Abstract
Background
Radiomics has emerged as a new approach that can help identify imaging information associated with tumor pathophysiology. We developed and validated a radiomics nomogram for preoperative prediction of microvascular invasion (MVI) in hepatocellular carcinoma (HCC).
Methods
Two hundred and eight patients with pathologically confirmed HCC (training cohort: n = 146; validation cohort: n = 62) who underwent preoperative gadoxetic acid-enhanced magnetic resonance (MR) imaging were included. Least absolute shrinkage and selection operator logistic regression was applied to select features and construct signatures derived from MR images. Univariate and multivariate analyses were used to identify the significant clinicoradiological variables and radiomics signatures associated with MVI, which were then incorporated into the predictive nomogram. The performance of the radiomics nomogram was evaluated by its calibration, discrimination, and clinical utility.
Results
Higher α-fetoprotein level (p = 0.046), nonsmooth tumor margin (p = 0.003), arterial peritumoral enhancement (p < 0.001), and the radiomics signatures of hepatobiliary phase (HBP) T1-weighted images (p < 0.001) and HBP T1 maps (p < 0.001) were independent risk factors of MVI. The predictive model that incorporated the clinicoradiological factors and the radiomic features derived from HBP images outperformed the combination of clinicoradiological factors in the training cohort (area under the curves [AUCs] 0.943 vs. 0.850; p = 0.002), though the validation did not have a statistical significance (AUCs 0.861 vs. 0.759; p = 0.111). The nomogram based on the model exhibited C-index of 0.936 (95% CI 0.895–0.976) and 0.864 (95% CI 0.761–0.967) in the training and validation cohort, fitting well in calibration curves (p > 0.05). Decision curve analysis further confirmed the clinical usefulness of the nomogram.
Conclusions
The nomogram incorporating clinicoradiological risk factors and radiomic features derived from HBP images achieved satisfactory preoperative prediction of the individualized risk of MVI in patients with HCC.
Keywords: Microvascular invasion, Hepatocellular carcinoma, Gadoxetic acid, Radiomics, Nomogram
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer and the third most frequent cause of cancer mortality worldwide [1]. Surgical resection and liver transplantation are potential curative treatments for HCC. However, tumor recurrence occurs in 70% of cases after hepatectomy and 25% after liver transplantation, with 5-year overall survival of around 10–20% [2, 3, 4]. Microvascular invasion (MVI) is associated with aggressive biological features of HCC [5], which has been established as a risk factor for early recurrence and poor outcome [6, 7, 8]. To improve prognosis of HCC with MVI, anatomical subsegmentectomy or partial hepatectomy with wide resection margin is recommended [6, 9]. Furthermore, given the scarcity of liver grafts and the possibility of tumor recurrence, some authors have proposed that liver transplantation is not suitable for the candidates with MVI [10, 11]. Therefore, preoperative knowledge of MVI can help stratify high-risk individuals for postoperative recurrence, thus aiding in treatment decision-making.
Currently, MVI is diagnosed by histopathology after surgery in most cases, which may hinder a rational treatment regimen. Fortunately, recent studies have shown that certain imaging modalities, particularly gadoxetic acid-enhanced magnetic resonance (MR) imaging, has great potential for predicting MVI. Radiological characteristics such as nonsmooth tumor margin [12, 13], arterial peritumoral enhancement [12, 13, 14], tumor hypointensity [15], and peritumoral hypointensity on hepatobiliary phase (HBP) images [13, 16] are noninvasive imaging biomarkers for MVI prediction. However, these qualitative findings suffer from limitations including interobserver variability and lack of external validation [11].
Radiomics performs the high-throughput mining and quantification of routinely acquired radiologic images, which provides important insights into cancer phenotype and tumor microenvironment [17]. In contrast to tissue biomarkers, which invasively evaluate regional tumor microscopic heterogeneity, radiomic biomarkers noninvasively interrogate the entire tumor at the millimeter scale [18]. Quantitative parameters retrieved from computed tomography (CT) and MR images have demonstrated improved diagnostic and prognostic precision in a variety of tumors including brain [19], nasopharynx [20], and lung [21] cancers. For MVI prediction, a recent investigation showed the promise of radiomic features extracted from preoperative CT images in HCCs less than 5 cm with an area under curve (AUC) of 0.8 [11]. Gadoxetic acid-enhanced MR imaging carries additional information on tumor microstructure [22, 23]. However, to the best of our knowledge, no studies published radiomic analysis of gadoxetic acid-enhanced MR images for MVI prediction.
In this study, we aimed to develop and validate a radiomics nomogram for preoperative prediction of MVI in HCC. This nomogram allows a preoperative prediction of the individualized risk of MVI in patients with HCC and is particularly helpful for the therapeutic stratification.
Materials and Methods
Patients
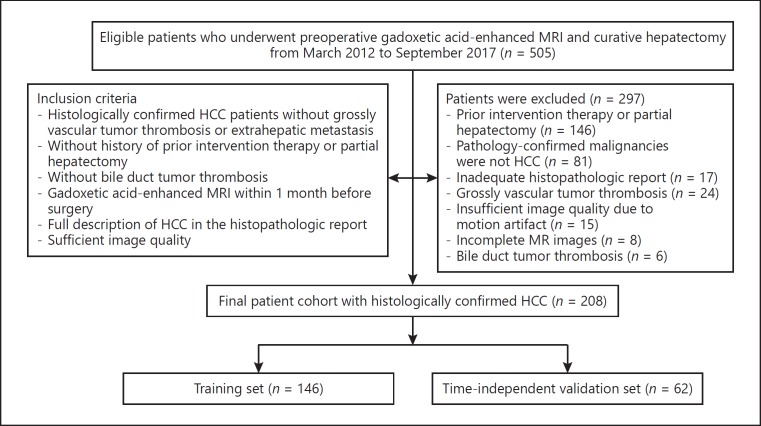
Our hospital ethics committee approved this retrospective study and waived patient informed consent. An institutional database was searched for all patients who underwent preoperative gadoxetic acid-enhanced MR imaging from March 2012 to September 2017 and found 505 patients. The final cohort consisted of 208 consecutive patients (183 men and 25 women; mean age, 55.5 ± 11.2 years) who met the following inclusion criteria (Fig. 1): (a) histologically confirmed HCC patients without evidence of grossly vascular invasion or extrahepatic metastasis at MR or CT imaging; (b) without history of prior intervention therapy or partial hepatectomy; (c) without bile duct tumor thrombosis; (d) gadoxetic acid-enhanced MRI within 1 month before surgery; (e) full description of HCC in the histopathologic report; (f) sufficient image quality. The cohort was divided into a training set (n = 146; 127 men and 19 women; mean age, 55.5 ± 10.9 years; from March 2012 to November 2016) and a time-independent validation set (n = 62; 56 men and 6 women; mean age, 55.5 ± 11.9 years; from December 2016 to September 2017) in a 7: 3 ratio according to the date of the MR examination.
Fig. 1.
Flow diagram of the study enrolment patients.
Laboratory Tests and Histology
Demographic, preoperative liver function tests and α-fetoprotein (AFP) levels were collected from medical records. Specimens from curative hepatectomy were sampled at the junction of the tumor and adjacent liver tissues in a 1: 1 ratio, at the 12, 3, 6, and 9 o'clock reference positions [24]. Pathological characteristics of tumor number, Edmondson-Steiner grade, MVI status, and cirrhosis of the noncancerous liver parenchyma were assessed in consensus by 2 dedicated pathologists from a team of experienced abdominal pathologists. MVI was defined as the presence of tumor in the portal vein, hepatic vein, or a large capsular vessel of the surrounding hepatic tissue lined with endothelium that was visible only on microscopy [24, 25].
MR Imaging
All study patients underwent gadoxetic acid-enhanced MR imaging using a 1.5T scanner (Magnetom Aera, Siemens Healthcare, Erlangen, Germany). Imaging sequences included axial T2-weighted imaging with fat suppression, diffusion-weighted imaging, in-phase and opposed-phase T1-weighted imaging, and pre-contrast and post-contrast dynamic three-dimensional T1-weighted volumetric-interpolated breath-hold examination (VIBE) at arterial phase (20–30 seconds), portal venous phase (60–70 s), delayed phase (180 s) and HBP (20 min) after injection of 0.025 mmol/kg of gadoxetic acid (Primovist, Bayer Schering Pharma, Berlin, Germany) into the cubital vein, followed by a 20-mL saline flush. Pre-contrast and HBP T1 mapping were also performed using a three-dimensional gradient-echo volumetric interpolated breath-hold examination with a dual flip-angle of 2 and 12°, and the quantitative precontrast and HBP T1 maps were automatically reconstructed on a voxel-by-voxel basis after data acquisition using the MapIt processing tool (MapIt software, Siemens Healthcare). Image acquisition parameters are shown in online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000494099).
Qualitative Analysis of MR Images
MR images were reviewed independently by 2 abdominal radiologists with 20 and 10 years of MR experience respectively. In case of any discrepancy, a consensus was reached after discussion. Both radiologists were aware that the lesions were HCCs but were blinded to all other clinical, laboratory, and histopathologic information. When patients had multiple tumors, the largest one was analyzed. The 2 radiologists assessed the following image features of HCC: (a) tumor size, defined as the maximum diameter on transverse HBP T1-weighted image; (b) tumor margin, categorized as smooth margin and nonsmooth margin on HBP images, round or oval tumors with smooth contour were identified as smooth margin, while irregular tumors with budding portion at the periphery were classified as nonsmooth tumor margin [13]; (c) arterial peritumoral enhancement, defined as detectable crescent or polygonal shaped enhancement surrounding the border on the arterial phase images, which becomes isointense during the delayed phase [13]; (d) enhancement pattern, classified as typical dynamic enhancement, with arterial hypervascularity, and portal washout, as well as atypical dynamic enhancement; (e) radiologic capsule appearance, defined as a hyper-enhanced structure encasing the tumor during the portal venous or delayed phase; (f) tumor hypointensity on HBP images, presenting as hypointense tumor on HBP images, when compared with the surrounding liver parenchyma; and (g) peritumoral hypointensity on HBP, defined as wedge-shaped or flame-like hypointense areas of the hepatic parenchyma located outside of the tumor margin on HBP images [16].
Radiomics Analysis of MR Images
Workflow
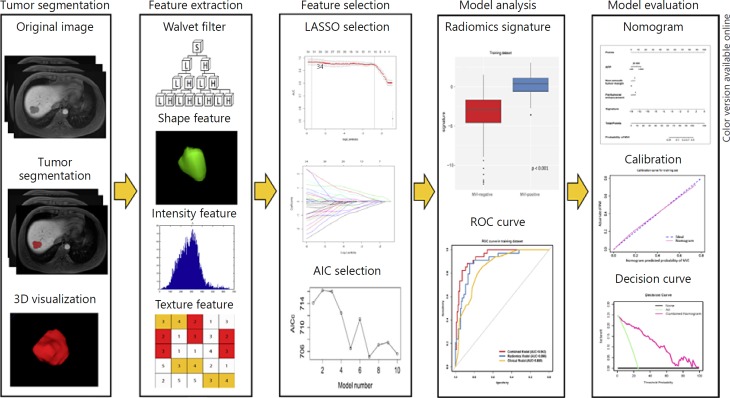
The workflow of the radiomics analysis included tumor segmentation, feature extraction, feature selection, and model construction and evaluation (Fig. 2).
Fig. 2.
Workflow of radiomics analysis. The radiomics workflow started with three-dimensional segmentation of tumor in MR images. After segmentation, radiomic features including shape, intensity and texture were extracted with or without wavelet filter of the images. Least absolute shrinkage and selection operator (LASSO) and Akaike information criteria (AIC) were used for the radiomic feature selection. Next, radiomics signature was built with the logistic regression model and receiver operating characteristic (ROC) curve was plotted. Finally, nomogram was developed and evaluated.
Image Segmentation
Three-dimensional segmentation of HCC was performed by a radiologist with an 8-year work experience using ITK-SNAP software (http://www.radiantviewer.com). Regions of interests were manually drawn on the T2-weighted images, diffusion-weighted images with b values of 500 s/mm2, unenhanced and enhanced arterial phase, portal venous phase, delayed phase, and HBP T1-weighted images, as well as pre-contrast T1 and HBP T1 maps, covering the whole tumor. The segmentation results were then validated by a senior radiologist with 18 years of work experience. A test-retest procedure was performed on a cohort of 20 randomly selected tumors not only to test the reproducibility of the extracted features from repeat segmentation, but also to exclude the features with intraclass correlation coefficients lower than 0.80.
Radiomics Feature Extraction
Image filtration was implemented on original image with an undecimated wavelet transform [26], which decomposed the original image into 8 decompositions. A set of 647 radiomic features were extracted from each segmented lesion on both the original and filtered images. These features are used to quantify tumor size (e.g., volume), shape (e.g., compactness, sphericity) and intensity (e.g., histogram-derived statistics of mean, SD, root mean square, median), as well as textural matrices including the gray level co-occurrence matrix [27], gray-level run-length matrix [28], gray-level size-zone matrix [27] and the neighborhood gray-tone difference matrix [29]. The detailed features are presented in online supplementary Table 2. All feature extraction was implemented using Matlab version 2014a (The MathWorks, Natick, MA, USA).
Clinicoradiological Risk Factors
Individual variables were analyzed for significant differences in the training and validate cohort using the Student t test, Mann Whitney U test, or Fisher exact tests, as appropriate. The univariate analysis was used to assess the single factor for discriminating MVI presence in the training cohort, and those significant variables at univariate analysis were entered into multivariate logistic regression analysis to determine potential risk factors of MVI.
Radiomic Feature Selection and Model Building
For each sequence, the extracted radiomic features were standardized into a normal distribution with z-scores to eliminate index dimension differences of the data. Features with intraclass correlation coefficients lower than 0.80 were excluded (online suppl. Fig. 1). The least absolute shrinkage and selection operator method was used to select features [30, 31] (online suppl. Fig. 2). Logistic regression analysis was then utilized to integrate the selected features with Akaike's information criterion as the stopping rule. Models with the minimum Akaike's information criterion value generated the final radiomics signature (online suppl. Tables 3, 4). The fusion radiomics signature combined the single MR sequence signature that showed satisfying predictive efficacy with an AUC greater than 0.7 in both the training and validation cohorts. A predictive model was built by incorporating the clinicoradiological risk factors and fusion radiomics signature with multivariable logistic regression modeling.
MVI Prediction Model Evaluation
The receiver operating characteristics curves were plotted and AUCs were used to quantify the discriminative efficacy for MVI prediction, and multiple comparisons of the curves were performed by the Delong test with Bonferroni-adjusted p values. The AUC with 95% CI, sensitivity, specificity, and accuracy were calculated.
Nomogram Construction and Evaluation
A radiomics nomogram was built on the predictive model as a graphical presentation. The discrimination performance of the nomogram was measured by Harrell's C-index [32]. Calibration curves were plotted to analyze the diagnostic performance of the nomogram in both the training and validation cohorts [33]. The Hosmer-Lemeshow test was used to assess the agreement between nomogram-predicted MVI and actual MVI from the calibration curves [34]. Decision curve analysis was conducted to determine the clinical usefulness of the nomogram by quantifying the net benefits at different threshold probabilities on the overall cohort [35].
Statistical Analysis
All the statistical analyses were performed using the SPSS (version 20, Chicago, IL, USA) and R software (version 3.4.1, Boston, MA, USA). A two-tailed p value less than 0.05 was considered statistically significant.
Results
Clinicoradiological Characteristics
Comparisons of clinicoradiological characteristics are shown in Table 1. No statistical difference was found in clinicoradiological factors between the training and validation cohorts (p = 0.070–0.987), except for enhancement pattern (p = 0.024) in MR imaging characteristics.
Table 1.
Comparisons of patient characteristics in training and validation datasets
| Characteristics | Training dataset (n = 146) |
Validation dataset(n =62) |
||||
|---|---|---|---|---|---|---|
| MVI present | MVI absent | p value | MVI present | MVI absent | p value | |
| Age, years* | 57 (51–60) | 55 (46–61) | 0.678 | 58 (45–64) | 55 (44–63) | 0.904 |
| Gender | 1.000 | 0.122 | ||||
| Male | 30 (88.2) | 97 (86.6) | 15 (78.9) | 41 (95.3) | ||
| Female | 4 (11.8) | 15 (13.4) | 4 (21.1) | 2 (4.7) | ||
| Etiology of liver disease | 0.626 | 0.860 | ||||
| HBV | 31 (91.2) | 104 (92.9) | 19 (100) | 41 (95.3) | ||
| HCV | 2 (5.9) | 3 (2.7) | 0 | 2 (4.7) | ||
| None or other | 1 (2.9) | 5 (4.5) | 0 | 0 | ||
| Total bilirubin | 0.613 | 0.860 | ||||
| <20.4 pmol/L | 32 (94.1) | 100 (89.3) | 19 (100) | 41 (95.3) | ||
| >20.4 pmol/L | 2 (5.9) | 12 (10.7) | 0 | 2 (4.7) | ||
| Alanine aminotransferase | 0.295 | 1.000 | ||||
| <40 U/L | 28 (82.4) | 80 (71.4) | 14 (73.7) | 32 (74.4) | ||
| >40 U/L | 6 (17.6) | 32 (28.6) | 5 (26.3) | 11 (25.6) | ||
| Aspartate aminotransaminase | 0.224 | 1.000 | ||||
| <35 U/L | 29 (85.3) | 82 (73.2) | 15 (78.9) | 33 (76.7) | ||
| >35 U/L | 5 (14.7) | 30 (26.8) | 4 (21.1) | 10 (23.3) | ||
| γ-Glutamyltransferase | 0.720 | 0.950 | ||||
| <60 U/L | 26 (76.5) | 80 (71.4) | 15 (78.9) | 32 (74.4) | ||
| >60 U/L | 8 (23.5) | 32 (28.6) | 4 (21.1) | 11 (25.6) | ||
| Platelets | 0.639 | 1.000 | ||||
| >125×109/L | 16 (47.1) | 60 (53.6) | 10 (52.6) | 22 (51.2) | ||
| <125×109/L | 18 (52.9) | 52 (46.4) | 9 (47.4) | 21 (48.8) | ||
| α-Fetoprotein | 0.003 | 0.114 | ||||
| <20 ng/mL | 9 (26.5) | 67 (59.8) | 7 (36.8) | 26 (60.5) | ||
| 20–400 ng/mL | 19 (55.9) | 32 (28.6) | 10 (52.6) | 11 (25.6) | ||
| >400 ng/mL | 6 (17.6) | 13 (11.6) | 2 (10.5) | 6 (14) | ||
| Edmondson-Steiner grade | 0.020 | 0.445 | ||||
| Grade I | 0 | 2 (1.8) | 0 | 2 (4.7) | ||
| Grade II | 16 (47.1) | 79 (70.5) | 8 (42.1) | 22 (51.2) | ||
| Grade III | 18 (52.9) | 31 (27.7) | 11 (57.9) | 19 (44.2) | ||
| Cirrhosis of background liver | 0.803 | 1.000 | ||||
| Absent | 11 (32.4) | 41 (36.6) | 8 (42.1) | 18 (41.9) | ||
| Present | 23 (67.6) | 71 (63.4) | 11 (57.9) | 25 (58.1) | ||
| Number of tumors | 0.720 | 0.068 | ||||
| Solitary | 27 | 93 | 13 | 39 | ||
| Multiple | 7 | 19 | 6 | 4 | ||
| MR imaging features | ||||||
| Tumor size, cm* | 2.4 (1.5–3.1) | 1.7 (1.3–2.7) | 0.049 | 2.2 (1.2–3.5) | 1.7 (1.0–2.7) | 0.144 |
| Tumor margin Smooth margin | 7 (20.6) | 73 (65.2) | <0.001 | 5 (26.3) | 30 (69.8) | 0.004 |
| Non-smooth margin | 27 (79.4) | 39 (34.8) | 14 (73.7) | 13 (30.2) | ||
| Peritumoral enhancement | <0.001 | 0.112 | ||||
| Absent | 11 (32.4) | 90 (80.4) | 9 (47.4) | 31 (72.1) | ||
| Present | 23 (67.6) | 22 (19.6) | 10 (52.6) | 12 (27.9) | ||
| Enhancement pattern | 0.460 | 0.590 | ||||
| Typical | 26 (76.5) | 94 (83.9) | 19 (100) | 40 (93) | ||
| Atypical | 8 (23.5) | 18 (16.1) | 0 | 3 (7) | ||
| Radiologic capsule | 0.612 | 0.434 | ||||
| Absent | 5 (14.7) | 23 (20.5) | 1 (5.3) | 7 (16.3) | ||
| Present | 29 (85.3) | 89 (79.5) | 18 (94.7) | 36 (83.7) | ||
| Tumor hypointensity on HBP | 1.000 | 1.000 | ||||
| Absent | 1 (2.9) | 4 (3.6) | 0 | 1 (2.3) | ||
| Present | 33 (97.1) | 108 (96.4) | 19 (100) | 42 (97.7) | ||
| Peritumoral hypointensity on HBP image | 0.006 | 0.068 | ||||
| Absent | 24 (70.6) | 102 (91.1) | 13 (68.4) | 39 (90.7) | ||
| Present | 10 (29.4) | 10 (8.9) | 6 (31.6) | 4 (9.3) | ||
Unless otherwise noted, data are shown as number of patients, with the percentage in parentheses.
Data are medians, with interquartile ranges in parentheses.
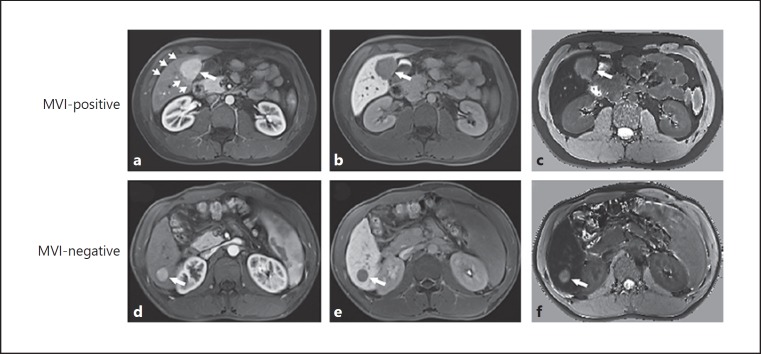
Univariate analysis showed serum AFP levels, Edmondson-Steiner HCC grade, tumor size, nonsmooth tumor margins, arterial peritumoral enhancement, and peritumoral hypointensity on HBP images were significantly related to MVI (p < 0.05). At the multivariate analysis, AFP levels (OR 1.903; 95% CI 1.018–3.644; p = 0.046), nonsmooth tumor margins (OR 4.817; 95% CI 1.781–14.412; p = 0.003), and arterial peritumoral enhancement (OR 5.322; 95% CI 2.033–14.551; p < 0.001) were independent predictors of MVI. The AUCs of combining the 3 predictors were 0.850 (95% CI 0.784–0.915) in the training cohort and 0.759 (95% CI 0.641–0.876) in the validation cohort (Table 2). Examples of typical radiological characteristics of MVI-positive and MVI-negative HCCs are shown in Figure 3.
Table 2.
Predictive efficacy of the clinicoradiological factors, radiomics signature and the predictive model
| Different models | Training dataset (n = 146) |
Validation dataset (n = 62) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| sensitivity, % | specificity, % | accuracy, % | AUC (95% CI) | sensitivity, % | specificity, % | accuracy, % | AUC (95% CI) | ||||
| Imaging traits +AFP | 85.3 | 71.4 | 74.7 | 0.850 | (0.784–0.915) | 73.7 | 67.4 | 69.4 | 0.759 | (0.641–0.876) | |
| HBP T1-w image | 88.2 | 68.8 | 73.3 | 0.754 | (0.668–0.840) | 63.2 | 65.1 | 64.5 | 0.705 | (0.570–0.840) | |
| HBP T1 map | 91.2 | 68.8 | 74.0 | 0.858 | (0.788–0.929) | 89.5 | 46.5 | 59.7 | 0.721 | (0.583–0.859) | |
| Fusion radiomics signature | 88.2 | 84.8 | 85.6 | 0.895 | (0.837–0.953) | 84.2 | 74.4 | 77.4 | 0.837 | (0.730–0.945) | |
| Predictive | model | 88.2 | 87.5 | 87.7 | 0.943 | (0.905–0.980) | 89.5 | 81.4 | 83.9 | 0.861 | (0.750–0.970) |
AUC, area under curve; HBP, hepatobiliary phase; T1-w, T1-weighted image; AFP, α-fetoprotein. Imaging traits refer to the non-smooth tumor margin and arterial peritumoral enhancement. Fusion radiomics signature represents the fusion radiomics signature of the HBP T1-weighted images and HBP T1 maps. The predictive model consists of radiomic features of HBP T1-weighted image and HBP T1 map, serum AFP level and imaging traits.
Fig. 3.
Representative gadoxetic acid-enhanced MR images of microvascular invasion (MVI)-positive and MVI-negative hepatocellular carcinoma. a Axial arterial phase image shows a hypervascular tumor (long arrow) with peritumoral enhancement (short arrows) in hepatic segment V, and (b) hepatobiliary phase (HBP) T1-weighted image and (c) HBP T1 map show nonsmooth tumor margin, suggestive of presence of MVI. d Axial arterial phase image displays a hypervascular mass (long arrow) without peritumoral enhancement in hepatic segment VI, and (e) HBP T1-weighted image and (f) HBP T1 map display smooth tumor margin, indicative of absence of MVI. Note HBP T1-weighted image shows hypointensity, while HBP T1 map displays hyperintensity, compared with surrounding liver parenchyma.
Performance of Radiomics Signature Using Single MR Sequence
The predictive performance of single radiomics signature on each MR sequence is summarized in online supplementary Table 5. For MVI prediction, the HBP T1-weighted image signature yielded an OR of 2.537 (95% CI 1.720–4.650, p < 0.001), and the HBP T1 map signature yielded an OR of 2.467 (95% CI 1.469–4.752; p < 0.001). Noticeably, radiomics signatures of HBP T1-weighted images and HBP T1 maps achieved satisfying performance, with AUCs of 0.754 (95% CI 0.668–0.840) and 0.858 (95% CI 0.788–0.929) in the training cohort, and AUCs of 0.705 (95% CI 0.570–0.840) and 0.721 (95% CI 0.583–0.859) in the validation cohort. Thus, HBP T1-weighted images and HBP T1 maps were further analyzed. The detailed feature information and the formulas of HBP T1-weighted images and HBP T1 maps radiomics signatures construction is provided in online supplementary Table 6 and online supplementary Formula.
Performance of Fusion Radiomics Signature Using Multi-Sequence
The fusion radiomics signature of the HBP T1-weighted images and HBP T1 maps was generated with the formula provided in the online Supplementary Formula. The distribution of the fusion radiomics signature is plotted in online supplementary Figure 3. There were significant differences of the fusion radiomic signatures between MVI-positive and MVI-negative HCCs in both the training (0.268 ± 1.485 vs. −3.447 ± 3.015; p < 0.001) and validation cohorts (0.197 ± 2.151 vs. −3.810 ± 5.006; p < 0.001).
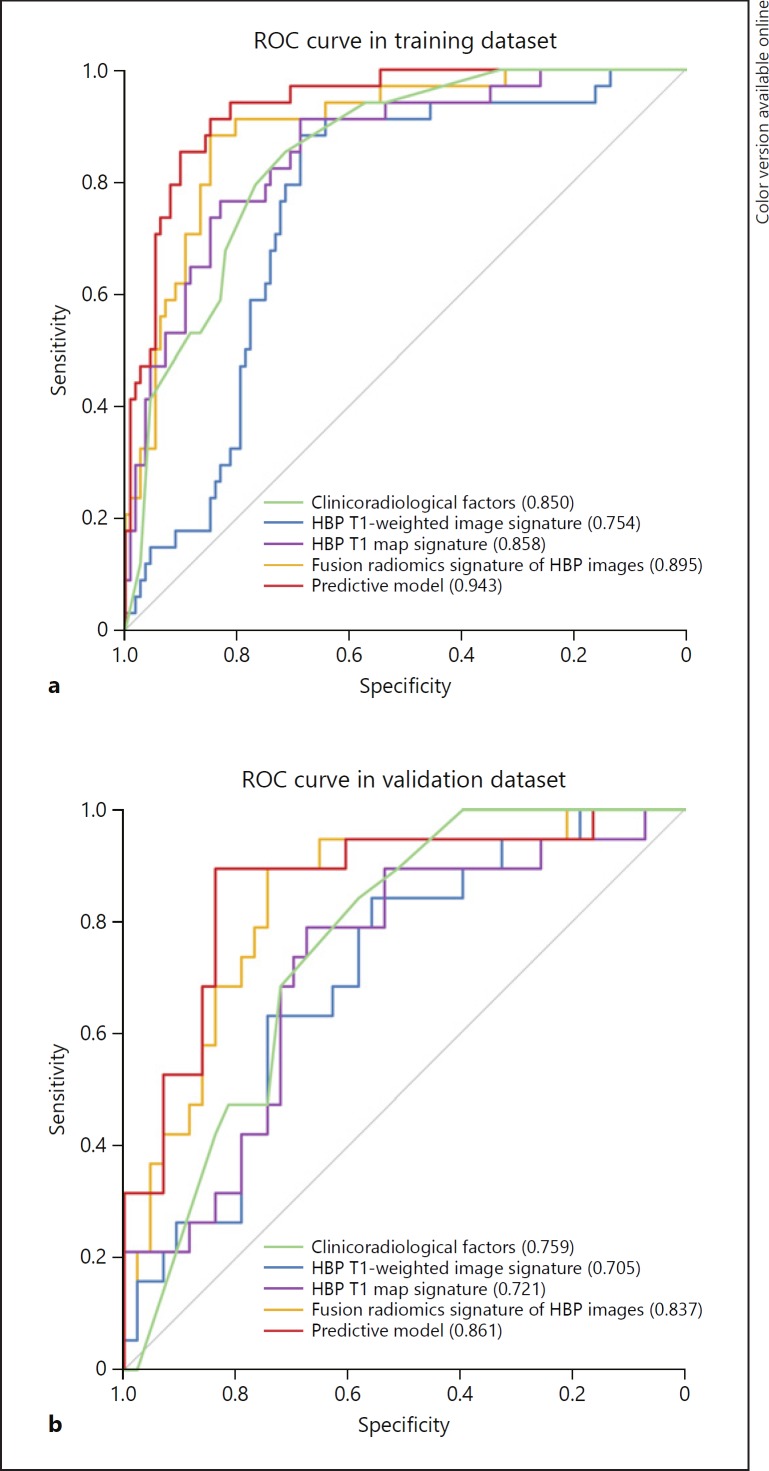
The fusion radiomics signature achieved better predictive efficacy for MVI than HBP T1-weighted image signature in both the training (AUCs 0.895 vs. 0.754; p = 0.002) and validation cohorts (AUCs 0.837 vs. 0.705; p = 0.040; Table 2; Fig. 4). The fusion radiomics signature performed better than the HBP T1 map signature in the validation cohort (AUCs 0.837 vs. 0.721; p = 0.037), but no statistical difference was found in the training cohort (0.895 vs. 0.858; p = 0.236).
Fig. 4.
Comparison of receiver operating characteristics (ROC) curves for prediction of microvascular invasion. ROC curves of clinicoradiological factors, radiomics signatures in hepatobiliary phase (HBP) T1-weighted images and HBP T1 map, the fusion radiomics signature of HBP images, and the predictive model that combines the clinicoradiological factors and the fusion radiomics signature in the (a) training and (b) validation dataset.
MVI Prediction Model
The MVI prediction model incorporated significant clinicoradiological factors with the fusion radiomics signature of HBP images. This model exhibited AUC of 0.943 (95% CI 0.905–0.980) in the training cohort with sensitivity, specificity, and accuracy of 88.2, 87.5, and 87.7% respectively. Applied in the validation cohort, the model yielded AUC of 0.861 (95% CI 0.750–0.970) with sensitivity, specificity, and accuracy of 89.5, 81.4, and 83.9% respectively. Moreover, the predictive model outperformed the combination of clinicoradiological factors (AUCs 0.943 vs. 0.850; p = 0.002) and fusion radiomics signature of HBP images (AUCs: 0.943 vs. 0.895; p = 0.031) in the training cohort. However, the performance of predictive model did not differ from that of the combination of clinicoradiological factors (AUCs 0.861 vs. 0.759; p = 0.111) and fusion radiomics signature of HBP images (AUCs 0.861 vs. 0.837; p = 0.548) in the validation cohort.
Development and Validation of the Nomogram
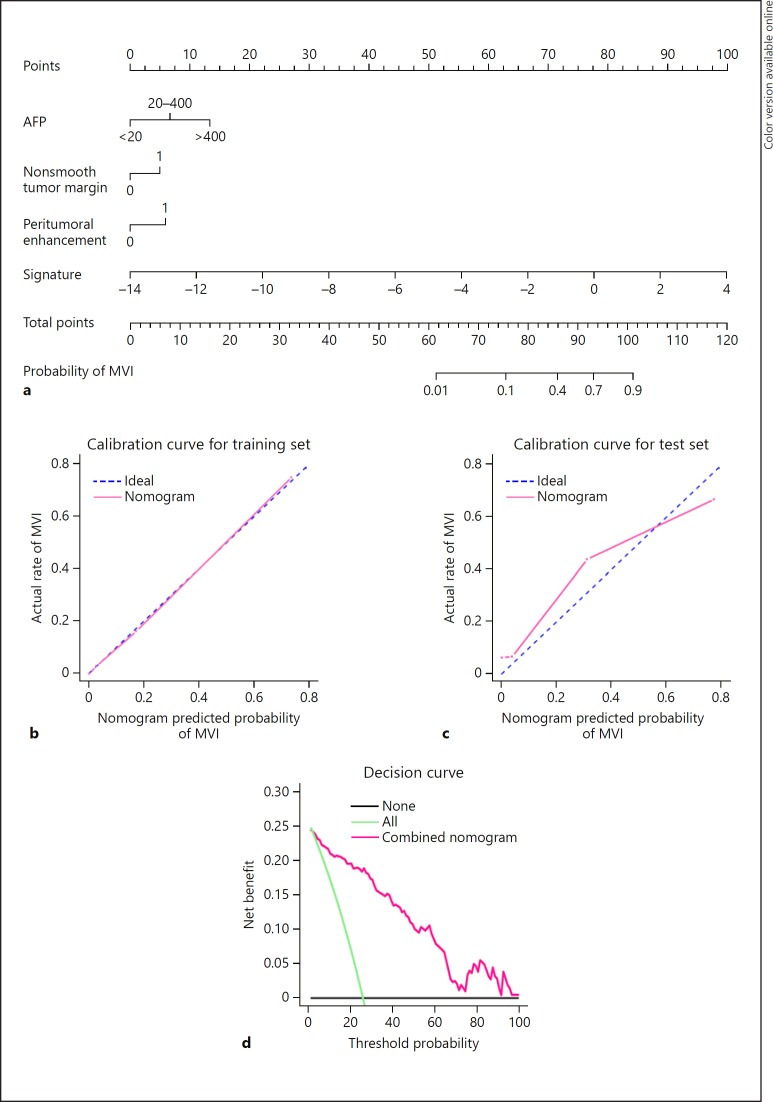
The nomogram based on the predictive model is presented in Figure 5a. Satisfactory predictive performances of the nomogram were obtained with a C-index of 0.936 (95% CI 0.895–0.976) in the training cohort and a C-index of 0.864 (95% CI 0.761–0.967) in the validation cohort. Calibration curves (Fig. 5b, c) showed that the predicted probabilities of the nomogram were closely aligned with the actual MVI estimates in both the training (p = 0.983) and validation cohort (p = 0.329). Decision curve for the nomogram is demonstrated in Figure 5d. The net benefit of the decision curve for the predictive nomogram is higher than that for assuming all patients have MVI when the threshold probability was greater than 2%. This suggests that basing therapy strategy on our nomogram will improve clinical outcome.
Fig. 5.
Nomogram for predicting microvascular invasion (MVI) probabilities, calibration of the nomogram and decision curve in the overall patients. a Nomogram for predicting microvascular invasion probabilities, calibration curves in the (b) training and (c) validation datasets, and (d) decision curve in the overall patients. a A radiomics nomogram integrated the fusion radiomics signature extracted from hepatobiliary phase images with clinicoradiological factors including serum AFP level, nonsmooth tumor margin and arterial peritumoral enhancement. b, c Calibration curves of the nomogram in the training and validation datasets; X-axis is nomogram-predicted probability of MVI. Y-axis is observed MVI, and the diagonal dashed line indicates the ideal prediction by a perfect model. d Decision curve for the nomogram predicting the MVI in the overall patients. The green line is the net benefit of assuming that all patients have MVI; the black line is the net benefit of assuming no patients have MVI; and the pink line is expected net benefit of per patient based on the predictive nomogram.
Discussion
In this work, we showed radiomics signatures of HBP T1-weighted images and HBP T1 maps were capable of predicting MVI in patients with HCC, and the fusion radiomics signature of HBP images could discriminate MVI-positive HCC with high sensitivity. Furthermore, we developed and validated a predictive model that incorporated serum AFP level, nonsmooth tumor margin, arterial peritumoral enhancement, and the fusion radiomics signature derived from HBP images, which exhibited high accuracy for preoperatively predicting MVI. Importantly, the conglomerate of the radiomics signature and clinicoradiological risk factors in the nomogram provides a straightforward, noninvasive and robust approach for personalized prediction of MVI before surgery.
This represents the first study to report the radiomics analysis on gadoxetic acid-enhanced MR imaging for preoperative prediction of MVI. Our results revealed that radiomics signatures of HBP T1-weighted images and HBP T1 maps achieved optimal performance among all the MR sequences, and the fusion radiomics signature of HBP images allowed MVI status to be stratified with favorable efficacy in both the training and validation cohorts. Previously, Zheng et al. [11] suggested that radiomics signature derived from preoperative CT images could be a potential predictor of MVI in HCC, with an AUC of 0.80 in tumors less than 5 cm. Predicting MVI with radiomic features in HBP images, we obtained an AUC of 0.943 and 0.861 in the training and time-independent validation cohorts. Noticeably, the fusion radiomics signature achieved a high sensitivity in both the training and validation cohorts. It was not surprising that the radiomics signature derived from HBP images performed better than that derived from CT in previous report because the intensity of HCC on HBP T1-weighted images was significantly correlated with MVI [15]. Besides HBP T1-weighted images, we conducted an additional radiomics analysis on HBP T1 map, as T1 map permitted more reliable quantitative measurement of signal intensity by overcoming some intrinsic limitations of T1-weighted image, and acquisitions with variable flip angles for T1 mapping can be easily incorporated into routine liver MR imaging [36]. In this study, the fusion radiomics signatures of HBP T1-weighted images and HBP T1 map achieved better efficacy than HBP T1-weighted image signature alone in both the training and validation cohort, and this indicated that HBP T1 map could serve as a complementary modality for predicting MVI.
During the construction of the fusion radiomics signature with 10 radiomic features, we discovered 3 features that were in concordance with the biological characteristics of HCC with MVI: sphericity (HBP T1 map_ori_Sph_dis), root mean square (HBP T1-w_Coif1_fos_root_ mean_square), and median of the intensity histogram (HBP T1 map_Coif4_fos_median). According to histology, MVI-positive HCC has an aggressive tendency to invade the tumor capsule and protrude into the noncancerous parenchyma, which leads to a higher frequency of irregular tumor margins [37]. The root mean square of the histogram represents the discrete degree of gray level in different tumor areas on the image, and the variance between the MVI-positive and MVI-negative HCC could be attributed to intratumoral heterogeneity (e.g., tumor cellularity, micro-necrosis, and inflammation) induced by MVI [38]. The presence of MVI contributed to a lower median of the intensity histogram because the lower signal intensity was found to be more frequent for MVI-positive HCCs than for MVI-negative HCCs [15]. The 7 other features derived from texture analysis also yielded significant information, which may further reflect macroscopic heterogeneities of MVI-positive HCCs.
In addition to the radiomics analysis, we also evaluated the preoperative clinicoradiological factors. Our results recapitulated previous findings that serum AFP levels, nonsmooth tumor margins and arterial peritumoral enhancement were independent variables associated with MVI. We observed the same trend of serum AFP levels increasing with the likelihood of MVI in HCC [25, 39]. Recent studies with gadoxetic acid also demonstrated that nonsmooth tumor margin on HBP images was a significant descriptor of MVI [12, 13]. Pathologic data showed that a single nodular type with extranodular growth or a confluent multinodular type was more frequently observed in MVI-positive HCC cases [40, 41], resulting in nonsmooth tumor margin as a feature of MVI-positive HCC on MR images. Additionally, prior studies [12, 13] are consistent with our results that arterial peritumoral enhancement on gadoxetic acid-enhanced MR imaging was a significant predictor of MVI. The explanation suggested by these reports was that compensatory arterial hyperperfusion occurs in the area of reduced portal flow due to minute portal branch occlusions caused by microscopic tumor thrombin in the adjacent nontumor hepatic parenchyma.
In the predictive model for MVI, we incorporated the fusion radiomics signature derived from HBP images and the significant clinicoradiological variables. The predictive model discriminated better than the combination of clinicoradiological risk factors in the training cohort. In this regard, the use of the fusion radiomics signature of HBP images could improve the predictive efficacy for MVI compared with clinicoradiological features alone.
Moreover, the nomogram based on the predictive model showed satisfactory predictive performance across the spectrum of MVI predictions in both the training cohort (C-index: 0.936) and validation cohort (C-index: 0.864) with good calibration. Lei et al. [25] first developed an MVI predictive nomogram based on clinicoradiological factors alone, yielding C-indexes of 0.81 and 0.80 in the training and validation cohort, respectively, which was less accurate than our nomogram. This also suggested the supplementary value of radiomic signature of HBP image in MVI prediction. Therefore, our nomogram represents an improvement on the basis of the radiomics signatures by extracting essential information related to MVI over the entire HCC in the HBP images. The use of our proposed nomogram may be an important method of assisting surgeon in therapeutic decision making, and this may promote personalized therapeutic regime in HCC patients.
There are some limitations to this study. On the one hand, this was a single-center retrospective study. Therefore, results from our database should be supplemented with further prospective validation by larger cohorts from other centers. On the other hand, we did not include genomic factors related with MVI. Banerjee et al. [42] mapped CT image features to 91 genes to predict histological MVI with high accuracy. However, technological complicity and high costs of multi-gene expression assays make radiogenomic venous invasion difficult to apply in routine clinical setting.
In conclusion, radiomic features of HBP T1-weighted images and HBP T1 maps are potential biomarkers for predicting MVI in patients with HCC. The predictive nomogram that incorporates clinicoradiological risk factors and fusion radiomics signatures of HBP images achieves satisfactory preoperative prediction of the individualized risk assessment of MVI.
Disclosure Statement
The authors declare that they have no conflicts of interest to disclose.
Supplementary Material
Supplementary data
Acknowledgment
This study was supported by the National Natural Science Foundation of China (81571661) and Zhongshan Hospital Science Foundation (2018ZSQN23).
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012 Mar;379((9822)):1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018 Mar;68((3)):526–49. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Schwartz M, Mazzaferro V. Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis. 2005;25((2)):181–200. doi: 10.1055/s-2005-871198. [DOI] [PubMed] [Google Scholar]
- 4.De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, et al. EUROCARE-5 Working Group Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE—5-a population-based study. Lancet Oncol. 2014 Jan;15((1)):23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 5.Mazzaferro V, Bhoori S, Sposito C, Bongini M, Langer M, Miceli R, et al. Milan criteria in liver transplantation for hepatocellular carcinoma: an evidence-based analysis of 15 years of experience. Liver Transpl. 2011 Oct;17((S2 Suppl 2)):S44–57. doi: 10.1002/lt.22365. [DOI] [PubMed] [Google Scholar]
- 6.Hwang S, Lee YJ, Kim KH, Ahn CS, Moon DB, Ha TY, et al. The Impact of Tumor Size on Long-Term Survival Outcomes After Resection of Solitary Hepatocellular Carcinoma: Single-Institution Experience with 2558 Patients. J Gastrointest Surg. 2015 Jul;19((7)):1281–90. doi: 10.1007/s11605-015-2849-5. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-Perálvarez M, Luong TV, Andreana L, Meyer T, Dhillon AP, Burroughs AK. A systematic review of microvascular invasion in hepatocellular carcinoma: diagnostic and prognostic variability. Ann Surg Oncol. 2013 Jan;20((1)):325–39. doi: 10.1245/s10434-012-2513-1. [DOI] [PubMed] [Google Scholar]
- 8.Sumie S, Nakashima O, Okuda K, Kuromatsu R, Kawaguchi A, Nakano M, et al. The significance of classifying microvascular invasion in patients with hepatocellular carcinoma. Ann Surg Oncol. 2014 Mar;21((3)):1002–9. doi: 10.1245/s10434-013-3376-9. [DOI] [PubMed] [Google Scholar]
- 9.Shi M, Guo RP, Lin XJ, Zhang YQ, Chen MS, Zhang CQ, et al. Partial hepatectomy with wide versus narrow resection margin for solitary hepatocellular carcinoma: a prospective randomized trial. Ann Surg. 2007 Jan;245((1)):36–43. doi: 10.1097/01.sla.0000231758.07868.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017 Jul;11((4)):317–70. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J, Chakraborty J, Chapman WC, Gerst S, Gonen M, Pak LM, et al. Hepatopancreatobiliary Service in the Department of Surgery of the Memorial Sloan Kettering Cancer Center. Research Staff in the Department of Surgery at Washington University School of Medicine Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma Using Quantitative Image Analysis. J Am Coll Surg. 2017 Dec;225((6)):778–788.e1. doi: 10.1016/j.jamcollsurg.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renzulli M, Brocchi S, Cucchetti A, Mazzotti F, Mosconi C, Sportoletti C, et al. Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology. 2016 May;279((2)):432–42. doi: 10.1148/radiol.2015150998. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Kim SH, Lee JE, Sinn DH, Park CK. Preoperative gadoxetic acid-enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J Hepatol. 2017 Sep;67((3)):526–34. doi: 10.1016/j.jhep.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Ahn SY, Lee JM, Joo I, Lee ES, Lee SJ, Cheon GJ, et al. Prediction of microvascular invasion of hepatocellular carcinoma using gadoxetic acid-enhanced MR and (18)F-FDG PET/CT. Abdom Imaging. 2015 Apr;40((4)):843–51. doi: 10.1007/s00261-014-0256-0. [DOI] [PubMed] [Google Scholar]
- 15.Kim JY, Kim MJ, Kim KA, Jeong HT, Park YN. Hyperintense HCC on hepatobiliary phase images of gadoxetic acid-enhanced MRI: correlation with clinical and pathological features. Eur J Radiol. 2012 Dec;81((12)):3877–82. doi: 10.1016/j.ejrad.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Kim KA, Kim MJ, Jeon HM, Kim KS, Choi JS, Ahn SH, et al. Prediction of microvascular invasion of hepatocellular carcinoma: usefulness of peritumoral hypointensity seen on gadoxetate disodium-enhanced hepatobiliary phase images. J Magn Reson Imaging. 2012 Mar;35((3)):629–34. doi: 10.1002/jmri.22876. [DOI] [PubMed] [Google Scholar]
- 17.Lambin P, Leijenaar RT, Deist TM, Peerlings J, de Jong EE, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017 Dec;14((12)):749–62. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 18.Bowen SR, Yuh WT, Hippe DS, Wu W, Partridge SC, Elias S, et al. Tumor radiomic heterogeneity: multiparametric functional imaging to characterize variability and predict response following cervical cancer radiation therapy. J Magn Reson Imaging. 2018 May;47((5)):1388–96. doi: 10.1002/jmri.25874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Götz M, Wick A, et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-oncol. 2018 May;20((6)):848–57. doi: 10.1093/neuonc/nox188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Tian J, Dong D, Gu D, Dong Y, Zhang L, et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin Cancer Res. 2017 Aug;23((15)):4259–69. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology. 2016 Dec;281((3)):947–57. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 22.Choi JW, Lee JM, Kim SJ, Yoon JH, Baek JH, Han JK, et al. Hepatocellular carcinoma: imaging patterns on gadoxetic acid-enhanced MR Images and their value as an imaging biomarker. Radiology. 2013 Jun;267((3)):776–86. doi: 10.1148/radiol.13120775. [DOI] [PubMed] [Google Scholar]
- 23.Kitao A, Matsui O, Yoneda N, Kozaka K, Kobayashi S, Koda W, et al. Hypervascular hepatocellular carcinoma: correlation between biologic features and signal intensity on gadoxetic acid-enhanced MR images. Radiology. 2012 Dec;265((3)):780–9. doi: 10.1148/radiol.12120226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cong WM, Bu H, Chen J, Dong H, Zhu YY, Feng LH, et al. Guideline Committee Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J Gastroenterol. 2016 Nov;22((42)):9279–87. doi: 10.3748/wjg.v22.i42.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei Z, Li J, Wu D, Xia Y, Wang Q, Si A, et al. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus-Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg. 2016 Apr;151((4)):356–63. doi: 10.1001/jamasurg.2015.4257. [DOI] [PubMed] [Google Scholar]
- 26.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014 Jun;5((1)):4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thibault G, Fertil B, Navarro C, Pereira S, Cau P, Levy N, et al. Shape and Texture Indexes Application to Cell Nuclei Classification. Int J Pattern Recognit Artif Intell. 2013;27((01)):1357002. [Google Scholar]
- 28.Galloway MM. Texture analysis using grey level run lengths. NASA STI/Recon Technical Report N. 1974. p. 75.
- 29.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern. 1989;19((5)):1264–74. [Google Scholar]
- 30.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997 Feb;16((4)):385–95. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Sauerbrei W, Royston P, Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med. 2007 Dec;26((30)):5512–28. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]
- 32.Uno H, Cai T, Pencina MJ, D'Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011 May;30((10)):1105–17. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015 Apr;16((4)):e173–e80. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med. 2007 Sep;35((9)):2052–6. doi: 10.1097/01.CCM.0000275267.64078.B0. [DOI] [PubMed] [Google Scholar]
- 35.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006 Nov-Dec;26((6)):565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haimerl M, Verloh N, Zeman F, Fellner C, Nickel D, Lang SA, et al. Gd-EOB-DTPA-enhanced MRI for evaluation of liver function: comparison between signal-intensity-based indices and T1 relaxometry. Sci Rep. 2017 Mar;7((1)):43347. doi: 10.1038/srep43347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu H, Zheng Q, Huang Y, Huang XW, Lai ZC, Liu J, et al. A non-smooth tumor margin on preoperative imaging assesses microvascular invasion of hepatocellular carcinoma: A systematic review and meta-analysis. Sci Rep. 2017 Nov;7((1)):15375. doi: 10.1038/s41598-017-15491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang WT, Yang L, Yang ZX, Hu XX, Ding Y, Yan X, et al. Assessment of Microvascular Invasion of Hepatocellular Carcinoma with Diffusion Kurtosis Imaging. Radiology. 2018 Feb;286((2)):571–80. doi: 10.1148/radiol.2017170515. [DOI] [PubMed] [Google Scholar]
- 39.Cucchetti A, Piscaglia F, Grigioni AD, Ravaioli M, Cescon M, Zanello M, et al. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: a pilot study. J Hepatol. 2010 Jun;52((6)):880–8. doi: 10.1016/j.jhep.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 40.Hui AM, Takayama T, Sano K, Kubota K, Akahane M, Ohtomo K, et al. Predictive value of gross classification of hepatocellular carcinoma on recurrence and survival after hepatectomy. J Hepatol. 2000 Dec;33((6)):975–9. doi: 10.1016/s0168-8278(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 41.Sumie S, Kuromatsu R, Okuda K, Ando E, Takata A, Fukushima N, et al. Microvascular invasion in patients with hepatocellular carcinoma and its predictable clinicopathological factors. Ann Surg Oncol. 2008 May;15((5)):1375–82. doi: 10.1245/s10434-008-9846-9. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015 Sep;62((3)):792–800. doi: 10.1002/hep.27877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data