Abstract
The emergence of rapidly evolving multidrug-resistant pathogens and a deficit of new compounds entering the clinical pipeline necessitate the exploration of alternative sources of antimicrobial therapeutics. Cyclotides revealed in Viola spp. are a class of highly stable, cyclic, and disulfide-bound peptides with diverse intrinsic bioactivities. Herein we have identified a novel complement of 42 putative cyclotide masses in the plant species Viola inconspicua. Cyclotide-containing fractions of a V. inconspicua peptide library revealed potent bioactivities against the Gram-negative bacteria Escherichia coli ATCC 25922 and the highly virulent and multidrug-resistant Klebsiella pneumoniae VK148. As such, six previously uncharacterized cyclotides, cycloviolacins I1–6 (cyI1–cyI6), were prioritized for molecular characterization. Cyclotides cyI3—cyI6 contain a novel “TLNGNPGA” motif in the highly variable loop six region, expanding the already substantial sequence diversity of this peptide class. Library fractions comprised of cyclotides cyI3–cyI6 exhibited MIC values of 18 and 35 μM against E. coli and K. pneumoniae, respectively, whereas isolated cyI3 killed ~50% of E. coli at 60 μM and isolated cyI4 demonstrated no killing at concentrations >60 μM against both pathogens. This work expands the repertoire of bioactive cyclotides found in Viola spp. and highlights the potential of these antibacterial cyclic peptides.
Graphical Abstract

As the world rapidly evolves into a postantibiotic era, it is necessary to examine the successes and limitations of standard approaches for new lead discovery, including reevaluation of previous and current traditional medicines. Botanical remedies derived from the >500-membered genus Viola (Violaceae) have been a mainstay of Chinese,1 Middle Eastern,2 Indian,3 and African3,4 ethnomedicine systems for generations. Viola essential oils, extracts, and poultices were used classically in the treatment of bacterial and viral infections (cough, cold, fever), in addition to inflammation (skin disease, migraine) and oxidative stress (cancer, hypertension).3 True to age-old beliefs, modern examination of the chemical constituents present in Viola spp. has revealed a wealth of bioactive secondary metabolites with broad-spectrum antibacterial activity,5,6 inhibition of inflammation pathways,7 antioxidant activity toward the treatment of hypertension8 and oxidative stress,9 as well as selective cytotoxic effects on drug-resistant10 and triple-negative11 cancers. The impressive biochemical repertoire presented by this genus highlights its clinical relevance and suggests that further exploration of its bioactive potential may unearth additional potent and diverse biological activities.
Numerous structurally and functionally distinct molecules orchestrate the biological activities characteristic of Viola spp.; however, a group of small ribosomally synthesized cyclic plant peptides, the cyclotides, are a primary driver of this bioactive powerhouse genus. Cyclotides are defined by their unique three-dimensional structure—the highly stable cyclic cysteine knot motif (CCK) generated via three conserved disulfides and a cyclic backbone forged through a standard peptide bond.12 Six “loops” or sequence regions exist between the six cysteine residues and are numbered consecutively from the first cysteine residue after the N-terminal cyclic ligation site to the C-terminus. Loops 1 and 4 are largely conserved among known cyclotide variants, while loop 6 has been subject to substantial evolutionary diversity, contributing to ~400 distinct cyclotide species (CyBase, the database of cyclic proteins, http://www.cybase.org.au), with innumerous variations thought to await discovery.13–15 Cyclotide expression, both in quantity and identity, is highly dynamic among different botanical species and even within the same species arising from seasonal variation, environmental conditions, geographical location, and external cues, dramatically increasing the sequence space sampled.16,17 Novel cyclotide motifs are often emerging,18 suggesting that current attempts to elucidate the extent of sequence diversity in this peptide class have barely scratched the surface of variations that may exist in nature.
As diverse as their sequences, cyclotides exhibit a range of robust biological activities. Discovery of the first cyclotide, kalata B1, followed the observation that African women consumed Oldenlandia affinis tea to speed childbirth, and the uterotonic activities of this concoction were later attributed to its cyclotide content.19 Subsequent characterization of numerous additional cyclotide species over the past three decades has illuminated a world of intrinsically bioactive compounds. Believed to participate in host defense,20,21 the antimicrobial22 and insecticidal activities demonstrated by cyclotide family have made these fascinating peptides obvious candidates for potential clinical and agricultural use. As membranolytic pore-formers, cyclotides achieve cytotoxicity via electrostatic-driven interactions and an amphipathic tertiary structure to facilitate target membrane disruption. Affecting cells with net negative membrane potentials, the largely nondiscriminatory cyclotide mechanism of action has led to additional clinically important anti-HIV23,24 and antineoplastic bioactivities.11,25–27
Viola inconspicua Blume is a weedy perennial prone to rapid dispersion through abundant cleistogamous flowers distributed among the grasslands, fields, and forests of Asia. This commonly hybridized Viola species is used in Chinese herbal remedies and was recently implicated in antifungal activity against the dermatophyte Trichophyton rubrum.28 As few as three cyclotide sequences (kalata S, vinc A, vinc B) have been identified in V. inconspicua,29,30 thus warranting further examination of this underexplored botanical species to reveal previously uncharacterized cyclotide sequences.
Herein, analysis of V. inconspicua revealed 42 uncharacterized cyclotide candidates and the known cyclotides viba 11, cyO8 and cyO9. To assess bioactivity of this cyclotide-rich species, a V. inconspicua peptide library was assayed against E. coli ATCC 25922 and the highly virulent and rifampin/streptomycin resistant pathogen Klebsiella pneumoniae VK148.31 Cyclotide-containing library fractions demonstrated robust antibacterial bioactivity, and the most abundant putative cyclotides masses in these fractions were prioritized for sequence characterization. Six novel cyclotide sequences were revealed (cycloviolacins I1–I6, cyI1–cyI6), four of which contain a previously unknown “TLNGNPGA” motif in the hypervariable loop 6 region.
Cyclotide sequences containing this novel motif were characterized for biological activity and secondary structure. Antibacterial activities of cyI3–cyI6 library fractions and isolated cyclotide species cyI3 and cyI4 were investigated. Determination of minimum inhibitory concentrations of cyclotide mixtures may begin to elucidate the synergistic effects among the numerous cyclotide species expressed in planta, likely enhancing the bioactive potencies of individual cyclotide members. Molecular modeling of the cyclotide cycloviolacin I3 (cyI3, 3378 Da) with Robetta32 suggests these novel-motif containing species maintain secondary and tertiary structure to known NMR-based cyclotide structures.
RESULTS AND DISCUSSION
Discovery of Novel V. inconspicua Cyclotides.
Identification of putative cyclotide masses in complex botanical extracts has been achieved routinely via mass spectrometry, where the reduction and alkylation of three conserved disulfides characteristic of cyclotides produces a telltale mass shift of +348.16 Da.33,34 This analysis of the V. inconspicua peptidome resulted in 42 putative cyclotide masses and three previously identified cyclotides, Viba 11 (Viola tricolor, Viola baoshanensis, Viola philippica), cyO8 (Viola odorata, Viola adunca), and cyO9 (Viola odorata, Viola biflora) (Table S1, Supporting Information). This novel complement of cyclotide-like masses suggests a much larger population of cyclotidyl species present in V. inconspicua than reported in the literature. Although the 42 masses identified via reduction alkylation mass shift analysis have not yet been mapped to specific cyclotides in V. inconspicua, 10 of these putative cyclotide masses are isobaric with cyclotides known in other botanical species (CyBase.org.au). Without full sequence characterization, however, it is not possible to confirm the identity of these masses.
Exploration of V. inconspicua Antibacterial Activities.
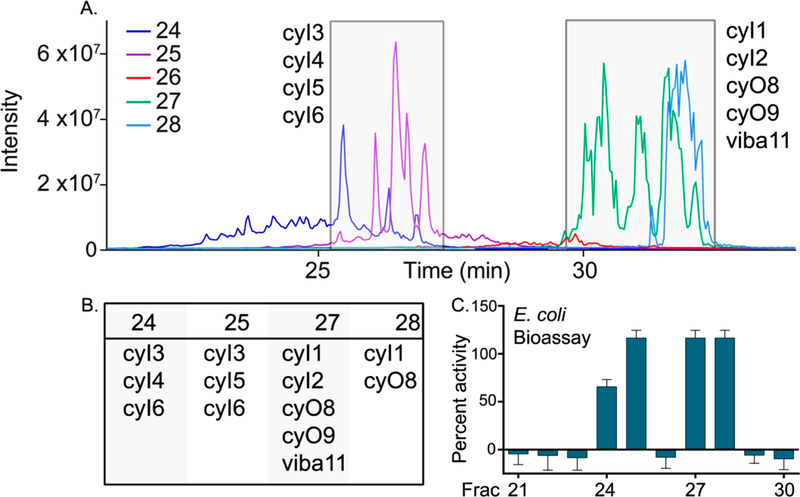
Sequence diversity within cyclotides is a likely contributor to the broad range of activities exhibited by this bioactive family of molecules. V. inconspicua reversed-phase peptide libraries produced regions of robust bioactivity (50–100%) against the Gram-negative bacteria E. coli ATCC 25922 and K. pneumoniae VK148 (Figure 1). LC–MS/MS analysis of this peptide library revealed bioactive fractions to be highly populated with both novel and known cyclotides (Figure 2), specifically library fractions 24, 25, 27, and 28. Six dominant and uncharacterized masses in these bioactive regions [3236.4 Da (cycloviolacin I1, cyI1), 3259.4 Da (cyI2), 3378.5 Da (cyI3), 3391.4 Da (cyI4), 3406.5 Da (cyI5), and 3433.5 Da (cyI6)] were thus prioritized for molecular characterization and validation of bioactivity.
Figure 1.
V. inconspicua peptide library exhibits robust bioactivity against E. coli ATCC 25922 (top) and K. pneumoniae VK148 (bottom).
Figure 2.
Bioactive V. inconspicua peptide library fractions contain abundant novel cyclotides (cyI1–cyI6). (A) Overlaid total ion chromatograms (TICs) of V. inconspicua reversed-phase library fractions 24–28. (B) Abundant cyclotide species present in each of the bioactive fractions. (C) V. inconspicua vs E. coli ATCC 25922 assay bioactivity region matches elution profile of novel cyclotide species cyI1–cyI6.
Sequence Characterization of Six Novel Cyclotides.
Multiple material harvests were required to amass sufficient material for characterization, and notably, abundances of putative and confirmed cyclotide sequences identified in this study were variable, as has been described in a previous study.35 The cause of these fluctuations, whether seasonal, temperature-related, or pathogen-induced, was indiscernible. This highlights the dynamic nature of cyclotide expression and suggests that cyclotides described in current literature are only a snapshot of possible sequences that exist in nature.
The six targeted masses (cyI1–cyI6) were sequenced via LC–MS/MS analysis of the reduced, alkylated, and Glu-C enzymatic digests via de novo sequence assignment and deposited on CyBase.org.au. Assigned cyO8/cyO9 MS/MS spectra aided the de novo sequencing of cyclotides cyI1–2 (Figure S1 and S2, Supporting Information), whereas MS/MS fragment ions of the highly abundant cyclotide cyI3 (Figure 3) were used to guide the sequence elucidation of cyclotides cyI4–6 (Figures S3–S5, Supporting Information). Low abundance MS/MS fragmentation within loop five required additional sequence verification with chymotrypsin digestion and LC–MS/MS analysis. Additionally, chymotrypsin selectivity for leucine residues and homology with known cyclotide sequences were used to discern isoleucine/leucine identities in several positions.
Figure 3.
V. inconspicua cyclotide cyI3 (3378.4 Da) CID MS/MS spectra of (A) gluC linearized cyI3 (+4), (B) cyI3 chymotrypsin fragment 2035.8 Da (+2), and (C) cyI3 chymotrypsin fragment 1218.5 Da (+2). Spectra are annotated with b and y ion series, and coverage is shown on sequence maps above each spectrum.
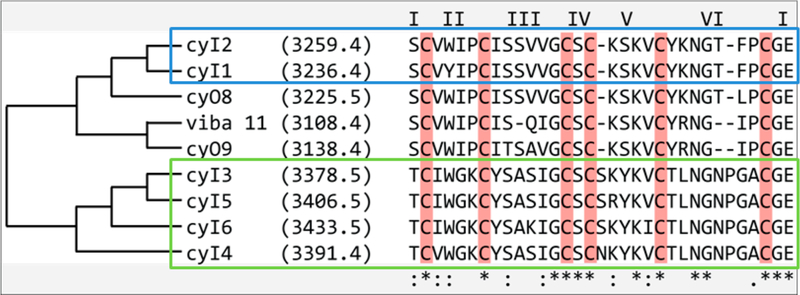
Four newly found sequences (cyI3–cyI6) demonstrated a novel “TLNGNPGAC” motif, deemed motif “Type II,” in the hypervariable loop 6 and were on the high end of the mass range typically exhibited by cyclotides (~2800–3700 Da, with most sequences clustering under 3400 Da33) (Table 1). The remaining two sequences (cyI1 and cyI2) were more homologous to known cyclotides cyO8/cyO9 based on cluster analysis, deemed motif “Type I” (Figure 4). Several parallels exist among the novel sequences identified in this work and the cytotoxic vibi cyclotides from the alpine violet, Viola biflora.36 CyI1 (3236.4 Da) was found to contain a tyrosine residue in loop two often occupied by a tryptophan residue (as in known cyclotides vibi H), a conserved mutation unlikely to affect activity. Cyclotides cyI1 and cyI2 contain a “GTXPCGE” motif, but contain a phenylalanine residue in the third position similar to only two other known cyclotides, vibi G and vibi J. Often occupied primarily by hydrophobic residues, loop three of cyclotide cyI6 contains a basic residue (lysine), common to only vibi J and ~10 other known cyclotides36 (CyBase.org.au). All cyclotides cyI1–cyI6 contain an asparagine residue in loop 6, a critical feature involved in the intracellular cyclization process and highly conserved throughout cyclic cyclotide variants.37–39 Interestingly, Type II cyclotides lack the loop 6 basic residue found in most cyclotide species; chemically masking this lysine or arginine residue decreases the activity of cyclotides cyO2 and varv A40 and may contribute to the weak activity of cyI3–cyI6.
Table 1.
Six Novel Cyclotide Sequences (cyI1–cyI6) Identified in Viola inconspicua
| identity | mass (Da) | sequence |
|---|---|---|
| cyI1 | 3236.4 | SCVYIPCISSVVGCSC-KSKVCYKNGT-FPCGE |
| cyI2 | 3259.4 | SCVWIPCISSVVGCSC-KSKVCYKNGT-FPCGE |
| cyI3 | 3378.5 | TCIWGKCYSASIGCSCSKYKVCTLNGNPGACGE |
| cyI4 | 3391.4 | TCVWGKCYSASIGCSCNKYKVCTLNGNPGACGE |
| cyI5 | 3406.5 | TCIWGKCYSASIGCSCSRYKVCTLNGNPGACGE |
| cyI6 | 3433.5 | TCIWGKCYSAKIGCSCSKYKICTLNGNPGACGE |
Figure 4.
Alignment of novel cyclotide sequences cyI1–cyI6 with previously characterized cyclotides viba 11, cyO8, and cyO9. Loops are indicated by roman numerals above sequences. Homology is indicated by tree on left. Asterisks on the bottom of sequences indicate high conservation, where colons indicate conservative mutations.
Examination of the tandem mass spectrometry data revealed that the two distinct sequence types, i.e., Type I and II motifs, detected in V. inconspicua are discernible via characteristic MS/MS fingerprint ions (Figure 5). The previously uncharacterized high molecular weight motifs represented by Type II cyclotides (cyI3–cyI6) were easily identified by the distinct C-terminus containing y-ions resulting from commonly used collision-induced dissociation (CID) fragmentation, m/z 590.2, 365.1, and 205.1, whereas Type I sequences are indicated by the highly abundant N-terminus containing b-ion series m/z 248.1, 347.1, 533.2, 646.3. Sample losses from reduction, alkylation, and enzymatic digestion often complicate sequence elucidation of novel cyclotide species. Using these highly abundant m/z fingerprints, MS/MS data can provide us with some information about mass identity and sequence type, though overlap in the MS1 may lead to ambiguity from “mixed” populations of Type I and II cyclotide fingerprints (Table S1, Supporting Information). Additionally, reversed-phase C18 retention times of cyO8/cyO9-like sequences cyI1 and cyI2 (Type I) were distinct from those containing the novel loop 6 motif, cyI3–cyI6 (Type II) (Figure 2). Interestingly, both types are similar in hydrophobic content and isoelectric point, however, the distribution of these residues may modify the tertiary structure. For example, a tyrosine residue found in position one of loop six in Type I sequences is in position one of loop three of type II cyclotides (Figure 4).
Figure 5.
CID MS/MS spectra of GluC-digested V. inconspicua cyclotides contain several highly abundant fragment ions characteristic of motif type, either Type I (left column) or the novel Type II motif reported herein (right column). Fragments are color-coded throughout spectra.
Bioactivity Assessment.
Due to the high sequence homology of cyclotides cyI3–cyI6, chromatographic isolation of quantities sufficient for bioactivity characterization of individual cyclotides was challenging. However, the early reversed-phase retention time of cyI4 allowed separation from other novel Type II cyclotides cyI3, 5, and 6 in enough quantity for bioactivity assays against both E. coli and K. pneumoniae, and the high abundance of cyI3 allowed for isolation of enough material for bioactivity testing solely against E. coli. Minimum inhibitory concentration of isolated cyI4 was not achieved at a concentration >60 μM against both E. coli and K. pneumoniae and cyI3 exhibited ~50% activity against E. coli at 60 μM. In contrast, fractions predominantly containing cyclotides cyI3–cyI6 demonstrated an MIC of 18 μM and 35 μM against E. coli ATCC and K. pneumoniae, respectively. As the library fractions generated in this work are representative of the relative concentrations found in physiological conditions of the botanical specimens used, fractions containing mixed cyclotide species allow for synergistic interactions potentially occurring in nature. Though it is posited that the ability of cyclotides to self-associate in solution may affect membranolytic activity,41 synergistic effects of different cyclotide sequences on bioactivity is largely unexplored.42,43 Regardless, cyclotide sequence diversity seen in planta is likely a multifaceted defense strategy evolved to combat diverse and seasonal stress conditions,33 supported by the observation that small sequence changes influence and tune cyclotide hemolytic ability, and thus target specificity.35 Interestingly, the observed bioactivity of Type II cyclotides is significantly less potent than the bactericidal cyclotide cyO2 (MIC 5 and <12.5 μM against E. coli and K. pneumoniae, respectively).
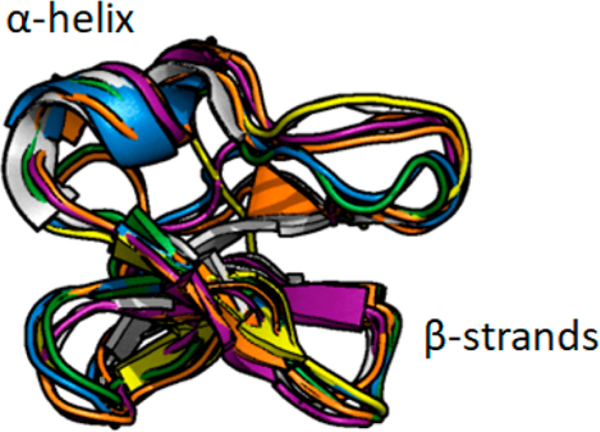
All new cyclotides were found to belong to the bracelet subfamily, defined by a high isoelectric point and the lack of a cis-proline in loop 5. Unlike nonribosomally synthesized bacterial-derived cyclic peptides, which tend to be smaller in size, cyclotides can form various elements of secondary structure. Molecular modeling of cyclotide cyI3 with Robetta generated five homology-based models and predicted an overall three-dimensional and secondary structure (a short α-helix in loop 3 and antiparallel β-sheets with loops 4 and 5) similar to that of cycloviolcin O2 (cyO2), the prototypical bracelet cyclotide (Figure 6).
Figure 6.
Overlay of Robetta predicted novel cyclotide cyI3 structures with prototypical bracelet cyclotide cyO2 (white structure, PDB 2KNM). Generated in PyMol (Schrödinger). CyI3 predicted structures conform to known bracelet subfamily secondary and tertiary structure.
Concluding Remarks.
New compounds with potent bioactivity and novel mechanisms of action are needed to supplement the increasingly ineffective antimicrobial therapeutics used in the modern clinic, particularly those with efficacy against Gram-negative bacterial species. Herein, we have established the antibacterial activity of a V. inconspicua peptide library against the Gram-negative pathogens E. coli ATCC 25922 and K. pneumoniae VK148. Forty-two putative cyclotide masses were identified in Viola inconspicua. Six novel sequences (cyI1–cyI6) highly abundant in bioactive library fractions were determined, expanding the already substantial sequence diversity of this highly stable and potent peptide class. Notably, each motif type was readily identifiable via characteristic MS/MS fingerprint ions that may be used in future analysis of cyclotide contents of botanical extracts. MIC values determined for library fractions containing novel cyclotides cyI3–cyI6 were in the low micromolar range, whereas MIC values of isolated cyclotides cyI3 and cyI4 were considerably higher. Due to the high sequence homology of cyI3–cyI6, the discrepancy in MIC values between isolated and complex cyclotide samples could indicate synergistic interactions among different sequences; however, it is possible that minor sequence variations in cyI5 and cyI6 could account for this increase in potency. Molecular modeling of a cyclotide with the novel loop six “Type II” motif provided evidence for standard cyclotide bracelet subfamily secondary structure elements. This work adds to the sequence diversity of the highly bioactive and stable cyclotide class.
EXPERIMENTAL SECTION
Plant Material.
Viola inconspicua seeds (Mountain Gardens, Burnsville, NC) were grown in a laboratory greenhouse at 17.5–20.3 °C on a 14/10 h light/dark cycle. Mature aerial tissue was harvested with immediate flash freezing and storage at −80 °C until extraction. V. inconspicua greenhouse specimens have been deposited at the UNC Herbarium under accession numbers NCU00303107 and NCU00303108. Specimens are publicly available for viewing on the SERNEC (Southeast Regional Network of Expertise and Collections) Web site (http://sernecportal.org/portal/index.php).
Peptide Library Preparation.
V. inconspicua strong cation exchange (SCX) peptide library was prepared as described previously.41 Briefly, peptidyl constituents were extracted from 100 g fresh weight material in 300 mL 10% acetic acid buffer and selected for by passing extracts through a high molecular weight cutoff filter (MWCO 30 kDa; Millipore) to remove large proteins and by dialyzing into 5 mM ammonium formate pH 2.7 to remove small-molecules (0.1–1 kDa cutoff; SpectrumLabs). A V. inconspicua SCX peptide library was generated with an SCX method (PolySulfethyl A column, 100 mm × 4.6 mm, 3 μm particles, PolyLC) using a linear salt gradient (mobile phase A: 5 mM ammonium formate, 20% acetonitrile, pH 2.7; mobile phase B: 500 mM ammonium formate, 20% acetonitrile, pH 3.0). SCX library fractions were desalted via vacuum centrifugation. A reversed-phase peptide library was subsequently prepared by injecting combined SCX library fractions 15–40 (to avoid collecting early eluting small molecules) onto a Phenomenex Jupiter C18 column (300 Å, 5 μm, 4.6 mm × 150 mm) with a flow rate of 0.5 mL/min and a 25 min linear ramp (mobile phase A, 95/5 water/acetonitrile with 0.1% TFA; mobile phase B, acetonitrile with 0.1% TFA). Reversed-phase library fractions were dried down via vacuum centrifugation and resuspended in water.
Mass Shift Analysis.
V. inconspicua SCX peptide library fractions 15–40 were combined, reduced with 10 mM dithiothreitol (45 °C, 850 rpm, 30 min) and alkylated with 100 mM iodoacetamide (25 °C, 850 rpm, 15 min). Salts were removed by solid phase extraction with Pierce C18 spin columns (Thermo Scientific) prior to LC–MS/MS analysis.
Antibacterial Bioactivity Assays.
The V. inconspicua reversed-phase peptide library was assayed against E. coli ATCC 25922 and clinical isolate Klebsiella pneumoniae VK148 (rifampin and streptomycin resistant derivative of ATCC 43816) in a 96-well plate format, as described previously.41 Bioassay plate wells each contained 30 μL Mueller–Hinton medium, 10 μL bacterial culture, and 10 μL V. inconspicua library fraction. Positive and negative controls consisted of antibiotic (E. coli: ampicillin, 100 μg/mL; K. pneumoniae and Staphylococcus aureus: erythromycin 100 μg/mL) and sterile water, respectively. Plates were incubated at 37 °C, 250 rpm for 4 h. Resazurin was added to each well to a final concentration of 1 mM and incubated for approximately 1 h. Cell viability was quantified using an excitation wavelength of 544 nm and emission wavelength of 590 nm. Assays were performed in triplicate.
Proteolysis for Sequence Characterization.
To avoid overly complex LC–MS/MS spectra, reduction and alkylation was performed as described above on individual SCX fractions. Reduced and alkylated material was incubated 1:50 enzyme:substrate with endoproteinase Glu-C enzyme (Sigma) or chymotrypsin (Sigma) at 37 °C for 3 h.
LC–MS/MS Analysis.
Approximately 1 μg of acidified reduced/alkylated, reduced/alkylated/glu-C digested, or reduced/alkylated/chymotrypsin digested V. inconspicua sample was injected onto a nano-LC-ESI-MS/MS platform consisting of a NanoAcquity (Waters, Milford, MA) coupled to a TripleTOF5600 MS (AB Sciex, Framingham, MA). Front-end UPLC separation of peptides was achieved on an HSS T3C18 column (100 Å, 1.8 μm, 75 μm × 250 mm, Waters), after passing a Symmetry C18 trap column (100 Å, 5 μm, 180 μm × 20 mm, Waters), with a flow rate of 0.3 μL/min and a 30 min linear ramp of 5%–50% B (mobile phase A, 1% formic acid in water; mobile phase B, 1% formic acid in acetonitrile). The TripleTOF5600 MS was operated in positive-ion, high-sensitivity mode with the MS survey spectrum using a mass range of 350–1600 Da in 250 ms. Targeted CID MS/MS data was acquired for the six novel cyclotide sequences using reduced/alkylated and digested samples and a collision energy (CE) of 40. Deisotoped peak lists for the intact and reduced/alkylated samples were generated using Progenesis QI for Proteomics software (Nonlinear Dynamics, v.2.0) with a retention time filter of 14–45 min and a maximum charge of +10. “Peptide ion data” was exported from Progenesis and analyzed for the characteristic reduction/alkylation mass shift corresponding to three disulfide bonds (348.16 Da) using Python.
Isolation of CyI3 and CyI4.
CyI3 and cyI4 isolation was achieved by collecting cyclotide-containing SCX fractions, desalting with SPE, and injecting onto an Agilent Zorbax extended C18 column (300 Å, 5 μm, 4.6 mm × 250 mm) with a flow rate of 2 mL/min and a linear ramp of 5–25% mobile phase B from 0 to 5 min and 25–50% mobile phase B from 5 to 20 min (mobile phase A, water with 0.1% formic acid; mobile phase B, acetonitrile with 0.1% formic acid). CyI3/cyI4 reversed-phase peaks from multiple runs were manually collected, dried down in a vacuum concentrator, resuspended in water, and combined for subsequent MIC testing. CyI3/cyI4 purity was analyzed by LC–MS analysis on a Thermo Scientific Q Exactive HF-X via direct infusion.
Determination of Minimum Inhibitory Concentration.
The cyI3–cyI6 concentration in V. inconspicua reversed-phase library fractions and isolated cyI3/cyI4 samples was determined using a NanoDrop (Thermo) and an extinction coefficient of 8855 M−1 cm−1. MIC values were assessed with a 2-fold serial dilution performed in a 96-well plate format. In each well, 30 μL minimal medium, 10 μL bacterial culture, and 10 μL library fraction was added, and the plate was incubated at 37 °C for 4 h, shaking at 250 rpm, and both OD600 and resazurin were used to establish MIC values. The assay was performed in triplicate for all concentrations (except the highest concentration of cyI3, in which availability of material limited replicates to two).
Molecular Modeling of Cyclotide CyI3.
The three-dimensional structure of cyI3 was modeled with the Robetta server,32 an automated structure prediction server based on the Rosetta software package (Rosetta Commons). Circulin B (PDB 2ERI) was chosen as the homology model template. The five resulting structure predictions were aligned in Pymol (Schrödinger) with the prototypical bracelet cyclotide, cyO2.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Carol Ann McCormick (NCU Herbarium, Chapel Hill, NC) and Harvey Ballard (Ohio University) for curation and identification of Viola specimens. NSF MRI (CHE-1726291) supported the purchase of the Q-Exactive HF-X mass spectrometer, and we thank Dr. Brandie Ehrmann for training on this instrument. This work was funded by NIH (1R01GM125814–01) to L.M.H., and N.C.P. acknowledges NIH Biophysics training grant support (T32 GM008570).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnat-prod.9b00359.
The authors declare no competing financial interest.
REFERENCES
- (1).Muluye RA; Bian Y; Alemu PN J. Trad. Complement. Med 2014, 4, 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Feyzabadi Z; Ghorbani F; Vazani Y; Zarshenas MM Phytother. Res 2017, 31, 1669–1675. [DOI] [PubMed] [Google Scholar]
- (3).Chandra D; Kohli G; Prasad K; Bisht G; Punetha VD; Khetwal KS; Devrani MK; Pandey HK Curr. Res. Chem 2015, 7, 44–52. [Google Scholar]
- (4).Gran L Lloydia 1973, 36, 174–178. [PubMed] [Google Scholar]
- (5).Witkowska-Banaszczak E; Bylka W; Matlawska I; Goslinska O; Muszynski Z Fitoterapia 2005, 76, 458–461. [DOI] [PubMed] [Google Scholar]
- (6).Gautam SS; Navneet; Kumar S. Proc. Natl. Acad. Sci., India, Sect. B 2012, 82, 567–572. [Google Scholar]
- (7).Jeong YH; Oh YC; Cho WK; Shin H; Lee KY; Ma JY BMC Complementary Altern. Med 2016, 16, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Siddiqi HS; Mehmood MH; Rehman NU; Gilani AH Lipids Health Dis 2012, 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Habibi E; Arab-Nozari M; Elahi P; Ghasemi M; Shaki F Appl. Physiol., Nutr., Metab 2019, 44, 521–527. [DOI] [PubMed] [Google Scholar]
- (10).Deng W; Hu B; Dai C; Wang Y; Chen H; Zito SW; Fu L; Chen ZJ Cancer Res. Updates 2013, 2, 87–94. [Google Scholar]
- (11).Alipanah H; Bigdeli MR; Esmaeili MA Iran. J. Pharm. Res 2018, 17, 276–291. [PMC free article] [PubMed] [Google Scholar]
- (12).Saether O; Craik DJ; Campbell ID; Sletten K; Juul J; Norman DG Biochemistry 1995, 34, 4147–4158. [DOI] [PubMed] [Google Scholar]
- (13).Zhang J; Li J; Huang Z; Yang B; Zhang X; Li D; Craik DJ; Baker AJ; Shu W; Liao BJ Plant Physiol 2015, 178, 17–26. [DOI] [PubMed] [Google Scholar]
- (14).Weidmann J; Craik DJ J. Exp. Bot 2016, 67, 4801–4812. [DOI] [PubMed] [Google Scholar]
- (15).Hellinger R; Koehbach J; Soltis DE; Carpenter EJ; Wong GK; Gruber CW J. Proteome Res 2015, 14, 4851–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Trabi M; Svangard E; Herrmann A; Goransson U; Claeson P; Craik DJ; Bohlin LJ Nat. Prod 2004, 67, 806–810. [DOI] [PubMed] [Google Scholar]
- (17).Narayani M; Chadha A; Srivastava SJ Nat. Prod 2017, 80, 1972–1980. [DOI] [PubMed] [Google Scholar]
- (18).Niyomploy P; Chan LY; Harvey PJ; Poth AG; Colgrave ML; Craik DJ J. Nat. Prod 2018, 81, 2512. [DOI] [PubMed] [Google Scholar]
- (19).Gran L Acta Pharmacol. Toxicol 1973, 33, 400–408. [DOI] [PubMed] [Google Scholar]
- (20).Slazak B; Kapusta M; Stromstedt AA; Slomka A; Krychowiak M; Shariatgorji M; Andren PE; Bohdanowicz J; Kuta E; Goransson U Front. Plant Sci 2018, 9, 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Slazak B; Kapusta M; Malik S; Bohdanowicz J; Kuta E; Malec P; Goransson U Planta 2016, 244, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pranting M; Loov C; Burman R; Goransson U; Andersson DI J. Antimicrob. Chemother 2010, 65, 1964–1971. [DOI] [PubMed] [Google Scholar]
- (23).Wang CK; Colgrave ML; Gustafson KR; Ireland DC; Goransson U; Craik DJ J. Nat. Prod 2008, 71, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ireland DC; Wang CK; Wilson JA; Gustafson KR; Craik DJ Biopolymers 2008, 90, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gerlach SL; Rathinakumar R; Chakravarty G; Goransson U; Wimley WC; Darwin SP; Mondal D Biopolymers 2010, 94, 617–625. [DOI] [PubMed] [Google Scholar]
- (26).Lindholm P; Goransson U; Johansson S; Claeson P; Gullbo J; Larsson R; Bohlin L; Backlund A Mol. Cancer Ther 2002,1, 365–369. [PubMed] [Google Scholar]
- (27).Burman R; Svedlund E; Felth J; Hassan S; Herrmann A; Clark RJ; Craik DJ; Bohlin L; Claeson P; Goransson U; Gullbo J Biopolymers 2010, 94, 626–634. [DOI] [PubMed] [Google Scholar]
- (28).Sun Z; Du Y; Cheng L; Zhu N Int.J. Clin. Exp. Med 2014, 7, 5004–5009. [PMC free article] [PubMed] [Google Scholar]
- (29).Ravipati AS; Poth AG; Troeira Henriques S; Bhandari M; Huang YH; Nino J; Colgrave ML; Craik DJJ Nat.Prod 2017, 80, 1522–1530. [DOI] [PubMed] [Google Scholar]
- (30).Ravipati, A. S. Ph.D. Dissertation, University of Queensland, Queensland, Australia, 2016.
- (31).Pechous RD; Broberg CA; Stasulli NM; Miller VL; Goldman WE mBio 2015, 6, No. e02302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kim DE; Chivian D; Baker D Nucleic Acids Res 2004, 32, W526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Burman R; Yeshak MY; Larsson S; Craik DJ; Rosengren KJ; Goransson U Front. Plant Sci 2015, 6, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gruber CW; Elliott AG; Ireland DC; Delprete PG; Dessein S; Goransson U; Trabi M; Wang CK; Kinghorn AB; Robbrecht E; Craik DJ Plant Cell 2008, 20, 2471–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ireland DC; Colgrave ML; Craik DJ Biochem. J. 2006, 400, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Herrmann A; Burman R; Mylne JS; Karlsson G; Gullbo J; Craik DJ; Clark RJ; Goransson U Phytochemistry 2008, 69, 939–952. [DOI] [PubMed] [Google Scholar]
- (37).Craik DJ; Malik U Curr. Opin. Chem. Biol 2013,17, 546–554. [DOI] [PubMed] [Google Scholar]
- (38).Nguyen GK; Wang S; Qiu Y; Hemu X; Lian Y; Tam JP Nat. Chem. Biol 2014, 10, 732–738. [DOI] [PubMed] [Google Scholar]
- (39).Saska I; Gillon AD; Hatsugai N; Dietzgen RG; Hara-Nishimura I; Anderson MA; Craik DJ J. Biol. Chem 2007, 282, 29721–29728. [DOI] [PubMed] [Google Scholar]
- (40).Troeira Henriques S; Craik DJ Biochemistry 2017, 56, 669–682. [DOI] [PubMed] [Google Scholar]
- (41).Rosengren KJ; Daly NL; Harvey PJ; Craik DJ Pept. Sci 2013, 102, 136–136. [Google Scholar]
- (42).Craik DJ Toxins 2012, 4, 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Kirkpatrick CL; Broberg CA; McCool EN; Lee WJ; Chao A; McConnell EW; Pritchard DA; Hebert M; Fleeman R; Adams J; Jamil A; Madera L; Stromstedt AA; Goransson U; Liu Y; Hoskin DW; Shaw LN; Hicks LM Anal. Chem 2017, 89, 1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.