Abstract
MicroRNAs (miRNAs or miRs) are small, noncoding RNAs that are implicated in the regulation of most biological processes. Global miRNA biogenesis is altered in many cancers, and RNA-binding proteins play a role in miRNA biogenesis, presenting a promising avenue for targeting miRNA dysregulation in diseases. miR-34a exhibits tumor-suppressive activities by targeting cell cycle regulators CDK4/6 and anti-apoptotic factor BCL-2, among other regulatory pathways such as Wnt, TGF-β, and Notch signaling. Many cancers exhibit down-regulation or loss of miR-34a, and synthetic miR-34a supplementation has been shown to inhibit tumor growth in vivo. However, the post-transcriptional mechanisms that cause miR-34a loss in cancer are not entirely understood. Here, using a proteomics-mediated approach in non-small-cell lung cancer (NSCLC) cells, we identified squamous cell carcinoma antigen recognized by T-cells 3 (SART3) as a putative pre-miR-34a–binding protein. SART3 is a spliceosome recycling factor and nuclear RNA-binding protein with no previously reported role in miRNA regulation. We found that SART3 binds pre-miR-34a with higher specificity than pre-let-7d (used as a negative control) and elucidated a new functional role for SART3 in NSCLC cells. SART3 overexpression increased miR-34a levels, down-regulated the miR-34a target genes CDK4/6, and caused a cell cycle arrest in the G1 phase. In vitro binding experiments revealed that the RNA-recognition motifs within the SART3 sequence are responsible for selective pre-miR-34a binding. Our results provide evidence for a significant role of SART3 in miR-34a biogenesis and cell cycle progression in NSCLC cells.
Keywords: microRNA (miRNA), microRNA biogenesis, RNA-binding protein, RNA-protein interaction, cancer, lung cancer, precursor microRNA (pre-miRNA), proteomics, squamous cell carcinoma antigen recognized by T-cells 3 (SART3), miR-34a, microRNA biology, cyclin-dependent kinase 4/6 (CDK4/6), cell cycle, non-small cell lung cancer (NSCLC), posttranscriptional regulation, small noncoding RNA, RNA-induced silencing
Introduction
RNA-binding proteins (RBPs)2 are proteins containing one or more RNA-binding domains and have been widely implicated in post-transcriptional regulation of gene expression (1–3). RBPs serve as mediators of RNA transcription (4), modification (5, 6), splicing (6, 7), transport (8, 9), and turnover (10). For many years, RBPs were almost exclusively studied with respect to mRNA (11, 12); however, more recently, an improved understanding of RNA biology and next-generation sequencing technologies have led to the discovery of new roles for RBPs as regulators of noncoding RNAs such as miRNAs (11, 13–17).
miRNAs are a class of small noncoding RNA that act as post-transcriptional regulators of gene expression. Briefly, miRNAs are transcribed as several-kb primary transcripts (pri-miR) by RNA polymerase II (18, 19). This pri-miR is processed by the nuclear Microprocessor complex, composed of Drosha and DiGeorge syndrome critical region 8 (DGCR8), resulting in a ∼60–80-nucleotide pre-miR hairpin (20–23). Following nuclear export by Exportin-5, the pre-miR is processed by Dicer to yield a mature miRNA duplex (24–27). The guide strand of the duplex is loaded onto an Argonaute (AGO) protein to form the RNA-induced silencing complex (RISC), whereas the passenger strand is degraded (28–30). RISC then facilitates silencing of miRNA target gene transcripts (31–33).
The canonical miRNA biogenesis pathway can be disrupted in cancers, and miRs with tumor-suppressive functions are often down-regulated in these diseased states (34–36). RBPs have recently garnered increased attention as modulators of miRNA processing and potential contributors to loss of miRNA activity (13–17). A prominent example is the let-7–Lin28 miRNA-protein interaction, which has been shown to promote several forms of human cancer (34, 37–40). The let-7 family of miRNA has been widely implicated in tumor suppression by targeting oncogenes such as RAS and Myc (34, 41). Lin28 protein binds the hairpin loop of pri- or pre-let-7 to inhibit processing by Drosha or Dicer, respectively, leading to let-7 degradation (42–44).
miR-34a is one of the most extensively characterized miRs and has been shown to mediate tumor suppression by targeting the Notch (45, 46), TGF-β (47), and Wnt signaling pathways (48), as well as influencing the cell cycle (49, 50), senescence (51, 52), and apoptosis. Like let-7, as a tumor suppressor, miR-34a is lost or down-regulated in multiple cancers (53–55). Transcriptional regulation of miR-34a has been widely studied; miR-34a is transcriptionally induced by the p53 tumor suppressor (50, 54, 56–58) and can also be inactivated by CpG methylation (59, 60). Previous work in non-small-cell lung cancer (NSCLC) demonstrated that exogenous miR-34a supplementation inhibited tumor growth in vitro and in vivo (55, 61). Remarkably, growth inhibition occurred independent of p53 status and endogenous expression levels of miR-34a (61). These findings, in conjunction with a growing interest in RBPs as modulators of miRNAs (13–16), led us to hypothesize that an RBP may be influencing the observed loss of miR-34a activity in NSCLC. Herein, we describe the use of a tandem MS-based approach to identify such protein-binding partners for miR-34a.
Results
SART3 is a putative pre-miR-34a–binding protein
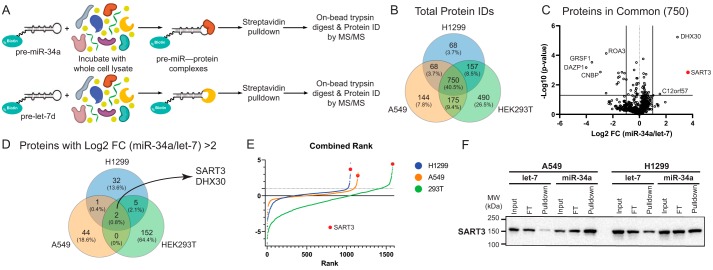
To identify proteins in the miR-34a interactome, we first established a method for miR-binding protein discovery in mammalian cells (Fig. 1A). In brief, a 5′ biotinylated pre-miR is used as bait and incubated with cell lysate, enabling subsequent pulldown of the RNA and its bound proteins on streptavidin-coated resin. After stringent washing, the resin is analyzed by Western blotting or LC-tandem MS (LC-MS/MS) to identify protein interactors. We optimized experimental conditions using the pre-let-7–Lin28 interaction as proof-of-concept, and our method led to the identification of Lin28 as a specific binding partner for pre-let-7d relative to pre-miR-21 in HEK293T and NTERA-2 cell lines by MS and Western blotting (Fig. S1).
Figure 1.
Proteomic analysis identifies SART3 as a pre-miR-34a–binding protein. A, pulldown scheme for MS-based proteomics using a biotinylated pre-miR probe as bait. B, Venn diagram of total proteins identified across each cell line. C, volcano plot of the 750 proteins commonly identified in all cell lines (one replicate per cell line, n = 3). D, Venn diagram of proteins with >2 FC across all cell lines. E, combined ranking of protein -fold changes. F, Western blotting validation of SART3 pulldown with pre-miR-34a (representative of n = 3 experiments).
Next, we used our pulldown strategy to identify RBPs for pre-miR-34a. With the aim of uncovering a p53-independent interaction, we opted to conduct experiments in two NSCLC cell lines: A549, which bears WT p53, and H1299, which carries mutant p53. Pre-let-7d was chosen as our negative control such that Lin28 enrichment could serve as an internal control for proteomics experiments. Pulldowns were performed in freshly lysed A549, H1299, and HEK293T cell lines for analysis by LC-MS/MS. 754 proteins were identified and quantified across all three cell lines (Fig. 1B). These proteins were visualized based on reproducibility and -fold change enrichment for pre-miR-34a (Fig. 1C). Only two proteins exhibited >2 log2 -fold change for pre-miR-34a/pre-let-7: SART3 and the DExH-box protein DHX30 (Fig. 1D). Interestingly, the interaction between SART3 and DHX30 has been observed in several cell lines (62), and both proteins have been shown to interact with AGO1 (63). As DHX30 belongs to a highly conserved family of RNA helicases broadly involved in most aspects of RNA biology (64), we turned our focus to SART3, which showed the highest pre-miR-34a enrichment by two methods of label-free relative quantification (Fig. S2). A combined ranking of all proteins highlighted SART3 as a top hit across all cell lines (Fig. 1E), a finding that was also validated by Western blotting (Fig. 1F). Conversely, we also found that miR-34a was enriched upon immunoprecipitation of SART3 (Fig. S3). In further support of our findings, we noted that SART3 had also been detected as a pre-miR-34a-BP via MS-based proteomics in a recent report, which identified this interaction across several cancer cell lines using a test set of 72 pre-miRs (15). From these results, we concluded that SART3 protein interacts with pre-miR-34a with specificity relative to pre-let-7d and several other pre-miRs.
C-terminal RNA-recognition motifs give rise to selective pre-miR-34a binding
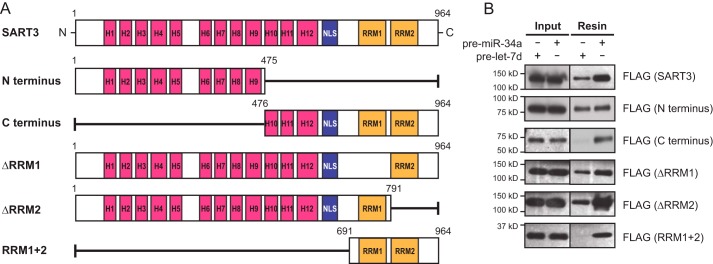
Having characterized a phenotype related to SART3 expression, we next sought to elucidate a potential mechanism for recognition of pre-miR-34a by SART3. SART3 is a 110-kDa nuclear RBP composed of multiple HAT (half-a-tetratricopeptide repeat) domains, two neighboring nuclear localization sequences (NLSs), and two RNA-recognition motifs (RRMs) (Fig. 2A, top) (65, 66). The HAT domain is a conserved helical motif that has been found in several proteins involved in RNA metabolism (67–69), and the RRM is a domain important for RNA recognition and binding (3, 70). Within SART3 specifically, the HAT domains have roles in pre-mRNA 3′-end processing, ubiquitin-specific protease (USP) recruitment, and spliceosome recycling (65, 66, 71–74). The NLS allows SART3 to function as a nuclear protein, and the RRMs contribute to RNA recognition, binding, and splicing activity (65, 74).
Figure 2.
C-terminal RRMs are necessary for specific pre-miR-34a binding. A, SART3 variants cloned into 3×FLAG-pcDNA3 for pre-miR pulldown assays. H, half-a-tetratricopeptide. B, assessment of in vitro binding of SART3 variants by Western blotting.
Recognizing that each of these regions of SART3 could be involved in miRNA regulation, we asked which of these domains were crucial for binding to pre-miR-34a. To this end, we generated 3×FLAG constructs to express different variants of SART3, each containing deletions or truncations of one or more domains of the protein (Fig. 2A). To assess the binding activity for each construct, we transiently transfected HEK293T cells with each plasmid, subsequently performed pre-miR-34a pulldown experiments as outlined in Fig. 1A, and visualized binding relative to pre-let-7d by Western blotting for the FLAG epitope tag.
We first compared the N and C termini of SART3 with the full-length protein using an empty 3×FLAG vector as a negative control. Interestingly, we found that the N terminus bound both pre-miR probes, whereas the C terminus was enriched with pre-miR-34a (Fig. 2B). We further investigated the C terminus of SART3 by testing constructs where either of the two RRMs was deleted, as well as a short fragment containing only the RRMs (Fig. 2A). The RRM 1 + 2 fragment appeared to bind pre-miR-34a with the highest specificity relative to pre-let-7d. Moreover, each of the individual RRM deletions showed a moderate enrichment of pre-miR-34a (Fig. 2B). These data suggest that the HAT domains participate in nonspecific RNA binding, whereas the RRMs of SART3 are important for specific binding of pre-miR-34a.
SART3 knockdown up-regulates miR-34a target genes CDK4 and CDK6
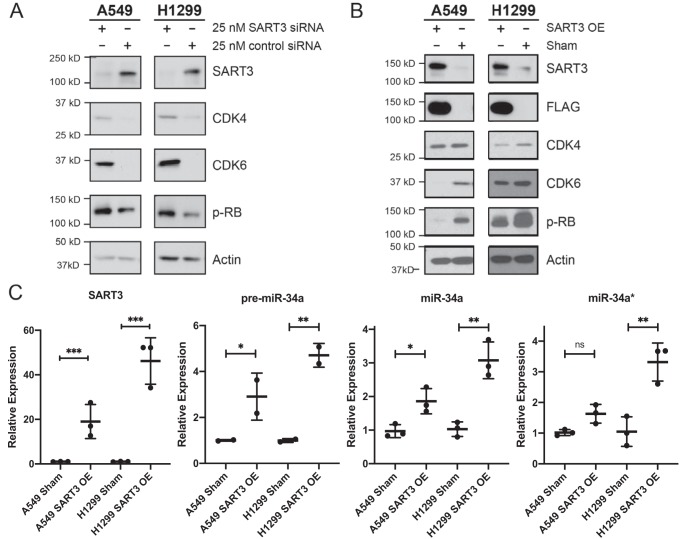
SART3, also referred to as Tip110, is annotated as a nuclear RBP and has been thoroughly characterized as a spliceosome recycling factor (65, 71, 73–75). SART3 has additional reported roles in pre-mRNA splicing (76), regulation of viral gene transcription (77, 78), histone chaperoning (66, 72), and stem cell growth and pluripotency (65, 79, 80). Moreover, SART3 has been investigated as an antigen for cancer immunotherapies (76, 81–85). Among these reported functions, we found no evidence that SART3 had previously been studied as a modulator of miRNA biogenesis or activity. As such, we first asked whether manipulation of SART3 levels would influence miR-34a expression or that of its target genes. To address this question, we depleted SART3 levels in A549 and H1299 cells via transfection with SART3-targeted siRNA. Robust knockdown of SART3, as well as significant increases in two well-established miR-34a target genes, CDK4 and CDK6 (54, 56), was observed relative to cells transfected with a noncoding siRNA control (Fig. 3A). Cyclin-dependent kinases 4 and 6 (CDK4/6) are cell cycle regulators that facilitate progression through the G1 phase via cyclin D–mediated phosphorylation of the retinoblastoma (RB) protein (86–88). Thus, to explore a functional consequence for up-regulation of CDK4/6, we performed additional knockdown experiments and observed that RB phosphorylation was also increased (Fig. 3A). These initial results suggested that, unlike Lin28-mediated regulation of let-7, SART3 may instead function to facilitate miR-34a maturation.
Figure 3.
SART3 levels influence CDK4/CDK6 expression and miR-34a levels. A, SART3 knockdown results in up-regulation of CDK4, CDK6, and phospho-RB (p-RB). B, stable overexpression of 1×FLAG-SART3 results in decreased CDK4, CDK6, and phospho-RB. C, SART3 mRNA, pre-miR-34a, miR-34a, and miR-34a* levels as determined by qRT-PCR. SART3 was normalized to UBB, and miRNAs were normalized to U6 snRNA (n = 2 for pre-miR-34a, n = 3 for all others). Two-tailed Student's t test was applied. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ns, not significant. Error bars, S.D.
SART3 overexpression increases miR-34a levels and decreases CDK4/6
To test the hypothesis that SART3 promotes miR-34a maturation, we generated stable cell lines overexpressing 1×FLAG-SART3. Following lentiviral transduction, levels of miR-34a, CDK4, and CDK6 were assessed by quantitative RT-PCR (qRT-PCR). Importantly, we found increased levels of pre-miR-34a, as well as the active and inactive strands of the mature miR-34a duplex (miR-34a and miR-34a*, respectively) upon SART3 overexpression (Fig. 3C). Despite our efforts to quantify or visualize pri-miR-34a by qPCR and Northern blotting, we were unable to detect the primary transcript in any of our cell lines. This was not surprising, as pri-miRNAs are often processed very rapidly or co-transcriptionally. Reciprocal to what was observed after SART3 knockdown, decreased levels of CDK4 and CDK6 were noted at the protein and mRNA levels (Fig. 3B and Fig. S4). Similarly, phosphorylation of RB protein was also decreased concomitant with CDK4/6 (Fig. 3B). These results indicate that SART3 overexpression leads to increased miR-34a levels and further support the hypothesis that SART3 has a role in aiding miR-34a biogenesis.
SART3 overexpression leads to G1 arrest
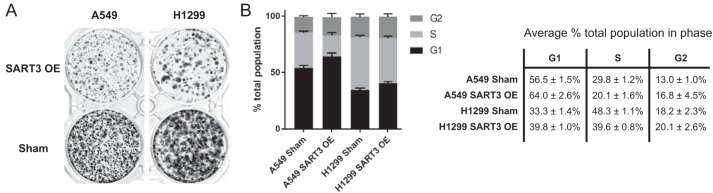
Prominent tumor-suppressive functions of miR-34a include induction of cell cycle arrest, senescence, and apoptosis (50–52, 54, 56–58, 89, 90). Due to the well-known roles of CDK4 and CDK6 as cell cycle regulators, our results prompted us to characterize growth and cell cycle distribution in our SART3 overexpressing cell lines. Consistent with the observed decrease in CDK4, CDK6, and phospho-RB upon SART3 overexpression, we found that these cells exhibited dramatic growth inhibition by colony formation assays (Fig. 4A). This slowed growth was in line with what has previously been observed in cancer cells treated with miR-34a, including NSCLC (53, 54, 56, 61, 91).
Figure 4.
Increased SART3 expression results in growth inhibition and cell cycle arrest in G1 phase. A, colony formation of SART3-overexpressing cell lines. B, cell cycle distribution upon SART3 overexpression in A549 and H1299 cells determined by flow cytometry (n = 3; error bars, S.D.). Representative histograms are shown in Fig. S5.
We next analyzed cell cycle distribution by flow cytometry with propidium iodide–stained cells. In both A549 and H1299 parental lines, substantial growth arrest in the G1 phase was observed upon overexpression of SART3 (Fig. 4B). In addition to cell cycle profiles, we analyzed apoptosis and cellular senescence in these populations. Only a modest increase in apoptosis was detected in SART3-overexpressing cells by annexin V staining (Fig. S6), and no indication of senescence by senescence-associated β-gal staining was found (data not shown). These experiments provide evidence that high SART3 expression levels induce G1 arrest via the miR-34a-CDK4/6 axis.
Discussion
Here we have described a novel miRNA-protein interaction between pre-miR-34a and SART3 in two NSCLC cell lines. Moreover, we have characterized what is, to our knowledge, a new function for the SART3 protein as a modulator of CDK4, CDK6, and cell cycle regulation. Interestingly, the phenotypes we observed indicate that SART3 possesses tumor-suppressive properties in NSCLC cells, which is in contrast to reports describing SART3 as an antigen in other cancers. This would suggest that the functions of SART3 are diverse and not yet fully understood. As such, our results warrant additional studies to probe the SART3 interactome in a more comprehensive fashion.
Although we report compelling evidence of a role for SART3 in the regulation of miR-34a biogenesis, the stage of biogenesis where this interaction occurs remains unclear. It is possible that the second protein identified, DHX30, also plays a role in miR-34a maturation. Notably, both proteins interact with AGO1, but neither has been reported to bind to Drosha, Exportin-5, or Dicer; however, DHX30 interacts with DGCR8 (62). Given these networks, it is conceivable that SART3 could act on pri- or pre-miR-34a as part of a larger complex containing DHX30 and Drosha/DGCR8. Based on the changes observed in pre- and mature miR-34a levels in response to SART3 overexpression, as well as the annotated roles for SART3 as a nuclear RBP, we hypothesize that this protein acts to facilitate miR-34a processing in the nucleus, likely at the precursor level. However, it is also possible that SART3 interacts with pri-miR-34a, as the precursor hairpin is embedded within the primary transcript (Fig. S7). Either hypothesis is supported by the trends observed in miR-34a target genes in response to altered SART3 expression. Future work will investigate the effects of SART3 on other miRNAs and cellular pathways to better understand the specificity of this RNA-protein interaction.
From a molecular recognition standpoint, our results suggest that the RNA-recognition motifs play an important role in forming a specific interaction with pre-miR-34a. The N terminus of SART3 containing several HAT domains binds RNA in a more promiscuous fashion; thus, we postulate that the RRMs contribute toward selectivity and recognition of miR-34a, whereas the N terminus supplies additional RNA-binding capacity. Future work will aim to elucidate a structural basis for this interaction. Such studies will provide added insight into how SART3 functions and will uncover any potential nucleotide sequence or secondary structures important for RNA recognition.
Although significant progress has been made in identifying RBPs as modulators of miRNA biogenesis, our results highlight the need for more detailed functional investigations of these interactions. Characterization of new miR-RBP binding events will improve our current understanding of miRNA regulation in disease, in addition to offering the potential to find new networks for selective therapeutic targeting of dysregulated miRNAs.
Experimental procedures
Cell culture
A549 (a gift from Dr. Beth Lawlor) and HEK293T cells (a gift from Dr. Carol Fierke) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and 2 mm l-glutamine. H1299 cells (a gift from Dr. Nouri Neamati) were cultured in RPMI 1640 medium supplemented with 10% FBS and 2 mm l-glutamine. NTERA-2 cells were purchased from ATCC and cultured in Dulbecco's modified Eagle's medium with 10% FBS, 2 mm l-glutamine, 1 mm sodium pyruvate, and 1% penicillin-streptomycin. Cells were grown at 37 °C with 5% CO2 in a humidified incubator. All cell lines were authenticated by STR profiling and regularly tested for mycoplasma contamination. For knockdown experiments, cells were reverse-transfected in triplicate with 25 nm SART3 SmartPool siRNA (Dharmacon) or siGENOME Nontargeting siRNA 2 (Dharmacon), using 7.5 μl of Lipofectamine RNAiMax (Invitrogen) in 500 μl of Opti-MEM (Gibco) per well of a 6-well dish. Lysates were harvested 48 h after transfection.
Pre-miR pulldown assays
Cells were grown to 80% confluence, washed with PBS, and harvested with a cell scraper. Cells were collected in 1 ml of lysis buffer (50 mm Tris, pH 7.6, 150 mm NaCl, 2 mm MgCl2, 10% glycerol, 0.5% Triton X-100, and freshly added protease inhibitor mixture) per 10-cm dish, kept on ice, and lysed by sonication. Lysate concentrations were normalized to 1 mg/ml by BCA assay (Thermo), and 200 μl of lysate was aliquoted for each pulldown. To the lysate aliquots, 100 μl of binding buffer (50 mm Tris, pH 7.6, 150 mm NaCl, 5% glycerol, 0.05% Tween 20, freshly added 2 mm ZnCl) was added, and a biotinylated pre-miR probe was added to a final concentration of 500 nm. Mixtures were incubated at room temperature for 30 min. To ensure an excess of miRNA relative to beads, 5 μl of streptavidin-coated magnetic resin (Roche Applied Science) per sample was aliquoted and washed with binding buffer. Lysate incubations were added to the streptavidin beads and incubated at room temperature for 1 h with constant agitation. Flow-throughs from each pulldown were collected, and resins were washed once with a stringent buffer (100 mm phosphate, pH 7, 200 mm NaCl, 0.25% Tween 20) and three times with PBS.
Protein identification by LC-MS/MS
Protocols for in-solution digestion are adapted from Yu et al. (92). Beads were resuspended in 50 μl of 0.1 m NH4HCO3 buffer (pH ∼8). Cysteines were reduced by adding 50 μl of 10 mm DTT and incubating at 45 °C for 30 min. Samples were cooled to room temperature, and cysteines were alkylated with 65 mm 2-chloroacetamide for 30 min at room temperature. Proteins were digested overnight with 1 μg of sequencing grade trypsin at 37 °C. Digestion was stopped by acidification, and peptides were desalted using SepPak C18 cartridges (Waters) and then dried using a Vacufuge concentrator (Eppendorf). Resulting peptides were dissolved in 8 μl of 0.1% formic acid, 2% acetonitrile solution, and 2 μl of peptide solution was resolved on a nanocapillary reverse-phase column (Acclaim PepMap C18, 2 μm, 50 cm, Thermo) over 180 min. Eluent was directly introduced into an Orbitrap Fusion mass spectrometer (Thermo) using an EasySpray source. MS1 scans were acquired at 120,000 resolution (AGC target = 1 × 106; maximum IT = 50 ms). Data-dependent collision-induced dissociation MS/MS spectra were acquired using the top speed method (3 s) following each MS1 scan (normalized collision energy ∼32%; AGC target 1 × 105; maximum IT = 45 ms). Protein identification and quantification was performed using MaxQuant (version 1.6.7.0) (93). MS/MS spectra were searched with Andromeda against the reference human database from Uniprot (February 2, 2014 download, 39,882 sequences) appended with common contaminants and the automatically generated reverse database for the decoy search, which was used to calculate the false discovery rate (FDR). Carbamidomethylation of cysteine (57.021464 Da) was set as a fixed modification; acetylation of protein N termini (42.010565 Da) and oxidation of methionine (15.994915 Da) were set as variable modifications. Other search parameters included fixed main-search MS1 error of 4.5 ppm, with 0.5-Da mass deviation allowed for fragment ions; minimum peptide length of 7; two missed cleavages were allowed. Match between runs was enabled for consecutively run samples within the same cell line, with a match time window of 0.5 min.
Data analysis
Relative quantification of proteins was achieved with the MaxLFQ algorithm using default settings. Proteins identified with an FDR of <1% were further filtered by removal of known contaminants and decoy proteins, as were proteins identified by <3 peptide-to-spectrum matches in all samples and those identified by only one peptide. This final list of proteins was loaded into Perseus (version 1.6.5.0), and LFQ intensities were log2-transformed before imputing missing values column-wise, based on a normal distribution (downshift of 1.8 and a width of 0.3). For proteins identified by multiple isoforms, LFQ intensities were reported or imputed for each isoform individually. For the volcano plot (Fig. 1C), -fold changes (LFQ miR34 versus LFQ Let7d control) for the 754 common proteins were calculated within each cell line (n = 3) and combined into a single file before importing into Perseus to perform a two-sided Student's t test, with pulldowns in the different cell lines serving as replicates. All replicates are biological. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (94) partner repository with the data set identifier PXD015278. Raw proteomics data is also shown in Table S1.
Lentivirus production and infection
1×FLAG-SART3 was amplified from A549 cDNA and cloned into pLentilox-IRES-Puro (obtained from University of Michigan vector core) by standard PCR using NheI and XhoI restriction enzymes (see Table S2 for primers). Lentiviruses were packaged, and stable cell lines were generated as described (95).
Western blotting
All lysates were prepared, and immunoblotting was performed as described previously (95). The antibodies used in this work are listed in Table S2.
RNA isolation and qRT-PCR
Total and small RNA were isolated using the mirVana miRNA isolation kit (Invitrogen). For mRNA quantification, cDNA was prepared from total RNA using the Superscript III first-strand synthesis kit (Invitrogen) according to the manufacturer's instructions. PowerUP SYBR Green master mix (Applied Biosystems) was used for gene expression analysis. For miRNA quantification, cDNA was prepared from small RNA using the miScript II RT kit (Qiagen) using the manufacturer's instructions. miScript SYBR Green PCR kit (Qiagen) was used for miRNA expression analysis. All qPCR was performed on a ViiA7 thermocycler using the fast qPCR protocol, and relative -fold change was calculated using the comparative threshold (CT) method. Primers used in this work are listed in Table S2.
Colony formation assays
Cells were seeded in 6-well plates at a density of 1,000 cells per well for A549 and 500 cells/well for H1299. After 9 days, cells were stained and imaged as described (95).
Flow cytometry
Cell cycle analysis was performed as described (94). For apoptosis analysis, cells were grown to 70% confluence, collected 96 h post-transduction, and stained with annexin V–Alexa FluorTM 488 (Invitrogen) according to the manufacturer's protocol. Fluorescence of stained cellular DNA content and/or annexin V conjugates were measured on a CytoFLEX Flow Cytometer (Beckman-Coulter). Cells were gated and analyzed using FlowJo software (version 10).
3×FLAG-SART3 variant cloning
Full-length SART3 and all variants were amplified from our pLentilox-1×FLAG-SART3 vector using standard PCR and cloned into pcDNA3 containing a 3×FLAG tag (95) using EcoRI and XbaI restriction enzymes. All primers used for cloning are listed in Table S2.
Author contributions
E. J. S. and A. L. G. conceptualization; E. J. S. data curation; E. J. S. and D. C. M. formal analysis; E. J. S. investigation; E. J. S. methodology; E. J. S. writing-original draft; E. J. S., D. C. M., and A. L. G. writing-review and editing; A. L. G. supervision; A. L. G. funding acquisition; A. L. G. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Venkatesha Basrur and Kevin Conlon (University of Michigan Proteomics Resource Facility) for assistance in generating proteomics data. We thank Prof. Johnny He's laboratory at the University of North Texas Health Science Center for gifting us with a SART3 antibody that was used in some preliminary experiments. Finally, we thank Dr. Dan Lorenz for intellectual contributions to this work and Dr. Pat O'Brien for input on the manuscript.
This work was supported by the Dr. Ralph and Marian Falk Medical Research Trust (to A. L. G.) and the National Science Foundation (to E. J. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1, S2, and Figs. S1–S7.
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD015278.
- RBP
- RNA-binding protein
- miRNA or miR
- microRNA
- DGCR8
- DiGeorge syndrome critical region 8
- AGC
- automatic gain control
- IT
- injection time
- RISC
- RNA-induced silencing complex
- NSCLC
- non-small-cell lung cancer
- HAT
- half-a-tetratricopeptide repeat
- NLS
- nuclear localization sequence
- RRM
- RNA-recognition motif
- CDK
- cyclin-dependent kinase
- RB
- retinoblastoma
- qPCR and qRT-PCR
- quantitative PCR and RT-PCR, respectively
- FBS
- fetal bovine serum
- FDR
- false discovery rate
- LFQ
- label-free quantitation.
References
- 1. Burd C. G., and Dreyfuss G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 10.1126/science.8036511 [DOI] [PubMed] [Google Scholar]
- 2. Glisovic T., Bachorik J. L., Yong J., and Dreyfuss G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bandziulis R. J., Swanson M. S., and Dreyfuss G. (1989) RNA-binding proteins as developmental regulators. Genes Dev. 3, 431–437 10.1101/gad.3.4.431 [DOI] [PubMed] [Google Scholar]
- 4. Das A. (1993) Control of transcription termination by RNA-binding proteins. Annu. Rev. Biochem. 62, 893–930 10.1146/annurev.bi.62.070193.004333 [DOI] [PubMed] [Google Scholar]
- 5. Nishikura K. (2010) Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rueter S. M., Dawson T. R., and Emeson R. B. (1999) Regulation of alternative splicing by RNA editing. Nature 399, 75–80 10.1038/19992 [DOI] [PubMed] [Google Scholar]
- 7. Fu X.-D., and Ares M. Jr. (2014) Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakielny S., and Dreyfuss G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell 99, 677–690 10.1016/S0092-8674(00)81666-9 [DOI] [PubMed] [Google Scholar]
- 9. Shyu A.-B., and Wilkinson M. F. (2000) The double lives of shuttling mRNA binding proteins. Cell 102, 135–138 10.1016/S0092-8674(00)00018-0 [DOI] [PubMed] [Google Scholar]
- 10. Parker R., and Song H. (2004) The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127 10.1038/nsmb724 [DOI] [PubMed] [Google Scholar]
- 11. Gerstberger S., Hafner M., and Tuschl T. (2014) A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dreyfuss G., Kim V. N., and Kataoka N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- 13. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.-C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., and Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nussbacher J. K., and Yeo G. W. (2018) Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol. Cell 69, 1005–1016.e7 10.1016/j.molcel.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Treiber T., Treiber N., Plessmann U., Harlander S., Daiss J.-L., Eichner N., Lehmann G., Schall K., Urlaub H., and Meister G. (2017) A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol. Cell 66, 270–284.e13 10.1016/j.molcel.2017.03.014 [DOI] [PubMed] [Google Scholar]
- 16. van Kouwenhove M., Kedde M., and Agami R. (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 11, 644–656 10.1038/nrc3107 [DOI] [PubMed] [Google Scholar]
- 17. Treiber T., Treiber N., and Meister G. (2019) Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- 18. Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., and Kim V. N. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cai X., Hagedorn C. H., and Cullen B. R. (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Denli A. M., Tops B. B. J., Plasterk R. H. A., Ketting R. F., and Hannon G. J. (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- 21. Gregory R. I., Yan K.-P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., and Shiekhattar R. (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- 22. Han J., Lee Y., Yeom K.-H., Kim Y.-K., Jin H., and Kim V. N. (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 10.1101/gad.1262504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Landthaler M., Yalcin A., and Tuschl T. (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162–2167 10.1016/j.cub.2004.11.001 [DOI] [PubMed] [Google Scholar]
- 24. Hutvágner G., McLachlan J., Pasquinelli A. E., Bálint E., Tuschl T., and Zamore P. D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- 25. Lund E., Güttinger S., Calado A., Dahlberg J. E., and Kutay U. (2004) Nuclear export of microRNA precursors. Science 303, 95–98 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- 26. Yi R., Qin Y., Macara I. G., and Cullen B. R. (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bohnsack M. T., Czaplinski K., and Gorlich D. (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10, 185–191 10.1261/rna.5167604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carmell M. A., Xuan Z., Zhang M. Q., and Hannon G. J. (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733–2742 10.1101/gad.1026102 [DOI] [PubMed] [Google Scholar]
- 29. Hammond S. M., Boettcher S., Caudy A. A., Kobayashi R., and Hannon G. J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150 10.1126/science.1064023 [DOI] [PubMed] [Google Scholar]
- 30. Kawamata T., and Tomari Y. (2010) Making RISC. Trends Biochem. Sci. 35, 368–376 10.1016/j.tibs.2010.03.009 [DOI] [PubMed] [Google Scholar]
- 31. Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., and Izaurralde E. (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898 10.1101/gad.1424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eulalio A., Huntzinger E., and Izaurralde E. (2008) Getting to the root of miRNA-mediated gene silencing. Cell 132, 9–14 10.1016/j.cell.2007.12.024 [DOI] [PubMed] [Google Scholar]
- 33. Rehwinkel J., Behm-Ansmant I., Gatfield D., and Izaurralde E. (2005) A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640–1647 10.1261/rna.2191905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin S., and Gregory R. I. (2015) MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., and Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 36. Zhang B., Pan X., Cobb G. P., and Anderson T. A. (2007) microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12 10.1016/j.ydbio.2006.08.028 [DOI] [PubMed] [Google Scholar]
- 37. King C. E., Cuatrecasas M., Castells A., Sepulveda A. R., Lee J.-S., and Rustgi A. K. (2011) LIN28B promotes colon cancer progression and metastasis. Cancer Res. 71, 4260–4268 10.1158/0008-5472.CAN-10-4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molenaar J. J., Domingo-Fernández R., Ebus M. E., Lindner S., Koster J., Drabek K., Mestdagh P., van Sluis P., Valentijn L. J., van Nes J., Broekmans M., Haneveld F., Volckmann R., Bray I., Heukamp L., et al. (2012) LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 44, 1199–1206 10.1038/ng.2436 [DOI] [PubMed] [Google Scholar]
- 39. Nguyen L. H., Robinton D. A., Seligson M. T., Wu L., Li L., Rakheja D., Comerford S. A., Ramezani S., Sun X., Parikh M. S., Yang E. H., Powers J. T., Shinoda G., Shah S. P., Hammer R. E., et al. (2014) Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 26, 248–261 10.1016/j.ccr.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O'Sullivan M., Lu J., Phillips L. A., Lockhart V. L., Shah S. P., Tanwar P. S., Mermel C. H., Beroukhim R., Azam M., et al. (2009) Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848 10.1038/ng.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roush S., and Slack F. J. (2008) The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516 10.1016/j.tcb.2008.07.007 [DOI] [PubMed] [Google Scholar]
- 42. Viswanathan S. R., Daley G. Q., and Gregory R. I. (2008) Selective blockade of microRNA processing by Lin28. Science 320, 97–100 10.1126/science.1154040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heo I., Joo C., Cho J., Ha M., Han J., and Kim V. N. (2008) Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32, 276–284 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- 44. Newman M. A., Thomson J. M., and Hammond S. M. (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 10.1261/rna.1155108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bu P., Chen K.-Y., Chen J. H., Wang L., Walters J., Shin Y. J., Goerger J. P., Sun J., Witherspoon M., Rakhilin N., Li J., Yang H., Milsom J., Lee S., Zipfel W., et al. (2013) A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell 12, 602–615 10.1016/j.stem.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Guessous F., Zhang Y., Dipierro C., Kefas B., Johnson E., Marcinkiewicz L., Jiang J., Yang Y., Schmittgen T. D., Lopes B., Schiff D., Purow B., and Abounader R. (2009) MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576 10.1158/0008-5472.CAN-09-0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Genovese G., Ergun A., Shukla S. A., Campos B., Hanna J., Ghosh P., Quayle S. N., Rai K., Colla S., Ying H., Wu C.-J., Sarkar S., Xiao Y., Zhang J., Zhang H., et al. (2012) MicroRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-β signaling in glioblastoma. Cancer Discov. 2, 736 10.1158/2159-8290.CD-12-0111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim N. H., Kim H. S., Kim N.-G., Lee I., Choi H.-S., Li X.-Y., Kang S. E., Cha S. Y., Ryu J. K., Na J. M., Park C., Kim K., Lee S., Gumbiner B. M., Yook J. I., and Weiss S. J. (2011) p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 4, ra71 10.1126/scisignal.2001744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carleton M., Cleary M. A., and Linsley P. S. (2007) MicroRNAs and cell cycle regulation. Cell Cycle 6, 2127–2132 10.4161/cc.6.17.4641 [DOI] [PubMed] [Google Scholar]
- 50. Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., Meister G., and Hermeking H. (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6, 1586–1593 10.4161/cc.6.13.4436 [DOI] [PubMed] [Google Scholar]
- 51. Ito T., Yagi S., and Yamakuchi M. (2010) MicroRNA-34a regulation of endothelial senescence. Biochem. Biophys. Res. Commun. 398, 735–740 10.1016/j.bbrc.2010.07.012 [DOI] [PubMed] [Google Scholar]
- 52. Zhao T., Li J., and Chen A. F. (2010) MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am. J. Physiol. Endocrinol. Metab. 299, E110–E116 10.1152/ajpendo.00192.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hermeking H. (2010) The miR-34 family in cancer and apoptosis. Cell Death Differ. 17, 193–199 10.1038/cdd.2009.56 [DOI] [PubMed] [Google Scholar]
- 54. He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., and Hannon G. J. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gallardo E., Navarro A., Viñolas N., Marrades R. M., Diaz T., Gel B., Quera A., Bandres E., Garcia-Foncillas J., Ramirez J., and Monzo M. (2009) miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 30, 1903–1909 10.1093/carcin/bgp219 [DOI] [PubMed] [Google Scholar]
- 56. Bommer G. T., Gerin I., Feng Y., Kaczorowski A. J., Kuick R., Love R. E., Zhai Y., Giordano T. J., Qin Z. S., Moore B. B., MacDougald O. A., Cho K. R., and Fearon E. R. (2007) p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307 10.1016/j.cub.2007.06.068 [DOI] [PubMed] [Google Scholar]
- 57. Chang T.-C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., and Mendell J. T. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 10.1016/j.molcel.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., and Oren M. (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26, 731–743 10.1016/j.molcel.2007.05.017 [DOI] [PubMed] [Google Scholar]
- 59. Lodygin D., Tarasov V., Epanchintsev A., Berking C., Knyazeva T., Körner H., Knyazev P., Diebold J., and Hermeking H. (2008) Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600 10.4161/cc.7.16.6533 [DOI] [PubMed] [Google Scholar]
- 60. Vogt M., Munding J., Grüner M., Liffers S.-T., Verdoodt B., Hauk J., Steinstraesser L., Tannapfel A., and Hermeking H. (2011) Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 458, 313–322 10.1007/s00428-010-1030-5 [DOI] [PubMed] [Google Scholar]
- 61. Wiggins J. F., Ruffino L., Kelnar K., Omotola M., Patrawala L., Brown D., and Bader A. G. (2010) Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70, 5923–5930 10.1158/0008-5472.CAN-10-0655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huttlin E. L., Bruckner R. J., Paulo J. A., Cannon J. R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M. P., Parzen H., Szpyt J., Tam S., Zarraga G., Pontano-Vaites L., Swarup S., et al. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 10.1038/nature22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Höck J., Weinmann L., Ender C., Rüdel S., Kremmer E., Raabe M., Urlaub H., and Meister G. (2007) Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep. 8, 1052–1060 10.1038/sj.embor.7401088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tanner N. K., and Linder P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8, 251–262 10.1016/S1097-2765(01)00329-X [DOI] [PubMed] [Google Scholar]
- 65. Whitmill A., Timani K. A., Liu Y., and He J. J. (2016) Tip110: physical properties, primary structure, and biological functions. Life Sci. 149, 79–95 10.1016/j.lfs.2016.02.062 [DOI] [PubMed] [Google Scholar]
- 66. Zhang Q., Harding R., Hou F., Dong A., Walker J. R., Bteich J., and Tong Y. (2016) Structural basis of the recruitment of ubiquitin-specific protease USP15 by spliceosome recycling factor SART3. J. Biol. Chem. 291, 17283–17292 10.1074/jbc.M116.740787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hammani K., Cook W. B., and Barkan A. (2012) RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. U. S. A. 109, 5651–5656 10.1073/pnas.1200318109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Preker P. J., and Keller W. (1998) The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 23, 15–16 10.1016/S0968-0004(97)01156-0 [DOI] [PubMed] [Google Scholar]
- 69. Champion E. A., Lane B. H., Jackrel M. E., Regan L., and Baserga S. J. (2008) A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol. Cell. Biol. 28, 6547–6556 10.1128/MCB.00906-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Birney E., Kumar S., and Krainer A. R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21, 5803–5816 10.1093/nar/21.25.5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bell M., Schreiner S., Damianov A., Reddy R., and Bindereif A. (2002) p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 21, 2724–2735 10.1093/emboj/21.11.2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Long L., Thelen J. P., Furgason M., Haj-Yahya M., Brik A., Cheng D., Peng J., and Yao T. (2014) The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J. Biol. Chem. 289, 8916–8930 10.1074/jbc.M114.551754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Medenbach J., Schreiner S., Liu S., Lührmann R., and Bindereif A. (2004) Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol. Cell. Biol. 24, 7392–7401 10.1128/MCB.24.17.7392-7401.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gu J., Shimba S., Nomura N., and Reddy R. (1998) Isolation and characterization of a new 110 kDa human nuclear RNA-binding protein (p110nrb). Biochim. Biophys. Acta 1399, 1–9 10.1016/S0167-4781(98)00082-7 [DOI] [PubMed] [Google Scholar]
- 75. Damianov A., Schreiner S., and Bindereif A. (2004) Recycling of the U12-type spliceosome requires p110, a component of the U6atac snRNP. Mol. Cell. Biol. 24, 1700–1708 10.1128/MCB.24.4.1700-1708.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Harada K., Yamada A., Yang D., Itoh K., and Shichijo S. (2001) Binding of a SART3 tumor-rejection antigen to a pre-mRNA splicing factor RNPS1: a possible regulation of splicing by a complex formation. Int. J. Cancer 93, 623–628 10.1002/ijc.1391 [DOI] [PubMed] [Google Scholar]
- 77. Liu Y., Li J., Kim B. O., Pace B. S., and He J. J. (2002) HIV-1 Tat protein-mediated transactivation of the HIV-1 long terminal repeat promoter is potentiated by a novel nuclear Tat-interacting protein of 110 kDa, Tip110. J. Biol. Chem. 277, 23854–23863 10.1074/jbc.M200773200 [DOI] [PubMed] [Google Scholar]
- 78. Zhao W., Liu Y., Timani K. A., and He J. J. (2014) Tip110 protein binds to unphosphorylated RNA polymerase II and promotes its phosphorylation and HIV-1 long terminal repeat transcription. J. Biol. Chem. 289, 190–202 10.1074/jbc.M113.529784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu Y., Lee M.-R., Timani K., He J. J., and Broxmeyer H. E. (2012) Tip110 maintains expression of pluripotent factors in and pluripotency of human embryonic stem cells. Stem Cells Dev. 21, 829–833 10.1089/scd.2011.0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu Y., Timani K., Mantel C., Fan Y., Hangoc G., Cooper S., He J. J., and Broxmeyer H. E. (2011) TIP110/p110nrb/SART3/p110 regulation of hematopoiesis through CMYC. Blood 117, 5643–5651 10.1182/blood-2010-12-325332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Suefuji Y., Sasatomi T., Shichijo S., Nakagawa S., Deguchi H., Koga T., Kameyama T., and Itoh K. (2001) Expression of SART3 antigen and induction of CTLs by SART3-derived peptides in breast cancer patients. Br. J. Cancer 84, 915–919 10.1054/bjoc.2000.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Furugaki K., Cui L., Kunisawa Y., Osada K., Shinkai K., Tanaka M., Kataoka K., and Nakano K. (2014) Intraperitoneal administration of a tumor-associated antigen SART3, CD40L, and GM-CSF gene-loaded polyplex micelle elicits a vaccine effect in mouse tumor models. PLoS One 9, e101854 10.1371/journal.pone.0101854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Koga M., Komatsu N., Kawamoto N., Shichijo S., Itoh K., and Yamada A. (2004) Analysis of cellular localization of SART3 tumor antigen by a newly established monoclonal antibody: heterotopic expression of SART3 on the surface of B-lineage leukemic cells. Oncol. Rep. 11, 785–789 10.3892/or.11.4.785 [DOI] [PubMed] [Google Scholar]
- 84. Miyagi Y., Imai N., Sasatomi T., Yamada A., Mine T., Katagiri K., Nakagawa M., Muto A., Okouchi S., Isomoto H., Shirouzu K., Yamana H., and Itoh K. (2001) Induction of cellular immune responses to tumor cells and peptides in colorectal cancer patients by vaccination with SART3 peptides. Clin. Cancer Res. 7, 3950–3962 [PubMed] [Google Scholar]
- 85. Yang D., Nakao M., Shichijo S., Sasatomi T., Takasu H., Matsumoto H., Mori K., Hayashi A., Yamana H., Shirouzu K., and Itoh K. (1999) Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res. 59, 4056–4063 [PubMed] [Google Scholar]
- 86. Morgan D. O. (1997) Cyclin-dependent Kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291 10.1146/annurev.cellbio.13.1.261 [DOI] [PubMed] [Google Scholar]
- 87. Weinberg R. A. (1995) The retinoblastoma protein and cell cycle control. Cell 81, 323–330 10.1016/0092-8674(95)90385-2 [DOI] [PubMed] [Google Scholar]
- 88. Malumbres M., and Barbacid M. (2001) To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer 1, 222–231 10.1038/35106065 [DOI] [PubMed] [Google Scholar]
- 89. Yamakuchi M., Ferlito M., and Lowenstein C. J. (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426 10.1073/pnas.0801613105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bandi N., and Vassella E. (2011) miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol. Cancer 10, 55 10.1186/1476-4598-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ji Q., Hao X., Zhang M., Tang W., Yang M., Li L., Xiang D., Desano J. T., Bommer G. T., Fan D., Fearon E. R., Lawrence T. S., and Xu L. (2009) MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One 4, e6816 10.1371/journal.pone.0006816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yu L., Jearawiriyapaisarn N., Lee M. P., Hosoya T., Wu Q., Myers G., Lim K.-C., Kurita R., Nakamura Y., Vojtek A. B., Rual J.-F., and Engel J. D. (2018) BAP1 regulation of the key adaptor protein NCoR1 is critical for γ-globin gene repression. Genes Dev. 32, 1537–1549 10.1101/gad.318436.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 94. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mitchell D. C., Menon A., and Garner A. L. (2019) Chemoproteomic profiling uncovers CDK4-mediated phosphorylation of the translational suppressor 4E-BP1. Cell Chem. Biol. 26, 980–990.e8 10.1016/j.chembiol.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.