Abstract
The Research Domain Criteria (RDoC) paradigm was launched 10 years ago as a superior approach for investigation of mental illness. RDoC conceptualizes normal human behavior, emotion, and cognition as dimensional, with mental illnesses as dimensional extremes. We suggest that RDoC may have value for understanding normal human psychology and some conditions plausibly construed as extremes of normal variation. By contrast, for the most serious of mental illnesses, including dementia, autism, schizophrenia, and bipolar disorder, we argue that RDoC is conceptually flawed. RDoC conflates variation along dimensional axes of normal function with quantitative measurements of disease phenotypes and with the occurrence of diseases in overlapping clusters or spectra. This moves away from the disease model of major mental illness. Further, RDoC imposes a top-down approach to research. We argue that progress in major mental illness research will be more rapid with a bottom-up approach, starting with the discovery of etiological factors, proceeding to investigation of pathogenic pathways, including use of cell and animal models, and leading to a refined nosology and novel, targeted treatments.
Keywords: Research Domain Criteria, Schizophrenia, Bipolar disorder, Autism, Psychosis, Dimension, Category, Diagnosis, Nosology, Genetics, Gene-environment interaction, National Institute of Mental Health, Spectrum
Introduction
The Research Domain Criteria (RDoC) [1] paradigm was introduced as a new and superior approach for investigation of psychiatric illness as part of the 2008 National Institute of Mental Health (NIMH) strategic plan [2, 3, 4]. The basic concept is to parse human behavior and brain function into neuropsychological “domains,” currently totaling six (“negative valence systems,” “positive valence systems,” “cognitive systems,” “social processes,” “arousal and regulatory systems,” and “sensorimotor”), and their component “constructs” (e.g., within the negative valence system domain: “acute threat,” “potential threat,” “sustained threat,” “loss,” and “frustrative nonreward”). This strategy arose from the argument that an “… important contributing factor to the slow rate of progress [in psychiatric research] is the widespread reliance of research projects on categorical, symptom-based diagnostic systems …” [5], meaning the Diagnostic and Statistical Manual of Mental Disorders (DSM) and the International Classification of Diseases classification schemes.
RDoC characterizes mental illnesses as extremes along prespecified neuropsychological dimensions. It attempts to describe “the full range of variation, from normal to abnormal, among the fundamental components [dimensions] to improve understanding of what is typical versus pathological” [6, 7]. Further, “… disorders are considered in terms of disruptions of the normal-range operation of these systems,” and “often both extremes of a dimension may be considered as ‘abnormal’ - for example, a complete lack of fear may be associated with aggressive or psychopathic behavior, and the opposite end of diminished reward-seeking may be mania” [7]. The RDoC scheme became a preferred mode for NIMH-sponsored research and at times a criterion for gauging fundability of research projects: “RDoC frees clinical investigators from the current diagnostic categories and encourages basic scientists to identify molecular or neural mechanisms of specific domains of a mental function rather than creating models of diseases” [8].
Now, 10 years after its dissemination, it is timely to reassess the intellectual impact of RDoC on psychiatric research. In brief, we contend that conceptualizing psychiatric disorders as extremes of normal variation has merit for some conditions. However, RDoC is not the optimal approach to major mental illnesses, such as dementia, autism, schizophrenia, and bipolar disorder – conditions best conceptualized using the disease model, the powerful paradigm behind much of contemporary medical research.
In this review, we will describe our view of strengths and weakness of the RDoC scheme, in part by comparison with examples from other areas of biomedical research. We suggest that RDoC is inconsistent with some of the most promising contemporary lines of psychiatric research into major mental illnesses, with an emphasis on recent genetic advances that demonstrate the effectiveness of the bottom-up approach to major mental illnesses. We conclude that RDoC should be abandoned as a guide for directing the course of future NIMH-funded research into major mental illnesses, in favor of more straightforward application of the disease model, so successful in many other fields of medicine.
Strengths and Accomplishments of RDoC
Perhaps the greatest potential strength of RDoC is its focus on systematizing normal human mental function and on guiding attempts to associate underlying psychological constructs of cognition and emotion with specific neural circuitry. As an example, circuitry related to the habenula, and associated behavior such as stress-induced anhedonia, has been described in terms of RDoC domains and constructs [9]. The correlation of neuroimaging findings and genetic variation with the initial responsiveness to reward attainment and approach motivation has been described, in RDoC terms, as aspects of positive valence systems [10]. RDoC has informed studies of social cognition [11], analyses of reward processing in relation to mood symptoms [12], conditioned fear learning [13], and some interpretations of working memory [14], to name only a few examples. A second potential strength has been to highlight that many psychiatric disorders have substantial phenotypic overlap, though this perspective was already emerging from genetic studies.
RDoC may be useful for those psychiatric disorders that appear to reflect too much or too little of a normal trait, such as personality disorders [15], certain anxiety disorders, including posttraumatic stress disorder, and perhaps some forms of major depression [16, 17]. For instance, amygdala function associated with fear-related behaviors may help elucidate the pathophysiology of posttraumatic stress disorder [18], and amygdala-insula connectivity has been associated with emotional dysregulation [19], including childhood anxiety [20]. The severity of social anxiety has been correlated with hypervigilance in response to facial expression [21]. It is not yet clear to what extent RDoC provides added value; however, more success may be possible as RDoC refines its domains and constructs and develops measurements for its dimensions [16].
A major strength of the RDoC perspective has been to encourage researchers to include a wider scope of patients in studies, for instance schizophrenia and bipolar disorder or all psychoses together, rather than focusing on individual disorders. However, we believe that this would have happened even in the absence of RDoC, given the many recent genetic studies demonstrating extensive overlap in risk loci.
In our view, the outcome of RDoC-related research to date has been of narrow scope and limited impact [22]. It is notable, for instance, that RDoC has had little or no impact on two of the most successful recent initiatives of the NIMH in relation to major mental illness, the B-SNIP and PsychENCODE consortia [23, 24].
Unfounded Claims of RDoC
Before a more general critique, we would like to refute two claims often made about the value of RDoC. First is the claim that RDoC represents an advance over the approach of the DSM because it – but not the DSM – focuses on underlying neurobiology [2, 3] rather than phenomenology. This is not correct. While the DSM has in the past avoided premature classification based on etiological hypotheses (e.g., those of psychoanalysis), the DSM approach, and particularly DSM-5, clearly focuses on neurobiology in those conditions for which a consensus has been reached that a disease model is appropriate or that neurobiological knowledge is sufficiently advanced to contribute to disease classification. For instance, neurocognitive disorders, including Alzheimer's disease, frontotemporal dementia, Huntington's disease (HD), and others, are defined by etiology, pathology, or pathogenesis. Substance abuse and posttraumatic stress disorder definitions are based on specific etiologies and relevant neurobiology. Our purpose here is not to defend the DSM, which certainly is beset by multiple conceptual, methodological, and scientific flaws (e.g., a lack of nosological hierarchy [25, 26] and long-standing struggles to classify personality disorders [27]), but simply to point out that the claim about its lack of consideration of neurobiology is incorrect.
While RDoC claims to focus on neurobiology and its dimensional manifestations, most RDoC domains and constructs are in fact defined not by neurobiology, but rather by phenomenology and behavior, associated with hypothesized physiology. We have previously suggested [28] that, with this focus, RDoC has formal resemblance to the Galenic humoral theory. Ghaemi previously noted the same resemblance between Galenic reasoning and pre-RDoC psychiatric nosology [25]. All told, it is difficult to make the argument that RDoC offers a conceptual advance over previous nosological strategies.
A second unfounded claim is that RDoC has fostered recent advances in psychiatric neuroscience, or that it has a special claim on approaches to “precision medicine” [2]. RDoC, however, has no monopoly on the use of neurobiological tools to understand cognition, emotion, and behavior, or on individualizing the treatment of disease. Furthermore, the RDoC scheme, as detailed in the NIMH strategic plan [8] and on the RDoC website (www.nimh.nih.gov/research-priorities/rdoc/index.shtml?), focuses on molecules and pathways that regulate normal brain function, rather than those related to disease.
The net effect is that RDoC maintains research focus on previously well-established pathways rather than directing attention to novel neurobiology based on emerging genetic and other risk factors. Specifically, in the current (spring 2019) RDoC matrix, dopamine or serotonin appear 33 times, GABA or glutamate 36 times, and oxytocin or vasopressin 18 times, while there is no mention of the many more recently implicated molecules conferring risk for major mental illness, as described below. We argue that RDoC thus has actually diverted attention away from the new neurobiology that is most relevant to psychiatric disease.
Weaknesses of RDoC: Conceptual Issues
Previous critiques of RDoC have addressed, among other issues, the problematic overall structure of the matrix, the lack of consideration of time course, the lack of consideration of the differential response to psychotropic medicines such as lithium, and the lack of benchmarks to indicate success or failure [29, 30, 31, 32, 33]. Many of these critiques might be addressable, at least in principle, by modifying RDoC. We believe, however, that RDoC is fundamentally flawed. Here we seek to provide a further conceptual view of these issues and to assess the RDoC approach to psychiatric research in contrast to approaches that have led to success in other areas of medicine.
Our underlying conceptual concern with RDoC relates to the nature of severe mental illness. RDoC assumes that all psychiatric disorders result from a deviation along dimensional axes of normal variation – too much or too little of a normal neuropsychological or neurobiological function – and hence that research should emphasize understanding normal variation. By contrast, we begin from the standpoint that syndromic psychiatric illnesses are the result of specific, though complex, pathogenic processes that fundamentally disrupt normal neurobiological function, and hence disrupt normal patterns of cognition, emotion, and behavior.
This is simply a restatement of the disease model of major mental illness – psychiatric diseases [25], like other diseases, have etiologies and pathogenic processes, leading to specific forms of pathology at the level of the cell and tissue, which in turn lead to abnormal pathophysiological states and clinical syndromes [15]. The state of disease is distinctly different from normal pathology and function. While some aspects of etiology, such as polygenetic vulnerability, or of pathology, such as the extent of ventricular enlargement, may have dimensional aspects, these features of the disease do not negate the concept of disease nor detract from the value of the disease model in guiding clinical care and research. Research into major mental illness should therefore focus less on dimensions of normal behavior and more on discovery of disease etiology and pathogenesis. Given this perspective, we believe there are at least four major conceptual flaws in the RDoC system:
(1) The RDoC Matrix
The structure and elements of the RDoC matrix are arbitrary and problematic. The domains appear capricious, jargon-laden, and incomplete. If the goal is to describe normal emotional function, where are universal experiences such as love, hatred, anger, jealousy, and hopefulness? If the goal is to capture clinical problems not well covered in the DSM, where are apathy, irritability, aggression, and anosognosia? If the goal is to supplant existing dimensional schemes, then why is there no systematic set of psychometric instruments measuring the constructs? Further, why are there no rigorous comparisons and cross-validations with the five-factor model or other dimensional systems?
The system is hermetic, with no rigorous path to external validation. There appears to be continued debate on definitions of the constructs [10] and whether they are independent or overlapping or redundant – without an objective route to resolve these debates. Even if the structure were to be changed, it is not clear what criteria would substantiate improvement.
(2) Dimensionality
RDoC's reliance on dimensionality moves away from the disease model. We would make the distinction between “quantitative” and “dimensional” analyses. Many phenomena in medicine are indeed quantitative and complex. However, that does not imply that clinical syndromes are simply extremes on dimensions anchored in normality. Some medical examples may be illustrative.
One simple example is respiratory rate. Normal individuals vary in rate and depth of breathing. However, while respiratory diseases, such as pneumonia, may be manifest in part by changes in rate and depth of breathing, the fundamental problem is not changes in pulmonary function along normal axes, but rather that the quantitative changes in pulmonary function reflect the onset of a qualitatively new condition. The goal of research then becomes discovering and correcting the underlying etiology (e.g., pneumococcus) and pathogenesis (e.g., infection and inflammation), rather than better understanding normal variation in the regulation of breathing.
Fever provides another example. We measure body temperature because fever indicates the onset of a qualitative shift from normal – an infectious, inflammatory, or other pathogenic disease process. The research goal is to elucidate this underlying disease process, not to study the range of normal variation of temperature regulation.
A third example is diabetes. Glucose is a quantitative measure, but diabetes, once established, causes dysfunction by many avenues no longer solely related to elevated glucose.
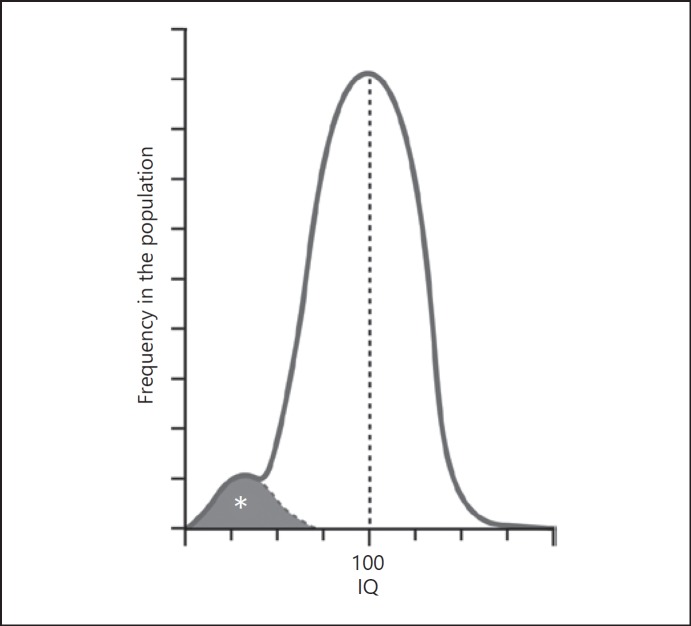
A neuropsychiatric example is intellectual function as measured by IQ. The distribution of IQ in the human population has the characteristic bell-shaped dimensional distribution overall, but at the lower end of the curve there is another small peak (Fig. 1) reflecting syndromal forms of intellectual disability, such as fragile X syndrome, Rett syndrome, Down syndrome, and others. Recognizing the syndromic nature of these disorders, each distinct from normal human variation and each distinct from each other, and in which intellectual disability is but one feature, was the first step in determining that they are caused by discrete genetic abnormalities, which in turn has guided research into pathogenesis and the detection of potential therapeutic targets.
Fig. 1.
Bimodality of the superficially unimodal Gaussian distribution of IQ in the population. In addition to the normal distribution of IQ in the general population, a second distribution, at the very low end, reflects the existence of a separate population of individuals with specific, etiologically defined, syndromes, such as fragile X syndrome, Rett syndrome, Angelman syndrome, Down syndrome (adapted from McHugh and Slavney [15]). Note that we are not claiming that other disorders (such as schizophrenia) have Mendelian origins, but simply that a quantitative trait with a superficially Gaussian “dimensional” distribution can conceal disparate subcategories and etiologies best seen as categorically distinct, and using the disease model. Illustrator: Joan M.K. Tycko.
We are not arguing that psychiatric disorders will turn out to be infectious or Mendelian in etiology, but rather that misinterpreting a quantitative measure as a dimensional axis can falsely lead to viewing phenotypes as simple variations along a dimension, rather than as qualitatively distinct clinical syndromes. In many cases (e.g., respiratory rate or temperature in infection, IQ in fragile X), the quantitative measures may be indirect markers of disease activity, but not fundamental to disease pathogenesis.
These are complex issues, and we do not wish to oversimplify. However, we would argue that it is better for research to start with phenomena present in disease and base quantification on such phenomena, rather than assume that variation of normal phenomena captures the essence of a disease phenotype.
A related issue arises in the nosology of clusters of complex and heterogeneous sets of syndromes. In many cases, based on their underlying etiology and pathobiology, such clusters may be considered a “spectrum” or “continuum” of disease, categorically distinct from normal, but not part of a dimension of normal variation. The key point is that in our view major mental illnesses are best conceptualized as categorically distinct from normal dimensional variation, and most effectively using the disease model.
We would add, however, that even categorically distinct disease entities typically have gradations of severity, just as the symptoms of pneumonia or diabetes may vary from subclinical to mild to severe. In the case of psychiatric disorders with complex etiology, this is manifest as heterogeneity of severity and disability and in subclinical manifestations of illness. Examples include the mild cognitive deficits and other “intermediate phenotypes” observed in relatives of individuals with schizophrenia [34] and the broad autism phenotype observed in family members of probands with strictly defined autism [35]. As in nonpsychiatric disorders, gradations complicate the conceptualization of separate disease categories, and suggest the potential value of investigation of specific dimensional properties underlying categorically defined psychiatric disorders, with the caveat that any one factor is not the disease itself.
A practical consideration arises in the essential actions of a physician caring for a patient – making a diagnosis and choosing a treatment; for instance, the decision about whether a patient with psychosis has bipolar disorder or schizophrenia, and therefore the decision to prescribe, or not to prescribe, lithium. This is inherently a binary decision process. Our categories at present are imperfect and may reflect a spectrum of phenotypes, but the process of diagnosis and treatment of major mental illness seems likely to retain a categorical aspect.
(3) Natural History
RDoC does not address the natural history of disease, which typically involves initiating, sustaining, and homeostatic processes that change with time, leading to different phenotypic manifestations of the same disease at different stages in its evolution. In a recent extended defense of RDoC, the “four key issues” discussed included neither natural history nor pathophysiology [36]. The RDoC perspective is fundamentally static. We would add that studies of normal neurodevelopment, for which RDoC does advocate [37], are not the same as investigations of disease course.
Diseases are dynamic, with a natural history in which evolving pathogenic changes lead to changes in clinical presentation. In the pneumonia example, fever and altered respiratory rate may reflect, sequentially, bronchitis, pneumonia, and sepsis. In diabetes, hyperglycemia may lead to neuropathy, stroke, myocardial infarction, and ketoacidosis, each with its own pathogenesis, time course, and therapeutic implications.
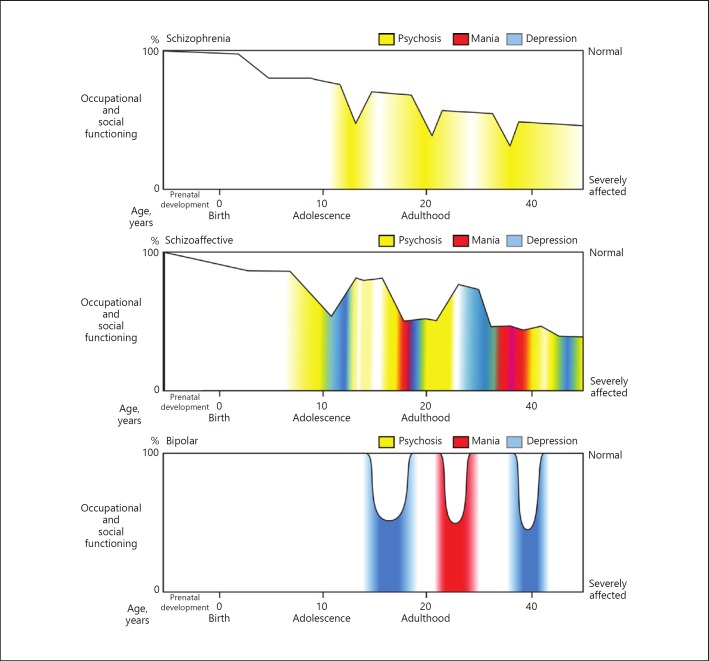
Autism, the dementias, schizophrenia, and bipolar disorder have distinct natural histories, which should be an integral part of their definition and need to be incorporated into research initiatives (see Fig. 2 for idealized diagrams of natural histories in the psychosis spectrum). By disregarding natural history, RDoC loses a perspective that has guided all other fields of medicine, and fails to direct psychiatric research toward the pathogenic processes that underlie different stages of major mental illness.
Fig. 2.
Idealized natural histories of forms of psychotic disorder. These schematics are based on simplified views of the natural histories of these disorders, originating with the ideas of Kraepelin, Bleuler, Baillarger, Falret, and Leonhard, and generally validated by longitudinal outcome studies. The broad distinction is between schizophrenia and bipolar illness. Schizophrenia is notable for subtle developmental abnormalities, detectible even in childhood. Manifest symptoms emerge in adolescence and young adulthood, with a functional and cognitive decline followed by a chronic course punctuated by episodes of more acute illness. By contrast, bipolar disorder, in its pure form, appears to involve fewer developmental abnormalities, and does not universally lead to cognitive and social decline. Instead, bipolar disorder, in its classic form, has an episodic course in which normality is interrupted by episodes of depression or mania. Many patients, however, have intermediate forms of illness, often termed schizoaffective disorder, characterized by a mixture of affective changes and psychotic phenomena, and a relatively chronic course. These three categories are oversimplifications of what is in reality a complex and heterogeneous group of diseases, which may be better described as a “spectrum” of related disorders. Nonetheless, the diagram emphasizes the importance of incorporating the natural history of the major mental illnesses, including the changes in pathophysiology that underlie changes in phenotype over the course of the disease. This concept that diseases are not static over time is one of the fundamental features of the biomedical approach to disease nosology and research, largely missing in RDoC. Illustrator: Joan M.K. Tycko.
(4) Top-Down versus Bottom-Up Approach to Research
RDoC dictates primarily a top-down approach to research – a methodology that begins by postulating how a system under investigation must function, and then breaking down the postulated function into increasingly smaller units [38]. The RDoC top-down approach to mental illness begins by postulating the existence of essential components of normal brain function (domains) that are divided into subsystems (constructs), which in turn are broken down into increasingly granular sub-subsystems (circuits, molecules, and eventually genes) that are hypothesized to underpin the entire set of domains (Fig. 3). There is little connection to human clinical syndromes. By contrast, progress in medicine usually comes from a bottom-up approach, a research methodology that begins with “base elements” that are linked together to form more complex subsystems, leading to a “top-level” system. In medicine, this means beginning with research into the etiological factors of a disease, determining how the combination and interaction of these factors leads to pathogenic pathways and pathology, and discovering how these abnormalities lead to clinical syndromes. The process is iterative and naturally leads to the development of increasingly refined diagnostic categories and to targets for rational treatment. Many disorders in many fields of medicine result from a complex interplay of genomic and environmental risk factors, with both heterogeneity (the same or similar phenotypes deriving from different genetic mechanisms) and pleiotropy (different phenotypes derived from variations at the same genetic locus). Complexity, therefore, is not a rationale for abandoning the medical model of disease reasoning.
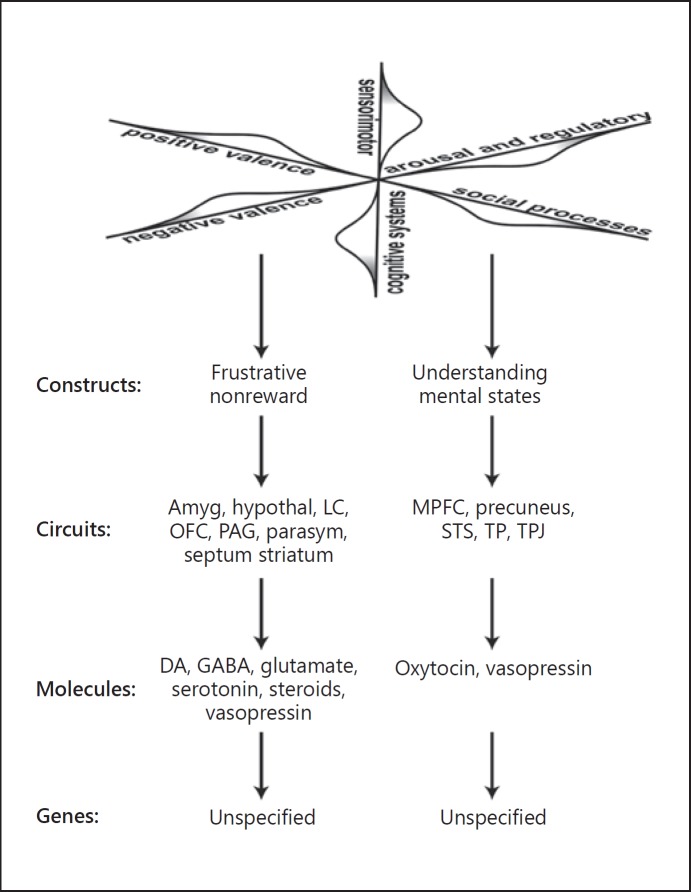
Fig. 3.
The top-down approach of RDoC. The RDoC approach to psychiatry begins with arbitrary domains, divided into constructs, which are the representations of one, and often many, circuits based on a few already known molecules. Depicted are one construct each from two different domains. Note the plethora of ill-defined “circuits,” the paucity of novel molecules, and the decision to exclude genes (as of 2019) for lack of evidence. The relevance to psychiatric diseases, as manifest in patients, is obscure. Amyg, amygdala; hypothal, hypothalamus; LC, locus coeruleus; MPFC, medial prefrontal cortex; OFC, orbital frontal cortex; PAG, periaqueductal gray; parasym, parasympathetic; STS, superior temporal sulcus; TP, temporal pole; TPJ, temporal parietal junction. All terms and relationships adapted from the NIMH RDoC matrix website (https://www.nimh.nih.gov/research/research-funded-by-nimh/rdoc/constructs/rdoc-matrix.shtml).
Cancer provides a classic example of the fruitfulness of a bottom-up approach in a complex disorder (which also has a natural history, typically staged by sequential dysplasia, carcinoma in situ, tumor growth, invasion, and metastasis). Progress in recent decades has emerged from identification of genetic and environmental etiological factors, the discovery of convergent pathogenic pathways, and the development of histological and molecular markers. Continued and iterative revisions of nosology, diagnosis, and prognosis, aided by cell and animal models, are now yielding rational individualized therapeutics [39].
A comparable situation holds for autoimmune disorders. For instance, a genome-wide association study (GWAS) of ankylosing spondylitis, Crohn's disease, psoriasis, and ulcerative colitis found substantial shared genetic risk among the diseases, yet “… despite the profound pleiotropy, clear demarcations of the genetic risk for the individual conditions exist” [40].
Diabetes, caused by a highly complex mixture of genetic and environmental factors, serves as another example. As in many disorders with complex genetics, familial risk does not follow a Mendelian pattern, and first-degree relatives may have subclinical phenotypes [41]. Diabetes also demonstrates the concept of a spectrum of disorders, in which multiple related syndromes are subject to quantitative measures, but are categorically distinct from normal [42]. Despite its complexity, application of the bottom-up disease approach, including the use of cell and animal models based on etiological factors [43], has led to substantial progress, which would have been difficult or impossible with a top-down approach focusing on normal mechanisms of glucose regulation.
Another aspect of the top-down approach is the imposition of RDoC upon the scientific community. The NIMH decreed it after relatively little discussion with the community, with no consideration of alternatives, and no mechanism for evaluation as to success or failure. Funding of extramural grants is determined in part by adherence to this conceptual scheme. This prescriptive approach to research is atypical of American biomedicine.
Conceptualization of Disease in the Central Nervous System
Movement Disorders
Movement disorders provide a striking example of the inadequacy of dimensional reasoning, the value of considering etiology, pathogenesis, and natural history, and the success of the bottom-up approach. Clinically, involuntary movements are not at extremes of a normal dimension of movements, but rather categorical abnormalities subject to quantitative measurements. For instance, the tremor of Parkinson's disease is quite different from the “physiological” tremor most people have with fatigue, and different again from cerebellar tremor or essential tremor, and all are distinct again from chorea and dystonia.
The recent reclassification of movement disorders with genetic etiologies [44] highlights the value of careful clinical-genetic correlation using a bottom-up approach. The classification scheme recognizes that mutations may have pleiotropic effects and that some mutations are causative while others are risk factors. HD provides an interesting example of quantitative – but not dimensional – reasoning [45]. The CAG repeat within the gene huntingtin varies in length from about 5 to 30 triplets in the normal population; variation in this range has no established clinical relevance. HD arises in individuals with repeat lengths greater than the threshold of 35 CAG triplets, with longer repeats resulting in earlier onset age and more rapid progression, again a quantitative but not a dimensional phenomenon.
Classification of movement disorders with complex genetics is also categorical rather than dimensional. For instance, the pathological classification of parkinsonian syndromes [46] as synucleinopathies or tauopathies, with protein inclusions in glia or neurons, has refined previous clinical diagnoses. Genetic distinctions promise additional improvements.
Dystonia [47] appears not to involve neurodegeneration, and thus may provide a model for investigation of complex psychiatric disorders. Reminiscent of diabetes, it can be described as a “broad clinical spectrum, from childhood-onset generalized dystonias to adult-onset focal PTD [primary torsion dystonia]” [47]. Mutations in single genes, such as DYT1, often with reduced penetrance, can cause early-onset primary dystonia. Late-onset dystonia, e.g., cervical dystonia, often appears sporadically, and like many psychiatric disorders is probably a consequence of complex genetic factors. Neurobiological mechanisms, emerging from exploration of etiology and pathogenesis using model organisms, may involve abnormal neuronal plasticity with an imbalance of excitatory and inhibitory motor circuitry.
Dementias
The bottom-up approach [48] has proven to be a powerful method for deciphering the dementias. We can now distinguish Alzheimer's disease, frontotemporal dementia, Lewy body dementia, vascular dementia, and others based in part on clinical course (including phenomenology), pathology, and, increasingly, etiology. Diagnostic precision has improved in an iterative fashion as both etiology and phenotype are refined. The discovery of genetic mutations has enabled the development of animal and cellular models, which, in turn, have stimulated fruitful hypotheses about pathogenesis and identification of potential therapeutic targets.
Autism
The molecular genetics approach to autism is making rapid progress [24, 49, 50, 51, 52] and will be just briefly discussed here. The discovery of rare de novo mutations of relatively large effect using sequencing and microarray strategies, in combination with GWAS findings of single nucleotide polymorphisms (SNPs) with small contributions to risk, has implicated variations in SCN2A, SHANK1, and NRXN1, ANK2, and CHD8, among many other genes, as etiological factors. Mutations in promotors are often in conserved regions of the distal promoter at known transcription binding sites [53]. Many of the variants also appear to contribute to the risk of intellectual dysfunction in the absence of autism. First-degree relatives of patients with autism often have subclinical features of the autism syndrome. Despite genetic heterogeneity and pleiotropy, common pathogenic mechanisms are emerging, in part through study of animal models, including chromatin remodeling, synaptic dysfunction, wnt signaling, neural-immune function especially involving NFkappaB, and activity-dependent transcription or translation [24, 44, 45, 46, 48, 49, 50].
Schizophrenia and Affective Disorders
The phenotypic and genetic overlap of the psychotic disorders as currently defined, including schizophrenia, schizoaffective disorder, bipolar disorder, and major depression with psychosis, have generated controversy in nosology [54, 55], suggesting the description “polymorphous polygenicity” [56]. This complexity appears to have been part of the impetus for the development of RDoC. We would argue that the available evidence supports viewing these disorders as a spectrum of disease phenotypes (or perhaps multiple spectra or clusters of related clinical syndromes), best approached using bottom-up research strategies that begin with the discovery of etiological factors.
In 2014, a GWAS identified 108 genome-wide-significant loci for schizophrenia [57]. Additional loci continue to emerge with larger sample sizes, methodological advances, and integration of GWAS findings with detailed analyses of transcript expression [24]. These discoveries are illuminating potential pathogenic pathways. For example, the strong GWAS signal on chromosome 6 implicated increased expression of C4a [58], a critical regulator of synaptic pruning. Pathway analyses of other genes associated with schizophrenia by SNPs or copy number variants suggest a pathogenic role for abnormalities in synaptic transmission (especially glutamate) and plasticity, activity-regulated cytoskeleton function, microRNAs, and FMRP-RNA targets [59, 60]. Coding variants detected in schizophrenia have also been implicated in developmental disorders [61], consistent with the concept of schizophrenia as a disorder of neurodevelopment [34].
The PsychENCODE Consortium has added enormous amounts of data and a sophisticated analytic approach to the genetics of schizophrenia and other major mental illnesses, especially at the level of gene expression and alternative splicing. Consortium investigations have provided new or additional evidence for many contributory factors. These include altered function of excitatory neurons (via differential expression of genes such as GRIN1 and NRXN1); the genetic targets of specific transcription factors (e.g., FMRP and RBFOX1); calcium signaling; the complement cascade (via expression variants of C4A, C4B, and CLU); and cell adhesion (via expression variation of Protocadherin) – among other pathways, systems, and cell types [24, 62].
The polygenic risk score, developed to measure the contribution to schizophrenia risk of many small common variants acting together [63], has emphasized the genetic complexity of schizophrenia. It has also clarified the role of interactions of environmental factors with genomic variants in increasing the risk of disease. For instance, the polygenic risk score for schizophrenia is greater in individuals with a complicated gestation or with birth trauma. Unexpectedly, this interaction appears to be mediated, at least in part, by a subset of risk genes expressed in the placenta [64].
GWASs of bipolar disorder are also now yielding replicated loci with genome-wide significance [65]. Polygenic risk scores demonstrate a genetic overlap between schizophrenia and bipolar disorder [65, 66], providing further evidence for conceptualizing these disorders as related along a spectrum, though some risk loci common to the two disorders may have opposite effects on gene expression or splicing [24, 51]. Variants in several subunits of voltage-gated calcium channels are prominent for both schizophrenia and bipolar disorder, as are variants in other genes coding for proteins involved in neuronal signaling [67]. However, distinctions are also emerging – for instance, the strong GWAS finding on chromosome 6, so prominent in schizophrenia, is not prominent in bipolar disorder, while an association with TRANK1 may be stronger in bipolar disorder. Genes involved in potassium ion response may distinguish between the two disorders, and variants detected in the two disorders may have an opposite effect on some genes (e.g., the transcriptional repressor GATAD2A). Rare variants have been detected in bipolar disorder and implicate a number of genes, consistent with GWAS findings, including ANK3, a gene whose protein product regulates axonal initial segment function, and a number of genes encoding calcium channels [68].
A high schizophrenia polygenic risk score in individuals with bipolar disorder predicts lack of response to lithium [69]. SNPs associated with genes expressed in excitatory neurons may be more relevant for schizophrenia, while SNPs associated with genes expressed in inhibitory neurons may be more relevant for bipolar disorder [24, 51, 70]. The PsychENCODE Consortium data associate fetally expressed genes more strongly with schizophrenia than with bipolar disorder [50], while processes including oxidative phosphorylation, ubiquitin-mediated proteolysis, RNA transport, and endocytosis and synaptic vesicle cycle are more strongly associated with bipolar disorder [24].
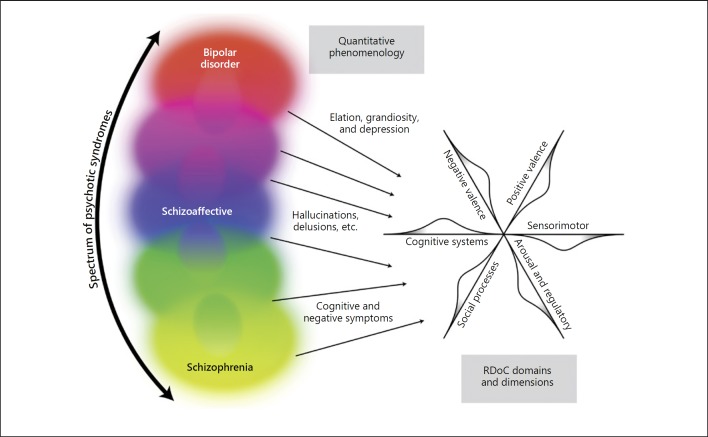
A persistent issue in the nosology of psychosis has been the status of syndromes that have features of both bipolar disorder and schizophrenia, often designated “schizoaffective disorder.” Recent data indicate that the schizophrenia polygenic risk score is correlated with the presence and severity of mood-incongruent psychotic symptoms in affective and schizoaffective disorders [71]. This is consistent with the view that the psychotic disorders form a “spectrum” of related diseases, with neurobiology and clinical phenotypes categorically distinct from normal human biology and function, not easily understood using the RDoC scheme of “dimensional” variations along an axis anchored in normal functions (Fig. 4).
Fig. 4.
Disjunction between phenomenology of psychosis and “dimensions” of RDoC. The “spectrum” of psychosis is shown using current terminology, recognizing that both the terminology and the relationship among the various disorders will inevitably evolve in the future. Certain phenomena of this spectrum of diseases, such as delusions, hallucinations, and thought disorder, appear qualitatively distinct from normal phenomenology. Other characteristic phenomena (e.g., mood and cognition) can be measured quantitatively, and may resemble extremes of normal phenomenology. However, we argue that despite this phenomenological resemblance to extremes of normal variation, these quantitatively measurable phenomena do not necessarily correspond to the domains and constructs of the RDoC scheme (illustrated schematically as axes to show the six current RDoC domains and their dimensional nature). In our view, the phenomena of major mental illnesses represent a shift off dimensional axes of normal variation, the consequence of a breakdown in normal brain function caused by complex etiological and pathophysiological processes, and are best understood using the disease model. The main point is that even though a clinical phenomenon is quantifiable, it does not follow that such a phenomenon can best be understood as an extreme of dimensional variation along a normal axis. This line of argument also leads to the conclusion that, for disease research and clinical practice, it is better to focus attention on the quantitative phenomena observed in patients, rather than to make the a priori assumption that variations in quantitative phenomena observed in the normal population will be of direct relevance. Illustrator: Joan M.K. Tycko.
Progress is emerging in major depression, a very difficult problem due to clinical heterogeneity. A recent GWAS involving more than 480,000 subjects identified variation at 44 loci as significant risk factors with evidence of partially shared, causal biological pathways with schizophrenia [72]. Indeed, the genetic overlap extends to other psychiatric disorders and to some personality traits [73].
Ironically, the era of remarkable progress in the discovery of the genetic basis of mental illness began about 10 years ago, following years of frustration, just about the same time that the NIMH was calling for the creation of RDoC. The frustration then was perhaps understandable – new pharmacological treatments had not emerged despite decades of progress in neuroscience and efforts such as the Decade of the Brain. Perhaps a comparison might be made to the state of oncology in the 1950s – surgical, radiation, and chemical treatments that were nonspecific, dangerous, and based on simple schemes of disease classification. Subsequent advances in the understanding of cancer etiology, pathogenesis, and pathology, including the mechanisms behind oncogenesis and metastasis, have led to remarkable improvements in classification schemes, diagnosis, and treatment. Though a few decades behind these efforts, we take the optimistic stand that advances in psychiatry can follow a comparable trajectory. Together, the rapid genetic advances in schizophrenia, the affective disorders, and other major mental illnesses over the past few years provide powerful support for a bottom-up approach to the study of the etiology and pathogenesis of these conditions, based on the discovery of the roles of specific genes and pathways in disease pathogenesis, some common to multiple diseases, others more specific.
Alternatives to RDoC for Future Research in Psychiatry
We believe that the alternative to the RDoC scheme for investigation of major mental illness is a straightforward application of the disease model, with a bottom-up focus on etiology and pathogenesis. Psychiatric etiologies, both genetic and environmental, are heterogeneous and pleiotropic. Family members of patients with major mental illnesses often share subclinical versions of some of the same phenotypes, presumably because of shared risk alleles [54], sometimes blurring the distinction between disease state and normal variation. However, even when describing subsyndromal psychiatric phenotypes, we believe that it is better to start from the disease phenotypes, guided by genotype-phenotype correlations in patients with the diseases, rather than attempting to use normal variation as a starting point. It is important not to mistake quantitative differences in measures of disease severity and genetic risk, or differences along a disease spectrum, with variations along a dimension of normal. We would anticipate an iterative process of redefining and refining clinical syndromes based on a progressive understanding of etiology and pathogenesis, exactly the process that has led to progress in many other areas of medicine.
We are still at an early stage of genetic research. For many GWAS loci, the actual allele conferring risk, and often even the relevant gene, remains uncertain, and the effect of the variant on gene expression is often unknown. For many loci, variation is likely to have complex pleiotropic effects. This is particularly relevant for loci associated with both schizophrenia and bipolar disorder. Abundant examples exist in other diseases, including disorders affecting brain function, in which different variations in the same gene lead to different clinical phenotypes. For instance, point mutations in SCN2A affect the structure of membrane-spanning domains in the encoded sodium channel protein (NaV1.2), potentially increasing channel function resulting in epilepsy. On the other hand, SCN2A mutations, such as those causing protein truncation, that lead to a loss of protein function may cause cognitive dysfunction [74].
Full understanding of genetic variants and their relationship with environmental insults will require detailed insight into the regulation of gene expression (e.g., repressors, enhancers, splice variants, epigenetics, three-dimensional chromosomal organization) and into developmental shifts in gene expression and splicing. Rare mutations with major impact on disease risk, and heritable copy number variations, may provide insight into the effect of common variations.
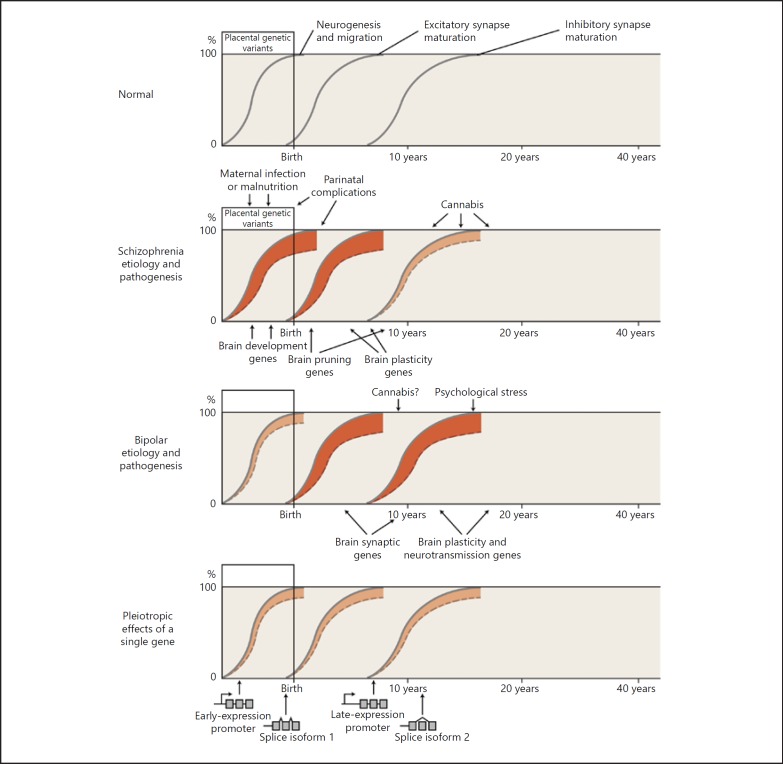
We would emphasize the importance, and feasibility, of using complex genetic information to construct testable and potentially clinically useful hypotheses. The potential use of genetic information to predict lithium response is one example. A more complex hypothesis might arise from distinctions between bipolar disorder and schizophrenia found in genetic association and gene expression studies. One possible broad hypothesis might be that genetic disturbances (splicing or expression changes) manifest earlier in development, especially at excitatory synapses, and might preferentially increase the risk for schizophrenia. By contrast, genetic disturbances in the same genes, but manifesting later in development, especially at inhibitory synapses, might have more of an effect on the risk for bipolar disorder (Fig. 5) [24, 51, 62, 70, 75]. Hypothesis building based on complex genetic data should take into account known aspects of neurobiology, such as developmental trajectories and cell subtype specificity; in this case, the key neurobiological insight is the relatively later development of inhibitory compared to excitatory neurons. We are well aware that these hypotheses involve oversimplifications and may turn out to be incorrect, but we believe that it is useful to develop and test concepts that attempt to correlate genetic, pathophysiological, and clinical information.
Fig. 5.
Relationship between genetic and environmental risk factors and clinical phenotypes. Genetic and environmental factors, acting in part on the differential expression of genes, disrupt the normal developmental processes, leading to abnormal brain structure and function. The y axis indicates the extent to which a developmental process has reached completion. The x axis indicates age, reflecting developmental status. A solid gray line indicates normal brain development (neurogenesis and migration, excitatory synapses, and inhibitory synapses). A dashed line below the solid gray line, with red shading between the lines, indicates a disruption of the process – a shift to an abnormal brain state (depiction modified from Insel [93]). Normal development involves multiple neurobiological processes; here we show just three as a simplification to highlight that different processes mature at different times. Phenotypic differences arise because of individual differences in exposure to genetic and environmental risk factors, in the timing of exposure to environmental risk factors, and in the interactions among genetic and environmental risk factors. We emphasize here that risk factor-induced changes in gene expression, including different isoforms of the same gene, may lead to changes in synaptic structure and function, though many other pathogenic mechanisms are also likely involved. We hypothesize (no doubt as an oversimplification) that risk alleles and environmental risk factors acting during earlier stages of development (such as placental stress occurring in an individual with a genetic vulnerability to such stress) tend to predispose to schizophrenia. By contrast, in general, alleles and environmental factors altering gene expression later in development predispose more to bipolar disorder. The earlier influences may preferentially affect excitatory synapses (which generally mature earlier), while the later influences may preferentially affect inhibitory synapses (which generally mature later). The bottom panel highlights how different alleles of the same gene can have effects at different time points in development via differing promotor usage or splice isoforms. Many other mechanisms (not shown), such as epigenetic changes or posttranslational regulation via microRNAs, could give similar results. We are well aware that these hypotheses involve oversimplifications and may turn out to be incorrect, but we believe that it is useful to develop and test concepts that attempt to correlate genetic, pathophysiological, and clinical information. Illustrator: Joan M.K. Tycko.
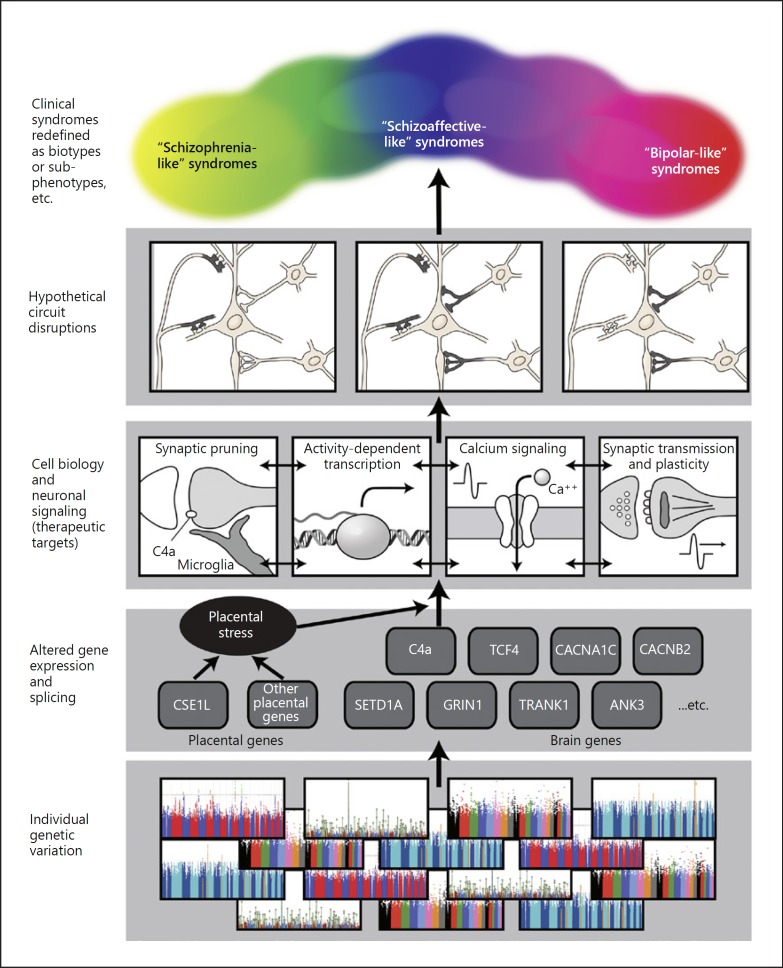
While psychiatric research must embrace the complications (and “big data” methods) of multiplex etiology, it can also seek simplification by determining the key points of convergence in the pathogenic processes that lead to disease. The neural network model described by the PsychENCODE group [24] provides a conceptual framework for considering multiple levels of the disease process, from genetic variant to gene expression to clinical phenotype. We would argue that the most relevant level for discovery of therapeutic targets will not be at the level of genes and gene expression, nor at the level of circuits, but rather at the intermediate level of intracellular pathways and cell-cell interactions (Fig. 6), corresponding to the “hidden” L2 layer described by Wang et al. [24]. This is also the most difficult level to approach with current methods. Progress will require intensive in vitro study of cell models derived from patients and in vivo study of genetically manipulated animals.
Fig. 6.
Hypothetical bottom-up scheme for conceptualizing the causal chain from genetic etiologies to clinical phenotypes. We hypothesize that the large number of genetic risk factors (also environmental factors, not shown) for major mental illness converge on a smaller number of molecular pathways, which in turn will selectively alter brain circuitry, leading to clinical phenotypes. This scheme stands in contrast to the top-down approach of RDoC in its emphasis on the etiology and pathogenesis of disease, rather than on normal biology. Furthermore, this depiction of the bottom-up approach corrects an imbalance in RDoC, which overemphasizes circuit-level analysis and underemphasizes analysis at the molecular and cellular levels. We include a representation of the hypotheses noted in the text that genetic risk variants affecting isoforms expressed earlier in development and in excitatory neurons might predispose more to schizophrenia-like phenotypes, while variants affecting isoforms expressed later in development and in inhibitory neurons may predispose more to bipolar disorder. A representation of genes predisposing to an abnormal response to placental stress, which we hypothesize are more relevant for schizophrenia-like syndromes than for bipolar-like syndromes, is provided as an example of gene-environment interaction. While these are no doubt oversimplifications (and particularly the circuit changes are not yet well defined, so are shown as hypothetical synaptic alterations), they may illustrate the possibility that general principles illuminating nosology may emerge even from very complex GWAS, transcriptome, and other datasets. We represent the clinical phenotypes as a “spectrum,” with the expectation that the clinical syndromes may well be redefined and subcategorized based on increasing understanding of etiology and pathophysiology. In general, etiologies and pathogeneses more relevant to schizophrenia-like syndromes are shown on the left side of the figure, while etiologies and pathogeneses more relevant to bipolar-like syndromes are shown on the right side. While no doubt oversimplifications, these and other hypotheses can be tested and validated or rejected, a critical strength of the bottom-up approach. Illustrator: Joan M.K. Tycko.
Cell models of both bipolar disorder and schizophrenia [62] have already provided suggestive initial data pointing to pathogenic processes, such as aberrant migration and increased oxidative stress in schizophrenia [76], and differential signaling responses in cells from lithium-responsive versus -nonresponsive bipolar subjects [77]. Mouse models using genetic variations associated with schizophrenia and bipolar disorder have also provided intriguing insights [78, 79]. Of course, the concept of “animal models” – especially mouse models – relevant to human disorders of cognition and emotion is fraught with difficulties. Nonhuman primates [80], such as recently developed marmoset models [81], may facilitate more refined genotype-phenotype interpretations [82]. Overall, despite recognition of limitations, there is growing consensus that animal models will prove critical in advancing knowledge of psychiatric disorders [83].
Work at the level of neuropathology and cellular protein may provide unexpected insights. In particular, increased levels of abnormal insoluble proteins have been detected in a subset of postmortem brains from schizophrenia patients [84]. A separate investigation reported increased protein ubiquitination in homogenates of brain tissue and red blood cells from patients with schizophrenia, suggestive of abnormal protein processing [85]. These results might be correlated with the study of Ursini et al. [64] mentioned above, which found that a subset of genes related to cell stress were enriched as risk factors in cases with “early-life complications” such as birth trauma. It might be hypothesized that patients without abnormal proteins have genetic liabilities more related to synapses and neuronal signaling, while cases with abnormal proteins have genetic liabilities more related to oxidative or other cell stress and altered proteostasis. A subset of patients marked by abnormal proteins may even have a phenotype reminiscent of Kraepelin's “dementia praecox.” These protein-based approaches may provide a novel means to subdivide and characterize schizophrenia and related disorders, to identify central and potentially peripheral biomarkers, and to suggest new therapeutic targets. The RDoC scheme has little room to incorporate such novel and unexpected discoveries.
Even after pathogenic pathways have been validated, it is a long process to reach the point of exploiting therapeutic targets; it has taken 25 years from the discovery of the HD gene for rational disease-modifying therapy (e.g., oligonucleotides to suppress Huntingtin protein expression) to reach human clinical trials [86]. Nonetheless, in autism and schizophrenia, genetic and pathogenic approaches are beginning to set the stage for rational therapeutics [87, 88], demonstrating the potential for the bottom-up approach even in complex psychiatric disorders.
Current guidance from the NIMH emphasizes study of genes emerging from GWAS, but – in our view counterproductively – discourages the generation of “models of diseases” [8]. We believe that the directive toward “pure” neurobiology, and the emphasis on normal behavior and normal neurobiology in the RDoC scheme, diverts attention from more direct and efficient studies of disease etiology and pathophysiology. We acknowledge that modeling disease phenotypes, and especially those disorders with polygenic etiology, is challenging. Nonetheless, even when models are limited to single components, careful exploration of the correlation between genotypes and phenotypes imposes direction and criteria for the success or failure of a program of investigation. (We agree that the phrase “animal model useful in the study of condition X” is more accurate than the phrase “animal model of condition X” as suggested in the ACNP 2018 animal research symposium [89], but use the latter phrase as shorthand – in any case, one must be aware of all the many caveats about animal models.) Furthermore, disease-relevant models are essential for testing candidate novel therapeutics (and conversely the action of known therapeutics can help validate candidate animal models).
Longitudinal efforts to define more closely the clinical courses of the major mental illnesses will be essential for the development and validation of models, and for testing the efficacy of therapeutic interventions that emerge from clinical models. The longitudinal studies that defined the clinical course of HD [90, 91] provide a precedent for the value of such efforts in developing strategies for clinical trials. It is remarkable that despite the scope and complexity of the major mental illnesses, there are relatively few long-term longitudinal studies of clinical course [92].
We conclude that RDoC may offer a few advantages for research into psychiatric disorders that are inherently dimensional. To be successful for these disorders, RDoC will need reformulation to include reliable measurements and a path to validation, with criteria for success or failure. However, for the major mental illnesses, RDoC deflects research away from studying disease etiology and pathogenesis, the path so valuable for much of medicine, just when this approach is yielding remarkable advances. As noted above, we believe the salutary change to research on broader sets of patients with different diagnoses would have happened in the absence of RDoC as a natural consequence of recent genetics research. We therefore propose the abandonment of the RDoC system as a method for directing the course of future NIMH-funded research.
As a guide to research in major mental illness, we favor approaches focused on the disease model, beginning with etiology, including genetics and environment, and proceeding to pathogenesis, including cell and animal models, human imaging and other biomarker studies, and additional studies of postmortem brains. Further development and study of cell and animal models relevant to disease will be necessary for experimental therapeutics. Basic and translational science can inform iterative refinement of nosology, and lead to clinical trials of rational therapeutics.
Disclosure Statement
Dr. Ross reports grant support from NIH and CHDI, previous support from JNJ/Janssen, Teva, and Raptor/Horizon, clinical trial support from Teva, Vaccinex, Roche/Genentech, and CHDI, and consulting for Teva, Sage, uniQure, Roche/Ionis, and HSG. Dr. Margolis reports grant support from the NIH, ABCD Charitable Trust, and Teva.
Funding Sources
This work was supported in part by the ABCD Charitable Trust, which had no role in the conceptualization, research, or writing of this paper.
Author Contributions
Both authors made a substantial contribution to the conception and design of the manuscript, participated in the drafting and revision of the manuscript, approved the final version for publication, and agree to be accountable for all aspects of the work.
Acknowledgments
This work is dedicated to the late Pamela Sklar, whose perspective guided many of our ideas. We thank Drs. Daniel Ebert, Fernando Goes, Mikhail Pletnikov, Jimmy Potash, Crystal Salcido, and Daniel Weinberger for their comments and insight, and Dr. Paul McHugh for intellectual inspiration.
References
- 1.Cuthbert BN, Workgroup NR, NIMH RDoC Workgroup The RDoC framework: continuing commentary. World Psychiatry. 2014 Jun;13((2)):196–7. doi: 10.1002/wps.20140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Insel TR. The NIMH Research Domain Criteria (RDoC) Project: precision medicine for psychiatry. Am J Psychiatry. 2014 Apr;171((4)):395–7. doi: 10.1176/appi.ajp.2014.14020138. [DOI] [PubMed] [Google Scholar]
- 3.Cuthbert BN. The RDoC framework: facilitating transition from ICD/DSM to dimensional approaches that integrate neuroscience and psychopathology. World Psychiatry. 2014 Feb;13((1)):28–35. doi: 10.1002/wps.20087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Institute of Mental Health Strategy 1.4 of the 2008 NIMH Strategic Plan. 2008:pp 1–60. [Google Scholar]
- 5.Morris SE, Cuthbert BN. Research Domain Criteria: cognitive systems, neural circuits, and dimensions of behavior. Dialogues Clin Neurosci. 2012 Mar;14((1)):29–37. doi: 10.31887/DCNS.2012.14.1/smorris. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Institute of Mental Health . A Strategic Plan for Research. 2015. p. p. 56. [Google Scholar]
- 7.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013 May;11((1)):126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Institute of Mental Health . A Strategic Plan for Research. 2015. p. p. 12. [Google Scholar]
- 9.Elmer GI, Brown PL, Shepard PD. Engaging Research Domain Criteria (RDoC): Neurocircuitry in Search of Meaning. Schizophr Bull. 2016 Sep;42((5)):1090–5. doi: 10.1093/schbul/sbw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hess JL, Kawaguchi DM, Wagner KE, Faraone SV, Glatt SJ. The influence of genes on “positive valence systems” constructs: A systematic review. Am J Med Genet B Neuropsychiatr Genet. 2016 Jan;171B((1)):92–110. doi: 10.1002/ajmg.b.32382. [DOI] [PubMed] [Google Scholar]
- 11.Gur RC, Gur RE. Social cognition as an RDoC domain. Am J Med Genet B Neuropsychiatr Genet. 2016 Jan;171B((1)):132–41. doi: 10.1002/ajmg.b.32394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nusslock R, Walden K, Harmon-Jones E. Asymmetrical frontal cortical activity associated with differential risk for mood and anxiety disorder symptoms: an RDoC perspective. Int J Psychophysiol. 2015 Nov;98((2 Pt 2)):249–61. doi: 10.1016/j.ijpsycho.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Glatt CE, Lee FS. Common Polymorphisms in the Age of Research Domain Criteria (RDoC): integration and Translation. Biol Psychiatry. 2016 Jan;79((1)):25–31. doi: 10.1016/j.biopsych.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz E, Tost H, Meyer-Lindenberg A. Working memory genetics in schizophrenia and related disorders: an RDoC perspective. Am J Med Genet B Neuropsychiatr Genet. 2016 Jan;171B((1)):121–31. doi: 10.1002/ajmg.b.32353. [DOI] [PubMed] [Google Scholar]
- 15.McHugh PR, Slavney PR. The Perspectives of Psychiatry. Baltimore: Johns Hopkins University Press; 1998. [Google Scholar]
- 16.Van Dam NT, O'Connor D, Marcelle ET, Ho EJ, Cameron Craddock R, Tobe RH, et al. Data-Driven Phenotypic Categorization for Neurobiological Analyses: beyond DSM-5 Labels. Biol Psychiatry. 2017 Mar;81((6)):484–94. doi: 10.1016/j.biopsych.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufman J. Unraveling the Genetics of Major Depression and Stress-Related Psychiatric Disorders: Is It Time for a Paradigm Shift? Biol Psychiatry. 2018 Jul;84((2)):82–4. doi: 10.1016/j.biopsych.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Stevens JS, Kim YJ, Galatzer-Levy IR, Reddy R, Ely TD, Nemeroff CB, et al. Amygdala Reactivity and Anterior Cingulate Habituation Predict Posttraumatic Stress Disorder Symptom Maintenance After Acute Civilian Trauma. Biol Psychiatry. 2017 Jun;81((12)):1023–9. doi: 10.1016/j.biopsych.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bebko G, Bertocci M, Chase H, Dwojak A, Bonar L, Almeida J, et al. Decreased amygdala-insula resting state connectivity in behaviorally and emotionally dysregulated youth. Psychiatry Res. 2015 Jan;231((1)):77–86. doi: 10.1016/j.pscychresns.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebowitz ER, Gee DG, Pine DS, Silverman WK. Implications of the Research Domain Criteria project for childhood anxiety and its disorders. Clin Psychol Rev. 2018 Aug;64:99–109. doi: 10.1016/j.cpr.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McTeague LM, Laplante MC, Bulls HW, Shumen JR, Lang PJ, Keil A. Face Perception in Social Anxiety: Visuocortical Dynamics Reveal Propensities for Hypervigilance or Avoidance. Biol Psychiatry. 2018 Apr;83((7)):618–28. doi: 10.1016/j.biopsych.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carcone D, Ruocco AC. Six Years of Research on the National Institute of Mental Health's Research Domain Criteria (RDoC) Initiative: A Systematic Review. Front Cell Neurosci. 2017 Mar;11:46. doi: 10.3389/fncel.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, et al. Identification of Distinct Psychosis Biotypes Using Brain-Based Biomarkers. Am J Psychiatry. 2016 Apr;173((4)):373–84. doi: 10.1176/appi.ajp.2015.14091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FC, et al. PsychENCODE Consortium Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018 Dec;362((6420)):eaat8464. doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaemi SN. Taking disease seriously in DSM. World Psychiatry. 2013 Oct;12((3)):210–2. doi: 10.1002/wps.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghaemi SN. After the failure of DSM: clinical research on psychiatric diagnosis. World Psychiatry. 2018 Oct;17((3)):301–2. doi: 10.1002/wps.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldham JM. DSM models of personality disorders. Curr Opin Psychol. 2018 Jun;21:86–8. doi: 10.1016/j.copsyc.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Ross CA, Margolis RL. Research Domain Criteria: Cutting Edge Neuroscience or Galen's Humors Revisited? Mol Neuropsychiatry. 2018 Dec;4((3)):158–63. doi: 10.1159/000493685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casey BJ, Craddock N, Cuthbert BN, Hyman SE, Lee FS, Ressler KJ. DSM-5 and RDoC: progress in psychiatry research? Nat Rev Neurosci. 2013 Nov;14((11)):810–4. doi: 10.1038/nrn3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilienfeld SO. The Research Domain Criteria (RDoC): an analysis of methodological and conceptual challenges. Behav Res Ther. 2014 Nov;62:129–39. doi: 10.1016/j.brat.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Lilienfeld SO, Treadway MT. Ann Rev Clin Psychol. 2016. Clashing Diagnostic Approaches: DSM-ICD Versus RDoC; pp. pp. 12435–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberger DR, Glick ID, Klein DF. Whither Research Domain Criteria (RDoC)?: The Good, the Bad, and the Ugly. JAMA Psychiatry. 2015 Dec;72((12)):1161–2. doi: 10.1001/jamapsychiatry.2015.1743. [DOI] [PubMed] [Google Scholar]
- 33.Frances A. RDoC is necessary, but very oversold. World Psychiatry. 2014 Feb;13((1)):47–9. doi: 10.1002/wps.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017 Dec;18((12)):727–40. doi: 10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- 35.Woodbury-Smith M, Paterson AD, O'Connor I, Zarrei M, Yuen RK, Howe JL, et al. A genome-wide linkage study of autism spectrum disorder and the broad autism phenotype in extended pedigrees. J Neurodev Disord. 2018 Jun;10((1)):20. doi: 10.1186/s11689-018-9238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark LA, Cuthbert B, Lewis-Fernández R, Narrow WE, Reed GM. Three Approaches to Understanding and Classifying Mental Disorder: ICD-11, DSM-5, and the National Institute of Mental Health's Research Domain Criteria (RDoC) Psychol Sci Public Interest. 2017 Sep;18((2)):72–145. doi: 10.1177/1529100617727266. [DOI] [PubMed] [Google Scholar]
- 37.National Institute of Mental Research Domain Criteria, Developmental and Environmental Aspects. 2018. https://www.nimh.nih.gov/research-priorities/rdoc/developmental-and-environmental-aspects.shtml.
- 38.Anonymous Difference between Top-down and Bottom-up Approach. https://techdifferences.com/difference-between-top-down-and-bottom-up-approach.html June 271, 2018.
- 39.Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer. 2017 Feb;17((2)):116–30. doi: 10.1038/nrc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. International IBD Genetics Consortium (IIBDGC) International Genetics of Ankylosing Spondylitis Consortium (IGAS) International PSC Study Group (IPSCSG) Genetic Analysis of Psoriasis Consortium (GAPC) Psoriasis Association Genetics Extension (PAGE) Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016 May;48((5)):510–8. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderwald C, Stadler M, Golay A, Krebs M, Petrie J, Luger A, RISC Investigators Impact of family history on relations between insulin resistance, LDL cholesterol and carotid IMT in healthy adults. Heart. 2010 Aug;96((15)):1191–200. doi: 10.1136/hrt.2009.177436. [DOI] [PubMed] [Google Scholar]
- 42.Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 2015 Mar;6((1)):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jayasimhan A, Mansour KP, Slattery RM. Advances in our understanding of the pathophysiology of Type 1 diabetes: lessons from the NOD mouse. Clin Sci (Lond) 2014 Jan;126((1)):1–18. doi: 10.1042/CS20120627. [DOI] [PubMed] [Google Scholar]
- 44.Marras C, Lang A, van de Warrenburg BP, Sue CM, Tabrizi SJ, Bertram L, et al. Nomenclature of genetic movement disorders: recommendations of the international Parkinson and movement disorder society task force. Mov Disord. 2016 Apr;31((4)):436–57. doi: 10.1002/mds.26527. [DOI] [PubMed] [Google Scholar]
- 45.Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014 Apr;10((4)):204–16. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 46.Fogel BL, Clark MC, Geschwind DH. The neurogenetics of atypical parkinsonian disorders. Semin Neurol. 2014 Apr;34((2)):217–24. doi: 10.1055/s-0034-1381738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phukan J, Albanese A, Gasser T, Warner T. Primary dystonia and dystonia-plus syndromes: clinical characteristics, diagnosis, and pathogenesis. Lancet Neurol. 2011 Dec;10((12)):1074–85. doi: 10.1016/S1474-4422(11)70232-0. [DOI] [PubMed] [Google Scholar]
- 48.42. Hinz FI, Geschwind DH. Molecular Genetics of Neurodegenerative Dementias. Cold Spring Harb Perspect Biol. 2017;9((4)) doi: 10.1101/cshperspect.a023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodbury-Smith M, Scherer SW. Progress in the genetics of autism spectrum disorder. Dev Med Child Neurol. 2018 May;60((5)):445–51. doi: 10.1111/dmcn.13717. [DOI] [PubMed] [Google Scholar]
- 50.Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. BrainSpan Consortium. PsychENCODE Consortium. PsychENCODE Developmental Subgroup Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018 Dec;362((6420)):eaat7615. doi: 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. PsychENCODE Consortium Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018 Dec;362((6420)):eaat8127. doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, et al. PsychENCODE Consortium Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 2018 Dec;362((6420)):eaat6720. doi: 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.An JY, Lin K, Zhu L, Werling DM, Dong S, Brand H, et al. Genome-wide de novo risk score implicates promoter variation in autism spectrum disorder. Science. 2018 Dec;362((6420)):eaat6576. doi: 10.1126/science.aat6576. 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015 Sep;349((6255)):1489–94. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawrie SM, O'Donovan MC, Saks E, Burns T, Lieberman JA. Improving classification of psychoses. Lancet Psychiatry. 2016 Apr;3((4)):367–74. doi: 10.1016/S2215-0366(15)00577-5. [DOI] [PubMed] [Google Scholar]
- 56.Sklar P. Schizophrenia. Neurosci Abst; 2017. Polymorphous Polygenicity: The Story of the Genome; p. p. 201715. [Google Scholar]
- 57.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014 Jul;511((7510)):421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium Schizophrenia risk from complex variation of complement component 4. Nature. 2016 Feb;530((7589)):177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ. Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biol Psychiatry. 2015 Jan;77((1)):52–8. doi: 10.1016/j.biopsych.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 60.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Psychosis Endophenotypes International Consortium. CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017 Jan;49((1)):27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh T, Walters JT, Johnstone M, Curtis D, Suvisaari J, Torniainen M, et al. INTERVAL Study. UK10K Consortium The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017 Aug;49((8)):1167–73. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casiño C, et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science. 2018 Dec;362((6420)):eaat4311. doi: 10.1126/science.aat4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009 Aug;460((7256)):748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ursini G, Punzi G, Chen Q, Marenco S, Robinson JF, Porcelli A, et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat Med. 2018 Jun;24((6)):792–801. doi: 10.1038/s41591-018-0021-y. [DOI] [PubMed] [Google Scholar]
- 65.Stahl E, Breen G, Forstner A, McQuillin A, Ripke S, Cichon S, et al. Genomewide association study identifies 30 loci associated with bipolar disorder. bioRxiv. 2018;••• doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JM, et al. Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 2018 Jun;173((7)):1705–1715.e16. doi: 10.1016/j.cell.2018.05.046. Electronic address: douglas.ruderfer@vanderbilt.edu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 Apr;381((9875)):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ament SA, Szelinger S, Glusman G, Ashworth J, Hou L, Akula N, et al. Bipolar Genome Study Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci USA. 2015 Mar;112((11)):3576–81. doi: 10.1073/pnas.1424958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016 Mar;387((10023)):1085–93. doi: 10.1016/S0140-6736(16)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50((4)):621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allardyce J, Leonenko G, Hamshere M, Pardiñas AF, Forty L, Knott S, et al. Association Between Schizophrenia-Related Polygenic Liability and the Occurrence and Level of Mood-Incongruent Psychotic Symptoms in Bipolar Disorder. JAMA Psychiatry. 2018 Jan;75((1)):28–35. doi: 10.1001/jamapsychiatry.2017.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. eQTLGen. 23andMe. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018 May;50((5)):668–81. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Brainstorm Consortium Analysis of shared heritability in common disorders of the brain. Science. 2018 Jun;360((6395)):eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanders SJ, Campbell AJ, Cottrell JR, Moller RS, Wagner FF, Auldridge AL, et al. Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends Neurosci. 2018 Jul;41((7)):442–56. doi: 10.1016/j.tins.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, et al. BrainSeq Consortium Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018 Aug;21((8)):1117–25. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roussos P, Guennewig B, Kaczorowski DC, Barry G, Brennand KJ. Activity-Dependent Changes in Gene Expression in Schizophrenia Human-Induced Pluripotent Stem Cell Neurons. JAMA Psychiatry. 2016 Nov;73((11)):1180–8. doi: 10.1001/jamapsychiatry.2016.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B, et al. Pharmacogenomics of Bipolar Disorder Study Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015 Nov;527((7576)):95–9. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010 Apr;464((7289)):763–7. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu S, Cordner ZA, Xiong J, Chiu CT, Artola A, Zuo Y, et al. Genetic disruption of ankyrin-G in adult mouse forebrain causes cortical synapse alteration and behavior reminiscent of bipolar disorder. Proc Natl Acad Sci USA. 2017 Sep;114((39)):10479–84. doi: 10.1073/pnas.1700689114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bauman MD, Schumann CM. Advances in nonhuman primate models of autism: Integrating neuroscience and behavior. Exper Neurol. 2018;299((Pt A)):252–265. doi: 10.1016/j.expneurol.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sasaki E. Prospects for genetically modified non-human primate models, including the common marmoset. Neurosci Res. 2015 Apr;93:110–5. doi: 10.1016/j.neures.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 82.Zhu Y, Sousa AM, Gao T, Skarica M, Li M, Santpere G, et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018 Dec;362((6420)):eaat8077. doi: 10.1126/science.aat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Monteggia LM, Heimer H, Nestler EJ. Meeting Report: Can We Make Animal Models of Human Mental Illness? Biol Psychiatry. 2018 Oct;84((7)):542–5. doi: 10.1016/j.biopsych.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nucifora LG, MacDonald ML, Lee BJ, Peters ME, Norris AL, Orsburn BC, et al. Increased protein insolubility in brains from a subset of patients with schizophrenia. Am J Psychiatry. 2019 doi: 10.1176/appi.ajp.2019.18070864. epub before print. [DOI] [PubMed] [Google Scholar]
- 85.Bousman CA, Luza S, Mancuso SG, Kang D, Opazo CM, Mostaid MS, et al. Elevated ubiquitinated proteins in brain and blood of individuals with schizophrenia. Sci Rep. 2019 Feb;9((1)):2307. doi: 10.1038/s41598-019-38490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Roon-Mom WM, Roos RA, de Bot ST. Dose-Dependent Lowering of Mutant Huntingtin Using Antisense Oligonucleotides in Huntington Disease Patients. Nucleic Acid Ther. 2018 Apr;28((2)):59–62. doi: 10.1089/nat.2018.0720. [DOI] [PubMed] [Google Scholar]
- 87.Lacivita E, Perrone R, Margari L, Leopoldo M. Targets for Drug Therapy for Autism Spectrum Disorder: Challenges and Future Directions. J Med Chem. 2017 Nov;60((22)):9114–41. doi: 10.1021/acs.jmedchem.7b00965. [DOI] [PubMed] [Google Scholar]
- 88.Millan MJ, Andrieux A, Bartzokis G, Cadenhead K, Dazzan P, Fusar-Poli P, et al. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov. 2016 Jul;15((7)):485–515. doi: 10.1038/nrd.2016.28. [DOI] [PubMed] [Google Scholar]
- 89.Bale TL, Abel T, Akil H, Carlezon WA, Jr, Moghaddam B, Nestler EJ, et al. The critical importance of basic animal research for neuropsychiatric disorders. Neuropsychopharmacology. 2019 Jul;44((8)):1349–53. doi: 10.1038/s41386-019-0405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paulsen JS, Long JD, Ross CA, Harrington DL, Erwin CJ, Williams JK, et al. PREDICT-HD Investigators and Coordinators of the Huntington Study Group Prediction of manifest Huntington's disease with clinical and imaging measures: a prospective observational study. Lancet Neurol. 2014 Dec;13((12)):1193–201. doi: 10.1016/S1474-4422(14)70238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tabrizi SJ, Scahill RI, Owen G, Durr A, Leavitt BR, Roos RA, et al. TRACK-HD Investigators Predictors of phenotypic progression and disease onset in premanifest and early-stage Huntington's disease in the TRACK-HD study: analysis of 36-month observational data. Lancet Neurol. 2013 Jul;12((7)):637–49. doi: 10.1016/S1474-4422(13)70088-7. [DOI] [PubMed] [Google Scholar]
- 92.Heilbronner U, Samara M, Leucht S, Falkai P, Schulze TG. The longitudinal course of schizophrenia across the lifespan: clinical, cognitive, and neurobiological aspects. Harv Rev Psychiatry. 2016 Mar-Apr;24((2)):118–28. doi: 10.1097/HRP.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Insel TR. Rethinking schizophrenia. Nature. 2010 Nov;468((7321)):187–93. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]