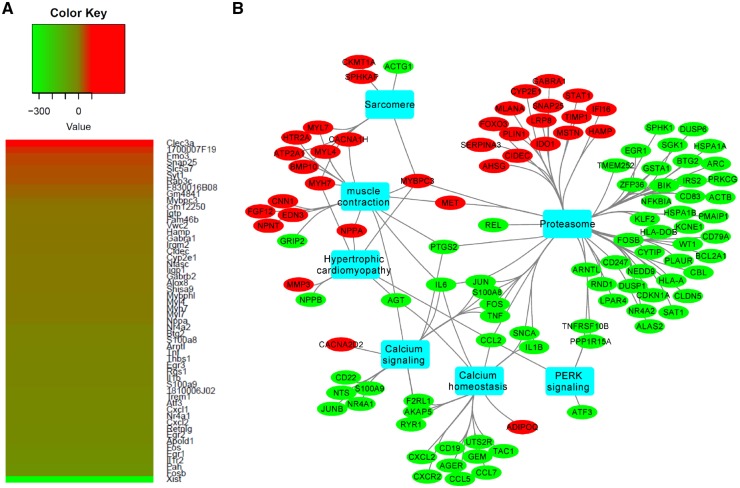
3.5 Gene pathway enrichment analyses and network visualization
To determine the global gene expression changes in cMyBP-CΔC10mut hearts compared with NTG controls, we performed RNA-seq analysis using total mRNA isolated from the hearts of these mice (Figure 6). In total, we observed 222 upregulated and 299 downregulated genes in cMyBP-CΔC10mut compared with NTG hearts. As expected, total Mybpc3 gene expression was highly up-regulated in cMyBP-CΔC10mut compared with NTG hearts by the TG overexpression of cMyBP-CΔC10mut. Supporting the observed increased heart size and wall thickness, we detected an upregulation of several hypertrophy genes, including Myh7 and Nppa, in cMyBP-CΔC10mut hearts. We next performed gene ontology analyses of this RNA-Seq dataset for biological processes and pathways that were overrepresented or underrepresented in cMyBP-CΔC10mut compared with NTG hearts, as we described previously.49 Selected enriched biological processes and pathways in cMyBP-CΔC10mut compared with NTG hearts are depicted in the network representation in Figure 6B.
Figure 6.
Differentially regulated genes in cMyBP-CΔC10mut compared with NTG hearts. (A) RNA-seq heat maps depicting clusters of genes differentially regulated in cMyBP-CΔC10mut vs. NTG hearts. Shown is a subset of the most up-regulated (red) and down-regulated (green) genes from the total gene set. The fold change, up or down, is represented in the key for the respective panels (n = 4 pooled samples per genotype). (B) Network of significantly enriched gene ontology terms for upregulated genes in cMyBP-CΔC10mut hearts. The red and green circles denote upregulated and downregulated genes, respectively, in cMyBP-CΔC10mut compared with NTG hearts. Blue rectangles represent significantly enriched (P < 0.05; FDR) biological processes using the ToppFun application of ToppGene Suite.
3.6 cMyBP-CΔC10mut protein mislocalizes at the Z-line
It has been established that the cMyBP-CΔC10mut leads to the replacement of 62 of the C-terminal amino acids with a non-native sequence of 55 residues. This includes the four out five of critical amino acids crucial for binding to the rod domain of myosin heavy chain.21 We and others have shown that this domain is crucial for incorporation into the sarcomere; therefore, the amount of cMyBP-C in the soluble fraction (i.e. non-sarcomere) was studied. Levels of cMyBP-C in the soluble fraction were increased 22.5 ± 6.8-fold in cMyBP-CΔC10mut mice vs. NTG controls (Figure 7A). The apparent mislocalization of cMyBP-C was confirmed with immunofluorescence staining of isolated cardiomyocytes from cMyBP-CΔC10mut and NTG hearts (Figure 7B and C). Staining with an antibody raised against the N-terminus of cMyBP-C resulted in the typical doublet staining between each actinin-labelled Z-line (Figure 7B). In contrast, localization of cMyBP-CΔC10mut, as specifically identified with an antibody raised against the c-Myc tag, was not restricted to the C-zone of the sarcomere, but was, instead, present at the Z-line with some diffuse, randomly distributed staining along the rest of the sarcomere (Figure 7C). This suggests that mislocalization of cMyBP-CΔC10mut at the Z-line in cMyBP-CΔC10mut cardiomyocytes could cause the contractile abnormalities, as observed in the hearts of these animals, and trigger myocardial remodelling leading to HCM.
Figure 7.
cMyBP-CΔC10mut protein is highly soluble and localizes at the Z-line in the cardiac sarcomere. (A) Western blot analysis depicting the levels of total soluble cMyBP-C (endogenous and/or cMyBP-CΔC10mut) from cMyBP-CΔC10mut and NTG hearts (n = 4 NTG, 5 cMyBP- *P < 0.027). (B) Single cardiomyocytes were isolated from cMyBP-CΔC10mut and NTG hearts and were stained with antibodies detecting cMyBP-C’s N-terminus (C0 domain) (green) and α-actinin (red). In both NTG and cMyBP-CΔC10mut myocytes, the classical staining pattern of two cMyBP-C bands between Z-lines (stained with α-actinin) was observed, indicating proper localization of endogenous cMyBP-C in cMyBP-CΔC10mut and NTG cardiomyoctyes. (C) Cardiomyocytes from cMyBP-CΔC10mut and NTG hearts were stained with antibodies to detect c-Myc (blue), which will show only the c-Myc-tagged cMyBP-CΔC10mut protein and α-actinin (red). The c-Myc signal was only observed in myocytes from cMyBP-CΔC10mut hearts. The presence of c-Myc staining at the Z-line with α-actinin suggests that the cMyBP-CΔC10mut protein aberrantly distributes within the cardiac sarcomere in cMyBP-CΔC10mut myocytes. Scale bar = 5 μm in B and C.
4. Discussion
The cMyBP-CΔC10mut variant is associated with the development of various forms of cardiomyopathy and HF, but its functional and molecular effects are unknown.19 Seventy percent of all MYBPC3 mutations, including cMyBP-CΔC10mut, are predicted to produce proteins that have premature stops or frame shifts that terminate or change the C’-terminal, which includes key myosin-binding residues. These truncated protein products are either not expressed or quickly degraded, as expression of the mutant protein cannot be found in cardiac tissue from HCM patients.23,28,50 This means that reduced cMyBP-C expression, i.e. haploinsufficiency, rather than detrimental effects of the mutant protein, i.e. poison polypeptide, is the predominant disease mechanism of MYBPC3 truncation mutants,51,52 but the aetiology of HCM in heterozygous MYBPC3 truncation mutation carriers may vary depending on the specific mutation.
The genotype–phenotype relationship in HCM is far from straightforward. The same mutation can lead to HCM development at young age in one person, while a family member could live symptom-free until late age. This was illustrated in a recent study in which genotype–positive/phenotype–negative subjects (family members of overt HCM patients) were monitored for almost two decades.38 The study determined that carrying the cMyBP-CΔC10mut does not mean that disease will develop, at least not at a young age.38 It is possible that some mutations are more penetrant than others, but this needs to be established. Studying the disease mechanisms of specific MYBPC3 mutations is complicated by the occurrence of mutations in other sarcomeric proteins, resulting in worse prognoses.34 For example, if carriers of cMyBP-CΔC10mut also carry a mutation in MYH7, this combination frequently results in sudden cardiac death.33 More shocking statistics indicate that South Asians carrying the cMyBP-CΔC10mut have 50% higher rates of morbidity and mortality after ischaemia–reperfusion injury compared with other ethnic groups.53–55 Supporting these data, Srivastava et al.36 showed that the presence of cMyBP-CΔC10mut leads to reduced contractile function following coronary artery disease, suggesting that cMyBP-CΔC10mut pathogenicity can exacerbate cardiac dysfunction in patients with myocardial infarction.
To the best of our knowledge, the pathological consequences of cMyBP-CΔC10mut expression have never been studied in vivo. Given its presence in and potential impact on 100 million people of South Asian descent worldwide, systematically studying the presumed expression of cMyBP-CΔC10mut at the whole-organ level would have clear translational value. The cMyBP-CΔC10mut mutation was initially described in a group of Indian HCM patients in 2003.34 In a more extensive follow-up study using a large cohort of cardiomyopathy cases and healthy controls, this mutation was shown to be a founder mutation highly prevalent among South Asians (6%).38 The mutation was overrepresented in the cardiomyopathy group, indicating that carrying the mutation confers a greater risk of developing cardiomyopathy. Of note, carrying the mutation could lead to HCM, but it could also lead to dilated cardiomyopathy to a similar degree, or to restrictive cardiomyopathy to a lesser degree.19 In a recent study, we found a similarly high mutation frequency in people of South Asian descent living in the USA (6.0%).38 The screening performed in this study was done on people attending community events, not a hospital setting. Therefore, most people were free of cardiac disease. In this group of subjects, we did not find an increase in systolic or diastolic dysfunction measured by echocardiography. This discrepancy could be explained if cMyBP-CΔC10mut causes late disease onset. Most people screened (84.5%) were under the age of 60.38 Founder mutations can subsist in the population if they are associated with a mild phenotype or a late disease onset.
We performed cardiomyopathy panel screening on this cohort and identified a second, previously undescribed, mutation on the same MYBPC3 allele, D389V. This mutation strongly correlated with hyperdynamic ventricular function and ventricular wall thickening.38 The D389V mutation sits at a critical regulatory region of cMyBP-C that may alter the ability of cMyBP-C to regulate actomyosin interactions, but future studies are needed to directly assess the function of this mutation. Because this missense mutation was strongly linked to HCM and occurred on the same allele as that of the cMyBP-CΔC10mut, it can be argued that this mutant protein must at least be translated to exert a pathophysiological function, whether by improper binding and resultant myofilament dysregulation, or by a protein aggregation mechanism. This information provides support for studying the TG expression of cMyBP-CΔC10mut in mice at the whole-heart level.
By examining its underlying molecular mechanism, the present study aimed to determine the sufficiency of cMyBP-CΔC10mut expression as a causative factor in the development of an HCM phenotype. We have demonstrated that TG expression of cMyBP-CΔC10mut in mouse hearts in vivo results in a phenotype of HCM with hypertrophy, diastolic dysfunction, and fibrosis. Indeed, this was supported by earlier in vitro findings showing that expression of cMyBP-CΔC10mut in adult rat cardiomyocytes leads to contractile dysfunction.37 Further in vitro analysis showed that the mutant C10 domain loses its ability to interact with the light meromyosin region of myosin heavy chain. The decreased thick filament affinity of the C’-terminal of cMyBP-CΔC10mut also explains the increased presence of cMyBP-C in the soluble fraction of cMyBP-CΔC10mut heart lysates. Since TG expression of wild-type cMyBP-C in mice did not result in a phenotype,56,57 we have evidence that the culprit is cMyBP-CΔC10mut, not high expression levels of cMyBP-C.
The apparent discrepancy between the high levels of non-symptomatic human mutation carriers and the TG mouse data presented in this manuscript, as well as the rat cardiomyocyte data published earlier,37 could be explained if high expression levels of cMyBP-CΔC10mut are needed to induce disease. In both the TG mice and adenoviral-based experiments in rat cardiomyocytes, high expression levels of cMyBP-CΔC10mut were induced. At present, the protein level of cMyBP-CΔC10mut in human heart tissue is unknown, but it could be that heterozygous cMyBP-CΔC10mut carriers express the mutant protein at a low level. If that is the case, the addition of a secondary stressor, whether by a co-mutation or other external factors, could be the trigger leading to cardiomyopathy. It has been shown that carrying cMyBP-CΔC10mut leads to greater cardiac dysfunction in post-myocardial infarction patients,53–55 supporting its role as a disease modifier.
The exact mechanism by which cMyBP-CΔC10mut expression leads to dysfunction is unknown. cMyBP-CΔC10mut did not localize to its usual position in the C-zone. At least part of the protein localized to the Z-line (Figure 7), and upon subcellular fractionation with high levels of detergent we observed increased levels of soluble cMyBP-CΔC10mut. While the fold change in soluble cMyBP-C between cMyBP-CΔC10mut and NTG is sizeable (22.5 fold), it should be noted that the level of soluble cMyBP-C in NTG is very low. So even in the cMyBP-CΔC10mut, the vast majority of cMyBP-C is likely to be myofilament associated. Our in vitro analysis also revealed Z-line staining.37 It is unknown if this mislocalization contributes to impaired relaxation. As we did not observe an increase of total cMyBP-C in the myofilament fraction (Supplementary material online, Figure S1), despite the Z-disc staining of cMyBP-CΔC10mut, it could be possible that endogenous cMyBP-C expression is lower. We did indeed observe a lower mRNA expression of endogenous cMyBP-C (Figure 1B, right panel). A lower level of cMyBP-C and cMyBP-CΔC10mut that localizes to the Z-line rather than C-zone could cause C-zone haploinsufficiency. From studies in mouse models and HCM patients, we know that haploinsufficiency of cMyBP-C could lead to increased calcium sensitivity.58–60 We did observe an increased calcium sensitivity of force development that was not caused by decreased cTnI phosphorylation. In a mouse model of a truncating cMyBP-C mutation, a bigger shift in calcium sensitivity was seen,58 but in another study using the same mouse model an increase of a similar magnitude as the present study was observed.59 This same shift is also seen in myectomy tissue from HCM patients with a MYBPC3 mutation.60 However, the mechanism by which cMyBP-CΔC10mut leads to increased calcium sensitivity is unknown. Recent studies have revealed another possible pathogenic effect of cMyBP-C haploinsufficiency: a reduction in the level of myosin that is in the super-relaxed state. The super-relaxed state of myosin is a low energy consuming state and is dependent on cMyBP-C.61 In HCM patients with a MYBPC3 mutation leading to haploinsufficiency, reduced levels of super-relaxed myosin were observed.62 This could lead to increased energy consumption that could contribute to HCM pathophysiology. Indeed, treatment of mouse cardiomyocytes with reduced cMyBP-C levels with a compound that restored the levels of super-relaxed myosin normalized contractile performance.63 Whether changes in the super-relaxed state are present and contribute to pathology in the cMyBP-CΔC10mut model remains to be studied.
Generally, truncating mutations in MYBPC3 appear to act via haploinsufficiency, although additional lines of inquiry have looked into the possibility that these truncated mutants can dysregulate the NMD or proteasomal pathways as a primary or secondary disease mechanism.6 While a clear relationship between MYBPC3 mutations and these degradation pathways has not emerged, data do suggest that altering the ubiquitin-proteasome system can alter disease progression.6 In our RNA-seq analysis, many genes associated with proteasome function were differentially expressed. Since the proteasome is involved in the degradation of proteins and is known to be involved in HCM pathophysiology,10,64 it is not surprising to find that it is differentially regulated in the cMyBP-CΔC10mut group. Whether or not the high expression of mutant protein overwhelms the proteasome system and whether activation of the system would be a therapeutic strategy6 requires further study.
In conclusion, we showed that TG expression of cMyBP-CΔC10mut leads to all the hallmarks of HCM in mice. However, recent data from people carrying the cMyBP-CΔC10mut mutation imply that a heterozygous cMyBP-CΔC10mut mutation causes late-onset HCM17–19 or functions as a disease modifier in HCM17–19 or may not be pathogenic under baseline normal conditions.38 Follow-up studies should focus on elucidating the effects of carrying heterozygous cMyBP-CΔC10mut mutation in humans and whether the 25-base pair deletion results in exon 33 skipping. If so, we should investigate the underlying mechanism and the likelihood of skipping, as these data could have clinical implications for the large group of mutation carriers living in the USA or abroad.
Supplementary Material
Acknowledgements
We acknowledge Suresh Govindan, PhD, for his help in cloning cMyBP-CΔC10mut, mouse genotyping, breeding, and colony management.
Conflict of interest: S.S. provided consulting and collaborative services to AstraZeneca, Merck, and Amgen unrelated to the content of this manuscript. All other authors declared no conflict of interest.
Funding
S.S. has received support from National Institutes of Health grants R01 HL130356, R56 HL139680, R01 AR067279, and R01 HL105826; American Heart Association Cardiovascular Genome-Phenome Study (15CVGPSD27020012) and Catalyst (17CCRG33671128) awards; and AstraZeneca, Merck and Amgen. D.Y.B. (11PRE7240022), D.W.D.K. (13POST17220009), and T.L.L. (15PRE22430028) were supported with American Heart Association fellowship training grants. R.C. was supported by NIH grants P01 HL059408 and R01 HL139883. R.N.-S. has received support from NHLBI grants 1RHL118067 and 2RHL118067.
Time for primary review: 32 days
References
- 1. Semsarian C, Ingles J, Maron MS, Maron BJ.. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 2015;65:1249–1254. [DOI] [PubMed] [Google Scholar]
- 2.Authors/Task Force members, Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, McKenna WJ, Mogensen J, Nihoyannopoulos P, Nistri S, Pieper PG, Pieske B, Rapezzi C, Rutten FH, Tillmanns C, Watkins H.. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the task force for the diagnosis and management of hypertrophic cardiomyopathy. Russ J Cardiol 2014;35:2733–2779. [DOI] [PubMed] [Google Scholar]
- 3. Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M.. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 2003;107:2227–2232. [DOI] [PubMed] [Google Scholar]
- 4. Gupta MK, Gulick J, Liu R, Wang X, Molkentin JD, Robbins J.. Sumo E2 enzyme UBC9 is required for efficient protein quality control in cardiomyocytes. Circ Res 2014;115:721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang X, Robbins J.. Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol 2014;71:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schlossarek S, Singh SR, Geertz B, Schulz H, Reischmann S, Hubner N, Carrier L.. Proteasome inhibition slightly improves cardiac function in mice with hypertrophic cardiomyopathy. Front Physiol 2014;5:484.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bahrudin U, Morikawa K, Takeuchi A, Kurata Y, Miake J, Mizuta E, Adachi K, Higaki K, Yamamoto Y, Shirayoshi Y, Yoshida A, Kato M, Yamamoto K, Nanba E, Morisaki H, Morisaki T, Matsuoka S, Ninomiya H, Hisatome I.. Impairment of ubiquitin-proteasome system by E334K cMyBPC modifies channel proteins, leading to electrophysiological dysfunction. J Mol Biol 2011;413:857–878. [DOI] [PubMed] [Google Scholar]
- 8. Sandri M, Robbins J.. Proteotoxicity: an underappreciated pathology in cardiac disease. J Mol Cell Cardiol 2014;71:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang W, Li S, Wang H, Li B, Shao L, Lai Y, Horvath G, Wang Q, Yamamoto M, Janicki JS, Wang XL, Tang D, Cui T.. Nrf2 enhances myocardial clearance of toxic ubiquitinated proteins. J Mol Cell Cardiol 2014;72:305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dorsch LM, Schuldt M, Knezevic D, Wiersma M, Kuster DWD, van der Velden J, Brundel B.. Untying the knot: protein quality control in inherited cardiomyopathies. Pflugers Arch 2019;471:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Pointu H, Mougenot N, Guiard J, Reimer R, Hohenberg H, Schwartz K, Vernet M, Eschenhagen T, Carrier L.. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res 2009;105:239–248. [DOI] [PubMed] [Google Scholar]
- 12. Carrier L. Too much of a good thing is bad: proteasome inhibition induces stressed hearts to fail. Cardiovasc Res 2010;88:389–390. [DOI] [PubMed] [Google Scholar]
- 13. Carrier L, Schlossarek S, Willis MS, Eschenhagen T.. The ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy. Cardiovasc Res 2010;85:330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, Maron BJ, Seidman JG, Seidman CE.. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet 1995;11:434–437. [DOI] [PubMed] [Google Scholar]
- 15. Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, Gautel M, Labeit S, James M, Beckmann J, Weissenbach J, Vosberg H-P, Fiszman M, Komajda M, Schwartz K.. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet 1995;11:438–440. [DOI] [PubMed] [Google Scholar]
- 16. Carrier L, Mearini G, Stathopoulou K, Cuello F.. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 2015;573:188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adalsteinsdottir B, Teekakirikul P, Maron BJ, Burke MA, Gudbjartsson DF, Holm H, Stefansson K, DePalma SR, Mazaika E, McDonough B, Danielsen R, Seidman JG, Seidman CE, Gunnarsson GT.. Nationwide study on hypertrophic cardiomyopathy in Iceland: evidence of a MYBPC3 founder mutation. Circulation 2014;130:1158–1167. [DOI] [PubMed] [Google Scholar]
- 18. Alders M, Jongbloed R, Deelen W, van den WA, Doevendans P, Ten CF, Regitz ZV, Vosberg HP, van LI, Wilde A, Dooijes D, Mannens M.. The 2373insG mutation in the MYBPC3 gene is a founder mutation, which accounts for nearly one-fourth of the HCM cases in the Netherlands. Eur Heart J 2003;24:1848–1853. [DOI] [PubMed] [Google Scholar]
- 19. Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K.. A common MYBPC3 (cardiac myosin binding protein-C) variant associated with cardiomyopathies in South Asia. Nat Genet 2009;41:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaffer JF, Kensler RW, Harris SP.. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem 2009;284:12318–12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuster DW, Sadayappan S.. MYBPC3's alternate ending: consequences and therapeutic implications of a highly prevalent 25 bp deletion mutation. Pflugers Arch 2014;466:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barefield D, Sadayappan S.. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol 2010;48:866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C, Watkins H.. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res 2009;105:219–222. [DOI] [PubMed] [Google Scholar]
- 24. Michels M, Soliman OI, Phefferkorn J, Hoedemaekers YM, Kofflard MJ, Dooijes D, Majoor-Krakauer D, Ten Cate FJ.. Disease penetrance and risk stratification for sudden cardiac death in asymptomatic hypertrophic cardiomyopathy mutation carriers. Eur Heart J 2009;30:2593–2596. [DOI] [PubMed] [Google Scholar]
- 25. Pezzoli L, Sana ME, Ferrazzi P, Iascone M.. A new mutational mechanism for hypertrophic cardiomyopathy. Gene 2012;507:165–169. [DOI] [PubMed] [Google Scholar]
- 26. Barefield D, Kumar M, Gorham J, Seidman JG, Seidman CE, de Tombe PP, Sadayappan S.. Haploinsufficiency of MYBPC3 exacerbates the development of hypertrophic cardiomyopathy in heterozygous mice. J Mol Cell Cardiol 2015;79:234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spudich JA. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J 2014;106:1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J.. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation 2009;119:1473–1483. [DOI] [PubMed] [Google Scholar]
- 29. Page SP, Kounas S, Syrris P, Christiansen M, Frank-Hansen R, Andersen PS, Elliott PM, McKenna WJ.. Cardiac myosin binding protein-C mutations in families with hypertrophic cardiomyopathy: disease expression in relation to age, gender, and long term outcome. Circ Cardiovasc Genet 2012;5:156–166. [DOI] [PubMed] [Google Scholar]
- 30. Blankenburg R, Hackert K, Wurster S, Deenen R, Seidman JG, Seidman CE, Lohse MJ, Schmitt JP.. Beta-Myosin heavy chain variant Val606Met causes very mild hypertrophic cardiomyopathy in mice, but exacerbates HCM phenotypes in mice carrying other HCM mutations. Circ Res 2014;115:227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tardiff JC, Carrier L, Bers DM, Poggesi C, Ferrantini C, Coppini R, Maier LS, Ashrafian H, Huke S, van der Velden J.. Targets for therapy in sarcomeric cardiomyopathies. Cardiovasc Res 2015;105:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Velden J, Ho CY, Tardiff JC, Olivotto I, Knollmann BC, Carrier L.. Research priorities in sarcomeric cardiomyopathies. Cardiovasc Res 2015;105:449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakthivel S, Waldmuller S, Saadi AV, Joseph PK, Rakesh PG, Padmakumar R, Tharakan JM, Richard P, Schwartz K, Rajamanickam C, Vosberg HP.. Novel mutations in MYH7 and MYBPC3 of an Indian family causing hypertrophic cardiomyopathy. J Mol Cell Cardiol 2001;33:A105 (abstract). [Google Scholar]
- 34. Waldmuller S, Sakthivel S, Saadi AV, Selignow C, Rakesh PG, Golubenko M, Joseph PK, Padmakumar R, Richard P, Schwartz K, Tharakan JM, Rajamanickam C, Vosberg HP.. Novel deletions in MYH7 and MYBPC3 identified in Indian families with familial hypertrophic cardiomyopathy. J Mol Cell Cardiol 2003;35:623–636. [DOI] [PubMed] [Google Scholar]
- 35. Simonson TS, Zhang Y, Huff CD, Xing J, Watkins WS, Witherspoon DJ, Woodward SR, Jorde LB.. Limited distribution of a cardiomyopathy-associated variant in India. Ann Hum Genet 2010;74:184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Srivastava A, Garg N, Mittal T, Khanna R, Gupta S, Seth PK, Mittal B.. Association of 25 bp deletion in MYBPC3 gene with left ventricle dysfunction in coronary artery disease patients. PLoS One 2011;6:e24123.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuster DW, Govindan S, Springer TI, Martin JL, Finley NL, Sadayappan S.. A hypertrophic cardiomyopathy-associated MYBPC3 mutation common in populations of South Asian descent causes contractile dysfunction. J Biol Chem 2015;290:5855–5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viswanathan SK, Puckelwartz MJ, Mehta A, Ramachandra CJA, Jagadeesan A, Fritsche-Danielson R, Bhat RV, Wong P, Kandoi S, Schwanekamp JA, Kuffel G, Pesce LL, Zilliox MJ, Durai UNB, Verma RS, Molokie RE, Suresh DP, Khoury PR, Thomas A, Sanagala T, Tang HC, Becker RC, Knoll R, Shim W, McNally EM, Sadayappan S.. Association of cardiomyopathy with MYBPC3 D389V and MYBPC3Δ25bp intronic deletion in South Asian descendants. JAMA Cardiol 2018;3:481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barefield D, Kumar M, de Tombe PP, Sadayappan S.. Contractile dysfunction in a mouse model expressing a heterozygous MYBPC3 mutation associated with hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol 2014;306:H807–H815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lynch TL, Sivaguru M, Velayutham M, Cardounel AJ, Michels M, Barefield D, Govindan S, dos Remedios C, van der Velden J, Sadayappan S.. Oxidative stress in dilated cardiomyopathy caused by MYBPC3 mutation. Oxid Med Cell Longev 2015;2015:424751.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sivaguru M, Fried G, Sivaguru BS, Sivaguru VA, Lu X, Choi KH, Saif MT, Lin B, Sadayappan S.. Cardiac muscle organization revealed in 3-D by imaging whole-mount mouse hearts using two-photon fluorescence and confocal microscopy. Biotechniques 2015;59:295–308. [DOI] [PubMed] [Google Scholar]
- 42. Kumar M, Govindan S, Zhang M, Khairallah RJ, Martin JL, Sadayappan S, de Tombe PP.. Cardiac myosin-binding protein C and troponin-I phosphorylation independently modulate myofilament length-dependent activation. J Biol Chem 2015;290:29241–29249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schneider CA, Rasband WS, Eliceiri KW.. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rundell VL, Manaves V, Martin AF, de Tombe PP.. Impact of beta-myosin heavy chain isoform expression on cross-bridge cycling kinetics. Am J Physiol Heart Circ Physiol 2005;288:H896–H903. [DOI] [PubMed] [Google Scholar]
- 45. Chandra M, Rundell VL, Tardiff JC, Leinwand LA, De Tombe PP, Solaro RJ.. Ca(2+) activation of myofilaments from transgenic mouse hearts expressing R92Q mutant cardiac troponin T. Am J Physiol Heart Circ Physiol 2001;280:H705–H713. [DOI] [PubMed] [Google Scholar]
- 46. Sumandea MP, Pyle WG, Kobayashi T, de Tombe PP, Solaro RJ.. Identification of a functionally critical protein kinase C phosphorylation residue of cardiac troponin T. J Biol Chem 2003;278:35135–35144. [DOI] [PubMed] [Google Scholar]
- 47. Schlossarek S, Mearini G, Carrier L.. Cardiac myosin-binding protein C in hypertrophic cardiomyopathy: mechanisms and therapeutic opportunities. J Mol Cell Cardiol 2011;50:613–620. [DOI] [PubMed] [Google Scholar]
- 48. Moravsky G, Ofek E, Rakowski H, Butany J, Williams L, Ralph-Edwards A, Wintersperger BJ, Crean A.. Myocardial fibrosis in hypertrophic cardiomyopathy: accurate reflection of histopathological findings by CMR. JACC Cardiovasc Imaging 2013;6:587–596. [DOI] [PubMed] [Google Scholar]
- 49. Lynch TLt, Ismahil MA, Jegga AG, Zilliox MJ, Troidl C, Prabhu SD, Sadayappan S.. Cardiac inflammation in genetic dilated cardiomyopathy caused by MYBPC3 mutation. J Mol Cell Cardiol 2017;102:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moolman JA, Reith S, Uhl K, Bailey S, Gautel M, Jeschke B, Fischer C, Ochs J, McKenna WJ, Klues H, Vosberg HP.. A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation 2000;101:1396–1402. [DOI] [PubMed] [Google Scholar]
- 51. McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG.. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest 1999;104:1771.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL.. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res 2002;90:594–601. [DOI] [PubMed] [Google Scholar]
- 53. Uppaluri CR. Heart disease and its related risk factors in Asian Indians. Ethn Dis 2002;12:45–53. [PubMed] [Google Scholar]
- 54. Jones DA, Gallagher S, Rathod KS, Redwood S, de Belder MA, Mathur A, Timmis AD, Ludman PF, Townend JN, Wragg A.. Mortality in South Asians and Caucasians after percutaneous coronary intervention in the United Kingdom: an observational cohort study of 279,256 patients from the BCIS (British Cardiovascular Intervention Society) National Database. JACC Cardiovasc Interv 2014;7:362–371. [DOI] [PubMed] [Google Scholar]
- 55. Maron BJ, Kalra A.. Hypertrophic cardiomyopathy in the developing world: focus on India. Eur Heart J 2014;35:2492–2495. [DOI] [PubMed] [Google Scholar]
- 56. Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J.. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res 2005;97:1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, Seidman CE, Seidman JG, Robbins J.. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc Natl Acad Sci USA 2006;103:16918–16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fraysse B, Weinberger F, Bardswell SC, Cuello F, Vignier N, Geertz B, Starbatty J, Kramer E, Coirault C, Eschenhagen T, Kentish JC, Avkiran M, Carrier L.. Increased myofilament Ca2+ sensitivity and diastolic dysfunction as early consequences of Mybpc3 mutation in heterozygous knock-in mice. J Mol Cell Cardiol 2012;52:1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Najafi A, Sequeira V, Helmes M, Bollen IA, Goebel M, Regan JA, Carrier L, Kuster DW, Van Der Velden J.. Selective phosphorylation of PKA targets after beta-adrenergic receptor stimulation impairs myofilament function in Mybpc3-targeted HCM mouse model. Cardiovasc Res 2016;110:200–214. [DOI] [PubMed] [Google Scholar]
- 60. Sequeira V, Wijnker PJ, Nijenkamp LL, Kuster DW, Najafi A, Witjas-Paalberends ER, Regan JA, Boontje N, Ten Cate FJ, Germans T, Carrier L, Sadayappan S, van Slegtenhorst MA, Zaremba R, Foster DB, Murphy AM, Poggesi C, Dos Remedios C, Stienen GJ, Ho CY, Michels M, van der Velden J.. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ Res 2013;112:1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McNamara JW, Li A, Smith NJ, Lal S, Graham RM, Kooiker KB, van Dijk SJ, Remedios CGD, Harris SP, Cooke R.. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J Mol Cell Cardiol 2016;94:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McNamara JW, Li A, Lal S, Bos JM, Harris SP, van der Velden J, Ackerman MJ, Cooke R, Dos Remedios CG.. MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One 2017;12:e0180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Toepfer CN, Wakimoto H, Garfinkel AC, McDonough B, Liao D, Jiang J, Tai AC, Gorham JM, Lunde IG, Lun M, Lynch TL, McNamara JW, Sadayappan S, Redwood CS, Watkins HC, Seidman JG, Seidman CE.. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci Transl Med 2019;11pii:eaat1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schlossarek S, Frey N, Carrier L.. Ubiquitin-proteasome system and hereditary cardiomyopathies. J Mol Cell Cardiol 2014;71:25–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.