Abstract
We investigated potential biosynthetic pathways of long chain alkenols (LCAs), long chain alkyl diols (LCDs), and long chain hydroxy fatty acids (LCHFAs) in Nannochloropsis oceanica and Nannochloropsis gaditana, by combining culturing experiments with genomic and transcriptomic analyses. Incubation of Nannochloropsis spp. in the dark for 1 week led to significant increases in the cellular concentrations of LCAs and LCDs in both species. Consistently, 13C-labelled substrate experiments confirmed that both LCA and LCD were actively produced in the dark from C14–18 fatty acids by either condensation or elongation/hydroxylation, although no enzymatic evidence was found for the former pathway. Nannochloropsis spp. did, however, contain (i) multiple polyketide synthases (PKSs) including one type (PKS-Clade II) that might catalyze incomplete fatty acid elongations leading to the formation of 3-OH-fatty acids, (ii) 3-hydroxyacyl dehydratases (HADs), which can possibly form Δ2/Δ3 monounsaturated fatty acids, and (iii) fatty acid elongases (FAEs) that could elongate 3-OH-fatty acids and Δ2/Δ3 monounsaturated fatty acids to longer products. The enzymes responsible for reduction of the long chain fatty acids to LCDs and LCAs are, however, unclear. A putative wax ester synthase/acyl coenzyme A (acyl-CoA): diacylglycerol acyltransferase is likely to be involved in the esterification of LCAs and LCDs in the cell wall. Our data thus provide useful insights in predicting the biosynthetic pathways of LCAs and LCDs in phytoplankton suggesting a key role of FAE and PKS enzymes.
Keywords: Alkenols, Bioproduct, Diols, Hydroxylated fatty acids, Nannochloropsis, Polyketide synthase
Introduction
Since phytoplankton do not require clean water for their growth and can encompass high levels of biomass productivities per area compared with terrestrial plants (Chisti 2007), microalgal mass culturing could contribute to the sustainable production of chemical products of interest for the biotechnological industry. Nannochloropsis species (Eustigmatophyceae) are considered among the most suitable candidates for biofuel development because of their high growth rate and lipid content with respect to other phytoplankters (Rodolfi et�al. 2009). Specifically, both free and ester-bound fatty acids from Nannochloropsis spp. are currently considered as potential candidates for biodiesel production (Chen et�al. 2012). The genomes of several Nannochloropsis species have been sequenced allowing the identification of major metabolic pathways for lipid biosynthesis (Radakovits et�al. 2012, Vieler et�al. 2012, Corteggiani Carpinelli et�al. 2014, Wang et�al. 2014). Transcriptomic analyses of Nannochloropsis cultures have contributed to the identification of the genes potentially involved in different lipid pathways including the biosynthesis of polyunsaturated fatty acids (PUFAs) (Vieler et�al. 2012) and triacylglycerols (Radakovits et�al. 2012, Li et�al. 2014).
Besides regular fatty acids and PUFAs, Eustigmatophyceae also produce long chain hydroxy fatty acids (LCHFAs), with an alkyl chain of 28–32 carbon and a hydroxyl group at a mid-chain position (Volkman et�al. 1992, Gelin et�al. 1997a). Plant hydroxy fatty acids, such as the 12-OH C18:1 from Ricinus communis and 14-OH C20:1 from Physaria fendleri were previously shown to act as lubricants when added to reference diesel fuel (Goodrum and Geller 2005). This suggests that the quality of Nannochloropsis oils for biodiesel development might also be improved if tiny amounts of LCHFAs (≤1%) are present in the lipid extract to be used for methanol transesterification. LCHFAs possess a combustion enthalpy slightly higher than that of C14–18 fatty acids (Table�1) and thus such addition would not affect the energy yield. Furthermore, hydroxylated aliphatic compounds are also under investigation for polymer development (Sharma and Kundu 2006, Mutlu and Meier 2010), hence their diversity and biosynthetic pathways have been partially elucidated (Buschhaus et�al. 2013, Busta and Jetter 2018, Li et�al. 2018).
Table 1.
Combustion enthalpies of the main fatty acids present in Nannochloropsis species
| Compound | Reaction | Combustion enthalpy | |
|---|---|---|---|
| KJ�mol−1 | KJ�g−1 | ||
| C14:0 FA | C14:0 FA + 20 O2 → 14 H2O + 14 CO2 | 8,300 | 36.0 |
| C16:1 FA | C16:1 FA + 22.5 O2 → 15 H2O +16 CO2 | 9,400 | 37.0 |
| C16:0 FA | C16:0 FA + 23 O2→ 16 H2O + 16 CO2 | 9,500 | 37.0 |
| C18:1 FA | C18:1 FA + 25.5 O2 → 17 H2O + 18 CO2 | 11,160 | 39.5 |
| C18:0 FA | C18:0 FA + 26 O2 → 18 H2O + 18 CO2 | 10,800 | 38.0 |
| C20:5 FA | C20:5 FA + 26.5 O2 → 15 H2O + 20 CO2 | 11,400 | 38.0 |
| C30:0 OH-FA | C30:0 OH-FA+ 43.5 O2 → 30 H2O + 30 CO2 | 18,000 | 39.0 |
| C32:0 OH-FA | C32:2 OH-FA + 46.5 O2 → 32 H2O + 32 CO2 | 19,200 | 39.0 |
Nannochloropsis spp. produce two other classes of hydroxylated compounds related to LCHFAs, in which the terminal carboxylic group is replaced with an alcohol group, i.e. long chain alkyl diols (LCDs) and long chain alkenols (LCAs). LCAs differ from LCDs because of an intermediate double bond instead of the secondary alcohol group. Similarly to bifunctional aliphatic compounds from plants, LCDs might also attract the interest of the polymer industry. For example, polyricinoleate diol, prepared from 12-OH-C18 fatty acid (ricinoleic acid) was tested for the synthesis of polyurethane, revealing faster degradation times than petrochemical polyurethanes (Petrovic et�al. 2010). Polyurethane synthesis requires highly hydroxylated compounds as starters and there is a common interest in screening natural products with a high number of hydroxyl groups (Petrovic 2008). LCDs from Nannochloropsis might thus be of interest for the polymer industry. However, it is crucial to identify the culturing conditions affecting the cellular concentrations of LCHFAs, LCAs, and LCDs as well as their biosynthetic pathways.
While the total lipid content of microalgae typically increases during the stationary phase of their growth (Dunstan et�al. 1993) as well as under high salinity (Martinez-Roldan et�al. 2014) or nitrogen deprivation (Pal et�al. 2011), such culture manipulations do not increase the cellular concentration of LCAs, LCDs, and LCHFAs significantly (Balzano et�al. 2017), suggesting that these compounds are unlikely to serve as storage lipids. Instead, their decrease under hydrogen peroxide-driven oxidative stress suggests a protective role for LCAs, LCDs, and LCHFAs in Nannochloropsis cells (Balzano et�al. 2017). Finally, LCAs and LCDs are thought to occur in the outer layer of the Nannochloropsis cell wall (Gelin et�al. 1997b, Scholz et�al. 2014, Zhang and Volkman 2017, Volkman 2018) as part of a polymer, termed algaenan.
Since LCAs, LCDs, and LCHFAs are structurally related among each other in terms of carbon number and position of the functional groups, common biosynthetic pathways have been long hypothesized for Nannochloropsis species (Volkman et�al. 1992, Versteegh et�al. 1997, Gelin et�al. 1997a). LCHFAs were suggested to originate from the elongation or condensation of C14–18 fatty acids (Gelin et�al. 1997a, Scholz et�al. 2014) and this was confirmed by the positive correlation recently found between the cellular concentrations of C14–16 fatty acids and two LCHFAs (13-hydroxy C30:0 and 15-hydroxy C32:0 fatty acids), respectively, in three Nannochloropsis spp. (Balzano et�al. 2017). However, the biosynthetic pathways of LCHFAs, LCAs, and LCDs are not fully understood and the enzymes potentially involved in the process are unknown.
Here, (i) we analyzed the genomes from different Nannochloropsis spp. to identify the enzymes potentially involved in the biosynthesis of LCAs and LCDs, (ii) searched for culturing conditions promoting the accumulation of LCAs, LCDs and LCHFAs in Nannochloropsis oceanica and Nannochloropsis gaditana, and (iii) performed transcriptomic analyses to identify genes potentially involved in their biosynthesis.
Results and Discussion
Dark incubation enhances LCA and LCD concentrations
To identify potential genes for LCA, LCD, and LCHFA synthesis, we first investigated conditions which stimulated the production of these compounds. A previous study showed that manipulations that typically promote the accumulation of storage lipids, such as nitrogen deprivation (<1 μM nitrate), exposure to high light irradiance (300 μE�m−2�s−1), and culturing at high (50 g�kg−1) salinity, did not increase the cellular concentrations of LCAs, LCDs and LCHFAs significantly (Balzano et�al. 2017). Unfavorable environmental conditions in the marine water column, such as prolonged exposure to dark conditions, are known to trigger the formation of resting stages in phytoplankton (McQuoid and Hobson 1996) for cell protection purposes. Resting forms of Nannochloropsis limnetica exhibit a thicker cell wall compared with active cells (Fietz et�al. 2005), and might thus contain higher amounts of LCAs and LCDs. We therefore attempted to enhance the production of these lipids by incubating living cultures of N. oceanica and N. gaditana in the dark for 1 week.
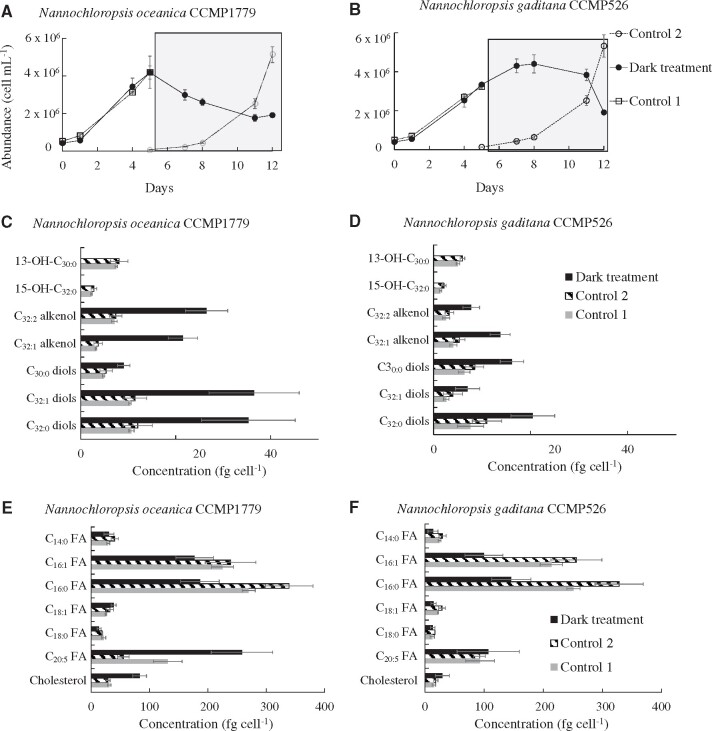
Before the incubation in the dark, cells from both N. oceanica and N. gaditana exhibited growth rates (Fig.�1A, B) comparable to those reported previously (Balzano et�al. 2017). Cell abundance decreased by ca. 50% in both species over 1 week under dark conditions. Cells were observed under transmission electron microscopy and the outer cell wall, which contains LCAs and LCDs (Scholz et�al. 2014), was extremely thin (approximately 10 nm) for the resolution of the instrument used and its thickness could not be measured reliably (Supplementary Fig. S1).
Fig. 1.
Growth curves depicting the cell abundance of N. oceanica CCMP1779 (A) and N. gaditana CCMP526 (B) during the experiments. The shaded areas correspond to the time interval during which the cultures were incubated under dark conditions. Cellular levels of LCAs, LCDs, LCHFAs (C, D), and major fatty acids (E, F) for N. oceanica CCMP1779 (C, E) and N. Gaditana CCMP526 (D, F).
Both N. oceanica and N. gaditana exhibited increased cellular concentrations of LCAs and LCDs after dark incubation. Cells were enumerated by flow cytometry before filtration and we did not observe dead cells or large debris (i.e. particles with comparable forward scatter but lower chlorophyll fluorescence compared with ordinary Nannochloropsis cells) in the cytograms of cells harvested from the dark treatment. This suggests that most of the material filtered contained viable cells, or at least dead cells with intact chloroplasts, while debris were likely to be smaller in size (i.e. with a forward scatter comparable to the background noise of the instrument) and were not retained by the filters. In spite of the significant decline in viable cells under dark conditions the LCAs and LCDs analyzed here are thus likely derived from intact cells. The cellular concentration of C32:1 alkenols increased by almost one order of magnitude (3.5 � 0.5 to 22 � 3 fg�cell−1), while C32:1 and C32:0 diols tripled in concentration reaching 26 � 4, 37 � 9 and 35 � 10 fg�cell−1, respectively (Fig.�1C), and the C30:0 diol nearly doubled in N. oceanica at the end of the dark incubation. Similarly, in N. gaditana, C32:2 and C32:1 alkenols increased from 2.9 � 0.8 to 7.8 � 1.7 fg�cell−1 and from 4.7 � 1.0 to 14 � 2.0 fg�cell−1, respectively, the C30:0 diols doubled (7.4 � 1.4 to 16 � 2.5 fg�cell−1) and the C32:1 and C32:0 diols almost tripled (3.3 � 1.3 to 7.0 � 2.5 and 9.3 � 2.9 to 20 � 4, respectively, Fig.�1D) in concentration. In contrast with LCAs and LCDs, the concentration of LCHFAs dropped dramatically, with the 13-OH-C30:0 fatty acid decreasing from 7.8 � 1.0 and 5.5 � 0.4 fg�cell−1 for N. oceanica and N. gaditana, respectively, to values below the detection limit. Similarly, the 15-OH-C32:0 fatty acid decreased from 2.5 � 0.4 and 1.7 � 0.4 fg�cell−1, for N. oceanica and N. gaditana, respectively, to below the detection limit (Fig.�1C, D). Furthermore, the concentration of C16:0 fatty acid decreased by nearly half for both species (Fig.�1E, F). In contrast, the other C14–20 fatty acids followed different dynamics with the concentration of C16:1 fatty acid decreasing under dark conditions and that of the C20:5 PUFA increasing for N. gaditana, whereas no significant changes were observed in N. oceanica (Fig.�1C, D). The decrease in C16:0 fatty acid under dark conditions is likely due to the consumption of storage lipids necessary to sustain cell metabolism. Storage lipids such as triacylglycerols are typically dominated by the C16:0 fatty acid in Nannochloropsis spp. (Alboresi et�al. 2016).
The incubation under dark conditions for 1 week thus promoted a substantial increase in the cellular concentrations of LCAs and LCDs (Fig.�1C, D), which is the first culture condition ever described shown to trigger an increase in the LCAs and LCDs content in Nannochloropsis spp. Seemingly, prolonged light deprivation affects the biosynthetic pathways of LCHFAs, LCAs and LCDs resulting in the complete removal of LCHFAs and an accumulation of both LCAs and LCDs. Since dark conditions are thought to promote the formation of resting stages, which can result in thicker cell walls as shown for N. limnetica (Fietz et�al. 2005), the dramatic decline in LCHFAs under dark conditions strongly suggests that these lipids are unlikely to be present in the cell wall, but rather form the precursors of LCAs and LCDs. In turn, the LCHFAs might derive from C14–18 fatty acids (Volkman et�al. 1992, Gelin et�al. 1997b, Balzano et�al. 2017).
Further clues were obtained by determination of the double-bond positions of unsaturated LCAs and LCDs in a replicate from the dark treatment of N. oceanica CCMP1779 using dimethyl disulfide derivatization. Consistent with previous findings (Gelin et�al. 1997b), the double bond in LCAs occurs at the same position as that of the mid-chain alcohol group in the corresponding LCDs and LCHFAs (Supplementary Fig. S2). For example, the C32:1 alkenol mostly consists of two isomers with double bonds at Δ14 and Δ15 which correspond to the position of the mid-chain alcohol group in the 15-OH C32:0 fatty acid and the C32:0 1,15-diol. Moreover, the C32:2 alkenol has a second double bond at the same position (Δ27) as that of the C32:1 diol (Supplementary Fig. S2). The excellent correspondence between the double-bond position of monounsaturated alkenols and the position of the intermediate hydroxyl group in diols and LCHFAs with the same carbon number, along with the presence of a double bond at Δ27 in both diunsaturated alkenols and monounsaturated diols, clearly indicates common biosynthetic pathways for LCAs, LCDs and LCHFAs (Supplementary Fig. S2).
13C-labelling indicates active biosynthesis of LCAs and LCDs in the dark
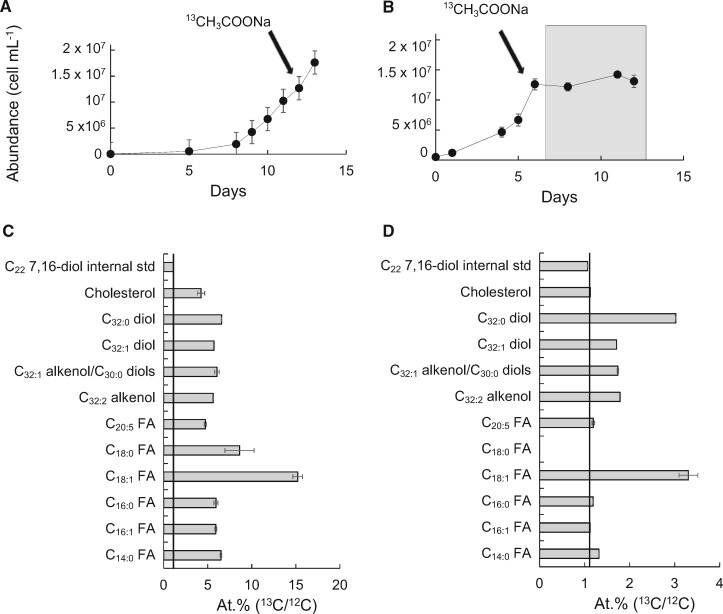
We incubated N. oceanica CCMP1779 with 13C-[2]-acetate under alternating 12/12 dark/light conditions to investigate the biosynthetic relationships among C14–18 fatty acids, LCHFAs, LCAs, and LCDs (Fig.�2A). All these lipids were significantly labelled with 13C (Fig.�2C), with atomic 13C percentages ranging from 4.2% (cholesterol) to 15.2% (C18:1 fatty acid). In a second experiment, cells were initially cultured under alternating 12/12 dark/light conditions, and during exponential growth, 13C-[2]-acetate was then added to the culture which was subsequently incubated under dark conditions for 1 week (Fig.�2B). As expected, the enrichment levels of lipids observed under dark conditions were significantly lower than those found under dark/light conditions due to the absence of growth. The C16:1 fatty acid showed no incorporation of 13C and the C14:0, C16:0, and the C20:5 were only slightly labelled (1.3%, 1.2% and 1.2%, respectively). However, the C18:1 fatty acid (3.2%), the C32:0 diols (3.0%) and to a lesser extent the other LCDs and the LCAs (1.7–1.8%) showed substantial incorporation of 13C label (Fig.�2D). Thus, our labelling experiments show that during regular growth under alternating dark/light conditions, the 13C-[2]-acetate was taken for de novo synthesis of C16:0 fatty acids as well as for the formation of LCHFAs, LCAs, and LCDs. In contrast, when labelled sodium acetate was supplied prior to incubation in the dark, de novo fatty acid synthesis was likely to be insignificant as the cell growth in the dark was nearly negligible (Fig.�2B); however, the lack of detection of C18:0 fatty acid along with the high atomic 13C percentage measured for the C18:1 fatty acid (Fig.�2D) suggests that an active synthesis of C18:1, probably via C16:0 elongation to C18:0 followed by desaturation to C18:1, was taking place. Importantly, both LCAs and LCDs were actively synthesized under dark conditions. The greater 13C content of LCAs and LCDs compared with C14–16 under dark conditions suggests that if the biosynthesis of LCD and LCA took place by condensation of two C14–16 fatty acids, such process would have rapidly taken up all 13C-labelled C14–16 fatty acids. Alternatively, and perhaps more likely, unlabelled C14–16 fatty acids were elongated with 13C-labelled sodium acetate to LCHFAs and subsequent reduction may have resulted in the formation of 13C-labelled LCAs and LCDs (Fig.�2D).
Fig. 2.
Incorporation of 13C-labelled sodium acetate in the biomass of N. oceanica CCMP1779 under dark/light (A, C), and dark (B, D) conditions. Growth curves (A, B) depicting the cell abundance of the culture during the experiment. The grey area denotes the time interval of dark incubation whereas arrows indicate the supply of 13C-labelled sodium acetate. Atomic 13C percentage (of 13C+12C) (C, D) measured by GC-IR-MS for selected lipids at the end of the experiment. Error bars represent the standard deviation calculated from three replicate measurements on the lipid extract (note that some error bars are too small to be visible in the graph). The straight line indicates the natural atomic 13C percentage.
Transcriptomic analyses and hypothetical biosynthetic pathway
To determine which genes were upregulated in the dark incubation experiments, and thus potentially involved in LCD and LCA biosynthesis, we extracted RNA and sequenced the transcriptomes of Nannochloropsis cultures harvested at the end of the experiments. We compared the gene expression level of Nannochloropsis spp. from the dark treatment (i.e. treatment leading to high concentrations of LCAs and LCDs) with the dark/light control. Overall we mapped 10,043 genes from N. oceanica CCMP1779 against the reference genome from the same strain (Vieler et�al. 2012) and 11,222 genes from N. gaditana CCMP526 (Table�2) against the reference genome from the strain N. gaditana B31 (Corteggiani Carpinelli et�al. 2014). In addition, we also carried out a functional analyses of all the proteins predicted from the genomes of N. oceanica CCMP1779 and N. gaditana B31 using Interproscan (Jones et�al. 2014) to identify putative catalytic domains for the elongation and hydroxylation of fatty acids, the dehydration of secondary alcohols, the reduction of their carboxylic groups, and the formation of ethers and esters.
Table 2.
Overview of the transcriptomic analyses of the strains analyzed in the present study
| N. gaditana CCMP526 | N. oceanica CCMP1779 | |
|---|---|---|
| Mapped genes | 10,043 | 11,222 |
| Upregulateda | 1,950 | 955 |
| Highly upregulatedb | 440 | 292 |
| Downregulateda | 2,067 | 1,855 |
| Highly downregulatedb | 612 | 1,133 |
| Not significant | 6,026 | 8,412 |
Gene expression of the dark treatment was compared with that of the dark/light controls.
A gene is considered upregulated or downregulated if its expression level changes by at least 2-fold in the dark treatment compared with the dark/light control and the change is associated with an FDR corrected P-value <0.01.
Number of downregulated and upregulated genes exhibiting an expression change of at least 8-fold.
About 60% of the genes from N. oceanica and 74% of the genes from N. gaditana did not change significantly in expression during the dark incubation (Table�2). Nannochloropsis oceanica exhibited a comparable number of upregulated and downregulated genes (1,950 and 2,067, respectively) whereas 1,855 genes from N. gaditana were downregulated and only 955 upregulated (Table�2). The expression changes of all the genes from both Nannochloropsis species are shown in detail in Supplementary Table S1. The results of our dark incubation experiments with and without stable isotope labelling (Figs.�1, 2), as well as previous findings (Volkman et�al. 1992, Gelin et�al. 1997a, Balzano et�al. 2017, Volkman 2018), suggest that LCHFAs originate from C14–18 fatty acids either by condensation of two fatty acids or elongation/in-chain hydroxylation. Subsequently LCHFAs are likely to be reduced to form LCDs. Similarly, LCAs might derive from the dehydration of the secondary alcohol groups in LCDs or LCHFAs.
We thus focused on genes potentially coding enzymes that catalyze (i) the condensation of two C14–18 fatty acids, (ii) the elongation and (iii) the in-chain hydroxylation of fatty acids ≥ C16, (iv) the reduction of fatty acids to alcohols, the (v) dehydration of secondary alcohol groups, and (vi) the formation of esters. We searched for these genes in Nannochloropsis genomes and compared their expression levels in the dark treatment with those found for the control treatment. We also searched publicly available genomes from other Nannochloropsis spp. to identify genes homologous to those potentially involved in the biosynthetic processes hypothesized here for N. gaditana and N. oceanica.
Condensation of two C14–18 fatty acids
The condensation of two fatty acids to form longer products has been rarely reported in literature. γ-Proteobacteria from the genera Xanthomonas and Photorabdus can perform head-to-head condensation of fatty acids mediated by oleA and photopyrone synthase enzymes, respectively (Kresovic et�al. 2015, Christenson et�al. 2017). Similarity analyses of oleA and photopyrone synthase sequences against N. oceanica and N. gaditana proteins did not yield significant results (Supplementary Table S2) suggesting Nannochloropsis spp. do not contain oleA or photopyrone homologs. Moreover, a head-to-head condensation would produce a mid-chain functionalized intermediate which would still require an additional ω-functionalization to yield a primary/secondary aliphatic compound such as LCDs, LCAs, or LCHFAs.
Acidobacteria are known to produce a C30 13,16-dimethyl dicarboxylic acid from a tail-to-tail condensation of two C15 iso fatty acids (Sinninghe Damst� et�al. 2011) but the enzymes involved in such process are unknown. Furthermore, a tail-to-tail condensation would yield an intermediate functionalized on both ends such as a dicarboxylic acid, and one of these ends would therefore require to be fully reduced to a methyl group. Both head-to-head and tail-to-tail condensations would thus form intermediates which need a further functionalization or reduction step to yield the LHCFAs observed in Nannochloropsis spp. Long chain aliphatic compounds resulting from head-to-head (i.e. mid-chain functionalized only) or tail-to-tail (functionalized on both ends) condensation have never been detected in Nannochloropsis.
The condensation of the carboxylic end of a fatty acid with the aliphatic end of another fatty acid (head-to-tail condensation) would instead require fewer reaction steps and the resulting biosynthetic pathway appears thus to be less energy demanding compared with both head-to-head and tail-to-tail condensations. For example, the reaction between the carboxylic end of a C18 fatty acid with the methyl end of a C14 fatty acid would yield a C32 product functionalized on the first and the 15th carbon such as the 15-OH-C32 fatty acid, the C32 1,15-diol, the Δ15 C32:1 alkenol and the Δ15, 27 C32:2 alkenol. Similarly, the condensation between a C12 and a C18 fatty acids would lead to C30 compounds with a secondary functionalization on the 13th carbon as well as a terminal carboxylic group. Although this pathway cannot be fully discarded, we did not find any evidence reported in literature for such a biosynthetic process.
Enzymes responsible for chain elongation in Nannochloropsis
Fatty acid elongation is based on stepwise additions of two carbon units to the growing acyl coenzyme A (CoA) chain (Leonard et�al. 2004), with each addition consisting of the (i) condensation of the acyl CoA with a malonyl group to form a 3-ketoacyl-CoA, (ii) reduction of 3-ketoacyl-CoA to 3-hydroxyacyl-CoA, (iii) dehydration to enoyl-CoA, and (iv) reduction to an elongated acyl chain (Leonard et�al. 2004). While 3-ketoacyl-CoA synthases (KCS) are typically substrate specific (Leonard et�al. 2004, Haslam and Kunst 2013), the other three enzymes required for the elongation are known to have a broad substrate preference being able to accept 3-ketoacyl, 3-hydroxyacyl, or 3-enoyl units of different lengths. Enzymes belonging to two different families, the elongation proteins (ELO) and the fatty acid elongases (FAEs) possess the KCS domain (Leonard et�al. 2004, Haslam and Kunst 2013). In addition, the polyketide synthases (PKSs) family consists in proteins known to contain ketoacyl-acyl carrier protein (ACP) synthase (KAS) and can also accept C16–18 fatty acids as substrates for elongation (Staunton and Weissman 2001).
Δ0-ELOs as elongators of fatty acids
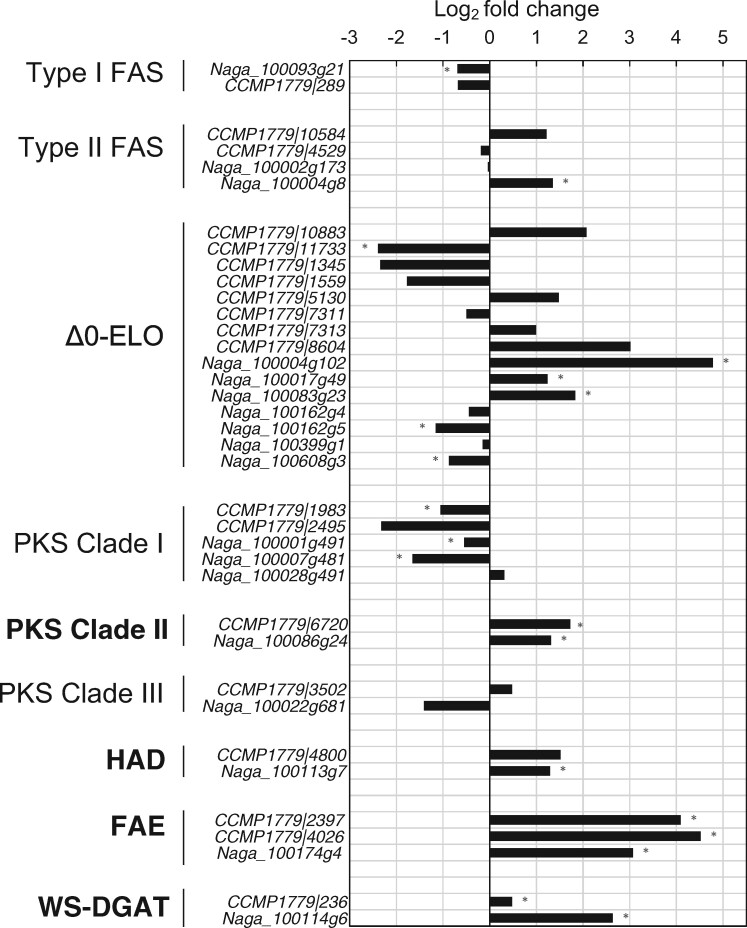
Seven ELOs that accept monounsaturated or saturated fatty acids as substrates (Δ0-ELO) have been previously identified in N. gaditana CCMP526 (Dolch et�al. 2017), and we found eight Δ0-ELOs in N. oceanica by similarity searches. Four Δ0-ELOs from N. gaditana and two Δ0-ELOs from N. oceanica can potentially accept fatty acids containing up to 28 carbons as substrates (Supplementary Data) as predicted by comparing their secondary structure (Supplementary Fig. S3) with that of known Δ0-ELOs from yeasts (Denic and Weissman 2007). Only two Δ0-ELOs from N. gaditana (Naga_100083g23 and Naga_100017g49) were upregulated under dark conditions (Fig.�3). One of these enzymes (Naga_100083g23) has been proven experimentally, by heterologous expression in yeasts, to catalyze the formation of saturated fatty acids containing up to 28 carbons (Dolch et�al. 2017). However, analysis of a mutant of N. gaditana in which the gene coding for the Δ0-ELO Naga_100083g23 has been knocked-out (Dolch et�al. 2017) exhibited a distribution of LCAs, LCDs and LCHFAs very similar to that of the wild type (CCMP526, Supplementary Fig. S4), indicating that Naga_100083g23 is not involved in the biosynthesis of these compounds in N. gaditana. This, along with the lack of upregulated Δ0-ELOs in N. oceanica under dark conditions (Fig.�3), suggests that Δ0-ELOs are not involved in the biosynthesis of LCHFAs in Nannochloropsis spp.. The intermediates required for the biosynthesis of LCAs, LCDs and LCHFAS might thus be formed by other enzymes.
Fig. 3.
Expression level of the genes potentially coding the enzymes catalyzing the different reactions involved in the biosynthesis of saturated C14–18 fatty acids, LCHFAs, LCAs, and LCDs as well as the formation of ester bonds within the cell wall biopolymer. The enzymes most likely involved in processing fatty acids ≥C18 and coded by genes upregulated under dark conditions are in bold. The horizontal axis indicates the log2 fold change in gene expression, between the dark treatment and the two control treatments (Control 1 and Control 2). Significant differences (P < 0.01) are indicated with an asterisk. The prefixes Naga and CCMP1779 denote transcripts from N. gaditana CCMP526 and N. oceanica CCMP1779, respectively. FAS, fatty acid synthase; Δ0-ELO, Saturated fatty acid elongases; PKS, polyketide synthase; HAD, hydroxyacyl-acyl carrier protein-dehydratase; FAE, fatty acid elongation enzyme; WS-DGAT, bifunctional wax ester synthase/diacylglycerol acyltransferase.
FAE enzymes as elongators of fatty acids
FAE enzymes are known to be involved in the biosynthesis of saturated and monounsaturated C20–28 fatty acids in plants (Joubes et�al. 2008, Haslam and Kunst 2013). Nannochloropsis gaditana contains one gene coding for FAE (Naga_100174g4) and we found in N. oceanica two amino acid (AA) sequences (CCMP1779|2397 and CCMP1779|4026) that align with two different regions of the gene product of Naga_100174g4 (Supplementary Fig. S5). The alignment of CCMP1779|2397 and CCMP1779|4026 with Naga_100174g4 as well as the similar expression level exhibited by CCMP1779|2397 and CCMP1779|4026 (increase by 18- to 22-fold in the dark treatment, Fig.�3), strongly suggest that these two AA sequences are two contiguous parts of the same protein. Phylogenetic analyses indicate that FAEs from Nannochloropsis spp. cluster with proteins from diatoms and Pelagophyceae forming a well-supported clade (Supplementary Fig. S6), which groups with known FAEs from higher plants (Joubes et�al. 2008). Nannochloropsis FAEs possess two trans-membrane helices (TMHs), two domains for KCSs, and a domain for chalcone/stilbene synthase (Supplementary Fig. S6).
Interestingly, the genes coding for FAEs in both N. oceanica and N. gaditana are upregulated by >10-fold in the dark treatment (Fig.�3) suggesting an enhanced enzymatic activity of FAEs under dark conditions. FAE enzymes are reported to elongate functionalized fatty acids at an intermediate position and also can accept substrates of variable length including C24–28 fatty acids (Haslam and Kunst 2013). For example, a FAE from the higher plant P. fendleri is known to catalyze the elongation of 12-OH-C18:1 to 14-OH-C20:1 fatty acid (Moon et�al. 2001) and the moss Funaria hygrometrica contains C32:0 1,7-diols, which have been suggested to originate from the elongation of 3-hydroxyacyl intermediates, catalyzed by FAE enzymes (Busta et�al. 2016). Furthermore, the 7-18-(OH)2-C24:1 fatty acid from Orychophragmus violaceus (Brassicaceae) has also been shown to derive from FAE-catalyzed elongation of a 3-OH-intermediate of the 12-OH-C18:1 fatty acid (Li et�al. 2018). Thus, the high expression level of genes coding FAEs in both N. oceanica and N. gaditana, along with the potential enzymatic capability of these proteins to elongate in-chain functionalized fatty acids, suggest that FAE enzymes might play a role in the formation of LCHFAs from C14–20 fatty acids.
Role of PKSs in fatty acid hydroxylation
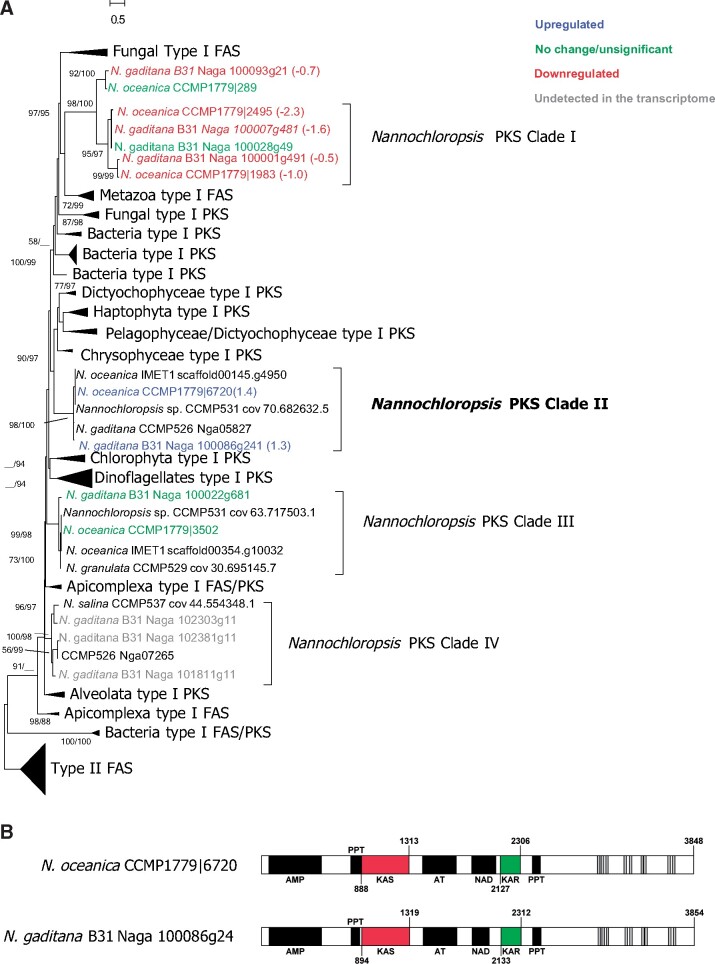
Since FAE enzymes can elongate not only fatty acids but also hydroxy fatty acids, the hydroxylation process required for the formation of mid-chain hydroxyl groups might occur before chain elongation takes place. The formation of both 13-OH-C30:0 and 15-OH-C32:0 fatty acids by FAE-based elongation would then require a 3-OH-C20:0 fatty acid as a starter (Fig.�4). Since 3-OH-C20:0 fatty acid has not been detected in Nannochloropsis spp. as well as other Eustigmatophyceae, it might be an intermediate in the chain elongation. The mid-chain functionalization of fatty acids can be catalyzed by PKS enzymes since they possess acyl transferase (AT) and KAS domains but might lack any or all of the other catalytic sites required to complete a fatty acid elongation cycle (Staunton and Weissman 2001, Jenke-Kodama et�al. 2005). Type I PKSs consist of single multifunctional enzymes possessing several catalytic domains and their distribution is scattered among different lineages since genes coding PKSs have not been found in ciliates and Rhizaria (Shelest et�al. 2015, Kohli et�al. 2016). Three genes from N. oceanica have been previously suggested to code for PKSs (Vieler et�al. 2012, Poliner et�al. 2015, Alboresi et�al. 2016) and two genetically distinct PKS clades were previously detected in N. oceanica and N. gaditana (Shelest et�al. 2015).
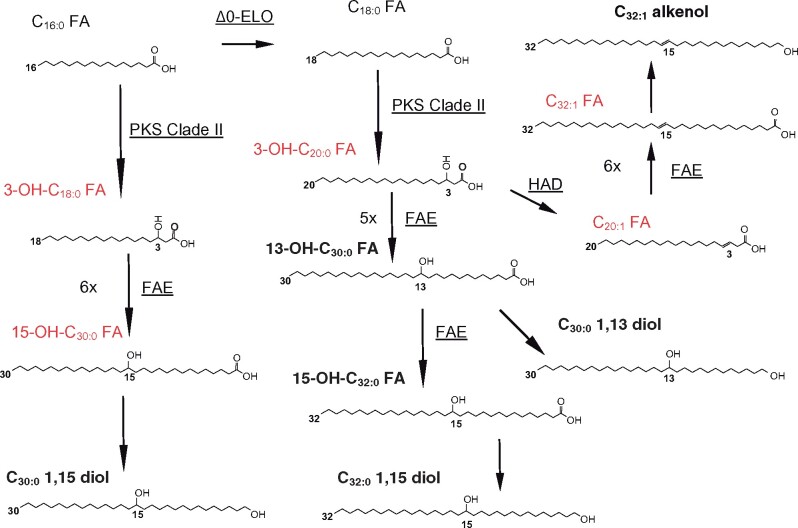
Fig. 4.
Putative pathways for the biosynthesis of LCHFAs, LCAs, and LCDs. For the biosynthetic steps in which the enzymes potentially involved have been predicted, the enzyme name is indicated next to the arrow, underlined. The lipids detected in Nannochloropsis spp. in this study or reported in previous studies are written in bold, whereas those that have not been observed are written in red.
We identified 22 genes coding for PKSs in the different Nannochloropsis spp. (Supplementary Table S3) and built a phylogenetic tree of the KAS domain (KAS-PKS). KAS-PKS phylogeny indicates that five gene products (PKS-Clade I) correspond to the iterative type I PKSs previously identified by Shelest et�al. (2015) and are closely related to two other PKSs from N. gaditana (Naga_100093g21) and N. oceanica (CCMP1779|289), respectively (Fig.�5A). Sequences from PKS-Clade I cluster with type I FAS/PKS from fungi and Metazoa, whereas 15 other gene products show only weak similarities with KAS-PKS from other species and form three distinct clades: PKS-Clade II, PKS-Clade III and PKS-Clade IV (Fig.�5A). Transcriptomic data from the dark incubation experiments of N. oceanica and N. gaditana indicate that the genes coding for PKS-Clade I and PKS-Clade III enzymes were downregulated or did not exhibit significant changes under dark conditions, while those coding for PKS-Clade II were upregulated (Fig.�3), and we did not detect genes coding for PKS-Clade IV in our transcriptomes. PKS-Clade II enzymes CCMP1779|6720 and Naga_100086g4 increased their expression in the dark treatment by 3.2- and 2.5-fold, respectively (Fig.�3) suggesting they can be potentially involved in the hydroxylation of fatty acids.
Fig. 5.
Phylogenetic and functional analyses of Nannochloropsis genes potentially coding for polyketide synthases (PKS) and potentially involved in the formation of 3-OH- intermediates. Phylogeny (A) of the deduced amino acid sequences from the ketoacyl ACP synthase (KAS) domain of the PKSs. Sequences from different Nannochloropsis spp. were compared with a pre-existing alignment from Kohli et al. 2016 and the phylogenetic tree was constructed using the Maximum Likelihood (ML) algorithm by RAxML. Sequences from N. oceanica CCMP1779 and N. gaditana B31 are colored according to their expression levels in the transcriptome and numbers in brackets after the sequences denote the expression levels of these genes in the transcriptome, indicated as log2-fold changes. Node labels indicate the bootstrap support based on ML and Neighbour Joining (NJ) algorithms, respectively; support values <50% are omitted. (B) Structural analyses of two putative PKS from N. oceanica and N. gaditana affiliated to Clade II. The domain structure of all putative PKSs from Nannochloropsis is shown in details in Supplementary Table S1. The domains likely to correspond to the catalytic sites for ketoacyl-acyl carrier protein (ACP)-synthase (KAS) and ketoacyl-ACP-reductase (KAR) are in red, and green, respectively. Transmembrane helices (TMHs) are in grey, other domains in black. AT, acyl transferase; HAD, hydroxyacyl dehydratase; NAD, Nicotinamide adenine dinucleotide-binding domain; ER, enoyl reductase; PPT, phosphopantetheine-binding domain; AMP, adenosine monophosphate-dependent synthetase/ligase.
Interestingly PKS-Clade II enzymes possess domains for PKS-KAS (IPR020841), AT (IPR020801) as well as an adenosine monophosphate (AMP) binding domain (IPR000873), a phosphopantetheine-binding ACP domain (PPT, IPR009081), and a ketoacyl-ACP-reductase (KAR, IPR013968) domain (Fig.�5B). The presence of catalytic domains for both KAS and KAR in PKS-Clade II enzymes and the lack of hydroxyacyl dehydratase (HAD) and enoyl reductase (ER) domains suggest that PKS-Clade II enzymes might catalyze an incomplete fatty acid elongation leading to the formation of 3-OH-fatty acids. C14-18 fatty acids might thus be elongated to form 3-OH-C16–20 fatty acid intermediates by PKS-Clade II enzymes. The incomplete elongation of the C18:0 fatty acid might lead to the formation of a 3-OH-C20:0 intermediate which, after five or six full elongation cycles, potentially catalyzed by the FAE enzymes, would form the 13-OH-C30:0 and 15-OH-C32:0 fatty acids, respectively, the two LCHFAs present in Nannochloropsis spp. (Gelin et�al. 1997a, Balzano et�al. 2017).
Reduction of LCHFAs to LCDs
LCDs and LCAs are likely formed from LCHFAs as evidenced by the depletion of LCHFAs and increase in LCAs and LCDs in the dark incubation experiments (Fig.�1). Furthermore, the presence of C14–24 alkanols (Volkman et�al. 1999) as well as C15–17 alkanes and the C15–31 alkenes (Gelin et�al. 1997b, Sorigue et�al. 2016, Zhou et�al. 2017), also suggests the occurrence of fatty acid reduction activities in Nannochloropsis (spp.). Odd-numbered alkanes and alkenes are typically formed from the reduction of even-numbered fatty acids to aldehydes followed by a decarbonylation step, as described for Arabidopsis thaliana (Bernard et�al. 2012) and Chlamydomonas reinhardii (Sorigue et�al. 2016). Similarly, fatty alcohols are also formed from the reduction of fatty acids catalyzed by alcohol-forming fatty acyl-CoA reductases (FAR) as shown in A. thaliana (Li-Beisson et�al. 2010), Apis mellifera (Teerawanichpan et�al. 2010), Calanus finmarchicus (Teerawanichpan and Qiu 2012), and Euglena gracilis (Teerawanichpan and Qiu 2010). However, we could not find any protein sequence containing the conserved motif [IVF]X[ILV]TGXTGF[MLV][GA] which corresponds to the FAR catalytic site (Hofvander et�al. 2011, Teerawanichpan and Qiu 2012), and none of the Nannochloropsis proteins belong to any FAR family (IPR026055, IPR008670, IPR016836 and IPR003157). Furthermore, similarity searches (blastp) of known FARs against the deduced amino acid sequences of Nannochloropsis genomes produced hits with low (bit score <50) similarity (data not shown). Indeed, a recent study also failed to detect genes coding for FARs in Nannochloropsis genomes (Sorigue et�al. 2016). This indicates that the enzymes involved in fatty acid reduction in Nannochloropsis are either unrelated or greatly divergent from known FARs.
We then searched within the genomes of N. oceanica and N. gaditana for genes coding for domains involved in the reduction of carboxylic acids. We found 44 genes coding for the short chain dehydrogenase/reductase (SDR) and eight genes that can code for the male sterility 2 (MS2) domain (Supplementary Table S4). Proteins with the male sterility (MS2) domain can catalyze the reduction of fatty acids in A. thaliana (Aarts et�al. 1997), and five of these proteins are annotated as PKS-Clade I (Supplementary Table S5, Fig.�5A), since they also possess the catalytic domains for fatty acid elongation. PKS-Clade I enzymes were previously suggested to be involved in the reduction of fatty acids in N. gaditana (Scholz et�al. 2014). Since the genes coding for PKS-Clade I as well as the other genes coding for the MS domain are not upregulated under dark conditions (Fig.�3, Supplementary Table S4) their products are unlikely to be involved in the reduction of LCHFAs to LCDs, although a role in other reduction processes cannot be discarded. Genes coding for SDR were also mostly downregulated under dark condition (Supplementary Table S5); only six of them were upregulated but blastp analyses revealed similarities with proteins from other species with very different functions (Supplementary Table S5).
Thus, although Nannochloropsis spp. contain a range of compounds (LCAs, LCDs, alkanes, alkenes and alkanols) that are very likely to originate from the reduction of fatty acids, we could not find any enzyme potentially involved in these reductive processes.
Δ2- and Δ3-C20:1 fatty acids as potential LCA precursors
LCAs contain a double bond at the same position where LCDs have the hydroxyl group (Supplementary Fig. S2, Gelin et�al. 1997b) suggesting that LCAs might be formed from the dehydration of the mid-chain alcohol group of LCDs. Thus, we searched for dehydratase domains and found 14 genes coding for different lipid dehydratase domains (Supplementary Table S7), and one of them (Naga_100113g71) was upregulated under dark conditions (Supplementary Table S8). Naga_100113g71, and its N. oceanica homolog (CCMP1779|4800) code for proteins possessing a HAD domain and cluster with HADs from other species in our phylogenetic analyses (Supplementary Fig. S7).
Alternatively, the dehydration of the secondary alcohol group may occur at an earlier stage, e.g. as a result from an incomplete fatty acid elongation process in which ER activity is missing, followed by several further elongation processes (Fig.�4). The higher expression levels of Naga_100113g71 under dark conditions (Fig.�3), in spite of a decrease in C14–16 fatty acids (Fig.�1F), suggest that these enzymes might have been catalyzing the dehydration of other compounds such as longer fatty acids. Since HAD enzymes are thought to have a broad substrate specificity (Heath and Rock 1996, Leonard et�al. 2004), potentially catalyzing the dehydration of 3-hydroxyacyl chains of different lengths, they might also accept 3-OH-C20:0 fatty acids as substrates to form the Δ2- and Δ3-C20:1 fatty acid, which, if further elongated and reduced, might lead to the formation of the C30–32 alkenols typically found in Nannochloropsis spp. (Fig.�4).
Incorporation of LCAs and LCDs in Nannochloropsis cell wall biopolymer
The presence of ether- and ester-bound LCAs and LCDs within the cell wall of Nannochloropsis spp. has been long hypothesized (Volkman et�al. 1992, Gelin et�al. 1996, Gelin et�al. 1997b, Volkman 2018). Fourier transform infrared spectroscopy on the cell wall of N. gaditana demonstrated the presence of ether bonds and also found some C=O stretches, but whether the latter are related to carboxylic, aldehyde, ketone or ester functional groups is not clear (Scholz et�al. 2014). Although the core of the cell wall polymer may be ether-bound, as they are resistant against base and acid hydrolysis (Gelin et�al. 1997b), some of the LCAs and LCDs present in Nannochloropsis spp. likely occur as ester-bound moieties to polymeric carboxyl groups (Volkman 2018). The formation of esters from alcohols and fatty acids is typically catalyzed by bifunctional wax ester synthase/acyl coenzyme A (acyl-CoA): diacylglycerol acyltransferase (WS-DGAT) (Kalscheuer and Steinbuchel 2003). A gene coding for WS-DGAT (Naga_100114g61) was previously predicted in N. gaditana (Cui et�al. 2018), and our phylogenetic analyses (Supplementary Fig. S8) indicate that the proteins encoded by Naga_100114g61 and its N. oceanica homolog (CCMP1779|236) are closely related to a WS-DGAT from Phaeodactylum tricornutum (PtWS-DGAT). PtWS-DGAT has been recently shown to catalyze the formation of esters from alcohols and fatty acids (Cui et�al. 2018) and has a domain structure (Fig.�1 in Cui et�al. 2018) similar to that found here for Nannochloropsis WS-DGATs (Naga_100114g61 and CCMP1779|236, Supplementary Fig. S8). Interestingly, putative WS-DGAT from N. gaditana and N. oceanica increased in expression by 6- and 1.5-fold, respectively (Fig.�3), under dark conditions, suggesting that an active production of esters was likely to take place during dark incubation. Thus, WS-DGATs in Nannochloropsis spp. might be involved in esterification of LCAs/LCDs to carboxyl groups to form the ester-bound structures which have been previously detected in the cell wall (Scholz et�al. 2014).
In contrast, we could not find any gene potentially catalyzing the formation of ether bonds within the cell wall biopolymers. Similarity (blastp) analyses of known ether synthases such as the 9-divinyl ether synthase from Solanum lycopersicum (tomato plant), the corvol ether synthase from Kisatasospora setae (bacterium), against the predicted proteins of N. oceanica CCMP1779 and N. gaditana B31 did not yield significant hits (Supplementary Table S9). The polymerization of LCAs and LCDs to form ether-bound structures in algaenans remains thus unclear.
Potential biosynthetic pathways for LCA, LCD and LCHFAs in Nannochloropsis spp.
As previously suggested (Gelin et�al. 1997a, Scholz et�al. 2014, Balzano et�al. 2017, Volkman 2018), our results from stable isotope experiments confirm that LCAs and LCDs derive from C14–18 fatty acids by either condensation or elongation.
If biosynthesis occurs via condensation the dominant pathway involves the interaction between the aliphatic end of a C14 fatty acid with the carboxylic end of a C18 fatty acid to produce a C32 compound functionalized on the 15th carbon which would be a precursor of the 15-OH-C32 fatty acid, the C32 1,15-diol and the C32 alkenols. Although we could not find evidence for such a pathway in literature nor could find genes potentially coding such biosynthetic processes within Nannochloropsis genomes, this pathway cannot be fully discarded.
In contrast, the elongation pathway is more likely to occur. The lack of elongation intermediates such as aliphatic compounds with a number of carbons comprised between 20 and 26 in Nannochloropsis spp. might be due to a rapid uptake of such compounds for the following steps of the pathway. Nevertheless, results from our genomic and transcriptomic analyses, combined with comparisons with biosynthetic pathways in plants, more likely suggest that LCHFAs are formed from C14–18 fatty acids via elongation. Specifically we found two key enzymes, PKS-Clade II and FAE, potentially involved in the elongation process. PKS-Clade II are likely to elongate and hydroxylate the C18:0 fatty acid and, to a lesser extent, the C16:0 fatty acid, to form the 3-OH-C20:0 and 3-OH-C18:0 fatty acids, respectively (Fig.�4). Subsequently, FAE enzymes can potentially catalyze the multiple elongation of the 3-OH-C20:0 fatty acid to 13-OH-C30:0 and 15-OH-C32:0 fatty acids after five or six complete elongation cycles, respectively (Fig.�4). Six complete elongation cycles of 3-OH-C18:0 fatty acid and subsequent reduction might form the C30:0 1,15-diol which is also present in Nannochloropsis spp. as well as other eustigmatophycean representatives (Rampen et�al. 2014).
Since saturated LCDs are functionalized at the same mid-chain position as their corresponding monounsaturated LCAs (Supplementary Fig. S2), both lipid classes are very likely to share a similar biosynthetic pathway and to originate from the same precursors, the 3-OH-C20:0 fatty acid and, to a lesser extent, the 3-OH-C18:0 fatty acid. The pathway leading to the formation of LCAs would start from the dehydration of 3-OH-C20:0 fatty acids to both Δ2 and Δ3 C20:1 fatty acid catalyzed by an HADs (Fig.�4). The Δ2 and Δ3C20:1 fatty acids would then undergo five complete FAE-catalyzed elongations and a reduction to form the Δ12 and Δ13 C30:1 alkenols or six elongations and a reduction to form Δ14 and Δ15 C32:1 alkenols (Fig.�4). The Δ12 and Δ13 C30:1 alkenols as well as Δ14 and Δ15 C32:1 alkenols have been detected here in both N. gaditana and N. oceanica (Supplementary Fig. S2) and were also found previously in Nannochloropsis salina and an unidentified Nannochloropsis strain (Gelin et�al. 1997b). The Δ14 and Δ15 C30:1 alkenols would instead derive from the dehydration of 3-OH-C18:0 fatty acid to Δ2 and Δ3 C18:1 fatty acids followed by six complete elongation cycles and the reduction of the carboxylic group to alcohol.
The formation of a double bond in LCDs and a second double bond in LCAs would originate at an early stage of the pathway, before the 3-OH C20:0 fatty acid is either elongated to form LCHFAs, or dehydrated to form LCA precursors. A double bond on a Δ27 position, for both the C32:2 alkenol and the C32:1 diol, might originate from a desaturation of the 13th carbon in C18:0 fatty acid or a desaturation of the 15th carbon in 3-OH-C20:1 fatty acid. The formation of Δ13 C18:1 or a Δ15 3-OH-C20:1 would potentially involve the activity of a stereospecific desaturase such as a Δ13stearoyl desaturase. Nannochloropsis spp. contains 29 proteins with domains for fatty acid desaturation and some of them are upregulated under dark conditions (Supplementary Table S9), it is unclear whether any of these enzymes exhibits Δ13stearoyl desaturase activity.
LCD production in other species
LCDs can also be produced by other phytoplankters (Sinninghe Damst� et�al. 2003, Rampen et al. 2011) as well as some plants (Buschhaus et�al. 2013). To evaluate the presence of FAEs and PKSs in LCD-producers other than Eustigmatophyceae we analyzed the proteins predicted from genomes or transcriptomes available to date. The diatom Proboscia alata can code for three putative PKSs as well as a FAE (Supplementary Fig. S9). Similarly to PKS-Clade II enzymes from Nannochloropsis spp. (Fig.�5B), PKSs from P. alata possess both KAS and KAR domains (Supplementary Fig. S9) being thus potentially able to catalyze the formation of hydroxylated products. Indeed, Proboscia species contain C28–30 1,14-diols and 12-OH C27–29 methyl alkanoates which were previously suggested to be formed from 12-OH-C26-28 fatty acids (Sinninghe Damst� et�al. 2003). The 12-OH-C26–28 fatty acids might originate after five full elongation cycles of 2-OH-C16–18 fatty acids, which would in turn derive from an incomplete elongation (and thus hydroxylation) of C14–16 fatty acids, with FAEs catalyzing the former reaction and PKSs the latter. The hydroxylation of C14–16 fatty acids should thus occur, in this case, on the second, rather than on the third carbon to eventually produce the LCD detected in Proboscia spp..
C26–32 aliphatic diols with a primary and a secondary alcohol group can also be present in the epicuticular waxes of aquatic ferns (Speelman et�al. 2009, Mao et�al. 2017), terrestrial ferns (Jetter and Riederer 1999) as well as other land plants such as mosses (Busta et�al. 2016), conifers (Wen and Jetter 2007) and flowering plants (Wen et�al. 2006, Racovita and Jetter 2016). Similarly to the biosynthetic pathways proposed here for Nannochloropsis spp., LCDs from plants could start with the formation of 3-hydroxyacyl compounds mediated by P450 hydroxylases or PKS enzymes, followed by FAE-catalyzed elongation of 3-hydroxyacyl intermediates as suggested for plants (Wen and Jetter 2007, Busta et�al. 2016).
Conclusions
LCHFAs are likely to originate from C14–18 fatty acids after either condensation of C14–18 fatty acids or an incomplete fatty acid elongation, forming 3-OH-fatty acids, followed by a further elongation. Enzymes potentially involved in the condensation of two fatty acids are not known to date. We identified instead two enzymes (PKS-Clade II and FAE) likely to be involved in the elongation of C14–18 fatty acids to LCHFAs which are then likely to be reduced to LCDs. HAD enzymes might play a role in the dehydration of secondary alcohols before, during, or after the elongation of 3-OH-FAs, forming the double bonds present in LCAs whereas WS-DGAT enzymes are potentially involved in the formation of the ester-bound structures present in the Nannochloropsis cell wall. Although the biosynthetic pathways for LCAs and LCDs have not been fully elucidated and the formation of ether bonds within cell wall polymers is still unclear, our work identifies a potential mechanism, similar to biosynthetic processes described in higher plants, for the formation of mid-chain functionalized aliphatic compounds in phytoplankton. Future challenges include the biochemical and functional characterization of the candidate enzymes predicted here. Eventually, if long chain aliphatic compounds are formed from the elongation of C14–18 fatty acids, genetic manipulations of PKS-Clade II and FAE enzymes might contribute to increase the productivity of both LCHFAs and LCDs in Nannochloropsis species.
Materials and Methods
Culturing and dark incubations
Nannochloropsis oceanica CCMP1779 and N. gaditana CCMP526 were cultured in batch using f/2 medium (Guillard 1975) under 12:12 dark/light conditions at 20�C and algal growth was regularly monitored using flow cytometry (Marie et�al. 1999). For the experiments each strain was grown in six replicate 1.5 L Erlenmeyer flasks (Supplementary Data) and cells were harvested from three flasks to assess the initial concentration of lipids and the background gene expression (Control 1). From each of the remaining six flasks (three per species) an aliquot (20 mL equals ∼7 � 107 cells) was transferred into new Erlenmeyer flasks prefilled with medium, incubated under dark/light conditions, and used as positive control (Control 2), whereas the initial flasks with the remaining volume (780 mL) were instead transferred under dark conditions (dark treatment). Both Control 2 and dark treatment were incubated for 1 week at 20�C. Cells were harvested from their culturing flasks by filtration through 0.7 μm GF/F filters (Whatman, Maidstone, UK). Cells were enumerated by flow cytometry before filtration and we did not observe dead cells or debris (i.e. particles with comparable forward scatter and lower chlorophyll fluorescence than ordinary Nannochloropsis cells) in our cytograms in any of the samples filtered. This suggests that most of the material filtered contained viable cells, or at worst dead cells with intact chloroplasts. Filters for lipid analyses were immediately rinsed in demineralized water and stored at −80�C, whereas filters for RNA extraction and further transcriptomic analyses were flash frozen in liquid nitrogen immediately and then stored at −80�C until analyses.
In addition, we analyzed the composition of LCAs, LCDs, and LCHFAs in mutant strains of N. gaditana CCMP526 in which the gene Naga_100083g23 coding for a Δ0-ELOs has been silenced (Dolch et�al. 2017). Three mutants (Clone 5, Clone 13, and Clone 15) were obtained from the Cell and Plant Physiology Laboratory (Grenoble, France), cultured under the same conditions as above (f/2 medium, 12/12 dark light cycle) along with the wild type (N. gaditana CCMP526) and harvested during the exponential phase of their growth.
Stable isotope labelling
To assess whether LCDs are formed from C14–16 fatty acids, we incubated N. oceanica CCMP1779 with sodium 13C-[2]-acetate (Sigma-Aldrich, 279315–1 G, Zwijndrecht, Netherlands) under both dark/light and dark conditions. We used an axenic culture of N. oceanica CCMP1779 to avoid any consumption of 13C-labelled acetate by heterotrophic bacteria typically present in phytoplankton cultures. The strain was cultured at 20�C under 12:12 dark/light conditions in a 5 L glass carboy and continuous air bubbling was provided by an aquarium pump connected through 0.2 �m pore size filters (Sartorius, G�ttingen, Germany) to dilute any formed 13C-labelled CO2. A volume of 10 mL of 0.5 M sodium 13C-[2]-acetate was added to the cultures when they reached cellular densities of approximately 1.5 � 107 cell mL−1. The dark/light treatment was then incubated at the same conditions for 2 d, whereas the dark treatment was incubated for 1 week in the dark. At the end of the experiments cells were harvested as described above.
Lipid extraction, gas chromatography-mass spectrometry and isotope ratio mass spectrometer
For the nonisotopically labelled dark incubation experiment, we extracted 18 filters in total, i.e. three replicates per treatment for two strains (N. oceanica CCMP1779 and N. gaditana CCMP526). The filters were freeze-dried using a Lyoquest (Telstart, Life Sciences) freeze-drier and then they were saponified and acid hydrolyzed as described previously (Rodrigo-G�miz et�al. 2015, Balzano et�al. 2017). The total lipid extract (TLE) was then dissolved in dichloromethane (DCM) and dried through anhydrous sodium acetate, and subsequently under nitrogen. After extraction, we added 20.4 �g C22:0 7, 16-diol as internal standard to the TLE, for quantification purposes. Subsequently, the extracts were methylated with diazomethane and cleaned over a small silica gel column using ethyl acetate as eluent. Prior to analysis, the TLEs were silylated by the addition of N,O-bis-(trimethylsilyl)-trifluoroacetamide (BSTFA) and pyridine, and heating at 60�C for 20 min.
An aliquot of one lipid extract from N. oceanica CCMP1779 (dark treatment) was used to determine the position of the double bond in alkenols and unsaturated LCDs, lipids were derivatized by adding 50 μL dimethyl disulfide and 5 μL iodine solution (60 mg iodine in 1 mL diethyl ether), eluted then in DCM and iodine was removed using 50–100 mL 5% sodium thiosulfate solutions.
Compounds were quantified using gas chromatography (GC) flame ionization detection using an Agilent 7890B GC (Agilent Technologies, Amstelveen, Netherlands) with a 25 m fused silica column diameter 0.32 mm, coated with CP Sil-5 (thickness 0.12 �m). Identification of the lipids was achieved using GC-mass spectrometry (GC-MS) with an Agilent 7890B GC coupled to an Agilent 5977 mass spectrometer (Agilent). Identification of the LCDs, LCAs, and LCHFAs was based on the fragmentation mass spectra obtained in full scan (m/z 50–800) as described by Versteegh et�al. (1997). To discriminate between C30:0 diols and the C32:2 alkenols, which co-elute on the GC, we determined the relative contributions of each compound to the total peak area in the GC-MS chromatogram (MassHunter software, Agilent) in which the two compounds are partially separated.
For the stable isotope experiment, we analyzed two filters corresponding to the two treatments of N. oceanica. The extraction set-up consisted in saponification followed by acid hydrolysis as described above. Subsequently, we added 19.2 �g C22:0 7,16-diol as internal standard to the extracts and we methylated the TLEs using boron trifluoride in methanol. The extracts were separated by column chromatography, using aluminum oxide as stationary phase, which was activated for 2 h at 150�C. Lipids were then extracted in three different solutions: hexane: DCM (9:1, v/v), hexane: DCM (1:1, v/v), and methanol: DCM (1:1, v/v). Fatty acids were mostly dissolved in the second hexane: DCM solution whereas LCAs, LCDs, and LCHFAs were dissolved in the methanol: DCM solutions. For the silylation and the methylation we used BSTFA and BF3/methanol, respectively, with known δ13C values of −32.2‰ and −25.7‰, respectively. Compounds were quantified by GC-Flame Ionization Detector (FID) chromatograms as described above, whereas identification was achieved using an Agilent 7890 A GC coupled to an Agilent 5975C MS. All GC-MS parameters were also identical, but the total run time was 60 min. The isotopic composition of the different compounds was analyzed using GC-isotope ratio mass spectrometry, using an Agilent 6890 GC coupled to a Thermo Delta Plus isotope ratio mass spectrometer (IR-MS, ThermoFisher, Landsmeer, Netherlands). Separation was achieved on a ZB-5MS column with a length of 60 m, a column diameter of 0.32 mm and a film thickness of 0.25 �m. Oven program was identical as that described for the GC-FID and GC-MS, but the end temperature of 320�C was held for 30 min (total run time of 80 min). The injection volume was 1 �L, the four fractions were all analyzed in triplicate, and the reported data represent averaged values. The isotopic compositions are reported in units of atom percent (At%). Values were corrected considering the δ13C values of both BSTFA and methanol.
Genomic analyses
To identify genes potentially involved in the biosynthesis of LCAs, LCDs, and LCHFAs in Eustigmatophyceae, we constructed a local blast database (Altschul et�al. 1990, Camacho et�al. 2009) using the predicted proteins from the genomes of eight Nannochloropsis strains (Nannochloropsis sp. CCMP531, N. gaditana B-31, N. gaditana CCMP526, Nannochloropsis granulata CCMP529, N. oceanica CCMP1779, N. oceanica IMET1, Nannochloropsis oculata CCMP525, and N. salina CCMP537, Supplementary Data). Conserved protein domains were searched using Interproscan (Jones et�al. 2014) or by manually enquiring for specific AA motifs. Specifically, we searched for proteins containing the motifs HWYHH, GMGCSAG and [D/E]TACSSS or H[G/A]TGT, which correspond to highly conserved regions of Δ0-ELOs (Hashimoto et�al. 2008), FAEs (Millar et�al. 1999) and PKSs (Shelest et�al. 2015) enzymes, respectively. Moreover we searched for genes potentially coding for the conserved motif [I/V/F]X[I/L/V]TGXTGF[M/L/V][G/A] which corresponds to the catalytic site of FARs (Hofvander et�al. 2011, Teerawanichpan and Qiu 2012). The presence and position of TMHs in Δ0-ELOs, FAEs, and PKSs proteins was assessed using TMHMM (Krogh et�al. 2001).
We carried out similarity searches of known proteins from other species against the locally built Nannochloropsis database as well as similarity searches of Nannochloropsis proteins potentially involved in the biosynthetic processes against the nonredundant (NR) (Pruitt et�al. 2005) and the Swissprot (Bateman et al. 2017) databases, using blastp (Camacho et�al. 2009).
To compare putative Nannochloropsis enzymes involved in the biosynthetic processes with known and unknown proteins from other species, we performed phylogenetic analyses on four protein families: FAEs, PKSs, the HADs and the WS-DGATs. We aligned putative Nannochloropsis FAEs with known FAE proteins from A. thaliana, Brassica napus (Joubes et�al. 2008) as well as a FAE known to elongate hydroxy fatty acids from P. fendleri (Moon et�al. 2001). Sequences were aligned using MAFFT-linsy (Katoh and Standley 2013) and poorly aligned regions (regions containing >50% gaps) were trimmed from the alignment which finally consisted of 50 sequences and 195 unambiguously aligned positions. We analyzed the KAS domain of PKSs (KAS-PKS) using Nannochloropsis proteins previously identified as PKSs (Shelest et�al. 2015, Alboresi et�al. 2016) as well as other proteins containing the KAS-PKS domain (IPR020841) and/or containing the conserved motifs [D/E]TACSSS and H[G/A]TGT. Sequences were then aligned to a pre-existing alignment of 92 KAS-PKS sequences (Kohli et�al. 2016) and trimmed as described above. The final alignment consisted of 138 sequences and 173 AA positions. For HADs, we extracted two AA sequences containing a domain for HAD (IPR010084) from N. oceanica and N. gaditana, respectively. We then searched for homologs in the Nannochloropsis and Swissprot databases, aligned and trimmed the sequences as described above and the final alignment included 50 AA sequences and 147 positions. For the WS-DGAT phylogeny, we downloaded known AA sequences from the Swissprot database, searched for homologs in Nannochloropsis, aligned the sequences and trimmed the alignment as described above. The alignment included 43 AA sequences and 245 positions. Phylogenetic trees were constructed using both maximum likelihood (ML) and neighbor joining (NJ) algorithms based on 1,000 bootstraps. ML phylogeny was inferred using RAxML with 1,000 bootstraps (Stamatakis 2014) and was used to build the phylogenetic trees, whereas NJ bootstrap support values were calculated using MEGA (Tamura et�al. 2007).
To evaluate the occurrence of similar biosynthetic processes in other LCD-producers (Balzano et�al. 2018), we downloaded a number of predicted proteins, obtained from transcriptomes of phytoplankton cultures (Keeling et�al. 2014) from iMicrobe (https://www.imicrobe.us/). The species used were Florenciella parvula (MMETSP1323), Florenciella sp. (MMETSP1324), Heterosigma akashiwo (MMETSP0292, MMETSP0294, MMETSP0295, MMETSP0296, MMETSP0409, MMETSP0410, MMETSP0411, MMETSP0414, MMETSP0415, MMETSP0416, MMETSP0894, MMETSP0895, MMETSP0896 and MMETSP0897), Phaeomonas parva (MMETSP1163), Florenciella parvula (MMETSP1323), Florenciella sp. (MMET SP1324), P. alata (MMETSP0174, MMETSP0176), Proboscia inermis (MMETSP0816) and Sarcinochrysi sp. (MMETSP1170). In addition, we downloaded the genomes of the plants Azolla filliculoides (www.fernbase.org) and Triticum aestivum (Kersey et�al. 2018) which are also known to produce LCDs (Speelman et�al. 2009, Racovita and Jetter 2016). We then analyzed the domain structure of these proteins using Interproscan (Jones et�al. 2014) and searched for PKSs coding KAS and KAR domains.
RNA extraction
To prevent RNA degradation, the extractions were carried out under sterile and cold (∼10�C) conditions in a clean laboratory; samples, tubing, and all other equipment used were kept in ice unless otherwise stated. RNA was extracted from each of three replicates of each of the three treatments (Control 1, Control 2, and dark treatment) from both N. oceanica CCMP1779 and N. gaditana CCMP526 for a total of 18 samples. Cells were disrupted using a combination of thermal, chemical, and mechanical lyses: from each sample about half of a GF/F filter was cut in many small pieces using sterile tweezers and scissors, drilled using disposable pellet pestles, and then transferred into 12 mL falcon tubes prefilled with 0.1 and 0.5 �m glass beads (Biospec, Bartlesville, Canada). Tubes were then rapidly submerged several times into liquid nitrogen to promote thermal cell lysis. One milliliter of RLT buffer (Qiagen, Venlo, Netherlands), 10 �L mercaptoethanol (Sigma-Aldrich), and 50 �L plant RNA isolation aid (Thermo Fisher Scientific) were then added to the tubes which were vortexed for 5 min, incubated for 5 min in ice, vortexed again for 5 min and finally centrifuged at 4,500�g. The supernatant was transferred into 2 mL tubes which were centrifuged again at 16,000�g and the supernatant removed. A solution of 35 �L lysozyme (Qiagen), 20 �L proteinase-K (Qiagen) and 100 �L sodium-dodecyl-sulfate (Ambion, Bleiswijk, Netherlands) were then added to the samples which were incubated at 37�C for 10 min. Tubes were then centrifuged for 15 min and the supernatant transferred into DNA spin column (DNAeasy blood and tissue kit, Qiagen) and centrifuged to remove most of the DNA. The lysate was transferred into RNAeasy spin columns (RNAeasy mini-kit, Qiagen) and the RNA was then isolated following the instructions provided by the supplier. Traces of DNA were removed from the RNA extract using Turbo DNAse (Thermo Fisher Scientific). RNA concentration and integrity were assessed using Qubit Fluorometric Quantitation (Thermo Fisher Scientific) and a Bioanalyzer (Agilent, Santa Cruz, USA) whereas a PCR using universal eukaryote primers (Stoeck et�al. 2010) was carried out to confirm the absence of DNA contamination within the RNA extracts.
RNA extracts were sent to Utrecht Sequencing Facility (www.useq.nl), where cDNA was generated, sequencing libraries prepared and sequencing carried out with two runs on a NextSeq500 with reads of 75 bp.
Gene expression analyses
A total of 614,537,691 raw fastq reads were obtained and processed locally on a bioinformatic cluster. Low-quality reads were trimmed or removed using Trimmomatic (Bolger et�al. 2014) with the maxinfo method (MAXINFO:40:0.6) and a minimal length of acceptable reads of 36 bp. Adapters were also removed using Trimmomatic with the Illuminaclip option. The quality of the trimmed reads was controlled with fastqc (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and reads were sorted and counted using the R library Rsamtools (Morgan et�al. 2017). Reads were mapped against the previously sequenced genomes of N. oceanica CCMP1779 (Vieler et�al. 2012) available at Joint Genome Institute (https://genome.jgi.doe.gov/pages/search-for-genes.jsf? organism=Nanoce1779) and N. gaditana B-31 (Corteggiani Carpinelli et�al. 2014) downloaded from www.nannochloropsis.org. Mapping was performed using the R library GenomicFeatures (Lawrence et�al. 2013), data were normalized using Deseq2 (Love et�al. 2014) and a gene expression table as well as log2-fold changes with corresponding P-values corrected with the false discovery rate (Benjamini and Hochberg 1995) was obtained. Expression changes were considered significant for P-values <0.01.
Supplementary Material
Acknowledgments
We thank M. Verweij for help in lipid analyses, M. Grego for support in algal culturing, S. Vreugdenhil for help in RNA extraction, A. Abdala for assistance in bioinformatic analyses and R. Langelaan from Naturalis for help in electron microscopy. We thank Utrecht Sequencing Facility (USEQ) for providing sequencing service and data. USEQ is subsidized by the University Medical Center Utrecht, Hubrecht Institute and Utrecht University.
Funding
This research was funded by the European Research Council (ERC) under the European Union’s Seventh Framework Program (FP7/2007–2013) ERC grant agreement [339206]. Financial support from the Netherlands Earth System Science Centre (NESSC) is provided to S.S. and J.S.S.D. The French National Research Agency supports J.L. and E.M. (ANR-10-LABEX-04; ANR-11-BTBR-0008; ANR-17-EURE-0003).
Disclosures
The authors have no conflicts of interest to declare.
References
- Aarts M.G.M., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., et al. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623. [DOI] [PubMed] [Google Scholar]
- Alboresi A., Perin G., Vitulo N., Diretto G., Block M., Jouhet J., et al. (2016) Light remodels lipid biosynthesis in Nannochloropsis gaditana by modulating carbon partitioning between organelles. Plant Physiol. 171: 2468–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Balzano S., Lattaud J., Villanueva L., Rampen S.W., Brussaard C.P.D., van Bleijswijk J., et al. (2018) A quest for the biological sources of long chain alkyl diols in the western tropical North Atlantic Ocean. Biogeosciences 15: 5951–5968. [Google Scholar]
- Balzano S., Villanueva L., de Bar M., Sinninghe Damst� J.S., Schouten S. (2017) Impact of culturing conditions on the abundance and composition of long chain alkyl diols in species of the genus Nannochloropsis. Org. Geochem. 108: 9–17. [Google Scholar]
- Bateman A., Martin M.J., O'Donovan C., Magrane M., Alpi E., Antunes R., et al. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45: D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate. A pratical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 57: 289–300. [Google Scholar]
- Bernard A., Domergue F., Pascal S., Jetter R., Renne C., Faure J.D., et al. (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhaus C., Peng C., Jetter R. (2013) Very-long-chain 1, 2-and 1, 3-bifunctional compounds from the cuticular wax of Cosmos bipinnatus petals. Phytochemistry 91: 249–256. [DOI] [PubMed] [Google Scholar]
- Busta L., Budke J.M., Jetter R. (2016) Identification of beta-hydroxy fatty acid esters and primary, secondary-alkanediol esters in cuticular waxes of the moss Funaria hygrometrica. Phytochemistry 121: 38–49. [DOI] [PubMed] [Google Scholar]
- Busta L., Jetter R. (2018) Moving beyond the ubiquitous: the diversity and biosynthesis of specialty compounds in plant cuticular waxes. Phytochem. Rev. 17: 1275–1304. [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009) BLAST plus: architecture and applications. BMC Bioinformatics 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu T.Z., Zhang W., Chen X.L., Wang J.F. (2012) Biodiesel production from algae oil high in free fatty acids by two-step catalytic conversion. Biores. Technol. 111: 208–214. [DOI] [PubMed] [Google Scholar]
- Chisti Y. (2007) Biodiesel from microalgae. Biotechnol. Adv. 25: 294–306. [DOI] [PubMed] [Google Scholar]
- Christenson J.K., Jensen M.R., Goblirsch B.R., Mohamed F., Zhang W., Wilmot C.M., et al. (2017) Active multienzyme assemblies for long-chain olefinic hydrocarbon biosynthesis. J. Bacteriol. 199: e00890–e008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corteggiani Carpinelli E., Telatin A., Vitulo N., Forcato C., D’Angelo M., Schiavon R., et al. (2014) Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant 7: 323–335. [DOI] [PubMed] [Google Scholar]
- Cui Y.L., Zhao J.L., Wang Y.C., Qin S., Lu Y.D. (2018) Characterization and engineering of a dual-function diacylglycerol acyltransferase in the oleaginous marine diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Weissman J.S. (2007) A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130: 663–677. [DOI] [PubMed] [Google Scholar]
- Dolch L.J., Rak C., Perin G., Tourcier G., Broughton R., Leterrier M., et al. (2017) A palmitic acid elongase affects eicosapentaenoic acid and plastidial monogalactosyldiacylglycerol levels in Nannochloropsis. Plant Physiol. 173: 742–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan G.A., Volkman J.K., Barrett S.M., Garland C.D. (1993) Changes in the lipid composition and maximization of the polyunsaturated fatty acid content of 3 microalgae grown in mass culture. J. Appl. Phycol. 5: 71–83. [Google Scholar]
- Fietz S., Bleiss W., Hepperle D., Koppitz H., Krienitz L., Nicklisch A. (2005) First record of Nannochloropsis limnetica (Eustigmatophyceae) in the autotrophic picoplankton from Lake Baikal. J. Phycol. 41: 780–790. [Google Scholar]
- Gelin F., Boogers I., Noordeloos A.A.M., Sinninghe Damst� J.S., Hatcher P.G., de Leeuw J.W. (1996) Novel, resistant microalgal polyethers: an important sink of organic carbon in the marine environment? Geochim. Cosmochim. Acta 60: 1275–1280. [Google Scholar]
- Gelin F., Boogers I., Noordeloos A.A.M., Sinninghe Damst� J.S., Riegman R., De Leeuw J.W. (1997a) Resistant biomacromolecules in marine microalgae of the classes Eustigmatophyceae and Chlorophyceae: geochemical implications. Org. Geochem. 26: 659–675. [Google Scholar]
- Gelin F., Volkman J.K., de Leeuw J.W., Sinninghe Damst� J.S. (1997b) Mid-chain hydroxy long-chain fatty acids in microalgae from the genus Nannochloropsis. Phytochemistry 45: 641–646. [Google Scholar]
- Goodrum J.W., Geller D.P. (2005) Influence of fatty acid methyl esters from hydroxylated vegetable oils in diesel fuel lubricity. Biores. Technol. 96: 851–855. [DOI] [PubMed] [Google Scholar]
- Guillard R.R.L. (1975). Culture of phytoplankton for feeding marine invertebrates. InCulture of Marine Invertebrate Animals. Edited by Smith W.L., Chanley M.H. (Eds.). pp. 29–60. Plenum Book Publication Corporation, New York. [Google Scholar]
- Hashimoto K., Yoshizawa A.C., Okuda S., Kuma K., Goto S., Kanehisa M. (2008) The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 49: 183–191. [DOI] [PubMed] [Google Scholar]
- Haslam T.M., Kunst L. (2013) Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210: 93–107. [DOI] [PubMed] [Google Scholar]
- Heath R.J., Rock C.O. (1996) Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271: 27795–27801. [DOI] [PubMed] [Google Scholar]
- Hofvander P., Doan T.T.P., Hamberg M. (2011) A prokaryotic acyl-CoA reductase performing reduction of fatty acyl-CoA to fatty alcohol. FEBS Lett. 585: 3538–3543. [DOI] [PubMed] [Google Scholar]
- Jenke-Kodama H., Sandmann A., M�ller R., Dittmann E. (2005) Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22: 2027–2039. [DOI] [PubMed] [Google Scholar]
- Jetter R., Riederer M. (1999) Long-chain alkanediols, ketoaldehydes, ketoalcohols and ketoalkyl esters in the cuticular waxes of Osmunda regalis fronds. Phytochemistry 52: 907–915. [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W.Z., McAnulla C., et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes J., Raffaele S., Bourdenx B., Garcia C., Laroche-Traineau J., Moreau P., et al. (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67: 547–566. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R., Steinbuchel A. (2003) A novel bifunctional wax ester synthase/acyl-CoA: diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278: 8075–8082. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.J., Burki F., Wilcox H.M., Allam B., Allen E.E., Amaral-Zettler L.A., et al. (2014) The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Bio. 12: e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., Allen J.E., Allot A., Barba M., Boddu S., Bolt B.J., et al. (2018) Ensembl genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 46: D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli G.S., John U., Van Dolah F.M., Murray S.A. (2016) Evolutionary distinctiveness of fatty acid and polyketide synthesis in eukaryotes. ISME J. 10: 1877–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresovic D., Schempp F., Cheikh-Ali Z., Bode H.B. (2015) A novel and widespread class of ketosynthase is responsible for the head-to-head condensation of two acyl moieties in bacterial pyrone biosynthesis. Beilstein J. Org. Chem. 11: 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., et al. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A.E., Pereira S.L., Sprecher H., Huang Y.S. (2004) Elongation of long-chain fatty acids. Prog. Lipid Res. 43: 36–54. [DOI] [PubMed] [Google Scholar]
- Li J., Han D., Wang D., Ning K., Jia J., Wei L., et al. (2014) Choreography of transcriptomes and lipidomes of Nannochloropsis reveals the mechanisms of oil synthesis in microalgae. Plant Cell. 26: 1645–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.J., Teitgen A.M., Shirani A., Ling J., Busta L., Cahoon R.E., et al. (2018) Discontinuous fatty acid elongation yields hydroxylated seed oil with improved function. Nat. Plants 4: 711–720. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y., Shorrosh B., Beisson F., Andersson M.X., Arondel V., Bates P.D., et al. (2010) Acyl-lipid metabolism. Arabidopsis Book 8: e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S.Y., Zhu X.W., Wu N.Y., Jia G.D., Sun Y.G., Guan H.X., Wu D.D. (2017) Alcohol compounds in Azolla imbricata and potential source implication for marine sediments. Sci. China Earth Sci. 60: 348–359. [Google Scholar]
- Marie D., Partensky F., Vaulot D., Brussaard C. (1999) Enumeration of phytoplankton, bacteria, and viruses in marine samples. InCurrent protocols in cytometry. Edited by Robinson P.J. Chapter 11. John Wiley & Sons, New York. [DOI] [PubMed] [Google Scholar]
- Martinez-Roldan A.J., Perales-Vela H.V., Canizares-Villanueva R.O., Torzillo G. (2014) Physiological response of Nannochloropsis sp. to saline stress in laboratory batch cultures. J. Appl. Phycol. 26: 115–121. [Google Scholar]
- McQuoid M.R., Hobson L.A. (1996) Diatom resting stages. J. Phycol. 32: 889–902. [Google Scholar]
- Millar A.A., Clemens S., Zachgo S., Giblin E.M., Taylor D.C., Kunst L. (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H., Smith M.A., Kunst L. (2001) A condensing enzyme from the seeds of Lesquerella fendleri that specifically elongates hydroxy fatty acids. Plant Physiol. 127: 1635–1643. [PMC free article] [PubMed] [Google Scholar]
- Morgan M., Pag�s H., Obenchain V., Hayden N. (2017) Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package version 1.30.0. http://bioconductor.org/packages/release/bioc/html/Rsamtools.html. (September, 2018, date last accessed).
- Mutlu H., Meier M.A.R. (2010) Castor oil as a renewable resource for the chemical industry. Eur. J. Lipid Sci. Technol. 112: 10–30. [Google Scholar]
- Pal D., Khozin-Goldberg I., Cohen Z., Boussiba S. (2011) The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl. Microbiol. Biotechnol. 90: 1429–1441. [DOI] [PubMed] [Google Scholar]
- Petrovic Z.S. (2008) Polyurethanes from vegetable oils. Polymer Revs. 48: 109–155. [Google Scholar]
- Petrovic Z.S., Xu Y.J., Milic J., Glenn G., Klamczynski A. (2010) Biodegradation of thermoplastic polyurethanes from vegetable oils. J. Polym. Environ. 18: 94–97. [Google Scholar]
- Poliner E., Panchy N., Newton L., Wu G.X., Lapinsky A., Bullard B., et al. (2015) Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles. Plant J. 83: 1097–1113. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. (2005) NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33: D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racovita R.C., Jetter R. (2016) Identification of in-chain-functionalized compounds and methyl-branched alkanes in cuticular waxes of Triticum aestivum cv. Bethlehem. PLoS One 11: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R., Jinkerson R.E., Fuerstenberg S.I., Tae H., Settlage R.E., Boore J.L., et al. (2012) Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat. Commun. 3: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampen S.W., Schouten S., Sinninghe Damst� J.S. (2011) Occurrence of long chain 1,14-diols in Apedinella radians. Org. Geochem. 42: 572–574. [Google Scholar]
- Rampen S.W., Willmott V., Kim J.-H., Rodrigo-Gamiz M., Uliana E., Mollenhauer G., et al. (2014) Evaluation of long chain 1, 14-alkyl diols in marine sediments as indicators for upwelling and temperature. Org. Geochem. 76: 39–47. [Google Scholar]
- Rodolfi L., Zittelli G.C., Bassi N., Padovani G., Biondi N., Bonini G., et al. (2009) Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102: 100–112. [DOI] [PubMed] [Google Scholar]
- Rodrigo-G�miz M., Rampen S.W., de Haas H., Baas M., Schouten S., Sinninghe Damst� J.S. (2015) Constraints on the applicability of the organic temperature proxies UK037, TEX86 and LDI in the subpolar region around Iceland. Biogeosciences 12: 6573–6590. [Google Scholar]
- Scholz M.J., Weiss T.L., Jinkerson R.E., Jing J., Roth R., Goodenough U., et al. (2014) Ultrastructure and composition of the Nannochloropsis gaditana cell wall. Eukaryot. Cell 13: 1450–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V., Kundu P.P. (2006) Addition polymers from natural oils—a review. Prog. Polym. Sci. 31: 983–1008. [Google Scholar]
- Shelest E., Heimerl N., Fichtner M., Sasso S. (2015) Multimodular type I polyketide synthases in algae evolve by module duplications and displacement of AT domains in trans. BMC Genomics 16: 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinninghe Damst� J.S., Rampen S., Irene W., Rijpstra C., Abbas B., Muyzer G., et al. (2003) A diatomaceous origin for long-chain diols and mid-chain hydroxy methyl alkanoates widely occurring in Quaternary marine sediments: indicators for high-nutrient conditions. Geochim. Cosmochim. Acta 67: 1339–1348. [Google Scholar]
- Sinninghe Damst� J.S., Rijpstra W.I.C., Hopmans E.C., Weijers J.W.H., Foesel B.U., Overmann J., et al. (2011) 13,16-Dimethyl octacosanedioic acid (iso-diabolic acid), a common membrane-spanning lipid of acidobacteria subdivisions 1 and 3. Appl. Environ. Microbiol. 77: 4147–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorigue D., Legeret B., Cuine S., Morales P., Mirabella B., Guedeney G., et al. (2016) Microalgae synthesize hydrocarbons from long-chain fatty acids via a light-dependent pathway. Plant Physiol. 171: 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speelman E.N., Reichart G.J., de Leeuw J.W., Rijpstra W.I.C., Sinninghe Damst� J.S. (2009) Biomarker lipids of the freshwater fern Azolla and its fossil counterpart from the Eocene Arctic Ocean. Org. Geochem. 40: 628–637. [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staunton J., Weissman K.J. (2001) Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18: 380–416. [DOI] [PubMed] [Google Scholar]
- Stoeck T., Bass D., Nebel M., Christen R., Jones M.D.M., Breiner H.W., et al. (2010) Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19: 21–31. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- Teerawanichpan P., Qiu X. (2010) Fatty Acyl-CoA reductase and wax synthase from Euglena gracilis in the biosynthesis of medium-chain wax esters. Lipids 45: 263–273. [DOI] [PubMed] [Google Scholar]
- Teerawanichpan P., Qiu X. (2012) Molecular and functional analysis of three fatty Acyl-CoA reductases with distinct substrate specificities in copepod Calanus finmarchicus. Mar. Biotechnol. 14: 227–236. [DOI] [PubMed] [Google Scholar]
- Teerawanichpan P., Robertson A.J., Qiu X.A. (2010) A fatty acyl-CoA reductase highly expressed in the head of honey bee (Apis mellifera) involves biosynthesis of a wide range of aliphatic fatty alcohols. Insect Biochem. Mol. Biol. 40: 641–649. [DOI] [PubMed] [Google Scholar]
- Versteegh G.J.M., Bosch H.J., De Leeuw J.W. (1997) Potential palaeoenvironmental information of C24 to C36 mid-chain diols, keto-ols and mid-chain hydroxy fatty acids; a critical review. Org. Geochem. 27: 1–13. [Google Scholar]
- Vieler A.G., Wu C.-H., Tsai B., Bullard A.J., Cornish C., Harvey I.-B., et al. (2012) Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 8: e1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman J.K. (2018). Lipids of geochemical interest in microalgae. InHydrocarbons, Oils and Lipids: Diversity, Origin, Chemistry and Fate. Edited by Wilkes H. pp. 1–34. Springer International Publishing, Cham. [Google Scholar]
- Volkman J.K., Barrett S.M., Blackburn S.I. (1999) Eustigmatophyte microalgae are potential sources of C-29 sterols, C22-C28n-alcohols and C28-C32n-alkyl diols in freshwater environments. Org. Geochem. 30: 307–318. [Google Scholar]
- Volkman J.K., Barrett S.M., Dunstan G.A., Jeffrey S.W. (1992) C30-C32 Alkyl diols and unsaturated alcohols in microalgae of the class Eustigmatophyceae. Org. Geochem. 18: 131–138. [Google Scholar]
- Wang D., Ning K., Li J., Hu J., Han D., Wang H., et al. (2014) Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 10: e1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen M., Au J., Gniwotta F., Jetter R. (2006) Very-long-chain secondary alcohols and alkanediols in cuticular waxes of Pisum sativum leaves. Phytochemistry 67: 2494–2502. [DOI] [PubMed] [Google Scholar]
- Wen M., Jetter R. (2007) Very-long-chain hydroxyaldehydes from the cuticular wax of Taxus baccata needles. Phytochemistry 68: 2563–2569. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Volkman J.K. (2017) Algaenan structure in the microalga Nannochloropsis oculata characterized from stepwise pyrolysis. Org. Geochem. 104: 1–7. [Google Scholar]
- Zhou L., Chen J., Xu J.L., Li Y., Zhou C.X., Yan X.J. (2017) Change of volatile components in six microalgae with different growth phases. J. Sci. Food Agric. 97: 761–769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.