Abstract
Piezo1 is a mechanosensitive cation channel with widespread physiological importance; however, its role in the heart is poorly understood. Cardiac fibroblasts help preserve myocardial integrity and play a key role in regulating its repair and remodeling following stress or injury. Here we investigated Piezo1 expression and function in cultured human and mouse cardiac fibroblasts. RT-PCR experiments confirmed that Piezo1 mRNA in cardiac fibroblasts is expressed at levels similar to those in endothelial cells. The results of a Fura-2 intracellular Ca2+ assay validated Piezo1 as a functional ion channel that is activated by its agonist, Yoda1. Yoda1-induced Ca2+ entry was inhibited by Piezo1 blockers (gadolinium and ruthenium red) and was reduced proportionally by siRNA-mediated Piezo1 knockdown or in murine Piezo1+/− cells. Results from cell-attached patch clamp recordings on human cardiac fibroblasts established that they contain mechanically activated ion channels and that their pressure responses are reduced by Piezo1 knockdown. Investigation of Yoda1 effects on selected remodeling genes indicated that Piezo1 activation increases both mRNA levels and protein secretion of IL-6, a pro-hypertrophic and profibrotic cytokine, in a Piezo1-dependent manner. Moreover, Piezo1 knockdown reduced basal IL-6 expression from cells cultured on softer collagen-coated substrates. Multiplex kinase activity profiling combined with kinase inhibitor experiments and phosphospecific immunoblotting established that Piezo1 activation stimulates IL-6 secretion via the p38 mitogen-activated protein kinase downstream of Ca2+ entry. In summary, cardiac fibroblasts express mechanically activated Piezo1 channels coupled to secretion of the paracrine signaling molecule IL-6. Piezo1 may therefore be important in regulating cardiac remodeling.
Keywords: ion channel, calcium, p38 MAPK, mitogen-activated protein kinase (MAPK), IL-6, fibroblast, heart, mechanotransduction, patch clamp, signal transduction, cardiac fibroblast
Introduction
Cardiac fibroblasts play important roles in the normal physiology of the heart and in its response to damage or stress. Altered environmental stiffness and mechanical stretching of cardiac fibroblasts, which occur secondary to fibrosis, cardiac dilatation, or changes in the hemodynamic burden in the heart, induce differentiation of fibroblasts into myofibroblasts with increased expression of pro-fibrotic cytokines, extracellular matrix (ECM)5 proteins, and ECM receptors (1–3). This preserves cardiac integrity and performance, but the response can become maladaptive; surplus deposition of cardiac ECM results in fibrosis, which increases the stiffness of the myocardium and reduces pumping capacity (4).
Much remains unknown regarding how mechanical forces are translated into transcriptional responses important for determination of the fibroblast phenotype and how this can lead to cardiac remodeling (3, 5). Stretch-activated ion channels have been identified as candidates for cardiac mechanotransducers (6). Intracellular Ca2+ controls many functions in cardiac fibroblasts, including ECM synthesis and cell proliferation (7, 8), and it is known that Ca2+ entry through ion channels is important for cardiac fibroblast responsiveness. For example, TRPV4 channel–mediated Ca2+ signaling has been shown to play a role in mechanosensation and differentiation of fibroblasts into myofibroblasts (9).
In 2010, the Piezo1 and Piezo2 proteins were identified as the long-sought molecular carriers of an excitatory mechanically activated current found in many cell types (10). Piezo1, a nonselective cation channel, can be activated by a synthetic small molecule called Yoda1 (11). Yoda1 was discovered in a screen of ∼3.25 million compounds aiming to identify activators of Piezo1 or Piezo2 using high-throughput Ca2+ imaging (11). Importantly, Yoda1 activates Piezo1 but not Piezo2 and can do so in artificial lipid bilayers without the need for other cellular components (11). More recently, Yoda1 has been used to explore the gating mechanism of Piezo1 (12). Hence, Yoda1 is a useful tool for activating the Piezo1 channel without the need for mechanical stimulation. Piezo1 mRNA has been detected in the murine heart, whereas Piezo2 mRNA was barely detected (10). Piezo1 has been shown recently to be expressed in cardiomyocytes and up-regulated in heart failure (13). However, there are currently no reports regarding the role of Piezo1 in cardiac fibroblasts. We hypothesized that Piezo1 plays an important role in cardiac fibroblast function by regulating Ca2+ entry and downstream signaling.
Our data provide evidence that Piezo1 acts as a functional Ca2+-permeable mechanosensitive ion channel in murine and human cardiac fibroblasts and that its activation by Yoda1 is coupled to secretion of IL-6, a cytokine important in the response to cardiac injury and hypertrophic remodeling. We further reveal that Piezo1-induced Ca2+ entry is coupled to IL-6 expression via activation of p38 MAPK.
Results
Piezo1 expression and activity in cardiac fibroblasts
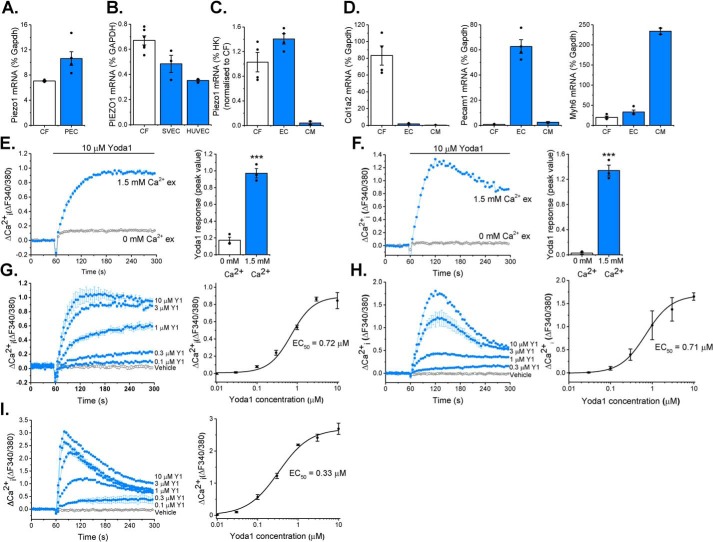
mRNA encoding Piezo1 was detected in cultured cardiac fibroblasts from both mouse and human hearts (Fig. 1, A and B). In comparison with endothelial cells, which are known to express high levels of this channel (14), Piezo1 mRNA expression levels in murine cardiac fibroblasts were similar to those observed in murine pulmonary endothelial cells, and those in human cardiac fibroblasts were similar to those in human saphenous vein endothelial cells and human umbilical vein endothelial cells (HUVECs) (Fig. 1, A and B). Using a magnetic antibody cell separation (MACS) technique (15), we confirmed that Piezo1 was expressed in the fibroblast-enriched (Col1a2-positive) fraction of freshly isolated cells from mouse heart at a level similar to that in the endothelial cell-enriched (Pecam-1-positive) fraction (Fig. 1, C and D). Piezo1 mRNA levels were 20 times higher in isolated cardiac fibroblasts than Myh6-positive cardiomyocytes when normalized to the average of three separate housekeeping genes (Fig. 1, C and D).
Figure 1.
Piezo1 is expressed by cardiac fibroblasts and forms a functional ion channel. A and B, RT-PCR analysis of Piezo1 mRNA expression in murine cardiac fibroblasts (CF, n = 3) compared with murine pulmonary endothelial cells (PEC, n = 5) (A) and human CF (n = 6) compared with human saphenous vein endothelial cells (SVEC, n = 3) and HUVECs (n = 3) (B). Expression was measured as percent of the housekeeping control (Gapdh/GAPDH). C, RT-PCR analysis of Piezo1 mRNA expression in fibroblast-enriched fraction 2 (CF) and endothelial cell–enriched fraction 1 (EC) isolated from murine heart using the MACS technique (n = 4). Cardiomyocytes (CM) were isolated from separate hearts (n = 2). Expression was measured relative to three housekeeping genes (Gapdh, Actb, and Hprt) and normalized to the CF sample. D, RT-PCR analysis of cell type-specific marker genes for CF (Col1a2), endothelial cells (Pecam1), and cardiomyocytes (Myh6) in MACS fractions as for C. E and F, representative Ca2+ traces and mean ± S.E. are shown. Ca2+ entry was evoked by 10 μm Yoda1 in murine (E) and human (F) cardiac fibroblasts in the presence or absence of extracellular Ca2+. ***, p < 0.001 (paired t test; n/N = 3/9). G–I, Ca2+ entry evoked by varying concentrations of Yoda1 application at 60 s, ranging from 0.1–10 μm in murine (G) and human (H) cardiac fibroblasts and HEK T-Rex-293 cells heterologously expressing mouse Piezo1 (I). Vehicle control is illustrated by the black trace. Mean ± S.E. are displayed as concentration–response curves, and fitted curves are plotted using a Hill equation, indicating the EC50 of Yoda1 (n/N = 3/9).
Having demonstrated that cardiac fibroblasts express Piezo1 mRNA, we investigated whether the Piezo1 protein was able to form a functional ion channel. Using the Fura-2 Ca2+ indicator assay, it was found that Yoda1, a Piezo1 agonist (11), elicited an increase in intracellular Ca2+ in murine and human cardiac fibroblasts (Fig. 1, E and F). Consistent with the Yoda1-induced increase in intracellular Ca2+ being due to influx of extracellular Ca2+ through an ion channel, the Ca2+ signal was reduced by more than 90% when extracellular Ca2+ was absent in human and mouse cardiac fibroblast cultures (Fig. 1, E and F).
Concentration–response data for Yoda1 in murine cardiac fibroblasts revealed a marked effect at 0.3 μm, and the maximal response was generated at 10 μm; the EC50 of Yoda1 was estimated to be 0.72 μm (Fig. 1G). This was almost identical to the EC50 observed in human cardiac fibroblasts in similar experiments (Fig. 1H) and comparable with that for mouse Piezo1 heterologously expressed in HEK T-RExTM-293 cells, where the EC50 of Yoda1 was 0.33 μm (Fig. 1I).
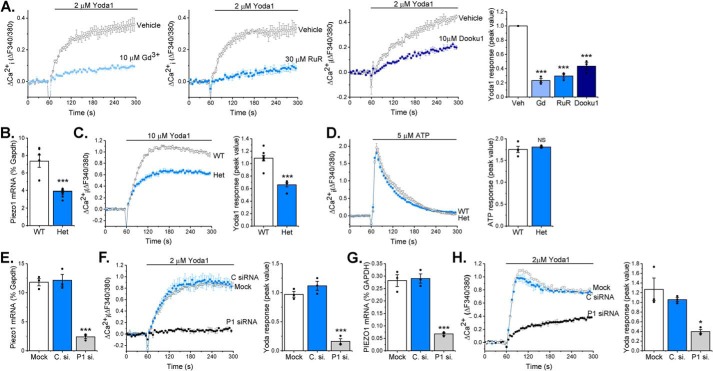
Gadolinium (Gd3+) and ruthenium red, both nonspecific inhibitors of mechanosensitive ion channels, including Piezo1 (10), were used to investigate the pharmacology of the channel. Murine cardiac fibroblasts preincubated with these inhibitors exhibited significantly reduced Yoda1-evoked Ca2+ entry (>70% reduction in both cases) (Fig. 2A). Additionally, Yoda1-evoked Ca2+ entry was significantly reduced by more than 50% by Dooku1 (Fig. 2A), an analog of Yoda1 that has antagonist properties against Yoda1 in Piezo1-overexpressing HEK293 cells and HUVECs (16). The data were similar in human cardiac fibroblasts, where again all three inhibitors significantly reduced Ca2+ influx in response to Yoda1 (Fig. S1). Ca2+ entry elicited by Yoda1 application was also investigated in cardiac fibroblasts isolated from a global heterozygous (Het) Piezo1+/− mouse line. RT-PCR analysis confirmed the predicted 50% reduction in Piezo1 mRNA expression in cardiac fibroblasts derived from Piezo1+/− hearts compared with the WT (Fig. 2B). The Ca2+ response to Yoda1 was reduced by 40% (Fig. 2C), whereas the response to ATP was similar in cells from WT and Piezo1+/− mice (Fig. 2D). Piezo1-specific siRNA, which decreased Piezo1 mRNA expression by 80% in murine cardiac fibroblasts (Fig. 2E), reduced Yoda1-evoked Ca2+ entry by a similar level (Fig. 2F), whereas control siRNA was without effect. Similar results were obtained with human cardiac fibroblasts (Fig. 2, G and H). Thus, the Yoda1 response depended on Piezo1 and was proportional to its expression level. Together, these data establish that Yoda1-induced Ca2+ entry in cardiac fibroblasts depends on Piezo1 expression and that the channel has the expected pharmacological properties.
Figure 2.
Yoda1-evoked Ca2+ entry depends on Piezo1 expression. A, representative intracellular Ca2+ traces and mean data after murine cardiac fibroblasts were exposed to 10 μm Gd3+, 30 μm ruthenium red (RuR), 10 μm Dooku1, or vehicle (Veh) for 30 min before activation of Piezo1 by application of 2 μm Yoda1. Data were normalized to vehicle-treated cells. Repeated measures one-way ANOVA: p < 0.0001, F = 114.1 (n/N = 3/9). Post hoc test: ***, p < 0.001 versus vehicle-treated cells. B, RT-PCR analysis of Piezo1 mRNA expression in cultured murine cardiac fibroblasts isolated from WT and Piezo1+/− (Het) mice. Expression is measured as percent of the housekeeping control Gapdh. ***, p < 0.001 (unpaired t test, n = 8). C, representative Ca2+ trace and mean data illustrating Ca2+ entry elicited by 10 μm Yoda in cardiac fibroblasts from WT (n/N = 8/24) and Piezo1+/− Het (n/N = 5/15) mice. ***, p < 0.001 (unpaired t test). D, representative Ca2+ trace and mean data illustrating Ca2+ entry evoked by 5 μm ATP in cardiac fibroblasts from WT (n/N = 4/12) and Piezo1+/− (Het, n/N = 3/9) mice. Unpaired t test: not significant (NS). E, RT-PCR analysis of Piezo1 mRNA expression following transfection of murine cardiac fibroblasts with Piezo1 siRNA, mock-transfected cells, and cells transfected with control siRNA. Expression is measured as percent of the housekeeping control Gapdh. Repeated measures one-way ANOVA: p = 0.0001, F = 61.1 (n = 3). Post hoc test: ***, p < 0.001 versus mock-transfected cells. F, representative Ca2+ trace and mean data showing the response to 2 μm Yoda1 in murine cardiac fibroblasts transfected with Piezo1-specific siRNA (P1 si.) compared with mock-transfected cells and cells transfected with control siRNA (C. si.). Repeated measures one-way ANOVA: p < 0.0001, F = 72.6 (n/N = 3/9). Post hoc test: ***, p < 0.001 versus mock-transfected cells. G, as for E but in human cardiac fibroblasts. Repeated measures one-way ANOVA: p = 0.0002, F = 50.8 (n/N = 3/9). Post hoc test: ***, p < 0.001 versus mock-transfected cells. H, as for F but in human cardiac fibroblasts. Repeated measures one-way ANOVA: p = 0.0108, F = 10.6 (n/N = 3/9). Post hoc test: *, p < 0.05 versus mock-transfected cells.
Cardiac fibroblasts contain mechanically activated currents
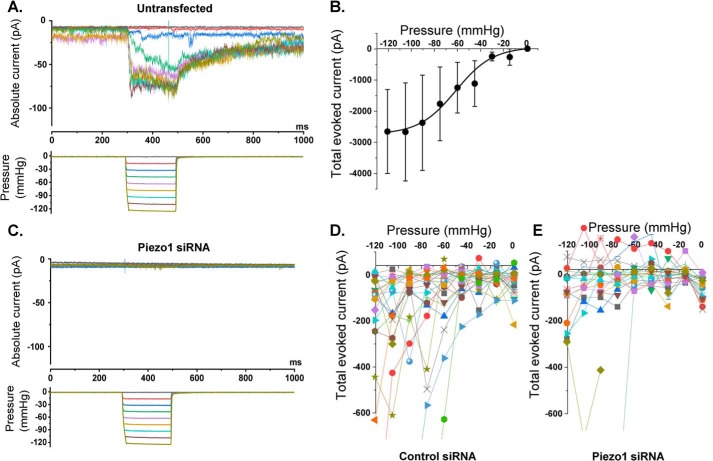
To investigate whether cardiac fibroblasts contain mechanically activated ion channels, we made cell-attached patch recordings from human cardiac fibroblasts. Mechanical force was applied to the patches using a fast pressure clamp system that generated calibrated suction pulses (pressure pulses) in the patch pipette and therefore increased membrane tension (Fig. 3A). Increased pressure caused increased inward currents up to a limit (Fig. 3, A and B). The currents were noisy macroscopic currents, suggesting the presence of multiple individual channels that summed to generate the overall current. The amplitude of the current was highly variable between patches, as seen by the large standard error in Fig. 3B. Despite this variability, we estimated that the pressure required for 50% activation was −61.3 mm Hg (Fig. 3B).
Figure 3.
Cardiac fibroblasts express mechanically activated ionic currents that depend on Piezo1. Recordings were made from cell-attached patches on human cardiac fibroblasts. A constant holding voltage of +80 mV was applied to the patch pipette to ensure a negative membrane potential across the patch (inside relative to outside). A rapid pressure clamp system applied 200-ms negative pressure (suction) steps of increasing magnitude to the patch pipette. A, example data from an untransfected cell. The color code of the pressure steps (bottom panel) matches the color code of the current traces in the top panel. B, mean ± S.E. data for experiments of the type shown in A, in which the total current during 180 ms of each pressure pulse was summed for n = 7–8 patches/data point. The fitted curve is the Boltzmann function, which gave a midpoint for 50% activation of −61.3 mm Hg. C, as for A but from a cell that was transfected with Piezo1 siRNA. D and E, each color shows the individual data for all recordings from cells transfected with control siRNA (D, n = 24 recordings) or Piezo1 siRNA (E, n = 21 recordings). As for B, the total current during 180 ms of each pressure pulse was summed. Two-way ANOVA: p = 0.0475, F = 3.95 (siRNA); p < 0.0001, F = 4.15 (pressure); p < 0.0001, F = 4.16 (interaction). In D and E, data values exceeding approximately −600 pA are clipped to maximize visibility of the majority of the data, but all data values were included in the statistical analysis. The two large current values in the Piezo1 siRNA group may have been from nontransfected cells because the transfection efficiency was estimated to be 90%.
The mechanically activated channel activity is Piezo1-dependent
To test whether the currents were Piezo1-dependent, we systematically compared two matched groups of human cardiac fibroblasts: one transfected with control siRNA and the other transfected with Piezo1 siRNA. Because the transfection efficiency was estimated to be 90% at best, we expected that at least one in every 10 recordings would be from a nontransfected cell. To compensate for this limitation, we made a large number of individual recordings before analyzing the data (45 in total). Our overall impression was that most Piezo1 siRNA–transfected cells showed no meaningful current in response to pressure steps (Fig. 3C). Data for all 45 recordings are shown in Fig. 3, D and E. The high variability was again observed, but it was also visually apparent that pressure-activated currents were more common in the control siRNA group than in the Piezo1 siRNA group (compare Fig. 3, D and E). In only two patches of the Piezo1 siRNA group were clear pressure-activated currents observed (Fig. 3E). We suspected that these latter two recordings were from nontransfected cells (assuming 90% transfection efficiency). Formal analysis using two-way ANOVA on all data in the two groups for all pressure steps (no data were excluded) indicated that currents in the Piezo1 siRNA group were statistically significantly different (smaller) than those in the control siRNA group. These data suggest that human cardiac fibroblasts express mechanically activated Piezo1 channels.
Yoda1-induced Piezo1 activation is coupled to increased Il6 gene expression
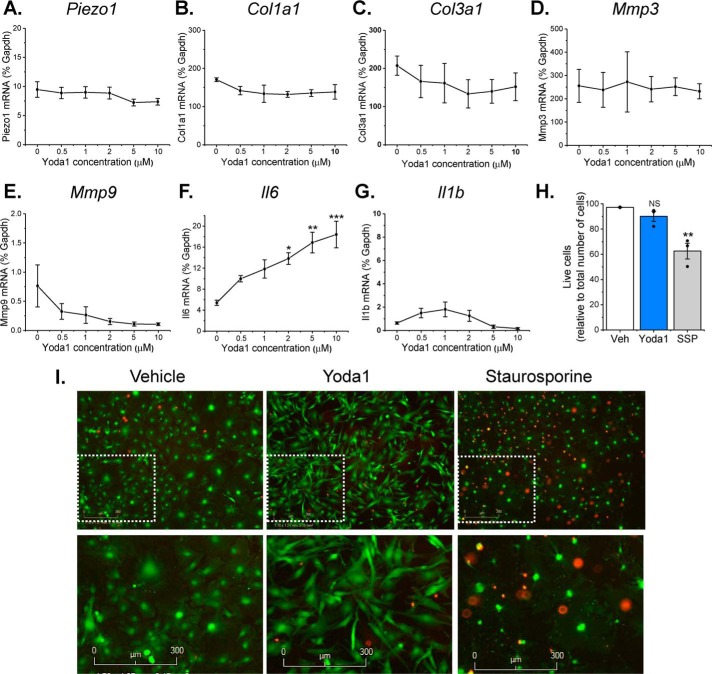
To gain insight into the functional role of Piezo1 activation in cardiac fibroblasts, the effect of 24-h treatment with 0.5–10 μm Yoda1 on the expression of selected remodeling genes in murine fibroblasts was investigated by RT-PCR (Fig. 4, A–G). Prolonged Yoda1 treatment did not modulate Piezo1 gene expression (Fig. 4A), nor did it affect the expression of genes involved in ECM turnover, including collagens I and III (Col1a1, Col3a1) or matrix metalloproteinase 3 and 9 (Mmp3, Mmp9) (Fig. 4, B–E). However, a concentration-dependent increase in mRNA expression of the inflammatory/hypertrophic cytokine IL-6 was observed, with significant 2- to 4-fold increases observed in response to 2–10 μm Yoda1 (Fig. 4F). This was not a generic inflammatory response because Il1b mRNA levels remained unaffected by Yoda1 treatment (Fig. 4G). Prolonged exposure to Yoda1 did not affect cell viability (Fig. 4, H and I), although it did appear to induce a morphological change in the cells toward a more spindle-like, less rhomboid shape (Fig. 4I).
Figure 4.
Effect of Yoda1 on gene expression and cell survival in cardiac fibroblasts. A–G, RT-PCR analysis of murine cardiac fibroblasts treated with concentrations of Yoda1 ranging from 0.5–10 μm or DMSO vehicle for 24 h (n = 3). All mRNA expression levels were normalized to those of the housekeeping gene Gapdh. Repeated measures one-way ANOVA: p = 0.0010, F = 8.9. Post hoc test: *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus vehicle-treated cells. H, live/dead cell assay performed on cultured murine cardiac fibroblasts treated with either vehicle (Veh), 10 μm Yoda1, or 1 μm staurosporine (SSP) for 24 h. The bar chart shows mean data for viable cells as a percentage of total cells. Repeated measures one-way ANOVA: p = 0.0028, F = 10.6 (n = 3). Post hoc test: **, p < 0.01; NS, not significant versus vehicle-treated cells. I, representative images from the live/dead cell assay. Green indicates live cells; red indicates dead cells. Bottom panels are magnified images of the indicated regions in the top panels.
Piezo1 activation stimulates IL-6 expression and secretion via p38 MAP kinase
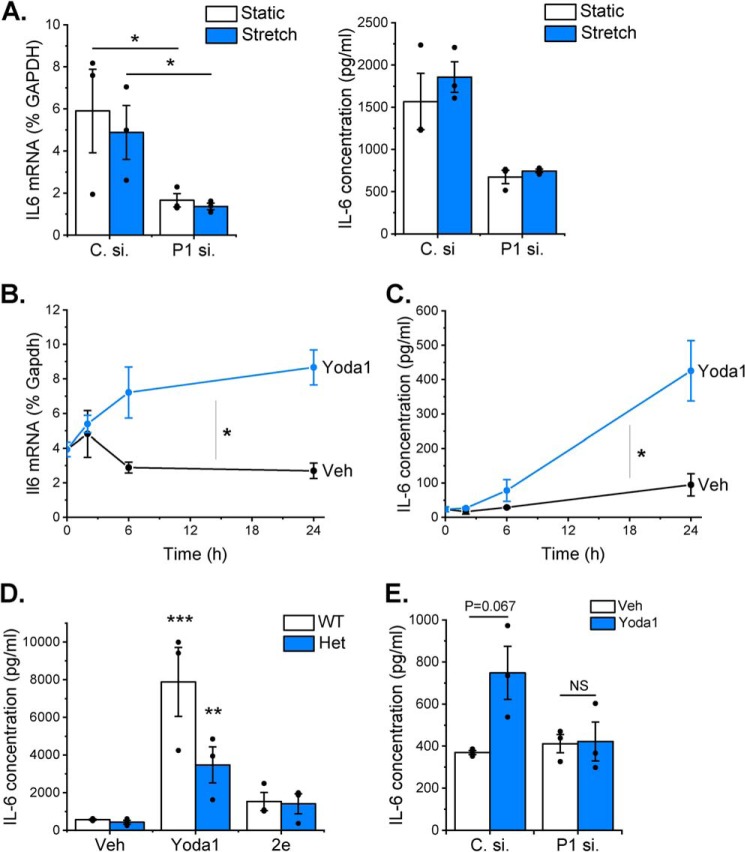
Given our recent findings regarding the potential importance of cardiac fibroblast-derived IL-6 in modulating cardiac hypertrophy (15) and the causative link between mechanical stimulation and cardiac hypertrophy (17), we proceeded to interrogate the mechanism by which Piezo1 activation was coupled to IL-6 expression. First, we explored whether mechanical activation of cardiac fibroblasts was coupled to IL-6 expression and whether this occurred via a Piezo1-dependent mechanism. Cyclic stretching (1 Hz, 10% stretch) of human cardiac fibroblasts for 24 h had no effect on IL6 mRNA levels or IL-6 protein secretion (Fig. 5A). However, during these experiments, it became apparent that PIEZO1 gene silencing significantly reduced basal IL-6 expression at both mRNA and protein levels (independently of stretch) when cells were grown on BioFlex plates (Fig. 5A) but not when they were maintained on regular tissue culture plastic (Fig. 5E). The BioFlex plates have a stiffness (Young's modulus) ∼1000 times less than standard tissue culture plates and were coated with type I collagen. Thus, activation of Piezo1 by chemical activation (Yoda1) or in response to altered substrate composition/stiffness was coupled to increased IL-6 expression in cardiac fibroblasts.
Figure 5.
Activation of Piezo1 is coupled to IL-6 expression. A, RT-PCR analysis of IL6 mRNA expression (left panel) and ELISA analysis of IL-6 protein secretion (right panel) after exposure of human cardiac fibroblasts to 24-h cyclical stretch (1 Hz, 10% stretch) on collagen-coated BioFlex plates compared with fibroblasts maintained in parallel under static conditions. Cardiac fibroblasts were transfected previously with either control siRNA (C. si.) or Piezo1-specific siRNA (P1 si.). RT-PCR data are expressed as percent of the housekeeping control GAPDH. Repeated measures two-way ANOVA for RT-PCR (n = 3): p = 0.0704, F = 12.7 (siRNA); p = 0.403, F = 1.1 (stretch); p = 0.754, F = 0.13 (interaction). Post hoc test: *, p < 0.05. Repeated measures two-way ANOVA for ELISA (n = 3): p = 0.0268, F = 35.8 (siRNA); p = 0.1545, F = 5.0 (stretch); p = 0.838, F = 0.05 (interaction). Post hoc test: not statistically significant. B and C, murine cardiac fibroblasts were exposed to vehicle (Veh) or 10 μm Yoda1 for 2–24 h before measuring Il6 mRNA levels by RT-PCR, where expression is measured as percent of the housekeeping control Gapdh (B) or analyzing conditioned medium for IL-6 levels by ELISA (C) (both n = 3). Comparison of the area under the curve by paired t test: *, p < 0.05 for the effect of Yoda1 on IL-6 expression at both mRNA and protein levels. D, cardiac fibroblasts from WT (n = 4) and Piezo1+/− (Het, n = 3) mice were treated with vehicle, 10 μm Yoda1, or compound 2e for 24 h before measuring IL-6 levels in conditioned medium using ELISA. Two-way ANOVA: p < 0.0001, F = 24.9 (compound); p = 0.1041, F = 3.1 (genotype); p = 0.615, F = 0.51 (interaction). Post hoc test: ***, p < 0.001; **, p < 0.01 for the effect of Yoda1 versus vehicle. All other changes were not statistically significant. E, mouse cardiac fibroblasts were transfected with control or Piezo1-specific siRNA and treated with either vehicle or 10 μm Yoda1 for 24 h before collecting conditioned media and measuring IL-6 levels by ELISA (n = 3). Repeated measures two-way ANOVA: p = 0.2349, F = 2.82 (siRNA); p = 0.2187, F = 3.13 (Yoda1); p = 0.064, F = 14.2 (interaction). Post hoc test: NS, not significant versus vehicle-treated cells.
We then probed the mechanism further in murine cardiac fibroblasts using Yoda1 as a stimulus. Il6 mRNA expression increased in a time-dependent manner over a 2- to 24-h period of Yoda1 treatment (Fig. 5B). This increase correlated with an increase in IL-6 protein secretion (Fig. 5C). Yoda1-induced IL-6 secretion was reduced by ∼50% in cardiac fibroblasts isolated from hearts of Piezo1+/− mice compared with those from WT hearts (Fig. 5D), in keeping with the reduction in Piezo1 expression and Ca2+ response in these cells (Fig. 2, B and C). IL-6 secretion was unchanged following treatment of cardiac fibroblasts with compound 2e (Fig. 5D), an inactive analogue of Yoda1 (16) that did not induce Ca2+ entry in murine or human cardiac fibroblasts or in HEK T-REx-293 cells heterologously expressing mouse Piezo1 (Fig. S2, A–C). The Yoda1-induced increase in IL-6 secretion depended on Piezo1, as evidenced by attenuation of the secretion in murine cardiac fibroblasts transfected with Piezo1-specific siRNA (Fig. 5E). The same was true in human cardiac fibroblasts, where the Yoda1-evoked increase in Il6 mRNA expression was attenuated by Piezo1-specific siRNA, and again the inactive Yoda1 analog 2e had no effect (Fig. S2, D and E). These data establish that Yoda1 induces IL-6 expression in cardiac fibroblasts via a Piezo1-dependent mechanism.
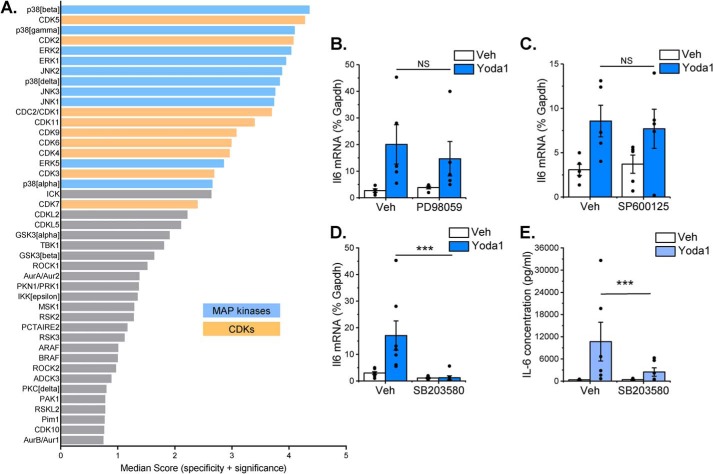
PamChip multiplex serine/threonine kinase activity profiling was used to assess differences in kinase activity following treatment of murine cardiac fibroblasts with Yoda1 for 10 min. Combinatorial analysis of phosphorylation of 140 peptide substrates revealed a hierarchical list of predicted serine/threonine kinases that were activated downstream of Piezo1 (Fig. 6A and Table S1). Within the top 20 hits, two major kinase families were identified: MAPKs (extracellular signal-regulated kinases (ERK1/2/5), c-Jun N-terminal kinases (JNK1/2/3), and p38 mitogen-activated protein kinases (p38α/β/γ/δ)) and cyclin-dependent kinases (CDK 1–7, 9, and 11) (Fig. 6A). The primary role of CDK family kinases is to phosphorylate cell cycle proteins and thereby regulate cell cycle progression, although it has also been reported that specific CDKs can regulate inflammatory gene expression through interaction with the NF-κB pathway (18). However, given that kinases within the NF-κB pathway (IKKα, IKKβ) were not activated in response to Yoda1 in our experiments (Table S1), we did not pursue the role of CDKs further. Subsequent experiments were therefore undertaken with selective pharmacological inhibitors of the ERK (PD98059), JNK (SP600125), and p38 MAPK (SB203580) pathways to identify which were required for expression of Il6 mRNA in response to Piezo1 activation in murine cardiac fibroblasts (Fig. 6, B–D). Of these, only SB203580 significantly reduced the Yoda1-induced increase in Il6 mRNA levels back to basal levels (Fig. 6D), suggesting an important role for the p38 MAPK pathway in Yoda1-induced Il6 gene expression. In agreement, SB203580 also inhibited Yoda1-induced IL-6 protein secretion by 80% (Fig. 6E).
Figure 6.
Piezo1 activation induces MAP kinase signaling, with p38 MAPK being coupled to IL-6 expression. A, mouse cardiac fibroblasts were stimulated with 10 μm Yoda1 for 10 min before analyzing Ser/Thr protein kinase activity with PamChip multiplex kinase activity profiling (n = 6). Kinases were identified based on peptide substrate phosphorylation and ranked by specificity and statistical significance (only the top 40 are shown). See Table S1 for the full dataset. The top kinase families predicted to be activated by Yoda1 were the MAPKs ERK1/2/5, JNK1/2/3, and p38α/β/γ/δ (blue) and CDKs 1–7, 9, and 11 (orange). B–D, RT-PCR analysis of Il6 mRNA expression after exposure of murine cardiac fibroblasts to 30 μm PD98059 (B, an ERK pathway inhibitor), 10 μm SP600125 (C, a JNK inhibitor), or 10 μm SB203580 (D, a p38 MAPK inhibitor) for 1 h, followed by treatment with vehicle (Veh) or 10 μm Yoda1 for a further 24 h. Expression is measured as percent of the housekeeping control Gapdh (n = 7 for SB203580, n = 5 for PD98059, and SP600125). Repeated measures two-way ANOVA for B: p = 0.7738, F = 0.133 (PD98059); p = 0.0529, F = 7.41 (Yoda1); p = 0.069, F = 6.08 (interaction). For C: p = 0.6133, F = 0.299 (SP600125); p = 0.0020, F = 51.9 (Yoda1); p = 0.475, F = 0.62 (interaction). For D: p = 0.0004, F = 52.1 (SB203580); p = 0.2942, F = 1.32 (Yoda1); p = 0.0028, F = 23.8 (interaction). Post hoc test: ***, p < 0.001; NS, not significant for the effect of inhibitor. E, murine cardiac fibroblasts were treated with vehicle or 10 μm SB203580 for 1 h and then treated with either vehicle or 10 μm Yoda1 for 24 h before collecting conditioned media and measuring IL-6 levels by ELISA (n = 6). Repeated measures two-way ANOVA: p = 0.0217, F = 10.8 (SB203580); p = 0.0283, F = 9.3 (Yoda1); p = 0.0002, F = 98.1 (interaction). Post hoc test: ***, p < 0.001 for the effect of SB203580.
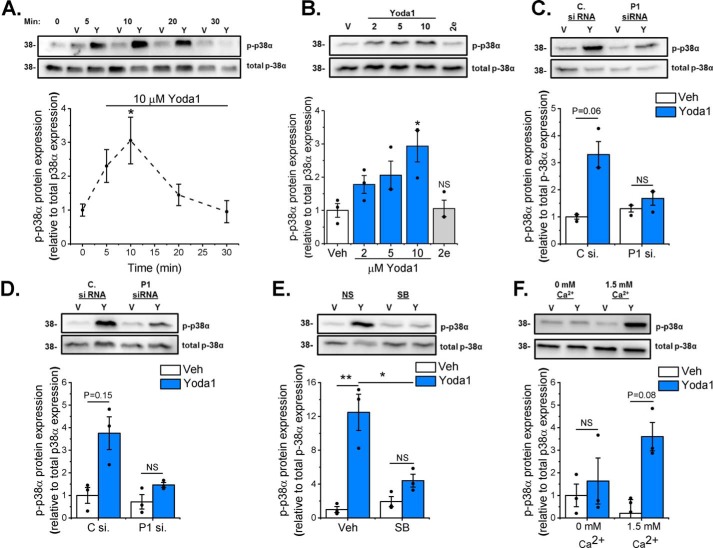
Western blotting was used to investigate whether the p38 MAPK pathway was stimulated following Piezo1 activation. Yoda1 increased p38 phosphorylation (activation) after 10 min before returning to basal levels after 30 min (Fig. 7A). Further studies investigating p38 activation revealed that Yoda1-induced p38 phosphorylation occurred in a concentration-dependent manner and that p38 was not activated in response to the inactive Yoda1 analog 2e (Fig. 7B). Piezo1-specific siRNA reduced Yoda1-induced activation of p38α in both murine (Fig. 7C) and human (Fig. 7D) cardiac fibroblasts, confirming the role of Piezo1 in Yoda1-induced p38 activation. The p38 inhibitor SB203580 significantly reduced the Yoda1-induced increase in p38 phosphorylation (Fig. 7E). Furthermore, p38 activation following Yoda1 treatment was shown to be dependent on the presence of extracellular Ca2+ (Fig. 7F), indicating that it is Ca2+ entry through the Piezo1 channel that is important for downstream signaling. Together, these data demonstrate that Ca2+-induced p38 MAPK activation is essential for inducing the expression and secretion of IL-6 in response to Piezo1 activation.
Figure 7.
Yoda1-induced p38 MAPK phosphorylation depends on Piezo1. Cellular protein samples were immunoblotted for p-p38α and reprobed with antibody for total p38α to confirm equal protein loading. The bar charts show mean densitometric data of p-p38α normalized to p38α expression. A, murine cardiac fibroblasts (n = 6) treated with DMSO vehicle (V) or 10 μm Yoda1 (Y) for 5–30 min. *, p < 0.05 versus vehicle-treated cells (repeated measures one-way ANOVA, p = 0.0085). B, murine cardiac fibroblasts (n = 3) treated for 10 min with varying concentrations of Yoda1 (2–10 μm) or 10 μm compound 2e. *, p < 0.05; NS, not significant versus vehicle-treated cells (repeated measures one-way ANOVA, p = 0.0122). C and D, murine (C) or human (D) cardiac fibroblasts (n = 3) transfected with either scrambled or Piezo1-specific siRNA before treatment with vehicle or 10 μm Yoda1 for 10 min. Repeated measures two-way ANOVA for C: p = 0.0202, F = 48.0 (Yoda1); p = 0.3045, F = 1.87 (siRNA); p = 0.0903, F = 9.6 (interaction). Repeated measures two-way ANOVA for D: p = 0.0282, F = 34.0 (Yoda1); p = 0.2669, F = 2.32 (siRNA); p = 0.478, F = 0.75 (interaction). Post hoc test: not significant. E, murine cardiac fibroblasts (n = 3) exposed to 10 μm SB203580 for 1 h before treatment with vehicle or 10 μm Yoda1 for 10 min. Repeated measures two-way ANOVA: p = 0.0312, F = 30.6 (Yoda1); p = 0.5898, F = 0.40 (SB203580); p = 0.0148, F = 65.9 (interaction). Post hoc test: **, p < 0.01; *, p < 0.05. F, murine cardiac fibroblasts (n = 3) treated for 10 min with vehicle or 10 μm Yoda1 in either standard DMEM or DMEM containing 1.75 mm EGTA to chelate free Ca2+. Repeated measures two-way ANOVA: p = 0.0123, F = 79.5 (Yoda1); p = 0.5739, F = 0.44 (Ca2+); p = 0.128, F = 6.34 (interaction). Post hoc test: not significant.
Discussion
The three main findings of this study are that human and mouse cardiac fibroblasts express functional Piezo1 channels coupled to a rapid rise in intracellular Ca2+, that these channels are mechanosensitive, and that Piezo1 activation is coupled to secretion of IL-6 via a p38 MAPK-dependent pathway. These data establish a link between cardiac fibroblast Piezo1 and secretion of paracrine signaling molecules that can modulate cardiac remodeling.
We established that murine and human cardiac fibroblasts express Piezo1 mRNA at levels similar to those found in endothelial cells from various sources, which are known to express high levels of functional Piezo1 (14). Piezo1 mRNA expression was 20 times higher in cardiac fibroblasts than in cardiomyocytes. The EC50 values of Yoda1 acting on endogenous Piezo1 in murine and human cardiac fibroblasts (0.72 μm and 0.71 μm, respectively) were comparable with our previous report of 0.23 μm in HUVECs (16). When artificially overexpressed in cell lines, our data on mouse Piezo1 in HEK T-REx-293 cells (EC50 = 0.33 μm) were broadly comparable with human Piezo1 expressed in HEK T-REx-293 cells (EC50 = 2.51 μm) (16) and somewhat lower than those originally reported in HEK293T cells transiently transfected with mouse (EC50 = 17.1 μm) or human (EC50 = 26.6 μm) Piezo1 (11). It is worth noting that we could only use Yoda1 at concentrations up to 10 μm because of solubility problems, so these EC50 values are estimates.
The ability of common inhibitors of mechanosensitive ion channels, gadolinium and ruthenium red (10), and the novel Yoda1 antagonist Dooku1 (16) to decrease the Yoda1-evoked Ca2+ entry in murine and human cardiac fibroblasts correlated with the expected properties of the channel. The mechanism by which Yoda1 modulates Piezo1 activation and gating has been described recently (12). However, it has also been suggested that Yoda1 may have some Piezo1-independent effects in endothelial cells (19). We further verified the importance of Piezo1 by showing that the Ca2+, p38 MAPK, and IL-6 responses in cardiac fibroblasts were all proportionally reduced by siRNA-mediated knockdown of Piezo1 or by use of cells from a global Piezo1+/− heterozygous knockout mouse model (14). Thus, the Yoda1 responses we observed in cardiac fibroblasts were unequivocally due to activation of Piezo1.
Our patch clamp data showed that increased pressure caused increased inward currents up to a limit, consistent with the presence of Piezo1 channels (10, 20). The amplitude of the current was highly variable between patches; we suspect that the variable amount of suction required to form the initial Giga-seal was a contributor to the variability in current amplitude (with greater suction apparently leading to smaller mechanically activated currents). Prior work has suggested that overexpressed human Piezo1 channels require −43 mm Hg for 50% activation (20), but the difference between this value and what we observed (−61.3 mm Hg) may reflect the influence of the native environment on channel properties at natural density. Although the kinetics of the currents were relatively slow and did not show inactivation, which contrasts with data for overexpressed Piezo1 channels (10), prior work has suggested that endogenous Piezo1 channels have slower kinetics and inactivate slowly or not at all (14, 21). These data suggest that Piezo1 channel activity most likely explains these mechanically activated currents in human cardiac fibroblasts.
Prolonged exposure to Yoda1 increased IL-6 mRNA and protein expression in a Piezo1-dependent manner in both human and mouse cardiac fibroblasts. IL-6 is a pleiotropic pro-inflammatory cytokine that promotes cardiac fibroblast proliferation and fibrosis (22) as well as cardiac hypertrophy through its actions on cardiomyocytes (23) and is readily secreted by cardiac fibroblasts in culture (24). Mechanical stretching of cardiac fibroblasts induces expression of several pro-fibrotic cytokines (1, 25), although IL-6 expression does not appear to be mechanically regulated in human cardiac fibroblasts (26), as we found in this study. This is in contrast to fibroblasts from various other noncardiac sources that have been shown to secrete IL-6 in response to mechanical stretch (27–29).
Although cyclic stretch did not modulate IL-6 expression in human cardiac fibroblasts (and no role for Piezo1 was found), it was apparent from the stretch experiments that Piezo1 siRNA could reduce basal IL-6 expression. This was evident only when cells were grown on the softer collagen-coated substrate of the BioFlex plates but not on the rigid plastic of regular tissue culture plates. Thus, Piezo1 may play a role in detecting altered substrate stiffness and/or composition and modulate IL-6 expression accordingly. It has been shown previously that stretch-induced expression of several cardiac fibroblast genes depends on matrix stiffness (3, 25); hence, there is a complex interplay of mechanosignaling pathways that convert these different mechanical stimuli into alterations in cellular phenotype. Further studies are required to more precisely determine the role of substrate stiffness in regulating IL-6 secretion in cardiac fibroblasts.
The p38 family of stress-activated MAPKs is known to play an important role in cardiac signaling and is activated in both acute and chronic cardiac pathologies (30). Recently, a key role for p38α in regulating multiple aspects of cardiac fibroblast function has emerged (31). For example, fibroblast-specific knockout of Mapk14, the gene encoding p38α, revealed a critical role for this kinase in driving cardiac myofibroblast differentiation and fibrosis in response to ischemic injury or chronic neurohumoral stimulation (32). In a similar model, we established a role for cardiac fibroblast p38α in stimulating cardiac hypertrophy after chronic β-adrenergic stimulation and uncovered a potential role for IL-6 acting as a paracrine inducer of cardiomyocyte hypertrophy in this context (15). We have also shown an important role for p38α in cytokine-induced IL-6 expression and secretion in cultured human cardiac fibroblasts (33). Thus, Piezo1-mediated activation of p38α in cardiac fibroblasts and subsequent secretion of IL-6 may be important in the cardiac remodeling process. Interestingly, hydrostatic pressure can stimulate p38 MAPK signaling through Piezo1 activation in mesenchymal stem cells (34).
In conclusion, our study establishes that Piezo1 is a functional mechanosensitive Ca2+-permeable ion channel in cardiac fibroblasts and is coupled to altered expression of genes important to the remodeling process. Specifically, activation of Piezo1 induces expression and secretion of IL-6, a pro-hypertrophic cytokine with important roles after cardiac injury. Furthermore, we revealed that p38 MAPK is activated downstream of Piezo1-mediated Ca2+ entry to cause increased IL-6 secretion. Functional studies and in vivo investigation into the role of Piezo1 in cardiac fibroblasts are warranted to provide additional information regarding its role in regulating cardiac remodeling.
Experimental procedures
Reagents
Yoda1 (Tocris), staurosporine (Sigma), PD98059 (Merck), SP600125 (Cambridge Bioscience), and SB203580 (Merck) were all solubilized in DMSO. ATP, gadolinium, and ruthenium red were all obtained from Sigma-Aldrich and dissolved in H2O. Dooku1 and compound 2e (16) were synthesized at the University of Leeds and solubilized in DMSO.
Mouse cardiac fibroblast culture
Adult C57BL/6J mice were euthanized according to guidelines of The UK Animals (Scientific Procedures) Act (1986). Cardiac fibroblast cultures were established from collagenase-digested hearts and cultured in DMEM supplemented with 10% fetal calf serum as described previously (15). All cells were kept at 37 °C and 5% CO2 throughout the study. Cells were studied at passage 1–2. Cells were kept in serum-free DMEM for 16 h prior to treatment to collect RNA, lysates, and conditioned media.
Human cell culture
Biopsies of human atrial appendages and long saphenous veins were obtained from patients undergoing elective coronary artery bypass grafting at the Leeds General Infirmary following local ethical committee approval and informed patient consent. The study complied with the principles of the Declaration of Helsinki. Cells were harvested and cultured as described previously for cardiac fibroblasts (35) and saphenous vein endothelial cells (36). Experiments were performed on human cardiac fibroblasts from passages 2–5. Serum-free DMEM was used for 1 h prior to treatment to collect RNA and conditioned media. HUVECs were purchased from Lonza and cultured in endothelial cell growth medium (EGM-2, Lonza), supplemented with 2% fetal calf serum and the EGM-2 SingleQuots Kit (Lonza). HUVECs were used at passage 2.
Mouse pulmonary endothelial cell culture
CD146+ pulmonary endothelial cells were isolated with immunomagnetic microbeads (Miltenyi) and cultured in MV2 medium (Promocell) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. Cells were studied at passage 1.
HEK T-Rex-293 cell culture
pcDNA3_mouse Piezo1_IRES_GFP, a kind gift from Ardem Patapoutian (10), was used as a template to clone the mouse Piezo1 coding sequence into pcDNA4/TO. Overlapping mouse Piezo1 (forward primer 5′-GTAACAACTCCGCCCCATTG-3′ and reverse primer 5′-GCTTCTACTCCCTCTCACGTGTC-3′) and pcDNA4/TO (forward primer 5′-GACACGTGAGAGGGAGTAGAAGCCGCTGATCAGCCTCGACTG-3′ and reverse primer 5′-CAATGGGGCGGAGTTGTTAC-3′) PCR products were assembled using Gibson Assembly (New England Biolabs) (37). This construct does not contain tetracycline operator sequences. HEK T-REx-293 cells (Invitrogen) were transfected with pcDNA4/TO-mPiezo1 using Lipofectamine 2000 (Invitrogen) and treated with 200 μg/ml zeocin (InvivoGen) to select stably transfected cells. Individual clones were isolated and analyzed for expression using Yoda1 and intracellular Ca2+ measurements. HEK T-REx-293 cells were maintained in DMEM supplemented with 10% fetal calf serum and 1% penicillin/streptomycin (Sigma-Aldrich). Nontransfected HEK T-Rex-293 cells were used as control cells.
Cardiac cell fractionation
Nonmyocyte cardiac cell fractions were prepared as described previously (15). Briefly, collagenase-digested heart tissue was filtered through a 30-μm MACS Smart Strainer (Miltenyi) to remove cardiomyocytes. Nonmyocytes were then separated into two fractions using a cardiac fibroblast magnetic antibody cell separation kit (Miltenyi). “Nonfibroblasts” (Pecam1-positive endothelial cells and leukocytes) were collected in fraction 1, and “fibroblasts” (Col1a1/Col1a2/Ddr2/Pdgfra-positive) were collected in fraction 2, as characterized previously (15). Separately, adult mouse cardiomyocytes were isolated from ventricles of 8-week-old WT mice. Hearts were cannulated through the aorta and perfused for 5 min with perfusion buffer (124.5 mm NaCl, 10 mm HEPES, 11.1 mm glucose, 1.2 mm NaH2PO4, 1.2 mm MgSO4, 4 mm KCl, and 25 mm taurine (pH 7.34)) containing 10 mm butanedione monoxime, followed by perfusion buffer containing 1 mg/ml type 2 collagenase, 0.05 mg/ml protease and 12.5 μm CaCl2 for 7–15 min until suitable digestion was observed. Ventricles were gently cut in perfusion solution containing 10% FBS and 12.5 μm CaCl2 and filtered. This step was repeated in perfusion solution containing 5% FBS and 12.5 μm CaCl2 and, finally, perfusion solution containing only 12.5 μm CaCl2. The filtrate was pelleted by gravity for 5 min before resuspension in perfusion buffer containing 0.5 mm CaCl2. This step was repeated with increasing concentrations of CaCl2 (1.5 mm, 3.5 mm, 8 mm, and 18 mm). Finally, the cell pellet was resuspended in 1 ml TRIzol. RNA was extracted from cardiac cell fractions, and quantitative RT-PCR was used to quantify gene expression.
Quantitative RT-PCR
For gene expression studies, cardiac fibroblasts were treated with Yoda1 or vehicle (DMSO) for the indicated time. RNA was extracted from cultured/fractionated cells with the Aurum RNA Extraction Kit (Bio-Rad). cDNA was synthesized using a reverse transcription system (Promega). Real-time RT-PCR was performed with the ABI-7500 system, gene expression master mix, and specific TaqMan primer/probe sets (Thermo Fisher Scientific): mouse Col1a1 (Mm01302043_g1), mouse Col3a1 (Mm01254476_m1), mouse Piezo1 (Mm01241545_g1), human PIEZO1 (Hs00207230_m1), mouse Il1b (Mm00434228_m1), mouse Il6 (Mm00446190_m1), human IL6 (Hs00174131_m1), mouse Mmp3 (Mm00440295_m1), and mouse Mmp9 (Mm00442991_m1). Data are routinely expressed as the percentage of mouse Gapdh (Mm99999915_g1) or human GAPDH (Hs99999905_m1) housekeeping gene mRNA expression using the formula 2−ΔCT × 100, in which CT is the cycle threshold number. For analysis of cardiac cell fractions, three housekeeping genes were used to mitigate the effects of any variation in expression of housekeeping genes between cell fractions: Gapdh (Mm99999915_g1), Hprt (Mm03024075_m1), and Actb (Mm00607939_s1). Data were expressed relative to the geometric mean of the pooled housekeeping gene expression using the formula 2−ΔCT.
Piezo1-modified mice
Animal use was authorized by the University of Leeds Animal Ethics Committee and by the UK Home Office under project license P144DD0D6. Animals were maintained in individually ventilated Optimice cages (Animal Care Systems) at 21 °C, 50–70% humidity, 12 h/12 h light/dark cycle, RM1 diet (Special Diet Services) ad libitum, and Pure'o Cell bedding (Datesand). C57BL/6 mice carrying global disruption of the Piezo1 gene with a lacZ insertion flanked by Flp recombinase target sites have been described previously (14). Identification of WT and Piezo1+/− mice was performed by genotyping using primers for LacZ: forward, AATGGTCTGCTGCTGCTGAAC; reverse, GGCTTCATCCACCACATACAG. Mice of varying ages and sexes were used for experiments.
Western blotting
Cells were treated with vehicle (DMSO), Yoda1, or compound 2e at the appropriate times prior to lysing. Cells were harvested in lysis buffer containing 10 mm Tris (pH 7.5), 150 mm NaCl, 0.5 mm EDTA, 0.5% NP-40, MiniComplete protease inhibitors (Roche), and PhosSTOP phosphatase inhibitors (Roche). A protein quantification assay was then performed using the DC Protein Assay (Bio-Rad). 25 μg of protein was loaded on a 10% polyacrylamide gel. After resolution by electrophoresis, samples were transferred to PVDF membranes, and Western blotting was performed as described previously (38) using a primary antibody for phospho-p38 MAPK (9215, Cell Signaling Technology). Membranes were reprobed with p38α antibody (9228, Cell Signaling Technology) to confirm equal protein loading. Species-appropriate secondary antibodies (GE Healthcare) and ECL Prime Western blotting detection reagent (GE Healthcare) were used for visualization. Syngene G:BOX Chemi XT4 was used for imaging alongside GeneSys image acquisition software for densitometric analysis.
Intracellular Ca2+ measurements
Changes in [Ca2+]i were measured using the ratiometric Ca2+ indicator dye Fura-2/AM and a 96-well fluorescence plate reader (FlexStationII384, Molecular Devices) controlled by SOFTmax PRO software v5.4.5. Cardiac fibroblasts and HUVECs were plated in clear 96-well plates (Corning) and HEK T-REx-293 cells in black, clear-bottomed 96-well plates (Grenier) at a confluence of 90% 24 h before experimentation. Cells were incubated for 1 h at 37 °C in standard bath solution (SBS; 130 mm NaCl, 5 mm KCl, 8 mm D-glucose, 10 mm HEPES, 1.2 mm MgCl2, and 1.5 mm CaCl2 (pH 7.4)) containing 2 μm Fura-2 in the presence of 0.01% pluronic acid (Thermo Fisher Scientific) to aid dispersion. During all Fura-2 assays involving cardiac fibroblasts, 2.5 mm probenecid (Sigma-Aldrich) was present to prevent extrusion of the Fura-2 indicator (39). Cells were washed with SBS and then incubated at room temperature for 30 min, during which time inhibitors were added when applicable. Stimuli were injected at 60 s of recording. The change in intracellular Ca2+ concentration (Δ[Ca2+]i) was measured as the ratio of Fura-2 emission (510 nm) intensity at 340 nm and 380 nm.
Gene silencing
Murine cardiac fibroblasts were grown to 80% confluence and transfected with 10 nm Piezo1-specific Silencer Select Pre-Designed siRNA (4390771, siRNA s107968, Life Technologies) or Silencer Select Negative Control No. 1 siRNA (4390843, Life Technologies) using Lipofectamine RNAiMAX reagent (Life Technologies) in Opti-MEM (Gibco) according to the manufacturer's instructions. Medium was replaced with full-growth medium 24 h later. For human cardiac fibroblasts, cells were grown to 90% confluence and transfected with 20 nm Piezo1-specific Silencer Select Pre-designed siRNA (4392420, siRNA s18891, Thermo Fisher Scientific) or ON-TARGETplus Nontargeting Pool siRNA (D-01810-10-20, Dharmacon) using Lipofectamine 2000 in Opti-MEM according to the manufacturer's instructions. Medium was replaced with full-growth medium after 4.5 h. Cells were used for experimentation 48 h after transfection.
Patch clamp electrophysiology
Ionic currents were recorded through cell-attached patches using a standard patch clamp technique in voltage clamp mode. Patch pipettes had a resistance of 4–6 megaohm when filled with pipette solution. An ionic solution composed of 145 mm CsCl, 2 mm MgCl2, 10 mm HEPES, 5 mm ATP, 0.1 mm GTP, and 1 mm EGTA (titrated to pH 7.2 using CsOH) was used in the pipette. The bath solution was SBS. Recordings were at a constant holding potential of +80 mV (applied to the patch pipette). 200-ms pressure steps were applied to the patch pipette with an interval of 10 s using a high-speed pressure clamp HSPC-1 system (ALA Scientific Instruments). All recordings were made with an Axopatch-200B amplifier (Axon Instruments, Inc.) equipped with Digidata 1550B and pClamp 10.6 software (Molecular Devices) at room temperature. Currents were filtered at 2 kHz and digitally sampled at 20 kHz. Data were analyzed using pClamp 10.6 and the MicroCal Origin 2018 (OriginLab Corp.) software packages. All recordings were made blind (i.e. without knowledge of which cells had been transfected with control or Piezo1 siRNA).
Cell viability assay
Cardiac fibroblasts were plated at 80% confluency and incubated overnight (37 °C, 5% CO2). Vehicle (DMSO), Yoda1 (10 μm), or staurosporine (1 μm) were applied the following day, and cells were incubated for 24 h prior to commencing the live/dead cell viability assay (Thermo Fisher Scientific) according to the manufacturer's instructions. Cells were imaged using the IncuCyte ZOOM Live Imaging System (Essen Bioscience). The total number of fluorescent cells in each well was calculated using built-in algorithms, using an average from 9 images/well of a 12-well plate. The mean data are shown as the number of live cells relative to the total number of cells.
Stretch experiments
Human cardiac fibroblasts were seeded at 1 × 105 cells/well on collagen-coated membranes (BioFlex 6-well culture plates). 72 h after transfection, cells were stretched while in serum-free medium using an FX-4000 or FX-5000 Flexercell Tension System (Flexcell International) to equibiaxially elongate the cell-seeded elastic membrane against a loading post. Elongation at 10% strain and 1 Hz was applied to the cells for 24 h. A 6-well stretching plate was housed in the incubator (37 °C, 5% CO2) alongside unstimulated cells adhered to BioFlex plates, which served as static controls. RNA was extracted, and conditioned medium was collected. Quantitative RT-PCR and ELISA were used to quantify gene and protein expression, respectively.
ELISA
Conditioned media were collected, centrifuged to remove cellular debris, and stored at −20 °C for subsequent analysis. The concentration of IL-6 in the media was measured by ELISA (M6000, R&D Systems) according to the manufacturer's instructions. Samples were diluted 1:5–1:30 prior to analysis.
Multiplex kinase activity profiling
PamGene serine-threonine kinase (STK) multiplex activity assays were used to investigate STK activity. Murine cardiac fibroblasts were treated with vehicle or Yoda1 for 10 min, collected, and lysed using mammalian protein extraction reagent lysis buffer (Thermo Fisher Scientific) containing Halt phosphatase and Halt protease inhibitor cocktails (Pierce) for 30 min on ice. A protein quantification assay was then performed as stated above, and samples were snap-frozen in liquid nitrogen. 1.2 μg of protein was loaded onto each STK PamChip array. Phosphorylation of PamChip peptides was monitored by PamStation 12 following the manufacturer's protocols as described previously (40). Signal intensities were analyzed using PamGene's BioNavigator software as Yoda1-treated versus DMSO treatment after 10 min. Permutation analysis resulted in a specificity score (mapping of peptides to kinases) and a significance score (difference between treatment groups) for each kinase. The combined score was used to rank and predict top kinase hits.
Statistical analysis
OriginPro 2015 (OriginLab) and GraphPad Prism version 7.05 (GraphPad Software) were used for data analysis, and OriginPro 2015 was used to prepare the charts and graphs. Averaged data are presented as mean ± S.E., where n represents the number of independent experiments and N indicates the total number of replicates within the independent experiments. Data were log-transformed prior to statistical analysis. For comparisons between two sets of data, paired or unpaired Student's t tests were used as appropriate. For multiple comparisons with a single factor, one-way ANOVA was used (p values in the figure legends) with Tukey post hoc test (p values in the figures). For analysis of two factors, regular or repeated measures two-way ANOVA was used (p values in the figure legends) with Sidak post hoc test (p values in the figures). p < 0.05 was considered statistically significant. For IC50 determination, data were normalized to vehicle, and curves were fitted using the Hill equation.
Author contributions
N. M. B., K. M., and V. S. data curation; N. M. B., M. J. L., V. S., H. T. G., J. S., J. L., F. A. v. N., and K. E. P. methodology; N. M. B. and N. A. T. writing-original draft; N. M. B., K. M., F. A. v. N., D. J. B., and N. A. T. writing-review and editing; M. J. L., H. T. G., E. L. E., K. C., R. F., J. S., J. L., F. A. v. N., K. E. P., D. J. B., and N. A. T. resources; J. L., M. J. D., F. A. v. N., D. J. B., and N. A. T. supervision; J. L., M. J. D., D. J. B., and N. A. T. funding acquisition; D. J. B. and N. A. T. conceptualization; N. A. T. project administration.
Supplementary Material
Acknowledgments
We thank Ardem Patapoutian (Scripps Research Institute) for providing the pcDNA3_mouse Piezo1_IRES_GFP plasmid. We also thank Savithri Rangarajan (PamGene) for assistance with analysis of data from the PamChip kinase activity arrays, David Eisner's group at the University of Manchester for supplying the mouse cardiomyocyte fraction, and the University of Leeds Alumni for a scholarship to N. M. B.
This work was supported by British Heart Foundation PhD Studentship FS/15/48/31665 (to N. M. B. (student), N. A. T., J. L., and D. J. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Table S1.
- ECM
- extracellular matrix
- MAPK
- mitogen-activated protein kinase
- HUVEC
- human umbilical vein endothelial cell
- MACS
- magnetic antibody cell separation
- Het
- heterozygous
- ANOVA
- analysis of variance
- ERK
- extracellular signal-regulated kinase
- CDK
- cyclin-dependent kinase
- JNK
- c-Jun N-terminal kinase
- SBS
- standard bath solution
- STK
- serine-threonine kinase
- CF
- cardiac fibroblast(s).
References
- 1. Deb A., and Ubil E. (2014) Cardiac fibroblast in development and wound healing. J. Mol. Cell. Cardiol. 70, 47–55 10.1016/j.yjmcc.2014.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turner N. A., and Porter K. E. (2013) Function and fate of myofibroblasts after myocardial infarction. Fibrogenesis Tissue Repair 6, 5 10.1186/1755-1536-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herum K. M., Lunde I. G., McCulloch A. D., and Christensen G. (2017) The soft- and hard-heartedness of cardiac fibroblasts: mechanotransduction signaling pathways in fibrosis of the heart. J. Clin. Med. 6, E53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan D., Takawale A., Lee J., and Kassiri Z. (2012) Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair 5, 15 10.1186/1755-1536-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saucerman J. J., Tan P. M., Buchholz K. S., McCulloch A. D., and Omens J. H. (2019) Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 16, 361–378 10.1038/s41569-019-0155-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reed A., Kohl P., and Peyronnet R. (2014) Molecular candidates for cardiac stretch-activated ion channels. Glob. Cardiol. Sci. Pract. 2014, 9–25 10.5339/gcsp.2014.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin T. P., Lawan A., Robinson E., Grieve D. J., Plevin R., Paul A., and Currie S. (2014) Adult cardiac fibroblast proliferation is modulated by calcium/calmodulin-dependent protein kinase II in normal and hypertrophied hearts. Pflugers Arch. 466, 319–330 10.1007/s00424-013-1326-9 [DOI] [PubMed] [Google Scholar]
- 8. Zhang X., Zhang T., Wu J., Yu X., Zheng D., Yang F., Li T., Wang L., Zhao Y., Dong S., Zhong X., Fu S., Xu C. Q., Lu F., and Zhang W. H. (2014) Calcium Sensing Receptor Promotes Cardiac Fibroblast Proliferation and Extracellular Matrix Secretion. Cellular Physiology and Biochemistry 33, 557–568 10.1159/000358634 [DOI] [PubMed] [Google Scholar]
- 9. Adapala R. K., Thoppil R. J., Luther D. J., Paruchuri S., Meszaros J. G., Chilian W. M., and Thodeti C. K. (2013) TRPV4 channels mediate cardiac fibroblast differentiation by integrating mechanical and soluble signals. J. Mol. Cell. Cardiol. 54, 45–52 10.1016/j.yjmcc.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coste B., Mathur J., Schmidt M., Earley T. J., Ranade S., Petrus M. J., Dubin A. E., and Patapoutian A. (2010) Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 10.1126/science.1193270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Syeda R., Xu J., Dubin A. E., Coste B., Mathur J., Huynh T., Matzen J., Lao J., Tully D. C., Engels I. H., Petrassi H. M., Schumacher A. M., Montal M., Bandell M., and Patapoutian A. (2015) Chemical activation of the mechanotransduction channel Piezo1. Elife 4, e07369 10.7554/eLife.07369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lacroix J. J., Botello-Smith W. M., and Luo Y. (2018) Probing the gating mechanism of the mechanosensitive channel Piezo1 with the small molecule Yoda1. Nat. Commun. 9, 2029 10.1038/s41467-018-04405-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang J., Huang B., Yuan G., Chen Y., Liang F., Zeng H., Zheng S., Cao L., Geng D., and Zhou S. (2017) Stretch-activated channel Piezo1 is up-regulated in failure heart and cardiomyocyte stimulated by AngII. Am. J. Transl. Res. 9, 2945–2955 [PMC free article] [PubMed] [Google Scholar]
- 14. Li J., Hou B., Tumova S., Muraki K., Bruns A., Ludlow M. J., Sedo A., Hyman A. J., McKeown L., Young R. S., Yuldasheva N. Y., Majeed Y., Wilson L. A., Rode B., Bailey M. A., et al. (2014) Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282 10.1038/nature13701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bageghni S. A., Hemmings K. E., Zava N., Denton C. P., Porter K. E., Ainscough J. F. X., Drinkhill M. J., and Turner N. A. (2018) Cardiac fibroblast-specific p38α MAP kinase promotes cardiac hypertrophy via a putative paracrine interleukin-6 signaling mechanism. FASEB J. 32, 4941–4954 10.1096/fj.201701455RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Evans E. L., Cuthbertson K., Endesh N., Rode B., Blythe N. M., Hyman A. J., Hall S. J., Gaunt H. J., Ludlow M. J., Foster R., and Beech D. J. (2018) Yoda1 analogue (Dooku1) which antagonizes Yoda1-evoked activation of Piezo1 and aortic relaxation. Br. J. Pharmacol. 175, 1744–1759 10.1111/bph.14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tarone G., and Lembo G. (2003) Molecular interplay between mechanical and humoral signalling in cardiac hypertrophy. Trends Mol. Med. 9, 376–382 10.1016/S1471-4914(03)00164-3 [DOI] [PubMed] [Google Scholar]
- 18. Schmitz M. L., and Kracht M. (2016) Cyclin-dependent kinases as coregulators of inflammatory gene expression. Trends Pharmacol. Sci. 37, 101–113 10.1016/j.tips.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 19. Dela Paz N. G., and Frangos J. A. (2018) Yoda1-induced phosphorylation of Akt and ERK1/2 does not require Piezo1 activation. Biochem. Biophys. Res. Commun. 497, 220–225 10.1016/j.bbrc.2018.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bae C., Gnanasambandam R., Nicolai C., Sachs F., and Gottlieb P. A. (2013) Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc. Natl. Acad. Sci. U.S.A. 110, E1162–E1168 10.1073/pnas.1219777110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Del Mármol J. I., Touhara K. K., Croft G., and MacKinnon R. (2018) Piezo1 forms a slowly-inactivating mechanosensory channel in mouse embryonic stem cells. Elife 7, e33149 10.7554/eLife.33149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banerjee I., Fuseler J. W., Intwala A. R., and Baudino T. A. (2009) IL-6 loss causes ventricular dysfunction, fibrosis, reduced capillary density, and dramatically alters the cell populations of the developing and adult heart. Am. J. Physiol. Heart Circ. Physiol. 296, H1694–H1704 10.1152/ajpheart.00908.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meléndez G. C., McLarty J. L., Levick S. P., Du Y., Janicki J. S., and Brower G. L. (2010) Interleukin 6 mediates myocardial fibrosis, concentric hypertrophy, and diastolic dysfunction in rats. Hypertension 56, 225–231 10.1161/HYPERTENSIONAHA.109.148635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LaFramboise W. A., Scalise D., Stoodley P., Graner S. R., Guthrie R. D., Magovern J. A., and Becich M. J. (2007) Cardiac fibroblasts influence cardiomyocyte phenotype in vitro. Am. J. Physiol. Cell Physiol. 292, C1799–C1808 10.1152/ajpcell.00166.2006 [DOI] [PubMed] [Google Scholar]
- 25. Herum K. M., Choppe J., Kumar A., Engler A. J., and McCulloch A. D. (2017) Mechanical regulation of cardiac fibroblast profibrotic phenotypes. Mol. Biol. Cell 28, 1871–1882 10.1091/mbc.e17-01-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ugolini G. S., Pavesi A., Rasponi M., Fiore G. B., Kamm R., and Soncini M. (2017) Human cardiac fibroblasts adaptive responses to controlled combined mechanical strain and oxygen changes in vitro. Elife 6, e22847 10.7554/eLife.22847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. House M., Tang P., Daniel J., Kaplan D., and Socrate S. (2012) 510: Mechanical stretch increases production of inflammatory cytokines from cervical fibroblasts in 2D and 3D culture. Am. J. Obstet. Gynecol. 206, S231 [Google Scholar]
- 28. Li J., Zhang Y., Cui L., Wang J., Pang X., Lai Y., Yao Y., Liu X., and Li Y. (2013) Mechanical stretch changes coronary artery fibroblasts function by upregulating HSF1 protein expression. Int. J. Biol. Macromol. 59, 105–110 10.1016/j.ijbiomac.2013.03.053 [DOI] [PubMed] [Google Scholar]
- 29. Skutek M., van Griensven M., Zeichen J., Brauer N., and Bosch U. (2001) Cyclic mechanical stretching enhances secretion of Interleukin 6 in human tendon fibroblasts. Knee Surg. Sports Traumatol. Arthrosc. 9, 322–326 10.1007/s001670100217 [DOI] [PubMed] [Google Scholar]
- 30. Zhang W., Elimban V., Nijjar M. S., Gupta S. K., and Dhalla N. S. (2003) Role of mitogen-activated protein kinase in cardiac hypertrophy and heart failure. Exp. Clin. Cardiol. 8, 173–183 [PMC free article] [PubMed] [Google Scholar]
- 31. Turner N. A., and Blythe N. M. (2019) Cardiac fibroblast p38 MAPK: a critical regulator of myocardial remodeling. J. Cardiovasc. Dev. Dis. 6, E27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molkentin J. D., Bugg D., Ghearing N., Dorn L. E., Kim P., Sargent M. A., Gunaje J., Otsu K., and Davis J. (2017) Fibroblast-specific genetic manipulation of p38 mitogen-activated protein kinase in vivo reveals its central regulatory role in fibrosis. Circulation 136, 549–561 10.1161/CIRCULATIONAHA.116.026238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sinfield J. K., Das A., O'Regan D. J., Ball S. G., Porter K. E., and Turner N. A. (2013) p38 MAPK α mediates cytokine-induced IL-6 and MMP-3 expression in human cardiac fibroblasts. Biochem. Biophys. Res. Commun. 430, 419–424 10.1016/j.bbrc.2012.11.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sugimoto A., Miyazaki A., Kawarabayashi K., Shono M., Akazawa Y., Hasegawa T., Ueda-Yamaguchi K., Kitamura T., Yoshizaki K., Fukumoto S., and Iwamoto T. (2017) Piezo type mechanosensitive ion channel component 1 functions as a regulator of the cell fate determination of mesenchymal stem cells. Sci. Rep. 7, 17696–17696 10.1038/s41598-017-18089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turner N. A., Porter K. E., Smith W. H., White H. L., Ball S. G., and Balmforth A. J. (2003) Chronic β2-adrenergic receptor stimulation increases proliferation of human cardiac fibroblasts via an autocrine mechanism. Cardiovasc. Res. 57, 784–792 10.1016/S0008-6363(02)00729-0 [DOI] [PubMed] [Google Scholar]
- 36. Bauer C. C., Boyle J. P., Porter K. E., and Peers C. (2010) Modulation of Ca2+ signalling in human vascular endothelial cells by hydrogen sulfide. Atherosclerosis 209, 374–380 10.1016/j.atherosclerosis.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 37. Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 38. Turner N. A., Ball S. G., and Balmforth A. J. (2001) The mechanism of angiotensin II-induced extracellular signal-regulated kinase-1/2 activation is independent of angiotensin AT1A receptor internalisation. Cell. Signal. 13, 269–277 10.1016/S0898-6568(01)00135-8 [DOI] [PubMed] [Google Scholar]
- 39. Di Virgilio F., Steinberg T. H., and Silverstein S. C. (1989) in Methods in Cell Biology (Tartakoff A. M., ed.) pp. 453–462, Academic Press, Cambridge, MA: [DOI] [PubMed] [Google Scholar]
- 40. Schwill M., Tamaskovic R., Gajadhar A. S., Kast F., White F. M., and Plückthun A. (2019) Systemic analysis of tyrosine kinase signaling reveals a common adaptive response program in a HER2-positive breast cancer. Sci. Signal. 12, eaau2875 10.1126/scisignal.aau2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.