Abstract
The angiotensin II (AngII) type 1 receptor (AT1R) is a member of the G protein–coupled receptor (GPCR) family and binds β-arrestins (β-arrs), which regulate AT1R signaling and trafficking. These processes can be biased by different ligands or mutations in the AGTR1 gene. As for many GPCRs, the exact details for AT1R–β-arr interactions driven by AngII or β-arr–biased ligands remain largely unknown. Here, we used the amber-suppression technology to site-specifically introduce the unnatural amino acid (UAA) p-azido-l-phenylalanine (azF) into the intracellular loops (ICLs) and the C-tail of AT1R. Our goal was to generate competent photoreactive receptors that can be cross-linked to β-arrs in cells. We performed UV-mediated photolysis of 25 different azF-labeled AT1Rs to cross-link β-arr1 to AngII-bound receptors, enabling us to map important contact sites in the C-tail and in the ICL2 and ICL3 of the receptor. The extent of AT1R–β-arr1 cross-linking among azF-labeled receptors differed, revealing variability in β-arr's contact mode with the different AT1R domains. Moreover, the signature of ligated AT1R–β-arr complexes from a subset of azF-labeled receptors also differed between AngII and β-arr–biased ligand stimulation of receptors and between azF-labeled AT1R bearing and that lacking a bias signaling mutation. These observations further implied distinct interaction modalities of the AT1R–β-arr1 complex in biased signaling conditions. Our findings demonstrate that this photocross-linking approach is useful for understanding GPCR–β-arr complexes in different activation states and could be extended to study other protein–protein interactions in cells.
Keywords: G protein-coupled receptor (GPCR), angiotensin II, arrestin, receptor, photoaffinity labeling, angiotensin II type 1 receptor, biased signaling, p-azido-L-phenylalanine, unnatural amino acid
Introduction
G protein–coupled receptors (GPCRs)4 comprise the largest class of membrane-bound proteins and are involved in regulating many physiological functions, making them important therapeutic targets. Agonist-bound GPCRs interact with heterotrimeric G proteins to transduce signals and are subsequently desensitized following receptor phosphorylation by GPCR kinases (GRKs). GRKs enhance the binding of arrestin proteins to receptors, including the visual arrestin interaction to rhodopsin, and the binding of the two nonvisual arrestins, also known as β-arrestins (β-arrs) 1 and 2, to nonvisual GPCRs, leading to a reduction in G protein–dependent signaling at the plasma membrane (1–3). In addition to this classical role, β-arrs promote internalization of receptors and act as signal transducers where the GPCR–β-arr complex serves as a scaffold to recruit other signaling effectors (4, 5). The angiotensin II (AngII) type 1 receptor (AT1R), a member of the GPCR family, binds β-arrs with high affinity and forms long-lived complexes in cells. β-Arrs interact with AngII-bound receptors' C-tails, which have previously been phosphorylated on specific serine/threonine clusters (6, 7), and promote AT1R internalization and trafficking into endosomes. The AT1R–β-arr complex also forms a signaling entity by recruiting effectors, such as kinases (8–10). β-Arr–mediated AT1R internalization and signaling can also be selectively directed by different ligands (e.g. biased ligands) and mutations, which induce conformational changes in both the receptor and β-arrs (9, 11–16).
Despite many years of research on the mechanisms of GPCR activation and on the functions of the receptor–β-arr complex in cells, little is known about the details of the complex formation following ligand-mediated activation of receptors. This is in part due to the paucity of structural information due to inherent difficulties in obtaining homogenous and/or stable in vitro ligand–GPCR–β-arr complexes. Moreover, once engaged by ligands, the receptor–β-arr complex adopts different conformations and arrangements. So far, only a complex of rhodopsin bound to visual arrestin has been crystalized with high resolution (17). Nonetheless, other approaches, such as negative-stain EM combined with the use of a chimeric GPCR, have proposed distinct conformational arrangements of β-arr binding to the receptor (18). β-Arr has been shown to interact with the GRK-phosphorylated C-tail of the agonist-bound receptor to form a “tail” arrangement. In addition to this tail interaction, β-arr interacts with the intracellular core domains within the receptor to form a “fully engaged” complex (18–20). A similar core arrangement has also been reported in the structure of the rhodopsin–visual arrestin complex (17). These approaches provided valuable insights for arrestins' engagement with receptors, but the precise determinants for arrestins' interaction with the C-tail (other than the already identified phosphorylation sites) and/or the core domain of receptors (i.e. ICLs and TMs), especially in the context of a native cellular environment, need to be further explored. Moreover, because biased ligands and mutations within the receptor affect GPCR–β-arr complex conformations (21, 22), such as in the case of AT1R (12, 14–16), intermolecular interactions within the complex may also vary in biased signaling conditions.
Incorporation of photoactivable unnatural amino acids (UAAs) in peptides, including in AngII, have been previously used to identify important contact points between GPCR ligands and receptors through photolysis and cross-linking (23–25). Recently, UAA mutagenesis methods have enabled site-specific modification of receptors with photoactivable UAAs, such as p-azido-l-phenylalanine (azF), to map ligand-binding sites on GPCRs (26–31). In these cases, receptors containing amber substitutions at distinct sites in their sequences are expressed along with an engineered suppressor tRNACUA and a specific aminoacyl-tRNA synthetase (aaRS) in cells, which are subsequently supplemented with the UAA to incorporate the photoactivable residue in receptors. UV-mediated photolysis of azF-incorporated mutant receptors allows insertion of nitrene into primary amines or aliphatic hydrogens of interacting partners lying within close proximity (i.e. in the radius of 3–4 Å), enabling different complex cross-linking formations and the identification of important contact sites between receptors and ligands (32).
Here, we used UAA mutagenesis of AT1R with the photoactive azF inserted at various positions in the receptor to map the residues in AngII-bound AT1R involved in β-arr binding. We investigated the effects of two β-arr–biased ligands, DVG and TRV027, and a biased signaling mutation in AT1R on the photolabeling patterns of the AT1R–β-arr complexes from different vantage points. Our study revealed important intermolecular contact points between β-arr and AT1R located not only in the C-tail but also in the ICLs of the receptor. In addition, we show differences in the interaction of β-arr with certain residues in AT1R in response to AngII versus biased ligands and an AT1R biased signaling variant, suggesting that such a targeted photocross-linking approach can be used to study the conformational arrangement of receptor–β-arr complexes in cells.
Results
AzF incorporation into receptors and expression of AT1R amber mutants
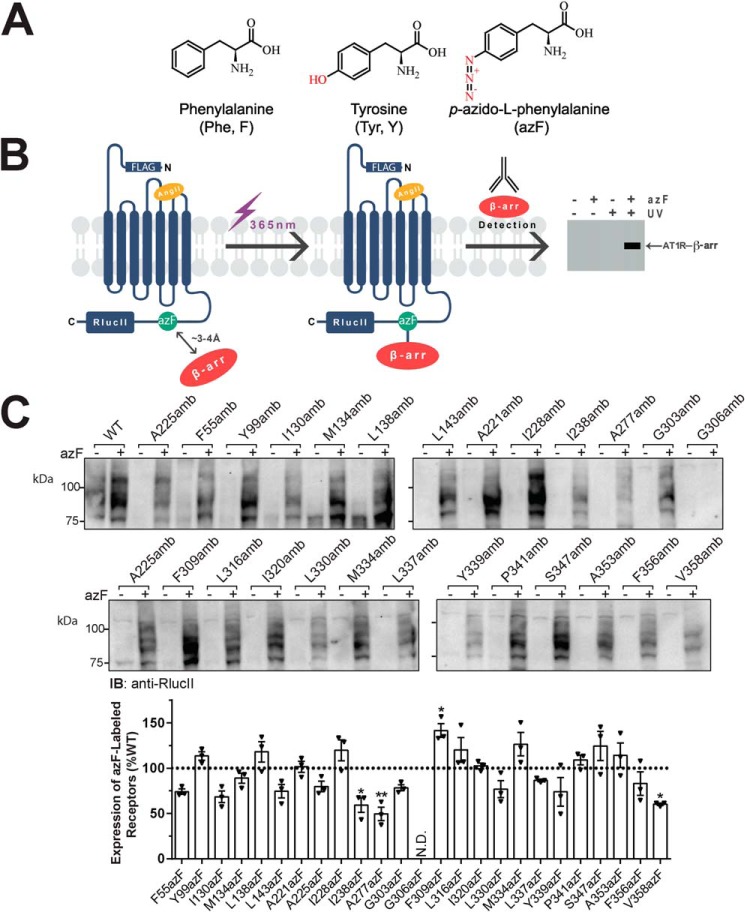
To uncover the binding sites of β-arr on AT1R, we labeled receptors that were expressed in HEK293T cells via site-specific incorporation of the photoreactive UAA azF (Fig. 1A) through introduction of amber (i.e. stop) codon mutations in AT1R at different sites. We used AT1R fused to RlucII in its C terminus, which was shown to be functional (15, 33, 34), to vet receptor expression and binding to β-arr. AzF-incorporated receptors were immunoprecipitated via the FLAG epitope on AT1R, and cross-linked receptor–β-arr complexes were detected after UV-mediated photolysis using an antibody against β-arr (Fig. 1B). To avoid disrupting receptor expression and/or their functionality as much as possible, we favored mutation of uncharged and nonpolar amino acids. We also considered residues located near important phosphorylation motifs in the C-tail of AT1R for the substitution (6, 7, 35, 36) because we rationalized that those could also form contact points with β-arr. To validate our strategy, two residues in the extracellular loops (ECLs) of AT1R were also selected as negative controls because these are not predicted to interact with β-arr. Amber receptors were expressed in cells along with the tRNACUA and the azF aaRS, and medium was supplemented with azF as done previously (26–29). We validated that receptors were fully translated only in the presence of azF through detection of the C-terminal RlucII epitope (Fig. 1C). Receptors were detected in total cell lysates in the presence of azF as shown by a broad band migrating between 75 and 100 kDa, which represents the full-length, glycosylated form of AT1R-RlucII (Fig. 1C). Quantification of receptor revealed that azF-incorporated receptors were expressed to comparable levels as the WT-AT1R except for F309azF, which showed significant increased expression, whereas I238azF, A277azF, and V358azF showed significant decreased expression (Fig. 1C). Of the 25 mutants, only G306azF-AT1R could not be detected. In all other cases, full-length amber mutant receptors were detected only in the presence of azF supplementation, indicating efficient amber codon suppression at various targeted sites and UAA incorporation in these receptor mutants.
Figure 1.
Chemical structures of natural and unnatural amino acids, photocross-linking approach, and azF-incorporated AT1R mutant expression. A, chemical structures of phenylalanine and tyrosine with three-letter and single-letter nomenclature and azF resonance structure (two double bonds). B, schematic of the photocross-linking approach. All experiments were conducted on AT1R containing an N-terminal FLAG tag and a C-terminal RlucII epitope. AzF is incorporated at site-specific amber (TAG) mutations using a heterologous cell system and the amber codon suppression technology as described under “Experimental procedures.” Cells expressing azF-incorporated AT1Rs along with β-arr1 are incubated with AngII followed by exposure to UV light (365 nm). Photoactivation of azF in AT1R promotes the formation of a covalent bond with primary amines or aliphatic hydrogens lying within its proximity (up to 3–4 Å), allowing capturing β-arr through cross-linking. The AT1R–β-arr complex is then immunologically detected to reveal its size. C, expression of each of the AT1R amber mutants transiently transfected into HEK293T cells in the absence (−) or presence (+) of 0.5 mm azF, as determined through antibody detection of the C-terminal RlucII epitope. Data are representative blots of three independent experiments (upper panel, including both sets of Western blots). Quantifications from blots are represented as mean ± S.E. (error bars) of the optical density of the band of three independent experiments and expressed as percentage of WT-AT1R (dashed line; lower panel). N.D., not detected. One-way ANOVA followed by Dunnett's multiple comparison tests was performed: *, p < 0.05; **, p < 0.01.
Functionality of azF-incorporated AT1R mutants
We next verified the functionality of the different azF-incorporated AT1Rs by assessing β-arr recruitment to receptors upon AngII stimulation. We quantified β-arr1-YFP binding to AT1R-RlucII (33, 34) using a bioluminescence resonance energy transfer (BRET) assay as done previously (12, 33, 37, 38). AT1R is a class B GPCR in terms of its trafficking behavior that shows high affinity for both β-arr1 and β-arr2 and forms long-lived complexes with β-arrs inside cells after prolonged AngII stimulation (i.e. 20 min; Fig. S1) (6, 7, 11, 33). To maximize the formation of receptor–β-arr complexes, cells expressing the azF-incorporated AT1R-RlucII mutants and β-arr1-YFP were stimulated for 20 min with AngII before BRET recording. AngII promoted β-arr binding to all expressed receptors with functional affinity (pEC50) comparable with that of the WT-AT1R-RlucII except for G303azF, which showed a 10-fold decrease in potency (Table 1 and Fig. S2). This reduction in potency was not explained by a decrease in receptor expression (Fig. 1C). In contrast, other AT1R mutants with expression levels comparable with that of G303azF, such as the F55azF, I130azF, L143azF A225azF, L330azF, and Y339azF, all recruited β-arr with similar potencies as WT-AT1R (Table 1 and Fig. S2). Moreover, A225azF also formed long-lived complexes inside the cells, analogous to that of WT and other well-expressed mutants (Fig. S1). The impact of azF incorporation was more readily observed on the functional efficacy (Emax) of β-arr complex formation with receptors, ranging from 11 to 76% of that promoted by WT-AT1R (Table 1 and Fig. S2). Overall, functionality of azF-incorporated AT1R mutants to recruit β-arr did not always correlate with their expression levels (Fig. 1C), revealing that azF incorporation at certain sites also differently affected β-arr binding to AT1R. Altogether, these data show that, in most cases, azF incorporation is well-tolerated and that these receptors are functional for binding and trafficking with β-arr.
Table 1.
Efficacies and potencies of β-arrestin binding to WT-AT1R and azF-incorporated receptor mutants
HEK293T cells transiently expressing RlucII-tagged receptor (WT-AT1R or amber mutants) along with β-arr1-YFP in the presence of 0.5 mm azF were stimulated with increasing concentrations of AngII. BRET signals were normalized to the maximal response of WT (%WT) and averaged. pEC50 and Emax were obtained from nonlinear regression curve of averaged data as described under “Experimental procedures.” Data represent means ± S.E. of three independent experiments. ND, not determined.
| pEC50 | Emax | |
|---|---|---|
| %WT | ||
| WT | −8.49 ± 0.13 | 100.0 ± 0.1 |
| F55azF | −8.56 ± 0.09 | 14.8 ± 5.5 |
| Y99azF | −8.33 ± 0.11 | 29.1 ± 7.2 |
| I130azF | −8.45 ± 0.02 | 12.5 ± 4.0 |
| M134azF | −8.38 ± 0.20 | 27.3 ± 10.4 |
| L138azF | −8.45 ± 0.12 | 39.1 ± 15.7 |
| L143azF | −8.29 ± 0.08 | 30.1 ± 13.6 |
| A221azF | −8.46 ± 0.17 | 35.0 ± 8.7 |
| A225azF | −8.23 ± 0.43 | 17.8 ± 1.8 |
| I228azF | −8.23 ± 0.30 | 32.2 ± 5.9 |
| I238azF | −8.55 ± 0.19 | 15.2 ± 3.8 |
| A277azF | −8.62 ± 0.22 | 16.0 ± 3.1 |
| G303azF | −7.50 ± 0.22 | 11.4 ± 2.8 |
| G306azF | ND | ND |
| F309azF | −8.34 ± 0.09 | 45.3 ± 13.4 |
| L316azF | −8.00 ± 0.10 | 45.7 ± 11.7 |
| I320azF | −8.00 ± 0.01 | 40.3 ± 11.2 |
| L330azF | −8.40 ± 0.12 | 22.7 ± 1.2 |
| M334azF | −8.30 ± 0.15 | 75.3 ± 23.9 |
| L337azF | −8.16 ± 0.10 | 33.2 ± 10.4 |
| Y339azF | −8.32 ± 0.03 | 71.5 ± 15.6 |
| P341azF | −8.43 ± 0.03 | 38.5 ± 25.2 |
| S347azF | −8.44 ± 0.18 | 76.4 ± 21.0 |
| A353azF | −8.33 ± 0.26 | 61.7 ± 18.8 |
| F356azF | −8.35 ± 0.10 | 57.2 ± 20.5 |
| V358azF | −8.31 ± 0.04 | 66.0 ± 25.2 |
AzF-mediated photocross-linking of AT1R–β-arr complexes and identification of critical intermolecular contacts
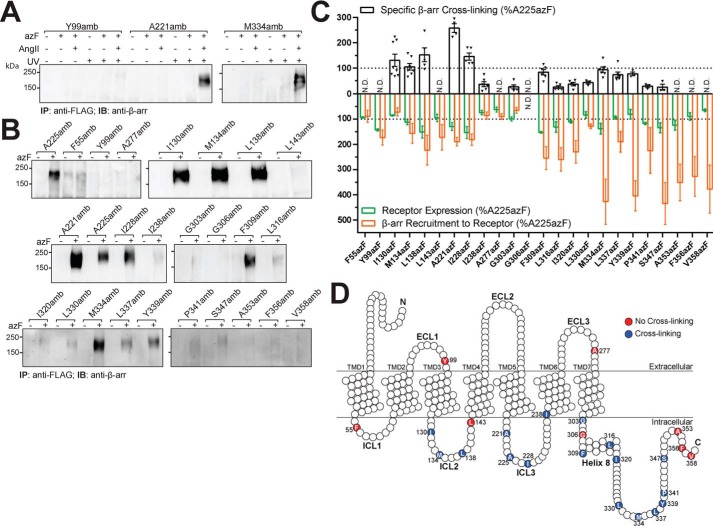
Photocross-linking experiments were carried out in HEK293T cells transiently expressing different site-specifically azF-labeled AT1R mutants and stimulated with AngII at concentrations achieving maximal receptor–β-arr complex formation as done in our BRET experiments. The interaction of β-arrs with agonist-bound GPCRs, including for AT1R, is labile in most detergent-solubilized conditions and can only be detected biochemically either through chemical cross-linking and immunoprecipitation of receptors (39–41) or here through immunoprecipitation of photocross-linked complexes. AzF-mediated cross-links between β-arr1 and AT1R after photolysis were determined from immunopurified FLAG-AT1R-RlucII–β-arr complex (bands migrating between 150 and 250 kDa; Fig. 2, A and B) using a specific antibody against β-arr1 C-terminal domain. Results showed that photocross-linking of β-arr to the A221amb and M334amb mutant receptors could only occur in the presence of azF, AngII stimulation, and photolysis (Fig. 2A). Expectedly, a mutation located in the ECL1 of AT1R (Y99amb; Fig. 2A), which was expressed similarly to WT-AT1R (Fig. 1C), was unable to covalently bind β-arr in similar conditions. WT-AT1R showed slight azF incorporation and AngII-mediated photocross-linking, although the level of receptor–β-arr complexes detected was marginal as compared with that of A225azF receptor (Fig. S3). In these conditions, WT-AT1R–β-arr complex detection was comparable with that obtained with photolysed A225azF in the absence of AngII stimulation; hence, such signal was considered as nonspecific. Photocross-linking of different AT1R amber mutants was compared with that of A225azF for quantification (Fig. 2, B and C). Mutant receptors showing photocross-linking signals similar to those obtained with WT-AT1R were considered as background and therefore interpreted as not detected (Fig. 2C, N.D.). Strong cross-linked complex formation was observed between β-arr and azF-AT1R mutants located in the ICL2 (I130azF, M134azF, and L138azF), ICL3 (A221azF, A225azF, and I228azF), and the C terminus of the receptor (F309azF and L337azF) (Fig. 2, B, C, and D), although their abilities to recruit β-arr ranged between 13 and 45% of that of WT-AT1R (Table 1 and Fig. S2). M334azF and Y339azF in the C-tail, which interacted well with β-arr, also formed strong cross-linked complexes. I238azF in ICL3; G303azF and L316az in helix 8; and I320azF, L330azF, P341azF, and S347azF in the C-tail region of the receptor all formed contacts with β-arr, although the extent of complexes was reduced as compared with that of A225azF. No interactions were detected between β-arr and AT1R mutants in the ECL1 and ECL3 (F99azF and A277azF, respectively) or with F55azF in ICL1; L143azF in ICL2; and A353azF, F356azF, and V358azF in the very distal C-terminal domain (Fig. 2, B, C, and D). The level of cross-linked AT1R–β-arr complexes observed did not always correlate with the expression of mutant receptors or with their ability to bind β-arr (Fig. S4) as well-illustrated by the different receptor C-tail mutants, which all recruited β-arr more efficiently than A225azF but formed cross-linked complexes to variable levels (Fig. 2C). These findings illustrate the sensitivity and selectivity of this approach for detecting discrete contact points between AT1R and β-arr1 and allowed identification of important residues in ICL2, ICL3, helix 8, and the C-tail of the receptor involved in this interaction (Fig. 2D).
Figure 2.
β-Arrestin binding on azF-incorporated AT1R mutants. A and B, AngII-mediated photocross-linking of azF-incorporated AT1Rs with β-arr1. HEK293T cells transiently expressing each of the AT1R amber mutants in the absence (−) and presence (+) of 0.5 mm azF were incubated with vehicle (−) or 1 μm AngII (+) followed by exposure (+) or not (−) to UV as described under “Experimental procedures.” Total cell lysates were then immunoprecipitated (IP) using an anti-FLAG antibody to isolate AT1Rs, and immunoprecipitated proteins were resolved by SDS-PAGE. Cross-linked complexes were detected with an anti-β-arr1 antibody (immunoblot (IB)). Shown are representative blots from three to seven independent experiments. C, quantification of β-arr cross-linking (black; above x axis), expression (green; below x axis), and β-arr1 recruitment to AT1R amber mutants (orange; below x axis). Receptors were transiently transfected in HEK293T cells in the presence of 0.5 mm azF, and their expression levels were determined through detection of C-terminal RlucII epitope. β-Arr binding to receptors was determined by BRET as described under “Experimental procedures” and represents the Emax of BRET ratio. Quantifications of optical density from the blots' bands and BRET signals are the mean ± S.E. (error bars), normalized to the A225azF mutant (dashed lines). N.D., not detected. D, schematic summary of AT1R contacts with β-arr.
Different binding modalities of the AT1R–β-arr complex as revealed by bias ligands and a bias AT1R variant
The variable levels of AT1R–β-arr cross-linked complexes obtained with different azF-labeled receptors highlight the importance of the relative position, distance, and/or orientation of the specific photochemical group in AT1R's residues involved in forming intermolecular interactions with β-arr, especially because the azido group in azF is sensitive to these parameters when forming covalent bounds upon UV activation (26, 42–44). Moreover, AngII-biased ligands have been shown to promote different conformations in both receptor and β-arr, which would suggest that there are also changes to some intermolecular interactions within the complex (11, 14, 16). We therefore reasoned that it would be possible to detect such biased ligand-mediated conformational changes in the AT1R–β-arr complex by assessing the variation in patterns and levels of photocross-linked complexes using different azF-labeled receptors. We used two AngII ligands, DVG and TRV027, which have been shown to engage β-arr binding to AT1R but are deficient in mediating Gαq signaling (e.g. β-arr–biased ligands) (11, 12, 45), and first verified their propensity to promote distinct changes in β-arr conformation as compared with AngII using a set of six β-arr-FlAsH sensors (F1–F6) as done previously (14). We show that these ligands promoted different changes in β-arr's conformation as compared with AngII when recruited to the ligand-bound receptor as illustrated by the Δnet BRET signal of individual sensors that produced a signature response that was characteristic for each ligand. As compared with AngII, DVG and TRV027 caused a significant decrease in the signal of the F2 sensor, whereas only TRV027 decreased and increased, respectively, the signals in the F4 and F6 sensors (Fig. 3A), suggesting differences in β-arr conformations promoted by specific ligand-bound AT1R complexes. We next assessed the effect of AngII-biased ligands on the photocross-linking patterns using five azF-AT1Rs located in distinct regions of the receptor (ICL2: I130azF and M134az; ICL3: A225azF; and the C-tail: M334azF and L337azF). AzF-labeled AT1R mutants were photolysed after 20 min of incubation with AngII, DVG, or TRV027, and cross-linked complexes were examined as described above. DVG stimulation of receptors resulted in significant decreases in cross-linked complexes for all azF-AT1R mutants as compared with AngII except for A225azF (Fig. 3B). Although DVG-mediated β-arr recruitment efficacies were reduced at all AT1R mutants (Fig. 3C), it promoted efficient A225azF-AT1R–β-arr complex formation comparable with that of AngII (Fig. 3B). In contrast, TRV027 produced a significant decrease in receptor–β-arr complexes only for I130azF, M134azF, and L337azF mutants, whereas levels of β-arr complexes with M334azF receptor were analogous to that promoted by AngII and significantly increased for the A225azF mutant despite reduction in TRV-mediated recruitment of β-arr to mutant receptors (Fig. 3C). Interestingly, a significant difference between DVG- and TRV027-mediated receptor–β-arr complexes for M334azF was observed despite similar efficacies of both biased ligands to promote β-arr recruitment to the mutant.
Figure 3.
Modulation of β-arrestin binding to AT1R using AngII-biased agonists. A, conformational signatures in β-arr binding to the AT1R. HEK293SL cells were transfected with WT-AT1R and β-arr-FlAsH reporters (F1–F6). Cells were stimulated with 1 μm AngII (black), DVG (red), or TRV027 (blue), and 5 min post-stimulation, six consecutive BRET measurements were taken every minute. Change in net BRET is reported, and data are represented as means ± S.E. (error bars) of triplicates from four to five independent experiments. Two-way ANOVA followed by Dunnett's multiple comparison tests was performed: *, p < 0.05. B, bias agonist–mediated photocross-linking. HEK293T cells transiently expressing each of the AT1R amber mutants in the presence of 0.5 mm azF were incubated with 1 μm AngII, DVG, or TRV027 followed by exposure to UV as described under “Experimental procedures.” Total cell lysates were then immunoprecipitated (IP) using an anti-FLAG antibody to isolate AT1Rs, and products were resolved by SDS-PAGE. Cross-linked complexes were detected with an anti-β-arr1 antibody (immunoblot (IB)). Shown are representative blots from three independent experiments (upper panel, including both Western blots). Quantifications represent the means ± S.E. (error bars) of optical density of the band, normalized to AngII (%AngII) (lower panel). Two-way ANOVA followed by Tukey's multiple comparison tests was performed: *, p < 0.05; **, p < 0.01; ***, p < 0.0001. C, β-arr1 recruitment to mutant AT1Rs, as assessed by BRET. HEK293T cells transiently expressing the RlucII-tagged receptor (WT-AT1R or amber mutants) along with the β-arr1-YFP in the presence of 0.5 mm azF were stimulated with 1 μm AngII, DVG, or TRV027. BRET signals were normalized to AngII (%Emax AngII). Data represent means ± S.E. (error bars) of three independent experiments. Two-way ANOVA followed by Tukey's multiple comparison tests was performed: ***, p < 0.0001.
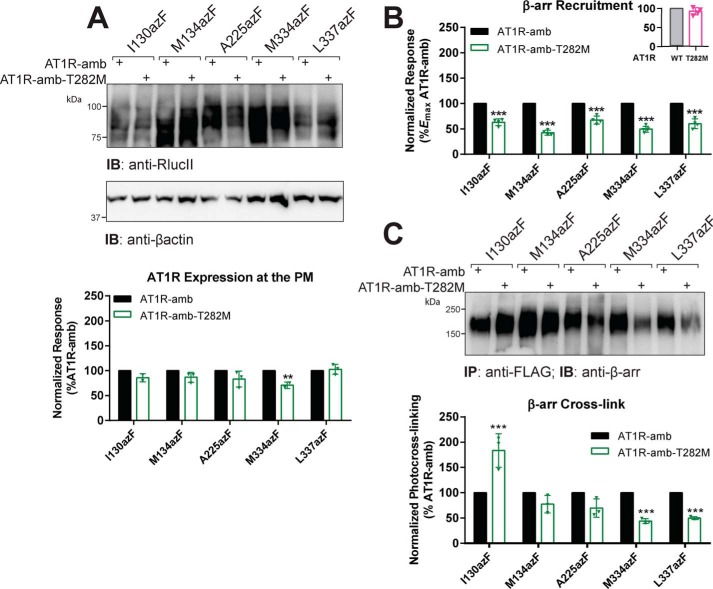
Naturally occurring variants can also impact AT1R signaling by altering the receptor's conformation (12, 46–48). Here, we focused on the nonsynonymous T7.33M AT1R (Ballesteros–Weinstein numbering (49); hereafter abbreviated T282M) in the TM7 of AT1R because we previously showed that this mutant biased receptor signaling. AT1R-T282M showed reduced efficiency to bind and cotraffic with β-arr into endosomes as compared with its ability to activate G proteins, which suggests different conformational arrangements of receptor–β-arr complexes between AT1R and AT1R-T282M mutant (12). At maximal AngII occupancy, AT1R and AT1R-T282M showed similar efficacy to recruit β-arr (Fig. 4B, inset), although we consistently observed a reduction of ∼10–15% in β-arr recruitment to AT1R-T282M as compared with AT1R. We introduced this variant in the same sets of amber AT1R mutants used to study biased ligands (Ile-130, Met-134, Ala-225, Met-334, and Leu-337) and assessed its impact on the extent of β-arr recruitment and photocross-linking to mutant receptors (Fig. 4). AzF-incorporated receptors bearing the T282M mutation (AT1R-amb-T282M) were all well-expressed, although M334azF-T282M was slightly reduced compared with its azF-labeled AT1R counterpart (Fig. 4A). Introduction of T282M in the different amber mutants reduced β-arr interaction with receptors as compared with their azF-labeled counterparts lacking this variant (Fig. 4B). Despite this, we observed a significant AngII-mediated increase in the receptor–β-arr complex formation for I130azF-T282M as compared with the same photoactive receptor lacking this variant (Fig. 4C). M134azF-T282M and A225azF-T282M showed similar levels of cross-linked receptor–β-arr complexes as their photoactive receptor counterparts lacking the T282M mutation, whereas we observed a significant reduction in complex formation for the M334azF-T282M and L337azF-T282M AT1R mutants (Fig. 4C). Although this reduction in AT1R–β-arr complexes between different azF-labeled T282M receptors and their azF-labeled receptor counterparts may result from the lower expression of the variant receptors (Fig. 4A) and/or a reduced ability of β-arr to bind these receptors (Fig. 4B), it cannot explain the significant 2-fold increase in the complex formation of the AT1R-I130azF-T282M mutant with β-arr as compared with AT1R-I130azF (Fig. 4C). Taken together, these data suggest that β-arr–biased ligands and a biased mutation in the receptor impose different binding conformations in the AT1R–β-arr complex, hence differentially impacting specific contacts within the complex.
Figure 4.
Modulation of β-arrestin photocross-linking on the AT1R variant T282M. A, plasma membrane (PM) expression of each AT1R-amber and AT1R-amber-T282M mutants transiently transfected into HEK293T cells in the presence of 0.5 mm azF, as determined through antibody detection of the C-terminal RlucII epitope after cell-surface membrane protein isolation. Shown are representative blots from three independent experiments. Quantifications represent the means ± S.E. (error bars) of optical density of the RlucII signal over the optical density of the β-actin (loading control), normalized to AT1R-amber (%AT1R-amb). Two-way ANOVA followed by Bonferroni's multiple comparison tests was performed: **, p < 0.001. B, β-arr1 recruitment to AT1R variant T282M amber mutants. HEK293T cells transiently expressing the RlucII-tagged receptor (WT-AT1R and variant T282M (inset) or amber mutants with or without the T282M variant (body)) along with β-arr1-YFP in the presence of 0.5 mm azF were stimulated with 1 μm AngII. BRET signals were normalized to AT1R-amber (%Emax AT1R-amb). Data represent means ± S.E. (error bars) of three independent experiments. Two-way ANOVA followed by Bonferroni's multiple comparison tests was performed: ***, p < 0.0001. C, AngII-mediated photocross-linking in the AT1R variant T282M amber mutants. HEK293T cells transiently expressing either the AT1R or AT1R variant T282M amber mutants in the presence of 0.5 mm azF were incubated with 1 μm AngII followed by UV exposure as described under “Experimental procedures.” Total cell lysates were then immunoprecipitated (IP) using an anti-FLAG antibody to isolate AT1Rs, and products were resolved by SDS-PAGE. Cross-linked complexes were detected with an anti-β-arr1 antibody (immunoblot (IB)). Shown are representative blots from three independent experiments (upper panel). Quantifications represent the means ± S.E. (error bars) of optical density of the band, normalized to AT1R-amber (%AT1R-amb; lower panel). Two-way ANOVA followed by Bonferroni's multiple comparison tests was performed: ***, p < 0.0001.
Discussion
Important gaps exist in our understanding of GPCRs' interaction with regulatory proteins, including with β-arrs, particularly in the formation of these complexes in their native cellular environment. We begin filling these gaps by resolving β-arr–binding sites within AT1R through the use of bioorthogonal labeling of receptor with UAAs followed by a photocross-linking approach in live mammalian cells. This allowed us to map critical residues in AT1R involved in β-arr1 binding, unveiling the importance of regions in AT1R for this interaction as well as differences in β-arr binding modalities to ligand-bound receptor in different bias signaling conditions.
We identified important residues for β-arr1 binding to AT1R in ICL2, ICL3, helix 8, and the C-tail of the receptor. Detection of numerous β-arr contact points within the receptor's C-tail is perhaps not surprising given that many of these residues (e.g. Leu-330, Met-334, Leu-337, Tyr-339, and Ser-347) are near phosphorylated serine/threonine clusters that are important for β-arr engagement with activated GPCRs (6, 7, 35, 36). Residues in helix 8, but not in the very distal region of the AT1R's C-tail, participated in β-arr binding. We also found critical residues in ICL2 and ICL3 involved in β-arr interaction. Interestingly, recent structural data on AT1R revealed important intramolecular changes between ligand-bound versus inactive receptor states, including reorganization of ICL2 and repositioning of helix 8 (50). Moreover, studies using EM on a chimeric complex of the β2-adrenergic receptor bound to β-arr1 and a resolved crystal structure of the rhodopsin–visual arrestin complex have, respectively, highlighted the importance of ICL3 and of ICL2 and ICL3 in this interaction with receptors (17, 18). These observations combined with our findings suggest that ligand-mediated structural changes in AT1R allow exposition of critical residues in ICL2, ICL3, and helix 8 for binding to β-arr1. We did not observe cross-linking between β-arr1 and the uniquely azF-labeled receptor in ICL1, which does not exclude that other residues within this domain may participate in this interaction.
Substitution of residues in AT1R by azF, including seemingly conserved residues (e.g. Phe or Tyr), had in some cases a negative impact on receptor expression and/or β-arr1 binding to receptors. We nonetheless observed that most of these replacements were well-tolerated for AT1R's functions and that azF-labeled receptors were efficiently cross-linked to β-arr although to different extents. These observations suggest that the molecular distance and orientation of the azido group in the azF of the different AT1R mutants in relation to their respective reactive chemical groups in β-arr (e.g. amines or aliphatic hydrogen in the interacting residues in the radius of 3–4 Å) (26, 42–44) importantly contributed to the variability in AT1R–β-arr1 cross-linked complexes detected.
Our findings also suggest that AT1R and β-arr form a fully engaged (18–20) complex whereby β-arr binds to both the tail and the core of the receptor when it is internalized in endosomes. Whether the modalities of AT1R interaction with β-arr differ in function of the cellular compartment where the complex assembles or between β-arr1 and β-arr2 binding remains to be determined. Indeed, β-arrs bind different effectors in distinct cellular compartments, for instance clathrin and AP-2 at the plasma membrane, which may differently modulate β-arrs' conformations in an allosteric manner, thus altering their interaction with AT1R. Moreover, in vitro data on β-arrs bound to a phosphopeptide derived from the C terminus of the V2 vasopressin receptor have revealed different conformations between β-arr1 and β-arr2 (51); hence, these two β-arrs may interact differently with AT1R. Such possibilities, as well as the interaction of AT1R with other intracellular effectors and the identification of critical residues in β-arrs involved in receptor binding, will be interesting to explore using bioorthogonal labeling and targeted photocross-linking approaches.
Biased agonism implies that GPCRs adopt multiple active conformations upon ligand binding in receptors, which lead to distinct signaling outputs in cells (52, 53). Recent structural data on AT1R using NMR and double electron–electron resonance spectroscopy have indeed revealed the existence of diverse conformational populations in the same ligand-bound receptor (16). These conformational populations of AT1R also differentially varied upon AngII versus biased ligand binding to the receptor. The photoreactive azido in the different UAA-labeled AT1Rs would be expected to covalently bind optimally distanced reactive groups in β-arr's residues, giving rise to changes in levels and patterns of complex formation, as the distance between the photoreactive group in the azF-labeled AT1R and its interacting partner in β-arr increases or decreases at some but not all positions for different ligand–AT1R–β-arr complexes. This is indeed what we observed with the two β-arr–biased ligands DVG and TRV027 as compared with AngII and with the T282M biased signaling variant of AT1R. Although our approach does not allow us to determine the conformations of the different ligand–receptor–β-arr complexes, it nonetheless provided an intermolecular signature of the arrangement of the complex that suggests the existence of distinctive AT1R–β-arr conformations driven by AngII versus biased signaling conditions. Our data also highlight the importance of different intracellular domains of AT1R, such as ICL2, ICL3, and helix 8, in binding β-arr1, which like the TMs in the receptor may also be susceptible to differential conformational intramolecular rearrangements driven by β-arr binding to ligand-bound receptors.
Our findings using bioorthogonal labeling of AT1R with UAAs and photocross-linking have allowed us to identify critical determinants within the receptor's domains for ligand-mediated AT1R–β-arr complex formation in cells. As our search to understand how ligand-bound GPCRs lead to distinct conformations and β-arr engagement and signaling continues, information gathered from the use of such approaches should inform structural, biophysical, and cell biological data. We anticipate that, given the usefulness of this approach to study AT1R–β-arr interaction, it will enable investigating the binding modalities of other GPCR-interacting partners in their native environment.
Experimental procedures
Materials
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, and other cell culture reagents were purchased from Gibco, Life Technologies. Polyethylenimine (PEI) was purchased from Polysciences (Warrington, PA). Human AngII, polyornithine, 1,2-ethanedithiol (EDT), 2,3-dimercapto-1-propanol, anti-FLAG M2 Affinity Gel®, and horseradish peroxidase–conjugated rabbit secondary antibody were purchased from Sigma-Aldrich. Horseradish peroxidase–conjugated mouse secondary antibody was purchased from Bio-Rad. Anti-RlucII antibody (MAB4400) was obtained from EMD Millipore. β-Arr1 A1CT antibody was obtained from Dr. Robert J. Lefkowitz (Duke University), and the polyclonal β-arr1 antibody (3978) was described elsewhere (54). The angiotensin ligands DVG and TRV027 were described previously (12). β-Actin antibody (C4) was obtained from Santa Cruz Biotechnology. AzF was purchased from Chem Impex International (Wood Dale, IL). FlAsH-EDT2 was synthesized at McGill University. Coelenterazine H was purchased from Nanolight Technology (Pinetop, AZ). Chemiluminescence reagents were purchased from PerkinElmer Life Sciences. Q5 high fidelity DNA polymerase, restriction enzymes, and Gibson Assembly Mix were obtained from New England Biolabs. Oligonucleotides were synthesized at Integrated DNA Technologies. All other reagents were obtained from Thermo Fisher Scientific (Waltham, MA) and were of analytical grade.
Constructs and receptor mutagenesis
Suppressor tRNA and azF aaRS plasmids were constructed as described previously (55). Amber codons (TAG) were introduced in the desired location of an N-terminal FLAG-tagged and C-terminal RlucII-labeled WT human AT1R or AT1R variant T282M in the pcDNA3.1 vector described previously (12, 33). The β-arr1 and its N-terminally tagged YFP version (β-arr1-YFP) were described previously (7). Rluc-β-arr2-FlAsH BRET sensors were described previously (14). High-throughput mutagenesis was carried out using a two-fragment PCR approach as described (56). All AT1R mutants were validated by automated sequencing at McGill University.
Transfections and cell culture
HEK293T cells and HEK293SL clonal cell line (described previously in Ref. 33) were cultured in DMEM supplemented with 10% fetal bovine serum and 20 μg/ml gentamycin at 37 °C in 5% CO2 and 90% humidity. Transient transfections were performed using the PEI method (PEI:DNA ratio, 3:1). WT-AT1R, suppressor tRNA, azF aaRS, and β-arr1 were typically transfected at a ratio of 0.1:1:0.1:0.1 (supplemented with pcDNA3.1 DNA to reach final amounts of 2.2 μg/6-well plate or 35-mm dish or 100 ng/96-well plate), whereas the amber mutant AT1R, suppressor tRNA, azF aaRS, and β-arr1 were typically transfected at a ratio of 1:1:0.1:0.1. For experiments with AT1R variant T282M, the AT1R variant T282M, suppressor tRNA, azF aaRS, and β-arr1 were typically transfected at a ratio of 0.1:1:0.1:0.1 (supplemented with pcDNA3.1 DNA to reach final amounts of 2.2 μg/6-well plate or 35-mm dish or 100 ng/96-well plate), whereas the amber mutant AT1R variant T282M (double mutant), suppressor tRNA, azF aaRS, and β-arr1 were typically transfected at a ratio of 1:1:0.1:0.1. 18 h post-transfection, the medium was replaced by DMEM with or without 0.5 mΜ azF for 48 h. For the BRET experiments, cells were transiently transfected with either β-arr1-YFP (8.3 ng) or β-arr2-FlAsH constructs along with the WT-AT1R-RlucII or amber mutants and tRNA pair. For the microscopy experiments, cells were transiently transfected with β-arr1-YFP (75 ng) construct along with the WT-AT1R-RlucII or amber mutants and tRNA pair.
Photocross-linking
HEK293T cells were seeded at 4 × 105 cells/well in 6-well culture plates, transiently transfected using PEI as detailed above, and incubated for 18 h at 37 °C in 5% CO2. ∼72 h post-transfection, medium was replaced with 1 μm AngII, DVG, or TRV027, each prepared in Tyrode's buffer (140 mm NaCl, 2.7 mm KCl, 1 mm CaCl2, 12 mm NaHCO3, 5.6 mm d-glucose, 0.5 mm MgCl2, 0.37 mm NaH2PO4, and 25 mm Hepes (pH 7.4)) and 0.1% (w/v) BSA and incubated for 20 min at room temperature (21 °C). Cells were then irradiated with a Blak-Ray B-100AP/R UV light (Analytik Jena) for 20 min on ice. Cells were then washed and resuspended in solubilization buffer (50 mm Tri-HCl, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40 (v/v), 0.5% sodium deoxycholate (w/v), and 0.1% SDS (w/v) (pH 7.4)) supplemented with protease inhibitors (20 μg/ml leupeptin, 10 μg/ml aprotinin, 2 μg/ml pepstatin A, and 10 mm phenylmethylsulfonyl fluoride). Solubilization was carried out for 1 h at 4 °C on a nutating mixer followed by centrifugation at 14,000 rpm (18,407 × g) for 20 min at 4 °C to isolate solubilized proteins (supernatant fraction) from cellular debris (pellet).
Immunoprecipitation
WT-AT1R, AT1R-T282M, or amber mutants with and without T282M mutations, tagged with an N-terminal FLAG epitope, were immunopurified from detergent-solubilized cell lysates using anti-FLAG M2 Affinity Gel. Briefly, anti-FLAG M2 Affinity Gel was incubated with cell lysates for 3 h at 4 °C on a nutating mixer. The samples were spun at 5,000 rpm (2,348 × g) for 1 min, and beads were then washed three times with solubilization buffer and incubated in Laemmli buffer for 1 h at 37 °C before being loaded on an SDS-polyacrylamide gel.
Immunoblotting
Total cell lysates or immunopurified proteins from the same experiment were individually separated on three to four SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and immunoblotted with primary antibody (diluted in blocking buffer 1:2,000 for anti-β-arr1 A1CT, 1:500 for anti-β-arr1 3978, 1:500 for anti-RlucII, and 1:1,000 for anti-β-actin) overnight at 4 °C on a nutating mixer. Secondary anti-rabbit or anti-mouse antibodies conjugated to horseradish peroxidase were used to detect bands by chemiluminescence (1:10,000). Chemiluminescence signals on blots from the same experiment were detected at once using the ChemiDoc Touch Imaging System (Bio-Rad) and the same exposure time. Densitometry analysis of immunoblots was performed using Image LabTM 6.0 software (Bio-Rad), and quantification of receptor expression or cross-linking of β-arr-receptor complexes was determined as a measure of optical density of chemiluminescence signals and was either normalized to that of AT1R (WT) or A225azF samples in the same experiment.
BRET experiments
For assessing β-arr1 recruitment to receptors, HEK293T cells were seeded at 2 × 104 cells/well in white 96-well polyornithine-coated culture plates and transiently transfected as detailed above. At ∼72 h after transfection, cells were washed once with 100 μl of Tyrode's buffer/well and then incubated with 90 μl of Tyrode's buffer for 30 min at 37 °C with 5% CO2. Cells were stimulated with various concentrations of ligand for 20 min at room temperature followed by BRET measurements. For β-arr2-FlAsH BRET experiments, FlAsH labeling was performed as described previously (14). Briefly, 1.75 μl of FlAsH-EDT2 stock reagent was mixed with 3.5 μl of 25 mm EDT solution in DMSO and left for 10 min at room temperature. 100 μl of Tyrode's buffer was then added to the mixture and left for 5 min at room temperature. The volume was then adjusted to 5 ml with Tyrode's buffer to complete the labeling solution. Cells were washed with Tyrode's buffer and incubated with 60 μl of labeling solution/well for 1 h at 37 °C. Cells were then washed twice with 2,3-dimercapto-1-propanol wash buffer followed by another wash with Tyrode's buffer and then incubated with 90 μl of Tyrode's buffer for 1h at 37 °C with 5% CO2. Cells were stimulated with ligand, and 5 min post-stimulation, six consecutive BRET measurements were taken every minute. For all BRET experiments, the cell-permeable substrate coelenterazine H was added to cells to a final concentration of 2 μm. BRET measurements were performed in triplicates using a PerkinElmer Life Sciences Victor X Light multilabel plate reader with filter sets (center wavelength/bandwidth) of 460/25 nm (donor) and 535/25 nm (acceptor). BRET signals were determined by calculating the ratio of the light emitted by the acceptor over the intensity of light emitted by the donor.
Cell-surface membrane protein purification
HEK293T cells were seeded at 4 × 105 cells/well in 6-well culture plates, transiently transfected using PEI as detailed above, and incubated for 18 h at 37 °C in 5% CO2. ∼72 h post-transfection, cells were washed with PBS and incubated with ice-cold hypotonic buffer (50 mm Tris-HCl (pH 7.4) and 5 mm EDTA) supplemented with protease inhibitors (20 μg/ml leupeptin, 10 μg/ml aprotinin, 2 μg/ml pepstatin A, and 10 mm phenylmethylsulfonyl fluoride). Cells were gently scraped from dishes and lysed using a Dounce homogenizer. The resultant mixture was centrifuged at 5,000 rpm (2,348 × g) for 5 min at 4 °C to remove nuclei. The postnuclear supernatant was subsequently centrifuged at 40,000 × g for 30 min at 4 °C, and the crude membrane pellets were resuspended in solubilization buffer (50 mm Tri-HCl, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40 (v/v), 0.5% sodium deoxycholate (w/v), and 0.1% SDS (w/v) (pH 7.4)) supplemented with protease inhibitors (20 μg/ml leupeptin, 10 μg/ml aprotinin, 2 μg/ml pepstatin A, and 10 mm phenylmethylsulfonyl fluoride). Solubilization was carried out for 1 h at 4 °C on a nutating mixer followed by centrifugation at 14,000 rpm (18,407 × g) for 20 min at 4 °C to isolate plasma membrane proteins (supernatant fraction) from cellular debris (pellet).
Confocal microscopy
HEK293SL cells were seeded in 35-mm glass-bottom dishes and transiently transfected as detailed above. Around 72 h after transfection, cells were serum-starved in DMEM for 1 h and imaged at 0 and 20 min following 1 μm AngII stimulation using a Zeiss LSM-510 Meta laser-scanning microscope. To detect YFP, a laser was used with 514-nm excitation and 530–600-nm bandpass emission filter. Images (1024 × 1024) were collected using a 63×/1.4 oil immersion lens.
Data analysis and statistics
All data were analyzed using Image LabTM 6.0 software and Prism 6.0 (GraphPad Software Inc., La Jolla, CA). Estimation of the Emax and pEC50 values for ligand-mediated β-arr1 recruitment to AT1R was calculated using the GraphPad Prism software best-fit curves. One- or two-way analysis of variance (ANOVA), Dunnett's post hoc multiple comparisons test, Tukey's post hoc multiple comparisons test, or Bonferroni's post hoc multiple comparisons test was performed as appropriate, and statistical significance was accepted as p < 0.05.
Author contributions
L. G. and S. A. L. conceptualization; L. G. and Y. C. data curation; L. G. and Y. C. formal analysis; L. G., T. P. S., and S. A. L. investigation; L. G., Y. C., A. C., D. S., T. H., T. P. S., and S. A. L. methodology; L. G. and S. A. L. writing-original draft; L. G., Y. C., A. C., D. S., T. H., T. P. S., and S. A. L. writing-review and editing; Y. C., A. C., and D. S. validation; Y. C. visualization; T. H., T. P. S., and S. A. L. supervision; T. P. S. resources; S. A. L. funding acquisition; S. A. L. project administration.
Supplementary Material
Acknowledgments
We are grateful to the members of the Sakmar laboratory for help with the targeted photocross-linking methodology and to the members of the Laporte laboratory for discussions. We thank Robert J. Lefkowitz for the generous gift of the A1CT β-arr1 antibody.
This work was supported by Canadian Institutes of Health Research (CIHR) Grant MOP-74603 (to S. A. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- GPCR
- G protein–coupled receptor
- AngII
- angiotensin II
- AT1R
- angiotensin II type 1 receptor
- β-arr
- β-arrestin
- UAA
- unnatural amino acid
- azF
- p-azido-l-phenylalanine
- ICL
- intracellular loop
- GRK
- GPCR kinase
- TM
- transmembrane
- aaRS
- aminoacyl-tRNA synthetase
- ECL
- extracellular loop
- BRET
- bioluminescence resonance energy transfer
- DMEM
- Dulbecco's modified Eagle's medium
- PEI
- polyethylenimine
- EDT
- 1,2-ethanedithiol
- ANOVA
- analysis of variance
- YFP
- yellow fluorescent protein
- Rluc
- Renilla luciferase
- DVG
- [Asp1, Val5, Gly8]-AngII.
References
- 1. Pierce K. L., and Lefkowitz R. J. (2001) Classical and new roles of β-arrestins in the regulation of G-protein-coupled receptors. Nat. Rev. Neurosci. 2, 727–733 10.1038/35094577 [DOI] [PubMed] [Google Scholar]
- 2. Lefkowitz R. J. (2004) Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends Pharmacol. Sci. 25, 413–422 10.1016/j.tips.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 3. Gurevich V. V., and Gurevich E. V. (2019) GPCR signaling regulation: the role of GRKs and arrestins. Front. Pharmacol. 10, 125 10.3389/fphar.2019.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shukla A. K., Xiao K., and Lefkowitz R. J. (2011) Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem. Sci. 36, 457–469 10.1016/j.tibs.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shenoy S. K., and Lefkowitz R. J. (2011) β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol. Sci. 32, 521–533 10.1016/j.tips.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., and Caron M. G. (1999) Association of β-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization. J. Biol. Chem. 274, 32248–32257 10.1074/jbc.274.45.32248 [DOI] [PubMed] [Google Scholar]
- 7. Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., and Caron M. G. (2001) Molecular determinants underlying the formation of stable intracellular G protein-coupled receptor-β-arrestin complexes after receptor endocytosis. J. Biol. Chem. 276, 19452–19460 10.1074/jbc.M101450200 [DOI] [PubMed] [Google Scholar]
- 8. Tohgo A., Pierce K. L., Choy E. W., Lefkowitz R. J., and Luttrell L. M. (2002) β-Arrestin scaffolding of the ERK cascade enhances cytosolic ERK activity but inhibits ERK-mediated transcription following angiotensin AT1a receptor stimulation. J. Biol. Chem. 277, 9429–9436 10.1074/jbc.M106457200 [DOI] [PubMed] [Google Scholar]
- 9. Wei H., Ahn S., Shenoy S. K., Karnik S. S., Hunyady L., Luttrell L. M., and Lefkowitz R. J. (2003) Independent β-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 100, 10782–10787 10.1073/pnas.1834556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fessart D., Simaan M., and Laporte S. A. (2005) c-Src regulates clathrin adapter protein 2 interaction with β-arrestin and the angiotensin II type 1 receptor during clathrin-mediated internalization. Mol. Endocrinol. 19, 491–503 10.1210/me.2004-0246 [DOI] [PubMed] [Google Scholar]
- 11. Zimmerman B., Beautrait A., Aguila B., Charles R., Escher E., Claing A., Bouvier M., and Laporte S. A. (2012) Differential β-arrestin-dependent conformational signaling and cellular responses revealed by angiotensin analogs. Sci. Signal. 5, ra33 10.1126/scisignal.2002522 [DOI] [PubMed] [Google Scholar]
- 12. Namkung Y., LeGouill C., Kumar S., Cao Y., Teixeira L. B., Lukasheva V., Giubilaro J., Simões S. C., Longpré J. M., Devost D., Hébert T. E., Piñeyro G., Leduc R., Costa-Neto C. M., Bouvier M., et al. (2018) Functional selectivity profiling of the angiotensin II type 1 receptor using pathway-wide BRET signaling sensors. Sci. Signal. 11, eaat1631 10.1126/scisignal.aat1631 [DOI] [PubMed] [Google Scholar]
- 13. Saulière A., Bellot M., Paris H., Denis C., Finana F., Hansen J. T., Altié M. F., Seguelas M. H., Pathak A., Hansen J. L., Sénard J. M., and Galés C. (2012) Deciphering biased-agonism complexity reveals a new active AT1 receptor entity. Nat. Chem. Biol. 8, 622–630 10.1038/nchembio.961 [DOI] [PubMed] [Google Scholar]
- 14. Lee M. H., Appleton K. M., Strungs E. G., Kwon J. Y., Morinelli T. A., Peterson Y. K., Laporte S. A., and Luttrell L. M. (2016) The conformational signature of β-arrestin2 predicts its trafficking and signalling functions. Nature 531, 665–668 10.1038/nature17154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Devost D., Sleno R., Pétrin D., Zhang A., Shinjo Y., Okde R., Aoki J., Inoue A., and Hébert T. E. (2017) Conformational profiling of the AT1 angiotensin II receptor reflects biased agonism, G protein coupling, and cellular context. J. Biol. Chem. 292, 5443–5456 10.1074/jbc.M116.763854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wingler L. M., Elgeti M., Hilger D., Latorraca N. R., Lerch M. T., Staus D. P., Dror R. O., Kobilka B. K., Hubbell W. L., and Lefkowitz R. J. (2019) Angiotensin analogs with divergent bias stabilize distinct receptor conformations. Cell 176, 468–478.e11 10.1016/j.cell.2018.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kang Y., Zhou X. E., Gao X., He Y., Liu W., Ishchenko A., Barty A., White T. A., Yefanov O., Han G. W., Xu Q., de Waal P. W., Ke J., Tan M. H., Zhang C., et al. (2015) Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 10.1038/nature14656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shukla A. K., Westfield G. H., Xiao K., Reis R. I., Huang L. Y., Tripathi-Shukla P., Qian J., Li S., Blanc A., Oleskie A. N., Dosey A. M., Su M., Liang C. R., Gu L. L., Shan J. M., et al. (2014) Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 10.1038/nature13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cahill T. J. 3rd, Thomsen A. R., Tarrasch J. T., Plouffe B., Nguyen A. H., Yang F., Huang L. Y., Kahsai A. W., Bassoni D. L., Gavino B. J., Lamerdin J. E., Triest S., Shukla A. K., Berger B., Little J. 4th, et al. (2017) Distinct conformations of GPCR-β-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl. Acad. Sci. U.S.A. 114, 2562–2567 10.1073/pnas.1701529114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gurevich V. V., and Gurevich E. V. (2004) The molecular acrobatics of arrestin activation. Trends Pharmacol. Sci. 25, 105–111 10.1016/j.tips.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 21. Reyes-Alcaraz A., Lee Y.-N., Yun S., Hwang J.-I., and Seong J. Y. (2018) Conformational signatures in β-arrestin2 reveal natural biased agonism at a G-protein-coupled receptor. Commun. Biol. 1, 128 10.1038/s42003-018-0134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ranjan R., Dwivedi H., Baidya M., Kumar M., and Shukla A. K. (2017) Novel structural insights into GPCR-β-arrestin interaction and signaling. Trends Cell Biol. 27, 851–862 10.1016/j.tcb.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 23. Fillion D., Cabana J., Guillemette G., Leduc R., Lavigne P., and Escher E. (2013) Structure of the human angiotensin II type 1 (AT1) receptor bound to angiotensin II from multiple chemoselective photoprobe contacts reveals a unique peptide binding mode. J. Biol. Chem. 288, 8187–8197 10.1074/jbc.M112.442053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laporte S. A., Boucard A. A., Servant G., Guillemette G., Leduc R., and Escher E. (1999) Determination of peptide contact points in the human angiotensin II type I receptor (AT1) with photosensitive analogs of angiotensin II. Mol. Endocrinol. 13, 578–586 10.1210/mend.13.4.0270 [DOI] [PubMed] [Google Scholar]
- 25. Servant G., Laporte S. A., Leduc R., Escher E., and Guillemette G. (1997) Identification of angiotensin II-binding domains in the rat AT2 receptor with photolabile angiotensin analogs. J. Biol. Chem. 272, 8653–8659 10.1074/jbc.272.13.8653 [DOI] [PubMed] [Google Scholar]
- 26. Grunbeck A., Huber T., Abrol R., Trzaskowski B., Goddard W. A. 3rd, Sakmar T. P. (2012) Genetically encoded photo-cross-linkers map the binding site of an allosteric drug on a G protein-coupled receptor. ACS Chem. Biol. 7, 967–972 10.1021/cb300059z [DOI] [PubMed] [Google Scholar]
- 27. Grunbeck A., Huber T., Sachdev P., and Sakmar T. P. (2011) Mapping the ligand-binding site on a G protein-coupled receptor (GPCR) using genetically encoded photocrosslinkers. Biochemistry 50, 3411–3413 10.1021/bi200214r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grunbeck A., Huber T., and Sakmar T. P. (2013) Mapping a ligand binding site using genetically encoded photoactivatable crosslinkers. Methods Enzymol. 520, 307–322 10.1016/B978-0-12-391861-1.00014-9 [DOI] [PubMed] [Google Scholar]
- 29. Koole C., Reynolds C. A., Mobarec J. C., Hick C., Sexton P. M., and Sakmar T. P. (2017) Genetically encoded photocross-linkers determine the biological binding site of exendin-4 peptide in the N-terminal domain of the intact human glucagon-like peptide-1 receptor (GLP-1R). J. Biol. Chem. 292, 7131–7144 10.1074/jbc.M117.779496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valentin-Hansen L., Park M., Huber T., Grunbeck A., Naganathan S., Schwartz T. W., and Sakmar T. P. (2014) Mapping substance P binding sites on the neurokinin-1 receptor using genetic incorporation of a photoreactive amino acid. J. Biol. Chem. 289, 18045–18054 10.1074/jbc.M113.527085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coin I., Katritch V., Sun T., Xiang Z., Siu F. Y., Beyermann M., Stevens R. C., and Wang L. (2013) Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell 155, 1258–1269 10.1016/j.cell.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu W., Brock A., Chen S., Chen S., and Schultz P. G. (2007) Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods 4, 239–244 10.1038/nmeth1016 [DOI] [PubMed] [Google Scholar]
- 33. Namkung Y., Le Gouill C., Lukashova V., Kobayashi H., Hogue M., Khoury E., Song M., Bouvier M., and Laporte S. A. (2016) Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat. Commun. 7, 12178 10.1038/ncomms12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beautrait A., Paradis J. S., Zimmerman B., Giubilaro J., Nikolajev L., Armando S., Kobayashi H., Yamani L., Namkung Y., Heydenreich F. M., Khoury E., Audet M., Roux P. P., Veprintsev D. B., Laporte S. A., et al. (2017) A new inhibitor of the β-arrestin/AP2 endocytic complex reveals interplay between GPCR internalization and signalling. Nat. Commun. 8, 15054 10.1038/ncomms15054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou X. E., He Y., de Waal P. W., Gao X., Kang Y., Van Eps N., Yin Y., Pal K., Goswami D., White T. A., Barty A., Latorraca N. R., Chapman H. N., Hubbell W. L., Dror R. O., et al. (2017) Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170, 457–469.e13 10.1016/j.cell.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayer D., Damberger F. F., Samarasimhareddy M., Feldmueller M., Vuckovic Z., Flock T., Bauer B., Mutt E., Zosel F., Allain F. H. T., Standfuss J., Schertler G. F. X., Deupi X., Sommer M. E., Hurevich M., et al. (2019) Distinct G protein-coupled receptor phosphorylation motifs modulate arrestin affinity and activation and global conformation. Nat. Commun. 10, 1261 10.1038/s41467-019-09204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Angers S., Salahpour A., Joly E., Hilairet S., Chelsky D., Dennis M., and Bouvier M. (2000) Detection of β2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. U.S.A. 97, 3684–3689 10.1073/pnas.060590697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao Y., Namkung Y., and Laporte S. A. (2019) Methods to monitor the trafficking of β-arrestin/G protein-coupled receptor complexes using enhanced bystander BRET. Methods Mol. Biol. 1957, 59–68 10.1007/978-1-4939-9158-7_3 [DOI] [PubMed] [Google Scholar]
- 39. Shenoy S. K., Drake M. T., Nelson C. D., Houtz D. A., Xiao K., Madabushi S., Reiter E., Premont R. T., Lichtarge O., and Lefkowitz R. J. (2006) β-Arrestin-dependent, G protein-independent ERK1/2 activation by the β2 adrenergic receptor. J. Biol. Chem. 281, 1261–1273 10.1074/jbc.M506576200 [DOI] [PubMed] [Google Scholar]
- 40. Luttrell L. M., Ferguson S. S., Daaka Y., Miller W. E., Maudsley S., Della Rocca G. J., Lin F., Kawakatsu H., Owada K., Luttrell D. K., Caron M. G., and Lefkowitz R. J. (1999) β-Arrestin-dependent formation of β2 adrenergic receptor-Src protein kinase complexes. Science 283, 655–661 10.1126/science.283.5402.655 [DOI] [PubMed] [Google Scholar]
- 41. Qian H., Pipolo L., and Thomas W. G. (2001) Association of β-arrestin 1 with the type 1A angiotensin II receptor involves phosphorylation of the receptor carboxyl terminus and correlates with receptor internalization. Mol. Endocrinol. 15, 1706–1719 10.1210/mend.15.10.0714 [DOI] [PubMed] [Google Scholar]
- 42. Xiang Z., Lacey V. K., Ren H., Xu J., Burban D. J., Jennings P. A., and Wang L. (2014) Proximity-enabled protein crosslinking through genetically encoding haloalkane unnatural amino acids. Angew. Chem. Int. Ed. Engl. 53, 2190–2193 10.1002/anie.201308794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lancia J. K., Nwokoye A., Dugan A., Joiner C., Pricer R., and Mapp A. K. (2014) Sequence context and crosslinking mechanism affect the efficiency of in vivo capture of a protein-protein interaction. Biopolymers 101, 391–397 10.1002/bip.22395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanaka Y., Bond M. R., and Kohler J. J. (2008) Photocrosslinkers illuminate interactions in living cells. Mol. Biosyst. 4, 473–480 10.1039/b803218a [DOI] [PubMed] [Google Scholar]
- 45. Violin J. D., DeWire S. M., Yamashita D., Rominger D. H., Nguyen L., Schiller K., Whalen E. J., Gowen M., and Lark M. W. (2010) Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J. Pharmacol. Exp. Ther. 335, 572–579 10.1124/jpet.110.173005 [DOI] [PubMed] [Google Scholar]
- 46. Hansen J. L., Haunsø S., Brann M. R., Sheikh S. P., and Weiner D. M. (2004) Loss-of-function polymorphic variants of the human angiotensin II type 1 receptor. Mol. Pharmacol. 65, 770–777 10.1124/mol.65.3.770 [DOI] [PubMed] [Google Scholar]
- 47. Zhang H., Unal H., Desnoyer R., Han G. W., Patel N., Katritch V., Karnik S. S., Cherezov V., and Stevens R. C. (2015) Structural basis for ligand recognition and functional selectivity at angiotensin receptor. J. Biol. Chem. 290, 29127–29139 10.1074/jbc.M115.689000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arsenault J., Lehoux J., Lanthier L., Cabana J., Guillemette G., Lavigne P., Leduc R., and Escher E. (2010) A single-nucleotide polymorphism of alanine to threonine at position 163 of the human angiotensin II type 1 receptor impairs losartan affinity. Pharmacogenet. Genomics 20, 377–388 10.1097/FPC.0b013e32833a6d4a [DOI] [PubMed] [Google Scholar]
- 49. Ballesteros J. A., and Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors, in Methods in Neurosciences (Sealfon S. C., ed) pp. 366–428, Academic Press, New York [Google Scholar]
- 50. Wingler L. M., McMahon C., Staus D. P., Lefkowitz R. J., and Kruse A. C. (2019) Distinctive activation mechanism for angiotensin receptor revealed by a synthetic nanobody. Cell 176, 479–490.e12 10.1016/j.cell.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nobles K. N., Guan Z., Xiao K., Oas T. G., and Lefkowitz R. J. (2007) The active conformation of β-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of β-arrestins1 and -2. J. Biol. Chem. 282, 21370–21381 10.1074/jbc.M611483200 [DOI] [PubMed] [Google Scholar]
- 52. Kenakin T. (1995) Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol. Sci. 16, 232–238 10.1016/S0165-6147(00)89032-X [DOI] [PubMed] [Google Scholar]
- 53. Costa-Neto C. M., Parreiras-E-Silva L. T., and Bouvier M. (2016) A pluridimensional view of biased agonism. Mol. Pharmacol. 90, 587–595 10.1124/mol.116.105940 [DOI] [PubMed] [Google Scholar]
- 54. Zimmerman B., Simaan M., Lee M. H., Luttrell L. M., and Laporte S. A. (2009) c-Src-mediated phosphorylation of AP-2 reveals a general mechanism for receptors internalizing through the clathrin pathway. Cell. Signal. 21, 103–110 10.1016/j.cellsig.2008.09.013 [DOI] [PubMed] [Google Scholar]
- 55. Ye S., Köhrer C., Huber T., Kazmi M., Sachdev P., Yan E. C., Bhagat A., RajBhandary U. L., and Sakmar T. P. (2008) Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 283, 1525–1533 10.1074/jbc.M707355200 [DOI] [PubMed] [Google Scholar]
- 56. Heydenreich F. M., Miljuš T., Jaussi R., Benoit R., Milić D., and Veprintsev D. B. (2017) High-throughput mutagenesis using a two-fragment PCR approach. Sci. Rep. 7, 6787 10.1038/s41598-017-07010-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.