Abstract
Cellular characteristics and their adjustment to a state of disease have become more evident due to recent advances in imaging, fluorescent reporter mice, and whole genome RNA sequencing. The uncovered cellular heterogeneity and/or plasticity potentially complicates experimental studies and clinical applications, as markers derived from whole tissue ‘bulk’ sequencing is unable to yield a subtype transcriptome and specific markers. Here, we propose definitions on heterogeneity and plasticity, discuss current knowledge thereof in the vasculature and how this may be improved by single-cell sequencing (SCS). SCS is emerging as an emerging technique, enabling researchers to investigate different cell populations in more depth than ever before. Cell selection methods, e.g. flow assisted cell sorting, and the quantity of cells can influence the choice of SCS method. Smart-Seq2 offers sequencing of the complete mRNA molecule on a low quantity of cells, while Drop-seq is possible on large numbers of cells on a more superficial level. SCS has given more insight in heterogeneity in healthy vasculature, where it revealed that zonation is crucial in gene expression profiles among the anatomical axis. In diseased vasculature, this heterogeneity seems even more prominent with discovery of new immune subsets in atherosclerosis as proof. Vascular smooth muscle cells and mesenchymal cells also share these plastic characteristics with the ability to up-regulate markers linked to stem cells, such as Sca-1 or CD34. Current SCS studies show some limitations to the number of replicates, quantity of cells used, or the loss of spatial information. Bioinformatical tools could give some more insight in current datasets, making use of pseudo-time analysis or RNA velocity to investigate cell differentiation or polarization. In this review, we discuss the use of SCS in unravelling heterogeneity in the vasculature, its current limitations and promising future applications.
Keywords: Heterogeneity, Single-cell sequencing, Vasculature, Atherosclerosis
1. Introduction
Atherosclerosis is a long process of lipid and inflammatory cell accumulation in the vessel wall, leading to plaque formation and ultimately plaque rupture. Clinical manifestations of cardiovascular diseases are still the leading cause of death worldwide, necessitating better, targeted treatment.1 Current therapies to reduce the clinical manifestations, myocardial infarction and stroke, have been aimed at one or multiple risk factors such as dyslipidaemia, hypertension, or inflammation.2–4 However, as many cell types are involved and/or dysfunctional, a fully effective therapy has not been developed. Pinpointing progression of a disease to a certain cell type is challenging because of strong heterogeneity and/or plasticity of cells not only inside the plaque but also the surrounding tissue. Hence, we see the need to define and address heterogeneity in the healthy and atherosclerotic vasculature, and highlight a new technology to capture this heterogeneity at an unprecedented level: single-cell sequencing (SCS).
Before discussing cellular heterogeneity and plasticity in detail, one has to consider the classical definition of a cell type. The distinction between classical vascular cell types, such as endothelial cells (ECs), vascular smooth muscle cells (vSMCs), macrophages, and fibroblasts, is based on embryonic germ line origin, anatomical or organ location, microscopic morphology, and phenotype/function. In the distant past, this was largely based on morphology, while it is currently also based on population averages of the transcriptome, the expression of classical cell type markers, i.e. cluster of differentiation (CD) molecules, and lineage reporters models using these classical markers. In this framework, one can distinguish differences within a cell type, and changes between cell types, i.e. heterogeneity and plasticity. These terms are used interchangeably, causing considerable confusion. Much knowledge can be gained from the stem cell field, where cell plasticity and heterogeneity are often discussed subjects. The presence of absence of marker genes linked to stem cells, for instance stem cell antigen-1 (Sca-1) or CD34, does not limit the cell in question as belonging to the stem cell population.5,6 It rather shows that a range of genes are linked to stem cells and the expression of these genes is possibly different between cells in this population, creating a very heterogeneous cell population. Therefore, in this review, we will describe heterogeneity as moderate changes in transcriptome and function, enabling adaptation to the micro-environment, organ or anatomical location. Importantly, this adaptation does not lead in loss or acquisition of classical cell identity markers and yields cellular subtypes. Often adaptation of a cell to its environment is also termed phenotypic plasticity, but if cell identity is not lost, we regard this as heterogeneity. Cellular plasticity, on the other hand, is used here to refer to complete changes in cell identity, upon changes in micro-environment. This process is accompanied by loss or acquisition of classical cell identity markers, and includes so called trans-differentiation and reversal of this. Taken together, plasticity and heterogeneity may be regarded as cell types versus subtypes. A schematic overview of vascular cell types and their heterogeneous phenotypes is depicted in Figure 1. Certainly, this definition is not always unambiguous, and we will discuss the potential benefit of SCS to aid in this distinction.
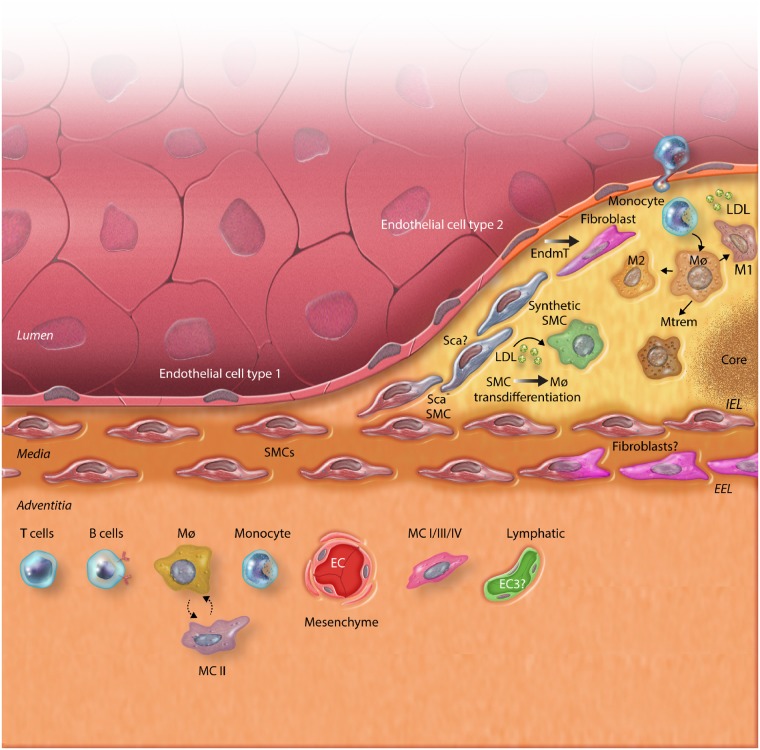
Figure 1.
The different layers of the vasculature (adventitia, media, and intima) and the development of atherosclerosis with all involved cell types. The graphical overview shows heterogeneity (indicated here by thick, black/white filled arrows and cell types in distinct colors) and plasticity (indicated here by single line, black arrows, and cell types in shades of the same color) of all these subsets and their capability to adjust their phenotype to the lipid-rich environment. Endothelial cell (EC) types are zonated46,55 and EC I and II can undergo endothelial-to-mesenchymal transition (EndMT) in hyperlipidemia. Smooth muscle cells (SMCs) can translocate to the cap and become more synthetic. Moreover, they can transdifferentiate into a macrophage-like cell upon lipid engulfing. Macrophages (Mϕ) are depicted with their different subsets according to certain gene expression profiles (M1, M2, MTrem61). They are located in the lipid-rich intima, just above the interna elastica lamina (IEL). In the adventitia, located underneath the externa elastica lamina (EEL), several mesenchymal subsets appear, indicated with I-II-III-IV.56 The adventitia is mostly inhabited by these subsets of mesenchymal cells (MCs), immune cells, and distinct EC subsets. These different subsets all have different functional profiles. Macrophages and MC II were shown to cross-talk as indicated by dotted arrow. The ? indicates new findings or unclarities that need further study.
2. Heterogeneity and plasticity in the vasculature: current evidence
The process of atherosclerosis starts with dysfunction of the ECs facing the vascular lumen. This leads to the extravasation of low-density lipoprotein (LDL) into the subendothelial space. Here LDL accumulates, is oxidized and will further trigger inflammation.7,8 Monocytes are attracted by inflammatory cytokines originating from the forming fatty streak and try to phagocytose the growing amount of LDL, leading to the formation of foamy macrophages. After extensive LDL uptake, these macrophages go into apoptosis and are cleared by other macrophages through efferocytosis.9 However, when plaque development progresses, the amount of apoptotic cells increases and clearance by other macrophages becomes ineffective. Post-apoptotic necrosis occurs, leading to the formation of a necrotic core in the plaque consisting of dead cells and cholesterol crystals. Macrophages release tissue factor, matrix proteases, and pro-angiogenic factors, which influence plaque stability and ultimately plaque rupture.10,11 Alongside the growing amount of macrophages and thus growing necrotic core, the amount of alpha smooth muscle actin (aSMA)+ vSMCs lining the atherosclerotic plaque will diminish. The role of vSMCs in atherosclerosis is already marked at the very beginning of the process, when intimal thickening is observed due to haemodynamic shear stress. Matrix proteoglycans, collagen and elastin fibres are secreted and a stable environment is created.7 While the plaque grows, vSMCs migrate from the medial layer towards the lumen forming the fibrous cap. This fibrous cap becomes thinner over the years due to smooth muscle cell (SMC) apoptosis and matrix degradation by macrophages. This can ultimately lead to cap rupture, exposing the plaques’ thrombogenic content, triggering thrombus formation and lumen occlusion, and consequently causing myocardial infarction or stroke.
In recent years evidence accumulates that most of the major cell types in atherosclerotic plaques, e.g. ECs, macrophages, T-cells, and vSMCs are heterogeneous and/or plastic to some extent. William Aird highlighted different concepts of endothelial heterogeneity in atherosclerosis with regards to anatomical location, activation, and dysfunction.12 He stipulated that EC heterogeneity and plasticity are dependent on multiple factors. The same is true for vSMCs, who undergo phenotypic switching upon lipid and cytokine exposure in the plaque. The vSMCs switch from a quiescent state to a proliferative, more migrative state, is also known as contractile-to-synthetic switch, which we classify as heterogeneity of subtypes.13 Lipid loading does not solely trigger contractile-to-synthetic switch, but also initiates trans-differentiation to macrophage-like smooth muscle cells and may be classified as plasticity of vSMCs. Multiple groups have now shown that expression of different macrophage markers, like galectin-3 (LGALS3) and CD68, increased during lipid-loading of vSMCs, while the vSMC markers alpha-actin-2 (ACTA2) and myosin heavy chain 11 (MYH11) decreased in expression.14–18
Immune cell heterogeneity is also widely discussed in atherosclerosis development, emphasizing different polarization states of macrophages. Polarization of macrophages into the pro-inflammatory M1 macrophage, via lipopolysaccharide or tumor necrosis factor-alpha, vs. the anti-inflammatory M2 macrophage, via interleukin (IL)-4 or IL-10, already shows a distinct phenotypic difference.19 However, we now know that the range of phenotypes is much more subtle and diverse than M1 vs. M2 and that stimuli and microenvironment are decisive for every subset of macrophages.20–22 Immune cell heterogeneity is not only restricted to macrophages but also occurs in other immune subsets. Activated macrophages can recruit T cells, and therefore further enhance inflammation. These T cells are not only activated by macrophages and their secreted cytokines but also by the vast amount of oxLDL in the plaque.23 This again yields a broad spectrum of differentially activated T cells. The diversity of macrophages and T cells opens up the possibility for drugs to tackle small subsets of immune cells with distinct phenotypes regarding plaque progression.24
Although ECs, macrophages, T cells, and vSMCs are the most discussed cells relating to disease progression, recent research in the field has shown that mesenchymal cells possibly also play a role. They may originate from a mesenchymal stem cell-like cell type, which can give rise to various cell types like (myo)fibroblasts or vSMCs.25 These cells have been reported to stem from the adventitial layer surrounding the vasculature and are positive for stem cell markers, like stem cell antigen 1 (Sca1) and GLI-Kruppel family member 1 (Gli1).26–28 Furthermore, evidence suggests that these cells originate from a process called endothelial-to-mesenchymal transition (EndMT), which can be triggered via various pathways.29,30 ECs exposed to different plaque traits, like hypoxia, oxidative stress, or transforming growth factor β (TGF-β), undergo this transformation where they lose gene and protein expression of endothelial markers like CD31, endothelial nitric oxide synthase (eNOS), while simultaneously gaining mesenchymal markers such as fibroblast activation protein (FAP), alpha-actin 2 (ACTA2), and regulatory transcription factors Snail and Slug, SNAI1&2, respectively.29 In human atherosclerotic plaques, EndMT is usually found in larger, unstable plaques and thus linking EndMT to plaque instability.
These examples already clearly illustrate the intricate complexity of atherosclerosis development and all cell types involved, with heterogeneity as a key concept. Heterogeneity of vSMCS, ECs, mesenchymal cells, and immune cells makes it difficult to study them in the context of healthy and diseased vasculature. However, an emerging technique might be able to give us more insight than ever before. The recent advances in the field of SCS are providing an unprecedented opportunity to unravel complex biological systems on multiple biological levels with single-cell resolution. The averaged data scientists have generated using bulk populations of cells or whole tissues can obscure relevant biological insight. Moreover, SCS enables researchers to zoom in on cell populations and investigate them in more depth. This potentially yields new cell phenotypes, uncovering subpopulations with different functions, and providing definitive answers to issues of cellular-trans-differentiation. In this review, we will discuss the use of SCS to unravel heterogeneity in healthy and disease vasculature. We will first summarize the principles and different methods for SCS, followed by discussion of published data on heterogeneity in healthy and diseased vasculature using SCS.
3. SCS technologies today
A decade ago, Tang et al.31 reported that it was possible to gain substantial transcriptomic information out of a single cell using next generation sequencing. This discovery soon sparked the development of technologies that now allow researchers to study large numbers of cell simultaneously. Although an overview of all types of SCS technologies goes beyond the scope of this review, there are certain landmarks that must be mentioned, as they also illustrate that the choice of single-cell technology is often depending on the experimental question.
In many experimental setups, a primary selection of the cells of interest from an organ or organism is necessary, and one of the most common tools to select cells of interest is Fluorescence Assisted Cell Sorting (FACS). This technology allows for single cells to be identified in a cell suspension and subsequent sorting of the cells in wells of a 96 or 384 well plate that contains lysis buffer and RNAse inhibitors. After this primary selection of cells, the chemistry by which a sequencing library is generated from the mRNA of a single cell can be chosen freely, with the Smart-Seq2 chemistry commonly used as being the most sensitive and accurate.32 Smart-seq2 allows for the recovery of sequencing information of the entire mRNA molecule, only limited by the efficiency of the reverse transcriptase used to create cDNA.33,34 However, Smart-Seq2 is sensitive to PCR-induced biased amplification noise. A solution to this bias is the inclusion of Unique Molecular Identifiers (UMI’s), a barcode that is unique to every cDNA molecule in the sequencing library. This allows for accurate counting of mRNA molecules expressed per cell, provided that each mRNA molecule is captured for reverse transcription only once.35
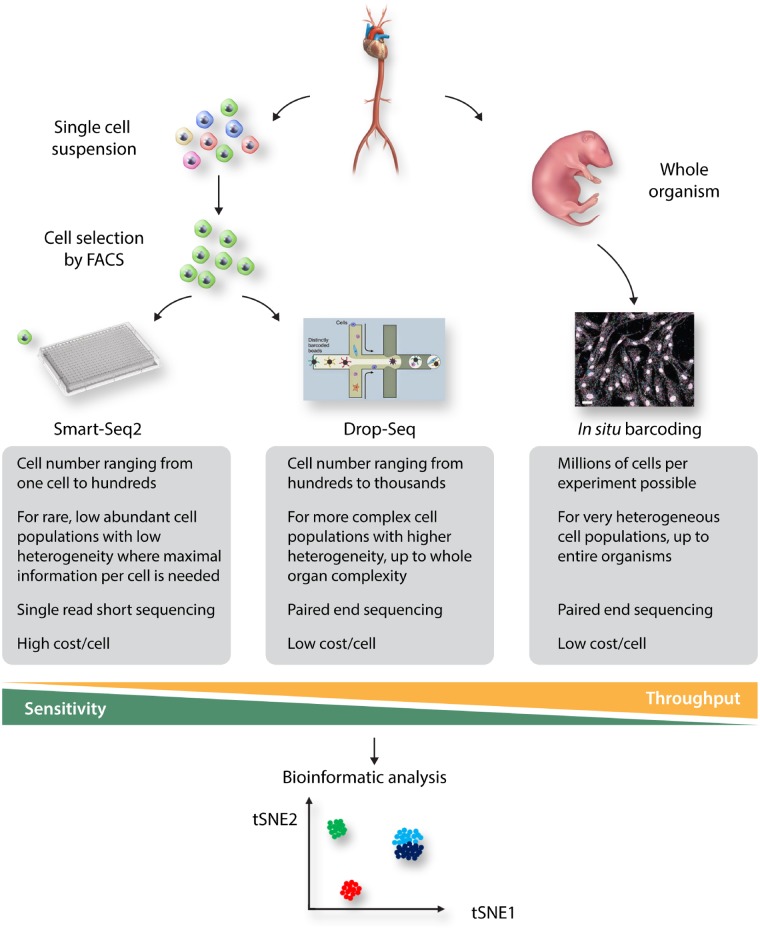
The selection of cells using FACS also provides drawbacks. The procedure of cell sorting by itself is already introducing bias, since large and/or delicate cells will not survive the fluidic shear stress induced by the FACS. But most importantly, selection of cells using FACS is limited in its throughput, as most FACS machines are only accurate enough to sort reliably in a 384 well format, thus limiting the liquid handling of the library preparation to a format in which only 384 cells can be analysed. This makes larger scale single-cell analysis using microwell plate based chemistry too inefficient to consider for experimental questions where a large heterogeneity is expected and thousands of cells need to be analysed. For these experiments, technologies based on droplet encapsulation,36 capture of cells in microwells,37 and in situ barcoding38,39 are the most prominent ones used today, with the drop-seq implementation commercialized by 10x Genomics being the most popular technology due to its ease of use and simple implementation in research environments. This technology allows the analysis of thousands of cells per sample at a decent gene recovery per cell. Finally, in situ barcoding allows for the analysis of millions of cells simultaneously, however, at a comparably low gene recovery per cell.40 For very small sample sizes, where every cell needs to be analysed in the highest detail, the depth of Smart-Seq2 is preferred, while for samples with enormous complexity (like whole organisms), the width of in situ barcoding or Drop-Seq is needed. This allows researchers, depending on the presence of cell populations in certain organs and pre-enriching techniques like FACS, to decide on which technique is most capable of answering a specific research question. A complete overview of the workflow, from tissue towards bioinformatical analysis, is depicted in Figure 2.
Figure 2.
Complete overview from tissue collection, processing, selection, sequencing method, and analysis. The advantages and disadvantages from all three sequencing methods are shown in a small diagram within Figure 1.
Today, the generation of single-cell data is widely accessible to researchers thanks to the plethora of available technologies and their various commercial implementations. However, the proper analysis of single-cell data is often not trivial due to the high complexity of the data that it provides. In a dataset, every cell is in essence a separate sample with quantitative information for every single gene, making the data several orders of magnitude richer compared to bulk transcriptomics. Most commonly, the data are visualized using a t-stochastic neighbour embedding algorithm (t-SNE).41 This algorithm takes the high-dimensional data points (i.e. the cells with gene expression information) and reduces this complexity to two dimensions (an X–Y graph). Data points (cells) with high similarity are placed in neighbouring positions, with different neighbourhoods (often called ‘clouds’ or ‘data clusters’) represented. However, one needs to be aware that t-SNE is a visualization foremost, and that it can easily be tuned to change the look of the data by changing the algorithm’s parameters. Also, it is important to remember that the distance between data clusters is not always a measure for difference between cell types, a common misconception.42 For this reason, many new algorithms are being developed. Recently, the Uniform Manifold Approximation and Projection (UMAP) algorithm was created, which is similar in its visualization style to t-SNE, but represents the relationship between cell types with higher fidelity.43 Another hurdle in single-cell data analysis is that the data is often a snapshot in time, while cells in a heterogeneous tissue are seldomly static. For example, in a diseased state like atherosclerosis, the vSMC are very plastic and to explore the dynamics of the cells, clustering of the cells while preserving the relationship between cell types is paramount. The RNA velocity algorithm allows prediction of future cell states by taking into account the ratio of unspliced vs. spliced RNA, which is a measurement of the ‘age’ of the RNA and the activity of the gene that produced it.44 Finally, the vasculature is difficult to classify into cell types since the ECs are zonated (i.e. their transcriptome gradually changes according to an anatomical axis).45,46 This gradual change in phenotype is well visualized with the Sorting Points Into Neighbourhoods (SPIN) algorithm, which sorts all cells on an X-axis according to similarity, while the Y-axis represents the expression level of a chosen zonated gene.47 A clustering variant of the SPIN algorithm, BackSPIN, can then be used to split the sorted cells into clusters, if desired.48 For a recent overview and discussion on clustering algorithms for single-cell data, we would like to refer the reader to an excellent recent review by Kiselev et al.49
4. Healthy vasculature
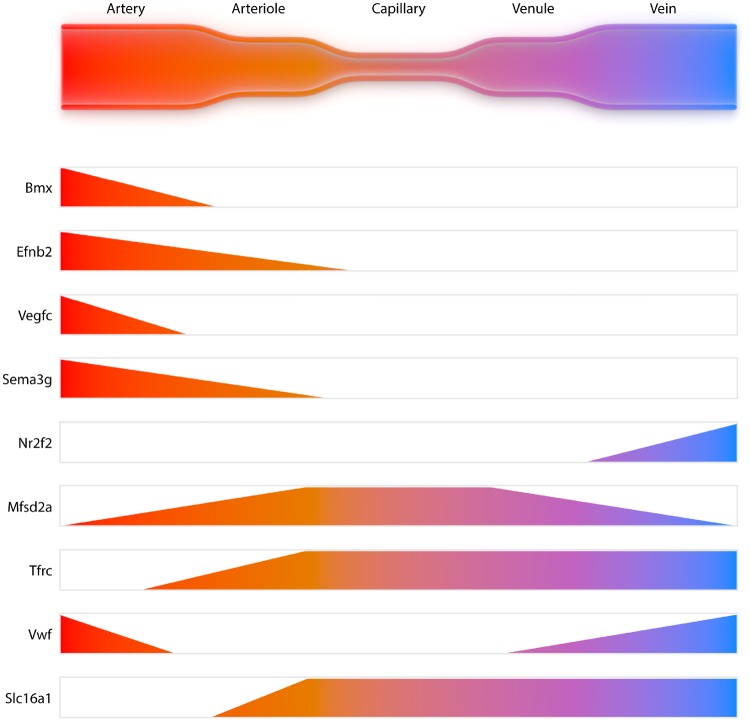
All organs have a specific vasculature dedicated to their relative function and this vascular organotypicity has long been recognized. Indeed, the vasculature can present itself as a strong barrier, a highly permeable fenestrated structure, or, as exemplified by the lungs, an interface for facilitated gas exchange. For an excellent overview of organotypic vasculature, we refer the reader to a recent review of Augustin and Koh.50 However, the heterogeneity of a vascular network within an organ has been studied poorly, until the use of SCS to molecularly define cell types in a vascular network. Recent SCS work has shown that brain and lung ECs are zonated, a term indicating that the transcriptome of cells gradually changes according to an anatomical axis. This thus relates to heterogeneity of cells, as phenotype adaptation does not result in full change or loss of EC identity.45,46 This term was first coined by Jungermann et al.,51 when describing metabolic zonation of the liver hepatocytes, which later has been extended towards molecular zonation by Halpern et al.52 using SCS. The zonation of brain ECs as shown by gradual changes in their transcriptome, related to their position in the vasculature, is schematically shown in Figure 3. In contrast to the ECs, mural cells (vSMC and pericytes) do not present a clear zonated pattern in the brain. While pericytes appear to be largely uniform in their transcriptome and mostly lack expression of genes required for contractility, vSMC differ between arteries and veins. Indeed, vSMCs on arteries stand out by the strong presence of gene programmes required for contractility, while vSMCs on veins are more similar to pericytes. Similar zonated patterns were also found in lung vasculature, although strong organotypicity of ECs and pericytes was found.46 Furthermore, Vanlandewijck et al. also described the presence of ‘fibroblast-like’ cells that sit outside of the smooth muscle cell layer, but under the astrocyte end-feet, of the larger arteries and veins. These cells have previously also been referred to as vascular leptomeningeal cells.53 Dobnikar et al.54 provided further support and detail of vSMCs heterogeneity using SCS on healthy mouse vessels and was able to show that a small portion of vSMCs are positive for stem cell marker Sca-1. They also showed that this specific subset of vSMCs is up-regulated during disease. Even more recently, Kalluri et al.55 and Gu et al.56 were able to describe vascular heterogeneity in both the healthy murine aorta and adventitia using an unbiased methods starting from all vascular cells. Kalluri isolated total aortic medial and intimal cells without FACS preselection, compared two different enzymatic digestion mixes, sequenced over 6000 cells with low and high sequencing depth (17 000 and 145 000 reads/cell, respectively) to define a complete aortic atlas.55 Eleven major cell populations were identified with both read depths and enzyme mixes, including SMCs, fibroblasts, monocytes, and ECs. They emphasized three phenotypically distinct EC subsets, revealing different functional aspects.55 The major EC subset was enriched for canonical EC markers, however, the other two would have been missed by pre-sorting for canonical markers. Differential genes for the second largest subset were involved in angiogenesis, lipid handling, and it was enriched in a tip cell gene signature. The third subset were marked as lymphatic ECs, which together with the large number of fibroblasts, raises the question if the dissection of adventitia from aorta was sufficient, as both cells are mainly thought to reside in the adventitia. However as in total 33% of all cells appear to be fibroblasts this contradicts the possibility of a small contamination. Validation of tissue RNA or protein localization of these cell types and subsets would add greatly to the biological insight, which is where most current reports are still lacking. However, this report adds considerable insight into the healthy murine cell atlas and shows that cell number is more important for discovery of new subsets than sequencing depth.
Figure 3.
Zonation of endothelial cells in the brain. Gene expression profiles differ along the anatomical axis of the vasculature and thus influence the functional profile of the endothelial cells.
The second recent report by Gu et al. made use of SCS to sketch an atlas of all cell types in the adventitia from ∼2000 total cells from healthy and ∼3000 cells from atherosclerotic mice. Gu et al. was able to obtain 15 different cell clusters, including T-cells, B-cells, natural killer cells, monocytes, macrophages and two clusters which they classify as non-immune cells. Despite the relatively small number of non-immune cells (∼800), they showed four different mesenchymal clusters, all linked to specific markers and functional aspects, specifically one cluster was linked to immune cells activation.56
Recently, the liver vasculature was investigated with SCS by Halpern et al.45 using paired-cell sequencing, a method where endothelial/hepatocyte cell pairs are deliberately selected. Thus, spatial information can be obtained from the zonated profile of the hepatocyte52 and endothelial specific gene signatures can be found by subtracting the hepatocyte transcriptome. Although the work has provided interesting insights in liver vascular heterogeneity, the authors also recognize that the dependence on specific paired cells (hepatocytes) is limiting in capturing the complete complexity of the liver vasculature. Further profiling of single cells of the entire vasculature of the liver is warranted.
In order to unify the efforts to create organism-wide single-cell atlases, international consortia are formed. Most prominently are the Tabula Muris Consortium and the Human Cell Atlas Consortium, aiming to profile all cell types of the mouse and the human, respectively. In large, organ-wide single-cell datasets, the vasculature is also represented, but often the lack of specific focus impedes a molecular characterization of the vasculature for several reasons. First, the separation of ECs from pericytes often requires special dissociation protocols, since they are embedded within the same basement membrane.57 As most organ-wide single-cell atlas projects are not specifically aiming for the dissociation of these two cell types, an artificial endothelial/pericyte hybrid is commonly described as a cell type of its own.37,58 Secondly, the vasculature is often underrepresented or insufficiently subclustered into separate cells, leading to annotation of all vascular cells as ‘endothelium’.59,60 For these reasons, specific SCS profiling of vascular beds of healthy, adult organs and the body’s main large arteries and veins (aorta, carotid artery, vena cava) is still paramount in establishing a molecular definition of vascular cell types across organs.
5. Diseased vasculature
While heterogeneity and zonation are already evident from recent SCS studies using healthy vasculature, heterogeneity is greatly amplified when looking at different disease models. Two reports emerging at the same time focused on immune cell heterogeneity in atherosclerosis.61,62 Cochain et al.61 dived into the immune aspect of atherosclerosis, using mice on a LDL receptor knock-out (LDLR−/−) background for SCS. CD45 positive cells from healthy and atherosclerotic tissue were used to investigate immune cell heterogeneity. In total, 13 clusters were found with distinct gene expression patterns, of which three clusters were only present in atherosclerotic tissue. These findings clearly show again the cellular adaptability within disease progression, emphasizing the importance of cellular heterogeneity and plasticity in the vasculature. SCS enabled them to find a new gene, triggering receptor expressed on myeloid cells 2 (TREM2), to be highly expressed on a subset of atherosclerotic macrophages, which had not been described before. This subset is involved in lipid metabolism, regulation of cholesterol efflux and oxidative stress, and was previously linked to osteoclasts and disease-associated microglia. Winkels et al.62 showed a diversity of 11 different clusters of leucocytes based on unsupervised clustering and validate these clusters by using a secondary technique, mass spectrometry cytometry of time of flight (CyTOF). Even though these papers are leading in the field of immune cell heterogeneity in context of diseased vasculature, there are still some limitations to these studies. Both papers only make use of CD45 positive cells, eliminating the option to look at their communication with other cells within the same tissue. Furthermore, the amount of cells used for analysis could be greatly enhanced.
Aforementioned papers were the first to use SCS as a new technique to investigate immune cell heterogeneity; however, heterogeneity and plasticity of other cells in diseased vasculature have already been described by others over the last years. Hao et al.63 already proposed vSMC heterogeneity in vascular disease back in 2003. With regards to arterial calcification, location seems to be key in the genetic and functional properties of the different SMCs.64 In atherosclerosis, Chappell et al.65 show that a small subset of very plastic vSMCS proliferate extensively, which results in accumulation of vSMCs that can gain macrophage markers, like CD107b (MAC3). The given is not only true for atherosclerotic vSMCs, but also for those involved in vascular injury.
In recent years, mesenchymal progenitor cells, a plastic and thus heterogeneous cell type by nature, have been getting more and more attention in vascular disease. These cells are thought to originate from the adventitia which is a progenitor niche, according to Majesky et al.26 This is supported by earlier data by Hu et al.27 who showed clusters of cells in the adventitia of aortic roots, positive for stem cell markers like Sca-1, CD34, and c-Kit. In addition to the detection of these cells, they also demonstrate their ability to differentiate into vSMCs upon PDGF-BB stimulation. Further support for the relevance of arterial progenitor cells, stems from a report showing that vascular endothelial growth factor stimulation of CD34+ isolated cells in vitro pushes them to an EC type with the ability to form small capillaries.66 These progenitor cells are not only important in maintaining normal vessel composition but also play a crucial role in vascular disease. The earlier mentioned study by Hu et al.27 showed that the transformation of Sca-1+ cells to SMCs is also happening in a murine vein graft in vivo. When combined with the hyperlipidaemic, apolipoprotein E knockout (ApoE−/−) atherosclerosis mouse model, they observed that ∼20% of SMCs were Sca-1+ and thus of progenitor origin. These findings are supported by multiple groups who also described the transition of adventitial progenitor cells to SMCs and ECs in the neointima, depending on the stimulus.67–69 Furthermore, these mesenchymal cells can generate myofibroblasts and therefore play a role in organ fibrosis, which is not only restricted to the vasculature of large arteries but also in kidneys, lungs, or liver.25 The aforementioned SCS study by Gu et al.56 mapping cells in the adventitia of healthy and hypercholesterolaemic ApoE-/- mice now confirms heterogeneity of adventitial mesenchyme. However, the relative contribution and function of the observed four mesenchymal clusters was not adapting drastically to the diseased situation. Possibly, the low number of cells (∼800) prevented full assessment of changes. Interestingly, cross-communication of an inflammatory mesenchymal subset was observed with activated macrophages in the diseased setting. The unbiased approach use to map all cells allowed this important new biological insight.56 Likewise, the whole aortic medial and intimal cell atlas resulting from the study by Kalluri et al. was derived from an unbiased approach. Here, the observed three EC subsets were conserved upon a high cholesterol diet, while induction of genes involved in collagen turnover suggested the presence of EndMt. Further the relative presence of the main subset was enhanced in diseased, while the opposite was true for the lipid/angiogenic EC subset. This seems rather contradictory to the current knowledge of angiogenic induction upon true hypercholesterolaemic disease-settings in double deficient apoE−/− to LDLr−/− mice.70 However the current study involved diet fed wildtype C57Bl6 mice representing possibly very early EC dysfunction, not an atherosclerosis model with overt hypercholesterolaemia and plaque development. It is, therefore, very interesting to compare the EC subsets in the—yet unavailable—total cell atlas of atherosclerotic plaques.
All these data together already stress the importance of heterogeneity and/or plasticity of ECs, vSMCs, mesenchymal cells, and immune cells in the vasculature and how this can affect vascular disease progression. Published SCS data on large, diseased arteries is currently limited to murine studies, only on atherosclerosis, with no data yet available on ECs in atherosclerotic models or on other large artery pathologies, such as pulmonary hypertension, and aneurysms. However, further support for disease-driven amplification of heterogeneity can be gained from a non-cardiovascular model, i.e. hyperpermeable tumour microvasculature. Zhao et al.71 used human xenografts implanted in mice and detected tumour heterogeneity in endothelial and mesenchymal cells, linked to Notch signalling. Also Lambrechts et al.72 made use of SCS in a lung cancer model in mice, separating stromal cells into 52 different subsets with their own gene signature. Bian et al.73 even combined single-cell transcriptome data with methylome and mutation data of human colorectal cancer samples, broadening the genetic fingerprint all the more so. Data from tumour microvasculature again confirms the heterogeneity in the vasculature and how SCS gave more insight in processes involved, e.g. the methylome.
To summarize, these data show the complexity of murine vasculature and how SCS enables us look at different cell types and their gene expression patterns on a deeper level than ever before. This could impact the identification of cell types and new subtypes, since SCS gives more depth to expression patterns belonging to different cell types and their subpopulations. However, few studies go beyond description of the subsets, and it is yet to be resolved if there are actual implications for functional heterogeneity. Further investigation of functional heterogeneity and cell-cell interactions in human atherosclerotic tissue can elucidate processes involved in disease and how the compares between physiology and pathology.
6. Biological implications
Thus far, the first groups confirmed the basal atherosclerotic plaque immune cell compositions and have described subtype heterogeneity thereof, and uncovered a potential new macrophage subtype, while SCS of ECs in healthy brain and lung vasculature revealed EC zonation, and arterial- and venous-specific vSMC types.45,46,61,62 Broadly, we see biological implications related to cell type identity and the pathogenesis of disease.
The introduction of SCS has challenged the classical definition of cell types, which was determined by morphology, tissue location and a few cell identity markers. Bulk transcriptomics and fluorescent reporter mice have already changed this simplified view, and uncovered new subtypes and trans differentiated cells. This distinction between heterogeneity and/or plasticity of cells is often ambiguous and open to errors. In the past, these errors have been made due to lack of high resolution microscopy in three dimensions, the lack of specific cell identity markers used for CRE reporters, and the analysis of population averages, obscuring individual differences and subpopulations. SCS has the potential to clearly distinguish between heterogeneity and plasticity. Grouping cells with similar transcriptomes will identify complete gene signatures of cell identity, validating classical identity markers and uncovering new ones. Clustering tools will enable dissecting major cell types with very distinct cell identity marker signatures, from subtypes whose signatures differ within the boundaries of a cell type signature. Although SCS may simplify the distinction, ambiguity may still exist when there is no real end-stage identity, such as in a dynamic and reversible process like EndMT. Hopefully, detailed pseudotime bioinformatics analysis of the temporal changes in the transcriptome in a controlled experimental setting, may further resolve these issues.
Upon consensus of cell type and subtype identity signatures, and the functional implications thereof, there may arise opportunities for improved resolution of disease. While general anti-inflammation therapy in humans has shown proof-of-concept, it only prevented the relative risk of clinical events by 15%.2 Speculating about the potential future advances this insight from SCS could bring the field, raises the possibility of new, subtype selective imaging targets and/or adaptable regulation of cell and subtypes. Adapting therapy to selectively inhibit immune cell subtypes with a detrimental function, or to trigger the conversion into a cell subtype with a more beneficial function could in theory be more effective to prevent clinical events. In future, the adaptable regulation of cell types and subtypes, potentially even in a personalized manner, is expected to have a durable effect on improving life expectancy, quality of life, and avoiding unnecessary treatments. Nevertheless, development and delivery of such subtype-specific inhibitors or reprogramming agents are far more clear and many hurdles need to be taken.
7. Future technical improvements
Although clearly important new insights are gained from latest SCS reports, several improvements can be made, both on the technological level, as well as bioinformatics. Here, we will discuss limitations and solutions to incomplete genome coverage, number of replicates and how to deal with stoichiometry, low throughput, loss of spatial information and cellular interaction, the need for fresh material, as well as highlight new technologies and analysis tools.
One major limitation of the current technologies is that not the entire transcriptome of individual cells can be mapped and thus, every single-cell transcriptome is but a stochastic sample of the pool of mRNA present in that cell In addition, only highly expressed non-coding RNAs can now be identified. However, the sensitivity of the methods is continuously improving allowing the detection of more and more genes in every individual cell, as well as non-coding RNAs. Recently, an optimized version of SCRB-Seq was developed called mcSCRB-Seq74 (molecular crowding single-cell RNA Barcoding and sequencing) using the molecular crowding agent PEG (polyethylenglycol), which increases the efficiency of the RT (reverse transcription) reaction in a concentration dependent manner. Thus, this protocol is at the moment the most sensitive plate-based single-cell RNA-Seq protocol (benchmarked using ERCCs). In addition, 10X genomics recently released a new version of the 3′ single-RNA-Seq assay with a higher capture capacity of polyadenlylated RNA, thus leading to more detected genes per cell.
The matter of biological replicates is also important to consider. Many published studies use a single sample, or a single pool of samples for the assessment of heterogeneity in an organ or disease condition. Although a single sample is already very informative in exploring cellular heterogeneity, it is often dangerous to extrapolate the result into a definite ‘atlas’. Taking into account current pricing of SCS, the trade-off between using more replicates, comparing different models or including more time-points is often hard to decide upon and is very specific to the experimental question.75,76
It is obvious that more replicates would increase the robustness of the data, yet one has to take batch effects into account that occur with every RNA-Seq reaction. To minimize batch effects, different experimental groups should ideally not be on separate days. With the use of barcoded oligos, different samples can be pooled in one reaction for droplet based assays, like 10X genomics, thus reducing batch effects. Sometimes, this may not be applicable due to low number of cells, but bioinformatic tools exist that allow the correction for batch effects.77
High-throughput analysis of multiple ‘omics’ on single-cell level will likely provide new biological insights into tissue heterogeneity and disease development. Single-cell RNA sequencing has evolved to a high-throughput technology with the development of technological advances like combinatorial indexing or droplet based technologies, which reduced costs and increased throughput to over 100 000 cells that can be analysed in one experiment.38,39,78,79 Indeed, early studies may have been hindered by low cell numbers studied, obscuring the identification of rare (sub)populations. Higher throughput of cells would allow inclusion of all cells in an organ, and with appropriate bioinformatics their interaction could then also be mapped. This could for instance be achieved by studying receptor-ligand interaction pairs as described by Skelly et al.80 in the mouse heart. In addition to cell–cell interactions, also spatial information can now be retrieved. Most single-cell analysis experiments start with the dissociation of single cells from tissues, so that spatial information is lost. One solution to regain spatial information has been demonstrated by Halpern et al.52 who used a panel of zonated landmark genes with smFISH to remap the single-cell transcriptomes of mouse liver cells to the zonation profile. Other approaches are osmFISH or huluFISH.81,82 Techniques for direct in situ transcriptomics have also been described (e.g. MERFISH, STARmap).83,84
The majority of studies report on murine material as proof of principle, due to need for fresh, homogeneous samples, necessitating fewer biological replicates. To speed up human discovery, the use of frozen, bio-archived material would make large sets of previously collected, frozen patient material available. The current use of fresh material for droplet-based technology, hinders the step to large scale collection of human samples, usually presenting one by one. In addition to larger heterogeneity compared with animal models, this adds potential batch effects and might obscure disease-related transcriptional changes. Although some reports claim that transcriptomics are comparable between fresh and frozen samples,85 recently also isolation protocols and studies of single nuclei have been evolved that allow the analysis of bio-archived tissues.86 These isolation protocols also reduce the isolation bias that comes with tissue dissociation protocols resulting in better isolation of some cell types compared to others. Furthermore, nuclear isolation might minimize transcriptional changes during the isolation process since the full isolation can be carried out at 4°C, as no enzymatic digestion is needed.87
The aforementioned technological advancements can overcome some of the current limitations. We will briefly highlight other developments allowing, i.e. simultaneous quantifications of protein levels, multiplexing of samples, and sequencing of the active transcriptome. The addition of oligonucleotide based barcoded antibodies to the single-cell suspension has added protein expression abundance on the cell surfaces to the sequencing data called Cite-Seq.88 This technique was also developed to combine more sample in one reaction, e.g. on the 10X Chromium to reduce batch effects and study more cells called cell-hashing. Another approach to multiplex several samples from different individuals in one single-cell experiment by using genetic variation of individuals has been recently described.89 Further developments to study intracellular proteins or phosphoproteins are being developed.90 Another exciting technology is the mapping of open chromatin regions in single cells.91 Using combinatorial indexing techniques or commercially kits, single ATAC sequencing has now become available for high throughput analysis.92 The additional DNA accessibility information in detected cell populations combined with mRNA-expression data from regular scRNA-seq will certainly help to identify novel cell populations and also validate the mRNA expression data on whether a detected population is truly distinct from the other cell-populations. An additional level has been recently added to this using FANS (fluorescent associated nuclei sorting) and single nuclear (sn)ATAC.92,93 snATAC allows the discovery of unique enhancer regions and regulatory logic in distinct cell types but, due to the nature of the data, does not allow the same accuracy of unsupervised clustering as with scRNA-Seq data. Analysis of both datasets scRNA-seq and scATAC-seq complements each other and allows among other things the identification of rare cell clusters. 94
In addition, bioinformatics tools are also evolving to accommodate current limitations. Much can be gained from in-depth bioinformatics such as pseudo-time trajectory analysis to study cellular trans-differentiation in detail. New exciting computational tools that allow pseudo-time analysis in single-cell data have been developed and refined.95,96 As further techniques are being developed, bioinformatical integration of multi-omics datasets of single-cell analysis represents a major challenge.97
8. Conclusions
This review emphasizes the importance of cell heterogeneity and plasticity in healthy vasculature and how this relates to atherosclerosis development and progression. We discussed SCS as a very useful technique in further investigating cell heterogeneity and plasticity. SCS has given the opportunity to link gene expression patterns to classical cell types and their subpopulations, but also how these patterns vary upon different environmental stimuli, challenging the plasticity of these cells. The depth in which SCS can offer genetic insight is dependent on the method chosen by researchers. Where Smart-Seq2 offers researchers the possibility to investigate expression of full-length RNA in cells, it limits the number processed cells per batch to 384. On the other hand, Drop-seq and in situ barcoding enable researchers to use larger quantities of cells or even complete embryos, but with lower gene recovery per cell. Development of new bioinformatic analysis tools is emerging and is allowing researchers analyse more information, such as RNA splicing or zonation. The latter seems of great importance regarding cell heterogeneity, which is proven in multiple organs in healthy state such as liver, lung, and the brain as shown for the latter by Vanlandewijck et al.46 Heterogeneity and plasticity of ECs, vSMCs, immune cells, and mesenchymal cells has shown to be present in healthy vasculature but is even more amplified in diseased vasculature. Current studies highlight this by using SCS in studying changes in cell populations and gene expression patterns in atherosclerotic mouse models. However, these studies are still only limited to murine models, since only fresh material can be used. Nowadays more advanced methods, such as Single Nucleus RNAseq, are broadening the field with the use of frozen tissue and thus also adding possibility of using human biopsies from tissue banks, expanding single-cell knowledge across species. This insight could help to identify novel therapeutic targets and pave the way towards urgently needed novel targeted therapeutics for the vast and growing patient population suffering from cardiovascular disease. However, latest and future advancements in technology and bio-informatics should be implemented to drive the insight from SCS data from mere description of existing and new subpopulations towards a full, in-depth insight into functional and spatial heterogeneity in vivo and cell–cell communication in healthy and diseased vasculature.
Conflict of interest: none declared.
Funding
This work has been supported by the Netherlands Scientific Organization [NWO Vidi 91718364 to J.C.S], the German Society of Internal Medicine (DGIM) as part of the clinician scientist program [to C.K.], the German Research Foundation [SFB TRR219 to R.K.], the Swedish Science Council [2015-00550 to M.V. and C.B.], the Swedish Cancer Society [150735 to M.V. and C.B.], the Knut and Alice Wallenberg Foundation [2015:0030 to M.V. and C.B.] and the Leducq Foundation [14CVD02 to M.V. and C.B., and 15Autophagy to J.C.S].
References
- 1. Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S.. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res 2016;118:535–546. [DOI] [PubMed] [Google Scholar]
- 2. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ, Group CT.. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 3. Ridker PM, MacFadyen JG, Everett BM, Libby P, Thuren T, Glynn RJ, Ridker PM, MacFadyen JG, Everett BM, Libby P, Thuren T, Glynn RJ, Kastelein J, Koenig W, Genest J, Lorenzatti A, Varigos J, Siostrzonek P, Sinnaeve P, Fonseca F, Nicolau J, Gotcheva N, Yong H, Urina-Triana M, Milicic D, Cifkova R, Vettus R, Anker SD, Manolis AJ, Wyss F, Forster T, Sigurdsson A, Pais P, Fucili A, Ogawa H, Shimokawa H, Veze I, Petrauskiene B, Salvador L, Cornel JH, Klemsdal TO, Medina F, Budaj A, Vida-Simiti L, Kobalava Z, Otasevic P, Pella D, Lainscak M, Seung K-B, Commerford P, Dellborg M, Donath M, Hwang J-J, Kultursay H, Flather M, Ballantyne C, Bilazarian S, Chang W, East C, Forgosh L, Harris B, Ligueros M.. Relationship of C-reactive protein reduction to cardiovascular event reduction following treatment with canakinumab: a secondary analysis from the CANTOS randomised controlled trial. Lancet 2018;391:319–328. [DOI] [PubMed] [Google Scholar]
- 4. Nicholls SJ, Puri R, Anderson T, Ballantyne CM, Cho L, Kastelein JJ, Koenig W, Somaratne R, Kassahun H, Yang J, Wasserman SM, Scott R, Ungi I, Podolec J, Ophuis AO, Cornel JH, Borgman M, Brennan DM, Nissen SE.. Effect of evolocumab on progression of coronary disease in statin-treated patients: the GLAGOV randomized clinical trial. JAMA 2016;316:2373–2384. [DOI] [PubMed] [Google Scholar]
- 5. Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res 2012;22:457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loeffler M, Roeder I.. Tissue stem cells: definition, plasticity, heterogeneity, self-organization and models—a conceptual approach. Cells Tissues Organs 2002;171:8–26. [DOI] [PubMed] [Google Scholar]
- 7. Insull W., Jr. The pathology of atherosclerosis: plaque development and plaque responses to medical treatment. Am J Med 2009;122:S3–S14. [DOI] [PubMed] [Google Scholar]
- 8. Weber C, Noels H.. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med 2011;17:1410–1422. [DOI] [PubMed] [Google Scholar]
- 9. Linton MF, Babaev VR, Huang J, Linton EF, Tao H, Yancey PG.. Macrophage apoptosis and efferocytosis in the pathogenesis of atherosclerosis. Circ J 2016;80:2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gonzalez L, Trigatti BL.. Macrophage apoptosis and necrotic core development in atherosclerosis: a rapidly advancing field with clinical relevance to imaging and therapy. Can J Cardiol 2017;33:303–312. [DOI] [PubMed] [Google Scholar]
- 11. Martinet W, Schrijvers DM, De Meyer GR.. Necrotic cell death in atherosclerosis. Basic Res Cardiol 2011;106:749–760. [DOI] [PubMed] [Google Scholar]
- 12. Aird WC. Endothelial cell heterogeneity and atherosclerosis. Curr Atheroscler Rep 2006;8:69–75. [DOI] [PubMed] [Google Scholar]
- 13. Doran AC, Meller N, McNamara CA.. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol 2008;28:812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R.. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res 2014;115:662–667. [DOI] [PubMed] [Google Scholar]
- 15. Rong JX, Shapiro M, Trogan E, Fisher EA.. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc Natl Acad Sci USA 2003;100:13531–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK.. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Allahverdian S, Chaabane C, Boukais K, Francis GA, Bochaton-Piallat ML.. Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc Res 2018;114:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bennett MR, Sinha S, Owens GK.. Vascular smooth muscle cells in atherosclerosis. Circ Res 2016;118:692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson HM. Macrophages heterogeneity in atherosclerosis—implications for therapy. J Cell Mol Med 2010;14:2055–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagenborg J, Goossens P, Biessen EAL, Donners M.. Heterogeneity of atherosclerotic plaque macrophage origin, phenotype and functions: implications for treatment. Eur J Pharmacol 2017;816:14–24. [DOI] [PubMed] [Google Scholar]
- 21. Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL.. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014;40:274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson JL, Newby AC.. Macrophage heterogeneity in atherosclerotic plaques. Curr Opin Lipidol 2009;20: 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tse K, Tse H, Sidney J, Sette A, Ley K.. T cells in atherosclerosis. Int Immunol 2013;25:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tabas I, Lichtman AH.. Monocyte-macrophages and T cells in atherosclerosis. Immunity 2017;47:621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El Agha E, Kramann R, Schneider RK, Li X, Seeger W, Humphreys BD, Bellusci S.. Mesenchymal stem cells in fibrotic disease. Cell Stem Cell 2017;21:166–177. [DOI] [PubMed] [Google Scholar]
- 26. Majesky MW, Dong XR, Hoglund V, Daum G, Mahoney WM Jr.. The adventitia: a progenitor cell niche for the vessel wall. Cells Tissues Organs 2012;195:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu Y, Zhang Z, Torsney E, Afzal AR, Davison F, Metzler B, Xu Q.. Abundant progenitor cells in the adventitia contribute to atherosclerosis of vein grafts in ApoE-deficient mice. J Clin Invest 2004;113:1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kramann R, Goettsch C, Wongboonsin J, Iwata H, Schneider RK, Kuppe C, Kaesler N, Chang-Panesso M, Machado FG, Gratwohl S, Madhurima K, Hutcheson JD, Jain S, Aikawa E, Humphreys BD.. Adventitial MSC-like cells are progenitors of vascular smooth muscle cells and drive vascular calcification in chronic kidney disease. Cell Stem Cell 2016;19:628–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evrard SM, Lecce L, Michelis KC, Nomura-Kitabayashi A, Pandey G, Purushothaman K-R, d’Escamard V, Li JR, Hadri L, Fujitani K, Moreno PR, Benard L, Rimmele P, Cohain A, Mecham B, Randolph GJ, Nabel EG, Hajjar R, Fuster V, Boehm M, Kovacic JC.. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat Commun 2016;7:11853.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Souilhol C, Harmsen MC, Evans PC, Krenning G.. Endothelial-mesenchymal transition in atherosclerosis. Cardiovasc Res 2018;114:565–577. [DOI] [PubMed] [Google Scholar]
- 31. Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA.. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6:377–382. [DOI] [PubMed] [Google Scholar]
- 32. Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W.. Comparative analysis of single-cell RNA sequencing methods. Mol Cell 2017;65: 631–643.e4. [DOI] [PubMed] [Google Scholar]
- 33. Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R.. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013;10: 1096–1098. [DOI] [PubMed] [Google Scholar]
- 34. Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R.. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014;9:171–181. [DOI] [PubMed] [Google Scholar]
- 35. Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lonnerberg P, Linnarsson S.. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 2014;11:163–166. [DOI] [PubMed] [Google Scholar]
- 36. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan GC, Chen M, Guo G.. Mapping the mouse cell atlas by microwell-Seq. Cell 2018;172:1091–1107.e7. [DOI] [PubMed] [Google Scholar]
- 38. Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J.. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 2017;357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, Pun SH, Sellers DL, Tasic B, Seelig G.. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 2018;360:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J.. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019;566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Maaten LH. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–2605. [Google Scholar]
- 42. Wattenberg M, Viégas F, Johnson I.. How to use t-SNE effectively. Distill 2016;http://distill.pub/2016/misread-tsne. [Google Scholar]
- 43. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW.. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018;37:38. [DOI] [PubMed] [Google Scholar]
- 44. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundstrom E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV.. RNA velocity of single cells. Nature 2018;560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Halpern KB, Shenhav R, Massalha H, Toth B, Egozi A, Massasa EE, Medgalia C, David E, Giladi A, Moor AE, Porat Z, Amit I, Itzkovitz S.. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol 2018;36:962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L, Sun Y, Raschperger E, Rasanen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C.. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018;554:475–480. [DOI] [PubMed] [Google Scholar]
- 47. Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O, Notterman DA, Domany E.. Sorting points into neighborhoods (SPIN): data analysis and visualization by ordering distance matrices. Bioinformatics 2005;21:2301–2308. [DOI] [PubMed] [Google Scholar]
- 48. Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S.. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015;347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 49. Kiselev VY, Andrews TS, Hemberg M.. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet 2019;20:310. [DOI] [PubMed] [Google Scholar]
- 50. Augustin HG, Koh GY.. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science 2017;357:eaal2379.. [DOI] [PubMed] [Google Scholar]
- 51. Jungermann K, Katz N.. Functional hepatocellular heterogeneity. Hepatology 2007;2:385–3395. [DOI] [PubMed] [Google Scholar]
- 52. Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I, Itzkovitz S.. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017;542:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Haring M, Hochgerner H, Romanov RA, Gyllborg D, Munoz Manchado A, La Manno G, Lonnerberg P, Floriddia EM, Rezayee F, Ernfors P, Arenas E, Hjerling-Leffler J, Harkany T, Richardson WD, Linnarsson S, Castelo-Branco G.. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 2016;352:1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jorgensen HF.. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun 2018;9:4567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kalluri AS, Vellarikkal SK, Edelman ER, Nguyen L, Subramanian A, Ellinor PT, Regev A, Kathiresan S, Gupta RM.. Single cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation 2019;140:147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gu W, Ni Z, Tan YQ, Deng J, Zhang SJ, Lv ZC, Wang XJ, Chen T, Zhang Z, Hu Y, Jing ZC, Xu Q.. Adventitial cell atlas of wt (wild type) and ApoE (apolipoprotein E)-deficient mice defined by single-cell RNA sequencing. Arterioscler Thromb Vasc Biol 2019;39:1055–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Armulik A, Genove G, Betsholtz C.. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 2011;21:193–215. [DOI] [PubMed] [Google Scholar]
- 58. Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S.. Molecular architecture of the mouse nervous system. Cell 2018;174: 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sohni A, Tan K, Song HW, Burow D, de Rooij DG, Laurent L, Hsieh TC, Rabah R, Hammoud SS, Vicini E, Wilkinson MF.. The neonatal and adult human testis defined at the single-cell level. Cell Rep 2019;26: 1501–1517.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018;562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A.. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res 2018;122:1661–1674. [DOI] [PubMed] [Google Scholar]
- 62. Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AAJ, Cochain C, Vafadarnejad E, Saliba AE, Zernecke A, Pramod AB, Ghosh AK, Anto Michel N, Hoppe N, Hilgendorf I, Zirlik A, Hedrick CC, Ley K, Wolf D.. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res 2018;122:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hao H, Gabbiani G, Bochaton-Piallat ML.. Arterial smooth muscle cell heterogeneity: implications for atherosclerosis and restenosis development. Arterioscler Thromb Vasc Biol 2003;23:1510–1520. [DOI] [PubMed] [Google Scholar]
- 64. Espitia O, Chatelais M, Steenman M, Charrier C, Maurel B, Georges S, Houlgatte R, Verrecchia F, Ory B, Lamoureux F, Heymann D, Goueffic Y, Quillard T.. Implication of molecular vascular smooth muscle cell heterogeneity among arterial beds in arterial calcification. PLoS One 2018;13:e0191976.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jorgensen HF.. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res 2016;119:1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pasquinelli G, Tazzari PL, Vaselli C, Foroni L, Buzzi M, Storci G, Alviano F, Ricci F, Bonafe M, Orrico C, Bagnara GP, Stella A, Conte R.. Thoracic aortas from multiorgan donors are suitable for obtaining resident angiogenic mesenchymal stromal cells. Stem Cells 2007;25:1627–1634. [DOI] [PubMed] [Google Scholar]
- 67. Chen Y, Wong MM, Campagnolo P, Simpson R, Winkler B, Margariti A, Hu Y, Xu Q.. Adventitial stem cells in vein grafts display multilineage potential that contributes to neointimal formation. Arterioscler Thromb Vasc Biol 2013;33:1844–1851. [DOI] [PubMed] [Google Scholar]
- 68. Tsai TN, Kirton JP, Campagnolo P, Zhang L, Xiao Q, Zhang Z, Wang W, Hu Y, Xu Q.. Contribution of stem cells to neointimal formation of decellularized vessel grafts in a novel mouse model. Am J Pathol 2012;181:362–373. [DOI] [PubMed] [Google Scholar]
- 69. Kokkinopoulos I, Wong MM, Potter CMF, Xie Y, Yu B, Warren DT, Nowak WN, Le Bras A, Ni Z, Zhou C, Ruan X, Karamariti E, Hu Y, Zhang L, Xu Q.. Adventitial SCA-1(+) progenitor cell gene sequencing reveals the mechanisms of cell migration in response to hyperlipidemia. Stem Cell Rep 2017;9: 681–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kampschulte M, Stockl C, Langheinrich AC, Althohn U, Bohle RM, Krombach GA, Stieger P, Churin Y, Kremer S, Dierkes C, Rath T, Roeb E, Roderfeld M.. Western diet in ApoE-LDLR double-deficient mouse model of atherosclerosis leads to hepatic steatosis, fibrosis, and tumorigenesis. Lab Invest 2014;94:1273–1282. [DOI] [PubMed] [Google Scholar]
- 71. Zhao Q, Eichten A, Parveen A, Adler C, Huang Y, Wang W, Ding Y, Adler A, Nevins T, Ni M, Wei Y, Thurston G.. Single-cell transcriptome analyses reveal endothelial cell heterogeneity in tumors and changes following antiangiogenic treatment. Cancer Res 2018;78:2370–2382. [DOI] [PubMed] [Google Scholar]
- 72. Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwe H, Pircher A, Van den Eynde K, Weynand B, Verbeken E, De Leyn P, Liston A, Vansteenkiste J, Carmeliet P, Aerts S, Thienpont B.. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 2018;24:1277–1289. [DOI] [PubMed] [Google Scholar]
- 73. Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, Wang W, Yan J, Hu B, Guo H, Wang J, Gao S, Mao Y, Dong J, Zhu P, Xiu D, Yan L, Wen L, Qiao J, Tang F, Fu W.. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science 2018;362:1060–1063. [DOI] [PubMed] [Google Scholar]
- 74. Bagnoli JW, Ziegenhain C, Janjic A, Wange LE, Vieth B, Parekh S, Geuder J, Hellmann I, Enard W.. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat Commun 2018;9:2937.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kleyman M, Sefer E, Nicola T, Espinoza C, Chhabra D, Hagood JS, Kaminski N, Ambalavanan N, Bar-Joseph Z.. Selecting the most appropriate time points to profile in high-throughput studies. eLife 2017;6:e18541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sefer E, Kleyman M, Bar-Joseph Z.. Tradeoffs between dense and replicate sampling strategies for high-throughput time series experiments. Cell Syst 2016;3: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Haghverdi L, Lun ATL, Morgan MD, Marioni JC.. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018;36:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang Y, Navin NE.. Advances and applications of single-cell sequencing technologies. Mol Cell 2015;58:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen X, Teichmann SA, Meyer KB.. From tissues to cell types and back: single-cell gene expression analysis of tissue architecture. Annu Rev Biomed Data Sci 2018;1: 29–51. [Google Scholar]
- 80. Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR.. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep 2018;22:600–610. [DOI] [PubMed] [Google Scholar]
- 81. Codeluppi S, Borm LE, Zeisel A, La Manno G, van Lunteren JA, Svensson CI, Linnarsson S.. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods 2018;15:932–935. [DOI] [PubMed] [Google Scholar]
- 82. Cheng YZY, Hartmann K, Zou P, Bekki G, Alter H, Liu HK. Autonomous combinatorial color barcoding for multiplexing single molecule RNA visualization. bioRxiv2017;127373. doi:10.1101/127373.
- 83. Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X.. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015;348:aaa6090.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, Deisseroth K.. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018;361:eaat5691.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu H, Kirita Y, Donnelly EL, Humphreys BD.. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 2019;30:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, Weitz DA, Rozenblatt-Rosen O, Zhang F, Regev A.. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 2017;14:955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nawijn MC, Vieira Brage F, Berg M, Brouwer S, Kar G, Teichmann S, Van den Berge M.. Novel cell types and altered cell states in asthma revealed by single-cell RNA sequencing of airway wall biopsies. Eur Respir J 2018;52. [Google Scholar]
- 88. Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P.. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ.. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 2018;36:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gerlach JP, van Buggenum JAG, Tanis SEJ, Hogeweg M, Heuts BMH, Muraro MJ, Elze L, Rivello F, Rakszewska A, van Oudenaarden A, Huck WTS, Stunnenberg HG, Mulder KW.. Combined quantification of intracellular (phospho-)proteins and transcriptomics from fixed single cells. Sci Rep 2019;9: 1469.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, Daza RM, McFaline-Figueroa JL, Packer JS, Christiansen L, Steemers FJ, Adey AC, Trapnell C, Shendure J.. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 2018;361:1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J.. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015;348:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen X, Miragaia RJ, Natarajan KN, Teichmann SA.. A rapid and robust method for single cell chromatin accessibility profiling. Nat Commun 2018;9:5345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Stuart TB, Hoffman P, Hafemeister C, Papalexi E, Mauck Iii WM, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single cell data. bioRxiv2018;460147. doi:10.1101/460147. [DOI] [PMC free article] [PubMed]
- 95. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C.. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017;14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Barkas NP, Nikolaeva D, Lozinsky Y, Demharter S, Khodosevich K, Kharchenko PV. Wiring together large single-cell RNA-seq sample collections. bioRxiv2018;460246. doi:10.1101/460246. [DOI] [PMC free article] [PubMed]