Abstract
Streptococcus pneumoniae is an opportunistic respiratory pathogen that can spread to other body sites, including the ears, brain, and blood. The ability of this bacterium to break down, import, and metabolize a wide range of glycans is key to its virulence. Intriguingly, S. pneumoniae can utilize several plant oligosaccharides for growth in vitro, including raffinose-family oligosaccharides (RFOs, which are α-(1→6)-galactosyl extensions of sucrose). An RFO utilization locus has been identified in the pneumococcal genome; however, none of the proteins encoded by this locus have been biochemically characterized. The enigmatic ability of S. pneumoniae to utilize RFOs has recently received attention because mutations in two of the RFO locus genes have been linked to the tissue tropism of clinical pneumococcal isolates. Here, we use functional studies combined with X-ray crystallography to show that although the pneumococcal RFO locus encodes for all the machinery required for uptake and degradation of RFOs, the individual pathway components are biochemically inefficient. We also demonstrate that the initiating enzyme in this pathway, the α-galactosidase Aga (a family 36 glycoside hydrolase), can cleave α-(1→3)-linked galactose units from a linear blood group antigen. We propose that the pneumococcal RFO pathway is an evolutionary relic that is not utilized in this streptococcal species and, as such, is under no selection pressure to maintain binding affinity and/or catalytic efficiency. We speculate that the apparent contribution of RFO utilization to pneumococcal tissue tropism may, in fact, be due to the essential role the ATPase RafK plays in the transport of other carbohydrates.
Keywords: glycoside hydrolase, host-pathogen interaction, Streptococcus, structure-function, X-ray crystallography, virulence factor, galactosidase, pneumococcus, raffinose-family oligosaccharide (RFO)
Introduction
Streptococcus pneumoniae is a frequent asymptomatic colonizer of the upper airways, where free carbohydrates are scarce (1); therefore, it acquires much of its carbon from the depolymerization and import of complex human glycans (2–5). As well as being a common respiratory commensal, the pneumococcus is also capable of migrating to the ear, where it causes otitis media, and it can produce life-threatening invasive disease, such as pneumonia, meningitis, and bacteremia (6). As it transitions through the human body and different tissue types, S. pneumoniae encounters and degrades a diverse range of human glycans. The purpose of this degradation is not only nutritional; carbohydrate depolymerization is an essential contributor to the virulence of this pathogen (7–10), for example through deglycosylation of immune system components and the uncovering of host cell receptors (11–13). The pneumococcal genome encodes for more than 40 known or predicted proteins that cleave glycosidic bonds and more than 20 predicted carbohydrate transport systems (5). Together, these proteins enable S. pneumoniae to metabolize more than 30 different carbohydrates (14). Many of these carbohydrates represent complete or constituent parts of abundant human glycans, such as glycogen, glycosaminoglycans, N- and O-linked glycans, and blood group antigens. However, the pneumococcus also possesses the ability to metabolize a number of other, nonhuman glycans derived from plant material. The oligosaccharides sucrose, melibiose, cellobiose, fructooligosaccharides (FOSs),3 and raffinose-family oligosaccharides (RFOs) can all support growth of S. pneumoniae in vitro (14–19), but exactly how, where, and when the bacterium would encounter these glycans is not known. In the case of sucrose, cellobiose, and FOSs (for which many of the uptake and degradation proteins have been biochemically characterized) (16, 17, 20–22), hypotheses include: the potential intersection of these oligosaccharides and the pneumococcus in the oropharynx (23); the interaction of S. pneumoniae with components of these glycans when present in the capsule of other bacteria (17); and/or the structural similarity between these plant-derived glycans and human glycans (17, 18, 23). This last hypothesis suggests that the “true” in vivo substrates/ligands of some pneumococcal carbohydrate-active proteins may be different from those determined in vitro.
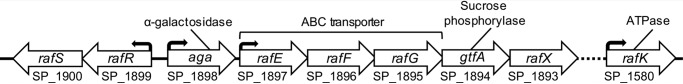
RFOs are a family of oligosaccharides possessing α-(1→6)-galactosyl extensions of sucrose (d-glucose-α-[1→2]-d-fructose) (24). The most common RFOs in plants are the trisaccharide raffinose, the tetrasaccharide stachyose, and the pentasaccharide verbascose. An RFO utilization locus has been identified in the genome of S. pneumoniae (25) and encodes for Aga, an α-galactosidase belonging to glycoside hydrolase (GH) family 36 (25); GtfA, a putative sucrose phosphorylase belonging to GH family 13; an ATP-binding cassette (ABC) transporter, consisting of the solute-binding protein (SBP) RafE (26) and the putative permeases RafF and RafG; RafR and RafS, two putative transcriptional regulators/repressors; and RafX, a putative protein of unknown function (Fig. 1). The ATP-binding protein RafK (also known as MsmK) that powers the ABC transporter is encoded elsewhere in the genome (15, 19).
Figure 1.
Organization of the core RFO utilization locus from S. pneumoniae. Genetic organization of a carbohydrate processing locus and an accessory ORF present in strains of S. pneumoniae whose protein products are known or putatively associated with raffinose utilization. The main locus encodes for an α-galactosidase (Aga) (25), an ABC transporter that consists of a solute-binding protein (RafE) (26) and two putative permeases (RafF and RafG), a putative sucrose phosphorylase (GtfA), two putative transcriptional regulators/repressors (RafR and RafS), and a putative protein of unknown function (RafX). Elsewhere in the genome is an ATP-binding protein (RafK) that powers the ABC transporter (15, 19). Arrows above ORFs indicate the presence of promoter sequences. Locus tags underneath ORFs correspond to the TIGR4 genome.
The complete RFO utilization locus is part of the “core” genome of S. pneumoniae (present in >98% of clinical isolates) (7, 27). In terms of virulence, both Aga and RafF have been identified as putative virulence factors in a signature-tagged mutagenesis screen (9) (this has not been validated in directed studies). Deletion mutants of rafK have also been shown to be outcompeted by WT in mouse models of pneumococcal colonization and infection (15, 19); however, RafK powers other carbohydrate transporters in addition to RafEFG (15). The most compelling evidence for the role of the RFO utilization locus in virulence comes from a recent study in which the ability of clinical pneumococcal isolates to utilize raffinose was correlated with their tissue tropism (28). Minhas et al. (28) compared the genomes of serotype- and sequence type–matched isolates from blood and ear infections and identified SNPs in rafK or rafR among the ear isolates. The ability of these isolates to grow on raffinose was significantly reduced compared with their paired blood isolates, and reversal of the rafR mutation restored full raffinose utilization competency. Expression of genes from each of the transcriptional units within the RFO locus (aga, rafG, and rafK) was also significantly lower in the ear isolates compared with the blood isolates. Finally, in a mouse model of infection, isolates bearing the rafR mutation were less able to persist in the lungs than rafR WT strains; however, the mutant strains exhibited significantly higher bacterial loads in the ear and, to a lesser extent, the brain. Therefore, the authors concluded that the capacity to utilize raffinose determines the ultimate site of infection following nasal challenge (28).
Exactly how expression of the RFO utilization locus affects the ability of S. pneumoniae to cause pneumonia or otitis media is not known. Evidence to indicate that the proteins encoded by this locus are involved in RFO processing is 3-fold. First, expression of aga is strongly induced by raffinose (25). Second, the SBP RafE has been crystallized in complex with raffinose (PDB code 2I58),4 and insertional inactivation of rafE abolishes raffinose uptake (19). Third, deletion of rafG abolishes growth of S. pneumoniae on raffinose and stachyose (14). However, none of the proteins encoded by the RFO utilization locus have been biochemically characterized to provide direct evidence of activity on RFOs.
The relationship between this locus, the processing of RFOs, and pneumococcal virulence is thus enigmatic. We therefore set out to biochemically and structurally characterize the key specificity determinants of the RFO utilization locus. Here we show that RafE binds RFOs, while Aga and GtfA sequentially degrade RFOs into free galactose, fructose, and glucose-1-phosphate (G1P). Overall, however, Aga has relatively poor activity on RFOs, and RafE demonstrates remarkably low affinities for these ligands, suggesting that this locus is not optimized for efficient RFO uptake and degradation. Given the apparent biochemical inefficiency of these proteins and the natural lifestyle of S. pneumoniae, we propose that this locus is a relic of streptococcal evolution that is not utilized by the pneumococcus in vivo. We also speculate that the reported link between the RFO locus and virulence may, in fact, be due to the promiscuous role RafK plays in the import of several human-derived carbohydrates.
Results
The RFO binding properties of RafE
SBPs are the specificity determinants of ABC transporters and, in turn, they dictate the array of glycans that are made available to downstream processing enzymes. To examine the ligand-binding properties of RafE, the protein lacking the native N-terminal secretion signal sequence and lipid-anchoring motif was produced recombinantly in Escherichia coli, and the binding properties of the purified protein were studied using isothermal titration calorimetry (ITC; Table 1). The ligands evaluated included RFOs (raffinose, stachyose, and verbascose), fragments of these glycans (i.e. galactose, melibiose, and sucrose), α-(1→3)-galactobiose, and the glucose trisaccharides isomaltotriose and panose (see Table 1 for chemical descriptions of each). The affinity of RafE for the tested mono- and disaccharides was too low to be detected, and the dissociation constants (Kd) for the trisaccharides and larger RFOs varied from ∼80 μm to ∼500 μm (Table 1). Notably, the measured affinities of RafE are significantly lower than those typically reported for SBPs, which are typically in the Kd range of ∼1 μm (4, 29–32).
Table 1.
Binding constants for RafE determined by isothermal titration calorimetry
The data shown are the means of three independent titrations.
| Ligand | Kd ± S.D. |
|---|---|
| μm | |
| Raffinose Gal-α-(1→6)-Glc-α-(1→2)-Fru | 478.6 ± 35.2 |
| Stachyose Gal-α-(1→6)-Gal-α-(1→6)-Glc-α-(1→2)-Fru | 198.5 ± 23.0 |
| Verbascose Gal-α-(1→6)-Gal-α-(1→6)-Gal-α-(1→6)-Glc-α-(1→2)-Fru | 368.8 ± 34.2 |
| Panose Glc-α-(1→6)-Glc-α-(1→4)-Glc | 80.2 ± 8.1 |
| Isomaltotriose Glc-α-(1→6)-Glc-α-(1→6)-Glc | 427 ± 20.1 |
Given the ability of RafE to accommodate glycans of different lengths and monosaccharide composition, we explored the molecular basis of its ligand binding by X-ray crystallography. Crystals that provided a diffraction data set to 2.35 Å resolution were initially obtained in the presence of stachyose in the space group P212121. This structure was solved by molecular replacement using the deposited coordinates of RafE in complex with raffinose (PDB code 2I58) as a search model; the final refined model comprised two molecules in the asymmetric unit. One of these monomers was used as a search model to solve the structures of RafE in complex with raffinose (to 2.65 Å resolution) and verbascose (to 2.45 Å resolution). In all three cases the electron density for the carbohydrates was clear, allowing unambiguous modeling of the ligand (Fig. S1).
RafE adopts a fold characteristic of the Cluster D-I (oligosaccharide-specific) SBPs (33), which are composed of two α/β domains joined together by a hinge region (Fig. 2A). Within the two α/β domains is a β-sheet core comprised of four and five β-strands (N and C terminus, respectively) surrounded by α-helices. Like other SBPs, the binding pocket is a deep cavity located in between the two α/β domains (Fig. 2B). The binding pocket is open to solvent, completely exposing the reducing end residue of bound glycans and therefore is consistent with the capacity to bind ligands of various lengths.
Figure 2.
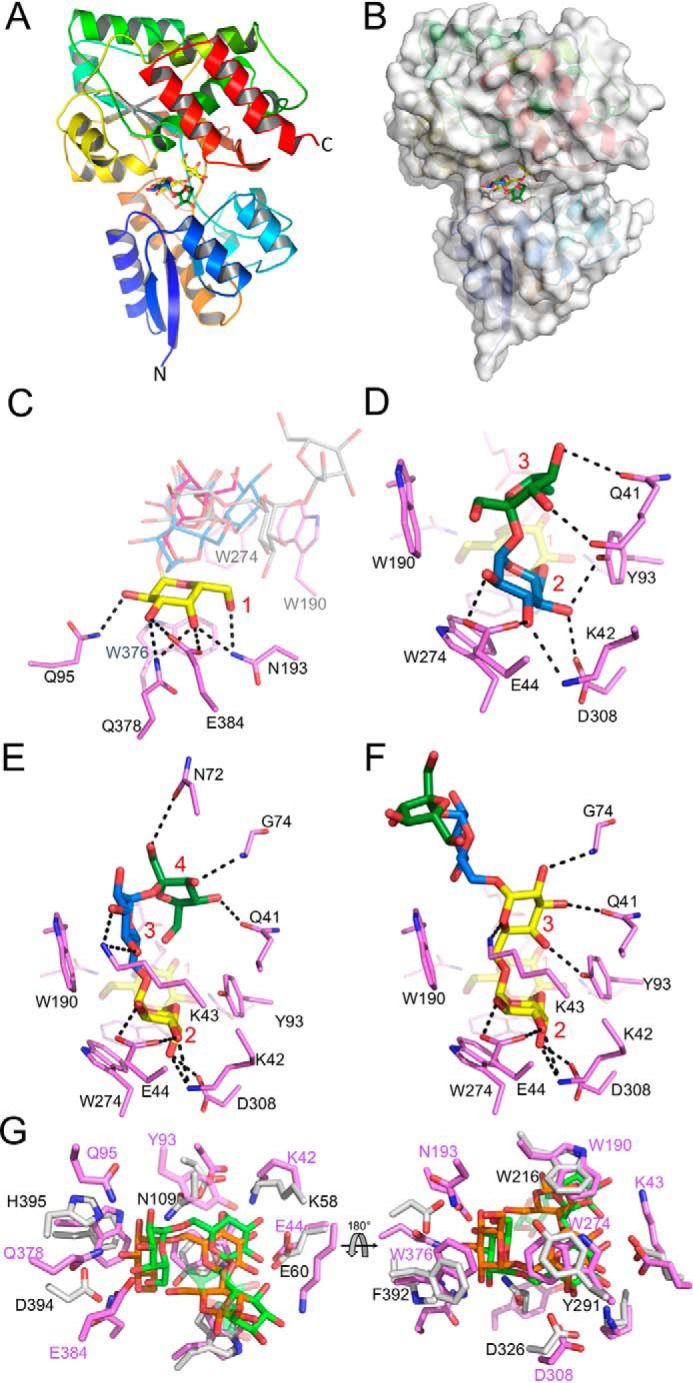
X-ray structure of RafE in complex with RFOs. A, global structure of RafE in complex with stachyose, showing secondary structure progression colored N to C termini. B, RafE surface representation of deep binding pocket accommodating the four sugar residues of stachyose. C, RafE RFO binding pocket overlay, displaying the highly conserved nonreducing end galactose residue(s) (yellow) direct hydrogen-bonding interactions (black dashes) with RafE side chains (magenta). The remaining residues of raffinose (red), stachyose (blue), and verbascose (gray) are shown as transparent lines. D–F, RafE binding pocket interactions with raffinose (D), stachyose (E), and verbascose (F), illustrating the binding pocket interactions between RafE side chains and fructose (green), glucose (blue), and galactose (yellow) residues. G, binding pocket overlay of RafE (magenta) bound to stachyose (green) and the B. animalis subsp. lactis SBP BlG16BP (light gray) bound to panose (orange) (PDB code 4ZZE). Residue numbering is shown in pink for RafE and black for BlG16BP. In C–F, the binding subsites are numbered in red.
An overlay of the three RafE complexes reveals a mode of binding whereby the nonreducing end galactose residues of the ligands are oriented toward the base of the binding pocket. In all three cases, the same set of interactions is made between this subsite, which we refer to as subsite 1, and the nonreducing end galactose (Fig. 2C). These comprise seven direct hydrogen bonds involving O2, O3, and O4 of the galactose residue. Trp376 provides a platform that interacts with the plane formed by O5-C1-C2. Although the axial O4 particular to galactose makes several hydrogen bonds with subsite 1, there appears to be nothing legislating against the presence of an equatorial O4, as in glucose, likely explaining the ability of this protein to also accommodate α-glucooligosaccharides as ligands.
Similar binding interactions are observed at subsite 2 with a mix of hydrophobic and polar interactions. When bound to stachyose or verbascose, subsite 2 of RafE contains a galactose residue; however, when bound to raffinose, this site is occupied by glucose (Fig. 2, D–F). Coordination is similar between both residues, with interactions between Trp274 and O5-C1-C2 plane of galactose/glucose. Direct hydrogen bonds are made between O4, O3, O2, and Asp308, Lys42, and Glu44, respectively.
In subsite 3, binding interactions are more diverse because of the presence of a fructose when bound to raffinose, a glucose when bound to stachyose, and a galactose when bound to verbascose. The binding interactions between the third residues of the RFOs (as well as the fourth residue of stachyose) and RafE are likely the key determinants underpinning the binding preferences of RafE. The galactose and fructose residues of verbascose and raffinose appear to have little or no van der Waal interactions with Trp190 in subsite 3, whereas the glucose residue of stachyose forms a close fitted C–H(π) interaction (Fig. 2E). Direct hydrogen bonds are limited for raffinose, with the main chain C=O and Oϵ of Gln41 being the only contributing atoms (to O3 and O4) (Fig. 2D). The third galactose residue of verbascose is flipped away from Trp190 and instead is coordinated by several hydrogen bonds between C4, C3, C2, and Tyr93, Gln41, and the N-H of Gly74 (Fig. 2F). The fourth and fifth residues of verbascose are not involved in any direct hydrogen bonding with RafE side chains. The glucose residue of stachyose only forms direct hydrogen bonding with Lys43, whereas its fructose residue makes several hydrogen bonds between O1, O3, O4, and Asn72, Gly74, and Gln41 (Fig. 2E).
A search for structural homologs of RafE using the Dali server (34) indicates that RafE shares the highest structural similarity (Z score 39.3; 23% sequence identity) with the raffinose- and panose-binding SBP BlG16BP from Bifidobacterium animalis subsp. lactis (PDB code 4ZZE) (35). Like RafE, BlG16BP exhibited the highest affinity for panose. A comparison of their structures reveals a conserved aromatic platform that complements the curvature of the ligands: Trp190, Trp274, and Trp376 in RafE, which are Trp216, Tyr291, and Phe392 in BlG16BP (Fig. 2G). Despite the similar specificity of the proteins, they have only ∼50% conservation of the residues involved in hydrogen bonding with none of the hydrogen bonding residues conserved in subsite 1, which accommodates the reducing end sugar (Fig. 2G).
The α-galactosidase activity of Aga
Given the membership of Aga in GH family 36 (GH36) and its previously postulated RFO activity, we anticipated the enzyme would have α-galactosidase activity (25). To examine this, we tested purified recombinant enzyme that was produced in E. coli against a panel of α- and β-configured synthetic substrates and found activity only on para-nitrophenyl-α-d-galactopyranoside (pNP-α-Gal; Fig. S2A). The pH optimum using pNP-α-Gal was between 6.0 and 6.4 (Fig. S2B), whereas the Km was 0.23 ± 0.02 mm and kcat was 1.8 ± 0.03 s−1 (at pH 6.5; Fig. S2C).
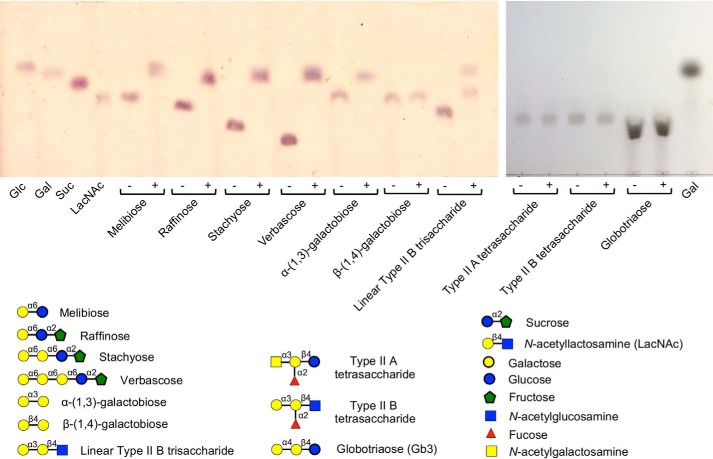
The range of α-galactoside substrates accepted by Aga was further screened using TLC (Fig. 3). Aga displayed clear activity against the RFOs raffinose, stachyose, and verbascose producing products with mobilities matching those of galactose and sucrose. Cleavage of melibiose produced products consistent with glucose and galactose. Aga also displayed activity on α-(1→3)-galactobiose but not β-(1→4)-galactobiose. Given the activity of Aga on α-(1→3)-galactobiose, we also tested glycans representing the major terminal α-linked galactose motifs present in mammals (36, 37). The type II blood group A and B tetrasaccharides, which terminate in α-(1→3)-linked GalNAc and galactose, respectively, and the Gb3 trisaccharide, which terminates in α-(1→4)-linked galactose, were not substrates for Aga (Fig. 3). However, the enzyme did have activity on the linear type II blood group B antigen, most likely because, in this case, the activity of the enzyme on the terminal α-(1→3)-linked Gal was not blocked by the presence of the fucosyl modification that defines the typical blood group B antigen.
Figure 3.
Activity screen of Aga against galactose-containing substrates. Substrates were incubated with Aga for 18 h prior to separation by TLC. Standards and substrates are indicated below each spot; ± indicates the presence/absence of Aga. Glc, glucose; Gal, galactose; Suc, sucrose; LacNAc, N-acetyllactosamine.
To better define the selectivity of the enzyme, we performed a kinetic analysis using raffinose, stachyose, melibiose, and α-(1→3)-galactobiose as substrates (Table 2). Aga displayed similar kcat/Km values for melibiose and α-(1→3)-galactobiose, which were ∼2-fold larger than the values obtained for raffinose and stachyose, thus suggesting the enzyme is better adapted to hydrolyze small substrates rather than RFOs.
Table 2.
Kinetic constants for Aga against α-galactoside substrates
The data shown are the means of three replicates.
| Substrate | Km ± S.E. | kcat ± S.E. | kcat/Km ± S.E. |
|---|---|---|---|
| mm | s−1 | s−1 mm−1 | |
| Melibiose Gal-α-(1→6)-Glc | 4.9 ± 0.4 | 162.0 ± 4.6 | 33.1 ± 2.9 |
| α-(1→3)-Galactobiose Gal-α-(1→3)-Gal | 2.2 ± 0.2 | 76.2 ± 2.3 | 34.6 ± 3.1 |
| Raffinose Gal-α-(1→6)-Glc-α-(1→2)-Fru | 5.8 ± 0.3 | 81.1 ± 1.3 | 14.0 ± 0.7 |
| Stachyose Gal-α-(1→6)-Gal-α-(1→6)-Glc-α-(1→2)-Fru | 11.0 ± 0.9 | 155.0 ± 4.3 | 14.1 ± 1.2 |
Substrate recognition by Aga
With the unexpectedly lower activity on RFOs, we attempted to pursue the molecular basis of this by X-ray crystallographic analysis of Aga structures. Crystals of Aga were initially obtained in the I222 space group and provided a diffraction data set to 2.10 Å resolution. The structure, comprising one molecule per asymmetric unit, was solved by molecular replacement using the structure of Geobacillus stearothermophillus GH36 α-galactosidase AgaA (PDB code 4FNU) (38) as the search model. Aga presents a three-domain fold characteristic of family GH36 members (Fig. 4A) (38, 39). The structure comprises a N-terminal domain composed of 20 β-strands forming a large twisted β-sandwich fold followed by an α-helix linking it to the catalytic domain. The latter domain is a classical (β/α)8-barrel fold harboring the catalytic machinery. The C-terminal domain corresponds to a small β-sandwich made of four antiparallel β-strands. The structure of Aga is highly similar to Lactobacillus acidophilus Mel36A and G. stearothermophilus AgaA and AgaB, with root-mean-square deviation values below 0.95 (over at least 542 residues). Also, Aga possesses all the sequence characteristics of GH36 subgroup I (39). Furthermore, most GH36 α-galactosidases belonging to subgroup I are known to form tetramers. This multimeric state is also observed in Aga through crystallographic symmetry (Fig. S3); this tetramer is predicted to be stable by PISA analysis (40).
Figure 4.
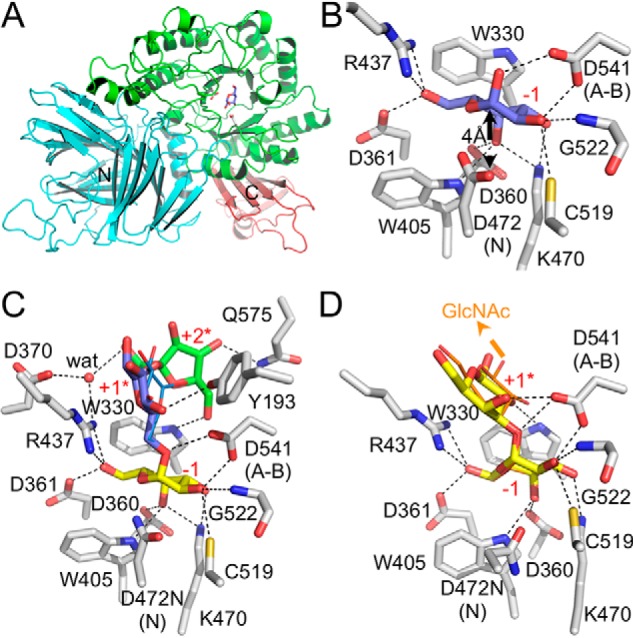
X-ray structure of Aga. A, cartoon representation of the overall fold of Aga in complex with its product galactose. Catalytic residues and galactose bound in the active site are shown as gray and blue sticks, respectively. The N and C termini are also indicated. B, Aga catalytic site in complex with galactose (represented as blue sticks). C, Aga D472N catalytic site in complex with raffinose (represented as sticks) and melibiose (represented as lines). D, Aga D472N catalytic site in complex with α-(1→3)-galactobiose (represented as sticks) and linear type II blood group B trisaccharide (represented as lines). The arrow pointing to GlcNAc indicates where GlcNAc should be located in the trisaccharide. Dashed lines denote hydrogen bonds. Residue numbering and subsites are indicated. N and A-B denote nucleophile and acid base catalytic residues, respectively. The water molecule (wat) is represented by a red sphere.
To gain further insight into the catalytic machinery, we solved the structure of Aga with its galactose product bound (Fig. 4B and Fig. S4A). The galactose product is accommodated within a −1 subsite in a manner identical to that described for other GH36s (38, 39). Specifically, the plane formed by C3 to C6 of the galactose ring packs against the indole group of Trp330. A series of hydrogen bonds are made between the sugar hydroxyl groups and Asp360, Asp361, Trp405, Arg437, Lys470, Cys519, Gly522, and Asp541 (also identified as the acid base catalytic residue) of the enzyme. Asp472, the second catalytic residue, sits 4 Å beneath the galactose C1 consistent with its role as a nucleophile.
To uncover the mode of substrate recognition by Aga, crystals of a catalytically inactive mutant of Aga, Aga D472N, were soaked with an excess (>50 mm) of melibiose or raffinose (Fig. 4C and Fig. S4, B and C), as well as with α-(1→3)-galactobiose or the linear blood group B type II trisaccharide (Fig. 4D and Fig. S4, D and E). The complexes of Aga D472N with unhydrolyzed substrates revealed that the galactose at the nonreducing end is bound via the same set of interactions as described above. In the raffinose complex, the glucose and fructose residues are positioned beyond the −1 subsite and make few interactions with Aga (Fig. 4C). Indeed, the glucose makes only one hydrogen bond between its O5 and Tyr193. The C6 and C4 hydroxyls of the fructose interact through direct hydrogen bonds with Trp330 and Gln575, respectively, whereas O1 makes a water-mediated hydrogen bond with Asp370 and Arg437. In the Aga D472N structure complexed with melibiose, the glucose aglycon is slightly shifted compared with the one in the raffinose complex and does not interact with any residue (Fig. 4C). Therefore, the glucose and fructose are located in what appear to be weak subsites, which could be described as pseudo +1 and +2 subsites (referred to as +1* and +2*). Attempts to trap a longer substrate, such as stachyose or verbascose, resulted in Aga structures displaying substrate density reduced solely to the galactose housed in the −1 subsite (data not shown). This agrees with the observation of weak or absent plus (+) subsites. The structure of Aga D472N in complex with α-(1→3)-galactobiose also reveals a +1* subsite where the galactose interacts through hydrogen bonding only with Asp541 and Trp330 (Fig. 4D). Interestingly, the galactose aglycon in the structure of Aga D472N with the linear blood group B type II trisaccharide possesses only a partial density (Fig. S3E), which still allowed modeling of the sugar in an orientation similar to the one observed for the galactobiose complex (Fig. 4D). However, the density for the linear blood group B type II trisaccharide GlcNAc is lacking, indicating disorder of this portion of the sugar (Fig. S3E). Furthermore, based on the latter two complexes, Aga would appear unable to accommodate the type II blood group B tetrasaccharide because the fucose α-(1→2) branched to the galactose at the +1* subsite would clash with Trp330 (data not shown), which explains the lack of activity toward such a substrate.
These structural data describe a catalytic machinery tuned toward the hydrolysis of disaccharides and potentially also trisaccharides. These findings converge toward the hypothesis that GH36s in a tetrameric organization are prone to accommodate only small unbranched carbohydrates such as di- and trisaccharides because of the narrowing of the catalytic pocket resulting from tetramerization (Fig. S3B) (39).
Complete depolymerization of RFOs
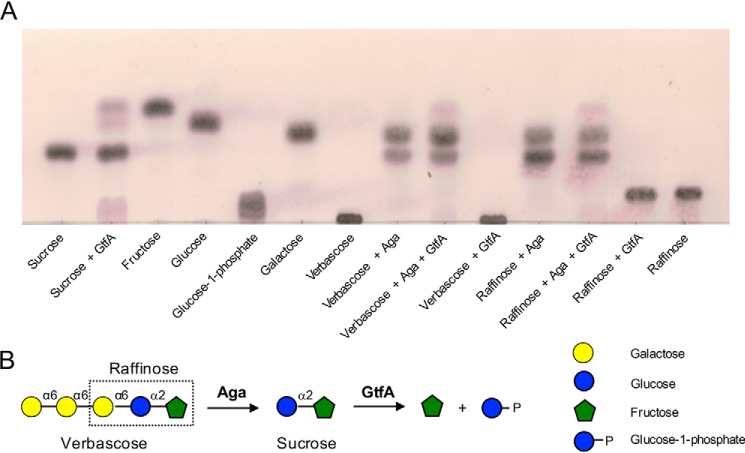
The degalactosylation of RFOs by Aga results in the production of sucrose. The raffinose utilization locus from S. pneumoniae encodes for a putative sucrose phosphorylase, GtfA (Fig. 1), that belongs to GH family 13 and that we hypothesized would act on RFOs after Aga to complete the degradation of RFOs into their constituent monosaccharides. To initially test GtfA for sucrose phosphorylase activity, recombinant GtfA was expressed and purified from E. coli. The enzyme failed to show activity on sucrose when reactions were performed in Tris-HCl buffer (not shown). However, GtfA exhibited partial activity against sucrose when reactions were performed in phosphate buffer, producing products with mobilities matching those of fructose, G1P, and glucose (Fig. 5). GtfA also exhibited activity against raffinose and verbascose, but only in the presence of Aga. Therefore, Aga and GtfA act sequentially to depolymerize RFOs into their constituent monosaccharides.
Figure 5.
The sequential depolymerization of RFOs by Aga and GtfA. A, the substrates sucrose, verbascose, and raffinose were incubated with Aga and/or GtfA for 18 h prior to separation by TLC. Standards and substrates are indicated below each spot. B, schematic depiction of the sequential breakdown of verbascose and raffinose by pneumococcal GHs. GHs are indicated in bold next to the arrow for the reaction they catalyze.
Discussion
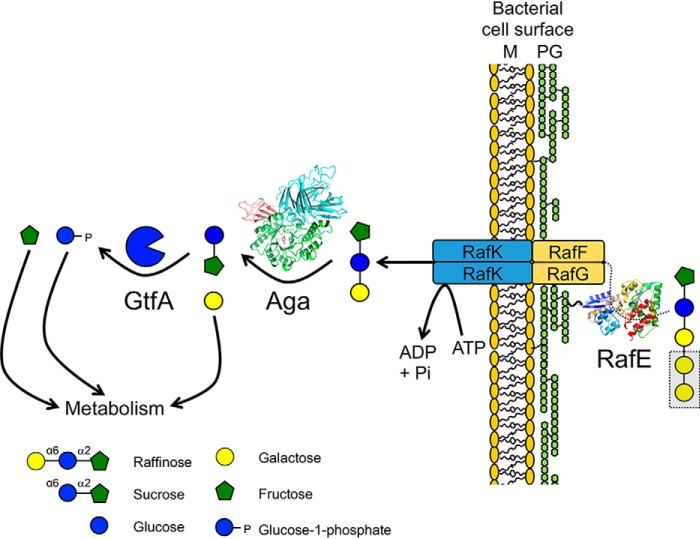
S. pneumoniae has the demonstrated ability to utilize RFOs for growth in vitro (14, 15, 28) and, as we have shown here biochemically, the pneumococcal genome encodes for the transporter and enzymes necessary to import RFOs and degrade them into their constituent monosaccharides (albeit with apparent inefficiency). Based on the data presented, we can assemble a model of a pathway that can be used by S. pneumoniae to utilize RFOs (Fig. 6). RafE binds RFOs outside of the cell and delivers them to the membrane components of the ABC transporter (RafF and RafG). The RFOs are then imported into the cytoplasm using energy provided by RafK. Once in the cytoplasm, Aga acts to sequentially degalactosylate the RFOs down to sucrose (we confirmed the intracellular localization of Aga in S. pneumoniae cells grown on raffinose; Fig. S5). Finally, the sucrose produced by Aga acts as a substrate for GtfA. The metabolism of free galactose in S. pneumoniae proceeds via either the Leloir or tagatose-6-phosphate pathway (41). The fructose released by GtfA is likely phosphorylated by a fructokinase, such as the previously characterized ScrK (16), prior to entering glycolysis. In addition to fructose, GtfA also generates G1P. G1P is interconverted to glucose-6-phosphate by a phosphoglucomutase (42), which can then enter either glycolysis or the pentose phosphate pathway (43). G1P itself is also an important intermediate in several pneumococcal anabolic pathways, including cell wall and capsule biosynthesis (42).
Figure 6.
Proposed pathway for RFO import and degradation in S. pneumoniae. The SBP RafE binds RFOs outside of the cell and delivers them to the membrane components of the ABC transporter (RafF and RafG). Powered by RafK (also known as MsmK), the RFOs are imported into the cytoplasm where they are sequentially degalactosylated by Aga. The released galactose can then enter metabolism either via the Leloir or tagatose-6-phosphate pathway (41). The remaining sucrose is cleaved by GtfA into fructose and G1P. Free fructose is likely phosphorylated by a fructokinase, such as ScrK (16), prior to entering glycolysis. G1P can be interconverted to glucose-6-phosphate by a phosphoglucomutase and enter either glycolysis or the pentose phosphate pathway (43). G1P is also an important intermediate in several pneumococcal anabolic pathways, including cell wall and capsule biosynthesis (42).
Although the data presented here indicate that S. pneumoniae possesses the ability to import and degrade RFOs, there are several lines of evidence to suggest that this capacity is unlikely to be biologically relevant to the normal lifestyle of this bacterium. First, RafE exhibits remarkably poor affinities for RFOs (Table 1). The Kd values determined for RafE with RFOs are ∼10-fold higher than that exhibited by BlG16BP with raffinose (35) and >100-fold higher than the Kd values that are typically reported for SBPs (4, 29–32). Therefore, RafE would need to encounter very high concentrations of RFOs for a significant proportion of this selectivity determinant of the ABC transporter to be engaged. Second, Aga exhibited relatively high Km values (>5 mm) and low kcat/Km values for RFOs (Table 2). In fact, the activity of Aga is not restricted to RFOs, and Aga exhibited a >2-fold lower Km and >2-fold higher kcat/Km against α-(1→3)-galactobiose compared with raffinose. Therefore, whereas RafE and Aga appear able to bind and degrade RFOs, the specific properties of these proteins suggest a pathway that would be quite inefficient. Finally, the question still remains as to how S. pneumoniae would encounter RFOs in its host given that they are plant oligosaccharides. The previous suggestion that small amounts of RFOs absorbed by the intestinal epithelium may be presented to S. pneumoniae on mucosal surfaces (28) is not congruent with the very low binding affinity of RafE.
These observations continue to highlight the unlikely relationship between RFO metabolism and pneumococcal virulence, which led to the suggestion that the locus targets alternate glycans. Indeed, Aga, which shows properties consistent with a strict α-galactosidase, demonstrates activity on a terminal α-linked galactose motif found in some mammalian glycans. However, we do not believe that Aga possesses activity on an in vivo substrate on the following basis. α-(1→6)-Linked galactose residues are not found on any human glycans, and although Aga also exhibits activity against α-(1→3)-linked galactose residues, it is inactive against the only known human glycan to bear this motif (the type II blood group B tetrasaccharide). Prior defucosylation of this tetrasaccharide into the linear form would allow Aga to act; however, the two characterized pneumococcal α-fucosidases, which are the only known fucosidases in this bacterium, lack this activity (44). Furthermore, we found Aga to have an intracellular location, which makes a potential role in host glycan processing even less likely. Thus, when interpreted in the wider context, our data do not support the alternate glycan substrate hypothesis.
Loci with similar genes and gene organization to the pneumococcal RFO locus are relatively common among the Lactobacillales (as determined by a STRING (45) gene neighborhood analysis using Aga as a search protein). Consistent with this, many other streptococcal species, including Streptococcus mutans, and lactic acid bacteria also have the ability to grow on raffinose (46–51). These bacteria reside in the oral cavity or gastrointestinal tract, and therefore, they would be expected to regularly encounter RFOs as part of the host diet. In the case of S. pneumoniae, however, a bacterium that is unlikely to encounter RFOs, we hypothesize that the RFO pathway may be an evolutionary relic that is not utilized and, as such, is not under strong selection pressure to maintain binding affinity and/or catalytic efficiency. Supporting this concept of divergence, the amino acid sequence conservation between components of the RFO pathway are as low as 55% within members of the Streptococcus genus. For example, RafE and Aga from the S. mutans locus share amino acid sequence identities of only 60 and 66%, respectively, with the pneumococcal homologs. However, it remains to be determined whether the biochemical efficiency of the RFO pathway components generally correlates with the niche the bacterium inhabits.
Despite the implications of our biochemical findings (as discussed above), there remains the enigmatic relationship between this locus and the tissue tropism and virulence of S. pneumoniae. In the recent study by Minhas et al. (28), isolates with a mutation in rafR were less able to persist in the murine lung than WT isolates but showed higher bacterial loads in the ear and brain. Isolates bearing a single amino acid substitution in rafK or a complete rafK deletion exhibited lower bacterial loads in all body sites. Given that RafK is a promiscuous ATPase known to power at least four different ABC transporters in S. pneumoniae (15, 17, 19) and that an effect of the reported rafR mutation was a significant reduction in rafK expression (28), we propose that the observed tissue tropism and virulence phenotype seemingly associated with the ability of S. pneumoniae to utilize raffinose may, in fact, be due to indirect effects arising from the lack or significantly reduced expression of RafK. RafK is known to be involved in the import of sialic acid, maltotetraose (derived from glycogen), FOSs, and RFOs by four different ABC transporters (15, 17, 19). It has also been proposed that RafK likely powers an additional two ABC transporters in S. pneumoniae, one of which we have shown imports N-glycans (4) (the other is uncharacterized). All of the characterized transporters have been associated with pneumococcal virulence and/or colonization in at least one animal model study (8, 9, 52–54). Furthermore, reduced bacterial loads in the lungs have been reported for ΔrafK pneumococci in two independent studies (19, 28). S. pneumoniae has the capacity to utilize more than 30 different carbohydrates, and these carbohydrates are likely differentially available at different human body sites (for example, gangliosides are most abundant in the brain (55)). Therefore, the essentiality of RafK for import of a subset of these carbohydrates has the potential to influence the tissue tropism of S. pneumoniae. Overall, the inefficiency of the RFO pathway suggests that it is not a key metabolic pathway in the pneumococcus, and previous virulence findings relating to this locus result from the more general context of carbohydrate uptake and the importance of this to the host–pneumococcus interaction.
Experimental procedures
Materials
Raffinose, stachyose, verbascose, and linear type II blood group B trisaccharide were purchased from Carbosynth Ltd. (Berkshire, UK). α-(1→3)-Galactobiose was from Dextra Laboratories Ltd. (Reading, UK). Type II blood group A and B tetrasaccharides and Gb3 were obtained from Elicityl (Crolles, France). All other materials were from Millipore–Sigma, unless otherwise stated.
Cloning and mutagenesis
The genes encoding for full-length Aga (locus tag SP_1898), RafE minus its secretion signal sequence and lipid-anchoring motif (amino acids 24–419; locus tag SP_1897), and full-length GtfA (locus tag SP_1894) were amplified by PCR from TIGR4 genomic DNA with the primers Aga-F and Aga-R, RafE-F and RafE-R, and GtfA-F and GtfA-R, respectively (Table S1). PCR products were cloned into pET28a between the NdeI and XhoI sites by In-Fusion® cloning (Takara Bio USA Inc., Mountain View, CA) to produce pET28a-Aga, pET28a-RafE, and pET28a-GtfA. Mutagenesis of pET28a-Aga to generate the Aga D472N mutation was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Mutagenic primers are listed in Table S1. The integrity of all constructs was confirmed by bidirectional sequencing (Sequetech, Mountain View, CA).
Protein expression and purification
Protein expression constructs were transformed into BL21(DE3). Expression of Aga, RafE, and GtfA was performed in LB broth with 0.5 mm isopropyl β-d-1-thiogalactopyranoside induction at 16 °C for 18 h. Standard procedures, as previously detailed (44), were used to lyse cells and purify the released proteins by immobilized metal affinity chromatography. Subsequent purification by size-exclusion chromatography, using either an S100 or S200 HiPrep 16/60 Sephacryl column (GE Healthcare) as appropriate, was performed using 20 mm Tris, pH 8.0, 500 mm NaCl. Protein purity was judged by SDS-PAGE analysis, and protein concentrations were determined using extinction coefficients calculated by ProtParam on the ExPASy server (56).
Isothermal titration calorimetry
ITC was performed as described previously (57) using a VP-ITC (MicroCal, Northampton, MA) in 50 mm potassium phosphate buffer, pH 6.6, at 25 °C. RafE was used at a concentration of 100 μm, and ligands at a concentration of 2.5 mm were titrated into protein. Ligand solutions were prepared using buffer saved from the last step of extensive dialysis of the protein solution. All solutions were filtered and degassed immediately before use. The data were fit to a one-site binding model. Because conditions were not sufficient to allow fitting of the stoichiometry (n), the n value was fixed at 1, which was justified based on the 1:1 interactions revealed by the crystallographic analyses of RafE.
α-Galactosidase assays
All Aga enzyme assays were performed at 37 °C. Aga was initially tested for activity against pNP-α-Gal and Y-β-Gal at 200 nm in 20 mm Tris, pH 8.0, with 1.5 mm substrate. Release of pNP was monitored at 405 nm in a SpectraMax M5 plate reader (Molecular Devices, San Jose, CA). The pH optimum of Aga (100 nm) was determined using 0.5 mm pNP-α-Gal in McIlvaine buffers, pH 2.4–8.0. Reactions in quadruplicate were incubated for 15 min, stopped by the addition of NaOH to 60 mm, and read at 405 nm. Kinetic constants for Aga (1 nm) against pNP-α-Gal were determined in Aga assay buffer (50 mm NaH2PO4/K2HPO4, pH 6.5) by following the release of pNP directly at 405 nm. The extinction coefficient of pNP in Aga assay buffer was experimentally determined to be 3707 m−1 cm−1. Aga was also screened for activity against a range of pNP substrates as described above for pNP-α-Gal and pNP-β-Gal. For all other substrates, kinetic constants were determined by quantifying the release of galactose in a stopped assay. Reactions (in triplicate) contained 40 nm Aga and varying concentrations of substrate in Aga assay buffer. Samples (30 μl) were taken every 3–5 min and stopped by heating to 95 °C for at least 10 min. Once cooled, 25 μl of each sample were mixed with components from the l-arabinose/d-galactose assay kit (Megazyme Inc., Chicago, IL), which contains a galactose mutarotase and a NAD+-dependent β-galactose dehydrogenase. Galactose detection reactions (100 μl) contained 0.5 μl of kit enzyme mix and 4 μl of NAD+ (kit supply) in kit buffer. Kit reactions were incubated at 25 °C and read at 340 nm every 10 s until the absorbance stabilized. Final absorbances were converted to galactose concentrations according to the manufacturer's instructions and accounting for the dilution factor of the original Aga reaction.
Thin-layer chromatography
TLC screening of Aga linkage specificity was performed with 1 mm substrate and 100 nm enzyme in Aga assay buffer at 37 °C for 18 h. Reactions and standards were spotted onto precoated POLYGRAM SIL G/UV254 TLC sheets (Thermo Fisher Scientific), separated in a solvent of 2:1:1 butanol:acetic acid:distilled H2O, and visualized with 0.2% (w/v) napthoresorcinol in acidified ethanol followed by heating at 90 °C. For GtfA-containing reactions, 10 mm substrate was digested with 30 μm GtfA and/or 100 nm Aga in 100 mm Aga assay buffer at 37 °C for 18 h. Spotted reactions and standards were separated in a solvent of 6:7:1 chloroform:acetic acid:distilled H2O, and visualized with acidified ethanol followed by heating at 90 °C.
General crystallography procedures
Crystals were obtained using sitting-drop vapor diffusion for screening and hanging-drop vapor diffusion for optimization at 18 °C. Prior to data collection, single crystals were flash-cooled with liquid nitrogen in crystallization solution supplemented with 20–25% (v/v) ethylene glycol as cryoprotectant. Diffraction data were collected on an “in-house” beam comprising a Pilatus 200K 2D detector coupled to a MicroMax-007HF X-ray generator with a VariMaxTM-HF Arc/Sec confocal optical system and an Oxford Cryostream 800. All diffraction data were processed using HKL2000 (58). Data collection and processing statistics are shown in Tables 3 and 4. All structures were solved by molecular replacement with PHASER (59). Initial models were built using BUCCANEER (60) followed by COOT (61). Refinement of atomic coordinates was performed with REFMAC (62). The addition of water molecules was performed in COOT with FINDWATERS and manually checked after refinement. In all data sets, refinement procedures were monitored by flagging 5% of all observations as “free” (63). Model validation was performed with MolProbity (64). Finally, the models obtained were represented using PyMOL (PyMOL Molecular Graphics System, version 1.6.0.0, Schrödinger, LLC).
Table 3.
X-ray data collection and structure statistics for RafE
The values for highest resolution shells are shown in parentheses. CIT, citric acid; EDO, ethylene glycol.
| RafE |
|||
|---|---|---|---|
| Raffinose | Stachyose | Verbascose | |
| Data collection | |||
| Beamline | Home Beam | Home Beam | Home Beam |
| Wavelength (Å) | 1.540 | 1.540 | 1.540 |
| Space group | P21 | P212121 | P21 |
| Cell dimensions (a, b, c) (Å) | 48.86, 145.82, 118.68 | 49.28, 118.84, 146.01 | 48.92, 145.88, 118.37 |
| Resolution (Å) | 30.00–2.65 (2.70–2.65) | 30.00–2.35 (2.39–2.35) | 30.00–2.40 (2.44–2.40) |
| Rmerge | 0.142 (0.565) | 0.084 (0.181) | 0.152 (0.530) |
| Rpim | 0.083 (0.369) | 0.036 (0.095) | 0.076 (0.344) |
| CC½ | 0.987 (0.729) | 0.995 (0.966) | 0.984 (0.706) |
| <I/σI> | 11.0 (2.0) | 20.2 (6.7) | 9.4 (2.2) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.8) | 99.6 (94.3) |
| Redundancy | 4.0 (3.6) | 5.0 (3.6) | 4.0 (3.1) |
| No. of reflections | 191,428 | 183,357 | 255,814 |
| No. unique | 47,919 | 36,605 | 64,353 |
| Refinement | |||
| Resolution (Å) | 2.65 | 2.35 | 2.40 |
| Rwork/Rfree | 0.24/0.29 | 0.22/0.24 | 0.23/0.26 |
| No. of atoms | |||
| Protein | 2968 (A), 2967 (B), 2972 (C), 2882 (D) | 3001 (A), 2986 (B) | 2991 (A), 2979 (B), 3000 (C), 2934 (D) |
| Ligand | 34 (RAFFINOSE-A), 34 (RAFFINOSE-B), 34 (RAFFINOSE-C), 34 (RAFFINOSE-D) | 45 (STACHYOSE-A), 45 (STACHYOSE-B), 26 (CIT), 16 (EDO) | 56 (VERBASCOSE-A), 56 (VERBASCOSE-B), 56 (VERBASCOSE-C), 56 (VERBASCOSE-D), 4 (EDO) |
| Water | 239 | 363 | 304 |
| B-factors | |||
| Protein | 36.4 (A), 40.0 (B), 39.3 (C), 49.0 (D) | 28.0 (A), 30.0 (B) | 32.8 (A), 36.6 (B), 34.9 (C), 47.5 (D) |
| Ligand | 31.3 (RAFFINOSE-A), 33.7 (RAFFINOSE-B), 37.3 (RAFFINOSE-C), 37.5 (RAFFINOSE-D) | 25.0 (STACHYOSE-A), 29.9 (STACHYOSE-B), 48.7 (CIT), 49.8 (EDO) | 33.4 (VERBASCOSE-A), 36.6 (VERBASCOSE-B), 34.6 (VERBASCOSE-C), 41.8 (VERBASCOSE-D), 32.1 (EDO) |
| Water | 31.3 | 32.2 | 31.4 |
| Root-mean-square deviation | |||
| Bond lengths (Å) | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.07 | 1.11 | 1.06 |
| Ramachandran (%) | |||
| Preferred | 96.9 | 97.8 | 96.7 |
| Allowed | 3.1 | 2.2 | 3.3 |
| Outliers | 0.0 | 0.0 | 0.0 |
Table 4.
X-ray data collection and structure statistics for Aga
The values for the highest resolution shells are shown in parentheses. EDO, ethylene glycol; TLA, l-tartaric acid; GLA, α-d-galactose; NA, sodium ion; TRS, Tris; MLB, melibiose; RAF, raffinose; ACT, acetate ion; Biose, α-(1→3)-galactobiose.
| Aga (apo) |
Aga |
Aga_D472N |
||||
|---|---|---|---|---|---|---|
| Native | Galactose | Melibiose | Raffinose | α-(1→3)-galactobiose | Linear type II blood group B trisaccharide | |
| Data Collection | ||||||
| Beamline | Home Beam | Home Beam | Home Beam | Home Beam | Home Beam | Home Beam |
| Wavelength (Å) | 1.540 | 1.540 | 1.540 | 1.540 | 1.540 | 1.540 |
| Space group | I222 | I222 | I222 | I222 | I222 | I222 |
| Cell dimensions (a, b, c) (Å) | 90.7, 127.2, 151.8 | 90.6, 127.3, 151.9 | 90.9, 127.2, 151.5 | 91.0, 127.5, 152.0 | 91.0, 127.2, 152.1 | 91.0, 127.3, 152.3 |
| Resolution (Å) | 30.00–2.20 (2.24–2.20) | 25.00–2.10 (2.14–2.10) | 25.00–2.20 (2.24–2.20) | 30.00–2.00 (2.03–2.00) | 25.00–2.00 (2.03–2.00) | 25.0–2.10 (2.14–2.10) |
| Rmerge | 0.152 (0.406) | 0.092 (0.316) | 0.134 (0.346) | 0.079 (0.289) | 0.072 (0.260) | 0.056 (0.093) |
| Rpim | 0.077 (0.244) | 0.042 (0.189) | 0.069 (0.184) | 0.036 (0.195) | 0.034 (0.186) | 0.026 (0.052) |
| CC½ | 0.986 (0.805) | 0.989 (0.901) | 0.991 (0.881) | 0.996 (0.926) | 0.997 (0.920) | 0.996 (0.991) |
| <I/σI> | 9.0 (2.1) | 14.8 (3.1) | 10.6 (3.5) | 17.0 (3.0) | 18.3 (3.1) | 24.0 (10.8) |
| Completeness (%) | 99.2 (98.1) | 97.9 (91.1) | 99.4 (99.8) | 99.0 (98.6) | 98.3 (87.2) | 98.7 (95.8) |
| Redundancy | 4.7 (3.7) | 3.5 (2.6) | 4.5 (4.2) | 4.6 (2.6) | 4.1 (2.2) | 4.3 (3.4) |
| No. of reflections | 209,955 | 178,337 | 199,826 | 273,621 | 240,313 | 219,887 |
| No. unique | 44,566 | 50,574 | 44,415 | 59,279 | 58,794 | 51,350 |
| Refinement | ||||||
| Resolution (Å) | 2.20 | 2.10 | 2.20 | 2.00 | 2.00 | 2.10 |
| Rwork/Rfree | 0.18/0.23 | 0.18/0.22 | 0.17/0.23 | 0.19/0.23 | 0.17/0.22 | 0.17/0.21 |
| No. of atoms | ||||||
| Protein | 5767 | 5760 | 5749 | 5762 | 5748 | 5773 |
| Ligand | 8 (EDO), 10 (TLA) | 28 (EDO), 12 (GLA), 1 (NA), 8 (TRS) | 23 (MLB), 20 (TLA), 36 (EDO) | 34 (RAF), 20 (TLA), 16 (EDO), 4 (ACT) | 23 (Biose_B), 20 (TLA) 23 (Biose_C), 24 (EDO) | 23 (Biose), 10 (TLA), 28 (EDO), 4 (ACT) |
| Water | 454 | 509 | 501 | 550 | 575 | 452 |
| B-factors | ||||||
| Protein | 34.9 | 27.3 | 33.3 | 28.6 | 26.9 | 26.1 |
| Ligand | 34.0 (EDO), 48.0 (TLA) | 36.3 (EDO), 26.4 (GLA), 24.3 (NA), 49.5 (TRS) | 36.6 (MLB), 50.1 (TLA), 50.1 (EDO) | 41.8 (RAF), 45.5 (TLA), 33.9 (EDO), 40.8 (ACT) | 31.3 (Biose_B), 42.4 (TLA) 47.9 (Biose_C), 40.9 (EDO) | 41.4 (Biose), 38.4 (TLA), 37.6 (EDO), 41.7 (ACT) |
| Water | 36.2 | 32.4 | 36.1 | 34.0 | 32.6 | 29.4 |
| Root-mean-square deviation | ||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.338 | 1.295 | 1.404 | 1.343 | 1.329 | 1.363 |
| Ramachandran (%) | ||||||
| Preferred | 95.1 | 96.0 | 95.6 | 96.1 | 96.2 | 95.7 |
| Allowed | 4.9 | 4.0 | 4.4 | 3.9 | 3.8 | 4.3 |
| Disallowed | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
RafE complex structure determinations
Cocrystals of RafE (28 mg ml−1) were obtained in 0.1 m Tris, pH 8.0, 0.2 mm CsCl, 20% (w/v) PEG 3350 with 10 mm raffinose, stachyose, or verbascose. The crystallization condition for the verbascose complex also contained 0.1 m MnCl2. The initial stachyose complex was solved by molecular replacement using an unpublished raffinose complex structure (PDB code 2I58) as the search model.
Aga Apo and complex structure determinations
Crystals of apo Aga (14 mg ml−1) were obtained in 0.1 m sodium acetate:acetic acid, pH 4.6, 1.1 m ammonium tartrate dibasic. The structure was solved by molecular replacement using the structure of G. stearothermophillus GH36 α-galactosidase AgaA (PDB code 4FNU) (38) as the search model. To obtain a galactose product complex, a crystal of Aga obtained in 1.0 m sodium phosphate monobasic/potassium phosphate dibasic, pH 5.6, was soaked with excess (>50 mm) raffinose for 45 min prior to cryoprotection. For all other complexes, crystals of Aga D472N (17 mg ml−1) obtained in 0.1 m sodium acetate:acetic acid, pH 4.6, 0.7–1.0 m ammonium tartrate dibasic were soaked with excess melibiose, raffinose, α-(1→3)-galactobiose, or linear type II blood group B trisaccharide for up to 30 min prior to cryoprotection.
Aga cellular localization
S. pneumoniae TIGR was grown in AGCH medium (65) containing 1% (w/v) raffinose at 37 °C in a candle jar to an A600 of 0.6, then fractionated as previously described (44). The presence of Aga in each cellular fraction was determined by adding 5 μl of fraction to 100 μl of 1 mm pNP-α-Gal in Aga assay buffer, incubating at 37 °C, and monitoring the absorbance at 405 nm in a SpectraMax M5 plate reader. Equal volumes of each fraction buffer were added to control wells containing substrate, and the data were subtracted from the corresponding test wells.
Author contributions
J. K. H. and A. B. B. conceptualization; J. K. H., E. P. W. M., B. P., M. A. M., and A. B. B. formal analysis; J. K. H., E. P. W. M., B. P., and M. A. M. investigation; J. K. H., E. P. W. M., B. P., and M. A. M. methodology; J. K. H., E. P. W. M., B. P., and A. B. B. writing-original draft; J. K. H., B. P., and A. B. B. writing-review and editing; A. B. B. supervision; A. B. B. funding acquisition; A. B. B. project administration.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Operating Grant PJT 159786. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S5.
N. G. Paterson et al., unpublished data.
- FOS
- fructooligosaccharide
- RFO
- raffinose family oligosaccharide
- GH
- glycoside hydrolase
- SBP
- solute-binding protein
- G1P
- glucose-1-phosphate
- ITC
- isothermal titration calorimetry
- pNP-α-Gal
- nitrophenyl-α-d-galactopyranoside
- PDB
- Protein Data Bank
- ABC
- ATP-binding cassette.
References
- 1. Siegel S. J., and Weiser J. N. (2015) Mechanisms of bacterial colonization of the respiratory tract. Annu. Rev. Microbiol. 69, 425–444 10.1146/annurev-micro-091014-104209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burnaugh A. M., Frantz L. J., and King S. J. (2008) Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190, 221–230 10.1128/JB.01251-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marion C., Limoli D. H., Bobulsky G. S., Abraham J. L., Burnaugh A. M., and King S. J. (2009) Identification of a pneumococcal glycosidase that modifies O-linked glycans. Infect. Immun. 77, 1389–1396 10.1128/IAI.01215-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robb M., Hobbs J. K., Woodiga S. A., Shapiro-Ward S., Suits M. D., McGregor N., Brumer H., Yesilkaya H., King S. J., and Boraston A. B. (2017) Molecular characterization of N-glycan degradation and transport in Streptococcus pneumoniae and its contribution to virulence. PLoS Pathog. 13, e1006090 10.1371/journal.ppat.1006090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hobbs J. K., Pluvinage B., and Boraston A. B. (2018) Glycan-metabolizing enzymes in microbe-host interactions: the Streptococcus pneumoniae paradigm. FEBS Lett. 592, 3865–3897 10.1002/1873-3468.13045 [DOI] [PubMed] [Google Scholar]
- 6. Henriques-Normark B., and Tuomanen E. I. (2013) The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 3, a010215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Obert C., Sublett J., Kaushal D., Hinojosa E., Barton T., Tuomanen E. I., and Orihuela C. J. (2006) Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect. Immun. 74, 4766–4777 10.1128/IAI.00316-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen H., Ma Y., Yang J., O'Brien C. J., Lee S. L., Mazurkiewicz J. E., Haataja S., Yan J.-H., Gao G. F., and Zhang J.-R. (2007) Genetic requirement for pneumococcal ear infection. PLoS One 3, e2950 10.1371/journal.pone.0002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hava D. L., and Camilli A. (2002) Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 10.1046/j.1365-2958.2002.03106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. King S. J. (2010) Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25, 15–24 10.1111/j.2041-1014.2009.00564.x [DOI] [PubMed] [Google Scholar]
- 11. Singh A. K., Pluvinage B., Higgins M. A., Dalia A. B., Woodiga S. A., Flynn M., Lloyd A. R., Weiser J. N., Stubbs K. A., Boraston A. B., and King S. J. (2014) Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA. PLoS Pathog. 10, e1004364 10.1371/journal.ppat.1004364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pluvinage B., Higgins M. A., Abbott D. W., Robb C., Dalia A. B., Deng L., Weiser J. N., Parsons T. B., Fairbanks A. J., Vocadlo D. J., and Boraston A. B. (2011) Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis. Structure 19, 1603–1614 10.1016/j.str.2011.08.011 [DOI] [PubMed] [Google Scholar]
- 13. Dalia A. B., Standish A. J., and Weiser J. N. (2010) Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78, 2108–2116 10.1128/IAI.01125-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bidossi A., Mulas L., Decorosi F., Colomba L., Ricci S., Pozzi G., Deutscher J., Viti C., and Oggioni M. R. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7, e33320 10.1371/journal.pone.0033320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marion C., Aten A. E., Woodiga S. A., and King S. J. (2011) Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect. Immun. 79, 4193–4200 10.1128/IAI.05290-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iyer R., and Camilli A. (2007) Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 66, 1–13 10.1111/j.1365-2958.2007.05878.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linke C. M., Woodiga S. A., Meyers D. J., Buckwalter C. M., Salhi H. E., and King S. J. (2013) The ABC transporter encoded at the pneumococcal fructooligosaccharide utilization locus determines the ability to utilize long- and short-chain fructooligosaccharides. J. Bacteriol. 195, 1031–1041 10.1128/JB.01560-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shafeeq S., Kloosterman T. G., and Kuipers O. P. (2011) CelR-mediated activation of the cellobiose-utilization gene cluster in Streptococcus pneumoniae. Microbiology 157, 2854–2861 10.1099/mic.0.051359-0 [DOI] [PubMed] [Google Scholar]
- 19. Tyx R. E., Roche-Hakansson H., and Hakansson A. P. (2011) Role of dihydrolipoamide dehydrogenase in regulation of raffinose transport in Streptococcus pneumoniae. J. Bacteriol. 193, 3512–3524 10.1128/JB.01410-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McKessar S. J., and Hakenbeck R. (2007) The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. J. Bacteriol. 189, 1342–1350 10.1128/JB.01170-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu W.-L., Jiang Y.-L., Pikis A., Cheng W., Bai X.-H., Ren Y.-M., Thompson J., Zhou C.-Z., and Chen Y. (2013) Structural insights into the substrate specificity of a 6-phospho-β-glucosidase BglA-2 from Streptococcus pneumoniae TIGR4. J. Biol. Chem. 288, 14949–14958 10.1074/jbc.M113.454751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Culurgioni S., Harris G., Singh A. K., King S. J., and Walsh M. A. (2017) Structural basis for regulation and specificity of fructooligosaccharide import in Streptococcus pneumoniae. Structure 25, 79–93 10.1016/j.str.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buckwalter C. M., and King S. J. (2012) Pneumococcal carbohydrate transport: food for thought. Trends Microbiol. 20, 517–522 10.1016/j.tim.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sengupta S., Mukherjee S., Basak P., and Majumder A. L. (2015) Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 6, 656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosenow C., Maniar M., and Trias J. (1999) Regulation of the alpha-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9, 1189–1197 10.1101/gr.9.12.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paterson N. G., Riboldi-Tunnicliffe A., Mitchell T. J., and Isaacs N. W. (2006) Purification, crystallization and preliminary X-ray diffraction analysis of RafE, a sugar-binding lipoprotein from Streptococcus pneumoniae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 676–679 10.1107/S1744309106021695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tettelin H., Nelson K. E., Paulsen I. T., Eisen J. A., Read T. D., Peterson S., Heidelberg J., DeBoy R. T., Haft D. H., Dodson R. J., Durkin A. S., Gwinn M., Kolonay J. F., Nelson W. C., Peterson J. D., et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 10.1126/science.1061217 [DOI] [PubMed] [Google Scholar]
- 28. Minhas V., Harvey R. M., McAllister L. J., Seemann T., Syme A. E., Baines S. L., Paton J. C., and Trappetti C. (2019) Capacity to utilize raffinose dictates pneumococcal disease phenotype. MBio. 10, e02596–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki R., Wada J., Katayama T., Fushinobu S., Wakagi T., Shoun H., Sugimoto H., Tanaka A., Kumagai H., Ashida H., Kitaoka M., and Yamamoto K. (2008) Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 283, 13165–13173 10.1074/jbc.M709777200 [DOI] [PubMed] [Google Scholar]
- 30. Abbott D. W., and Boraston A. B. (2007) Specific recognition of saturated and 4,5-unsaturated hexuronate sugars by a periplasmic binding protein involved in pectin catabolism. J. Mol. Biol. 369, 759–770 10.1016/j.jmb.2007.03.045 [DOI] [PubMed] [Google Scholar]
- 31. Quiocho F. A., Spurlino J. C., and Rodseth L. E. (1997) Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure 5, 997–1015 10.1016/S0969-2126(97)00253-0 [DOI] [PubMed] [Google Scholar]
- 32. Higgins M. A., Abbott D. W., Boulanger M. J., and Boraston A. B. (2009) Blood group antigen recognition by a solute-binding protein from a serotype 3 strain of Streptococcus pneumoniae. J. Mol. Biol. 388, 299–309 10.1016/j.jmb.2009.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berntsson R. P., Smits S. H., Schmitt L., Slotboom D.-J., and Poolman B. (2010) A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 10.1016/j.febslet.2010.04.043 [DOI] [PubMed] [Google Scholar]
- 34. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ejby M., Fredslund F., Andersen J. M., Vujičić Žagar, Henriksen J. R., Andersen T. L., Svensson B., Slotboom D. J., and Abou Hachem M. (2016) An ATP binding cassette transporter mediates the uptake of α-(1,6)-linked dietary oligosaccharides in Bifidobacterium and correlates with competitive growth on these substrates. J. Biol. Chem. 291, 20220–20231 10.1074/jbc.M116.746529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schnaar R. L., and Kinoshita T. (2017) Glycosphingolipids. In Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., and Seeberger P. H., eds) 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 37. Stanley P., and Cummings R. D. (2015) Structures common to different glycans. In Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L., and Seeberger P. H., eds) 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 38. Merceron R., Foucault M., Haser R., Mattes R., Watzlawick H., and Gouet P. (2012) The molecular mechanism of thermostable α-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies. J. Biol. Chem. 287, 39642–39652 10.1074/jbc.M112.394114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fredslund F., Hachem M. A., Larsen R. J., Sørensen P. G., Coutinho P. M., Lo Leggio L., and Svensson B. (2011) Crystal structure of α-galactosidase from Lactobacillus acidophilus NCFM: insight into tetramer formation and substrate binding. J. Mol. Biol. 412, 466–480 10.1016/j.jmb.2011.07.057 [DOI] [PubMed] [Google Scholar]
- 40. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 41. Paixão L., Oliveira J., Veríssimo A., Vinga S., Lourenço E. C., Ventura M. R., Kjos M., Veening J.-W., Fernandes V. E., Andrew P. W., Yesilkaya H., and Neves A. R. (2015) Host glycan sugar-specific pathways in Streptococcus pneumoniae: galactose as a key sugar in colonisation and infection. PLoS One 10, e0121042 10.1371/journal.pone.0121042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hardy G. G., Caimano M. J., and Yother J. (2000) Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182, 1854–1863 10.1128/JB.182.7.1854-1863.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Willenborg J., and Goethe R. (2016) Metabolic traits of pathogenic streptococci. FEBS Lett. 590, 3905–3919 10.1002/1873-3468.12317 [DOI] [PubMed] [Google Scholar]
- 44. Hobbs J. K., Pluvinage B., Robb M., Smith S. P., and Boraston A. B. (2019) Two complementary α-fucosidases from Streptococcus pneumoniae promote complete degradation of host-derived carbohydrate antigens. J. Biol. Chem. 294, 12670–12682 10.1074/jbc.RA119.009368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., and von Mering C. (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Russell R. R., Aduse-Opoku J., Sutcliffe I. C., Tao L., and Ferretti J. J. (1992) A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267, 4631–4637 [PubMed] [Google Scholar]
- 47. Barrangou R., Azcarate-Peril M. A., Duong T., Conners S. B., Kelly R. M., and Klaenhammer T. R. (2006) Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 103, 3816–3821 10.1073/pnas.0511287103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jarvis G. N., Kurtovic A., Hay A. G., and Russell J. B. (2001) The physiological and genetic diversity of bovine Streptococcus bovis strains. FEMS Microbiol. Ecol. 35, 49–56 10.1111/j.1574-6941.2001.tb00787.x [DOI] [PubMed] [Google Scholar]
- 49. Mao B., Tang H., Gu J., Li D., Cui S., Zhao J., Zhang H., and Chen W. (2018) In vitro fermentation of raffinose by the human gut bacteria. Food Funct. 9, 5824–5831 10.1039/C8FO01687A [DOI] [PubMed] [Google Scholar]
- 50. van den Bogert B., Erkus O., Boekhorst J., de Goffau M., Smid E. J., Zoetendal E. G., and Kleerebezem M. (2013) Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol. Ecol. 85, 376–388 10.1111/1574-6941.12127 [DOI] [PubMed] [Google Scholar]
- 51. Liébana J., Parejo E., Castillo A., and Gutiérrez J. (1993) Phenotypic characterization of oral streptococci by classical methods. Microbios. 76, 7–18 [PubMed] [Google Scholar]
- 52. Polissi A., Pontiggia A., Feger G., Altieri M., Mottl H., Ferrari L., and Simon D. (1998) Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marion C., Burnaugh A. M., Woodiga S. A., and King S. J. (2011) Sialic acid transport contributes to pneumococcal colonization. Infect. Immun. 79, 1262–1269 10.1128/IAI.00832-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Orihuela C. J., Radin J. N., Sublett J. E., Gao G., Kaushal D., and Tuomanen E. I. (2004) Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72, 5582–5596 10.1128/IAI.72.10.5582-5596.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu R. K., Tsai Y.-T., Ariga T., and Yanagisawa M. (2011) Structures, biosynthesis, and functions of gangliosides: an overview. J. Oleo Sci. 60, 537–544 10.5650/jos.60.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., and Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (Walker John M., ed) pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 57. van Bueren A. L., Morland C., Gilbert H. J., and Boraston A. B. (2005) Family 6 carbohydrate binding modules recognize the non-reducing end of β-1,3-linked glucans by presenting a unique ligand binding surface. J. Biol. Chem. 280, 530–537 10.1074/jbc.M410113200 [DOI] [PubMed] [Google Scholar]
- 58. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 59. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- 61. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 10.1038/355472a0 [DOI] [PubMed] [Google Scholar]
- 64. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lacks S. (1968) Genetic regulation of maltosaccharide utilization in Pneumococcus. Genetics 60, 685–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.