Abstract
Background and Aims
Diabetes mellitus is a risk factor for hepatocellular carcinoma (HCC) in patients with nonalcoholic steatohepatitis (NASH). Dipeptidyl peptidase-4 inhibitor (DPP4i), an antidiabetic agent, is reported to affect cell proliferation. We aimed to investigate the effects of DPP4i on the progression of NASH-related HCC and its metabolic pathway in a mouse model.
Methods
A mouse model of NASH-related HCC was used in this study. Eight-week-old mice were administered either DPP4i (sitagliptin 30 mg/kg/day; DPP4i group; n = 8) or distilled water (control group; n = 8) for 10 weeks. Then, HCC progression was evaluated by computed tomography. Changes in metabolites of HCC tissue were analyzed by metabolomic analysis. The localization and expression of p62, Keap1, Nrf2, and MCM7 were evaluated by immunostaining and immunoblotting, respectively.
Results
The number and volume of HCC were significantly lower in the DPP4i group than in the control group (1.8 ± 1.2 vs. 4.5 ± 1.7/liver, p < 0.01; 11.2 ± 20.8 vs. 37.5 ± 72.5 mm<sup>3</sup>/tumor, p < 0.05). Metabolome analysis revealed that DPP4i significantly increased 6-phosphogluconic acid and ribose 5-phosphate levels and decreased the AMP-to-adenine and GMP-to-guanine ratios (AMP-to-adenine ratio 0.7 ± 0.2 vs. 2.0 ± 1.2, p < 0.01; GMP-to-guanine ratio 0.6 ± 0.3 vs. 1.5 ± 0.7, p < 0.01). Immunostaining showed that p62 was localized in the cytoplasm of HCC in the DPP4i group, while p62 was localized in the nucleus of HCC in the control group. Keap1, Nrf2, and MCM7 expression decreased significantly in the DPP4i group compared to that in the control group.
Conclusions
We demonstrated that DDP4i prevented the progression of NASH-related HCC in a mouse model. Furthermore, metabolome analysis revealed that DDP4i downregulated the pentose phosphate pathway with suppression of the p62/Keap1/Nrf2 pathway. Thus, DDP4i may prevent tumor progression through inhibition of metabolic reprogramming in NASH-related HCC.
Keywords: Hepatoma, Nonalcoholic fatty liver disease, Dipeptidyl peptidase-4, Antidiabetic agent, Pentose phosphate shunt
Introduction
Along with an increase in the global prevalence of nonalcoholic steatohepatitis (NASH), the number of patients with NASH-related hepatocellular carcinoma (HCC) is increasing worldwide [1]. The incidence of HCC is lower among patients with NASH than among patients with hepatitis C virus-related chronic liver disease [2]; however, liver cirrhosis, patatin-like phospholipase domain-containing protein 3 rs738409 single-nucleotide polymorphism, male sex, and obesity are associated with an increased risk of HCC in patients with NASH [3, 4]. In addition, diabetes mellitus is an independent risk factor for hepatocarcinogenesis in patients with NASH [5].
Dipeptidyl peptidase-4 inhibitor (DPP4i) is an antidiabetic agent that inhibits degradation of glucagon-like peptide-1 and glucose-dependent insulinotropic peptide, which leads to stimulation of insulin secretion and inhibition of glucagon secretion [6]. Furthermore, DPP4 inhibition is reported to improve hepatic steatosis by downregulation of sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase-1, and fatty acid synthase and by upregulation of peroxisome proliferator-activated receptor-α and adenosine monophosphate (AMP)-activated protein kinase in the liver [7, 8]. Thus, DPP4i is thought to be beneficial for diabetic patients with NASH.
In addition to its effects on the metabolism, DPP4 exerts various biological activities such as immune stimulation and degradation of the extracellular matrix [9]. DPP4i also inhibits proliferation of lymphocytes and smooth vascular muscles [10, 11]. Moreover, DPP4i has been reported to reduce the risk of colon cancer and lung metastases of colorectal cancer in in vitro and in vivo studies [12, 13, 14, 15]. In patients with HCC, DPP4 expression is known to be upregulated in hepatoma cells [16], and DPP4i is frequently administered to patients with chronic liver disease including NASH [17]; however, the effects of DPP4i on HCC proliferation remain unclear. Furthermore, the mechanisms for DPP4i-induced alteration in tumor cell proliferation have never been investigated by global analyses including metabolomic analysis.
Thus, this study aimed to investigate the effects of DPP4i on the progression of NASH-related HCC in a mouse model. In addition, we performed a metabolomic analysis to investigate mechanisms for DPP4i-induced alteration in the proliferation of HCC.
Materials and Methods
Materials
All reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan) unless otherwise indicated.
Animals
We employed a NASH-related HCC mouse model (STAMTM mouse; SMC Laboratories, Inc., Tokyo, Japan) in this study. The mice were developed as described previously [18]. Briefly, 2-day-old male C57BL/6J mice were injected with streptozotocin (200 μg/mouse) and fed a high-fat diet (HFD-32; Clea, Tokyo, Japan) from the age of 4 weeks, and NASH and HCC occurred at 8 and 18 weeks of age, respectively.
Study Design
The mice were housed in a pathogen-free animal facility at 23°C under a controlled 12-h light/dark cycle and had access to autoclaved water ad libitum. The 8-week-old mice were orally administered either DPP4i (sitagliptin 30 mg/kg/day; DPP4i group; n = 8) or distilled water (control group; n = 8) for 10 weeks. Their body weight was evaluated every day, and the amount of food intake was evaluated every other day.
At 18 weeks of age, contrast-enhanced computed tomography was performed under pentobarbital anesthesia. HCC was defined as a washout lesion, and the number and size of HCC lesions were evaluated. Then, blood and liver samples were taken under ether anesthesia. The samples were rapidly frozen by liquid nitrogen and stored at −80°C until analysis.
Histology and Nonalcoholic Fatty Liver Disease Activity Score
Systematic random sampling was applied throughout this study [19]. For histological examination, the liver samples were fixed in 10% buffered formalin overnight and embedded in paraffin. All sections were sectioned 5 μm thick and stained with hematoxylin-eosin. Nonalcoholic fatty liver disease (NAFLD) activity was evaluated by the NAFLD activity score, in which the following findings were evaluated semiquantitatively: steatosis (0–3 points), lobular inflammation (0–2 points), hepatocellular ballooning (0–2 points), and fibrosis (0–4 points) [20].
DNA Microarray
A DNA microarray was performed using HCC tissue from the control and DPP4i groups (n = 5 in each group). Cy3 (cyanine-3)-labeled cRNA was prepared from 100 ng total RNA using the Low Input Quick Amp Labeling Kit® (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions, followed by RNeasy® column purification (QIAGEN, Venlo, The Netherlands). Dye incorporation and cRNA yield were evaluated by a spectrophotometer (NanoDrop ND-2000, Thermo Fisher Scientific Inc., Yokohama, Japan). Following the manufacturer's instructions, 0.6 μg of Cy3-labeled cRNA was fragmented at 60°C for 30 min in a reaction volume of 25 μL containing a fragmentation buffer and blocking agent.
Upon completion of the fragmentation reaction, 25 μL of hybridization buffer was added to the fragmentation mixture and hybridized to a SurePrint G3 Mouse Gene Expression 8×60K Microarray (Agilent Technologies) for 17 h at 65°C in a rotating Agilent hybridization oven. After hybridization, the microarrays were washed for 1 min at room temperature with a wash buffer (Agilent Technologies). The microarray slides were scanned immediately after washing on the Agilent SureScan Microarray Scanner (G2600D) using one color scan setting for the 8×60K array slides. The scanned images were analyzed with Feature Extraction Software 11.5.1.1 (Agilent Technologies) using the default parameters to subtract the background and obtain spatially detrended processed signal intensities.
Metabolomic Analysis
Fifty milligram of frozen HCC tissue (n = 5 in each group) was plunged into 1,500 µL of 50% acetonitrile/deionized water containing internal standards (H3304-1002; Human Metabolome Technologies, Inc., Tsuruoka, Japan) at 0°C in order to inactivate the enzymes. The tissue was homogenized 3 times at 1,500 rpm for 120 s, and then the homogenate was centrifuged at 2,300 g for 5 min at 4°C. Subsequently, 800 µL of the upper aqueous layer was centrifugally filtered through a 5-kDa cutoff filter (Merck KGaA, Darmstadt, Germany) at 9,100 g for 120 min at 4°C to remove proteins. The filtrate was centrifugally concentrated and resuspended in 50 µL of deionized water for capillary electrophoresis (CE)-mass spectrometry (MS) analysis.
A metabolomic analysis was performed using C-Scope (Human Metabolome Technologies Inc., Yamagata, Japan) as previously described [21]. Briefly, cationic compounds were measured in the positive mode of CE-time-of-flight MS (TOFMS) (CE-MS Agilent CE-TOFMS System; Agilent Technologies), and anionic compounds were measured in the positive and negative modes of CE-tandem MS (MS/MS) (CE Agilent CE System; Agilent Technologies). Peaks detected by CE-TOFMS and CE-MS/MS were extracted using the automatic integration software (MasterHands, Keio University, Tsuruoka, Japan [22], and MassHunter Quantitative Analysis B.04.00, Agilent Technologies, respectively) to obtain peak information including m/z, migration time, and peak area. The peaks were annotated with putative metabolites from the metabolite database (Human Metabolome Technologies Inc.) based on their migration times in CE and m/z values determined by TOFMS and MS/MS.
Immunohistochemistry
The paraffin-embedded liver samples were sectioned 6 μm thick. The tissue sections were subjected to antigen retrieval by placing them in Dako Target Retrieval Solution (pH 6.0; Dako, Glostrup, Denmark) for 20 min in a microwave oven and incubating them with primary antibodies for phospho-p62 (Ser351) (MBL Co. Ltd., Nagoya, Japan), Keap-1 (Abcam plc., Cambridge, UK), or Nrf2 (MBL Co. Ltd.) diluted 1: 100 in phosphate-buffered saline (PBS) for 60 min at room temperature. The tissue sections were also stained with anti-minichromosome maintenance protein 7 (MCM7) antibody (Cell Signaling Technology, Inc., Danvers, MA, USA). After several washes with PBS, the sections were incubated with the secondary antibodies (EnvisionTM FLEX/HRP; Agilent Technologies) at room temperature for 30 min. Subsequently, the sections were washed with PBS and developed with 3,3′-diaminobenzidine.
Immunoblotting
Liver tissue was homogenized on ice in 1 mmol/L NaHCO3 containing protease inhibitors and stored at −80°C as previously described [23]. Equal amounts of protein (40 µg) from the liver homogenates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 7.5% acrylamide gel. The resolved proteins were transferred electrophoretically onto polyvinylidene difluoride membranes. The membranes were incubated with an anti-phospho-p62 antibody (Ser351) (MBL Co. Ltd.), an anti-Keap1 antibody (Abcam plc.), or an anti-Nrf2 antibody (MBL Co. Ltd.) and were subsequently incubated with an HRP-conjugated goat anti-rabbit IgG (Cell Signaling Technology, Inc.). Proteins were visualized by a luminol-based enhanced chemiluminescence reagent (SuperSignalTM West Dura Luminol/Enhancer Kit; Thermo Fisher Scientific Inc., Waltham, MA, USA).
Statistical Analysis
Differences between the DPP4i group and the control group were analyzed by the Wilcoxon rank-sum test. Time course differences between the DPP4i group and the control group were analyzed by two-way repeated-measures analysis of variance. p values of < 0.05 were considered significant.
For DNA microarray analysis, statistical analysis was performed using the Aqua t test according to the manufacturer's instruction (Takara Bio Inc., Kusatsu City, Japan). Briefly, the Student t test was performed on genes, which were obtained by quality control and showed a log2 ratio greater than 1 or a log2 ratio of less than −1 [24]. The false discovery rate was adjusted by the Benjamini-Hochberg method, and q values of < 0.05 were defined as statistically significant [25].
In metabolome analysis, hierarchical cluster analysis and principal component analysis were performed by PeakStat and SampleStat software (Human Metabolome Technologies), respectively. Detected metabolites were plotted on metabolic pathway maps using VANTED (Visualization and Analysis of Networks containing Experimental Data) [26]. Welch's t test was applied to assess differences between the two groups. p values of < 0.05 were considered significant.
Results
Effects of DPP4i on Body Weight and Food Intake
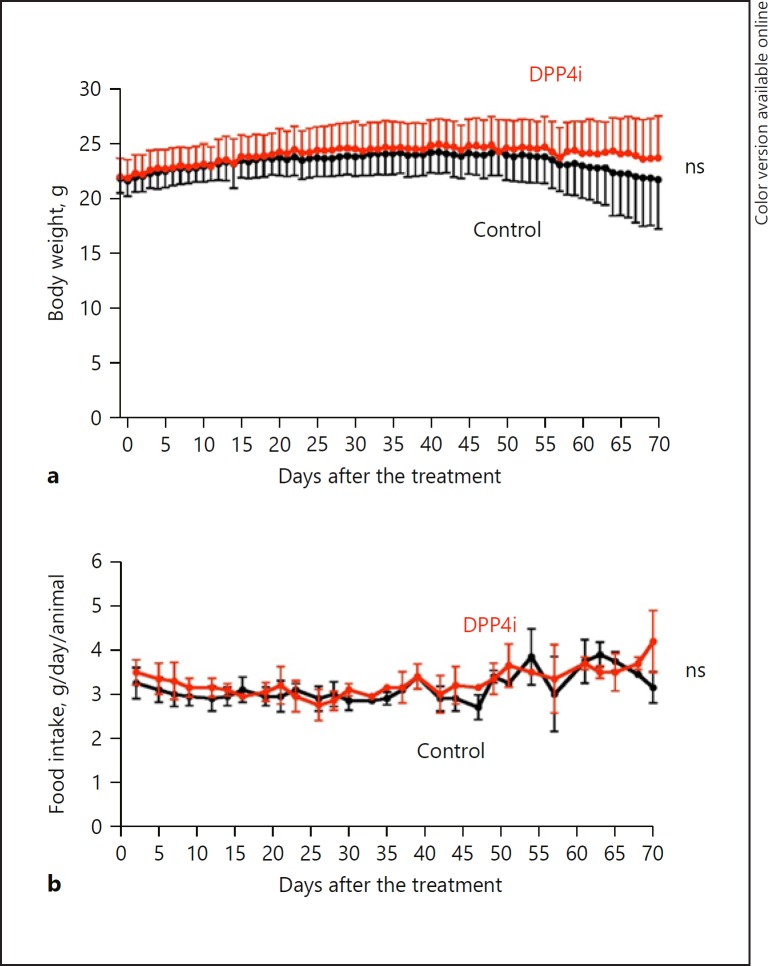
All mice in both groups survived until the end of this study. No significant difference was seen in body weight (Fig. 1a) or the amount of food intake (Fig. 1b) between the DPP4i group and the control group during the course of this study.
Fig. 1.
Effect of DPP4i on body weight and food intake in a mouse model of nonalcoholic steatohepatitis-related hepatocellular carcinoma. a Time course of body weight in the DPP4i group (red; n = 8) and in the control group (black; n = 8). b Time course of food intake in the DPP4i group (red; n = 8) and in the control group (black; n = 8). Statistical differences between the DPP4i group and the control group were analyzed by two-way repeated-measures analysis of variance. p values of < 0.05 were considered significant. DPP4i, dipeptidyl peptidase-4 inhibitor; ns, not significant.
Effects of DPP4i on Biochemical Examination Results
DPP4i significantly reduced the fasting blood glucose level and HbA1c value (Table 1). The serum insulin level was low in the control group, and DPP4i did not alter its level, since pancreatic beta-cells were damaged by streptozotocin treatment in this mouse model (Table 1). The serum levels of AST, ALT, and triglycerides were significantly reduced in the DPP4i group compared to those in the control group (Table 1).
Table 1.
Effects of DPP4i on biochemical examination results
| Control group (n = 8) | DPP4i group (n = 8) | p | |
|---|---|---|---|
| Fasting blood glucose, mg/dL | 452±75 | 366±84 | <0.05 |
| HbA1c, % | 6.1±0.5 | 5.6±0.6 | <0.05 |
| Fasting insulin, ng/mL | 0.16±0.04 | 0.17±0.05 | 0.67 |
| AST, IU/L | 101±26 | 77±21 | <0.05 |
| ALT, IU/L | 113±32 | 92±18 | <0.05 |
| Total cholesterol, mg/dL | 158±18 | 144±20 | 0.34 |
| Triglycerides, mg/dL | 425±124 | 310±101 | <0.05 |
Data are expressed as mean ± SD. Differences between the two groups were analyzed using the Wilcoxon rank-sum test.p values <0.05 were considered significant. DPP4i, dipeptidyl peptidase-4 inhibitor; HbA1c, hemoglobin A1c; AST, aspartate aminotransferase; ALT, alanine aminotransferase.
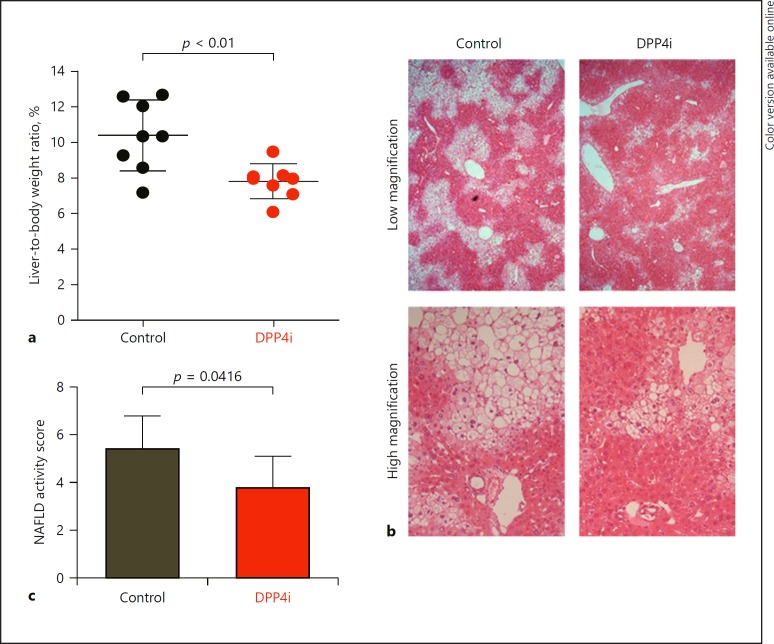
Effects of DPP4i on the Liver-to-Body Weight Ratio and NAFLD Activity Score
The liver-to-body weight ratio was significantly lower in the DPP4i group than in the control group (Fig. 2a). Microscopic images showed that hepatic steatosis was markedly decreased in the DPP4i group compared to the control group (Fig. 2b). The NAFLD activity score was also significantly lower in the DPP4i group than in the control group (Fig. 2c).
Fig. 2.
Effect of DPP4i on liver-to-body weight ratio and NAFLD activity score in a mouse model of nonalcoholic steatohepatitis-related hepatocellular carcinoma. a Liver-to-body weight ratio at 18 weeks of age in the DPP4i group (red; n = 8) and in the control group (black; n = 8). b Microscopic images for hematoxylin-eosin staining of the liver in the DPP4i group and in the control group. Upper row: low magnification. Lower row: high magnification. c NAFLD activity score in the DPP4i group (n = 8) and in the control group (n = 8). Statistical differences between the DPP4i group and the control group were analyzed by the Wilcoxon rank-sum test. p values of < 0.05 were considered significant. DPP4i, dipeptidyl peptidase-4 inhibitor; NAFLD, nonalcoholic fatty liver disease.
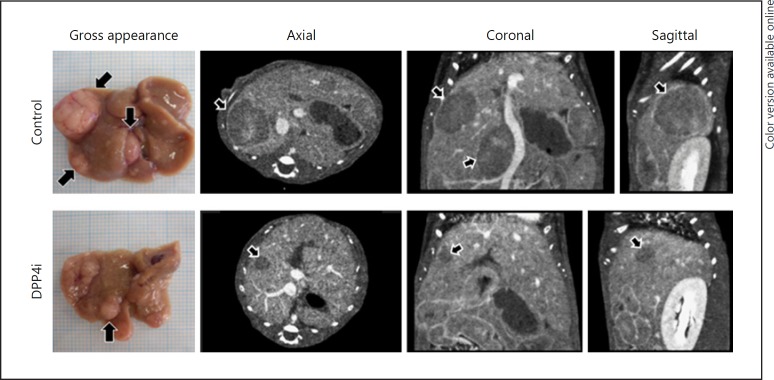
Effects of DPP4i on the Size and Number of HCC Lesions
In the control group, multiple large tumors were grossly visible in the liver (Fig. 3, left, upper row). However, the tumors were smaller in size and fewer in number in the DPP4i group (Fig. 3, left, lower row). Identical to the gross findings, contrast-enhanced computed tomography scan images showed that there were multiple large washout lesions, indicating HCCs, throughout the liver in the control group (Fig. 3, middle and right, upper row). On the other hand, the tumors were smaller and fewer in number in the DPP4i group than in the control group (Fig. 3, middle and right, lower row).
Fig. 3.
Effect of DPP4i on the size and number of nonalcoholic steatohepatitis-related hepatocellular carcinomas. Representative images of gross hepatic appearance (left) and contrast-enhanced computed tomography scans of the control group (upper row) and the DPP4i group (lower row). DPP4i, dipeptidyl peptidase-4 inhibitor.
Although there was no significant difference in the incidence of HCC between the DPP4i group and the control group, the number and volume of HCCs were significantly lower in the DPP4i group than in the control group (Table 2).
Table 2.
Effects of DPP4i on the incidence, number, and volume of HCCs
| Control group | DPP4i group | p | |
|---|---|---|---|
| Incidence of HCC | 100% (8/8) | 87.5% (7/8) | ns |
| Number of HCCs | 4.5±1.7/liver | 1.8±1.2/liver | <0.01 |
| Volume of HCCs | 37.5±72.5 mm3/liver | 11.2±20.8 mm3/liver | <0.05 |
Data are expressed as mean ± SD. Differences between the two groups were analyzed using the Wilcoxon rank-sum test.p values<0.05 were considered significant. HCC, hepatocellular carcinoma; DPP4i, dipeptidyl peptidase-4 inhibitor; ns, not significant.
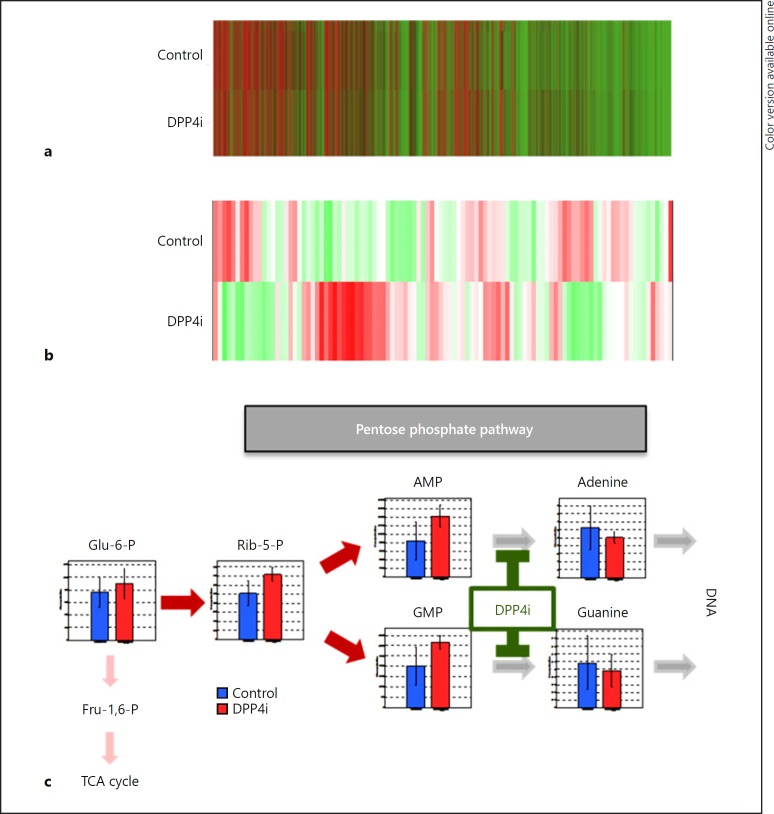
Effects of DPP4i on DNA Expression in HCC
The effects of DPP4i on the expression of 59,306 genes in HCC tissue were evaluated by DNA microarray analysis. In a hierarchical clustering analysis, the DNA expression pattern in the DPP4i group was similar to that in the control group, and there was no significant change in cluster classification between the two groups (Fig. 4a). In fact, gene expression was not significantly different between the DPP4i group and the control group (see online suppl. Table 1; for all online suppl. material, see www.karger.com/doi/10.1159/000491763).
Fig. 4.
Effect of DPP4i on gene expression and metabolite levels in hepatocellular carcinoma. a Heat map generated from DNA microarray data of the DPP4i group (n = 5) and the control group (n = 5). b Heat map generated from metabolomic analysis data of the DPP4i group (n = 5) and the control group (n = 5). c A scheme of DPP4i-caused changes in the metabolomic pathway. Statistical differences between the DPP4i group and the control group were analyzed by the Wilcoxon rank-sum test. p values of < 0.05 were considered significant. DPP4i, dipeptidyl peptidase-4 inhibitor; Glu, glucose; P, phosphate; Fru, fructose; TCA, tricarboxylic acid cycle; Rib, ribose; AMP, adenosine monophosphate; GMP, guanosine monophosphate; DNA, deoxyribonucleic acid.
Effects of DPP4i on Metabolites in HCC
The effects of DPP4i on 116 metabolites including 64 anions and 52 cations in HCC tissue were evaluated by CE-TOFMS and CE-MS/MS. In a hierarchical clustering analysis, the heat map of the metabolite expression pattern of the DPP4i group was different from that of the control group (Fig. 4b). In a comparative analysis, 7 anions (metabolites 2, 21, 23, 36, 43, 44, and 46 in online suppl. Table 2) and 2 cations (metabolites 80 and 88 in online suppl. Table 2) were significantly altered by DPP4i treatment, and most of these metabolites were associated with purine metabolism pathways, especially with the pentose phosphate pathways.
Metabolic pathway maps of the pentose phosphate pathways showed that intracellular levels of glucose 6-phosphate, ribose 5-phosphate, AMP, and GMP were higher in the DPP4i group than in the control group. However, intracellular adenine and guanine levels were lower in the DPP4i group than in the control group (Fig. 4c). DPP4i significantly decreased the AMP-to-adenine and GMP-to-guanine ratios (AMP-to-adenine ratio 0.7 ± 0.2 vs. 2.0 ± 1.2, p < 0.01; GMP-to-guanine ratio 0.6 ± 0.3 vs. 1.5 ± 0.7, p < 0.01), indicating inhibition of the final enzymatic reaction in adenine and guanine production.
Effects of DPP4i on the Differentiation of HCC and Expression of phosho-p62, Keap1, and Nrf2 in HCC Tissue
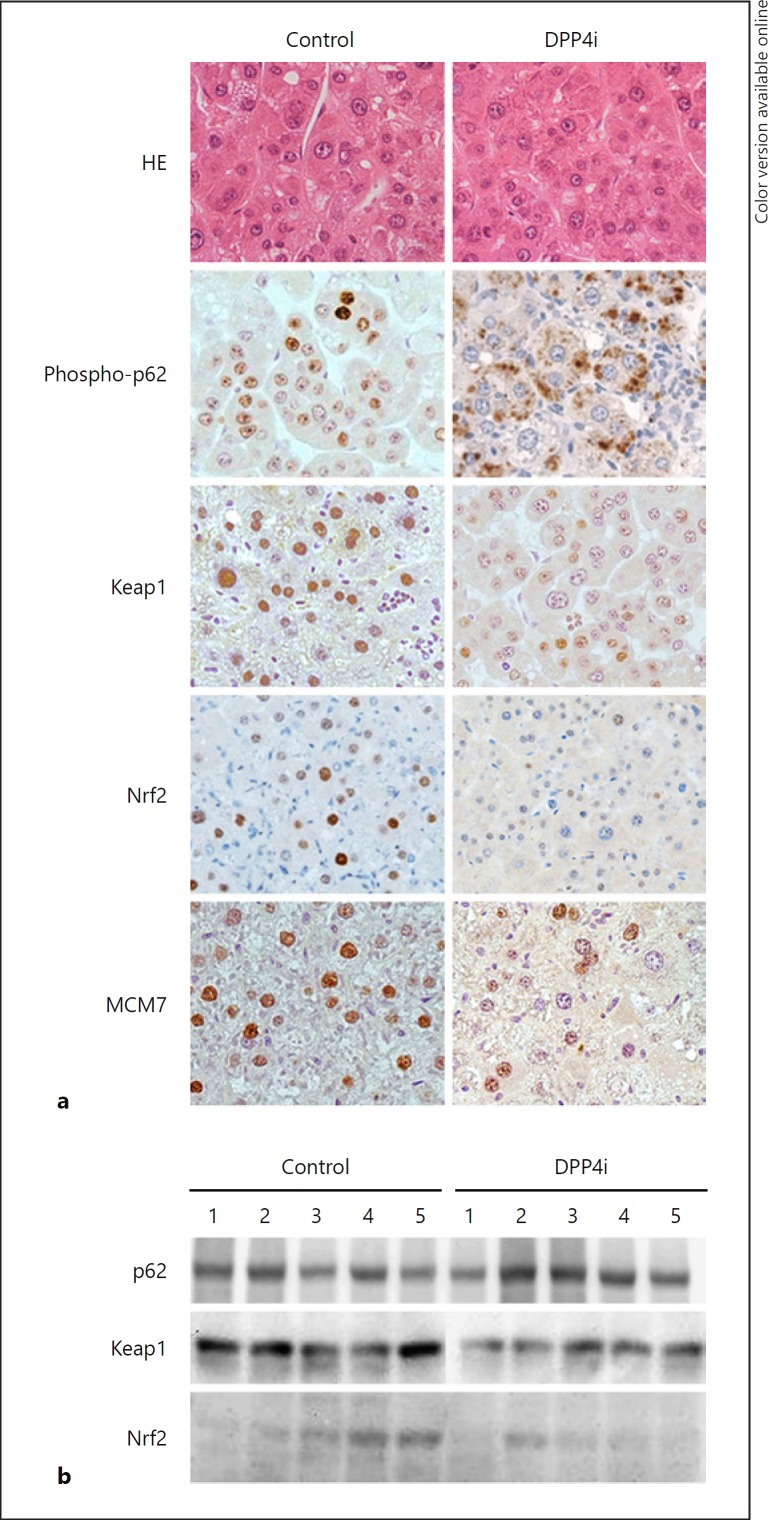
The HCC was a well-differentiated adenocarcinoma in both the control and the DPP4i group (Fig. 5a). Effects of DPP4i on the expression of phospho-p62, Keap1, Nrf2, and MCM7 in HCC tissue were evaluated. Immunostaining showed that phospho-p62, Keap1, and Nrf2 were localized in the nucleus of tumor cells in the control group (Fig. 5a, left column), whereas phospho-p62 was localized in the cytoplasm of tumor cells in the DPP4i group (Fig. 5a, right column). In the DPP4i group, expression of Keap1 and Nrf2 was weak (Fig. 5a, right column). Nuclear expression of MCM7 was also weak in the DPP4i group compared to that in the control group (Fig. 5a, right column).
Fig. 5.
Effect of DPP4i on the differentiation of hepatocellular carcinoma and expression of p62, Keap1, Nrf2, and MCM7 in hepatocellular carcinoma. a The differentiation of hepatocellular carcinoma was evaluated by HE staining in the DPP4i group (n = 5) and the control group (n = 5). Immunohistochemistry for p62, Keap1, Nrf2, and MCM7. The expression of each protein is visualized by 3,3′-diaminobenzidine (brown color) in the DPP4i group (n = 5) and the control group (n = 5). b Immunoblotting for p62, Keap1, and Nrf2. Each protein is visualized by a luminol-based enhanced chemiluminescence reagent in the DPP4i group (n = 5) and the control group (n = 5). DPP4i, dipeptidyl peptidase-4 inhibitor; HE, hematoxylin-eosin.
On immunoblotting, there was no significant change in phospho-p62 expression between the control group and the DPP4i group. Keap1 and Nrf2 expression was decreased in the DPP4i group compared to the control group (Fig. 5b).
Discussion
In this study, we demonstrated that DPP4i treatment prevented the progression of NASH-related HCC in a mouse model. A metabolomic analysis revealed that DPP4i regulated metabolites associated with the pentose phosphate pathway in HCC tissue. Furthermore, DPP4i inhibited the nuclear localization of p62 and suppressed Keap1 and Nrf2 expression in HCC. These data indicate that DPP4i prevented the progression of NASH-related HCC through downregulation of the pentose phosphate pathway via inhibition of the p62/Keap1/Nrf2 pathway.
Treatment with DPP4i significantly improved steatohepatitis in this study. Similarly, DPP4i has been reported to improve steatohepatitis in various types of NASH mouse models [7, 27, 28]. Akiyama et al. [29] reported that deletion of Nrf2 results in the development of NASH. However, Nrf2 expression was not seen in noncancerous liver tissue of either the control group or the DPP4i group in this study. Thus, the Nrf2 pathway may not be involved in the development of NASH in our model. Although the mechanisms for the improvement of steatohepatitis are unclear in this study, Shirakawa et al. [7] reported that DPP4i increases the expression of peroxisome proliferator-activated receptor-α and decreases the expression of sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase-1, and fatty acid synthase in the liver, leading to improvement of hepatic steatosis. Ideta et al. [8] also reported that DPP4i increases hepatic expression levels of phosphorylated AMP-activated protein kinase protein, resulting in suppression of de novo lipogenesis. Thus, our findings are in good agreement with those of previous reports, and the effect of DPP4i on lipid metabolism in hepatocytes could be the mechanism for improving steatohepatitis.
Although DPP4 expression is known to be upregulated in human HCC [16], the effect of DPP4i on the progression of HCC has never been reported. Here, we first demonstrated that DPP4i treatment prevented the progression of NASH-related HCC. In general, differentiation of HCC is closely associated with tumor progression. However, in this study, DPP4i did not affect the differentiation of HCC in this study, while it improved NASH. Therefore, we cannot deny the possibility that DDP4i improved the liver histological changes, which delayed the occurrence of HCC, not hepatocarcinogenesis. Thus, it is still unclear whether the preventive mechanism is due to an improvement of NASH or direct antitumor activity. Insulin enhances tumor progression, and the serum insulin level is significantly elevated in patients with NASH [23, 30]. Since insulin is associated with progression of HCC [30, 31], one would think that DPP4i improves hyperinsulinemia, resulting in prevention of tumor progression. However, the pancreatic beta-cells were damaged by streptozotocin treatment, and the serum insulin levels were low in the DPP4i group, suggesting that insulin-associated tumor growth might not be involved in the DPP4i-induced tumor prevention in this study. Recently, Yorifuji et al. [15] showed that DPP4i prevents colon carcinogenesis in mice with type 2 diabetes through suppression of mucosal IL-6 expression. Ye et al. [32] also showed that knockdown of DPP4i decreased the growth and liver metastasis of pancreatic cancer in vivo by using xenograft animal models. In addition, they showed that knockdown of DPP4 expression inhibited cell growth, migration, invasion, and colony formation and increased cell apoptosis of pancreatic cancer cells. Similar findings have been reported in thyroid tumor [33], endometrial carcinoma [34], and breast cancer [35]. Thus, DPP4i may have directly prevented the progression of HCC in this study.
DPP4 exerts various biological activities, such as inactivation of glucagon-like peptide-1, immune stimulation, binding to and degradation of the extracellular matrix, and resistance to anticancer agents [9]. In order to investigate the mechanisms for DPP4i-induced inhibition of tumor progression, we performed global analyses. First, we performed a DNA microarray analysis; however, none of the genes were significantly altered. Then, we performed a metabolome analysis and found that 9 metabolites were significantly altered by DPP4i treatment. These metabolites are associated with the pentose phosphate pathway, indicating a DPP4i-induced alteration of the pentose phosphate pathway. The pentose phosphate pathway is associated with cell proliferation via the production of nucleotide precursors of DNA from glucose 6-phosphate and is crucial for the “Warburg effect” of cancer cells [36, 37]. The pentose phosphate pathway is reported to be upregulated in HCC [38]. DPP4i is known to suppress DNA synthesis in keratinocytes [39]. In this study, we also demonstrated that nuclear expression of MCM7, which is associated with DNA replication [40] and progression of HCC [41], was low in the DPP4i group. Taken together, DPP4i might inhibit the pentose phosphate pathway, leading to suppression of DNA replication and subsequent prevention of HCC progression in this study.
The pentose phosphate pathway is known to be regulated by Nrf2-asccociated enzymes in cancer cells [42, 43]. Moreover, activation of Nrf2 is regulated by p62 and Keap1 in cancer cells [44]. In this study, we demonstrated that DPP4i suppressed the nuclear localization of p62 and inhibited Keap1 and Nrf2 expression in tumor cells. In previous studies, accumulation of p62 was reported to be associated with Nrf2 activation in human HCC cell lines, and upregulation of Nrf2 was related to progression of HCC, including metastasis [45, 46, 47]. In addition, the persistent activation of p62-induced Nrf2 contributes to the proliferation of human HCC cell lines [44]. Meanwhile, inhibition of the p62/Keap1/Nrf2 pathway significantly enhanced the erastin- and sorafenib-induced suppression of HCC [46]. Although the mechanisms for DPP4i-induced suppression of the p62/Keap1/Nrf2 pathway remain unclear, previous studies support that DPP4i suppresses the p62/Keap1/Nrf2 pathway, leading to the prevention of HCC progression in this study (Fig. 6). In contrast, Wang et al. [47] recently reported that DPP4i upregulates Nrf2 expression and enhances cancer cell migration and the promotion of metastasis in xenograft mouse models. Thus, DPP4i may not universally prevent tumor progression, and large-scale epidemiological observations from patients with diabetes treated with DPP4i are required.
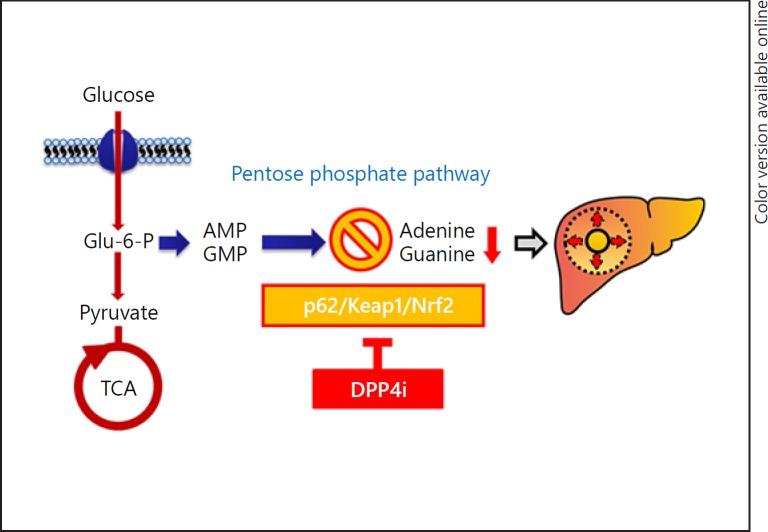
Fig. 6.
A scheme of possible mechanisms for DPP4i-induced HCC prevention. DPP4i may downregulate the p62/Keap1/Nrf2 pathway in HCC, leading to suppression of the pentose phosphate pathway. Lower adenine and guanine levels, end products of the pentose phosphate pathway, may inhibit HCC progression through the suppression of nucleic acid production. DPP4i, dipeptidyl peptidase-4 inhibitor; HCC, hepatocellular carcinoma; Glu, glucose; P, phosphate; TCA, tricarboxylic acid cycle; AMP, adenosine monophosphate; GMP, guanosine monophosphate.
Besides DPP4i, other antidiabetic agents such as metformin and sodium-glucose cotransporter 2 inhibitors are reported to exert preventive effects on NASH-related HCC. Metformin prevents HCC through the activation of AMP-activated protein kinase in a high-fat diet-fed mouse model [48]. Sodium-glucose cotransporter 2 inhibitor prevents NASH-related HCC through healthy adipose expansion in Western diet-fed melanocortin 4 receptor-deficient mice [49]. Thus, the suppressive mechanisms for NASH-related HCC differ with antidiabetic agents, and a combination therapy of different antidiabetic agents may have potent suppressive effects on NASH-related HCC.
A limitation of this study is that the STAM mouse is an insulin-deficient animal model. By using STAM mice, we could reveal that insulin-independent mechanisms might be involved in DPP4i-induced tumor prevention. However, insulin resistance is a feature of patients with NASH and is one of the main promoters of NASH-related HCC [30, 31]. Thus, further study is required to investigate the efficacy of DPP4i regarding HCC progression in insulin-resistant animal models for NASH.
In conclusion, we showed that DPP4i prevented NASH-related HCC in a mouse model. A metabolomic analysis revealed that DPP4i inhibited the pentose phosphate pathway in HCC tissue. Furthermore, DPP4i inhibited the nuclear localization of p62 and suppressed Keap1 and Nrf2 expression in tumor cells. Thus, our findings suggest that DPP4i prevents the progression of NASH-related HCC through downregulation of the pentose phosphate pathway via inhibition of the p62/Keap1/Nrf2 pathway.
Statement of Ethics
This animal study was approved by the Institutional Animal Care and Use Committee at Kurume University School of Medicine.
Disclosure Statement
All authors disclose no conflicts.
Funding Sources
This research was supported by AMED under Grants No. JP17fk0210304 and JP18fk0210040.
Author Contributions
T.K. participated in the study's conception and design, the acquisition and interpretation of the data, and drafting of the manuscript. D.N. participated in the analysis and interpretation of the data. H.K. and T.T. participated in the critical revision of the manuscript.
Supplementary Material
Supplementary data
Supplementary data
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016 Jul;64((1)):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology. 2010 Jun;51((6)):1972–8. doi: 10.1002/hep.23527. [DOI] [PubMed] [Google Scholar]
- 3.White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol. 2012;10:1342–1359. doi: 10.1016/j.cgh.2012.10.001. e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singal AG, Manjunath H, Yopp AC, Beg MS, Marrero JA, Gopal P, et al. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: a meta-analysis. Am J Gastroenterol. 2014 Mar;109((3)):325–34. doi: 10.1038/ajg.2013.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawamura Y, Arase Y, Ikeda K, Seko Y, Imai N, Hosaka T, et al. Large-scale long-term follow-up study of Japanese patients with non-alcoholic Fatty liver disease for the onset of hepatocellular carcinoma. Am J Gastroenterol. 2012 Feb;107((2)):253–61. doi: 10.1038/ajg.2011.327. [DOI] [PubMed] [Google Scholar]
- 6.Gallwitz B. Sitagliptin: profile of a novel DPP-4 inhibitor for the treatment of type 2 diabetes (update) Drugs Today (Barc) 2007 Nov;43((11)):801–14. doi: 10.1358/dot.2007.43.11.1157620. [DOI] [PubMed] [Google Scholar]
- 7.Shirakawa J, Fujii H, Ohnuma K, Sato K, Ito Y, Kaji M, et al. Diet-induced adipose tissue inflammation and liver steatosis are prevented by DPP-4 inhibition in diabetic mice. Diabetes. 2011 Apr;60((4)):1246–57. doi: 10.2337/db10-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ideta T, Shirakami Y, Miyazaki T, Kochi T, Sakai H, Moriwaki H, et al. The Dipeptidyl Peptidase-4 Inhibitor Teneligliptin Attenuates Hepatic Lipogenesis via AMPK Activation in Non-Alcoholic Fatty Liver Disease Model Mice. Int J Mol Sci. 2015 Dec;16((12)):29207–18. doi: 10.3390/ijms161226156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itou M, Kawaguchi T, Taniguchi E, Sata M. Dipeptidyl peptidase-4: a key player in chronic liver disease. World J Gastroenterol. 2013 Apr;19((15)):2298–306. doi: 10.3748/wjg.v19.i15.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro MM, Stoppa CL, Valduga CJ, Okuyama CE, Gorjão R, Pereira RM, et al. Sitagliptin inhibit human lymphocytes proliferation and Th1/Th17 differentiation in vitro. Eur J Pharm Sci. 2017 Mar;100:17–24. doi: 10.1016/j.ejps.2016.12.040. [DOI] [PubMed] [Google Scholar]
- 11.Ervinna N, Mita T, Yasunari E, Azuma K, Tanaka R, Fujimura S, et al. Anagliptin, a DPP-4 inhibitor, suppresses proliferation of vascular smooth muscles and monocyte inflammatory reaction and attenuates atherosclerosis in male apo E-deficient mice. Endocrinology. 2013 Mar;154((3)):1260–70. doi: 10.1210/en.2012-1855. [DOI] [PubMed] [Google Scholar]
- 12.Femia AP, Raimondi L, Maglieri G, Lodovici M, Mannucci E, Caderni G. Long-term treatment with Sitagliptin, a dipeptidyl peptidase-4 inhibitor, reduces colon carcinogenesis and reactive oxygen species in 1,2-dimethylhydrazine-induced rats. Int J Cancer. 2013 Nov;133((10)):2498–503. doi: 10.1002/ijc.28260. [DOI] [PubMed] [Google Scholar]
- 13.Jang JH, Baerts L, Waumans Y, De Meester I, Yamada Y, Limani P, et al. Suppression of lung metastases by the CD26/DPP4 inhibitor Vildagliptin in mice. Clin Exp Metastasis. 2015 Oct;32((7)):677–87. doi: 10.1007/s10585-015-9736-z. [DOI] [PubMed] [Google Scholar]
- 14.Amritha CA, Kumaravelu P, Chellathai DD. Evaluation of Anti Cancer Effects of DPP-4 Inhibitors in Colon Cancer- An Invitro Study. J Clin Diagn Res. 2015 Dec;9((12)):FC14–6. doi: 10.7860/JCDR/2015/16015.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yorifuji N, Inoue T, Iguchi M, Fujiwara K, Kakimoto K, Nouda S, et al. The dipeptidyl peptidase-4 inhibitor sitagliptin suppresses mouse colon tumorigenesis in type 2 diabetic mice. Oncol Rep. 2016 Feb;35((2)):676–82. doi: 10.3892/or.2015.4429. [DOI] [PubMed] [Google Scholar]
- 16.Stecca BA, Nardo B, Chieco P, Mazziotti A, Bolondi L, Cavallari A. Aberrant dipeptidyl peptidase IV (DPP IV/CD26) expression in human hepatocellular carcinoma. J Hepatol. 1997 Aug;27((2)):337–45. doi: 10.1016/s0168-8278(97)80180-8. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi T, Kohjima M, Ichikawa T, Seike M, Ide Y, Mizuta T, et al. The morbidity and associated risk factors of cancer in chronic liver disease patients with diabetes mellitus: a multicenter field survey. J Gastroenterol. 2015 Mar;50((3)):333–41. doi: 10.1007/s00535-014-0968-5. [DOI] [PubMed] [Google Scholar]
- 18.Fujii M, Shibazaki Y, Wakamatsu K, Honda Y, Kawauchi Y, Suzuki K, et al. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med Mol Morphol. 2013 Sep;46((3)):141–52. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi T, Sakisaka S, Sata M, Mori M, Tanikawa K. Different lobular distributions of altered hepatocyte tight junctions in rat models of intrahepatic and extrahepatic cholestasis. Hepatology. 1999 Jan;29((1)):205–16. doi: 10.1002/hep.510290115. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005 Jun;41((6)):1313–21. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimatsu H, Yonezawa A, Yamanishi K, Yao Y, Sugano K, Nakagawa S, et al. Disruption of Slc52a3 gene causes neonatal lethality with riboflavin deficiency in mice. Sci Rep. 2016 Jun;6((1)):27557. doi: 10.1038/srep27557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6((1)):78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaguchi T, Yoshida T, Harada M, Hisamoto T, Nagao Y, Ide T, et al. Hepatitis C virus down-regulates insulin receptor substrates 1 and 2 through up-regulation of suppressor of cytokine signaling 3. Am J Pathol. 2004 Nov;165((5)):1499–508. doi: 10.1016/S0002-9440(10)63408-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002 Dec;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001 Nov;125((1-2)):279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.Rohn H, Junker A, Hartmann A, Grafahrend-Belau E, Treutler H, Klapperstück M, et al. VANTED v2: a framework for systems biology applications. BMC Syst Biol. 2012 Nov;6((1)):139. doi: 10.1186/1752-0509-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung YA, Choi YK, Jung GS, Seo HY, Kim HS, Jang BK, et al. Sitagliptin attenuates methionine/choline-deficient diet-induced steatohepatitis. Diabetes Res Clin Pract. 2014 Jul;105((1)):47–57. doi: 10.1016/j.diabres.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, Fukunishi S, Yokohama K, Ohama H, Tsuchimoto Y, Asai A, et al. A long-lasting dipeptidyl peptidase-4 inhibitor, teneligliptin, as a preventive drug for the development of hepatic steatosis in high-fructose diet-fed ob/ob mice. Int J Mol Med. 2017 Apr;39((4)):969–83. doi: 10.3892/ijmm.2017.2899. [DOI] [PubMed] [Google Scholar]
- 29.Akiyama K, Warabi E, Okada K, Yanagawa T, Ishii T, Kose K, et al. Deletion of both p62 and Nrf2 spontaneously results in the development of nonalcoholic steatohepatitis. Exp Anim. 2017 doi: 10.1538/expanim.17-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaguchi T, Izumi N, Charlton MR, Sata M. Branched-chain amino acids as pharmacological nutrients in chronic liver disease. Hepatology. 2011 Sep;54((3)):1063–70. doi: 10.1002/hep.24412. [DOI] [PubMed] [Google Scholar]
- 31.Kawaguchi T, Taniguchi E, Morita Y, Shirachi M, Tateishi I, Nagata E, et al. Association of exogenous insulin or sulphonylurea treatment with an increased incidence of hepatoma in patients with hepatitis C virus infection. Liver Int. 2010 Mar;30((3)):479–86. doi: 10.1111/j.1478-3231.2009.02191.x. [DOI] [PubMed] [Google Scholar]
- 32.Ye C, Tian X, Yue G, Yan L, Guan X, Wang S, et al. Suppression of CD26 inhibits growth and metastasis of pancreatic cancer. Tumour Biol. 2016 Oct;37((12)):15677–86. doi: 10.1007/s13277-016-5315-4. [DOI] [PubMed] [Google Scholar]
- 33.Lee JJ, Wang TY, Liu CL, Chien MN, Chen MJ, Hsu YC, et al. Dipeptidyl Peptidase IV as a Prognostic Marker and Therapeutic Target in Papillary Thyroid Carcinoma. J Clin Endocrinol Metab. 2017 Aug;102((8)):2930–40. doi: 10.1210/jc.2017-00346. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Zhang X, Wu R, Huang Q, Jiang Y, Qin J, et al. DPPIV promotes endometrial carcinoma cell proliferation, invasion and tumorigenesis. Oncotarget. 2017 Jan;8((5)):8679–92. doi: 10.18632/oncotarget.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi HJ, Kim JY, Lim SC, Kim G, Yun HJ, Choi HS. Dipeptidyl peptidase 4 promotes epithelial cell transformation and breast tumourigenesis via induction of PIN1 gene expression. Br J Pharmacol. 2015 Nov;172((21)):5096–109. doi: 10.1111/bph.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi T, Veech RL, Uyeda K. Regulation of energy metabolism in macrophages during hypoxia. Roles of fructose 2,6-bisphosphate and ribose 1,5-bisphosphate. J Biol Chem. 2001 Jul;276((30)):28554–61. doi: 10.1074/jbc.M101396200. [DOI] [PubMed] [Google Scholar]
- 37.Stincone A, Prigione A, Cramer T, Wamelink MM, Campbell K, Cheung E, et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc. 2015 Aug;90((3)):927–63. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nwosu ZC, Megger DA, Hammad S, Sitek B, Roessler S, Ebert MP, Meyer C, Dooley S. Identification of the Consistently Altered Metabolic Targets in Human Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol. 2017;4:303–323. doi: 10.1016/j.jcmgh.2017.05.004. e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetter R, Reinhold D, Bühling F, Lendeckel U, Born I, Faust J, et al. DNA synthesis in cultured human keratinocytes and HaCaT keratinocytes is reduced by specific inhibition of dipeptidyl peptidase IV (CD26) enzymatic activity. Adv Exp Med Biol. 2000;477:167–71. doi: 10.1007/0-306-46826-3_19. [DOI] [PubMed] [Google Scholar]
- 40.Moreno SP, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014 Oct;346((6208)):477–81. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- 41.Qu K, Wang Z, Fan H, Li J, Liu J, Li P, et al. MCM7 promotes cancer progression through cyclin D1-dependent signaling and serves as a prognostic marker for patients with hepatocellular carcinoma. Cell Death Dis. 2017 Feb;8((2)):e2603. doi: 10.1038/cddis.2016.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cha YJ, Jung WH, Koo JS. Differential Site-Based Expression of Pentose Phosphate Pathway-Related Proteins among Breast Cancer Metastases. Dis Markers. 2017;2017:7062517. doi: 10.1155/2017/7062517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012 Jul;22((1)):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013 Sep;51((5)):618–31. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010 Mar;12((3)):213–23. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 46.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016 Jan;63((1)):173–84. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Liu X, Long M, Huang Y, Zhang L, Zhang R, et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci Transl Med. 2016;8:334ra351. doi: 10.1126/scitranslmed.aad6095. [DOI] [PubMed] [Google Scholar]
- 48.Tajima K, Nakamura A, Shirakawa J, Togashi Y, Orime K, Sato K, et al. Metformin prevents liver tumorigenesis induced by high-fat diet in C57Bl/6 mice. Am J Physiol Endocrinol Metab. 2013 Oct;305((8)):e987–e98. doi: 10.1152/ajpendo.00133.2013. [DOI] [PubMed] [Google Scholar]
- 49.Shiba K, Tsuchiya K, Komiya C, Miyachi Y, Mori K, Shimazu N, et al. Canagliflozin, an SGLT2 inhibitor, attenuates the development of hepatocellular carcinoma in a mouse model of human NASH. Sci Rep. 2018 Feb;8((1)):2362. doi: 10.1038/s41598-018-19658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data